Open Access Article
Open Access ArticleLysine-based non-cytotoxic ultrashort self-assembling peptides with antimicrobial activity†
Nagihan Özbeka,
Eugenio Llorens Vilarrocha *b,
Begonya Vicedo Jover
*b,
Begonya Vicedo Jover b,
Eva Falomir Ventura
b,
Eva Falomir Ventura c and
Beatriu Escuder
c and
Beatriu Escuder *a
*a
aInstitute of Advanced Materials (INAM), Universitat Jaume I, 12071 Castelló, Spain. E-mail: escuder@uji.es
bDepartment of Biology, Biochemistry and Natural Sciences, Universitat Jaume I, 12071 Castelló, Spain
cDepartment of Inorganic and Organic Chemistry, Universitat Jaume I, 12071 Castelló, Spain
First published on 8th May 2024
Abstract
Peptide-based molecules and their hydrogels are useful materials for biomedical applications due to the reversible nature of their self-assembly as well as the diversity of nanostructures that can be created starting from low-molecular weight compounds. In this study, we have focused on comprehending the characteristics of fibrillar networks of L-lysine-based self-assembled dipeptide hydrogels with a focus on their antibacterial properties. For that purpose, L-lysine has been complemented with hydrophobic aromatic moieties coming from L-phenylalanine and benzyloxyxarbonyl N-capping. In addition, the peptide C-terminus is blocked with alkylamides of different chain lengths which introduces additional dispersive interactions and hydrophobicity. These materials were well characterized by transmission electron microscopy, scanning electron microscopy, wide-angle powder X-ray diffraction and oscillatory rheology. Finally, biocompatibility and antimicrobial tests were performed showing that these hydrogels are compatible with HEK 293 cells and present a remarkable antibacterial activity against both Gram positive (S. aureus) and Gram negative (E. coli) bacteria.
Introduction
A pressing reality in today's world is the alarming rise of bacterial resistance caused by the excessive and improper use of antibiotics. This escalating issue poses a significant threat not only to human health but also to animals, agriculture, and the environment as well. The global consequences of microbial resistance have reached a critical stage, as indicated by a comprehensive report prepared by experts. It is estimated that over half a million individuals worldwide lose their lives annually due to infections caused by drug-resistant bacteria. Moreover, if effective treatment options are not developed and implemented swiftly, the number of fatalities could rise to millions in the near future, emphasizing the urgent need for immediate action.1,2 To handle this growing menace, researchers have been focused on innovative and targeted approaches in therapeutic research. Highly promising advancements include the utilization of liposomes, diverse polymeric structures, peptide-based hydrogels and nanoparticles, which are cutting-edge methods that hold great potential in the battle against microbial resistance.3,4 As biologically substantial molecules, supramolecular hydrogels have been identified as one of the most promising soft materials for modern biomedical applications.5–8 Thanks to their reversible gel–sol transition capability, which is specific to their non-covalent cross-links, supramolecular hydrogels show great potential as biomaterial scaffolds for diagnosis and therapy.9,10 For instance, the development of new supramolecular hydrogelators and the variety of their structures and functions are possible as a result of the non-covalent interactions.11–15 In addition, in many cases the inspiration for these optimizations is nature, leading to biomimetic or bioinspired materials.16–18 In this context, short and ultrashort peptides, and even single amino acid derivatives, have been reported as effective hydrogelators.19 Particularly, phenylalanine-based short peptides have attracted considerable attention due to their high tendency to aggregate by intermolecular π–π interactions.20–22 Moreover, additional aromatic moieties have been included at the N-terminus of peptides with the aim of increasing those π-stacking interactions. For instance, the introduction of the fluorenylmethyloxycarbonyl (Fmoc) group to the N-terminus of the diphenylalanine sequence (Fmoc-FF) promotes to form hydrogels by self-assembly whereas the diphenylalanine sequence (i.e., NH2-Phe-Phe-OH) is not able to form hydrogels alone and tends to form crystalline nanotubes.23–25 Nevertheless, due to the sensitivity of the Fmoc group to cleavage at pH values above 10, which increases when Fmoc-based peptide gelators dissolve in basic aqueous solutions to initiate gelation, and some unidentified toxic Fmoc-FF degradation products by releasing the highly reactive substance, dibenzofulvalene, scientists considered using alternative capping groups such as naphthalene-based or benzyloxycarbonyl (Z) groups among others.26 Naphthalene-based capping groups are, in general, not base-labile and present several sites for additional functionalization. Benzyloxycarbonyl compounds are typically metabolized in the body through hydrolysis, which is catalyzed by enzymes such as carboxylesterases and peptidases. This process releases the desired functional peptide and produces a benzyloxycarbonyl derivative, which can undergo further metabolism through oxidation, reduction, and conjugation reactions to form benzoic acid derivatives. These derivatives can then be excreted through conjugation with glucuronic acid or sulfate. The metabolism of benzyloxycarbonyl compounds can vary based on factors such as the compound's specific structure and the presence of other functional groups. Additionally, the use of compounds bearing the Z (benzyloxycarbonyl) group is prevalent in both peptide chemistry and pharmaceutical research. These compounds function as protecting groups for amino acids during the process of peptide synthesis, and they play a critical role in the development of peptide-based drugs and therapeutics.27,28 On the other hand, the use of Z group requires hydrophobic peptide sequences such as diphenylalanine to form hydrogels owing to their reduced aromatic surface.29,30 Following the previous results from our research group,31,32 different forms of F-based peptides protected with benzyloxycarbonyl (Z) group adjust the self-assembly characteristics to obtain amyloid aggregates. Indeed, the FF combination is able to create well-formed supramolecular networks by intermolecular interactions and intrinsic hydrophobicity as driving forces.33On the other hand, antimicrobial peptides (AMPs), have become very attractive tools nowadays due to their broad-spectrum antibacterial effects, stability against bacterial antibiotic resistance mechanisms unlike most conventional antibiotics and extraordinary biocompatibility features. Particularly, thanks to easy synthesis and lower cost conditions compared to long-chain peptides, short peptide sequences show a great potential.34–37 Briefly, the antibacterial mechanism might be foreseen by the interaction of positively charged amino acid residues with the negatively charged bacterial cell membranes. However, studies on antimicrobial peptides effective on both Gram-negative and Gram-positive bacterial types are very limited in the literature.38–44 A recent example proved that the antimicrobial effect of Fmoc-KnF, could not be increased by increasing the number of L-lysine residues in the structure.45 Also, there are still some difficulties with the solubility in water and limited knowledge about lysine-based dipeptide's antibacterial effect.
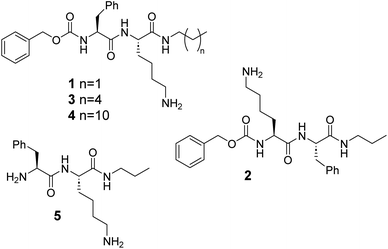
Here, we report on the self-assembly behaviour of Z-capped dipeptides 1–4 based on L-lysine and L-phenylalanine bearing alkylamide groups of different length at C-terminus (Fig. 1). The presence of the benzyloxycarbonyl (Z) group mimics the presence of an additional aromatic residue without the need of an additional F amino acid in the sequence. On the other hand, the length of the alkylamide fragment introduces additional control of the hydrophobicity and van der Waals interactions. These compounds and their hydrogels have been examined for antimicrobial activity with antibacterial assays, live/dead tests and cytotoxicity studies on human cell viability. Altogether, the combination of a remarkable antimicrobial activity with a simple and cost-effective molecular design makes these hydrogels promising candidates to fight against microbial infections.46,47
Experimental
Peptide synthesis
The designed peptides used in this study, compounds 1–5, were synthesized in gram scale by solution phase peptide synthesis and stored as hydrochloride salts (see ESI†).Aggregation studies
The hydrogels were prepared as the following methodology: the desired amount of each compound was weighted in cylindrical screw-capped vials with an internal diameter of 1.2 cm and 4.5 cm in length. Due to the lack of solubility of compounds 1–4 in water, the samples were dissolved by heating at concentrations of mM range in the aqueous medium and cooled down to r.t.Nuclear magnetic resonance spectroscopy studies
NMR spectra were recorded at 400 MHz 1H NMR and 101 MHz 13C NMR in a Bruker Ascend™ 400 NMR spectrometer in different deuterated solvents at 30 °C with the solvent signals as an internal reference.Mass spectrometry
The mass spectra were measured with a triple-quadrupole mass spectrometer with an electrospray source (waters). A capillary voltage of 3.5 kV was used in the positive scan mode. The observed isotopic pattern of each compound perfectly matched the theoretical isotope pattern calculated from their elemental composition using the MassLynx 4.0 program.FT-IR
Prepared hydrogels according to the protocol were lyophilized and FT-IR spectra were collected in a JASCO FTIR-6200 spectrometer – ATR PRO ONE at a wavenumber range of 400–4000 cm−1.Transmission electron microscopy
Samples were applied directly onto Carbon support film on 200 mesh copper grids. Excess amount of sample was drained gently, and extra salts from the buffer were washed with 5 μL of water once and the residue of water removed. Transmission electron microscopy (TEM) images were recorded in a JEM 2100plus Transmission Electron Microscope.Scanning electron microscopy
Indicated freeze-dried gels were placed on top of an aluminium specimen mount stub and sputtered with Pt. Scanning electron microscopy (SEM) analysis was performed in a Leica-Zeiss LEO 440 microscope.Oscillatory rheology
Rheological measurements were conducted on a Discovery RH-3 rheometer from TA instruments using a steel parallel plate-to-plate geometry (40 mm diameter). The gap distance was fixed at 500 μm. The hydrogels were prepared under the desired conditions and the hot solution of samples transferred to the rheometer plate, well-covered with the instrumental metal cover to avoid evaporation and stabilized for 5 minutes before analysing. Viscoelastic characteristics were studied under oscillatory experiments. Frequency and strain sweeps were examined at 25 °C. The experimental conditions were arranged by running a stress sweep step (0.1–100 Pa at 1 Hz) and a frequency sweep step (0.1–100 Hz at 1 Pa).Wide angle X-ray diffraction
Data were collected at r.t. with a Bruker D4 Endeavor X-ray powder diffractometer by using Cu-α radiation. Freeze-dried powders of hydrogels were placed on a sample holder and data were collected for 2θ values between 2° and 40° with a step size of 0.03° and a time step of 10 s.Bacterial culture and antibacterial assay
The compounds 1–4 were used to test their antimicrobial properties as glass coating. In detail, hydrogels 1–3 at their mgc (1; 10 mM, 2; 2.85 mM; 3; 0.5 mM) and compound 4 (1.6 mM; above cac; see ESI†), were applied to 15 mm diameter glass slides, 500 microliters for each slide, and dried by air for 24 hours. The assays were performed in Luria-Bertani Broth (LB) medium diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50. A blank control containing bacteria incubated in LB medium diluted 50 times was also added. Before testing the antimicrobial properties of the materials, the glass slides coated with different compounds were sterilized under UV light for 30 minutes on each side. The antimicrobial effect of the compounds was tested against E. coli strain NCIMB 9484 and S. aureus strain ATCC 29213 obtained from the Spanish Type Culture Collection (CECT). One day before antimicrobial testing, the bacteria were activated by inoculating a small amount of the suspension from a frozen stock onto Luria-Bertani agar medium and incubated for 24 h at 37 °C. From the obtained culture, a suspension was prepared in 10 mM MgSO4 for adjusted to a final concentration of 105 cfu mL−1 in LB medium diluted 50 times. Three samples of each compound-coated glass were placed in sterile 12-well plates. Then, 100 μL aliquots of the LB1/50 bacterial suspension was added to each glass. Three blank controls were also added in the same plate. The plates were incubated at 37 °C for 24 h. After 24 h of exposure to the compound-coated glasses the antibacterial activities of the compounds were evaluated by the drop plate counting method. For this purpose, each glass was washed with 2 mL of sterile MgSO4 (10 mM), 200 μL suspension aliquots were collected and four serial decimal dilutions were prepared. 5 μL of each dilution was inoculated on a solid LB plate. After 24 h incubation, the individual colonies were counted. Three independent experiments were performed with three samples for each compound and each bacterium.
50. A blank control containing bacteria incubated in LB medium diluted 50 times was also added. Before testing the antimicrobial properties of the materials, the glass slides coated with different compounds were sterilized under UV light for 30 minutes on each side. The antimicrobial effect of the compounds was tested against E. coli strain NCIMB 9484 and S. aureus strain ATCC 29213 obtained from the Spanish Type Culture Collection (CECT). One day before antimicrobial testing, the bacteria were activated by inoculating a small amount of the suspension from a frozen stock onto Luria-Bertani agar medium and incubated for 24 h at 37 °C. From the obtained culture, a suspension was prepared in 10 mM MgSO4 for adjusted to a final concentration of 105 cfu mL−1 in LB medium diluted 50 times. Three samples of each compound-coated glass were placed in sterile 12-well plates. Then, 100 μL aliquots of the LB1/50 bacterial suspension was added to each glass. Three blank controls were also added in the same plate. The plates were incubated at 37 °C for 24 h. After 24 h of exposure to the compound-coated glasses the antibacterial activities of the compounds were evaluated by the drop plate counting method. For this purpose, each glass was washed with 2 mL of sterile MgSO4 (10 mM), 200 μL suspension aliquots were collected and four serial decimal dilutions were prepared. 5 μL of each dilution was inoculated on a solid LB plate. After 24 h incubation, the individual colonies were counted. Three independent experiments were performed with three samples for each compound and each bacterium.
Live/dead test
The proportion of living vs. dead cells was quantified using the fluorescent LIVE/DEAD BacLight Bacterial Viability Kit, L13152 (Molecular Probes, Invitrogen, Paisley, UK). For live and dead cell quantification, 50 μL of bacterial suspension was mixed with 25 μL of each of the two components of the LIVE/DEAD BacLight kit and incubated in the dark for 20 min. Then, the ratio of live/dead cells was determined by flow cytometric analysis. Samples were analyzed on a Becton Dickinson FACScan flow cytometer using the CellQuest software, which was also used to determine the percentage of live/dead cells. SYTO9 (live) was excited at 480 nm and fluorescence was analyzed at 500 nm, whereas propidium iodide (dead) was excited at 490 nm and fluorescence was analyzed at 635 nm.Cell culture
Cell culture media were purchased from Gibco (Grand Island, NY). Fetal bovine serum (FBS) was obtained from Harlan-Seralab (Belton, U.K). Supplements and other chemicals not listed in this section were obtained from Sigma Chemical Co. (St. Louis, MO). Plastics for cell culture were supplied by Thermo Scientific BioLite. For tube formation assay, an IBIDI μ-slide angiogenesis (IBIDI, Martinsried, Germany) were used. All tested compounds were dissolved in DMSO at a concentration of 20 mM and stored at −20 °C until use. HEK-293 cell line was maintained in Dulbecco's modified Eagle's medium (DMEM) containing glucose (1 g L−1), glutamine (2 mM), penicillin (50 μg mL−1), streptomycin (50 μg mL−1), and amphotericin B (1.25 μg mL−1), supplemented with 10% FBS.Cell proliferation assay
In 96-well plates, 5 × 103 cells per well were incubated with serial dilutions of the tested compounds in a total volume of 100 μL of their respective growth media. The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma Chemical Co.) dye reduction assay in 96-well microplates was used. After 2 days of incubation (37 °C, 5% CO2 in a humid atmosphere), 10 μL of MTT (5 mg mL−1 in phosphate-buffered saline, PBS) was added to each well, and the plate was incubated for a further 3 h (37 °C). After that, the supernatant was discarded and replaced by 100 μL of DMSO to dissolve formazan crystals. The absorbance was then read at 550 nm by spectrophotometry. For all concentrations of compound, cell viability was expressed as the percentage of the ratio between the mean absorbance of treated cells and the mean absorbance of untreated cells. Three independent experiments were performed, and the IC50 values (i.e., concentration half inhibiting cell proliferation) were graphically determined using GraphPad Prism 4 software.Results and discussion
Synthesis
Compounds 1–4 were prepared by solution peptide synthesis and contain L-phenylalanine (F) and L-lysine (L) residues in different positions (see the ESI† for details). The Z group was used at the N-terminus as an additional aromatic fragment, reminiscent of F side chain, which is able to promote aggregation through π-stacking interactions,48,49 and C-terminus was designed with different alkyl chain lengths. Compound 1 (ZFKC3) is an N-capped FK derivative, while compound 2 (ZKFC3) is a reverse of the peptide sequence order. Compound 5 (FKC3), as an unprotected analogue of 1, was synthesized to observe the Z fragment effect on aggregation behaviour. Further, compounds 3 (ZFKC6) and 4 (ZFKC12) have the same molecular concept with a longer alkyl chain at C-terminus, to compare them in terms of the hydrophobicity effect on the self-assembly behaviour.Aggregation studies and characterization
The aggregation behaviour of compounds 1–4 was studied in terms of their ability to form hydrogels at physiological pH. For that purpose, the hydrochloride salts were dissolved in PBS (0.1 M, pH = 7.4) by heating and then cooled down to r.t. followed by aging for 24 h. The formation of hydrogels was qualitatively assessed by the vial inversion test and their apparent minimum gel concentrations (mgc) determined by sequential dilution-jellification experiments. As can be seen in Fig. 2, while 1 forms an opaque gel in PBS (mgc: 10 mM), 2 and 3 lead to transparent gels (mgc: 2.8 mM and 0.5 mM, respectively). In contrast, 4 was not able to form a gel in this medium.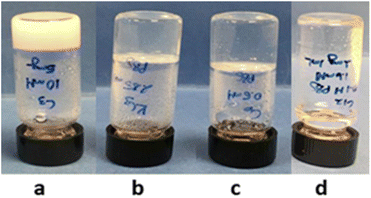 | ||
| Fig. 2 The macroscopic aspect of the hydrogels in PBS (0.1 M, pH = 7.4). (a) 1 (10 mM), (b) 2 (2.85 mM), (c) 3 (0.5 mM) and (d) 4 (1.6 mM). | ||
It has been previously reported in the literature that the self-assembly of diphenylalanine and its derivatives can lead to a wide variety of nanostructures. Gazit et al., for instance, examined how different metastable morphologies transform from monomers to spheres and then from fibrils to stable tubes, in terms of kinetics and thermodynamics of aggregation.50 As known, comprehending the kinetics of self-assembly allows the development of strategies to guide the formation of specific nanostructures. Indeed, controlling the formation of nanostructures is possible by directing factors such as concentration, temperature and solvent composition, which can influence the rate and pathway of aggregation.
Morphology analysis was performed by electron microscopy (TEM and SEM) to examine the characteristic network structures in PBS. As can be seen in Fig. 3, the three hydrogel-forming compounds (1–3) showed fibrillar networks of high aspect ratio with slight differences in fiber width (for additional images see Fig. S1†). Furthermore, SEM images of the three hydrogels support the observations made by TEM in terms of fibrillar network density and length differences between 1, 2 and 3 at their mgc. (Fig. 4). On the other hand, compound 4, which fails to form hydrogels within the studied concentration range (1 to 15 mM), precisely forms regular spherical particles of ca. 36 nm of diameter as well as few short fibers that did not make a self-sustainable hydrogel network (Fig. 3D) at 1.6 mM, which is higher than its critical aggregation concentration (cac) (see Table S1†). Additionally, samples of compound 4 at a higher concentration (10 mM) showed the formation of rods of different lengths that failed to form hydrogels in PBS (Fig. S3†). Therefore, this dipeptide-amphiphile is forming micelles at low concentration and micellar rods upon increasing concentration. In contrast, compound 4 was able to form transparent hydrogels at concentrations above 1 mM in distilled water, in the absence of buffer salts. Salts are known to play a remarkable effect on the self-assembly of amphiphiles in water.
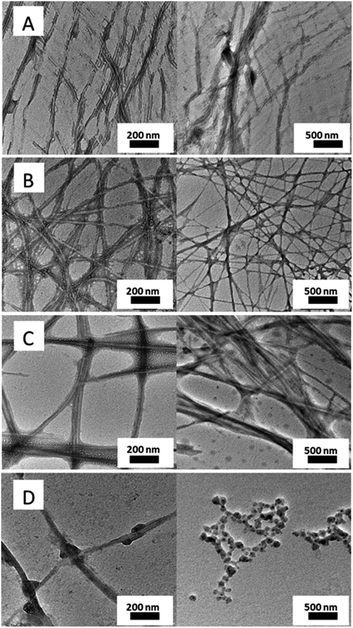 | ||
| Fig. 3 TEM micrographs of (A) 1 (10 mM), (B) 2 (2.85 mM), (C) 3 (0.5 mM) and (D) 4 (1.6 mM) in PBS (0.1 M, pH = 7.4). | ||
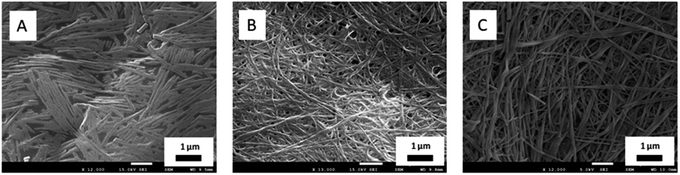 | ||
| Fig. 4 SEM micrographs of (A) 1 (10 mM), (B) 3 (0.5 mM) and (C) 2 (2.85 mM) in PBS (0.1 M, pH = 7.4). | ||
To monitor the mechanical behavior of hydrogels, we analyzed them by oscillatory rheology using a plate-to-plate geometry. The hot solution of pre-gels was transferred to rheometer plates and after 5 min., frequency and oscillatory strain sweeps were examined (see Fig. S4–S6†). Although, hydrogels presented a viscoelastic behavior typical of a gel with G′ > G′′; G′ values were below 1000 Pa, indicative that the hydrogels were very weak.
In order to get a structural insight at the supramolecular level, the hydrogels were further studied by fluorescence spectroscopy. Particularly, to monitor the effect of Z fragment and the presence of a ZF hydrophobic block on aggregation, fluorescence was evaluated. As can be seen in Fig. 5, the maximum of emission, λem, for hydrogels 1 and 2 at mgc, has been shifted to 296 and 285 nm respectively, compared to soluble analogue 5 (10 mM, λem 281 nm). Fluorescence studies for hydrogels revealed a common shift of 4–15 nm overall, suggesting the presence of strong π-stacking interactions (Fig. 5).51,52 In addition, in the presence of ZF block, the λem shows the biggest shift together to an additional broad band (350–500 nm) that has been previously assigned to a strong aromatic π–π stacking interaction.53
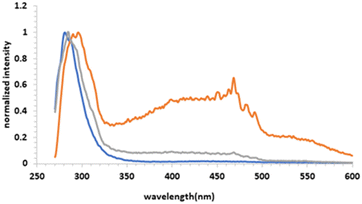 | ||
| Fig. 5 Fluorescence emission spectra (λex = 260 nm) of hydrogel 1 (orange line), hydrogel 2 (grey line) at mgc and solution of 5; 10 mM (blue line) in PBS (0.1 M, pH = 7.4). | ||
FT-IR and WAXD were performed to get insight into the connection between molecular structure and morphology of aggregated samples with the lyophilized samples. WAXD of lyophilized gels of compounds 1–3 in PBS were used to get information about the crystallinity of the aggregates (Fig. 6). As can be seen, only compound 1 showed some diffraction peaks with a low-angle reflection corresponding to a distance of 16.3 Å which could fit with the extended molecular size. Samples of compounds 2 and 3 were amorphous. These results are in agreement with the macroscopic and microscopic observations. Xerogel of compound 1, which forms an opaque material composed by rigid rod-like fibers as seen by SEM, is more crystalline than xerogels of compounds 2 and 3, which form transparent hydrogels with a more flexible fibrillar network.
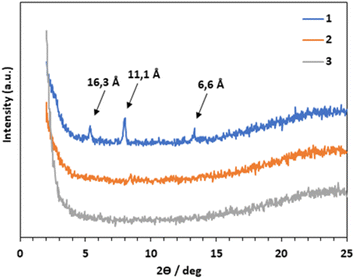 | ||
| Fig. 6 WAXD patterns of aggregates formed by 1 (blue line), 2 (orange line) and 3 (grey line). Samples were prepared from the lyophilized hydrogels of each compound at their mgc in PBS. | ||
Moreover, significant information about H-bonding interactions between amide groups can be obtained from the FT-IR of peptides. All aggregates present an amide I stretching band in the range 1622–1647 cm−1 assignable to a β-sheet structure as the significative feature of these aggregates54 (see Fig. S7†). Briefly, the self-assembly of F–K dipeptide derivatives results from a balanced combination of intermolecular interactions including H-bonding, π–π stacking of aromatic residues and the hydrophobic effect.
Overall, it seems that compounds 1–3 form stable fibrillar networks in PBS whereas compound 4, although it self-assembles into spherical particles and thin rods, it is not able to form a self-sustained network. It is known that the ability to form fibrillar networks effective for hydrogelation depends of a finely tunned balance between solubility and hydrophobicity.
Antibacterial studies
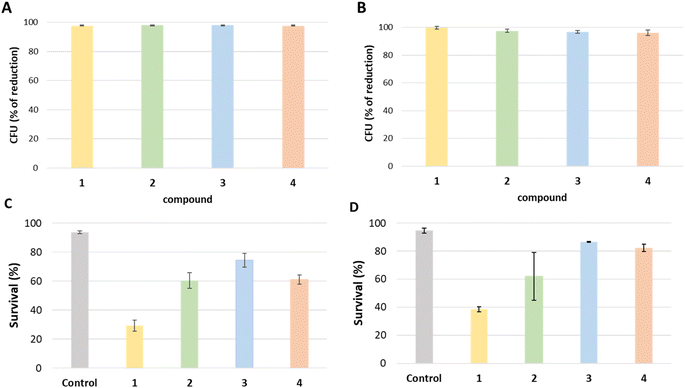
One of the main problems we are currently facing is the control of multi-resistant bacteria that are emerging in hospital environments. To solve this problem there we need to find alternatives to reduce the pressure on current antibiotics on these environments.55,56 As part of the response to this situation, here we analyzed the antimicrobial effect of 1, 2, 3 and 4 against Gram-negative (Escherichia coli) and Gram-positive (Staphylococcus aureus) by exposing two bacterial cultures to a glass surface covered with each compound for 24 hours. All tested compounds show strong antibacterial effects against E. coli and S. aureus whereas the antibacterial effect on E. coli was slightly higher for each sample and the growth rate of the same microorganisms on the mentioned surface was considerably low. In detail, the determined reduction in bacterial count against E. coli experiment was higher than 97% for all four samples compared to the control group, 97.5%, 98.1%, 98.0% and 97.6% respectively (Fig. 7A). On the other hand, 1 has the highest reduction at 99.9% in bacterial count against S. aureus compared to the control group whereas 2, 3 and 4 have relatively lower reductions with 97.5%, 96.8% and 96.1% respectively (Fig. 7B).To check whether the tested compounds had a bactericidal (kills the bacteria) or bacteriostatic (suppress the growth of bacteria) effect, live and dead cell quantification was performed (Fig. 7C and D). Consequently, 1 was the most effective in reducing the number of live bacteria. The results for E. coli obtained from stained cells that were counted with a flux cytometer showed the control group has an average percentage of live cells of 93.6% while treatments 1, 2, 3 and 4 show an average percentage of live cells of 29.3%, 60.3%, 74.3%, and 61% respectively. Hence, treatment 1 showed a 68.6% reduction of live cells compared to the control, being the most effective in reducing the number of live bacteria, while treatment 2, 3 and 4 showed 35.6%, 20.5% and 34.7% reduction respectively, as indicated in Fig. 7. Otherwise, the live/dead test result from the control group for S. aureus has an average percentage of live cells of 94.6%, while treatments 1, 2, 3, and 4 show an average rate of live cells of 38.6%, 62%, 86.6%, and 82.3% respectively. Overall, treatment 1 showed a 59.3% reduction of live cells compared to the control, while treatments 2, 3 and 4 showed 34.3%, 8.4% and 13.1% reduction respectively.
The obtained results reveal that compounds 1, 2, 3, and 4 show robust antimicrobial properties against both E. coli and S. aureus in comparison to the control group. Specifically, against E. coli, all tested compounds demonstrate a remarkable reduction in bacterial count, higher than 97% when contrasted with the control. Similarly, in the case of S. aureus, all compounds exhibit a reduction higher than 96%. This notable decrease suggests that besides the bactericidal effect observed in the live/dead test, the compounds inhibit the bacterial growth.
The majority of antibacterial have the potential to exhibit both bactericidal and bacteriostatic properties, and their main effect is to reduce or slow bacterial growth to a level that impedes the infection or can be easily controlled by the immune system.57 Bactericidal compounds kill bacteria, while a bacteriostatic compound stops the growth and reproduction of bacteria but does not necessarily kill them. Results proved that more than 90% of cells are alive in control samples, whereas this number is reduced in all the compounds tested depending on the bacteria. In general, all the compounds showed a higher bactericidal effect against E. coli than against S. aureus. 1 showed the higher bactericidal effect reducing the number of viable cells to 29% and 38% respective for E. coli and S. aureus. 2, 3 and 4 only reduced the number of live bacteria in a range between 60% and 80%, suggesting that the main effect of these compounds is bacteriostatic. The quantitative results for the live/dead test are the ones obtained with the cytometer confirmed successfully by confocal microscopy images that the cells are effectively stained in green; live or red; dead (see ESI†).
The results show different effects of the compounds on the analyzed bacteria types. This could indicate that each compound acts on the bacteria in a different way at a physical or physiological level. These results can lead to the combination of several treatments in order to avoid the acquisition of resistance.58 Regarding the reported studies, the presence of certain positively charged amino acids, such as lysine or arginine, in the molecule designed based on disrupting the negatively charged membranes of bacterial cells through electrostatic interactions, is essential due to the enhancement of interaction between positively charged AMPs and negatively charged components. This original work offers a new perspective for advanced antimicrobial studies with demanded efficiency and cost-effectiveness and a deeper glimpse into the use of AMPs by using a dipeptide sequence, which is distinguished from similar examples.
Cytotoxicity studies
Although peptide-based materials use to be biocompatible and highly biodegradable, here we complement the antimicrobial studies with biosafety assays. The effect of compounds 1–4 on human cell viability (HEK-293) was studied by MTT assay after 48 hours of treatment. This assay allowed us to establish the corresponding IC50 values (expressed as the concentration, in μM, at which 50% of cell viability compared to non-treated cells, is achieved). The data shown in Table 1 were obtained as the average of four experiments. In detail, we found that monomeric dipeptides were not cytotoxic because they do not cause any stress in cells and their proliferation rate in the presence of high micromolar concentration of the dipeptides was similar to the one observed for control cells that were no-treated. When cells are seeded in the presence of hydrogels, we observed no effect on cell proliferation after 48 h of incubation. So we decided to study the effect of monomeric dipeptides on cell viability. For these in vitro studies, we prepared a stock solution of each compound solubilized in DMSO (20 mM), in which the compounds do not form hydrogels. And we checked the effect of our compounds on human cell viability (HEK-293) in doses ranging from 400 μM to 200 nM in cell media by MTT assay after 48 h of treatment.| Compound | HEK-293 |
|---|---|
| 1 | >400 |
| 2 | >400 |
| 3 | >100 |
| 4 | >200 |
The results in Table 1 show that none of the compounds have any effect on human cell viability after 48 h of treatment at doses below 100 μM. In fact, 1 and 2 have no effect at all even at higher concentrations than 400 μM. 4 provoke some changes on the cell morphology at concentrations above 200 μM (Fig. S8†) while 3 was innocuous below 100 μM. We observed that cells preserved a morphology related to epithelial nature of HEK-293 though increased and brighter cytosolic granulation appeared (see Fig. S8†).
Conclusions
In this study, we investigated how the self-assembly of ultrashort peptide molecules with a benzyloxycarbonyl (Z) N-capping group and phenylalanine residues respond to different parameters such as hydrophobic interactions and C-terminus alkyl chain length. Indeed, thanks to the longer alkyl chain contribution of high van der Waals and hydrophobic interactions, 3 and 4 tend to aggregate at lower concentrations compared to short alkyl chain analogues. Self-assembly of lysine derivative dipeptides can be defined as a combination of intermolecular interactions including H- bonding, π–π stacking of aromatic residues and the hydrophobic effect. Not only increase hydrophobicity but also include additional intermolecular interactions N-capping group has been used and π–π interaction at the N-terminus seems that is effective on fibrillar network growth and leading to hydrogelation. Additionally, the aromatic and alkyl fragments' balance, which is driven by the hydrophobic effect, determines the final energetic state of the system. Overall, performed antibacterial activity and cytotoxicity tests proved the great antimicrobial effect of N-capped FK derivative 1 with a short alkyl chain on Gram-negative and Gram-positive bacteria among four designed peptides. Furthermore, 2 as a reverse sequence, KF, represented a remarkable antimicrobial activity against E. coli and S. aureus as well as longer alkyl chain analogs, 3 and 4, showed exciting results at precisely lower concentrations. According to the literature, super long chains were used e.g. NK-2 27 peptides studied by Andrä et al., which is not fairly comparable with our ultrashort peptides.38 Moreover, Lysine tryptophan (KW) long peptide sequences -4–10 peptides examined by Gopal et al.39 and Cao et al. were proposed antimicrobial peptides with a longer sequence of lysine, arginine and phenylalanine amino acid residues whereas would show negative effects on the antimicrobial activity in case of strong crosslinked structure of the hydrogel.42 Another example, Fmoc-F and Fmoc-AA were studied by Gahane et al. but there are effective on Gram-positive bacteria and do not include to evaluate comprehensive toxicological analysis.41 A recent example, the acylated tripeptides and pentapeptides reported by Glossop et al. do not show a strong antimicrobial effect as much as our originally designed ultrashort dipeptides.43 NapFFKK hydrogels studied by Laverty et al. and the dipeptide derivative NapFF displays less percentage of reduction against Escherichia coli-around 84%- compared to our peptides-over 96% against E.coli and S.aureus for all four peptide molecules.44 A very recent study of Hansda et al. reported a one histidine-based amphiphilic peptide with three amino acids, without any alternative derivatives to compare, an a potential antimicrobial agent but again it needs a peptide sequence which contains more than two amino acids.37 Consequently, our study demonstrates the great potential of simple dipeptide materials for antimicrobial application. Our compounds are ultrashort peptides of high biocompatibility that can be easily fabricated in a multigram scale. Moreover, they form weak hydrogels that can be simply spread over surfaces and protect them from bacterial infections. This work is an example on how supramolecular self-assembly converts low molecular weight compounds into high performance soft materials.Author contributions
The original idea was designed by BE with the input from ELV and BVJ. The materials were prepared and fully characterised by NÖ as a part of her PhD. The antimicrobial studies were performed by NÖ with the help and supervision of ELV and BVJ. The cytotoxicity studies were performed by EFV. All authors were involved in data analysis. The paper was written by NÖ under the supervision of BE, with contributions from all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Grant PID2019-110892RB-I00 funded by MCIN/AEI/10.13039/501100011033 and Grant UJI-B2020-21 funded by Universitat Jaume I are acknowledged. N. Ö. thanks the Grisolia Program of the Generalitat Valenciana for a predoctoral fellowship (GRISOLIAP/2020/040). The support given for the TEM studies by Maruxa Peiró Álvarez is sincerely acknowledged.References
- K. H. Luepke, K. J. Suda, H. Boucher, R. L. Russo, M. W. Bonney, T. D. Hunt and J. F. Mohr III, Pharmacotherapy, 2017, 37, 71–84 CrossRef PubMed.
- E. Tacconelli, E. Carrara, A. Savoldi, S. Harbarth, M. Mendelson, D. L. Monnet, C. Pulcini, G. Kahlmeter, J. Kluytmans, Y. Carmeli, M. Ouellette, K. Outterson, J. Patel, M. Cavaleri, E. M. Cox, C. R. Houchens, M. L. Grayson, P. Hansen, N. Singh, U. Theuretzbacher, N. Magrini, A. O. Aboderin, S. S. Al-Abri, N. Awang Jalil, N. Benzonana, S. Bhattacharya, A. J. Brink, F. R. Burkert, O. Cars, G. Cornaglia, O. J. Dyar, A. W. Friedrich, A. C. Gales, S. Gandra, C. G. Giske, D. A. Goff, H. Goossens, T. Gottlieb, M. Guzman Blanco, W. Hryniewicz, D. Kattula, T. Jinks, S. S. Kanj, L. Kerr, M.-P. Kieny, Y. S. Kim, R. S. Kozlov, J. Labarca, R. Laxminarayan, K. Leder, L. Leibovici, G. Levy-Hara, J. Littman, S. Malhotra-Kumar, V. Manchanda, L. Moja, B. Ndoye, A. Pan, D. L. Paterson, M. Paul, H. Qiu, P. Ramon-Pardo, J. Rodríguez-Baño, M. Sanguinetti, S. Sengupta, M. Sharland, M. Si-Mehand, L. L. Silver, W. Song, M. Steinbakk, J. Thomsen, G. E. Thwaites, J. W. M. van der Meer, N. Van Kinh, S. Vega, M. V. Villegas, A. Wechsler-Fördös, H. F. L. Wertheim, E. Wesangula, N. Woodford, F. O. Yilmaz and A. Zorzet, Lancet Infect. Dis., 2018, 18, 318–327 CrossRef PubMed.
- E. Meade, M. A. Slattery and M. Garvey, Antibiotics, 2020, 9, 32 CrossRef CAS PubMed.
- M. Nambiar and J. P. Schneider, J. Pept. Sci., 2022, 28, e3377 CrossRef CAS PubMed.
- I. W. Hamley, Bioconjugate Chem., 2021, 32, 1472–1490 CrossRef CAS PubMed.
- M. Yi, W. Tan, J. Guo and B. Xu, Chem. Commun., 2021, 57, 12870–12879 RSC.
- M. E. Afami, I. El Karim, I. About, S. M. Coulter, G. Laverty and F. T. Lundy, Materials, 2021, 14, 2237 CrossRef CAS PubMed.
- R. G. Weiss, J. Am. Chem. Soc., 2014, 136, 7519–7530 CrossRef CAS PubMed.
- S. Das and D. Das, Front. Chem., 2021, 9, 770102 CrossRef CAS PubMed.
- T. Sarkar, M. Chetia and S. Chatterjee, Front. Chem., 2021, 9, 691532 CrossRef CAS PubMed.
- R. Dong, Y. Pang, Y. Su and X. Zhu, Biomater. Sci., 2015, 3, 937–954 RSC.
- A. K. Das and P. K. Gavel, Soft Matter, 2020, 16, 10065–10095 RSC.
- W. Y. Seow and C. A. E. Hauser, Mater. Today, 2014, 17, 381–388 CrossRef CAS.
- W. T. Truong, L. Lewis and P. Thordarson, in Functional Molecular Gels, 2013, pp. 157–194 Search PubMed.
- J.-H. Kim and M. Benelmekki, in Nanostructured Thin Films, 2019, vol. 14, pp. 103–120 Search PubMed.
- J. Y. C. Lim, Q. Lin, K. Xue and X. J. Loh, Mater. Today Adv., 2019, 3, 100021 CrossRef.
- S. Mondal, S. Das and A. K. Nandi, Soft Matter, 2020, 16, 1404–1454 RSC.
- D. B. Amabilino, D. K. Smith and J. W. Steed, Chem. Soc. Rev., 2017, 46, 2404–2420 RSC.
- J. F. Miravet, B. Escuder, M. D. Segarra-Maset, M. Tena-Solsona, I. W. Hamley, A. Dehsorkhi and V. Castelletto, Soft Matter, 2013, 9, 3558–3564 RSC.
- V. Jayawarna, A. Smith, J. E. Gough and R. V. Ulijn, Biochem. Soc. Trans., 2007, 35, 535–537 CrossRef CAS PubMed.
- S. Burattini, B. W. Greenland, D. H. Merino, W. Weng, J. Seppala, H. M. Colquhoun, W. Hayes, M. E. Mackay, I. W. Hamley and S. J. Rowan, J. Am. Chem. Soc., 2010, 132, 12051–12058 CrossRef CAS PubMed.
- C. Berdugo, J. F. Miravet and B. Escuder, Chem. Commun., 2013, 49, 10608–10610 RSC.
- L. Adler-Abramovich and E. Gazit, Chem. Soc. Rev., 2014, 43, 6881–6893 RSC.
- A. M. Smith, R. J. Williams, C. Tang, P. Coppo, R. F. Collins, M. L. Turner, A. Saiani and R. V. Ulijn, Adv. Mater., 2008, 20, 37–41 CrossRef CAS.
- R. F. Silva, D. R. Araujo, E. R. Silva, R. A. Ando and W. A. Alves, Langmuir, 2013, 29, 10205–10212 CrossRef CAS PubMed.
- W. T. Truong, Y. Su, D. Gloria, F. Braet and P. Thordarson, Biomater. Sci., 2015, 3, 298–307 RSC.
- M. Bodanszky, Principles of Peptide Synthesis, Springer Science & Business Media edn, 2012 Search PubMed.
- K. Isobe, Nat. Biotechnol., 2010, 27, 751–754 CAS.
- K. McAulay, B. Dietrich, H. Su, M. T. Scott, S. Rogers, Y. K. Al-Hilaly, H. Cui, L. C. Serpell, A. M. Seddon, E. R. Draper and D. J. Adams, Chem. Sci., 2019, 10, 7801–7806 RSC.
- D. M. Ryan, S. B. Anderson, F. T. Senguen, R. E. Youngman and B. L. Nilsson, Soft Matter, 2010, 6, 475–479 RSC.
- R. Martí-Centelles and B. Escuder, ChemNanoMat, 2018, 4, 796–800 CrossRef.
- M. Tena-Solsona, S. Alonso-de Castro, J. F. Miravet and B. Escuder, J. Mater. Chem. B, 2014, 2, 6192–6197 RSC.
- M. Tena-Solsona, J. F. Miravet and B. Escuder, Chemistry, 2014, 20, 1023–1031 CrossRef CAS PubMed.
- A. Moretta, C. Scieuzo, A. M. Petrone, R. Salvia, M. D. Manniello, A. Franco, D. Lucchetti, A. Vassallo, H. Vogel, A. Sgambato and P. Falabella, Front. Cell. Infect. Microbiol., 2021, 11, 668632 CrossRef CAS PubMed.
- S. Marchesan, Y. Qu, L. J. Waddington, C. D. Easton, V. Glattauer, T. J. Lithgow, K. M. McLean, J. S. Forsythe and P. G. Hartley, Biomaterials, 2013, 34, 3678–3687 CrossRef CAS PubMed.
- S. Santos, I. Torcato and M. A. Castanho, Biopolymers, 2012, 98, 288–293 CrossRef CAS PubMed.
- B. Hansda, J. Majumder, B. Mondal, A. Chatterjee, S. Das, S. Kumar, R. Gachhui, V. Castelletto, I. W. Hamley, P. Sen and A. Banerjee, Langmuir, 2023, 39, 7307–7316 CrossRef CAS PubMed.
- J. Andrä, D. Monreal, G. Martinez de Tejada, C. Olak, G. Brezesinski, S. S. Gomez, T. Goldmann, R. Bartels, K. Brandenburg and I. Moriyon, J. Biol. Chem., 2007, 282, 14719–14728 CrossRef PubMed.
- R. Gopal, C. H. Seo, P. I. Song and Y. Park, Amino Acids, 2013, 44, 645–660 CrossRef CAS PubMed.
- D. Neubauer, M. Jaskiewicz, M. Bauer, K. Golacki and W. Kamysz, Molecules, 2020, 25, 257 CrossRef CAS PubMed.
- A. Y. Gahane, P. Ranjan, V. Singh, R. K. Sharma, N. Sinha, M. Sharma, R. Chaudhry and A. K. Thakur, Soft Matter, 2018, 14, 2234–2244 RSC.
- F. Cao, G. Ma, M. Song, G. Zhu, L. Mei and Q. Qin, Soft Matter, 2021, 17, 4445–4451 RSC.
- H. D. Glossop, G. H. De Zoysa, Y. Hemar, P. Cardoso, K. Wang, J. Lu, C. Valery and V. Sarojini, Biomacromolecules, 2019, 20, 2515–2529 CrossRef CAS PubMed.
- G. Laverty, A. P. McCloskey, B. F. Gilmore, D. S. Jones, J. Zhou and B. Xu, Biomacromolecules, 2014, 15, 3429–3439 CrossRef CAS PubMed.
- Y. Wu, Q. He, X. Che, F. Liu, J. Lu and X. Kong, Biochem. Biophys. Res. Commun., 2023, 648, 66–71 CrossRef CAS PubMed.
- J. K. Tripathi, S. Pal, B. Awasthi, A. Kumar, A. Tandon, K. Mitra, N. Chattopadhyay and J. K. Ghosh, Biomaterials, 2015, 56, 92–103 CrossRef CAS PubMed.
- Z. Azoulay, P. Aibinder, A. Gancz, J. Moran-Gilad, S. Navon-Venezia and H. Rapaport, Acta Biomater., 2021, 125, 231–241 CrossRef CAS PubMed.
- A. D. Martin and P. Thordarson, J. Mater. Chem. B, 2020, 8, 863–877 RSC.
- E. R. Draper, E. G. Eden, T. O. McDonald and D. J. Adams, Nat. Chem., 2015, 7, 848–852 CrossRef CAS PubMed.
- A. Levin, T. O. Mason, L. Adler-Abramovich, A. K. Buell, G. Meisl, C. Galvagnion, Y. Bram, S. A. Stratford, C. M. Dobson, T. P. Knowles and E. Gazit, Nat. Commun., 2014, 5, 5219 CrossRef CAS PubMed.
- U. Anand and M. Mukherjee, Langmuir, 2013, 29, 2713–2721 CrossRef CAS PubMed.
- J. Li, X. Du, S. Hashim, A. Shy and B. Xu, J. Am. Chem. Soc., 2017, 139, 71–74 CrossRef CAS PubMed.
- Y. Liu, G. Zhan, X. Zhong, Y. Yu and W. Gan, Liq. Cryst., 2011, 38, 995–1006 CrossRef CAS.
- J. Kong and S. Yu, Acta Biochim. Biophys. Sin., 2007, 39, 549–559 CrossRef CAS PubMed.
- P. Ferrinho, M. Viveiros and I. Fronteira, One Health, 2023, 16, 100512 CrossRef CAS PubMed.
- M. Balouiri, M. Sadiki and S. K. Ibnsouda, J. Pharm. Anal., 2016, 6, 71–79 CrossRef PubMed.
- G. A. Pankey and L. D. Sabath, Clin. Infect. Dis., 2004, 38, 864–870 CrossRef CAS PubMed.
- P. S. Ocampo, V. Lazar, B. Papp, M. Arnoldini, P. Abel zur Wiesch, R. Busa-Fekete, G. Fekete, C. Pal, M. Ackermann and S. Bonhoeffer, Antimicrob. Agents Chemother., 2014, 58, 4573–4582 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra08883a |
| This journal is © The Royal Society of Chemistry 2024 |