Open Access Article
Open Access ArticleSurface-enhanced Raman spectroscopy for characterization of filtrates of blood serum samples from patients with tuberculosis obtained by 50 kDa filtration devices†
Ali Kamran‡
 a,
Abdul Naman‡a,
Muhammad Irfan Majeed
a,
Abdul Naman‡a,
Muhammad Irfan Majeed *a,
Haq Nawaz
*a,
Haq Nawaz *a,
Najah Alwadie*b,
Noor ul Hudaa,
Umm-e- Habibaa,
Tania Tabussam
*a,
Najah Alwadie*b,
Noor ul Hudaa,
Umm-e- Habibaa,
Tania Tabussam a,
Aqsa Banoa,
Hawa Hajaba,
Rabeea Razaqa,
Ayesha Ashrafa,
Saima Aziza,
Maria Asghara and
Muhammad Imran
a,
Aqsa Banoa,
Hawa Hajaba,
Rabeea Razaqa,
Ayesha Ashrafa,
Saima Aziza,
Maria Asghara and
Muhammad Imran c
c
aDepartment of Chemistry, University of Agriculture Faisalabad, Faisalabad 38000, Pakistan. E-mail: irfan.majeed@uaf.edu.pk; haqchemist@yahoo.com
bDepartment of Physics, College of Science, Princess Nourah bint Abdulrahman University, P. O. Box 84428, Riyadh 11671, Saudi Arabia. E-mail: nhalwadie@pnu.edu.sa
cDepartment of Chemistry, Faculty of Science, King Khalid University, P. O. Box 9004, Abha 61413, Saudi Arabia
First published on 13th March 2024
Abstract
The ability of surface-enhanced Raman spectroscopy (SERS) to generate spectroscopic fingerprints has made it an emerging tool for biomedical applications. The objective of this study is to confirm the potential use of Raman spectroscopy for early disease diagnosis based on blood serum. In this study, a total of sixty blood serum samples, consisting of forty from diseased patients and twenty (controls) from healthy individuals, was used. Because disease biomarkers, found in the lower molecular weight fraction, are suppressed by higher molecular weight proteins, 50 kDa Amicon ultrafiltration centrifugation devices were used to produce two fractions from whole blood serum consisting of a filtrate, which is a low molecular weight fraction, and a residue, which is a high molecular weight fraction. These fractions were then analyzed, and their SERS spectral data were compared with those of healthy fractions. The SERS technique was utilized on blood serum, filtrate and residue of patients with tuberculosis to identify characteristic SERS spectral features associated with the development of disease, which can be used to differentiate them from healthy samples using silver nanoparticles as a SERS substrate. For further analysis, the effective chemometric technique of principal component analysis (PCA) was used to qualitatively differentiate all the analyzed samples based on their SERS spectral features. Partial least squares discriminant analysis (PLS-DA) accurately classified the filtrate portions of healthy and tuberculosis samples with 97% accuracy, 97% specificity, 98% sensitivity, and an area under the receiver operating characteristic (AUROC) curve of 0.74.
1. Introduction
Tuberculosis (TB) is a rapidly spreading contagious disease that is caused by Mycobacterium tuberculosis, and it remains an important public health concern. A recent survey of the World Health Organization and the United Nations General Assembly indicated that the death rate from TB has significantly increased up to 18% this year due to the global COVID-19 pandemic.1 The death rate from TB in Pakistan is ranked 5th worldwide due to poverty and inability to diagnose the disease at early stages.2 Mycobacterium tuberculosis enters the bloodstream through the airway and infects the breathing organ in the human body. TB is an airborne disease and is referred to as pulmonary TB when it mainly affects the lungs, but it also attacks other body tissues, such as kidneys, intestines, lymph nodes, joints and skin; when it exists in the body in a location other than the lungs, it is referred to as extra-pulmonary tuberculosis.3 Tuberculosis is considered to be a public health problem because its resistance against drugs is great.4The tuberculin skin test (TST) and interferon gamma release assay (IGRA) are used as medical tests for the diagnosis of tuberculosis disease. Different biomedical tests are used to detect Mycobacterium tuberculosis, including sputum smear microscopy,5 nucleic acid amplification test (NAAT),6 enzyme-linked immunosorbent assay (ELISA),7 chest radiograph,8 biosensor technique,9 acid-fast bacilli test,10 different DNA biomarkers and polymerase chain reaction.11 These tests are time-consuming, laborious and require costly consumables.
Raman spectroscopy is used in the early detection and identification of molecular components related to the disease. Vibrational spectroscopy has been widely used to analyze and understand processes in disease development. Raman spectroscopy of bodily fluids, notably blood serum, can be used to identify the chemical fingerprints of samples.12,13 Raman spectroscopy is an efficient qualitative and quantitative analytical tool for rapidly obtaining critical information.14 Although weak signal intensity often occurs with Raman spectroscopy, it is rapid, inexpensive, and precise. A few difficulties also exist when attempting to identify low concentrations of biofluids such as serum, plasma, joint submucosa, and lymph.15 Raman spectroscopy provides low-intensity Raman signals that are enhanced by nanoparticles as substrates, which enhance the ability to detect lower concentrations of biofluids.16 SERS has been previously used to characterize and compare the different types of TB disease, including pulmonary and extra-pulmonary TB, using healthy and diseased blood samples.2,17
Blood serum contains high molecular weight fraction (HMWF) and low molecular weight fraction (LMWF) proteins. The LMW protein fractions are thought to be tuberculosis disease biomarkers, but the analysis of human whole blood serum for the detection of TB disease-specific biomarkers is difficult because human blood serum contains large molecular weight proteins such as albumin and globulin that hinder the detection and identification of smaller molecular weight proteins.18
The current study focuses on the SERS analysis of filtrate samples obtained from healthy patients and patients with TB that were purified using 50 kDa filtration devices. This can assist in identifying characteristic SERS spectral signatures linked to specific protein molecules smaller than the filter size of 50 kDa and hence bypass the features related to proteins larger than this pore size. Thus, the blood serum samples from patients with TB can be identified and differentiated based on their distinct SERS spectral features. Notably, it was found that thus far, no studies have been published on this particular topic.
2. Materials and methods
2.1. Separation of serum from the blood of individuals affected by tuberculosis
Blood from healthy patients and patients with tuberculosis was collected in vials that were centrifuged to remove red blood cells. Serum samples were obtained that contained proteins, lipids, carbohydrates and other biomolecules related to infection of the human body. Sixty samples were obtained that included 40 from diseased patients and 20 from healthy individuals. Filtrate portions were obtained after centrifugal filtration of blood serum samples using Amicon Ultra 50 kDa centrifugal filtration devices. This process resulted in two fractions of human blood serum samples, including the filtrate, which contained the low molecular weight fraction and residue, which contained the high molecular weight fraction. The filtrate samples were analyzed, and their SERS spectral data from patients with TB and healthy individuals were compared.All blood serum samples were collected from PINUM Hospital, Faisalabad. All serum samples were collected from male patients 45 to 50 years of age, and healthy samples were collected from males without any co-morbidity or participation in any therapy. The mean patient age for all samples was calculated as 47.71, and the calculated standard deviation for age was 1.01 years. Thus, the controls and patients were of the same age group and sex. A total number of 60 samples were centrifuged, consisting of 20 healthy samples and 40 disease samples. This study was approved by the Bioethical Committee of the University of Agriculture Faisalabad, Pakistan.
2.2. SERS spectral measurements
Silver nanoparticles were used as a SERS substrate and were prepared following a previously published chemical reduction method.19 The average size of the silver nanoparticles was 53 nm.20 For SERS measurements, the nanoparticles were mixed with each sample using 40 μL of both, and the samples were then incubated for 30 minutes to ensure optimal sample–nanoparticle interaction. This incubation time was used for all measurements because it resulted in SERS spectra with strong intensity. SERS measurements were accomplished by analyzing 40 μL of the incubation mixture of healthy and TB disease samples one by one on an aluminum slide using a Raman spectrometer (ATR8300BS, Optosky, China). This instrument was equipped with an air-cooled charge-coupled device (CCD) detector, and for this study, a 785 nm diode laser served as the excitation source, delivering a laser power of 90 mW, and a lens of 40×/0.6 (NA) with a spot size of 2 μm was used. A total number of 15 SERS spectra were collected from all serum samples, within the spectral range of 200 to 1800 cm−1, and with a 10 seconds acquisition time for each spectrum.2.3. Pre-processing of the SERS spectral data
The SERS spectral data were pre-processed to convert raw spectral data from tuberculosis and healthy samples to a valuable data form using MATLAB 7.8 and recognized protocols. Various steps are involved in the pre-processing of the raw data including baseline correction, vector normalization, smoothing and substrate removal. The rubber band method and polynomial parameters were used for the rubber-band baseline removal. Smoothing of all spectra was accomplished by the Savitzky–Golay smoothing method (13 peak-point window, 3rd order polynomial). Normalization of the SERS spectral data was performed using vector normalization. Background (aluminum substrate) removal was performed by subtracting the SERS spectra of aluminum from all SERS spectra.212.4. SERS spectral data analysis
For the analysis of the SERS spectral data from the filtrate portions of samples from healthy and TB-positive patients, principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA) were used as multivariate data analysis techniques. PCA was employed to transform a large number of correlated variables into a smaller set of uncorrelated variables. An unsupervised method using PCA loadings was constructed that will assist in differentiating between various clusters of Raman spectral data with their positive and negative loadings.PLS-DA is a supervised methodology that plays a vital role in statistical modeling. PLS-DA is utilized to examine the interrelationship between two distinct sets of variables consisting of a set of independent variables and a set of dependent variables. By employing PLS-DA, the SERS spectral data can effectively be analyzed. PLS-DA was used to find the linear combinations of the variables that best explain the variation in the data, so that the data can be classified into different groups or classes. Monte Carlo simulation is a technique that can be used to estimate the uncertainty in PLS-DA results by generating many random samples from the data. PLS-DA is then performed, and this can provide a range of possible outcomes and the likelihood of certain results.
The receiver operating characteristic (ROC) curve is a visual representation of the performance of a binary classifier system as the decision threshold is adjusted. It illustrates how the true positive rate (sensitivity) and false positive rate (1-specificity) of the classifier model changes with varying thresholds. The effectiveness of the ROC curve is quantified by the area under the curve (AUC), which assesses the overall effectiveness of the classifier. A perfect classifier would have an AUC of 1, indicating ideal discrimination ability, while an AUC of 0.5 signifies a classifier model that performs no better than random guessing.
3. Results and discussion
3.1. Mean SERS spectra of uncentrifuged and centrifuged TB serum samples
Fig. 1 shows the mean SERS spectra of the uncentrifuged serum, filtrate and residue for patients who are tuberculosis-positive. This plot helps to distinguish the characteristic features between uncentrifuged and centrifuged disease-positive serum that are related to changes in biomolecules such as DNA/RNA, proteins, carbohydrates, and lipids. The SERS characteristic peaks and peak assignments along with references are listed in Table 1.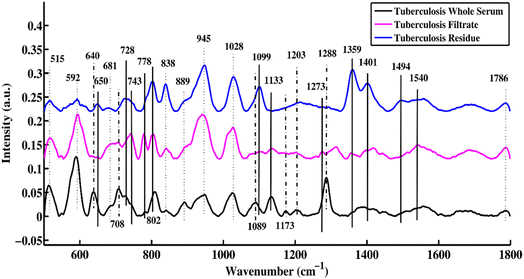 | ||
| Fig. 1 SERS mean spectra of uncentrifuged TB serum, and the filtrate and residue portions of TB serum. | ||
| SERS bands | SERS peak assignment | Components | References |
|---|---|---|---|
| 515–592 cm−1 | Phosphatidylinositol | Lipids | 22 and 23 |
| 640 cm−1 | Uric acid | Uric acid | 24 |
| 650 cm−1 | Tyrosine C–C twisting mode | Proteins | 23 |
| 681 cm−1 | Resonant ring vibrations within the DNA bases | DNA/RNA | 25 |
| 708 cm−1 | Cholesterol ester | Lipids | 26 |
| 728 cm−1 | Hypoxanthine | Hypoxanthine | 24 |
| 743 cm−1 | Symmetric breathing of tryptophan | Protein | 23 |
| 778 cm−1 | Cytosine/uracil ring breathing (nucleotides) | DNA/RNA | 27 |
| 802 cm−1 | Thymine-based DNA/RNA bases exhibiting ring-breathing mode | DNA/RNA | 23 |
| 815 cm−1 | C–O–C stretching vibration | Protein | 28 |
| 838 cm−1 | Glucose saccharide α-anomers exhibiting an α-saccharide or α-band | Carbohydrates | 16 |
| 889 cm−1 | Uric acid | Uric acid | 24 |
| 918 cm−1 | Proline, hydroxyproline, glycogen, and lactic acid | Proteins | 25 |
| 945 cm−1 | Stretching vibrations of single bonds in amino acids, including proline and valine | Proteins | 4 |
| 1008 cm−1 | Pectin, phenylalanine | Proteins | 13 |
| 1028 cm−1 | Stretching of methoxy groups (O–CH3) | DNA/RNA | 29 |
| 1089 cm−1 | Phospholipids | Lipids | 29 |
| 1099 cm−1 | Phospholipids | Lipids | 30 |
| 1133 cm−1 | C–C stretching mode of lipids | Lipids | 27 |
| 1173 cm−1 | C–H bending mode of tyrosine | DNA | 31 |
| 1203 cm−1 | Uric acid | Uric acid | 24 |
| 1273 cm−1 | Adenine ring-breathing mode | DNA | 32 |
| 1288 cm−1 | Cytosine ring-breathing mode in RNA | DNA | 32 |
| 1314 cm−1 | Guanine base | DNA/RNA | 33 and 34 |
| 1359 cm−1 | Guanine (N7, B, Z-marker) | DNA | 16 |
| 1401 cm−1 | Thymine | DNA | 35 |
| 1448 cm−1 | C–H vibration | Proteins | 27 and 36 |
| 1494 cm−1 | DNA base ring-breathing modes | DNA | 37 |
| 1540 cm−1 | Aromatic hydrogen, amide carbonyl (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) vibrations O) vibrations |
Proteins | 38 |
| 1689 cm−1 | Amide I (non-hydrogen bonded; disordered structure) | Proteins | 39 |
| 1786 cm−1 | Lipid content | Lipids | 40 |
In Fig. 1, three different types of SERS peak features are identified, which are denoted by solid, dotted, and dotted-dashed lines. These characteristic peaks, which are solely present in uncentrifuged/whole serum, are denoted by dotted-dashed lines, while the features that are present in only the filtrate and residue portions are represented by solid lines. The features common to both (filtrate and residue portions and whole serum) are shown by dotted lines. These SERS spectral features are the biomarkers that differentiate between filtrate and residue portions and uncentrifuged/whole serum samples. The SERS peak features, which are only present in uncentrifuged serum, are denoted by dotted-dashed lines and include 640, 708, 1089, 1173, 1203 and 1288 cm−1. The SERS peaks observed in the filtrate and residue portions of the serum are denoted by solid lines and include 650, 728, 743, 778, 802, 1099, 1133, 1273, 1359, 1401, 1494 and 1540 cm−1. There are other features that are common in the filtrate and residue portions and uncentrifuged serum samples, and they are shown with dotted lines and include 515, 592, 681, 838, 889, 945, 1028 and 1786 cm−1.
The bands that appear at 640 are associated with uric acid, 708 (cholesterol ester), 1089 (phospholipids), 1173 (C–H bending mode of tyrosine), 1203 (uric acid) and 1288 cm−1 (cytosine ring-breathing mode in RNA). After centrifugation of serum samples, some biomolecules with molecular sizes larger than 50 kDa were removed from the sample. The SERS peaks of these remaining biomolecules in the filtrate portion were compared with biomolecules of uncentrifuged samples to identify the differentiating SERS features.
3.2. SERS mean spectral peaks of healthy and tuberculosis filtrates
Fig. 2 shows a mean SERS plot between healthy and TB-positive blood serum filtrates. The peaks that are only present in the filtrate portions of healthy samples are denoted by dashed lines and were assigned to biomarkers only present in healthy samples, including 547, 743, 815, 1008 and 1099 cm−1. The SERS peaks denoted with solid lines are only present in the disease-positive filtrate portion and include 515, 681, 708, 802, 838, 945, 1028, 1314, 1540 and 1786 cm−1.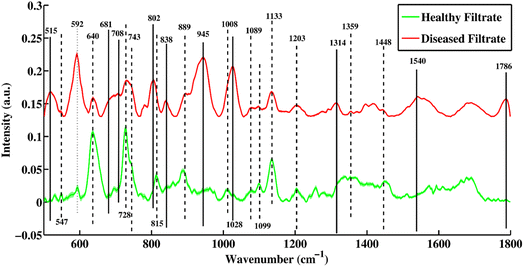 | ||
| Fig. 2 Mean SERS spectra of filtrate portions of healthy and tuberculosis positive blood serum samples with standard deviation. | ||
Some peaks present in the filtrate portions of healthy and disease samples denoted by dotted and dotted-dashed lines are linked with increasing and decreasing peak intensities, respectively. The SERS spectral peak at 592, which is associated with lipids (phosphatidylinositol) is present at a higher intensity in the disease filtrate as compared to the healthy filtrate portion, while peaks 640, 728, 1089, 1099, 1133, 1359 and 1448 cm−1 were higher in intensity in healthy filtrates as compared to disease filtrate portions.
The SERS spectral peaks of the tuberculosis filtrate include 515 (phosphatidylinositol), 681 (ring-breathing patterns in DNA bases), 708 (cholesterol ester), 802 (thymine-based ring-breathing mode of DNA/RNA base), 838 (glucose saccharide α-anomers exhibiting an α-saccharide or α-band), 945 (stretching vibrations of amino acids), 1028 (stretching of O–CH3), 1314 (guanine base of DNA), 1540 (aromatic hydrogen, amide carbonyl with (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) vibrations) and 1786 cm−1 (C–C stretching of lipids). These are the major biomarkers of the filtrate portion from patients with tuberculosis. Other SERS peaks that are only present in healthy filtrates were observed at 547 (lipids), 743 (symmetric breathing of tryptophan), 815 (C–O–C stretching vibration), 1008 (pectin, phenylalanine) and 1099 cm−1 (palmitic acid). SERS peaks at 889 and 1203 cm−1 are associated with uric acid.
O) vibrations) and 1786 cm−1 (C–C stretching of lipids). These are the major biomarkers of the filtrate portion from patients with tuberculosis. Other SERS peaks that are only present in healthy filtrates were observed at 547 (lipids), 743 (symmetric breathing of tryptophan), 815 (C–O–C stretching vibration), 1008 (pectin, phenylalanine) and 1099 cm−1 (palmitic acid). SERS peaks at 889 and 1203 cm−1 are associated with uric acid.
Some SERS spectral features were present with higher intensity in the disease filtrate portions (which are denoted by dotted-dashed lines) as compared to healthy filtrates of the serum, such as 592 cm−1, which is associated with lipids. Other features of the SERS spectra with higher intensity of peaks in the healthy filtrates as compared to disease filtrates are observed at 640 (uric acid), 728 (hypoxanthine), 889 (uric acid), 1089 (phospholipids), 1099 (palmitic acid), 1133 (palmitic acid), 1203 (uric acid), 1359 (guanine) and 1448 cm−1 (C–H vibration). The original SERS spectra of some diseased and healthy/control samples are shown in Fig. S1 and S2,† respectively.
3.3. Principal component analysis (PCA)
Surface-enhanced Raman spectroscopy is very efficient in differentiating between healthy and tuberculosis-positive filtrate samples. PCA is an unsupervised method that reduces the dimensionality of a complex dataset into new variables called principal components (PCs). PCA is employed for the differentiation between same groups as well as different groups/classes. The score plot, loadings and eigenvalues are extracted from the PCA model by applying a singular value decomposition (SVD) algorithm. PC values or eigenvalues are arranged in descending order, showing the values of explained variables.Fig. 3(a) shows scatter plots of the SERS spectra of filtrates of tuberculosis and healthy samples. Green dots represent the spectra of healthy filtrate portions, while pink dots show the spectra of filtrates of disease samples. The clusters of green dots on the positive side of the x-axis represent healthy filtrate samples, while clusters of pink dots observed on the negative side of the x-axis represent disease filtrates. PC-1 (first principal component) on the x-axis separating two different groups shows a maximum variability value of 70.92%. PC-2 (second principal component) shows 9.78% variability in the dataset. These results show that the diseased and healthy samples are separately clustered from each other, which indicates distinct and significant SERS spectral differentiation for both. PCA provides information only to visualize the data but does not provide any information regarding the variation in data that are separately clustered.
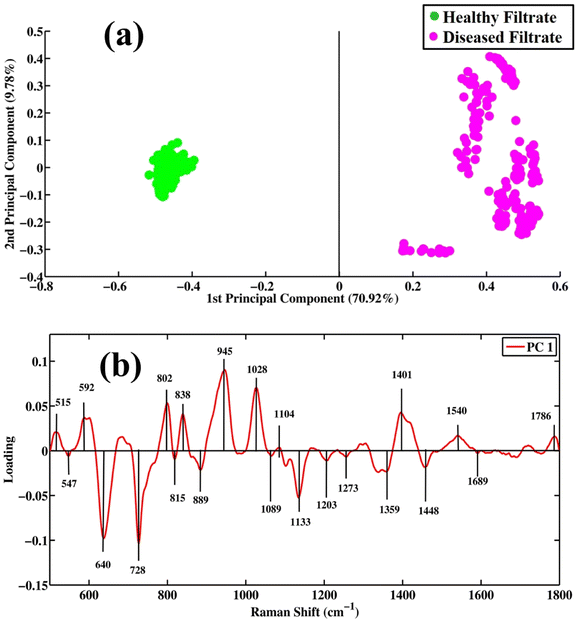 | ||
| Fig. 3 Pair-wise PCA analysis. (a) Scatter plot and (b) loadings between SERS spectral datasets of filtrate portions of healthy and tuberculosis disease samples. | ||
Fig. 3(b) shows loadings between SERS spectral bands of filtrates of disease and healthy samples. The negative loading shows the spectra in the scatter plot clustered on the negative side of the axis, while positive loading shows the spectra clustered on the positive axis of the scatter plot. The negative loadings include 547 (lipids), 640 (uric acid), 728 (hypoxanthine), 815 (C–O–C stretching vibration), 889 (uric acid), 1089 (phospholipids), 1133 (palmitic acid), 1203 (uric acid), 1273 (ring-breathing mode of adenine), 1359 (guanine (N7, B, Z marker)), 1448 (C–H vibration) and 1689 cm−1 (amide-I). The loadings of TB filtrates on the positive side of PC-1 include the SERS bands of 515 (phosphatidylinstole), 592 (phosphatidylinositol), 802 (thymine-based ring-breathing mode (DNA/RNA)), 838 (α-saccharide, α-anomers of glucose), 945 (stretching vibrations of single bonds in amino acids, including proline and valine), 1028 (stretching of (O–CH3)), 1401 (thymine), 1540 (aromatic hydrogen, amide carbonyl vibrations) and 1786 cm−1 (C–C stretching of lipids).
SERS spectral bands are extremely influenced by the elimination of larger proteins from biological samples, such as blood serum, through filtration with a 50 kDa filter. By separating major proteins with high molecular weights from smaller molecular weight proteins using a cellulose membrane, the efficiency of SERS spectroscopic detection is increased, which leads to greater differentiation of the SERS spectral bands associated with healthy and tuberculosis samples.
The presence of phosphatidylinositol in disease filtrates and its absence in healthy filtrates, as indicated by peaks observed at 519, 708, and 1786 cm−1, suggests that bacteria causing tuberculosis produce this molecule, and it could potentially play a pathogenetic role. Phosphatidylinositol (PI) is a phospholipid that is commonly found in the plasma membranes of eukaryotic cells, as well as in some bacteria that play a role in a range of cellular processes, including signal transduction, membrane trafficking, and autophagy. PI(4)P is a specific form of PI that is involved in regulating these processes. The peak assignments of 889, 1089, 1099 and 1133 cm−1 were present in the filtrate portion of serum samples from healthy and diseased patients. A common peak at 592 cm−1 was observed, but it was higher in intensity in the disease filtrate portion due to the presence of phosphatidylinositol.
The SERS peaks at 945 and 1540 cm−1 are associated with proline (protein) were only present in the disease filtrate portion and were absent in healthy filtrate samples. The tuberculosis bacterium secretes various proteins that assist in bacterial survival and replication within host cells. These proteins are involved in various processes such as nutrient acquisition, virulence, and immune evasion. For example, one protein called ESAT-6 is known to assist the tuberculosis bacterium in escaping from immune cells and establishing infection in the host. The SERS peak appearing at 815 cm−1 is specific to healthy serum filtrate and is related to the proteins. The other peaks are present in healthy and disease filtrate portions, but are higher in intensity in healthy samples. These peaks correspond to various protein assignments and vibrations, such as the symmetric breathing of tryptophan (743 cm−1) and the C–H vibration (1448 cm−1). Some SERS spectral features of DNA/RNA that appear in the filtrate portion of disease serum include 681, 862, 1028, and 1314 cm−1.
3.4. Partial least squares discriminant analysis (PLS-DA)
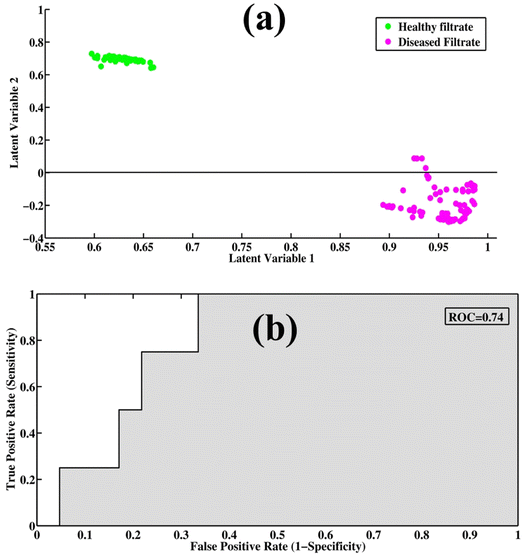
PLS-DA is a supervised model for the qualitative data analysis of filtrate portions of healthy and disease samples. The PCA model provides only visualization regarding SERS spectral datasets, but PLS-DA provides quantitative information regarding the dataset. It utilizes previous information for the datasets and provides quantitative discrimination of the SERS spectra of samples. The spectral datasets of the filtrates were randomized and split into 60% training datasets, with test datasets up to 40% to remove the bias from the model. PLS-DA works by discriminating the spectral datasets into two classes (TB and healthy), which can be differentiated through scatter score plot. The least prediction error of variables was predicted through optimum latent variables to find the maximum level of accuracy. The model calibration was performed with leave-one-out cross-validation (LOOCV) to determine the optimal number of latent variables (LVs). Three latent variables were determined to be optimum for the development of the classification model.Fig. 4(a) shows the PLS-DA score plot between SERS spectra of healthy and disease filtrate samples. The PLS-DA model was used to classify the samples into two different categories based on their characteristic SERS spectral features. In the score plot, the cluster of green dots represents healthy filtrate samples on the positive axis, while the disease filtrate samples are represented by pink dots and are present on the negative axis. Fig. 4(b) shows a graphical depiction of the (ROC) curve, which is a measure of the performance of the PLS-DA model. The area under the curve (AUC) value of 0.74 indicates very good performance of the model. An AUC value close to 1 represents high accuracy and validity of the model, while an AUC value below 0.5 indicates that the model is not fit, and the results may be invalid or false.
Table 2 provides the values of specificity, sensitivity, precision, and accuracy, which are important metrics for evaluating the performance of a classification model. These metrics provide insights into the ability of the model to correctly classify the SERS spectra of different samples. A PLS-DA scatter score plot of 60 samples (by averaging 10 spectra for each sample: 1 spectrum = 1 sample), where one dot represents one sample, is shown in Fig. S3.†
| Parameters of the PLS-DA model | Values |
|---|---|
| Sensitivity | 0.9874 |
| Specificity | 0.9795 |
| Precision | 0.9481 |
| Accuracy | 0.9711 |
| AUC | 0.74 |
4. Conclusion
Surface-enhanced Raman spectroscopy (SERS) with silver nanoparticles as the substrate was proven valuable in characterizing the blood serum samples of patients with tuberculosis and healthy individuals. The use of a 50 kDa centrifugal filtration device enabled the identification of distinct SERS spectral features that were specifically associated with tuberculosis disease diagnosis in patients, and thus allowed for clear differentiation from healthy controls. The key differentiating features of the SERS spectra are primarily related to samples from patients who are TB-positive, which exhibit peaks at 515, 681, 708, 802, 838, 945, 1028, 1314, 1540, and 1786 cm−1, and their comparison to the filtrate portion from healthy individuals.To demonstrate the efficiency of SERS, advanced multivariate techniques such as principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA) were employed. PCA is used to qualitatively distinguish between SERS datasets containing healthy and tuberculosis samples. The quantitative approach enables precise discrimination of different tuberculosis-positive samples based on the SERS spectral data. The establishment of the PLS-DA model further enhances the categorization of filtrate fractions from healthy individuals and patients tuberculosis on the basis of their SERS spectra. The model exhibits impressive performance with high accuracy (0.9711), precision (0.9481), sensitivity (0.9874), and specificity (0.9795), and thus enables precise disease diagnosis based on the SERS data.
Conflicts of interest
The authors have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University, Saudi Arabia, for funding this work through the research group program under grant number R.G.P. 2/574/44. This study was funded by the Princess Nourah bint Abdulrahman University Researchers Supporting Project (No. PNURSP2024R478), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.References
- J. Chakaya, E. Petersen, R. Nantanda, B. N. Mungai, G. B. Migliori, F. Amanullah, P. Lungu, F. Ntoumi, N. Kumarasamy and M. Maeurer, Int. J. Infect. Dis., 2022, 124, S26–S29 CrossRef PubMed.
- G. Dastgir, M. I. Majeed, H. Nawaz, N. Rashid, A. Raza, M. Z. Ali, M. Shakeel, M. Javed, U. Ehsan and S. Ishtiaq, Photodiagn. Photodyn. Ther., 2022, 38, 102758 CrossRef CAS PubMed.
- B. Acharya, A. Acharya, S. Gautam, S. P. Ghimire, G. Mishra, N. Parajuli and B. Sapkota, Mol. Biol. Rep., 2020, 47, 4065–4075 CrossRef CAS PubMed.
- S. Khan, R. Ullah, S. Shahzad, N. Anbreen, M. Bilal and A. Khan, Photodiagn. Photodyn. Ther., 2018, 24, 286–291 CrossRef PubMed.
- P. Desikan, Indian J. Med. Res., 2013, 137, 442 Search PubMed.
- C. f. D. Control and Prevention, Morb. Mortal. Wkly. Rep., 2009, 58, 7–10 Search PubMed.
- S. D. Gan and K. R. Patel, J. Invest. Dermatol., 2013, 133, e12 CrossRef CAS PubMed.
- C. C. Leung, Int. J. Tuberc. Lung. Dis., 2011, 15, 1279 CrossRef PubMed.
- S. K. Srivastava, C. J. Van Rijn and M. A. Jongsma, RSC Adv., 2016, 6, 17759–17771 RSC.
- M. Behr, S. Warren, H. Salamon, P. Hopewell, A. P. De Leon, C. Daley and P. Small, Lancet, 1999, 353, 444–449 CrossRef CAS PubMed.
- S. Negi, S. Khan, S. Gupta, S. Pasha, S. Khare and S. Lal, Indian J. Med. Microbiol., 2005, 23, 29–33 CrossRef CAS PubMed.
- U. Ehsan, H. Nawaz, M. I. Majeed, N. Rashid, Z. Ali, A. Zulfiqar, A. Tariq, M. Shahbaz, L. Meraj and I. Naheed, Spectrochim. Acta, Part A, 2023, 293, 122457 CrossRef CAS PubMed.
- M. Akram, M. I. Majeed, H. Nawaz, N. Rashid, M. R. Javed, M. Z. Ali, A. Raza, M. Shakeel, H. M. ul Hasan and Z. Ali, Photodiagn. Photodyn. Ther., 2022, 40, 103199 CrossRef CAS PubMed.
- H. J. Butler, L. Ashton, B. Bird, G. Cinque, K. Curtis, J. Dorney, K. Esmonde-White, N. J. Fullwood, B. Gardner and P. L. Martin-Hirsch, Nat. Protoc., 2016, 11, 664–687 CrossRef CAS PubMed.
- J. Smolsky, S. Kaur, C. Hayashi, S. K. Batra and A. V. Krasnoslobodtsev, Biosensors, 2017, 7, 7 CrossRef PubMed.
- S. Farquharson, C. Shende, F. E. Inscore, P. Maksymiuk and A. Gift, J. Raman Spectrosc., 2005, 36, 208–212 CrossRef CAS.
- K. Shahzad, H. Nawaz, M. I. Majeed, R. Nazish, N. Rashid, A. Tariq, S. Shakeel, A. Shahzadi, S. Yousaf and N. Yaqoob, Anal. Lett., 2022, 55, 1731–1744 CrossRef CAS.
- N. L. Anderson, M. Polanski, R. Pieper, T. Gatlin, R. S. Tirumalai, T. P. Conrads, T. D. Veenstra, J. N. Adkins, J. G. Pounds and R. Fagan, Mol. Cell. Proteomics, 2004, 3, 311–326 CrossRef CAS PubMed.
- M. Shakeel, M. I. Majeed, H. Nawaz, N. Rashid, A. Ali, A. Haque, M. U. Akbar, M. Tahir, S. Munir and Z. Ali, Photodiagn. Photodyn. Ther., 2022, 40, 103145 CrossRef CAS PubMed.
- N. Mehmood, M. W. Akram, M. I. Majeed, H. Nawaz, M. A. Aslam, A. Naman, M. Wasim, U. Ghaffar, A. Kamran and S. Nadeem, RSC Adv., 2024, 14, 5425–5434 RSC.
- S. Shakeel, H. Nawaz, M. I. Majeed, N. Rashid, M. R. Javed, A. Tariq, B. Majeed, A. Sehar, S. Murtaza and N. Sadaf, Photodiagn. Photodyn. Ther., 2022, 39, 102949 CrossRef CAS PubMed.
- R. J. Lakshmi, V. Kartha, C. Murali Krishna, J. R. Solomon, G. Ullas and P. Uma Devi, Radiat. Res., 2002, 157, 175–182 CrossRef CAS.
- W. T. Cheng, M. T. Liu, H. N. Liu and S. Y. Lin, Microsc. Res. Tech., 2005, 68, 75–79 CrossRef CAS PubMed.
- E. Avci, H. Yilmaz, N. Sahiner, B. G. Tuna, M. B. Cicekdal, M. Eser, K. Basak, F. Altıntoprak, I. Zengin and S. Dogan, Cancers, 2022, 14, 5021 CrossRef CAS PubMed.
- J. W. Chan, D. S. Taylor, T. Zwerdling, S. M. Lane, K. Ihara and T. Huser, Biophys. J., 2006, 90, 648–656 CrossRef CAS PubMed.
- C. Krafft, L. Neudert, T. Simat and R. Salzer, Spectrochim. Acta, Part A, 2005, 61, 1529–1535 CrossRef PubMed.
- Z. Cheng, H. Li, C. Chen, X. Lv, E. Zuo, X. Xie, Z. Li, P. Liu, H. Li and C. Chen, Photodiagn. Photodyn. Ther., 2023, 41, 103284 CrossRef CAS PubMed.
- T. Nguyen, C. Gobinet, J. Feru, S. B. Pasco, M. Manfait and O. Piot, J. Spectrosc., 2012, 27, 421–427 CrossRef CAS.
- Z. Huang, A. McWilliams, H. Lui, D. I. McLean, S. Lam and H. Zeng, Int. J. Cancer, 2003, 107, 1047–1052 CrossRef CAS PubMed.
- Y. Wei, Y.-y. Zhu and M.-l. Wang, Optik, 2016, 127, 7902–7907 CrossRef CAS.
- N. Stone, C. Kendall, J. Smith, P. Crow and H. Barr, Faraday Discuss., 2004, 126, 141–157 RSC.
- A. C. S. Talari, Z. Movasaghi, S. Rehman and I. U. Rehman, Appl. Spectrosc. Rev., 2015, 50, 46–111 CrossRef CAS.
- A. Tariq, M. R. Javed, M. I. Majeed, H. Nawaz, N. Rashid, S. Yousaf, A. Ijaz, N. u. Huda, H. Tahseen and A. Naman, Anal. Lett., 2023, 1–14 CAS.
- Y. Zhang, X. Lai, Q. Zeng, L. Li, L. Lin, S. Li, Z. Liu, C. Su, M. Qi and Z. Guo, Laser Phys., 2018, 28, 035603 CrossRef.
- F. Liu, H. Gu, Y. Lin, Y. Qi, X. Dong, J. Gao and T. Cai, Spectrochim. Acta, Part A, 2012, 85, 111–119 CAS.
- L. Silveira Jr, S. Sathaiah, R. A. Zângaro, M. T. Pacheco, M. C. Chavantes and C. A. Pasqualucci, Lasers Surg. Med., 2002, 30, 290–297 CrossRef PubMed.
- Z. Huang, A. McWilliams, S. Lam, J. English, D. I. McLean, H. Lui and H. Zeng, Int. J. Oncol., 2003, 23, 649–655 Search PubMed.
- B. R. Wood, M. A. Quinn, B. Tait, M. Ashdown, T. Hislop, M. Romeo and D. McNaughton, Biospectroscopy, 1998, 4, 75–91 CrossRef CAS.
- G. Shetty, C. Kendall, N. Shepherd, N. Stone and H. Barr, Br. J. Cancer, 2006, 94, 1460–1464 CrossRef CAS PubMed.
- D. Kaushik and B. Michniak-Kohn, AAPS PharmSciTech, 2010, 11, 1068–1083 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra00420e |
| ‡ The first two authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |