 Open Access Article
Open Access ArticleIdentification, screening and taste mechanisms analysis of two novel umami pentapeptides derived from the myosin heavy chain of Atlantic cod (Gadus morhua)†
Xu Zhao‡
a,
Wenpei Qiu‡a,
Xian-guang Shaoab,
Baifeng Fuc,
Xinyu Qiaoc,
Zhen Yuanc,
Meilian Yangc,
Pan Liu cd,
Ming Duc and
Maolin Tu
cd,
Ming Duc and
Maolin Tu *abc
*abc
aKey Laboratory of Animal Protein Food Deep Processing Technology of Zhejiang Province, Zhejiang-Malaysia Joint Research Laboratory for Agricultural Product Processing and Nutrition, College of Food Science and Engineering, Ningbo University, Ningbo 315832, China
bState Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Ningbo University, Ningbo, Zhejiang 315211, China
cSchool of Food Science and Technology, National Engineering Research Center of Seafood, Collaborative Innovation Center of Seafood Deep Processing, Dalian Polytechnic University, Dalian, Liaoning 116034, China
dCollege of Modern Agriculture, Neijiang Vocational & Technical College, Neijiang, Sichuan 641100, China
First published on 27th March 2024
Abstract
Umami peptides are new ingredients for the condiment and seasoning industries, with healthy and nutrition characteristics, some of which were identified from aquatic proteins. This study aims to further explore novel umami peptides from Atlantic cod (Gadus morhua) by combining in silico, nano-HPLC-MS/MS, sensory evaluation, and electronic tongue analysis. Two novel peptides, Leu-Val-Asp-Lys-Leu (LVDKL) and Glu-Ser-Lys-Ile-Leu (ESKIL), from the myosin heavy chain of Atlantic cod (Gadus morhua), were screened and confirmed to have strong umami tastes with the thresholds of 0.427 mM and 0.574 mM, respectively. The molecular docking was adopted to explore the interactions between the umami peptides and the umami taste receptor T1R1/T1R3, which showed that the umami peptides interacted with T1R1/T1R3 mainly by electrostatic interaction, hydrogen bond interaction, and hydrophobic interaction. Furthermore, the physicochemical properties of the peptides were investigated by in silico methods and cell viability experiments. This study will provide a better understanding of the umami taste in Atlantic cod and will promote the development of condiments and seasonings.
1. Introduction
Exploring the “blue food” is promising for supplying nutrition and flavor to humans. Atlantic cod (Gadus morhua), as one source of blue food, belongs to the Gadidae (Cod) family and is a type of fish well-known around the world to have great nutritional value and a delicious taste. The umami taste is recognized as the fifth taste, which can provide a pleasant sensory experience and is a special character of Atlantic cod. Some studies showed that umami substances can be used as a substitute for salt, which may be able to help people reduce their salt intake thereby decreasing the risk of people suffering from non-communicable diseases (NCD), such as cardiovascular disease, chronic kidney disease, and stomach cancer.1,2 The substances that can exert umami flavor contain nucleotides, free amino acids, peptides, organic acid, and their derivatives,3 and among which umami peptides have attracted a great deal of interest from food-related scientists and entrepreneurs as a consequence of their natural and valuable features.Compared with the widely used umami taste food additives, such as monosodium glutamate solution (MSG), umami peptides have the merits of having umami-enhancing effect on MSG and sodium chloride (NaCl) solution,4 with good nutrition value and bioactivity, as well as easily being metabolized by the human body. So far, over 400 umami peptides have been identified from diverse sources, such as chicken soup, beef soup, mushrooms, fish, beans, and oysters, most of which have been fully analyzed in terms of the relationships between the umami intensity and their primary structures.5 For example, the peptide PPQEAAQF reported by Zhang et al.6 screened from chicken soup by combining molecular docking and python script invocation, has the threshold of 0.12 mM. The umami peptide VEAL separated and identified from soy sauce was reported that it can enhance the umami taste of soy sauce,7 and the umami peptide HLQLAIR from Takifugu rubripes was analyzed and had the umami threshold of 0.24 mM.4 Moreover, several umami peptides were also identified from the Atlantic cod myosin protein by in silico hydrolysis, such as the peptides INKPEL,8 GGR, AGCD, and SGDAW.9 Although there are already many umami peptides that have been identified from various food raw materials, there is still some dispute about the taste of umami peptides,4,10 more umami peptides still need to be identified for better analyzing their taste mechanisms and structure characteristics. Myosin is the main protein found in muscle,11 which consists of heavy chains and light chains. Due to the special function and the abundant content in fish, many studies have been conducted on myosin.12
To screen the umami peptide conveniently and highly efficiently, increasing bioinformatics means appeared and have been used in some studies. Such as the iUmami-SCM web server (https://camt.pythonanywhere.com/iUmami-SCM), which is a predictor with the ability to predict the umami taste of peptides according to the unique sequence of a peptide.13 The Expasy-Compute pI/MW (https://web.expasy.org/compute_pi/) is able to evaluate the molecular weight (MW) and iso-electric point (pI) of the peptides. Meanwhile, the ProtParam (https://web.expasy.org/protparam/) and ToxinPred (http://crdd.osdd.net/raghava/toxinpred/) can be used to predict the instability index and the toxicity of peptides, respectively. Moreover, the water solubility of the peptide can be predicted by the Innovagen website (https://www.innovagen.com). Pepdraw (https://www.tulane.edu/%7Ebiochem/WW/PepDraw/) is with the function of predicting the net charge and hydrophobicity of the peptides.
At present, molecular docking is popularly used to screen the potential functional substance and analyze the active mechanisms, as well as used in the field of exploring the umami substances, which is based on the fact that ligands bind to T1R1–T1R3 heterodimers of taste receptor type 1 (T1R) to exhibit the umami taste.14 Therefore, the receptor T1R1/T1R3 was always chosen to analyze the interaction between the umami peptide and T1R1/T1R3.
To screen and develop umami peptides from Atlantic cod (Gadus morhua) is meaningful for the deep use of blue proteins. This study aims to identify the novel umami peptides from Atlantic cod (Gadus morhua). Using nano-HPLC-MS/MS to identify the peptides that existed in the Atlantic cod enzymatic hydrolysis. In silico methods were used to screen the umami peptides and illustrate the mechanisms of the peptide with umami taste, as well as the basic physicochemical information of the peptides was analyzed. This study will promote a deep understanding of the umami taste present in Atlantic cod-related cuisines, and provide a substitution choice for MSG used in food products to meet the requirements of customers pursuing more healthy and high-quality food products. In summary, identifying umami peptides in food is not only scientifically innovative but also helps to improve food quality and promote the development of the food industries, which is of great significance to the field of food science and the food market.
2. Materials and methods
2.1 Materials and reagents
Atlantic cod (Gadus morhua) peptides powder with a purity of 84.92% was obtained from Qingdao Yihexing Food Co., Ltd (Qingdao, Shandong, China). Acetonitrile (ACN) and methanol were obtained from Honeywell Burdick & Jackson (Muskegon, MI, USA). Monosodium glutamate was purchased from Ajinomoto Co., Inc. (Tokyo, Japan). Dulbecco's Modified Eagle Medium (DMEM) was obtained from Shanghai XP Biomed Ltd (Shanghai, China). Dimethyl sulfoxide (DMSO, 99.9%) and thiazolyl blue tetrazolium bromide (MTT, 98%) were from Shanghai Macklin Biochemical Technology Co., Ltd (Shanghai, China). Desalted peptides LVDKL (97.16%) and ESKIL (97.22%) were synthesized by ChinaPeptides Co., Ltd (Shanghai, China). Food-grade sucrose and citric acid were from Henan Wanbang Chemical Technology Co. Ltd (Henan, China), and sodium chloride was from Henan Tianma Chemical Technology Co. Ltd (Henan, China).2.2 Preparation of Atlantic cod hydrolysate (ACH)
The Atlantic cod (Gadus morhua) peptides powder was provided by Qingdao Yihexing Food Co., Ltd. The preparation method was described in the study reported by Yuan et al.15 Briefly, Atlantic cod (Gadus morhua) was cut into pieces (about 5 cm) and grounded with adding water at the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5 (meat
1.5 (meat![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water; w
water; w![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) w). Then, the Atlantic cod (Gadus morhua) meat solutions were adjusted to 58 °C and with 8.0 of pH. The enzymatic hydrolysis reaction started by adding 0.2% alkaline protease. Two hours later, the hydrolysis reaction was stopped by boiling the Atlantic cod (Gadus morhua) solutions for 15 min. Finally, the supernatant was spray-dried to obtain Atlantic cod peptides after the hydrolysate was sieved by a 120 mesh sieve. As described by Yuan et al.,15 83.42% and 1.74% of the Atlantic cod hydrolysate (ACH) were with the molecular weight <1000 Da and >3000 Da, respectively. The Atlantic cod (Gadus morhua) peptides powder was dissolved in ultrapure water for any further tests.
w). Then, the Atlantic cod (Gadus morhua) meat solutions were adjusted to 58 °C and with 8.0 of pH. The enzymatic hydrolysis reaction started by adding 0.2% alkaline protease. Two hours later, the hydrolysis reaction was stopped by boiling the Atlantic cod (Gadus morhua) solutions for 15 min. Finally, the supernatant was spray-dried to obtain Atlantic cod peptides after the hydrolysate was sieved by a 120 mesh sieve. As described by Yuan et al.,15 83.42% and 1.74% of the Atlantic cod hydrolysate (ACH) were with the molecular weight <1000 Da and >3000 Da, respectively. The Atlantic cod (Gadus morhua) peptides powder was dissolved in ultrapure water for any further tests.
2.3 Electronic tongue analysis
Electronic tongue analysis was conducted according to the reported studies by Fu et al.16 and Zhang et al.17 The TS-5000Z taste sensor system (Insent, Tokyo, Japan) equipped with 5 taste sensors, was used to analyze the taste properties of the samples. The taste sensors CT0, CA0, C00, AE1, and AAE corresponded to analyzing the saltiness, sourness, bitterness, astringency, and umami, respectively. Before acquiring the data, two reference probes conducted self-checking with reference solution (2.2365 mg mL−1 KCl and 0.045 mg mL−1 tartaric acid). Samples were dissolved in ultrapure water with different concentrations.2.4 Sensory evaluation
The sensory evaluation of the samples was conducted, according to the reported method with several modifications,17,18 by 10 panelists (4 females and 6 males) with the age range from 25–32 years old, without taste impairments or disorders, in a standard sensory evaluation laboratory. Panelists were first trained with the taste reference solutions. Monosodium glutamate solution (0.35%, w/v), sucrose solution (1%, w/v), sodium chloride solution (0.35%, w/v), L-isoleucine solution (0.25%, w/v), and citric acid solution (0.08%, w/v) were used as reference standards for umami, sweet, salty, bitter, and sour tastes, respectively. Then the panelists tasted the samples and gave the taste scores (ranging from 0 to 5). During the test, the panelists were required to drink one sip of the sample and keep it in their mouths for 10 s before spitting it out. Meanwhile, the panelists were asked to rinse their mouths with drinking water 2–3 times between different tests. For testing, Atlantic cod (Gadus morhua) peptides powder (5 mg mL−1) was prepared by dissolved in ultrapure water. Synthesized peptides LVDKL and ESKIL were prepared at 2 mg mL−1 in ultrapure water.2.5 Identification of peptides by nano-HPLC-MS/MS
The ethanol-soluble Atlantic cod (Gadus morhua) peptides mixture used for nano-HPLC-MS/MS analysis, which is reported by our previous study,15 and the nano-HPLC-MS/MS analysis methods were described as follows. The peptides in the sample were identified by using the Q Exactive™ coupled to an EASY-nano LC 1200 system (Thermo Fisher Scientific, MA, USA). Samples were first desalted with MonoSpin C18 column by following the manufacturer's protocol. Then the desalted sample was dried by using a centrifugal vacuum concentrator (Thermofisher, SAVANT SPD1010). Subsequently, samples were re-dissolved in 0.1% formic acid in water (buffer A) and loaded onto an analytical column (Acclaim PepMap C18, 25 cm, 75 μm i.d.) for nano-HPLC-MS/MS analysis. The analyzing parameters were set as: a total of 60 min washing gradient, starting at 2% buffer B (80% ACN with 0.1% FA) followed by a stepwise increase to 35% in 47 min, from 35% to 100% within 1 min and stayed for 12 min; 300 nL min−1 of flow rate; 40 °C of column temperature, and 2 kV of the electrospray voltage. Other mass spectrometer parameters were set as: data dependent acquisition (DDA) mode, automatically switched between MS and MS/MS mode, 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 of MS/MS resolution, 3 × 106 of automatic gain control target. Finally, the data were analyzed by PEAKS Studio version 10.6 (Bioinformatics Solutions Inc., Waterloo, Canada).
500 of MS/MS resolution, 3 × 106 of automatic gain control target. Finally, the data were analyzed by PEAKS Studio version 10.6 (Bioinformatics Solutions Inc., Waterloo, Canada).
2.6 In silico analysis
The possibility of the peptide with an umami taste was evaluated by the web server iUmami-SCM (https://camt.pythonanywhere.com/iUmami-SCM), which can predict the umami peptide by giving the peptide a unique score. Normally, the peptide with a score no less than 588 is recognized as with high umami intensity.13 Moreover, the admetSAR (https://lmmd.ecust.edu.cn/admetsar2) was used to analyze the ADMET (absorption, distribution, metabolism, excretion, and toxicity) properties of the peptides, by following the tutorial.19 Expasy-ProtParam tool (https://web.expasy.org/protparam/)20 and Innovagen peptide property calculator (https://www.innovagen.com/proteomics-tools) were adopted to predict the stability, water solubility, and net charge of peptides.2.7 Molecular docking
The molecular docking programs were performed by using Discovery Studio 2017 R2 software (NeoTrdent Technology Ltd, Beijing, China). Firstly, a 3-dimensional structure of the taste receptor (T1R1/T1R3) was established through the “Create homology models” module in the Discovery Studio software by following the method described by Dang et al.21 The protein sequences were downloaded from the UniProt database (https://www.uniprot.org/). The homology model template metabotropic glutamate receptor (PDB ID: 1EWK) was obtained from the Protein Data Bank (https://www.rcsb.org/). Then, the molecular docking of the potential umami peptides with the T1R1/T1R3 were conducted based on the CDOCKER program, and the “CDOCKER_ENERGY” scores were used to assess the affinity of the peptides with T1R1/T1R3 receptor.2.8 Cytotoxicity assay
In this work, the cell toxicity of the umami peptides was analyzed on the mouse macrophages (RAW 264.7 cells), which were purchased from Procell Life Science & Technology Co., Ltd (Wuhan, China). The RAW 264.7 cells were cultured in DMEM containing 10% FBS, 100 U mL−1 penicillin, and 100 μg mL−1 streptomycin. For MTT analysis, 5000 cells were seeded in each well in 96-well plate, and the old medium was replaced by different concentration peptide solutions (dissolved in DMEM) after the cells were cultured 24 h. The treatment time of peptides to RAW 264.7 cells was set as 24 h. Then, MTT was added to the wells and incubated for 4 h. Subsequently, the old medium was removed and added 150 μL DMSO. Following shaking the 96-well plate (10 min), the OD value was obtained by using a Miroplate reader under 570 nm. Finally, the cell viability was calculated by the following equation:| Cell viability (%) = OD(sample)/OD(control) × 100% |
2.9 Data analysis
In this study, all the experiments were conducted at least in triplicate. The data were presented as mean ± SD or mean ± SEM. Origin Pro 9.0 software (OriginLab Corporation, Northampton, MA, USA) and GraphPad Prism 6 (Dotmatics, Boston, MA, USA) were used to analyze the data. The heatmap in Fig. 3 was drawn by using the online server Hemi (https://hemi.biocuckoo.cn/).223. Results and discussion
3.1 Sensory of Atlantic cod hydrolysate and LC-MS/MS analysis
The flavor of the Atlantic cod hydrolysate was evaluated by the electronic tongue combined with the sensory evaluation, and the results were shown in Fig. 1. As results from the electronic tongue analysis (Fig. 1A), the Atlantic cod hydrolysate showed an umami taste from the concentration of 2 mg mL−1, and showed a strong umami taste at 4 mg mL−1 which is almost equal to 0.35% of MSG. Moreover, sensory evaluation results also verified the ACH possesses an umami taste characteristic (Fig. 1B). Therefore, the hydrolysate should contain some umami peptides. As the Atlantic cod hydrolysate was hydrolyzed by alkaline protease and is about 83.42% of which with molecular weight below 1000 Da, many kinds of peptides may exist in the mixture. To decrease the huge of work for analyzing identified peptides in the mixture, we choose the part of 80% ethanol soluble mixture for further LC-MS/MS analysis and focused on the “myosin heavy chain fast skeletal muscle [Gadus morhua]” as which is one of the major proteins existed in Atlantic cod. The method for preparing an 80% ethanol soluble Atlantic cod hydrolysate mixture, as well as the identification of the peptide was reported by our previous studies.16 Here, we deeply analyzed some of the data by a focus on the myosin”. Results as shown in Fig. S1,† a total of 208 peptides were identified from the “myosin heavy chain fast skeletal muscle [Gadus morhua]”, with total identification coverage in protein is approximately 31%, containing 3 to 21 amino acid residues, with a molecular weight ranging from 387.2481 to 2521.2034 Da. Wang et al.23 reported that most of the umami peptides are with peptide length <10, and among 208 identified peptides, 65.9% (137 peptides) of which meet the characteristics. However, the umami taste profile remain needs to be further verified.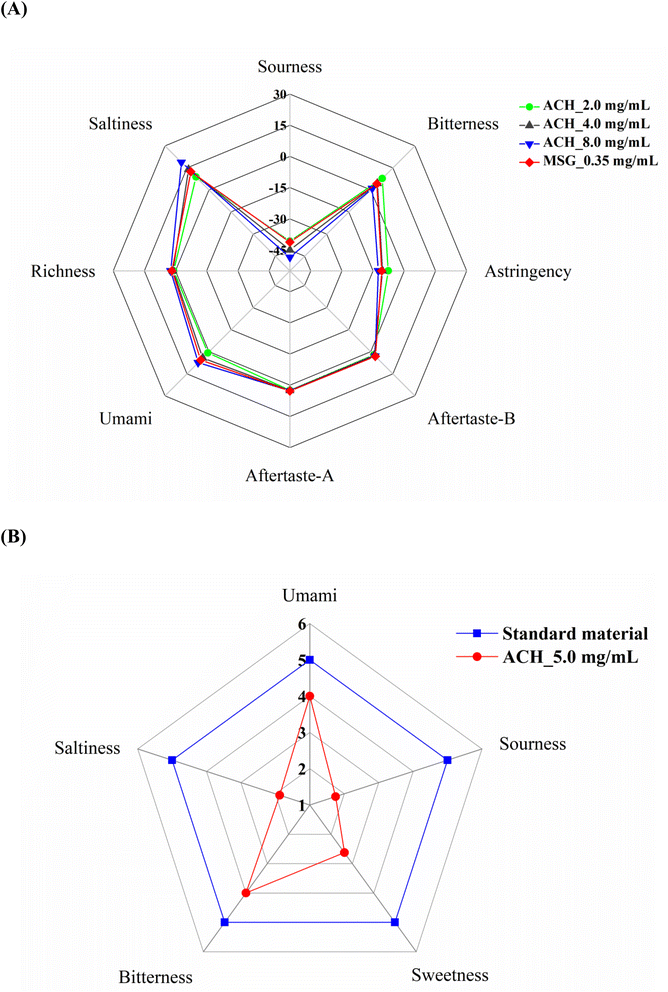 | ||
| Fig. 1 The taste properties of the hydrolysate of Atlantic cod (Gadus morhua). (A) Evaluated by the electronic tongue; (B) evaluated by sensory evaluation. | ||
3.2 Screening and synthesizing the umami peptide
As mentioned in section “3.1”, more than a hundred peptides were the umami peptide candidates, and it's hard to find the umami peptides among them accurately and easily. Therefore, the in silico methods were adopted to screen the umami peptides due to their high efficiency. In this study, the umami peptides were screened by combining the molecular docking and iUmami-SCM server (https://camt.pythonanywhere.com/iUmami-SCM), and the results were shown in Table 1. As most of the umami peptides with small molecular weight, a total of 47 peptides with 3–5 amino acid residues were selected for further consideration. Then, iUmami-SCM was used to predict the probability of peptides with umami taste, the scores <588 were considered to be non-umami taste, and a total of 15 peptides left were predicted as umami peptides. Then, 15 peptides were conducted molecular docking analysis. For molecular docking analysis, we first established the 3D-structure of T1R1/T1R3 receptor as indicated in our previous study.16 Subsequently, the umami taste of the peptides was evaluated by the molecular docking results of the “CDOCKER_ENERGY” index. Finally, the peptides LVDKL and ESKIL were selected to synthesize for further studies.| No. | Peptide sequence | iUmami-SCM | iUmami-SCM index | CDOCKER_ENERGY (kcal mol−1) | CDOCKER_INTERACTION_ENERGY (kcal mol−1) | Mass | m/z | z | Length |
|---|---|---|---|---|---|---|---|---|---|
| 1 | LVDKL | 601.50 | Umami | −102.39 | −70.92 | 586.369 | 587.3746 | 1 | 5 |
| 2 | ESKIL | 698.50 | Umami | −100.36 | −61.97 | 588.3483 | 589.3561 | 1 | 5 |
| 3 | VDDLE | 653.50 | Umami | −98.86 | −69.64 | 589.2595 | 590.2663 | 1 | 5 |
| 4 | TLDDL | 734.75 | Umami | −93.26 | −69.57 | 575.2803 | 576.2862 | 1 | 5 |
| 5 | AIDIL | 600.25 | Umami | −90.22 | −64.13 | 543.3268 | 544.3324 | 1 | 5 |
| 6 | KLQQF | 597.75 | Umami | −87.59 | −79.40 | 662.3751 | 663.3823 | 1 | 5 |
| 7 | ITDAA | 601.75 | Umami | −84.98 | −63.07 | 489.2435 | 490.2507 | 1 | 5 |
| 8 | KIEDM | 664.75 | Umami | −84.81 | −70.55 | 634.2996 | 635.3064 | 1 | 5 |
| 9 | LVDAS | 622.75 | Umami | −83.99 | −66.31 | 503.2591 | 504.266 | 1 | 5 |
| 10 | MIYTY | 605.00 | Umami | −81.35 | −75.92 | 689.3094 | 690.3148 | 1 | 5 |
| 11 | RVTF | 612.00 | Umami | −74.21 | −58.72 | 521.2961 | 261.6555 | 2 | 4 |
| 12 | TVNPY | 604.25 | Umami | −71.47 | −72.98 | 592.2856 | 593.2926 | 1 | 5 |
| 13 | AAMM | 589.33 | Umami | −68.32 | −53.81 | 422.1658 | 423.1732 | 1 | 4 |
| 14 | AVAK | 616.33 | Umami | −68.20 | −50.65 | 387.2481 | 388.2553 | 1 | 4 |
| 15 | SRVTF | 621.50 | Umami | −43.54 | −51.15 | 608.3282 | 305.1708 | 2 | 5 |
3.3 Umami taste analysis of the peptide
To explore the taste profiles of synthesized peptides LVDKL and ESKIL, the electronic tongue and sensory evaluation experiments were carried out, and the results were shown in Fig. 2 and Table S1.† Both peptides LVDKL and ESKIL possess umami flavor and the taste intensity of peptide LVDKL is a little stronger than ESKIL. Meanwhile, the umami taste intensity of peptides LVDKL and ESKIL are both higher than 0.07% MSG under the concentration of 0.5 mg mL−1. Although the umami taste of peptides LVDKL and ESKIL is lower than the standard umami taste compound (MSG) at the same concentration. However, with safety considerations, some people do not like MSG, but on the contrary, peptides are popularlly accepted as their variety of bioactivites are verified by plenty of experiments. Therefore, it's also meaningful for the peptides LVDKL and ESKIL with lower umami taste intensity than MSG. Meanwhile, the thresholds of the two peptides LVDKL and ESKIL are 0.427 mM and 0.574 mM, respectively, which is similar to the microbiota-derived umami peptide DFE (0.45 mM),24 yeast extract-derived IDVDGKPQIQ (0.56 mM),25 VQPEDNKGVFQ (0.61 mM),25 and lower than the peanut protein isolate hydrolysate derived umami peptide DQR (1.11 mM),26 and stronger than soy sauce derived peptide AQALQAQA (1.25 mM)27 as well as peanut protein isolate hydrolysate derived peptide EP (1.49 mM).28 Moreover, the electronic tongue analysis results indicated that the peptides LVDKL and ESKIL have slight saltiness and bitterness, which is a normal property for umami peptides, similar with peptides EEFLK and FLNQDEEAR.16 | ||
| Fig. 2 The taste properties of the hydrolysate of synthetic peptides. (A) Evaluated by the electronic tongue; (B) evaluated by sensory evaluation, with peptide concentration of 2 mg mL−1. | ||
3.4 Umami mechanisms of the peptides
To explore the flavor mechanisms of the umami peptides, molecular docking methodology was applied to analyze the interactions between the peptide and the umami taste receptor T1R1/T1R3, which is a popular studied receptor for analyzing the interaction between umami peptide and receptor.29 Meanwhile, the docking energy reflects the stability of the peptide-receptor complex, and more stable complex with lower docking energy.29 Here, the molecular docking results indicated that the peptides LVDKL and ESKIL have a strong affinity with T1R1/T1R3, with “CDOCKER_ENERGY” of −102.39 and −100.36 kcal mol−1, and “CDOCKER_INTERACTION_ENERGY” of −70.92 and −61.97 kcal mol−1. The docking results indicated that the peptide LVDKL formed a more stable complex with T1R1/T1R3 compared with the peptide ESKIL, which is consistent with the results obtained by the above mentioned electronic tongue and sensory evaluation analysis. The intermolecular forces existed between the umami peptides and T1R1/T1R3 were further explored, and as shown in Fig. 3 and Table S2,† LVDKL interacted with T1R1/T1R3 by electrostatic, hydrogen bond, and hydrophobic interactions on the residues ARG712, HIS915, GLN853, ARG854, GLY911, HIS914, LEU852, LEU912 and PHE851 of the receptor. Peptide ESKIL interacted with T1R1/T1R3 by forming 6 hydrogen bonds and 4 hydrophobic interactions with the receptor residues of GLN853, PRO715, HIS914, VAL714, LEU852 and HIS672, the distance of the hydrogen bond varied from 1.8658 Å to 3.0124 Å. Our results are similar to the peptide FSGLDGSK interacting with the T1R1/T1R3 receptor via hydrogen bond, electrostatic interactions, and hydrophobic interactions.30It's reported that the structure of the peptides decides their bioactivities and properties, as well as taste characteristics. In this work, the optimal 3D-structure conformations of the peptides LVDKL and ESKIL docked with T1R1/T1R3 receptor were displayed by the Discovery studio software (Fig. S2†). Studies showed that the Arg (R), Asp (D), Asn (N), Glu (E), Gln (Q), and Lys (K) in the umami peptide showed significant meaning for interacted with T1R1/T1R3.6 Meanwhile, no matter where (N-terminal, C-terminal, or middle) the amino acids residues Asp (D) and Glu (E) existed in the peptides, which will be the key active site exerting umami taste.23 Notably, the peptides LVDKL and ESKIL contain either ‘D’ or ‘E’ amino acid residues that are expected to contribute to the umami taste of those two peptides. Meanwhile, Wang et al.23 concluded that the ‘L’ with high frequency occurred in the N-terminal/C-terminal of umami peptides consisting of 4–7 amino acid residues, which may be one of the reasons that both peptide LVDKL and ESKIL possess umami taste.
3.5 Analysis of physicochemical characteristics of umami peptides
Here, 2 peptides from Atlantic cod hydrolysate are identified as umami peptides. The characteristics of peptides are critical to their bioactivities, taste profiles, and bioavailabilities. It's reported that the bioavailability of most compounds (64%) is mainly controlled by the intestinal absorption process.31The ADMET characteristics of the peptides LVDKL and ESKIL were predicted by the admetSAR web server, which will give the human intestinal absorption (HIA) and acute oral toxicity information of those 2 peptides. The admetSAR server will give HIA value by ‘−’/‘+’ of target compounds, which means poor-absorption (HIA−) and good-absorption (HIA+). As shown in Table S3.† The LVDKL was predicted with III toxicity and HIA−, HIA− means HIA% is less than 30%. The peptide ESKIL was predicted III toxicity and HIA+, meanwhile the “instability index” indicated that unstable. Toxicity predicted as category III indicated the peptide LVDKL and ESKIL with LD50 values among 500 mg kg−1 to 5000 mg kg−1,32 concluded them with low toxicity and high safety, which is same as the potential α-glucosidase inhibitory peptides TMMMLLP, FFLPQ, AMFLPGA, and FMLPQ.33 Moreover, both of the 2 umami peptides were predicted with good water solubility. However, all of those were predicted by the in silico method, therefore further in vitro and in vivo studies should be conducted to verify those characteristics.
The safety of the peptides is of great significance for its further application. To further analyze the safety of the peptides, the cell viability analysis was conducted. In this study, the murine macrophage cell line RAW 264.7 was chosen to conduct toxicity assessment experiments as it is one of the cell lines popularly used for predicting the potential human de novo responses and evaluating the bioactivity of test samples.34 As shown in Fig. 4, both peptides LVDKL and ESKIL showed no influence on the cell viability of RAW 264.7 cell after being treated for 24 h with the peptide concentrations ranging from 100 μg mL−1 to 750 μg mL−1, which means it is safe as predicted by admetSAR web server. However, more safety tests are expected to be done to verify its safety characteristics.
 | ||
| Fig. 4 Cell viability of the umami peptides on the RAW 264.7 cells. The influence of the peptides on the cell was tested by treated raw 264.7 cells for 24 h. | ||
4. Conclusions
The results of this study indicated the possibility of identifying umami peptide from the Atlantic cod (Gadus morhua) and verified the methodology of combining in silico methods and sensory evaluation experiments to screen umami peptide is highly efficient. In this work, two novel umami peptides were identified from the myosin heavy chain of Atlantic cod (Gadus morhua), which showed a strong intensity of umami taste. The umami taste mechanisms of the umami peptides were evaluated by molecular docking methodologies. Meanwhile, the stability, human intestinal absorption, and cell toxicity of the peptides were evaluated by in silico methods and MTT experiments.Data availability
Data will be made available on request.Author contributions
Xu Zhao: software, writing; Wenpei Qiu: data analysis, visualization; Xian-Guang Shao: writing-review & editing; Baifeng Fu: data analysis; Xinyu Qiao: data analysis; Zhen Yuan: methodology; Meilian Yang: writing-review & editing; Pan Liu: writing-review & editing; Ming Du: supervision, funding acquisition; Maolin Tu: writing, supervision, funding acquisition.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This study was funded by the Ministry of Science and Technology of the People's Republic of China [National Key R&D Program of China: investigate the mechanism of formation and control technologies of Chinese traditional and ethnic food quality (2021YFD2100100)], 2022 Ningbo Yongjiang Talent Introduction Programme (Young talent subproject), and the Research Startup Fund of Ningbo University (Grant Number ZX2022000432).References
- M. Adachi, A. Eguchi, H. Hayabuchi, T. Kawashima, S. Koganemaru, K. S. Lwin, H. Nakamura, T. Nishimura, S. Nomura, K. Shibuya, B. Sigel, S. Tanaka, Y. Tanoe, H. Uneyama, L. Yamasaki and D. Yoneoka, Public Health Nutr., 2023, 26, 488–495 CrossRef PubMed.
- H. Nakamura, T. Kawashima, L. Yamasaki, K. S. Lwin, A. Eguchi, H. Hayabuchi, Y. Tanoe, S. Tanaka, D. Yoneoka, C. Ghaznavi, H. Uneyama, K. Shibuya and S. Nomura, Food Sci. Nutr., 2023, 11, 872–882 CrossRef CAS PubMed.
- W. Wang, X. Zhou and Y. Liu, TrAC, Trends Anal. Chem., 2020, 127, 115876 CrossRef CAS.
- Z. Liu, Y. Zhu, W. Wang, X. Zhou, G. Chen and Y. Liu, Food Chem., 2020, 330, 127204 CrossRef CAS PubMed.
- Y. Gu, J. Zhang, Y. Niu, B. Sun, Z. Liu, X. Mao and Y. Zhang, Food Chem., 2024, 434, 137386 CrossRef CAS PubMed.
- J. Zhang, J. Zhang, L. Liang, B. Sun and Y. Zhang, Food Chem., 2023, 404, 134414 CrossRef CAS PubMed.
- X. Zhu, D. Sun-Waterhouse, J. Chen, C. Cui and W. Wang, J. Sci. Food Agric., 2021, 101, 158–166 CrossRef CAS PubMed.
- W. Zhu, W. He, F. Wang, Y. Bu, X. Li and J. Li, Int. J. Food Sci. Technol., 2021, 56, 402–412 CrossRef CAS.
- Y. Wang, J. Luan, X. Tang, W. Zhu, Y. Xu, Y. Bu, J. Li, F. Cui and X. Li, Food Funct., 2023, 14, 1510–1519 RSC.
- Y. Zhang, C. Venkitasamy, Z. Pan, W. Liu and L. Zhao, J. Food Sci., 2017, 82, 16–23 CrossRef CAS PubMed.
- S. Riebroy, S. Benjakul, W. Visessanguan, U. Erikson and T. Rustad, Food Chem., 2008, 109, 42–53 CrossRef CAS PubMed.
- P. A. Wang, I. Martinez and R. L. Olsen, Food Chem., 2009, 115, 1228–1233 CrossRef CAS.
- P. Charoenkwan, J. Yana, C. Nantasenamat, M. M. Hasan and W. Shoombuatong, J. Chem. Inf. Model., 2020, 60, 6666–6678 CrossRef CAS PubMed.
- X. Li, L. Staszewski, H. Xu, K. Durick, M. Zoller and E. Adler, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 4692–4696 CrossRef CAS PubMed.
- Z. Yuan, M. Yang, D. Zhu, D. Wu, S. Cheng, C. Wu, H. R. El-Seedi and M. Du, Food Sci. Hum. Wellness, 2023, 12, 1192–1203 CrossRef CAS.
- B. Fu, D. Wu, S. Cheng, X. Xu, L. Zhang, L. Wang, H. R. El-Seedi, H. Liu and M. Du, Food Sci. Hum. Wellness, 2024, 13, 146–153 CrossRef CAS.
- L. Zhang, D. Pu, J. Zhang, Z. Hao, X. Zhao, B. Sun and Y. Zhang, J. Agric. Food Chem., 2023, 71, 7803–7811 CrossRef CAS PubMed.
- S. Zhao, S. Ma, Y. Zhang, M. Gao, Z. Luo and S. Cai, Food Chem., 2024, 433, 137331 CrossRef CAS PubMed.
- H. Yang, C. Lou, L. Sun, J. Li, Y. Cai, Z. Wang, W. Li, G. Liu and Y. Tang, Bioinformatics, 2019, 35, 1067–1069 CrossRef CAS PubMed.
- E. Gasteiger, C. Hoogland, A. Gattiker, S. E. Duvaud, M. R. Wilkins, R. D. Appel and A. Bairoch, in The Proteomics Protocols Handbook, ed. J. M. Walker, Humana Press, Totowa, NJ, 2005, pp. 571–607, DOI:10.1385/1-59259-890-0:571.
- Y. Dang, X. Gao, A. Xie, X. Wu and F. Ma, Cell Biochem. Biophys., 2014, 70, 1841–1848 CrossRef CAS PubMed.
- W. Ning, Y. Wei, L. Gao, C. Han, Y. Gou, S. Fu, D. Liu, C. Zhang, X. Huang, S. Wu, D. Peng, C. Wang and Y. Xue, Nucleic Acids Res., 2022, 50, W405–w411 CrossRef CAS PubMed.
- W. Wang, Z. Cui, M. Ning, T. Zhou and Y. Liu, Biomaterials, 2022, 281, 121338 CrossRef CAS PubMed.
- F. An, K. Cao, S. Ji, Y. Wang, G. Pan, Y. Ma, Y. Zhao, J. Wu and R. Wu, Food Chem., 2023, 404, 134583 CrossRef CAS PubMed.
- H. Wang, W. Wang, S. Zhang, Z. Hu, R. Yao, H. Hadiatullah, P. Li and G. Zhao, Food Chem., 2023, 429, 136807 CrossRef CAS PubMed.
- J. Zhang, M. Zhao, G. Su and L. Lin, Food Chem., 2019, 278, 674–682 CrossRef CAS PubMed.
- M. Zhuang, L. Lin, M. Zhao, Y. Dong, D. Sun-Waterhouse, H. Chen, C. Qiu and G. Su, Food Chem., 2016, 206, 174–181 CrossRef CAS PubMed.
- J. Zhang, D. Sun-Waterhouse, Y. Feng, G. Su, M. Zhao and L. Lin, Food Res. Int., 2019, 120, 895–903 CrossRef CAS PubMed.
- R. Jia, Y. He, G. Liao, Z. Yang, D. Gu, Y. Pu, M. Huang and G. Wang, Food Res. Int., 2023, 172, 113208 CrossRef CAS PubMed.
- L. Liang, C. Zhou, J. Zhang, Y. Huang, J. Zhao, B. Sun and Y. Zhang, Food Chem., 2022, 387, 132870 CrossRef CAS PubMed.
- N.-N. Wang, C. Huang, J. Dong, Z. J. Yao, M.-F. Zhu, Z. K. Deng, B. Lv, A. P. Lu, A. F. Chen and D. S. Cao, RSC Adv., 2017, 7, 19007–19018 RSC.
- A. M. Faisal, S. H. Imtiaz, T. Zerin, T. Rahman and H. U. Shekhar, Bioinformation, 2017, 13, 417–429 CrossRef PubMed.
- Y. Fu, Z. Liu, H. Wang, F. Zhang, S. Guo and Q. Shen, Food Chem., 2023, 411, 135378 CrossRef CAS PubMed.
- L. Merly and S. L. Smith, Immunopharmacol. Immunotoxicol., 2017, 39, 55–58 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra00890a |
| ‡ The authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |

