Open Access Article
Open Access ArticleSynthesis of molecularly imprinted polymer with a methacrylate derivative monomer for the isolation of ethyl p-methoxycinnamate as an active compound from Kaempferia galanga L. extracts†
Marisa Dwi Ariania,
Ade Zuhrotunb,
Panagiotis Manesiotis c and
Aliya Nur Hasanah
c and
Aliya Nur Hasanah *ad
*ad
aPharmaceutical Analysis and Medicinal Chemistry Department, Faculty of Pharmacy, Universitas Padjadjaran, Sumedang, Bandung 45463, West Java, Indonesia. E-mail: aliya.n.hasanah@unpad.ac.id
bPharmacy Biology Department, Faculty of Pharmacy, Universitas Padjadjaran, Sumedang, Bandung 45463, West Java, Indonesia
cSchool of Chemistry and Chemical Engineering, Queens University of Belfast, Belfast, BT9 5 AG, UK
dDrug Development Study Centre, Faculty of Pharmacy, Universitas Padjadjaran, Sumedang, Bandung 45463, West Java, Indonesia
First published on 25th April 2024
Abstract
Kaempferia galanga rhizome is traditionally used as a treatment for various diseases. Ethyl p-methoxycinnamate (EPMC), which constitutes up to 31.77% of the total essential oil, is the main/marker compound. EPMC is responsible for various pharmacological activities of Kaempferia galanga rhizome. According to the existing research, the isolation yield of EPMC is still meager, namely 0.50–2.50%; thus, a new EPMC isolation method is needed to produce better results. In this study, after determining the association constant and obtaining the Jobs plot between methacrylate derivative monomers and EPMC, a molecularly imprinted polymer for solid phase extraction (MI-SPE) was synthesized through bulk polymerization with EPMC as a template, methacrylic acid as a monomer, TRIM/EDGMA as a crosslinker in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 (MIP1) or 1
20 (MIP1) or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7
7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
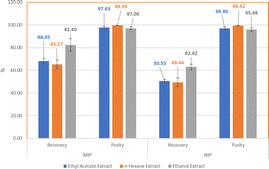
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 (MIP2). BPO was used as an initiator and n-hexane was used as a porogen. The synthesis of the NIP was also conducted using the same ratio but without the template. The MIPs were then characterized using Fourier transform infrared (FTIR) spectroscopy, scanning electron microscopy (SEM), and Brunauer–Emmett–Teller (BET) measurements, and their analytical performance was evaluated through adsorption capacity and selectivity. The results indicate that MIP2 exhibits better analytical performance with an adsorption capacity value of 0.0813 mg g−1. The selectivity of MIP2 was tested using EPMC analog compounds, namely ethyl cinnamic (EC), cinnamaldehyde (CD), and kaempferol (KF), with imprinting factor (IF) values of 17.436, 1.539, and 0.06, respectively. Lastly, MIP2 was applied to the SPE cartridge for the isolation of EPMC from Kaempferia galanga rhizome extract, and showed a percentage recovery of 82.40% for the ethanol extract, 68.05% for the ethyl acetate extract, and 65.27% for the n-hexane extract. MI-SPE 2 gives high purity results for the ethanol, ethyl acetate, and n-hexane extracts, with purities of 97.00%, 97.63%, and 99.59%, respectively. These results indicate that the MI-SPE technique shows great potential as a new method for isolating EPMCs with high yield and purity.
20 (MIP2). BPO was used as an initiator and n-hexane was used as a porogen. The synthesis of the NIP was also conducted using the same ratio but without the template. The MIPs were then characterized using Fourier transform infrared (FTIR) spectroscopy, scanning electron microscopy (SEM), and Brunauer–Emmett–Teller (BET) measurements, and their analytical performance was evaluated through adsorption capacity and selectivity. The results indicate that MIP2 exhibits better analytical performance with an adsorption capacity value of 0.0813 mg g−1. The selectivity of MIP2 was tested using EPMC analog compounds, namely ethyl cinnamic (EC), cinnamaldehyde (CD), and kaempferol (KF), with imprinting factor (IF) values of 17.436, 1.539, and 0.06, respectively. Lastly, MIP2 was applied to the SPE cartridge for the isolation of EPMC from Kaempferia galanga rhizome extract, and showed a percentage recovery of 82.40% for the ethanol extract, 68.05% for the ethyl acetate extract, and 65.27% for the n-hexane extract. MI-SPE 2 gives high purity results for the ethanol, ethyl acetate, and n-hexane extracts, with purities of 97.00%, 97.63%, and 99.59%, respectively. These results indicate that the MI-SPE technique shows great potential as a new method for isolating EPMCs with high yield and purity.
1. Introduction
Zingiberaceae is a diverse plant family consisting of 52 genera and approximately 1587 plant species. One of the best-known genera among the Zingiberaceae family is Kaempferia, which consists of 126 species that are widespread worldwide with wide distribution throughout tropical Southeast Asian countries such as Indonesia, Malaysia, Myanmar, Cambodia, Thailand, and even the South Asian region, namely India.1,2 Kaempferia galanga L., which is known in Indonesia as kencur, has been used empirically by approximately 109 ethnic groups. In Indonesia, Kaempferia galanga is present in several regions, such as Sumatra, Java, Kalimantan, East Nusa Tenggara, Sulawesi, and Maluku.3 It is ranked 16th as the most widely used medicinal plant.4 Kaempferia galanga rhizome has traditionally been used as an anti-inflammatory, analgesic, antimicrobial, antioxidant, nematicidal, and vasorelaxant.5–14 The rhizome and leaves of Kaempferia galanga L. have properties for the treatment of wounds, headaches, ulcers, the common cold, coughs, asthma, and breast cancer.15–17 In 2014, Kumar reported that the rhizome of Kaempferia galanga L. contains up to 50 volatile oil constituents (97.19% oil content), including ethyl-p-methoxycinnamate, ethyl cinnamate, γ-carene, γ-cadinene, 1,8-cineole, camphene, borneol, linoleoyl chloride and pinene.18,19Ethyl p-methoxycinnamate (EPMC) is the main component of the essential oil of Kaempferia galanga L., comprising 31.77% of the total essential oil (24.06% of the total metabolite content). EPMC is an ester compound responsible for the pharmacological activity and scent of Kaempferia galanga L.20 EPMC is responsible for various pharmacological activities, such as nematicidal, mosquito repellent, antimicrobial, analgesic, wound healing, antiangiogenic, and antineoplastic activity.7,11,12,21,22 The isolation yields of EPMC range from 0.50–2.50% using various isolation methods ranging from conventional column chromatography to CO2 supercritical chromatography.8,22–25 This isolation yield is still meager considering that EPMC is the dominant component of the essential oil of Kaempferia galanga L.; thus, an EPMC isolation method is needed to produce better results. Chanvimalueng et al. (2022) isolated EPMC from Kaempferia galanga in several stages, namely, maceration, fractionation, initial sub-fractionation, and advanced sub-fractionation, and obtained an EPMC yield of 2.68%.26 On the other hand, using the Molecularly Imprinted Polymer-Solid Phase Extraction (MI-SPE) technique for EPMC isolation requires only two steps in the process, which are maceration and SPE.
The molecularly imprinting technique (MIT) is one of the most promising techniques for preparing polymers with high selectivity and specific binding sites in molecularly synthesized polymers. Molecular imprinted polymers (MIPs) have advantages, because the use of cross-linkers will result in relatively stable binding sites on the polymer, allowing them to withstand high pressures without being damaged and remain stable under various pH and extreme chemical conditions. MIPs are currently widely used to isolate active compounds from natural materials because of their good selectivity and high reusability.27–31 Saad et al. (2021)31 conducted a study to develop a robust and selective method for the extraction of rosmarinic acid from the plant Rosmarinus officinalis L. using the molecularly imprinted solid phase extraction (MI-SPE) technique. The results showed that the MI-SPE technique was able to selectively extract rosmarinic acid from a very complex extract of Rosmarinus officinalis L. with a high yield and purity of 81.96 ± 6.33% and 80.59 ± 0.30%, respectively.31 Zhu et al., (2010)28 conducted a study for the isolation of vinblastine using the MIP technique. The results showed that the MIP cartridge had a high-capacity factor of 750 g g−1 in toluene, 625 g g−1 in chloroform, and 250 g g−1 in methanol. In addition, the resulting MIP sorbent has a high selectivity, which is indicated by the almost complete lack of selective binding of the sorbent to vincristine (an analog of vinblastine). The isolation yield of vinblastine from the extract of Catharanthus roseus applied to the MIP cartridge was also very high at 93.8%.28
Based on existing research results related to the isolation of secondary metabolites from this natural product, the MIP technique produces MIP sorbents with high selectivity and high yield. The MIP technique has not previously been reported for the isolation of EPMC compounds from Kaempferia galanga L. Therefore, the MIP technique can be used as a novel and promising method to improve the EPMC results from Kaempferia galanga L. This study will synthesize a molecularly imprinted functional polymer material with bulk polymerization. The synthesis is carried out by screening methacrylate monomer derivatives with the best template interactions and synthesizing them using bulk polymerization. The resulting material is characterized and used to isolate ethyl p-methoxycinnamate from a Kaempferia galanga L. rhizome extract that was extracted using two different solvents.
2. Materials and methods
2.1. Chemicals and reagents
Kaempferia galanga L. rhizome extract was provided by Herbal Study Center, Faculty of Pharmacy (Universitas Padjadjaran). Ethyl p-methoxycinnamate (EPMC), cinnamaldehyde (CD), and kaempferol (KF) were purchased from MarkHerb (Bandung, Indonesia). Benzoyl peroxide (BPO), 2-hydroxyethyl methacrylate (HEMA), methacrylic acid (MAA), benzoyl peroxide (BPO), ethyl cinnamate (EC), methyl methacrylate (MMA), and trifluoroacetic acid (TFA) were purchased from Sigma-Aldrich (Missouri, USA). 2-(Trifluoromethyl)acrylic acid (TFMAA), ethylene glycol dimethacrylate (EGDMA), and trimethylol propane trimethacrylate (TRIM) were purchased from Tokyo Chemical Industry (Tokyo, Japan). Acetic acid, ethanol, ethyl acetate, HPLC-grade acetonitrile, isopropanol, and n-hexane were purchased from Merck (Darmstadt, Germany). All materials other than those described were of pro analysis grades.2.2. Instruments
The instrumentation that was used in this research consisted of an analytical balance (Ohaus Pioneer), Fourier transform infrared (FTIR) spectrometer (IR Prestige-21 Shimadzu), JSM-6510 LA scanning electron microscope (JEOL), ultrasonicator (NEY 19H), ultra-high-performance liquid chromatography acquity H-Class PLUS core system with PDA detector dual pump with quartenery solvent manager (Waters), UV-Visible spectrophotometer (Analytical Jena Specord 200 using a 1.0 cm quartz cell), and ordinary glassware in the laboratory.2.3. Ultra-performance liquid chromatography (UPLC)
EPMC content was analyzed using a Waters ultra-performance liquid chromatography (UPLC) system coupled with a PDA detector. The chromatographic system used a C-18 (150 × 4.6 nm, 5 μm) column (Luna from Phenomenex). The mobile phase was acetonitrile and water (55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
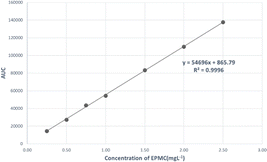
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45) with 0.2% trifluoroacetic acid at a flow rate 1.0 mL min−1. The wavelength was set to 310 nm and the injection volume was 10 μL. EPMC was measured at concentrations of 0.25, 0.50, 0.75, 1.00, 1.50, 2.00, and 2.50 mg L−1. A calibration curve was then created, which can be seen in Fig. 1, and the linear equation y = 546
45) with 0.2% trifluoroacetic acid at a flow rate 1.0 mL min−1. The wavelength was set to 310 nm and the injection volume was 10 μL. EPMC was measured at concentrations of 0.25, 0.50, 0.75, 1.00, 1.50, 2.00, and 2.50 mg L−1. A calibration curve was then created, which can be seen in Fig. 1, and the linear equation y = 546![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 96x + 865.79 was obtained, with the LOD and LOQ values being 0.07 mg L−1 and 0.20 mg L−1, respectively.
96x + 865.79 was obtained, with the LOD and LOQ values being 0.07 mg L−1 and 0.20 mg L−1, respectively.
2.4. Kaempferia galanga L. extract sample collection
Kaempferia galanga L. extract was obtained by the maceration method using three different solvents, namely, ethanol, ethyl acetate, and n-hexane. For each solvent, 250 grams of Kaempferia galanga L. dried rhizome were used. After extraction, the filtrate was evaporated using a rotary evaporator. The residue (crude extract) was collected and stored at 18–20 °C until use.24 The EPMC contents of all extracts were determined using UPLC. The calibration curve was prepared by the appropriate dilution of the EPMC standard in ethanol aliquots to obtain concentrations of 5.0, 10.0, 20.0, 70.0, 100.0 mg L−1. The sample extract was prepared using as much as 100.0 mg crude extract, then dissolved in a 100 mL volumetric flask. It was diluted by piping 1 mL of the solution into a 10 mL volumetric flask up to the mark; this was repeated three times. The extract samples were then filtered and analyzed using UPLC with a mobile phase of acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (55
water (55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 v/v) 0.2% TFA using C-18 (150 × 4.6 nm, 5 μm) column with a 1.0 mL min−1 flow rate, and a UV detector at 310 nm.32
45 v/v) 0.2% TFA using C-18 (150 × 4.6 nm, 5 μm) column with a 1.0 mL min−1 flow rate, and a UV detector at 310 nm.32
2.5. Determination of the template–functional monomer association constant using a UV-Vis spectrophotometer
Prior to synthesis, the functional monomer (FM) with the best interaction with the template (EPMC) was selected from among several methacrylic acid derivatives, namely, methacrylic acid (MAA), 2-hydroxyethyl methacrylate (HEMA), 2-(trifluoromethyl)acrylic acid (TFMAA), and methyl methacrylate (MMA). The structure of each monomer can be seen in Fig. S1 in the ESI.† The EPMC-FM interaction was studied using the UV titration method, referring to the research of Hasanah et al. (2019) with modification of the concentration of template molecules.33 1.5 × 10−4 M EPMC was dissolved in two solvents, namely n-hexane and isopropanol. Then, 0.01 M FM solution was gradually added from 0 μL to 300 μL (until the absorbance remained constant). Absorbance measurements were carried out using a microplate reader and UV detector with a single wavelength (310 nm) (the UV spectrum for determining the maximum wavelength of EPMC is shown in Fig. S2†). The absorbance at each addition was recorded and plotted on a curve of the FM concentration and the absorptive delta. The association constant was calculated using the Benesi–Hildebran equation.2.6. Stoichiometry reaction analysis (Job plot)
A stoichiometric reaction analysis was carried out to determine the equilibrium reaction between EPMC and the monomer to compare the composition of the best synthetic formula. A Job plot analysis was constructed by systematically varying the molar ratio of EPMC and monomer in the solvent, and was monitored using UV-Vis spectrophotometry referring to the study of Suryana et al., 2021 with modification of the solvent and the initial concentration of the template and functional monomer.34 The initial values of EPMC and the monomer were 2 × 10−5 and 2 × 10−4 M, respectively. The total volume was 3 mL and all absorbance was recorded at 310 nm; the delta of absorbance was plotted against the molar fraction of EPMC.2.7. Preparation of molecularly imprinted polymers (MIPs) and non-imprinted polymers (NIPs)
The MIPs were synthesized via bulk polymerization using two template![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FM
FM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) crosslinker (T
crosslinker (T![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FM
FM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cl) ratios, namely, 1
Cl) ratios, namely, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 (the most common ratio used in the literature),35,36 and the ratio obtained from the Job plot analysis. The MIPs were synthesized using 0.25 mmol of EPMC and 1 mmol of the selected FM. The EPMC and FM were dissolved in 5 mL of the selected solvent in a vial and then sonicated until fully dissolved. Then, 5 mmol of the crosslinker (EGDMA/TRIM) was added to the vial and sonicated until dissolved. Then, 0.206 mmol of the initiator BPO was added to the solution and sonicated for 5 minutes. The vial was sealed and transferred to an oven at 70 °C for 18 hours. The obtained polymer was ground and sieved using an 80-mesh sieve. The polymer was then rinsed with 20 mL of methanol to remove the remaining unreacted reagent and dried in an oven at 50 °C for 18 hours. The non-imprinted polymer (NIP) was prepared simultaneously under the same conditions without the addition of the template. Each polymer was synthesized twice. The composition of each MIP and NIP can be seen in Table 1.
20 (the most common ratio used in the literature),35,36 and the ratio obtained from the Job plot analysis. The MIPs were synthesized using 0.25 mmol of EPMC and 1 mmol of the selected FM. The EPMC and FM were dissolved in 5 mL of the selected solvent in a vial and then sonicated until fully dissolved. Then, 5 mmol of the crosslinker (EGDMA/TRIM) was added to the vial and sonicated until dissolved. Then, 0.206 mmol of the initiator BPO was added to the solution and sonicated for 5 minutes. The vial was sealed and transferred to an oven at 70 °C for 18 hours. The obtained polymer was ground and sieved using an 80-mesh sieve. The polymer was then rinsed with 20 mL of methanol to remove the remaining unreacted reagent and dried in an oven at 50 °C for 18 hours. The non-imprinted polymer (NIP) was prepared simultaneously under the same conditions without the addition of the template. Each polymer was synthesized twice. The composition of each MIP and NIP can be seen in Table 1.
| Polymer | Template (T) | Functional monomer (FM) | Crosslinker (Cl) | Ratio T![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FM FM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cl (mmol) Cl (mmol) |
|---|---|---|---|---|
| MIP 1 | EPMC | MAA | TRIM | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20 |
| NIP 1 | — | MAA | TRIM | 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20 |
| MIP 2 | EPMC | MAA | TRIM | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20 |
| NIP 2 | — | MAA | TRIM | 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20 |
| MIP 3 | EPMC | MAA | EGDMA | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20 |
| NIP 3 | — | MAA | EGDMA | 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20 |
| MIP 4 | EPMC | MAA | EGDMA | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20 |
| NIP 4 | — | MAA | EGDMA | 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20 |
A soxhlation method was used to remove EPMC from the resulting MIP. The soxhlation was carried out for 24 hours using a mixed methanol–acetic acid solution (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). The extracted polymer was rinsed with 20 mL of methanol and water and then dried at 50 °C for 18 hours. Monitoring of the completeness of the template removal from the MIP was done using the EPMC peak in adsorption studies.
1, v/v). The extracted polymer was rinsed with 20 mL of methanol and water and then dried at 50 °C for 18 hours. Monitoring of the completeness of the template removal from the MIP was done using the EPMC peak in adsorption studies.
2.8. Characterization of prepared MIP and NIP with Fourier transform infrared (FTIR) spectroscopy, scanning electron microscopy (SEM), a particle size analyzer (PSA) and Brunauer–Emmet–Teller (BET) analysis
Polymer characterization was performed to determine the properties of each polymer. Characterization was carried out using FTIR, SEM, PSA, and BET instruments. A total of 2 mg of the polymer was placed into a mortar, to which 198 mg of potassium bromide was then added. They were then mixed until homogeneous and formed into pellets. The pellets were then analyzed using FTIR in the 4000–400 cm−1 wavenumber range to confirm the synthesis process.33 SEM was carried out to determine the morphology of the resulting polymer. The polymers were coated with a thin layer of the gold film under vacuum and placed in the SEM instrument.37 The particle size of the polymer was analysed using a particle size analyzer (PSA). A test using a BET instrument was carried out to obtain a quantitative isothermic data curve, which was obtained by plotting the total volume of nitrogen gas adsorbed and desorbed against pressure.382.9. Adsorption capacity evaluation
The evaluation of adsorption capacity was based on the report of Suryana et al. (2021)34 with several modifications. EPMC solutions with several concentrations, namely 0.25, 0.50, 0.75, 1.00, 1.50, 2.00, and 2.50, 5, 7.5 mg L−1, were prepared for the adsorption capacity evaluation. 1.5 mL of EPMS was added to a vial containing 20 mg of the MIP or NIP sorbent and allowed to stand for 24 hours. The mixture was then filtered and measured using UPLC. The amount of EPMC absorbed was calculated based on the difference between the initial and final concentrations of EPMC in the filtrate. The results were plotted using the Freundlich and Langmuir isotherm adsorption models.342.10. Selectivity evaluation
Sorbent selectivity evaluation of EPMC was conducted towards analogs of EPMC, namely, the compounds EC, CD, and KF. Standards of EPMC, CD, EC, and KF with a concentration of 2 mg L−1 each were placed into a vial containing 20 mg of the sorbent and then shaken for 5 minutes. The concentration of each compound was measured using UPLC. Selectivity was calculated according to the values of the distribution, selectivity, and relative selectivity coefficients using the equation below.| Kd = (Ci − Cf)v/CfW |
| k = Kd1/Kd2 |
| k′ = kMIP/kNIP |
2.11. Application to real samples
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetic acid, 9
acetic acid, 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v). Each stage is given a contact time of 5 minutes. The collected fractions from each stage were analyzed by UPLC with a mobile phase of acetonitrile
1 v/v). Each stage is given a contact time of 5 minutes. The collected fractions from each stage were analyzed by UPLC with a mobile phase of acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (55
water (55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 v/v) with 0.2% TFA using a C-18 (150 × 4.6 nm, 5 μm) column with a 1.0 mL min−1 flow rate, and the UV detector set at 310 nm.32
45 v/v) with 0.2% TFA using a C-18 (150 × 4.6 nm, 5 μm) column with a 1.0 mL min−1 flow rate, and the UV detector set at 310 nm.32The test was carried out by first preparing a 100 mg L−1 extract solution. Each of the standard compounds EPMC, EC, CD, and KF was spiked into the n-hexane extract solution, and the solution was then extracted using MI-SPE and non-imprinted polymer-solid phase extraction (NI-SPE).
3. Results and discussion
3.1. Kaempferia galanga L. extract sample collection
Kaempferia galanga L. rhizome dried simplicia was extracted by maceration using various solvents, namely ethanol, ethyl acetate, and n-hexane. The selection of extraction solvents was based on the solvents generally used at each stage of the conventional EPMC isolation method, namely the extraction, fractionation, sub-fractionation, and isolation stages.42,43 Thus, this extraction process is expected to cover the EPMC withdrawal of Kaempferia galanga rhizome extract from the extraction process to sub-fractionation. In addition, the solvents used also match the solubility profile of EPMC.The maceration process was carried out three times for each solvent. First, 250 grams of Kaempferia galanga L. dried rhizome were used for each solvent. From the maceration of Kaempferia galanga L. rhizome, an average viscous extract of 65.40 grams was obtained for the ethanol extract, 71.18 grams for the ethyl acetate extract, and 80.15 grams for the n-hexane extract with extraction yields of 26.16 ± 0.31%, 28.48 ± 0.15%, and 32.06 ± 0.09%, respectively. Furthermore, the EPMC levels were analyzed in the extract using UPLC. The obtained EPMC levels in each extract are shown in Table 2.
| Extraction solvent | % assay average | % RSD |
|---|---|---|
| Ethyl acetate | 85.83 | 3.49% |
| Ethanol | 71.00 | 1.85% |
| n-Hexane | 96.10 | 1.11% |
From Table 2, it can be seen that EPMC compounds accumulate more in the n-hexane extract, with an assay average of 96.10 ± 1.11%. This is in accordance with the nature of EPMC, which tends to be non-polar;43 this, it is contained in many n-hexane extracts. The calibration curve for the EPMC content in the extract, along with the detailed experimental results, are presented in Fig. S3 and Table S1.†
3.2. Determination of the template–functional monomer association constant using a UV-Vis spectrophotometer
The association constant value represents the ability of a monomer to bind to a template molecule to form a stable complex before the polymerization process. The higher the association constant value, the more stable and rapid the formation of the monomer–template complex during the polymerization process, thus resulting in a high imprinting factor.44,45 In this study, the association constant value was determined using the monomers methacrylic acid (MAA), 2-hydroxyethyl methacrylate (HEMA), trifluoromethyl acrylic acid (TFMAA), and methyl methacrylate (MMA) on the EPMC template, with isopropanol and n-hexane as the porogen using the UV titration method. The association constant (Ka) value was calculated using the Benesi–Hildebrand equation using the slope and intercept values in the linear regression.46 The resulting association constant values can be seen in Table 3 below.| Monomer | Solvent | Ka value (M−1) | R2 |
|---|---|---|---|
| MAA | n-Hexane | 6569 | 0.997 |
| Isopropanol | 359 | 0.997 | |
| HEMA | n-Hexane | 264 | 0.997 |
| Isopropanol | 97 | 0.960 | |
| TFMAA | n-Hexane | 3378 | 0.993 |
| Isopropanol | 192 | 0.991 | |
| MMA | n-Hexane | 259 | 0.998 |
| Isopropanol | 89 | 0.998 |
The results showed that a higher association constant value was obtained using n-hexane than isopropanol as the solvent for each monomer in the test. This is because n-hexane (Kd 1.88) has a lower dielectric constant than isopropanol (19.92), so n-hexane is more non-polar than isopropanol. Polar solvents can disrupt the hydrogen bonds formed between the monomer and the template; thus, the porogen n-hexane will not interfere with the interaction between the monomer and the EPMC template.47,48 Using the methacrylic acid monomer with n-hexane as the solvent gives a higher Ka value of 6569 M−1 and R2 = 0.9970 compared to other monomers, so it can be considered to produce the most robust and stable complex to the EPMC template. Therefore, methacrylic acid was used as the functional monomer in the EPMC MIP polymerization synthesis process. The linear regression curve can be seen in Fig. S4.†
The Benesi–Hildebrand method has several limitations in determining association constants from UV spectrum titrations, such as the existence of zero and negative intercepts, which can cause the calculations to be invalid. Variations in the values obtained from the Benesi–Hildebrand method calculations can also be caused by unbonded guests in the complex bonds, which can affect the instrument signal. This contribution from the unbound guest response will cause potential errors in the slope and intercept of the Benesi–Hildebrand plot.49
3.3. Stoichiometry reaction analysis (Job plot)
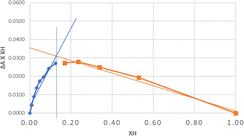
The Job plot is a chemical method used to determine the stoichiometric proportion of bonds, from which we can determine the number of monomer molecules (guests) that will bind to the template (host).50 In this study, various additions of monomer and template were carried out from 0 to 3 mL, which were then measured using UV spectrophotometry. A curve is then made between the template mole fraction and the absorbance delta multiplied by the template mole fraction. The template and monomer will form a complex, and when the reaction equilibrium point is reached, the concentration of the template–monomer complex will be at the maximum position, and the concentration of each template and monomer will be at a minimum approaching zero, so that the template and monomer will have equations that will be combined into a new equation to extrapolate the ratio of the template and monomer required to reach the reaction equilibrium point.51 Based on the calculations in Fig. S5 of the ESI,† the maximum peak value of the host fraction (XH) is 0.129, and the monomer template concentration ratio is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7. Moreover, the maximum value can also be seen using a tangent line, as shown in Fig. 2.
7. Moreover, the maximum value can also be seen using a tangent line, as shown in Fig. 2.
3.4. Preparation of MIPs and NIPs
The MIP and NIP were synthesized by bulk polymerization using the following two T![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FM
FM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cl ratios. MIP 1 and MIP 3 were synthesized in a ratio of 1
Cl ratios. MIP 1 and MIP 3 were synthesized in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, which is a common ratio in MIP synthesis.52 Meanwhile, MIP 2 and MIP 4 were synthesized using the ratio of 1
20, which is a common ratio in MIP synthesis.52 Meanwhile, MIP 2 and MIP 4 were synthesized using the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7
7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 based on the results of the Job plot analysis. The bulk polymerization method is simple, relatively fast, and does not require specific instruments.53 The MIP synthesis in this study required EPMC as a template, printed in a polymer matrix of methacrylic acid functional monomers, and used EGDMA or TRIM as the crosslinkers and benzyl peroxide (BPO) as the initiator in the solvent n-hexane.
20 based on the results of the Job plot analysis. The bulk polymerization method is simple, relatively fast, and does not require specific instruments.53 The MIP synthesis in this study required EPMC as a template, printed in a polymer matrix of methacrylic acid functional monomers, and used EGDMA or TRIM as the crosslinkers and benzyl peroxide (BPO) as the initiator in the solvent n-hexane.
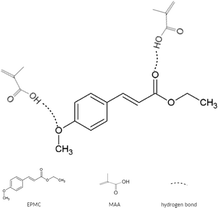
TRIM and EGDMA, which act as crosslinkers, play a role in controlling the morphology and stabilizing the bonding sites on the formed polymer so that the polymer can be chemically and physically stable. In this study, two different crosslinkers were used to determine which crosslinker would provide a better adsorption capacity on the EPMC MIP. The polymerization process was carried out at 70 °C, the temperature at which the initiator BPO decomposes.54 The template–monomer interactions expected to form during polymerization are non-covalent, such as hydrogen bonds. Interactions through hydrogen bonds, such as between the –OH from methacrylic acid and –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O from EPMC (Fig. 3), will form the basis for the specific binding in the MIP. Non-covalent interactions tend to be weaker than covalent interactions; thus, template release and the rebinding process in the MIP can easily occur.55 NIP synthesis was performed with the same procedure as MIP synthesis but without adding the EPMC template. The NIP was used to compare the results obtained from the MIP and ensures that the interactions that occur are molecular interactions, not non-specific interactions.
O from EPMC (Fig. 3), will form the basis for the specific binding in the MIP. Non-covalent interactions tend to be weaker than covalent interactions; thus, template release and the rebinding process in the MIP can easily occur.55 NIP synthesis was performed with the same procedure as MIP synthesis but without adding the EPMC template. The NIP was used to compare the results obtained from the MIP and ensures that the interactions that occur are molecular interactions, not non-specific interactions.
After the polymer is formed, the template is removed or extracted from the MIP by the soxhlation method. Extraction aims to remove the EPMC template from the MIP polymer matrix so that specific empty cavities remain. Under the right conditions, the cavity can re-bind the template molecule.56
3.5. Characterization of the prepared MIP and NIP using the Fourier transform infrared (FTIR) spectroscopy using attenuated reflectance, scanning electron microscopy (SEM), a particle size analyzer (PSA), and Brunauer–Emmet–Teller (BET) analysis
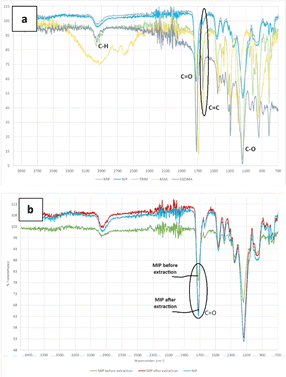
The FTIR analysis results in Fig. 4a show the spectra of MAA, TRIM, EGDMA, MIP, and NIP. In the MAA FTIR spectrum, the peak at 2927 cm−1 corresponds to the C–H stretch, the peaks at 1680 cm−1 and 1201 cm−1 correspond to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–O groups, respectively, and the peak at 1632 cm−1 corresponds to the C
O and C–O groups, respectively, and the peak at 1632 cm−1 corresponds to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching of the vinyl functional group. The FTIR spectra of EGDMA and TRIM show peaks at similar positions. In the spectra of the MIP and NIP, peaks for several groups remained, but no peak was observed in the area around 1630–1640 cm−1, which indicates the absence of the C
C stretching of the vinyl functional group. The FTIR spectra of EGDMA and TRIM show peaks at similar positions. In the spectra of the MIP and NIP, peaks for several groups remained, but no peak was observed in the area around 1630–1640 cm−1, which indicates the absence of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C from the vinyl group. The absence of this peak indicates that the polymerization process has run perfectly.57
C from the vinyl group. The absence of this peak indicates that the polymerization process has run perfectly.57
Fig. 4b presents a comparison of the MIP after and before soxhlation and the NIP. In general, the MIP before soxhlation, MIP after soxhlation, and NIP show similar spectra, but for the peak around 1700 cm−1, which is the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O band, there is a difference in intensity for the MIP before soxhlation and MIP after soxhlation, where the intensity for the MIP before soxhlation is lower than that for MIP after soxhlation. A plausible reason for this phenomenon is that the template molecule (EPMC) is assembled with the monomer (HEMA) via hydrogen bonding with C
O band, there is a difference in intensity for the MIP before soxhlation and MIP after soxhlation, where the intensity for the MIP before soxhlation is lower than that for MIP after soxhlation. A plausible reason for this phenomenon is that the template molecule (EPMC) is assembled with the monomer (HEMA) via hydrogen bonding with C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O during the preparation of the MIP prior to washing.
O during the preparation of the MIP prior to washing.
The particle size measurements in Table 4 show that all the MIPs have smaller particle sizes than the NIPs. MIP 2 has the smallest particle size (0.782 μm) compared to the other polymers. Smaller particle sizes lead to higher surface areas. This increased surface area will provide more binding sites for the template molecule, leading to increased adsorption capacity.58
| Polymer | Average particle size (μm) |
|---|---|
| MIP 1 | 0.828 |
| NIP 1 | 1.038 |
| MIP 2 | 0.782 |
| NIP 2 | 1.128 |
| MIP 3 | 1.021 |
| NIP 3 | 1.488 |
| MIP 4 | 0.815 |
| NIP 4 | 0.978 |
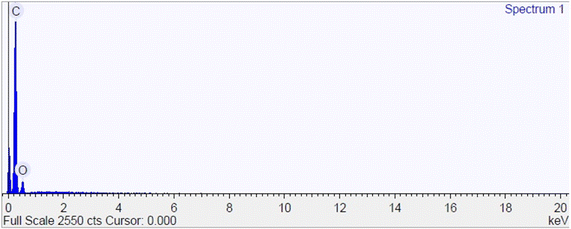
Quantitative energy dispersive X-ray (EDS) analysis was performed to observe the main atomic components that make up the polymers, such as carbon (C) and oxygen (O). Fig. 5 shows that the atomic components in MIP 2 are carbon and oxygen, with amounts of 83.49% and 16.51%, respectively. EDS for other polymers can be seen in ESI Fig. S6.†59
The SEM results in Fig. 6 show that all the polymer morphologies are spherical and non-uniform, with particle sizes of around 500–900 nm. From the SEM results, it can also be concluded that the MIP and NIP polymers with the TRIM crosslinker (Fig. 6a–d) produce larger and more uniform particle sizes than the MIP and NIP polymers with the EGDMA crosslinker (Fig. 6e–h). TRIM has a three-branched chain containing three vinyl groups that can participate in polymer formation, while EGDMA has two vinyl groups. Therefore, the TRIM crosslinker will produce polymers with larger particle sizes and greater rigidity compared to EGDMA.60
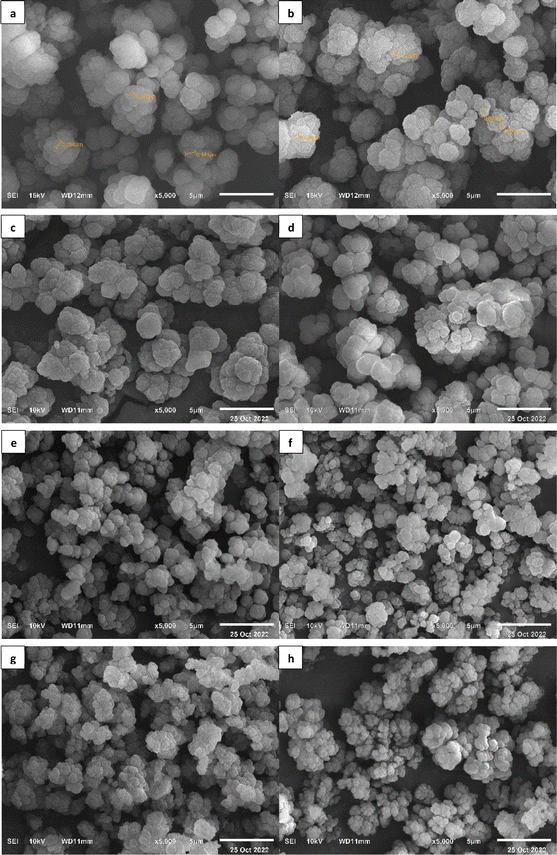 | ||
| Fig. 6 Polymer morphology using SEM at 5000× magnification: (a) MIP 1; (b) NIP 1; (c) MIP 2; (d) NIP 2; (e) MIP 3; (f) NIP 3; (g) MIP 4; (h) NIP 4. | ||
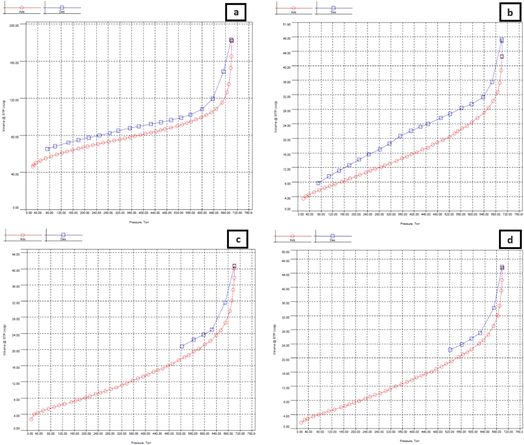
The BET instrument test results provided a quantitative isothermic data curve, which was obtained by plotting the total volume of nitrogen gas adsorbed and desorbed against pressure. Based on the IUPAC classification, the obtained polymer isothermic adsorption curve belongs to the type-V category.38 Type-V isotherms represent adsorption on mesoporous surfaces, which has a tendency to occur through the formation of multiple layers followed by capillary condensation. The type-V isotherm results from low interaction between the adsorbent and the adsorbate, which causes the absorption of the adsorbate to be small at low pressure and results in an increase in exchange with the rise in relative pressure. The curves can be seen in Fig. 7. A type-V isothermic adsorption curve is also usually shown by a mesoporous adsorbent solid (2–50 nm).38,61 In MIP 3 and MIP 4, desorption does not start from zero. This shows that a certain amount of adsorbate remains in the adsorbent, even at low pressure. This could be because the adsorbate may have a strong affinity for the adsorbent, which makes it difficult for the adsorbate to desorb, so desorption does not occur at low pressure.62
3.6. Adsorption capacity evaluation
The adsorption capacity is used to determine the mechanism of the rebinding interactions between template molecules and polymers. The adsorption capacity test was determined using an adsorption isotherm model that describes the characteristics of the MIP and calculates the relationship between the binding parameters and the affinity distribution.63,64 Isotherm adsorption analysis was carried out using the Freundlich and Langmuir isotherm models. The adsorption capacity results show that the tested MIP and NIP polymers are better fitted by the Freundlich isotherm model because the linearity value (R2) of the Freundlich isotherm model is greater than that of the Langmuir isotherm model.63 The Freundlich isotherm model shows that adsorption occurs on the surface of a heterogeneous multilayer with a weak bond affinity.65,66The Freundlich adsorption model has two parameters: 1/n and KF. The KF value indicates the affinity of the sorbent; the higher the affinity obtained, the greater the capacity of the sorbent to adsorb the target analyte.33 The 1/n value describes the degree of bond heterogeneity (homogeneity index) with a value between 0 and 1.66 A value of 1/n close to 1 indicates that the adsorption occurs homogeneously, while a value approaching 0 means that the adsorption occurs heterogeneously.67 A value of 1/n greater than 1 (as observed for NIP 3, MIP 3, and MIP 4) indicates a value of n < 1. This can be interpreted as meaning that adsorption energies are exponentially distributed. When n < 1, the Freundlich isotherm indicates a greater proportion of sorption sites with lower energies than those with higher energies. This leads to a decreasing rate of adsorption with increasing solute concentration.68,69
Table 5 shows that the MIP 2 polymer had a higher bond affinity (KF) value of 0.0813 compared to MIP 1, MIP 3, and MIP 4. The Kruskal–Wallis test was carried out to determine whether MIP 1, MIP 2, MIP 3, and MIP 4 affect the adsorption capacity. The Kruskal–Wallis test revealed that there is a significant difference between MIP 1, MIP 2, MIP 3, and MIP 4 on the dependent variable, adsorption capacity, with a p-value of 0.036 (α = 0.05). Thus, with the available data, the null hypothesis is rejected.
| Polymer | Freundlich | Langmuir | ||||
|---|---|---|---|---|---|---|
| R2 | 1/n | KF (mg g−1) | R2 | KL | Qm (mg g−1) | |
| MIP 1 | 0.9967 | 0.9702 | 0.0637 | 0.9996 | 0.8360 | 0.0923 |
| NIP 1 | 0.9701 | 0.3212 | 0.0545 | 0.8773 | 0.9070 | 0.0851 |
| MIP 2 | 0.9965 | 0.6367 | 0.0813 | 0.6796 | 0.5831 | 0.2198 |
| NIP 2 | 0.9762 | 0.9830 | 0.0400 | 0.1076 | 0.2751 | 0.4659 |
| MIP 3 | 0.9933 | 1.3314 | 0.0260 | 0.9520 | −0.4784 | −0.0297 |
| NIP 3 | 0.7722 | 1.9227 | 0.0035 | 0.0110 | 1.2972 | 0.0110 |
| MIP 4 | 0.9941 | 0.9394 | 0.0247 | 0.7004 | 0.1211 | 0.2314 |
| NIP 4 | 0.9816 | 1.4737 | 0.0192 | 0.5386 | 0.4786 | 0.0586 |
Furthermore, the bond affinity value of MIP 2 was higher than that of NIP 2 (0.0400). A Wilcoxon signed-rank test showed that MIP 2 and NIP 2 had statistically significant differences in adsorption capacity, with a p-value of 0.028 (α = 0.05). Therefore, MIP 2 was selected as the sorbent to be packaged in an SPE cartridge and applied for EPMC extraction from Kaempferia galanga L. rhizome extract. The Freundlich and Langmuir isotherms are presented in Fig. S7 in the ESI.†
3.7. Selectivity evaluation
The MIP selectivity was obtained from the calculation of the imprinting factor (IF), as mentioned in Section 2.10. KD is the ratio of the amount of adsorbed analyte to the concentration of the analyte in the solution, while the IF describes the quality of the binding sites formed on the MIP sorbent.67 The higher the IF value, the greater the difference between the analytes adsorbed on MIP and NIP. However, this high IF value does not prove conclusively that the template site formed is truly selective for the template compound.70 Therefore, the selectivity was determined by comparing the KD values of EPMC and analogous compounds, including ethyl cinnamate (EC), cinnamaldehyde (CD), and kaempferol (KF). These three compounds are also present in the Kaempferia galanga L. rhizome extract.18 The differences in the chemical structures of these three compounds can be seen in Fig. 8.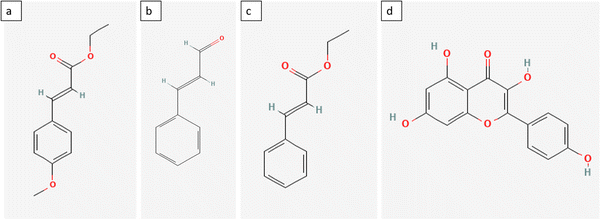 | ||
| Fig. 8 Chemical structure of compounds: (a) EPMC; (b) ethyl cinnamate; (c) cinnamaldehyde; (d) kaempferol. | ||
Based on the results of the study, MIP 2 gave the highest KD value of 8.821 compared to other MIPs, while NIP 2 gave a KD value of 3.138; thus, the EPMC imprinting factor on MIP 2 was 2.811. Therefore, the value of the MIP 2 imprinting factor proves that the MIP 2 polymer is more selective toward NIP than other MIP polymers. This is also supported by the particle size analysis results, which show that MIP 2 has a smaller particle size than MIP 1, 3, and 4. The smaller the particle size, the more binding sites there are, so the IF value will be greater. The KD value for each polymer can be seen in Table 6.
| Polymer | KD MIP | KD NIP | IF |
|---|---|---|---|
| MIP–NIP 1 | 3.440 | 7.976 | 0.431 |
| MIP–NIP 2 | 8.821 | 3.138 | 2.811 |
| MIP–NIP 3 | 1.090 | 1.925 | 0.566 |
| MIP–NIP 4 | 6.825 | 5.374 | 1.270 |
In addition to the imprinting factor, the value of the selectivity coefficient (k) also affects the selectivity of the MIP. The selectivity coefficient was obtained by comparing the KD of the EPMC template compound with the KD of other analogous or interfering compounds.70,71 The values of the selectivity coefficients can be seen in Table 7.
| Polymer | k (selectivity coefficient) | ||
|---|---|---|---|
| EC | CD | Kaempferol | |
| MIP 1 | 1.906 | 0.336 | N/A |
| MIP 2 | 17.436 | 1.539 | 0.061 |
| MIP 3 | 7.526 | 54.548 | 0.009 |
| MIP 4 | 0.536 | 3.659 | 0.092 |
A greater selectivity coefficient value indicates more significant ability of the MIP polymer to distinguish the EPMC template compound from its analog compounds.70,72 If the value of the selectivity coefficient is equal to 1, it indicates no difference in recognition or binding ability between the template and its analogous compounds. The Kruskal–Wallis test was used to determine whether MIP 1, MIP 2, MIP 3, and MIP 4 exhibit significant differences in terms of the selectivity coefficient for each analogue compound. The Kruskal–Wallis test revealed that significant differences between MIP 1, MIP 2, MIP 3, and MIP 4 in terms of the selectivity coefficient of the dependent variable, with p-values of 0.12, 0.12, and 0.18 (α = 0.05) for EC, CD, and kaempferol, respectively. Thus, with the available data, the null hypothesis is rejected. The obtained results demonstrated that MIP 2 could significantly distinguish EPMC from EC with a selectivity coefficient value of 17.436, followed by cinnamaldehyde with a value of 1.539 and kaempferol at 0.061.
As can be seen in Fig. 8, ethyl cinnamate and cinnamaldehyde have similar structures to EPMC with differences in several functional groups, namely the methoxy group in cinnamaldehyde and the ethoxy group in the ethyl cinnamate. This may allow EC and CD to enter the pores more easily and compete with EPMC for binding sites. However, the lack of active groups on EC causes EC to be less competitive than EPMC. On the other hand, kaempferol has a very different structure than EPMC, but this compound is also present in Kaempferia galanga extract. The more rigid structure of kaempferol with its three benzene rings may cause steric hindrance on the polymer surface. This can also be seen from the EPMC recovery results on NI-SPE2 spiked with kaempferol (Section 3.8.)
3.8. Application to real samples
The percentage recovery for EPMC from ethyl acetate extract was 68.05% ± 2.22 for MIP 2 and 50.55% ± 1.47 for the NIP with an IF of 1.35; the percentage recovery of EPMC from n-hexane extract was 65.27% ± 3.78 for the MIP and 49.44% ± 3.74 for the NIP, with an IF of 1.32; and the percentage recovery of EPMC from ethanol extract was 82.40% ± 5.52 for the MIP and 62.92% ± 2.49 for the NIP, with an IF value of 1.31. The IF values were obtained by comparing the amount or percentage of the analyte that binds to the MIP with the amount or percentage of the analyte that binds to the NIP. An IF value greater than 1 indicates that the MIP has better imprinting sites than the NIP.65 This is also confirmed by the results of the adsorption capacity evaluation, which show that the of adsorption capacity MIP 2 is twice that of NIP 2, indicating that there are more imprinting sites on MIP 2. Based on the obtained results, the three extract samples demonstrated better analytical performance of MIP 2 than NIP 2. The analytical performance for EPMC using MIP 2 and the ethanol extract showed the highest percentage recovery, even though the EPMC content in the ethanol extract was smaller compared to those in the ethyl acetate and n-hexane extracts; however, all three extracts give similar IF values.
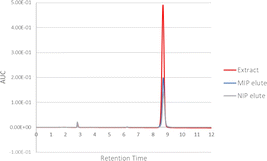
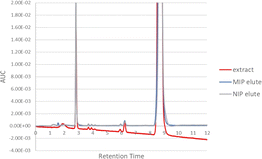
MIPs are known to be able to adsorb the target compound in at least in the same amounts as NIPs, so the NIP can be used as a comparison of the minimum recovery that the MIP can obtain.73 The NIP sorbent can also adsorb the EPMC template molecule because it is also composed of monomers and crosslinkers, which can provide non-specific interactions between the test sample passed through the NIP sorbent through non-covalent bonds such as hydrogen bonds and hydrophobic bonds. Adding a template to the MIP composition provides a cavity based on the template molecule, and thus the MIP sorbent will provide better recovery than the NIP due to the specific interaction between the EPMC and its particular cavity.74 From the chromatogram it can be seen that the extract elution results from the MIP contain more EPMC than those from the NIP. A visualization of the comparison of the EPMC peak chromatograms in the ethanol extracts, MIP elute, and NIP elute can be seen in Fig. 10.
The percentage purity of the SPE elution results of the Kaempferia galanga L. extract can be determined by comparing the AUC peak of EPMC with the total AUC peak that appears on the chromatogram. As can be seen in Fig. 11, other peaks appear at around 2.8 and 6.3 minutes. The percentage purity results for the SPE of the ethyl acetate extract are 97.63% ± 1.03% for MIP 2 and 96.90% ± 1.50% for the NIP; the percentage purity for the n-hexane extract is 99.59% ± 0.10% for the MIP and 99.42% ± 0.18% for the NIP; and the percentage purity for the ethanol extract is 97.00% ± 1.39% for the MIP and 95.98% ± 1.72% for the NIP.
The n-hexane extract has the highest purity value compared to the other extracts. Based on the figure above, it can be seen that the elutes resulting from the extraction process using MI-SPE have fewer impurity peaks compared to the extract samples. This shows that the sample extraction process using the MI-SPE technique can purify the extract sample to produce the template compound in high purity.71
Previously, other researchers had isolated EPMC using the column chromatography method. Chanvimalueng obtained an EPMC yield of 27.78% using four isolation stages.26 Based on the data above, it can be concluded that the EPMC isolation method using MI-SPE with two isolation stages can increase the EPMC yield compared to previous isolation methods.
| Compound | EPMC-, EC-, CD-, and KF-spiked extract | KF-spiked extract | EC-spiked extract | CD-spiked extract | ||||
|---|---|---|---|---|---|---|---|---|
| MI-SPE 2 | NI-SPE 2 | MI-SPE 2 | NI-SPE 2 | MI-SPE 2 | NI-SPE 2 | MI-SPE 2 | NI-SPE 2 | |
| a Abbreviations: CD: cinnamaldehyde; EC: ethyl cinnamate; EPMC: ethyl p-methoxycinnamate; KF: kaempferol. | ||||||||
| CD | 44.50 | 37.36 | — | — | — | — | 94.61 | 82.64 |
| EC | 27.62 | 26.77 | — | — | 13.27 | 16.07 | — | — |
| EPMC | 31.53 | 28.74 | 26.87 | 17.77 | 81.91 | 63.99 | 84.21 | 61.40 |
| KF | 72.87 | 52.90 | 44.15 | 34.10 | — | — | — | — |
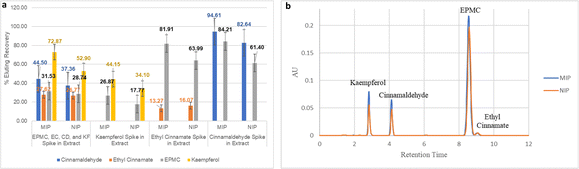 | ||
| Fig. 12 (a) % elution recovery of EPMC, EC, CD, and KF in kencur hexane extract (n = 3). (b) Chromatogram of elute from selectivity testing using MI-SPE and NI-SPE. | ||
The results show that the percent elution recovery value for EPMC from the sample solutions in the form of an extract and spiked with the four compounds is higher than that for EC, namely 31.53% for EPMC and 27.62% for EC, with a selectivity value of 1.14. On the other hand, the IF value for EPMC decreases from 1.3 to 1.1. However, there is a considerable difference in the results of the SPE sample solution which is spiked with only EC. This can be seen from the EPMC percent elution recovery value, which increased to 81.91% (IF 1.3) and EC percent elution recovery, which decreased to 13.27%, with a selectivity value of 6.17. A similar phenomenon occurred with the EPMC elution recovery in NI-SPE 2, which increased from 28.74% to 63.99%, and the EC elution recovery decreased from 26.77% to 16.07%. However, the IF EC value in the solution with four compounds (IF 1.03) and two compounds (0.83) does not have an imprinted site, as seen from the IF value of ≤1. From these data, it can be concluded that MI-SPE 2 is more selective for EPMC compared to EC. However, the presence of other compounds such as CD and KF can reduce the rebinding affinity of EPMC to MI-SPE 2. This can be observed from the 48% decrease in EPMC recovery on the MIP, from 65.27% to 31.53%, and the 58% decrease on the NIP, from 49.44% to 28.74%, in the extract spiked with the three analogue compounds. Since templates and interferents typically compete for the binding site,70 it can be assumed that this happens because CD and KF are more competitive for binding to MI-SPE 2. This can also be observed from the spiking test data with only CD or KF in the extract solution, as the % elution recovery for CD and KF are still higher than that of EPMC. Moreover, in the KF-spiked extract, there was also a decrease in the EPMC recovery for both the MIP and the NIP, namely 41% and 36%, respectively. The decrease in recovery that also occurs in the NIP can also be assumed to mean that KF covers the binding sites on the polymer surface.
Despite the relatively good results of MIP 2 for the separation of EPMC from Kaempferia galanga extract, this study has several limitations. For instance, we did not optimize the amount of crosslinker or the number or types of initiators used. We only examined the use of two different solvents as porogens without optimizing the solvent volume.
4. Conclusion
In this study, a novel molecularly imprinted polymer was prepared for EPMC using methacrylic acid as a functional monomer through bulk polymerization. The MIPs obtained were characterized using FTIR, SEM, and BET methods. According to the evaluation results, the MIP synthesized using TRIM as a crosslinker exhibited better adsorption capacity and selectivity values than that obtained using EGDMA. Moreover, these MIPs were applied as a sorbent combined with the SPE separation technique to isolate EPMC from Kaempferia galanga L. rhizome extract. A fairly good yield was obtained for the isolation of EPMC with a high purity value. This MI-SPE method is believed to be a novel and promising method for EPMC isolation from the extract. However, the analytical performance of the MIPs could be improved by screening functional monomers other than methacrylic acid derivatives. In addition, other polymerization methods to produce MIPs and NIPs could also be attempted in order to obtain MIP polymers with better adsorption capacity and selectivity.Author contributions
M. D. A. – data curation, investigation, visualization, writing – original draft; A. Z. – conceptualization, methodology, supervision; P. M. – conceptualization, methodology, supervision; A. N. H. – conceptualization, methodology, supervision, funding acquisition, writing – review & editing.Conflicts of interest
There are no conflicts to be declare.Acknowledgements
Directorate of Research and Community Engagement Universitas Padjadjaran for Research Funding through Beasiswa Unggulan Pascasarjana Padjadjaran (BUPP) 2021–2022.References
- J. Techaprasan, S. Klinbunga, C. Ngamriabsakul and T. Jenjittikul, Genet. Mol. Res., 2010, 9, 1957–1973 CrossRef CAS PubMed.
- The Plant List, A Working List of All Plant Species, https://www.theplantlist.org/ Search PubMed.
- Eisai, Medicinal Herb Index in Indonesia, P.T. Eisai Indonesia, Jakarta, 1st edn, 1986 Search PubMed.
- D. Subositi, N. Kurnianingrum, R. Mujahid and Y. Widiyastuti, Agrivita, 2020, 42, 45–52 Search PubMed.
- M. I. Umar, M. Z. Bin Asmawi, A. Sadikun, R. Altaf and M. A. Iqbal, Afr. J. Pharm. Pharmacol., 2011, 5, 1638–1647 CrossRef CAS.
- W. Ridtitid, C. Sae-wong, W. Reanmongkol and M. Wongnawa, J. Ethnopharmacol., 2008, 118, 225–230 CrossRef PubMed.
- D. Kanjanapothi, A. Panthong, N. Lertprasertsuke, T. Taesotikul, C. Rujjanawate, D. Kaewpinit, R. Sudthayakorn, W. Choochote, U. Chaithong, A. Jitpakdi and B. Pitasawat, J. Ethnopharmacol., 2004, 90, 359–365 CrossRef CAS PubMed.
- D. Lakshmanan, J. Werngren, L. Jose, K. P. Suja, M. S. Nair, R. L. Varma, S. Mundayoor, S. Hoffner and R. A. Kumar, Fitoterapia, 2011, 82, 757–761 CrossRef CAS PubMed.
- N. Rao and D. Kaladhar, World J. Pharm. Pharm. Sci., 2014, 3, 1180–1189 Search PubMed.
- I. H. Choi, J. Y. Park, S. C. Shin and I. K. Park, Nematology, 2006, 8, 359–365 Search PubMed.
- T. K. Hong, J. K. Lee, J. W. Heo, S. Il Kim, D. R. Choi and Y. J. Ahn, Nematology, 2010, 12, 775–782 Search PubMed.
- T. K. Hong, S. Il Kim, J. W. Heo, J. K. Lee, D. R. Choi and Y. J. Ahn, Nematology, 2011, 13, 235–244 CAS.
- R. Othman, H. Ibrahim, M. A. Mohd, K. Awang, A. U. H. Gilani and M. R. Mustafa, Planta Med., 2002, 68, 655–657 CrossRef CAS PubMed.
- R. Othman, H. Ibrahim, M. A. Mohd, M. R. Mustafa and K. Awang, Phytomedicine, 2006, 13, 61–66 CrossRef CAS PubMed.
- S. Tara V, S. Chandrakala, A. Sachidananda, B. L. Kurady, S. Smita and S. Ganesh, Indian J. Physiol. Pharmacol., 2006, 50, 384–390 Search PubMed.
- R. Mitra, J. Orbell and M. S. Muralitharan, Asia-Pac. BioTech News, 2007, 11, 105–110 CrossRef.
- H. L. Koh, T. K. Chua and C. H. Tan, A Guide to Medicinal Plants – An Illustrated, Scientific and Medicinal Approach, World Scientific Publishing Co. Pte. Ltd, 2010 Search PubMed.
- A. Kumar, Int. J. Pharma Bio Sci., 2014, 5, 225–231 CAS.
- A. P. Raina and Z. Abraham, J. Essent. Oil Res., 2016, 28, 29–34 CrossRef CAS.
- I. Komala, S. Nurhasni, O. S. Betha, E. Putri, S. Mufidah, M. F. Awaludin, M. Fahmi, M. Reza and N. P. Indriyani, Indones. J. Chem., 2018, 18, 60–65 CrossRef CAS.
- N. J. Kim, S. G. Byun, J. E. Cho, K. Chung and Y. J. Ahn, Pest Manage. Sci., 2008, 64, 857–862 CrossRef CAS PubMed.
- B. Liu, F. Liu, C. Chen and H. Gao, Nat. Prod. Res., 2010, 24, 1927–1932 CrossRef CAS PubMed.
- B. Elya, I. M. Kusuma, M. Jufri and R. Handayani, Res. J. Med. Plant, 2016, 10, 426–434 CrossRef CAS.
- N. S. S. Ambarwati, B. Elya, A. Malik and M. Hanafi, J. Young Pharm., 2017, 9, S56–S59 CrossRef CAS.
- A. Hakim, Y. Andayani and B. Deana Rahayuan, J. Phys.: Conf. Ser., 2018, 1095, 4–7 CrossRef.
- W. Chanvimalueng, A. Itharat, P. Thongdeeying, W. Pipatrattanaseree, K. Phumlek, S. Naknarin and N. Mukkasombut, Sci. Technol. Asia, 2022, 27, 134–143 Search PubMed.
- W. Peng-Ju, Y. Jun, S. Qing-De, G. Yun, Z. Xiao-Lan and C. Ji-Bao, Chin. J. Anal. Chem., 2007, 35, 484–488 Search PubMed.
- Q. Zhu, D. Huang, L. Li and Y. Yin, Sci. China: Chem., 2010, 53, 2587–2592 CrossRef CAS.
- T. Renkecz, K. László and V. Horváth, Mol. Imprinting, 2014, 2, 1–17 CAS.
- W. Ji, M. Zhang, H. Yan, H. Zhao, Y. Mu, L. Guo and X. Wang, Anal. Bioanal. Chem., 2017, 409, 7087–7096 CrossRef CAS PubMed.
- E. M. Saad, N. A. El Gohary, M. Abdel-Halim, H. Handoussa, R. Mohamed El Nashar and B. Mizaikoff, Molecularly Imprinted Polymers for Selective Extraction of Rosmarinic Acid from Rosmarinus officinalis L., Elsevier Ltd, 2021, vol. 335 Search PubMed.
- N. Srivastava, S. Mishra, H. Iqbal, D. Chanda and K. Shanker, J. Ethnopharmacol., 2021, 271, 113911 CrossRef CAS PubMed.
- A. N. Hasanah, T. N. D. Utari and R. Pratiwi, J. Anal. Methods Chem., 2019, 2019, 1–7 CrossRef PubMed.
- S. Suryana, M. Mutakin, Y. Rosandi and A. N. Hasanah, Polym. Adv. Technol., 2021, 1–14 Search PubMed.
- K. M. Wright, M. C. Bowyer, A. McCluskey and C. I. Holdsworth, Int. J. Mol. Sci., 2022, 24, 5117–5136 CrossRef PubMed.
- P. G. Arias, H. Martínez-Pérez-Cejuela, A. Combès, V. Pichon, E. Pereira, J. M. Herrero-Martínez and M. Bravo, J. Chromatogr. A, 2020, 1626, 461346 CrossRef CAS PubMed.
- Y. Wan, M. Wang, Q. Fu, L. Wang, D. Wang, K. Zhang, Z. Xia and D. Gao, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2018, 1097–1098, 1–9 CrossRef CAS PubMed.
- M. Thommes, K. Kaneko, A. V. Neimark, J. P. Olivier, F. Rodriguez-Reinoso, J. Rouquerol and K. S. W. Sing, Pure Appl. Chem., 2015, 87, 1051–1069 CrossRef CAS.
- A. Mehdinia, M. Ahmadifar, M. O. Aziz-Zanjani, A. Jabbari and M. S. Hashtroudi, Analyst, 2013, 20, 1345–1352 Search PubMed.
- Y. K. Lv, X. H. Liu, S. L. Yan, Y. Zhang and H. W. Sun, J. Porous Mater., 2013, 20, 1345–1352 CrossRef CAS.
- N. Li, T. B. Ng, J. H. Wong, J. X. Qiao, Y. N. Zhang, R. Zhou, R. R. Chen and F. Liu, Food Chem., 2013, 139, 1161–1167 CrossRef CAS PubMed.
- L. P. Dwita, N. P. E. Hikmawanti, Yeni and Supandi, J. Tradit. Complementary Med., 2021, 11, 563–569 CrossRef CAS PubMed.
- M. I. Umar, M. Z. Asmawi, A. Sadikun, I. J. Atangwho, M. F. Yam, R. Altaf and A. Ahmed, Molecules, 2012, 17, 8720–8734 CrossRef CAS PubMed.
- S. N. N. S. Hashim, R. I. Boysen, L. J. Schwarz, B. Danylec and M. T. W. Hearn, J. Chromatogr. A, 2014, 1359, 35–43 CrossRef CAS PubMed.
- K. F. Lim and C. I. Holdsworth, Molecules, 2018, 23, 1–19 Search PubMed.
- P. Thordarson, Chem. Soc. Rev., 2011, 40, 1305–1323 RSC.
- A. Korytkowska-Walach, Polym. Bull., 2013, 70, 1647–1657 CrossRef CAS.
- A. N. Hasanah, D. Fauzi, B. Z. Witka, D. Rahayu and R. Pratiwi, Mediterr. J. Chem., 2020, 10, 277–288 CrossRef CAS.
- S. M. Hoenigman and C. E. Evans, Anal. Chem., 1996, 68, 3274–3276 CrossRef CAS.
- X. Fu, Q. Yang, Q. Zhou, Q. Lin and C. Wang, Open J. Org. Polym. Mater., 2015, 05, 58–68 CrossRef CAS.
- R. Khattak and I. I. Naqvi, J. Res. (Sci.), 2007, 18, 219–235 Search PubMed.
- M. M. Qronfla, B. Jamoussi and R. Chakroun, Polymers, 2023, 15, 2398–2419 CrossRef CAS PubMed.
- C. Alvarez-lorenzo, Handbook of Molecularly Imprinted Polymers Handbook of Molecularly, Smithers Rapra Technology Ltd, Shawbury, 2013 Search PubMed.
- A. S. Carreira, R. F. A. Teixeira, A. Beirão, R. Vaz Vieira, M. M. Figueiredo and M. H. Gil, Eur. Polym. J., 2017, 93, 33–43 CrossRef CAS.
- H. Zhang, H. Song, X. Tian, Y. Wang, Y. Hao, W. Wang, R. Gao, W. Yang, Y. S. Ke and Y. Tang, Microchim. Acta, 2021, 188, 17 CrossRef CAS PubMed.
- A. N. Hasanah, G. Andriana, R. Pratiwi and S. Megantara, Eurasian J. Anal. Chem., 2018, 13, 36–43 CAS.
- R. Fernández-Cori, J. C. Morales Gomero, B. C. Huayhuas-Chipana, M. d. P. Taboada Sotomayor and J. G. Ruiz Montoya, ECS Trans., 2015, 66, 33–41 CrossRef.
- Z. Wang, T. Qiu, L. Guo, J. Ye, L. He and X. Li, React. Funct. Polym., 2018, 126, 1–8 CrossRef CAS.
- S. Bakhtiar, S. A. Bhawani and S. R. Shafqat, Chem. Biol. Technol. Agric., 2019, 6, 1–10 CrossRef CAS.
- M. H. Loghmani, A. F. Shojaie and S. A. Hosseini, J. Ind. Eng. Chem., 2021, 96, 98–108 CrossRef CAS.
- Z. A. Alothman, Materials, 2012, 5, 2874–2902 CrossRef CAS.
- F. Mikšík, T. Miyazaki and K. Thu, Energies, 2020, 13, 4247–4277 CrossRef.
- Y. Cui, Z. He, Y. Xu, Y. Su, L. Ding and Y. Li, Chem. Eng. J., 2021, 405, 126608 CrossRef CAS.
- A. N. Hasanah, M. Suherman, I. Susanti, I. Pitaloka and R. Mustarichie, Rasayan J. Chem., 2019, 12, 1269–1278 CrossRef CAS.
- S. Amin, S. Damayanti and S. Ibrahim, Jurnal Ilmu Kefarmasian Indonesia, 2018, 16, 12–19 CrossRef.
- L. Aljerf, J. Environ. Manage., 2018, 225, 120–132 CrossRef CAS PubMed.
- M. A. Décima, S. Marzeddu, M. Barchiesi, C. Di Marcantonio, A. Chiavola and M. R. Boni, Sustainability, 2021, 13, 11760–11809 CrossRef.
- K. V. Kumar, S. Gadipelli, B. Wood, K. A. Ramisetty, A. A. Stewart, C. A. Howard, D. J. L. Brett and F. Rodriguez-Reinoso, J. Mater. Chem. A, 2019, 7, 10104–10137 RSC.
- Y. Keren, M. Borisover and N. Bukhanovsky, Chemosphere, 2015, 138, 462–468 CrossRef CAS PubMed.
- G. Becskereki, G. Horvai and B. Tóth, Polymers, 2020, 13, 1781–1797 CrossRef PubMed.
- S. Zahara, M. A. Minhas, H. Shaikh, M. S. Ali, M. I. Bhanger and M. I. Malik, React. Funct. Polym., 2021, 166, 104984 CrossRef CAS.
- Q. Luo, N. Yu, C. Shi, X. Wang and J. Wu, Talanta, 2016, 161, 797–803 CrossRef CAS PubMed.
- Z. Dorkó, A. Szakolczai, T. Verbic and G. Horvai, J. Sep. Sci., 2015, 38, 4240–4247 CrossRef PubMed.
- N. A. Yusof, S. K. A. Rahman, M. Z. Hussein and N. A. Ibrahim, Polymers, 2013, 5, 1215–1228 CrossRef.
- A. Kumar, J. Ethnopharmacol., 2020, 253, 112667 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra01018c |
| This journal is © The Royal Society of Chemistry 2024 |