 Open Access Article
Open Access ArticleDesign strategies and biological applications of β-galactosidase fluorescent sensor in ovarian cancer research and beyond
Liangliang Li ,
Feifei Jia,
Yunxiu Li and
Yan Peng*
,
Feifei Jia,
Yunxiu Li and
Yan Peng*
Shenzhen Longhua District Central Hospital, Guangzhou 518000, People’s Republic of China. E-mail: 853312523@qq.com
First published on 18th January 2024
Abstract
Beta-galactosidase (β-galactosidase), a lysosomal hydrolytic enzyme, plays a critical role in the catalytic hydrolysis of glycosidic bonds, leading to the conversion of lactose into galactose. This hydrolytic enzyme is used as a biomarker in various applications, including enzyme-linked immunosorbent assays (ELISAs), gene expression studies, tuberculosis classification, and in situ hybridization. β-Galactosidase abnormalities are linked to various diseases, such as ganglioside deposition, primary ovarian cancer, and cell senescence. Thus, effective detection of β-galactosidase activity may aid disease diagnoses and treatment. Activatable optical probes with high sensitivity, specificity, and spatiotemporal resolution imaging capabilities have become powerful tools for visualization and real time tracking in vivo in the past decade. This manuscript reviews the sensing mechanism, molecular design strategies, and advances of fluorescence probes in the biological application of β-galactosidase, particularly in the field of ovarian cancer research. Current challenges in tracking β-galactosidase and future directions are also discussed.
1. Introduction
The glycoside hydrolase, beta-galactosidase (β-galactosidase), is specifically involved in cleaving the β-glycosidic bond between galactose and its organic moiety. The enzyme provides organisms with essential energy and carbon compounds by the hydrolysis of lactose into glucose and galactose. β-Galactosidase can lead to galactose denaturation (a lysosomal storage disease) or Morquio B syndrome.1 In 1994, β-galactosidase was identified as a homotetramer of 464 kD with a 2,2,2-point symmetry structure (Fig. 1a). Each of the subunits has five regions: the first forms a jelly-roll barrel, the second and fourth adopt a fibronectin type III-like barrel structure, the fifth is a β-sandwich type, and the central third region forms a TIM-type barrel.2,3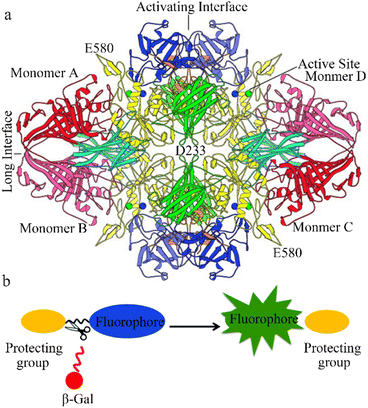 | ||
| Fig. 1 (a) View of the β-galactosidase tetramer looking down one of the two-fold axes. Coloring is by domain: complementation peptide, orange; domain 1, blue; domain 2, green; domain 3, yellow; domain 4, cyan; domain 5, red. Lighter and darker shades of a given color are used to distinguish the same domain in different subunits. The metal cations in each of the four active sites are shown as spheres: Na+, green; Mg+, blue. Reproduced with permission from ref. 4, Copyright 2000, John Wiley and Sons. (b) Design strategy for fluorescence β-galactosidase probes, based on grafting an enzyme active trigger onto a controllable emissive chromophore; that is, introducing a protective group (such as galactose) into the fluorophore. Reproduced with permission from ref. 32 Copyright 2021, Royal Society of Chemistry. | ||
β-Galactosidase is commonly used as a marker of gene expression, exhibiting a phenomenon known as alpha-complementation in blue/white screening of recombinant replicates. The enzyme has two peptides, LacZ gene alpha and LacZΩ, that are non-functional on their own but can spontaneously reassemble into a functional enzyme. Importantly, when a DNA fragment is inserted into a vector, the construction of LacZα is disrupted, preventing β-galactosidase activity.4,5
β-Galactosidase is also a significant biomarker of cell senescence and primary ovarian cancer (OC).6–8 Mutations in the enzyme are associated with Morquio B syndrome and β-galactosialidosis, and abnormal β-galactosidase accumulation is a major contributor to cellular senescence.9,10 β-Galactosidase is also more highly expressed in primary ovarian cancer cells (SHIN-3, SKOV-3, OVK-18, OVCAR-3, OVCAR-4, OVCAR-5, and OVCAR-8) than normal cells. Elevated β-galactosidase activity can cause separation of the amino polysaccharide side chain from the core protein by breaking the glycosidic bond. This, in turn, can induce the breakdown of large proteoglycan molecules and harm the basement membrane,11–13 promoting cancer cell infiltration and metastasis.14,15 These studies highlight the importance of accurate, rapid, and efficient detection and monitoring of β-galactosidase, particularly in fundamental biological research and early cancer diagnosis.
Several methods of measuring β-galactosidase activity, including colorimetric assays, electrochemical assays, single-photon emission computed tomography (SPECT), positron emission tomography (PET), and magnetic resonance imaging (MRI), have been developed.16 However, none have achieved real-time, non-invasive β-galactosidase detection in biological systems.17,18 Novel commercially accessible colorimetric methods, including the X-gal (5-bromo-4-chloro-3-indolyl β-D-galactopyranoside) substrate and Promega's assay kit utilizing o-nitrophenyl β-D-galactopyranoside, have also been used to assess β-galactosidase activity.19 However, these methods do not work for in vivo imaging due to their poor cell permeability and requirement for cell fixation and sacrificing of organisms.20 In recent years, optical imaging methods with non-invasive capability, high sensitivity, quick response time, precise spatial resolution, and the ability to conduct real-time imaging have surfaced as valuable instruments for biological investigation. Fluorescent probes play a critical role in optical imaging techniques, benefiting from their ease of acquisition, small size, and ease of modification for cellular staining and ex vivo detection.8,21 Several fluorescent probes have been developed that enable β-galactosidase visualization in biological systems.22,23
Ovarian cancer (OC) is one of three malignant gynecological tumors. Approximately 225![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 new OC cases occur globally each year, with a patient survival rate of about 30%. OC has the second highest incidence rate of malignant gynecological tumors and the highest mortality rate.24 At stage I, the 5 year survival rate is 90%. However, over 70% of OC cases are not diagnosed until the disease has progressed to stage III or IV, emphasizing the importance of early diagnosis.25 The most common clinical treatment is a combination of surgery and platinum-based chemotherapy. However, for patients who develop drug resistance or experience recurrence, single therapies often fail to achieve the desired result. Thus, there is an urgent need for more effective treatment strategies to control the disease and improve clinical prognosis.26,27 P53, the most frequently mutated gene in OC, plays a critical role in inducing cell senescence and death, DNA repair, regulating cell metabolism, activating the immune response, and maintaining stem cells.28 Identifying changes in methyltransferase-like protein 14 (METTL14)29 expression on the proliferation, invasion, and migration of the OC cell Lines, A2780 and SKOV3.30 There is a close association between the aberrant expression of β-galactosidase and tumor occurrence and metastasis in the OC cell lines, SKOV3 and SHIN3. Thus, this enzyme serves as a biomarker for OC clinical diagnosis and treatment.31 In recent years, several excellent fluorescent probes have been developed to aid diagnosis and OC tumor resection. However, few studies have reviewed the association between β-galactosidase and OC.8,16,32
000 new OC cases occur globally each year, with a patient survival rate of about 30%. OC has the second highest incidence rate of malignant gynecological tumors and the highest mortality rate.24 At stage I, the 5 year survival rate is 90%. However, over 70% of OC cases are not diagnosed until the disease has progressed to stage III or IV, emphasizing the importance of early diagnosis.25 The most common clinical treatment is a combination of surgery and platinum-based chemotherapy. However, for patients who develop drug resistance or experience recurrence, single therapies often fail to achieve the desired result. Thus, there is an urgent need for more effective treatment strategies to control the disease and improve clinical prognosis.26,27 P53, the most frequently mutated gene in OC, plays a critical role in inducing cell senescence and death, DNA repair, regulating cell metabolism, activating the immune response, and maintaining stem cells.28 Identifying changes in methyltransferase-like protein 14 (METTL14)29 expression on the proliferation, invasion, and migration of the OC cell Lines, A2780 and SKOV3.30 There is a close association between the aberrant expression of β-galactosidase and tumor occurrence and metastasis in the OC cell lines, SKOV3 and SHIN3. Thus, this enzyme serves as a biomarker for OC clinical diagnosis and treatment.31 In recent years, several excellent fluorescent probes have been developed to aid diagnosis and OC tumor resection. However, few studies have reviewed the association between β-galactosidase and OC.8,16,32
This review begins by systematically summarizing recent developments in fluorescent probe-based reactive probes for β-galactosidase, including synthesis strategies and applications based on their respective luminescence mechanisms. Both resolved and unresolved issues are explored that inform potential research directions.
2 β-Galactosidase sensing mechanisms
β-Galactosidase specifically recognizes β-galactosyl bonds, enabling the construction of β-galactosidase reaction-activated fluorescent probes. The activation of fluorescence or its activity signal has extremely high sensitivity due to its efficient catalytic effect. The most common strategy involves grafting an enzyme-active trigger onto a controllable emissive chromophore to introduce a protective group (such as galactose) into the fluorophore. Enzyme activity induces the controlled emission chromophores, generating a specific fluorescence change (Fig. 1b) that can be visualized.32 These fluorescent probes primarily involve two common mechanisms: intramolecular charge transfer (ICT) and Förster resonance energy transfer (FRET). Due to the significant charge change those results from O-cleavage before and after the reaction, the ICT mechanism predominates, leading to a noticeable alteration in the charge of the fluorescent probe. ICT-based probes involve the conjugation of an electron-donating unit in a molecule with an electron-accepting unit to produce a “push–pull” π-electron system in the excited state.33 The electron-donating character of the probe decreases when this region interacts with the analyte, resulting in a blue shift in the absorption spectrum (Fig. 2a). Conversely, when the ICT becomes more developed through the interaction between the analyte and the electron acceptor, a significant red shift is observed.34,35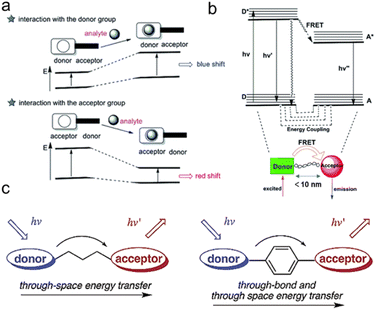 | ||
| Fig. 2 Schematic representation of (a) ICT process, (b) FRET and (c) TSET or TBET process. (a) and (b) Were reproduced with permission from ref. 34 Copyright 2018, Royal Society of Chemistry. (c) Was reproduced with permission from ref. 59 Copyright 2003, American Chemical Society. | ||
When distinct dual fluorophores are present in the probe design, the FRET mechanism becomes dominant. This typically occurs when the donor and acceptor can be reasonably approximated as point dipoles (Fig. 2b). While FRET doesn't require physical contact, the donor and acceptor must be close enough to interact, typically within a range of ≤10 nm (Förster distance).36–38 In addition to their proximity, the donor and acceptor must undergo resonance transition, and the energy transfer must compete with other pathways to allow the donor to relax to its ground state. These non-resistive conditions are important design considerations for the donor–acceptor configuration. In qualitative terms, a greater spectral overlap between the donor emission and acceptor absorption benefits a larger Förster distance, higher donor quantum yield, and a greater acceptor molar absorption coefficient. Thus, FRET is the energy transfer process between the luminescent donor (D) and the light-absorbing acceptor (A), which must be in very close proximity and energy resonance, as indicated by the spectral overlap integral.39,40
2.1 Design strategies using the ICT mechanism
Molecules capable of displaying ICT characteristics are inherently dipolar due to the presence of π-electron systems that connect D and A groups with electron-withdrawing and -donating properties. Various D–A combinations, such as D–π–A, D–A, D–A–D, A–π–D–A, or A–D–π–D–A, exist in their structures. Here, “π” represents unsaturated spacers of different lengths, including simple alkenes, phenyl derivatives, thiophene or thiazole functionalities. The π-spacer promotes constant stretching of the electron cloud between the donor and acceptor to maintain electronic transfer during reactions. Within π-conjugated systems, the electron donor typically involves lone pair electrons from heteroatoms present in heterocycles, which constitute the fluorophore unit of the probe. The acceptor unit requires electron deficiency, often found in aromatic groups or main group elements with strong electron-withdrawing substituents.41 Thus, the fusion of heterocycles with electron acceptor units is one of the most effective strategies for constructing ICT compounds. Small molecular compounds are typically used for ICT due to easy synthesis and modification. These are typically composed of chromophores such as anthracene, pyrene, anthraquinone, fluorene, perylene bisimide, carbazole, triphenylamine, and their derivatives.42Using the anion recognition ICT sensor as an example, this review explores the mechanism used to efficiently identify anionic sensing probes. Previous reports on chemical sensors for anion recognition can be categorized into four types:43–46 (1) the acidic-H in the probe binds with anions, enhancing the ICT process and making electron transfer more facile; (2) extraction of acidic protons from the probe allows anions to bind with the acidic-H in the probe, leading to deprotonation, increasing charge density and consequently promoting the ICT process; (3) anions undergo nucleophilic addition reactions with electrophilic sites on the probe, disrupting the conjugated system and weakening the ICT process (CN− is a notable example); (4) anions induce the destruction/elimination of certain functional groups within the molecular structure, enhancing the ICT process by increasing electron density on the sensor.
In contrast with anion-recognizing ICT sensors, fluorescence sensing of β-galactosidase focuses on its reaction substrate, the galactose molecule. β-Galactosidase's presence results in a significant charge variation that manifests as wavelength shifts governed by the ICT mechanism.47 In recent years, β-galactosidase fluorescent probes based on ICT have been developed, however, future research will be required to assess their efficacy under different conditions.
Photoacoustic imaging (PA) offers high resolution and deep tissue penetration but has limited sensitivity. FL (fluorescent imaging)/PA dual-modal imaging not only captures highly sensitive functional images but also provides high-resolution structural information for in vivo study. Hai et al. created an FL/PA probe (Gal-HCy-biotin) with tumor-targeting capabilities, allowing the imaging of tumor senescence through β-galactosidase activation (Fig. 3a).48 The Gal-HCy-biotin probe is composed of three components: β-D-galactose, a distinct substrate designed for β-galactosidase, a near-infrared (NIR) hydrocyanine dye (HCy) that serves as a responsive FL/PA imaging signal source, and biotin for targeting. The hydroxyl group in the HCy moiety on the Gal-HCy-Biotin probe is caged, inhibiting ICT and leading to FL/PA signal quenching. Thus, when β-galactosidase is overexpressed in the lysosomes of senescent tumor cells, Gal-HCy-biotin is hydrolyzed, releasing free HCy and restoring FL/PA signals. Gal-HCy-biotin or Gal-HCy can effectively image senescent tumor cells with a 4.6- or 3.5-fold FL enhancement, respectively, and a 4.1- or 3.3-fold PA enhancement, respectively. Gal-HCy-biotin or Gal-HCy can image tumor senescence with 2.9- or 1.7-fold FL enhancement, respectively, and 3.8- or 1.3-fold PA enhancement, respectively. This allows for specific imaging of senescent tumors, holding promise for the clinical application of Gal-HCy-biotin for tumor and senescence detection.
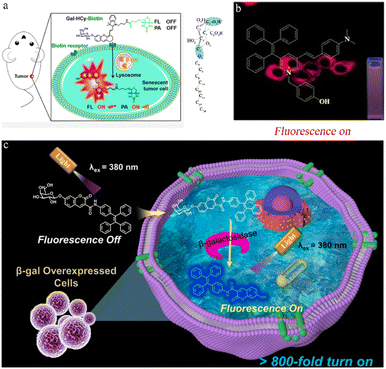 | ||
| Fig. 3 (a) Left: β-galactosidase-activatable fluorescent and photoacoustic imaging of tumor senescence. Biotin could help Gal-HCy-Biotin enter and accumulate in tumor cells with higher FL/PA signal. Right: the configuration of the galactoside carbon atoms. Reproduced with permission from ref. 48, Copyright 2023, American Chemical Society. (b) Imaging in living cells via a novel AIE fluorescent probe for β-galactosidase detection and its reaction products. Reproduced with permission from ref. 49, Copyright 2022, Elsevier. (c) Illustration of the sensing process of TC-gal for endogenous β-galactosidase detection. Biocompatible probe TC-gal is successfully applied to detect endogenous β-galactosidase in ovarian cancer cells and distinguish senescent cells. Reproduced with permission from ref. 50, Copyright 2021, Elsevier. | ||
The use of probes with aggregation-induced emission (AIE) helps to avoid the fluorescent dye aggregation-caused quenching (ACQ) of small molecules. Jia et al. designed a novel AIE probe with an ICT effect (Fig. 3b) that enables the selective and sensitive detection of β-galactosidase.49 Due to its low cytotoxicity, the AIE probe is suitable for β-galactosidase imaging in cells. Feng et al. carried out further optimization and design to improve its sensitivity. This group achieved ultra-sensitive β-galactosidase detection by designing a “turn-on” fluorescent probe by modulating twisted intramolecular charge transfer (TICT) effects in the fluorescent moiety (Fig. 3c).50 The tetraphenylethene segment reduced the TICT effect, while the hydroxyl group increased the ICT effect by donating electrons. This design significantly lowers background fluorescence and enhances the signal-to-noise ratio. The TC probe was successfully used to detect endogenous β-galactosidase expression in OC cells and senescent cell differentiation.
2.2 Design strategies of the FRET mechanism
Compared to single-emission fluorescent probes, ratiometric fluorescent probes can perform qualitative over quantitative detection using the ratio of two wavelengths, effectively avoiding interference by some environmental factors.51 FRET, which uses the energy transfer between donor and acceptor fluorophores, has gradually become the primary method for designing ratio-fluorescence sensors.52 Influenced by external factors, the intensity ratio between the FRET donor and acceptor fluorophores is highly dependent on FRET efficiency.53 The efficacy of FRET is dependent on a spectral overlap between the emission spectrum of the energy donor and the absorption spectrum of the energy acceptor, along with donor–acceptor distances of 1–10 nm. When these conditions are met, the FRET process can be toggled ON/OFF, switching fluorescence between donor and acceptor emissions.54 This ON/OFF mechanism of the FRET process forms the basis of FRET-based detection.During the fluorescent sensing of β-galactosidase, the fluorophore excited by β-galactosidase (donor), uses FRET, transferring excitation energy non-radiatively to an absorbing chromophore (acceptor).55 This energy transfer is primarily due to dipole–dipole interactions between the fluorophore and the absorbing chromophore. Using the dual-dipole response, a fluorophore in an excited state non-radiatively transfers energy to an adjacent absorbing chromophore, distinguishing this mechanism from ICT. However, designing FRET fluorescent probes requires stringent conditions, including dipole orientation and spectral overlap between the donor emission and the acceptor excitation.52 To address these challenges, researchers have successfully developed various FRET-based fluorescent β-galactosidase probes in recent years, establishing the groundwork for their advancement and theoretical investigation.
Yan et al. used FRET to develop a highly selective and sensitive fluorescent probe (FTR-βgal) for the specific detection of β-galactosidase (Fig. 4a).56 FTR uses naphthalene derivatives as energy donors and visual rhodamine derivatives as energy acceptors, with recognition sites introduced onto Np-Rhod. This probe demonstrates a notable emission shift, swift responsiveness, and minimal cytotoxicity, allowing it to effectively detect in vivo β-galactosidase activity with high sensitivity. The FTR-βgal probe also demonstrates enhanced tissue penetration and reduced cytotoxicity, making it suitable for in vivo imaging. These findings suggest that the probe has great potential for detecting endogenous β-galactosidase and being used within intricate biological systems. Dennis et al. successfully designed and synthesized a photosensitizer comprised of a galactose-based substrate, a photosensitive moiety based on boron-dipyrromethene (BODIPY), and a quencher 2 connected via an AB2 self-immolative linker, that responds to β-galactosidase (Fig. 4b).57 The BODIPY photosensitizer can be selectively activated by β-galactosidase, generating a singlet oxygen and restoring fluorescence emission, which is used to detect β-galactosidase activity in senescent cells (Fig. 4b). Nagano et al. used an amino-formate leaving group to synthesize a β-galactosidase-tagging probe (CMFβ-gal), on quinone methide amide.58 The CMFβ-gal probe operates using FRET, with a labeling reaction used to monitor changes in the fluorescence wavelength (Fig. 4c). FRET efficiency varies with changes in the labeling reaction process, and the fluorescence emitted by the labeled protein is noticeably distinct from that of the unreacted probe allowing for immediate detection of the target protein.
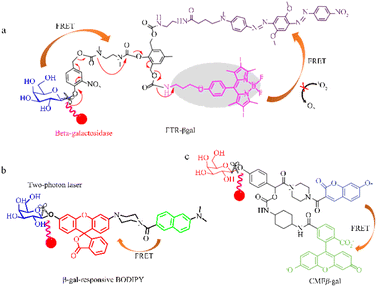 | ||
| Fig. 4 Quantum tracking through a proportional structure. (a) FTR-βgal, FTR-βgal was successfully applied in TP imaging of β-gal with the ratio signal rapidly transform from 0.15 to 5.9 (∼40-fold) between 540 nm and 450 nm. Reproduced with permission from ref. 57, Copyright 2020, Elsevier. (b) β-gal-responsive BODIPY β-gal-responsive BODIPY can activate a galactosyl distyryl boron dipyrromethene (BODIPY)-based photosensitiser inside the cells in a highly specific manner, effectively causing cell death and tumour eradication in nude mice. Reproduced with permission from ref. 58, Copyright 2023, Royal Society of Chemistry. (c) CMFβ-gal, reproduced with permission from ref. 59, Copyright 2006, American Chemical Society. | ||
2.3 Design strategies using other mechanisms
In addition to the two common mechanisms (ICT and FRET) mentioned above, various other methods and materials have been used to design fluorescent β-galactosidase probes. These include through-bond energy transfer (TBET),59,60 recognition between the host and guest, coupled with a specific signal transduction mechanism induced by static quenching,61 the inner filter effect (IFE),62 and ICT-FRET.63Lin et al. designed a coumarin-rhodamine TBET Box, in which the acceptor and donor units are connected by a conjugated spacer.60 The configuration allows large stokes and emission shifts, minimizing constraints on the strong spectral overlaps between acceptor absorption and donor emission. This represents a new paradigm in probe design that involves the energy transfer of dual fluorescent dyes using small molecules. The ratio fluorescence TBET-pH probe (Fig. 5a), which demonstrates significant emission shifts and intensity changes under different pH conditions along with excellent cell permeability, holds potential for further application in research and medicine.
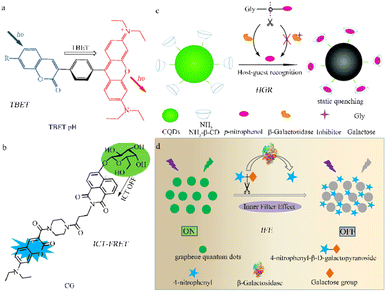 | ||
| Fig. 5 Fluorescence probe regulation strategies under different mechanisms. (a) Through-bond energy transfer (TBET), reproduced with permission from ref. 60, Copyright 2010, John Wiley and Sons. (b) Photoinduced electron transfer (PET), reproduced with permission from ref. 61, Copyright 2019, American Chemical Society. (c) ICT-FRET, reproduced with permission from ref. 62, Copyright 2017, Royal Society of Chemistry. (d) Inner filter effect (IFE), reproduced with permission from ref. 63, Copyright 2020, Elsevier. | ||
Feng et al. devised a comprehensive detection strategy using functional carbon quantum dots (CQDs) to screen glycosidases and inhibitors and a mechanism involving host-guest recognition and the specific signal transduction induced by static quenching (Fig. 5b).61 The nano-probe (β-CD-CQDs) was structured with a hydrophobic and appropriately sized β-cyclodextrin domain, promoting host-guest recognition and specific binding to p-nitrophenol. The carbon quantum dots within the probe also form an inclusion complex with p-nitrophenol which induces a static quenching mechanism. This proposed strategy holds potential for use in the detection of other glycosidases.
Lin et al. devised a convenient and label-free sensing strategy for detecting β-galactosidase activity using GQDs designed with IFE.62 When β-galactosidase is present, 4-nitrophenyl β-D-galactopyranoside (NPGal) can hydrolyze to 4-nitrophenol (4-NP) and quench the fluorescence (Fig. 5c). Compared to other strategies, this design requires only one type of fluorescent nanomaterial, eliminating the need for any modifications and avoiding time-consuming processing steps. This IFE-based fluorescence GQD sensing strategy has the potential to identify disease-relevant enzyme activities.
The ICT-FRET contrast rate type β-galactosidase fluorescence probe uses both ICT and FRET, effectively increasing interaction with the signal output after the β-galactosidase reaction to increase the ratio of emission intensity and achieve high sensitivity. Lin et al. constructed a highly sensitive fluorescent probe, CG, based on ICT-FRET that allows for visual detection of β-galactosidase (Fig. 5d).63 The CG probe can also be used to image in both overexpressing cell lines and those with transient expression, showcasing its versatility. This probe also demonstrates excellent selectivity for β-galactosidase in vitro and has the potential to become an ultra-sensitive tool for better understanding physiological and pathological processes in living organisms.
3 Application of fluorescent β-galactosidase probes in ovarian cancer research
This article introduces various strategies for designing fluorescent β-galactosidase probes using the sensing mechanism as a starting point. In recent years, fluorescent β-galactosidase probes have emerged as highly sensitive and specific tools for real-time tracking and in vivo visualization (Table 1 and Fig. 6).64–81 Recent advancements in the biological application of β-galactosidase-activatable probes based on spectral characteristics and responsive modulation, including probe mechanism categorization and synthesis strategies, have been discussed in other reviews.56 Despite the broad attention that these probes have received, however, there are few systematic summaries of their biological application.38 To address this gap, the current study provides a comprehensive overview of the potential biological applications of these probes by focusing on their use in OC diagnosis and treatment. In the subsequent sections, different probe applications, including single cancer detection probes, “multi-functional” detection probes, and targeted fluorescent probes, are used as classification criteria. While these criteria may not fully differentiate between these probe applications in a biological context, this simplified classification will help to summarize recent progress in β-galactosidase probe research to inform further studies and biological uses.| Name of the probe | λex/λem (nm) | Detection approach | Cells | Cytotoxicity | Quantum yield | LOD | Ref. |
|---|---|---|---|---|---|---|---|
| DDAOG | 456/608 | Turn on | 9L-LacZ cells | — | — | 70 | |
| HBT-gal | 415/492 | Turn on | HepG2 cells | 10 μL per well | — | 0.19 mU mL−1 | 72 |
| DCDHF-βgal | 580/665 | Ratiometric | HepG2, A549 and KB cells | 10 μL | — | — | 75 |
| NI-βGal | 395/545 | Ratiometric | C6/lacZ7 cells | — | — | — | 76 |
| PhoTO-gal | — | Turn on | HEK293 cells | — | — | — | 77 |
| NI–βgal | 440/545 | Ratiometric | C6/lacZ7 cells | 10 mM | — | — | 78 |
| SG1 | 376/490 | Turn on | DT2 and DT20 cells | — | 0.16 | — | 79 |
| 6SqGal | — | Ratiometric | HEK293 cells | — | 0.32 | — | 80 |
| DCMC | 660/690 | Turn on | HEK293 cells | — | — | — | 81 |
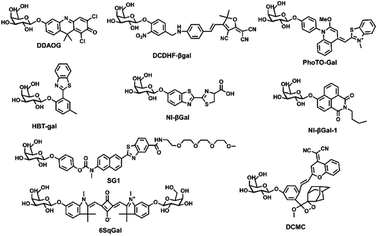 | ||
| Fig. 6 Corresponding to the structure of each probe in Table 1. | ||
3.1 Single-response probe
Single-response β-galactosidase fluorescence probes have been designed with high sensitivity or specificity and used to detect individual cancer cells. This section explores the benefits of these probes, including sensitivity enhancement, increased specificity, and improved visualization imaging. Lin et al. used the excited-state intramolecular proton transfer (ESIPT) mechanism to construct a novel fluorescence probe, ESIPT-GAL, for the detection of β-galactosidase that has low background signal and high sensitivity. This probe, which uses an HBT derivative as a signal reporter and D-galactose as a reaction moiety, displays high sensitivity in both living cells and tissues (Fig. 7a).82 ESIPT-GAL has good water solubility and biocompatibility, allowing fluorescence visualization up to 90 μm in tumor sections. This finding suggests that the probe has the potential for use as an in vivo imaging tool. Shi et al. designed a sensitive.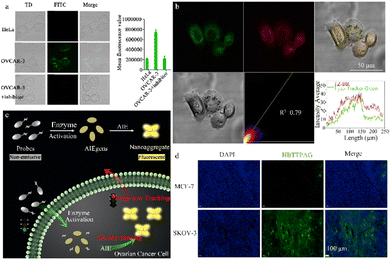 | ||
| Fig. 7 (a) Fluorescence images of HeLa and OVCAR-3 cells stained with the probe ESIPT-GAL reproduced with permission from ref. 82, Copyright 2021, Elsevier. (b) Fluorescence images of OVCAR-3 cells stained with the probe TZ-Br and Lyso-Tracker Green DND-26. Reproduced with permission from ref. 84, Copyright 2022, Elsevier. (c) Schematic illustration of an enzyme-regulated liberation strategy for on-site sensing and long-term tracking. By virtue of in situ enzyme-catalyzed conversion and intracellular retention characteristics of nanoaggregates, QM-βgal is able to perform on-site sensing and long-term trapping of β-gal activity in ovarian cancer cells. Reproduced with permission from ref. 85, Copyright 2019, Royal Society of Chemistry. (d) Fluorescence imaging of different cells incubated with HBTTPAG (10 μM) for 30 min. HBTTPAG can track β-galactosidase in ovarian cells for a long time, and can achieve specific imaging of β-galactosidase in ovarian cancer tissues. Reproduced with permission from ref. 87, Copyright 2021, Elsevier. | ||
β-Galactosidase -activatable probe (DCMCA), which is used for NIR imaging of OC tumors. Activation of β-galactosidase releases the NIR chromophore, DCM-NH2.83 The highly sensitive DCMCA probe enables real-time and visual imaging of β-galactosidase activity in an OC mouse model, offering significant clinical potential for early diagnosis and timely treatment. Hou et al. devised and synthesized a NIR emission fluorescent probe, TZ-Br, for β-galactosidase detection in senescent and cancer cells (Fig. 7b).84 After specific recognition of β-galactosidase by the TZ-Br probe, the glucoside bond is cleaved, releasing the phenol/phenolic acid of the fluorophore and enabling detection of endogenous β-galactosidase in senescent cells, cancer cells, and zebrafish. TZ-Br was successfully used to evaluate the curative effect of masitinib and quercetin on senescent cells. Thus, this probe could help to identify the pathological and physiological functions of β-galactosidase in biological systems.
To avoid the ACQ phenomenon of dyes, Guo et al. used a AIE probe (QM-βgal), which consists of a hydrophilic β-galactosidase triggered lactose fraction and a hydrophobic AIE active fluorescent group, QM-OH (Fig. 7c).85 This probe emits very little fluorescence in aqueous media and yet becomes highly emissive upon activation by endogenous β-galactosidase in living cells, demonstrating excellent detection performance. Moreover, the nanoaggregate structure of this probe enhances the intracellular retention of QM-βgal, enabling the long-term observation and detection of β-gal-overexpressing OC cells. This is critically important for biomedical applications and diagnostics. Gu et al. developed an activatable fluorescence probe (BOD-M-βgal) with an extended emission wavelength that can monitor the in vivo activity of β-galactosidase and investigated its potential implications in OC diagnosis.86 The luminescence mechanism of this probe involves the use of a self-quenching linker to introduce a β-galactose residue into an NIR fluorescent group based on boron dipyrromethene (BODIPY). An ethylene unit is simultaneously extended at position 3 of the BODIPY core to provide NIR-II fluorescence responsiveness. This probe can detect β-galactosidase in different states (buffer solution and SKOV3 cells) and image ovarian tumors in nude mice.
Xie et al. was the first to create a fluorescent probe, HBTTPAG, with AIE properties, achieving species-selective detection of β-galactosidase (from Escherichia coli and Aspergillus oryzae) (Fig. 7d).87 In the presence of β-galactosidase, the probe is hydrolyzed to HBTTPA, which activates the emission by restricting intramolecular rotation in aggregate states and restores intramolecular hydrogen bond transfer. HBTTPAG successfully identified heightened β-galactosidase activity in SKOV-3 cells, allowing for the long-term visualization and tracking of this enzyme in OC cells. Thus, HBTTPAG has the potential to achieve precise imaging of β-gal within OC tissue and offers a promising method for the species-selective detection of β-galactosidase.
3.2 Multiple-response probe
Abnormal β-galactosidase activity in eukaryotic cells promotes the formation, proliferation, and metastasis of certain cancers, including OC.88–90 The complexity of dynamic biological systems during cancer onset and progression makes it a challenge to track and visualize in vivo enzyme activity.91,92 Consequently, β-galactosidase is considered a more reliable biomarker for the early diagnosis of various cancers. While various fluorescent probes designed for β-galactosidase detection have been reported, their limited sensitivity restricts their use to cancers, such as OC, that have high β-galactosidase expression, and excludes those with comparatively lower expression, including colon or breast cancer. Advancements in the design of β-galactosidase fluorescence probes have informed the development of highly sensitive probes for detecting endogenous β-galactosidase activity and imaging various cancer cell types. These probes will not only aid in the detection of cancers with lower β-galactosidase expression but also significantly enrich the arsenal of effective tools for in situ tumor diagnosis and detection (Table 2).| Name of the probe | λex/λem (nm) | Detection approach | Cells | Cytotoxicity | Quantum yield | LOD | Ref. |
|---|---|---|---|---|---|---|---|
| β-CD-CQDs | 420/570 | Turn on | OVCAR-3 cells | 200 μg mL−1 | 0.02 | 0.6 U L−1 | 61 |
| CG | 370/475 | Ratiometric | — | 5 mg mL−1 | 0.95 | 0.081 U mL−1 | 63 |
| DCM-β-gal-UCNPs | 543/800 | Ratiometric | SKOV-3, 293 T and zebrafish | 1 mg mL−1 | — | 3.1 × 10−4 U mL−1 | 64 |
| Cyclometalated iridium(III) complexes | 282/550 | Turn on | Ovarian carcinoma cell and normal cell | 79.4 and 63.1 μM | — | 0.51 U mL−1 | 65 |
| GNPN | 475/545 | Ratiometric | MDBK and OVCAR-3 cells | 80 mM | 0.17 | 0.17 U L−1 | 66 |
| MC-βGal | 493/554 | Turn on | OVCAR-3 and HeLa cells | 100 μM | — | 0.019 U mL−1 | 67 |
| TMG | 580/650 | Turn on | MDBK and OVCAR-3 cells | 5 mg mL−1 | — | 0.86 U L−1 | 68 |
| FLM | 450/550 | Ratiometric | MDBK and OVCAR-3 cells | 5 mg mL−1 | 0.36 | 0.19 U L−1 | 69 |
| DMC-βgal | 725/770 | Turn on | SKOV-3 cells | 10 μM | — | 0.298 U L−1 | 71 |
| DCM-CHO-βgal | 525/665 | Turn on | HepG2 and SKOV-3 cells | 10 μM | 0.07 | 3.15 × 10−5 U mL−1 | 73 |
| AM-RP-G | 490/530 | Ratiometric | SKOV-3 cells | 10 mg mL−1 | — | 1.4 × 10−3 U mL−1 | 74 |
| ESIPT-GAL | 406/502 | Turn on | HeLa and OVCAR-3 cells | 30 μM | 0.0045 | 3.9 × 10−5 U mL−1 | 82 |
| ESIPT-GAL | 452/526 | Turn on | SKOV-3 cell | — | — | 1.26 × 10−3 U mL−1 | 83 |
| TZ-Br | 405/670 | Ratiometric | OVCAR-3 cells | 30 μM | 0.17 in PBS | 1.9 × 10−3 U mL−1 | 84 |
| QM-βgal | 355/434 | Turn on | SKOV-3 cells | 10 μM | 0.005 | 1.0 × 10−3 U mL−1 | 85 |
| BOD-M-βgal | 533/565 | Turn on | SKOV-3 cells | 100 μM | — | — | 86 |
| HBTTPAG | 488/574 | Turn on | SKOV-3 cells | 20 μM | 0.54 | 3.7 × 10−3 U mL−1 | 87 |
| CCGal1 | 620/662 | Turn on | SKOV-3 cells | 20 μM | 0.18 | 0.24 U mL−1 | 93 |
| Lyso-gal | 533/565 | Turn on | SKOV-3 cells | 50 μM | < 0.1% | 0.022 U mL−1 | 98 |
| HMRef-βGal | 600/725 | Ratiometric | SKOV-3 cells | 100 μM | — | — | 99 |
| DXM-βgal | 570/764 | Ratiometric | OVCAR 3 cells | 30 μM | 0.16 | 6.7 × 10−3 U mL−1 | 107 |
| DXM-βgal | 520/605 | Turn on | SKOV-3 cells | 10 μM | — | 2.92 × 10−4 U mL−1 | 108 |
| Gal-Br-NO2 | 415/670 | Turn on | MDBK, HeLa, and OVCAR-3 cells | 30 μM | 0.32 | 0.0035 U mL−1 | 109 |
Kim et al. designed a highly stable ratio fluorescence probe (CCGal1) characterized by tissue staining capability, excellent water solubility, high sensitivity, minimal cytotoxicity, and remarkable stability (Fig. 8a). This probe enables the quantitative monitoring of β-galactosidase enzyme activity in living cells and tissues, including the OC cell line, SKOV-3, and the colorectal cancer cell line, HT-29.93 The hydrolysis of β-galactosidase trigged by CCGal1 enhances the electron loading capacity of the probe and increases ICT efficiency resulting in a red shift in fluorescence. Thus, the use of CCGal1 to detect β-galactosidase enzyme activity is an effective tool to diagnose, prognose, and treat ovarian and colorectal cancers.
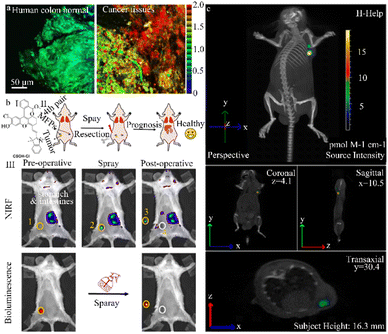 | ||
| Fig. 8 (a) Ratiometric images (Fred/Fyellow) of human colon normal and cancer tissues stained with CCGal1 (20 μM) for 3 h. Reproduced with permission from ref. 93, Copyright 2021, American Chemical Society. (b) (I) schematic diagram of the construction and surgery of the orthotopic breast tumor model. (II) NIRF and bioluminescence images for preoperative and postoperative stages after spraying of NIR-βgal-2. Reproduced with permission from ref. 94, Copyright 2022, American Chemical Society. (c) Tumor-bearing mice were not pretreated with avidin-β-gal before injection of DCM-βgal acting as the control. DCM-βgal manifests significantly ratiometric and turn-on NIR fluorescent signals simultaneously in response to β-gal concentration, which makes it favourable for monitoring dynamic β-gal activity in vivo with self-calibration in fluorescent mode. Reproduced with permission from ref. 95, Copyright 2016, American Chemical Society. | ||
Lin et al. successfully exploited a β-galactosidase-activated NIR fluorescence probe, NIR-βgal-2, to accurately monitor β-galactosidase activity in solid tumors and cancer cells, including the SKOV-3 and 4T1 cell lines (Fig. 8b).94 The CSOH–Cl dye (a NIR fluorescent dye) in this probe acts as a stable NIR fluorescent reporter, while β-galactosidase serves as an enzyme-activating trigger and the two are connected by a self-cleaving linker. In the presence of β-galactosidase, the NIR-βgal-2 probe is converted to CSOH–Cl, strongly enhancing the fluorescence signal. Importantly, since CSOH–Cl penetrates deeper into the tissue, NIR-βgal-2 can be used for “in situ spraying” to allow image-guided tumor resection in a murine breast tumor model, promoting favorable postoperative outcomes. These enzyme-activated NIR fluorescence probes have the potential to improve cancer diagnosis and image-guided surgeries.
Zhu et al. created a ratio emissive NIR fluorescence probe (DCM-βgal) that was used both in vivo and in situ to assess real-time fluorescence quantification and capture β-galactosidase activity in murine and human cancer cell lines (293 T and OVCAR-3) (Fig. 8c).95 In the DCM-βgal fluorescence probe, the dicyanomethyl-4H-pyran (DCM) chromophore serves as the NIR fluorescent reporter while the β-galactosidase-cleavable unit triggers enzyme activity. Upon recognition of β-galactosidase and subsequent elimination of the responsive group, the emission wavelength undergoes a significant shift, leading to the appearance of a new wide emission band with the NIR region. DCM-βgal has a high signal-to-noise ratio and directly and accurately monitors the distribution of endogenous β-galactosidase in transfected live cells. This probe also enables real-time in vivo bioluminescence imaging of β-galactosidase activity in nude mice with colorectal cancer. Notably, the NIR DCM-βgal probe was the first to use high-resolution three-dimensional in vivo fluorescence imaging and in situ monitoring of β-galactosidase activity in tumor-bearing nude mice. This probe offers the quantitative measurement and capture of in vivo β-galactosidase activity to aid human cancer diagnosis.
The use of poorly water-soluble probes may have limited use in biological applications. He et al. developed an enzyme-responsive photochromic fluorescent probe/human serum albumin assembly (NpG@HSA) that not only addressed the issue of poor water solubility but also allowed β-galactosidase imaging in both OC and senescent cells. Fluorogenic naphthalimide unit binding to HSA increases the fluorescence emission at 520 nm. When mediated by β-galactosidase, cleavage of the NpG@HSA lactose unit results in the formation of NpM@HSA and an increase in red fluorescence emission at 620 nm. The generated acrylonitrile fluorogenic unit then undergoes photoisomerization to facilitate stochastic optical reconstruction microscopy (STORM) imaging. NpG@HSA can be used to assess the subcellular distribution of β-galactosidase activity in different cell lines with nanoscale precision. This probe assembly provides a versatile imaging platform for the development of photochromic fluorescent probes which could be used to precisely locate disease-specific biomarkers.96
3.3 Targeted-response probe
β-Galactosidase is a representative lysosomal glycosidase, and the study of fluctuating levels of β-galactosidase in lysosomes, which are major subcellular organelles in the cell, is even more relevant.97 However, detecting and visualizing endogenous β-galactosidase in lysosomes continues to pose significant challenges, especially the development of targeted lysosomal fluorescence probes, which have received limited attention.98–100 This review describes representative β-galactosidase targeted fluorescence probes, with the assumption that additional probes will be developed to improve the precision of OC cell imaging.Wang et al. designed a specialized two-photon probe, FC-β-gal, that uses a 1,8-naphthalimide fluorophore with a morpholine ring as the lysosome targeting component. This innovative design allows endogenous β-galactosidase activity to be detected in cancerous SKOV-3 cells (Fig. 9a and b).98 FC-β-gal is strongly reactive to β-galactosidase, with glycosidic bond cleavage leading to a change in fluorescence from blue to green. Notably, using both two-photon and single-photon confocal imaging, this probe demonstrates ultra-low cytotoxicity and excellent endogenous β-galactosidase cell imaging in SKOV-3 cell line. The targeted two-photon probe design significantly expands its potential use in identifying pathologies, biological functions, and diseases associated with endogenous β-galactosidase expression in lysosomes.
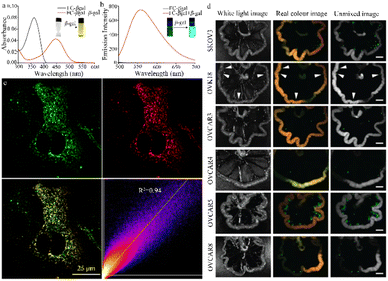 | ||
| Fig. 9 (a) and (b) The absorption and fluorescent spectra of the probe FC-βgal in the absence or presence of β-gal. Reproduced with permission from ref. 98, Copyright 2017, Elsevier. (c) CLSM imaging to investigate lysosome-targeting ability of LysoTracker R in SKOV-3 cells. Reproduced with permission from ref. 99, Copyright 2020, American Chemical Society. (d) Fluorescence spectral imaging of several mouse models of peritoneal metastasis at 1 h post administration of HMRef-βgal. In vivo, HMRef probe visualizes metastases as small as <1 mm in diameter in seven mouse models of disseminated human peritoneal ovarian cancer (SHIN3, SKOV3, OVK18, OVCAR3, OVCAR4, OVCAR5 and OVCAR8). Reproduced with permission from ref. 100, Copyright 2020, Spring Nature. | ||
Liu et al. created a lysosome-targeted NIR fluorescence probe, Lyso-gal, to detect and visualize endogenous β-galactosidase in SKOV-3 cell line (Fig. 9c).99 Lyso-gal consists of a lysosome-targeting core that connects anthocyanin and lactose by a linker. The galactopyranoside group of Lyso-gal is cleaved in the presence of β-galactosidase, promoting the removal of the blocking group on the phenolic hydroxyl and restoring the bright NIR fluorescence from the hydrolyzed β-galactosidase product. Lyso-gal offers the advantages of high selective imaging, sensitive detection, rapid detection time (1 min), and enhanced fluorescence intensity. This probe also has low toxicity and high sensitivity and selectivity, holding immense promise for the detection of endogenous β-galactosidase in primary OC cells.
Additional specialized β-galactosidase probes have been developed for OC imaging and detection. For example, Nobuyuki et al. designed a membrane-permeable HMRef-βGal probe capable of sensitively detecting endogenous β-galactosidase activity in living cells (Fig. 9d).100 This optimized intramolecular spiral functional probe provided >1400-fold fluorescence enhancement upon activation and successfully imaged intraperitoneal tumor metastasis in seven different mouse models. These findings provide strong confirmation that β-galactosidase can serve as a molecular target for visualizing peritoneal metastasis. There is also evidence that the HMRef-βgal probe could be used for other diseases, similar to how β-N-acetylglucosaminidase is used for lung cancer and gliomas, α-mannosidase is used for colon and breast cancer, α-fucosidase is used for thyroid cancer, and β-N-acetyl-glucosaminidase is used for stomach cancer. Thus, the HMRef-βgal probe has clinical promise for the fluorescence-guided diagnosis of OC peritoneal metastasis.
4 Application of fluorescent β-galactosidase probes in other fields
While this review has primarily discussed the use of fluorescent β-galactosidase probes in OC, studies indicate that this hydrolytic enzyme is widely present in animals, plants, and microorganisms (bacteria, fungi, yeast, etc.). Its primary sources include animal organs and plants such as apples, apricots, and peaches.101,102 In plant biology, β-galactosidase has been used to soften various fruits and vegetables, such as apples, pears, and potatoes, and ripen others, including peppers, cherries, and avocados.103,104 Thus, detecting β-galactosidase activity holds significant importance in the harvesting, processing, and storage of fruits, as well as in early disease prediction and disease monitoring. This section will emphasize the development and use of fluorescent β-galactosidase probes in diverse fields, including fruit flies, zebrafish, and fruits, to further understand their potential application in these areas.4.1 Detection of β-galactosidase activity in Drosophila
Urano et al. designed and synthesized a fresh β-galactosidase fluorescent probe (HMDER-βgal) that enables the rapid and clear observation of β-galactosidase activity in Drosophila melanogaster tissues and cultured cells (Fig. 10a).105 In a physiological environment, the spirocyclization structure of the hydroxymethyl in this probe is effectively used to modulate fluorescent emission before and after it reacts with the target enzyme. The use of the rhodopsin scaffold also increases the cellular accumulation of the HMDER-βGal probe, enabling distinct in situ visualization of β-galactosidase activity in Drosophila melanogaster tissues and living cells. Due to the functionality of the phenolic hydroxyl group, this simple and effective probe may aid the design of fluorescent probes capable of targeting a diverse range of molecules.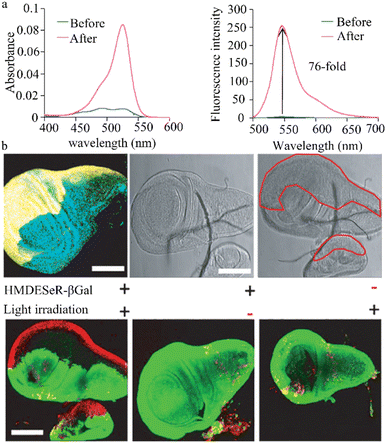 | ||
| Fig. 10 (a) Enzymatic reaction of HMDER-βGal with β-galactosidase. Reproduced with permission from ref. 105, Copyright 2011, American Chemical Society. (b) Immunostaining of β-galactosidase in an en-lacZ Drosophila larval wing disk. Reproduced with permission from ref. 106, Copyright 2014, John Wiley and Sons. | ||
This research group also successfully engineered a non-phototoxic spirocyclic structure and activatable HMDESeR-βgal photosensitizer (Fig. 10b),106 which can selectively induce cell death upon light irradiation of cells expressing β-galactosidase. In the presence of this enzyme, HMDESeR-βgal is converted into phototoxic HMDESeR, primarily in its open purine form. When applied to the posterior region of Drosophila larvae that express β-galactosidase in their wing discs, HMDESeR-βgal promotes light-induced cell death with significant specificity in the β-galactosidase-expressing region. The use of HMDESeR-βgal photosensitizers provides a new strategy for targeting tumor-selective photodynamic therapy (PDT) using enzymes that are overexpressed in tumors.
4.2 Detection of β-galactosidase activity in zebrafish
Ma et al. devised a distinctive NIR fluorescent probe, DXM-βgal, that is designed to identify and measure β-galactosidase activity in living cells. This probe features a significant stokes shift, which has effectively addressed challenges such as spectral overlap, low signal-to-noise ratio, self-absorption, and interference in imaging live systems (Fig. 11a).107 Due to its excellent biocompatibility, broad pH range, and strong tissue penetration, the DXM-βgal probe has successfully tracked β-galactosidase activity in living organisms and OC cells. Notably, the DXM-βgal probe was the first to accurately trace endogenous β-galactosidase from the embryonic to the juvenile stage of zebrafish, showcasing its potential for use in dynamic biological applications. This probe is a valuable tool for studying β-galactosidase activity in cancer cells and living organisms, informing cancer diagnostics and biological research.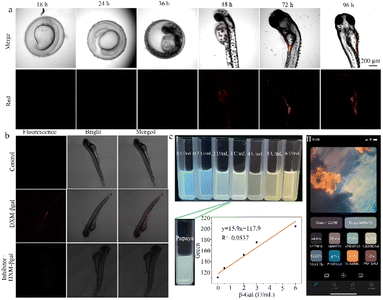 | ||
| Fig. 11 (a) Imaging of endogenous β-gal in zebrafish at different stages. XM can not only detect β-gal content in cancer cells but also track the changes of β-gal content in zebrafish at different developmental period. Reproduced with permission from ref. 107, Copyright 2022, Elsevier. (b) Fluorescence images of β-gal in zebrafish incubated with DXM-βgal (10 μM) after pretreated with inhibitor (5 mM). Reproduced with permission from ref. 108, Copyright 2020, Elsevier. (c) (I) Color changes of CB-FR with different concentrations of β-Gal, color changes of probe CB-FR with papaya juice, plot of the R value differences with 0–6.0 U mL−1 β-Gal. (II) Color removal software on smartphone. Reproduced with permission from ref. 110, Copyright 2023, Elsevier. | ||
Li et al. successfully designed a NIR fluorescent probe (DXM-βgal) to detect β-galactosidase activity both in vivo and in vitro (Fig. 11b).108 In the presence of β-galactosidase, the DXM-β-gal at 640 nm exhibits remarkable fluorescence enhancement, accompanied by a red to purple color change in solution. The reaction involves the β-galactosidase-catalyzed cleavage of the beta-galactosidic bond in DXM-βgal, causing its conversion into DXM-OH. Due to its excellent selectivity, sensitivity, and low detection limit, this probe has been successfully used for in vivo imaging in cells and zebrafish. The design of this probe and its strong biocompatibility suggest that it has the potential to become a valuable tool for studying β-galactosidase activity in the biomedical field.
Hou et al. achieved the successful creation of an enzyme-activated ratio NIR probe (Gal-Br-NO2), which can precisely identify endogenous β-galactosidase in OC cells, senescent cells, and zebrafish.109 Gal-Br-NO2 offers many advantages, including a short response time, large stokes shift, and NIR fluorescence emission, enabling accurate β-galactosidase imaging in zebrafish. This probe also has great potential as a tool for investigating β-galactosidase activity in aging and cancer research studies.
4.3 Detection of β-galactosidase activity in fruits
Hou et al. designed an intelligent FRET-based ratio fluorescent probe (CB-FR) to detect β-galactosidase activity in fruits and organisms (Fig. 11c).110 The CB-FR probe includes a classic FRET naphthalimide (acceptor) and pair coumarin (donor). Meanwhile, β-D-galactoside acts as a hydrolysis site for β-galactosidase and links the enzyme to naphthalimide. Due to its excellent water solubility, high affinity, rapid response time, and low detection limit, this probe can both detect overexpressed β-galactosidase in cancer cells and quantitatively analyze enzyme expression in fruits. The CB-FR probe enables the rapid semi-quantification of Gal activity through changes in pixel value pairs. Pixel value data can be extracted using color extraction software installed on a smartphone (Yiyu Qian Co., Ltd; APP). There is a strong linear relationship between the red channel value and β-galactosidase activity (0–6.0 U mL−1). Based on this relationship, the β-galactosidase activity in papaya can be accurately presented as 2.8951 U mL−1. Thus, the CB-FR system can be seamlessly integrated with smartphones to quickly and qualitatively evaluate β-Gal activity in actual fruit samples. This probe is a potent instrument for the instantaneous and swift identification of β-galactosidase activity in both fruits and living organisms, providing important insight into the physiological and pathological processes.5 Summary and outlook
In the past decade, there have been substantial advancements in the creation of β-galactosidase fluorescent probes used to image OC. These probes can directly identify and visualize aberrant enzyme levels in live OC cells, offering a new method for diagnosing this disease and informing the development of effective treatments. This review summarizes recent advances in OC-related fields, including the sensing mechanisms of β-galactosidase fluorescent probes and rational probe design strategies using various sensing mechanisms. The focus was on β-galactosidase fluorescent probes in the context of OC research. These probes are mainly categorized into single cancer detection, multiple cancer detection, and targeted fluorescent probes, highlighting their versatility and potential application in different fields. The current study provides examples of these designs to illustrate their significance and potential applications. While remarkable advancements have been achieved in the development of β-galactosidase enzyme-triggered fluorescence probes for OC imaging, transferring these sensing platforms from fundamental research to more clinical and practical applications has several challenges:(1) In early-stage cancers or cancer types with low β-galactosidase expression, the accurate detection of β-galactosidase enzyme activity at low concentrations requires fluorescence probes with ultra-high sensitivity. Developing novel β-galactosidase fluorescent probes can effectively increase the signal-to-noise ratio and thereby enhance sensitivity. For example, while NIR-II (1000–1700 nm) fluorescent probes can be used, their organic fluorophores often require complex synthesis processes with lower quantum yields and limited coverage in the NIR-II window. These significant limitations hinder the in vivo application of these probes. Thus, it is critical to devise simple and easily modifiable strategies to enhance their sensitivity.
(2) Effective and accurate in situ in vivo imaging requires emission probes, such as NIR-11, with deeper tissue penetration. However, these probes often contain extended π-conjugated structures that increase their hydrophobicity and reduce their water solubility. The design of such probes should involve appropriate functional groups and allow for both excellent water solubility and the capacity to detect deep cancer cells.
(3) NIR-II fluorescence imaging, due to its deep tissue penetration, high imaging resolution, and low autofluorescence, holds vast potential for a wide range of applications. NIR-II-emitting fluorophores are currently composed of both inorganic materials (quantum dots, rare-earth materials, gold nanoparticles) and organic molecules. Small molecule self-assembling nanoparticles can combine the simple chemical formulations found in small molecules with the tumor-targeting advantages of traditional nanoparticles. While effective self-assembling or encapsulated NIR-II probes have not yet been designed, they have exceptional imaging performance potential.111,112 The design of responsive and pH-sensitive self-assembling or encapsulated small molecules within a narrow physiological pH range (6.8–7.4) can be challenging. Self-assembling or encapsulated NIR-II small molecule dyes also often suffer from low quantum yields, limited chemical stability, and photostability in vivo. It is difficult to design reactive or targeted moieties that enhance the quantum yield and stability of these probes. This process requires intricate chemical processes and the development of activatable probes can be challenging. Furthermore, amphiphilic self-assembling or encapsulated NIR-II small molecules can disintegrate when diluted with hydrophobic biological molecules, potentially leading to false signals during in vivo imaging, which limits the receipt of accurate information in situ.
(4) β-Galactosidase expression by tumor cells varies by cancer type. For example, breast and colon cancers have less β-galactosidase expression than OC. As a result, β-galactosidase fluorescent probes are effective at detecting overexpressed β-galactosidase in OC but have limited accuracy at detecting breast or colon cancer. Thus, there is a need to develop β-galactosidase fluorescent probes capable of quantitatively measuring enzyme activity, allowing for accurate differentiation of enzyme activity in different cancer cells or variations in enzyme activity in a specific cancer type.
(5) When β-galactosidase fluorescent probes are released in situ, they tend to spread from the reaction site, decreasing the accuracy of detection. Approaches used to address this diffusion issue have often involved chemical attachments and lysosomal trafficking effects. Multifunctional β-galactosidase fluorescent probes capable of simultaneously detecting the activities of multiple enzymes in a single assay could increase diagnostic accuracy and provide more comprehensive biological information.
(6) There are distinctions between human endogenous β-galactosidase and β-galactosidase found in other species. While Escherichia coli β-galactosidase is often used as a substitute for this enzyme in humans, there are structural differences between the two. Effectively designing fluorescence probes that target human β-galactosidase holds great practical value for the accurate detection of this enzyme in humans.
(7) The development of organelle-targeted fluorescent probes has greatly improved subcellular imaging. Of the current subcellular localization fluorescence tools, ROS, RNS, and RSS (RONSS) probes are particularly effective at identifying the physiological and pathological functions of highly reactive, interacting, and interconvertible molecules associated with various biological events. Due to their distinct physicochemical properties, probes with specific targeting moieties can be localized to each organelle, including the nucleus, mitochondria, endoplasmic reticulum, lysosomes, and Golgi apparatus, and targeted to specific proteins.113 Nuclear targeting utilizes nuclear localization signals that bind with input proteins, actively transporting them into the nucleus through nuclear pores. Mitochondria, given their highly negative transmembrane potential (MMP, −180 mV), are targeted using ligands such as triphenylphosphonium (TPP), positively charged pyridine and quinoline derivatives, cyanines, and rhodamines. Some functional groups, including peptides and ketones, are used to construct mitochondrial-targeted probes because of their affinity for specific mitochondrial proteins. N,N-dimethylethylamine and 4-(2-hydroxyethyl) morpholine are commonly used to create lysosome-targeted fluorescent probes. These compounds are membrane-impermeable, protonated amines. Protein targeting involves the transfection and expression of a specific protein of interest (POI) with a fusion tag/enzyme. Once probes containing relevant substrates are introduced, they can effectively and spontaneously connect with the tag through highly specific biochemical reactions. This approach allows specific proteins within cells to be targeted. However, very little research has been conducted on organelle-targeted β-galactosidase fluorescence probes. Different targeting sites and their respective advantages could be used to design β-galactosidase fluorescence probes and further improve subcellular imaging.
Considering these research challenges, this review sought to inform the design and development of more sensitive and selective β-galactosidase enzyme fluorescence probes that can be used for the in situ detection and in vivo imaging of OC. This summary should offer guidance to improve these probes and foster their broad application in biosensors and imaging.
Author contributions
Liangliang Li: conceptualization, investigation, writing – original draft. Feifei Jia: supervision, conceptualization, writing. Yunxiu Li: investigation. Yan Peng: supervision, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
All authors are grateful for the support of Shenzhen Longhua District Central Hospital for the described work.Notes and references
- K. Ishikawa, M. Kataoka, T. Yanamoto, M. Nakabayashi, M. Watanabe, S. Ishihara and S. Yamaguchi, FEBS J., 2015, 282, 2540–2552 CrossRef CAS PubMed
.
- D. H. Juers, R. H. Jacobson, D. Wigley, X. J. Zhang, R. E. Huber, D. E. Tronrud and B. W. Matthews, Protein Sci., 2000, 9, 1685–1699 CrossRef CAS PubMed
.
- W. Tempel, Z. J. Liu, P. S. Horanyi, L. Deng, D. Lee, G. N. Newton, J. P. Rose, H. Ashida, S. C. Li, Y. T. Li and B. C. Wang, Proteins, 2005, 59, 141–144 CrossRef CAS PubMed
.
- D. H. Juers, R. H. Jacobson, D. Wigley, X. Zhang, R. E. Huber, D. E. Tronrud and B. W. Matthews, Protein Sci., 2000, 9, 1685–1699 CrossRef CAS PubMed
.
- D. H. Juers, T. D. Heightman, A. Vasella, J. D. McCarter, L. Mackenzie, S. G. Withers and B. W. Matthews, Biochemistry, 2001, 40, 14781–14794 CrossRef CAS PubMed
.
- D. H. Juers, B. W. Matthews and R. E. Huber, Protein Sci., 2012, 21, 1792–1807 CrossRef CAS PubMed
.
- M. Sakabe, D. Asanuma, M. Kamiya, R. J. Iwatate, K. Hanaoka, T. Terai, T. Nagano and Y. Urano, J. Am. Chem. Soc., 2012, 135, 409–414 CrossRef PubMed
.
- J. Zhang, P. Cheng and K. Pu, Bioconjug. Chem., 2019, 30, 2089–2101 Search PubMed
.
- P. Sosińska, J. Mikuła-Pietrasik, M. Ryżek, E. Naumowicz and K. Książek, Biogerontology, 2014, 15, 407–413 CrossRef PubMed
.
- Y. Wei, D. Cheng, T. Ren, Y. Li, Z. Zeng and L. Yuan, Anal. Chem., 2016, 88, 1842–1849 CrossRef CAS PubMed
.
- A. M. Konijn, R. Levy, G. Link and C. Hershko, J. Immunol. Methods, 1982, 54, 297–307 CrossRef CAS PubMed
.
- D. Baruch, H. Glickstein and Z. I. Cabantchik, Exp. Parasitol., 1991, 73, 440–450 Search PubMed
.
- G. P. Nolan, S. Fiering, J. F. Nicolas and L. A. Herzenberg, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 2603–2607 CrossRef CAS PubMed
.
- C. I. L. Justino, A. C. Freitas, R. Pereira, A. C. Duarte and T. A. P. R. Santos, Trac-Trend Anal. Chem., 2015, 68, 2–17 CrossRef CAS
.
- Y. Waerzeggers, P. Monfared, T. Viel, A. Winkeler, J. Voges and A. H. Jacobs, Methods, 2009, 48, 146–160 CrossRef CAS PubMed
.
- Y. Yao, Y. Zhang, C. Yan, W.-H. Zhu and Z. Guo, Chem. Sci., 2021, 12, 9885–9894 Search PubMed
.
- Y. Urano, M. Kamiya, K. Kanda, T. Ueno, K. Hirose and T. Nagano, J. Am. Chem. Soc., 2005, 127, 4888–4894 CrossRef CAS PubMed
.
- C. Wang, W. Chi, Q. Qiao, D. Tan, Z. Xu and X. Liu, Chem. Soc. Rev., 2021, 50, 12656–12678 RSC
.
- L. C. Xiao, L. J. Zhao, T. Li, D. K. Hartle, O. I. Aruoma and E. W. Taylor, Biofactors, 2006, 27, 157–165 Search PubMed
.
- A. Pal, M. Karmakar, S. R. Bhatta and A. Thakur, Coordin. Chem. Rev., 2021, 448, 214167 CrossRef CAS
.
- C.-H. Wu, P. Q. Nhien, T. T. K. Cuc, B. T. B. Hue and H.-C. Lin, Topics Curr. Chem., 2022, 381, 2 CrossRef PubMed
.
- N. S. M. Kamal, S. Safuan, S. Shamsuddin and P. Foroozandeh, Eur. J. Cell Biol., 2020, 99, 151108 CrossRef PubMed
.
- M. Safir Filho, P. Dao, M. Gesson, A. R. Martin and R. Benhida, Analyst, 2018, 143, 2680–2688 Search PubMed
.
- H. A. Kenny, P. C. Hart, K. Kordylewicz, M. Lal, M. Shen, B. Kara, Y. J. Chen, N. Grassl, Y. Alharbi, B. R. Pattnaik, K. M. Watters, M. S. Patankar, M. Ferrer and E. Lengyel, Cancers, 2021, 13, 3931 CrossRef CAS PubMed
.
- E. A. Pietilä, J. Gonzalez-Molina, L. Moyano-Galceran, S. Jamalzadeh, K. Zhang, L. Lehtinen, S. P. Turunen, T. A. Martins, O. Gultekin, T. Lamminen, K. Kaipio, U. Joneborg, J. Hynninen, S. Hietanen, S. Grénman, R. Lehtonen, S. Hautaniemi, O. Carpén, J. W. Carlson and K. Lehti, Nat. Commun., 2021, 12, 3904 CrossRef PubMed
.
- I. N. G. Budiana, M. Angelina and T. G. A. Pemayun, J. Turk.-Ger. Gynecol. Assoc., 2019, 20, 47–54 CrossRef PubMed
.
- J. Pfisterer and J. A. Ledermann, Semin. Oncol., 2006, 33, S12–S16 CrossRef CAS PubMed
.
- X. H. Chen, T. T. Zhang, W. Su, Z. H. Dou, D. P. Zhao, X. D. Jin, H. W. Lei, J. Wang, X. D. Xie, B. Cheng, Q. Li, H. Zhang and C. X. Di, Cell Death Dis., 2022, 13, 974 CrossRef CAS PubMed
.
- Y. C. Hou, X. T. Zhang, H. Yao, L. D. Hou, Q. W. Zhang, E. W. Tao, X. Q. Zhu, S. S. Jiang, Y. M. Ren, X. L. Hong, S. Y. Lu, X. X. Leng, Y. L. Xie, Y. Q. Gao, Y. Liang, T. Zhong, B. H. Long, J. Y. Fang and X. J. Meng, EMBO Rep., 2023, 24, e56325 CrossRef CAS PubMed
.
- Z. G. Xiao, J. Shen, L. Zhang, M. X. Li, W. Hu and C. H. Cho, Oncol. Lett., 2018, 15, 3395–3402 Search PubMed
.
- H. D. Li, D. Kim, Q. C. Yao, H. Y. Ge, J. Chung, J. L. Fan, J. Y. Wang, X. J. Peng and J. Yoon, Angew. Chem., Int. Ed., 2021, 60, 17268–17289 CrossRef CAS PubMed
.
- M. Li, M. Q. Yang and W. H. Zhu, Mater. Chem. Front., 2021, 5, 763–774 RSC
.
- B. Valeur and I. Leray, Coord. Chem. Rev., 2000, 205, 3–40 CrossRef CAS
.
- M. Liu, X. Yu, M. Li, N. Liao, A. Bi, Y. Jiang, S. Liu, Z. Gong and W. Zeng, RSC Adv., 2018, 8, 12573–12587 RSC
.
- Z. R. Grabowski and J. Dobkowski, Pure Appl. Chem., 1983, 55, 245–252 CrossRef CAS
.
- C. Curutchet, A. Franceschetti, A. Zunger and G. D. Scholes, J. Phys. Chem. C, 2008, 112, 13336–13341 CrossRef CAS
.
- E. Petryayeva, W. R. Algar and I. L. Medintz, Appl. Spectrosc., 2013, 67, 215–252 CrossRef CAS PubMed
.
- G. Beane, K. Boldt, N. Kirkwood and P. Mulvaney, J. Phys. Chem. C, 2014, 118, 18079–18086 CrossRef CAS
.
- T. Pons, I. L. Medintz, K. E. Sapsford, S. Higashiya, A. F. Grimes, D. S. English and H. Mattoussi, Nano Lett., 2007, 7, 3157–3164 CrossRef CAS PubMed
.
- T. Pons, I. L. Medintz, X. Wang, D. S. English and H. Mattoussi, J. Am. Chem. Soc., 2006, 128, 15324–15331 Search PubMed
.
- C. S. Abeywickrama, Chem. Commun., 2022, 58, 9855–9869 RSC
.
- H. Martin, L. R. Lázaro, T. Gunnlaugsson and E. M. Scanlan, Chem. Soc. Rev., 2022, 51, 9694–9716 RSC
.
- A. Belyaev, Y. H. Cheng, Z. Y. Liu, A. J. Karttunen, P. T. Chou and I. O. Koshevoy, Angew. Chem., Int. Ed., 2019, 58, 13456–13465 CrossRef CAS PubMed
.
- F. V. Bright, Anal. Chem., 1988, 60, 1031A–1039A CrossRef CAS PubMed
.
- F. C. Grozema, M. Swart, W. Robert, J. Zijlstra, J. J. Piet, D. Laurens, A. Siebbeles and P. T. V. Duijnen, J. Am. Chem. Soc., 2005, 127, 11019–11028 CrossRef CAS PubMed
.
- S. Paul, R. S. Fernandes and N. Dey, New J. Chem., 2022, 46, 18973–18983 Search PubMed
.
- S. K. Sharma, S. Poudel Sharma and R. M. Leblanc, Enzyme Microb. Technol., 2021, 150, 109885 CrossRef CAS PubMed
.
- F. Wu, J. Liu, M. Tao, M. Wang, X. Ren and Z. Hai, Anal. Chem., 2023, 95, 10481–10485 Search PubMed
.
- S. Zhang, X. Wang, X. Wang, T. Wang, W. Liao, Y. Yuan, G. Chen and X. Jia, Anal. Chem. Acta, 2022, 1198, 339554 CrossRef CAS PubMed
.
- Y. Niu, H. Wang, Y. Wang and L. Feng, Microchem. J., 2021, 166, 106205 CrossRef CAS
.
- R. M. Clegg, Fret and Flim Techniques, 2009, pp. 1–57 Search PubMed
.
- R. B. Sekar and A. Periasamy, J. Cell Biol., 2003, 160, 629–633 CrossRef CAS PubMed
.
- L. Wu, C. Huang, B. P. Emery, A. C. Sedgwick, S. D. Bull, X.-P. He, H. Tian, J. Yoon, J. L. Sessler and T. D. James, Chem. Soc. Rev., 2020, 49, 5110–5139 Search PubMed
.
- A. Kaur, P. Kaur and S. Ahuja, Anal. Method, 2020, 12, 5532–5550 RSC
.
- Y. Duo, G. H. Luo, W. T. Zhang, R. Z. Wang, G. G. Xiao, Z. H. Li, X. M. Li, M. L. Chen, J. Yoon and B. Z. Tang, Chem. Soc. Rev., 2023, 52, 1024–1067 RSC
.
- X. Wei, X.-X. Hu, L.-L. Zhang, J. Li, J. Wang, P. Wang, Z. Song, J. Zhang, M. Yan and J. Yu, Microchem. J., 2020, 157, 105046 CrossRef CAS
.
- J. Xiong, Y.-K. Cheung, W.-P. Fong, C. T. T. Wong and D. K. P. Ng, Chem. Commun., 2023, 59, 3471–3474 Search PubMed
.
- T. Komatsu, K. Kikuchi, H. Takakusa, K. Hanaoka, T. Ueno, M. Kamiya, Y. Urano and T. Nagano, J. Am. Chem. Soc., 2006, 128, 15946–15947 CrossRef CAS PubMed
.
- G. Jiao, L. H. Thoresen and K. Burgess, J. Am. Chem. Soc., 2003, 125, 14668–14669 Search PubMed
.
- W. Lin, L. Yuan, Z. Cao, Y. Feng and J. Song, Angew. Chem., Int. Ed., 2010, 49, 375–379 CrossRef CAS PubMed
.
- X. Kong, M. Li, B. Dong, Y. Yin, W. Song and W. Lin, Anal. Chem., 2019, 91, 15591–15598 CrossRef CAS PubMed
.
- C. Tang, J. Zhou, Z. Qian, Y. Ma, Y. Huang and H. Feng, J. Mater. Chem. B, 2017, 5, 1971–1979 Search PubMed
.
- X. Xie, Y. Lian, L. Xiao and L. Wei, Spectrochim. Acta A, 2020, 240, 118594 Search PubMed
.
- D. Jiang, Q. Tan, Y. Shen, M. Ye, J. Li and Y. Zhou, Spectrochim. Acta A, 2023, 292, 122411 CrossRef CAS PubMed
.
- W. Wang, K. Vellaisamy, G. Li, C. Wu, C.-N. Ko, C.-H. Leung and D.-L. Ma, Anal. Chem., 2017, 89, 11679–11684 Search PubMed
.
- X. Chen, X. Ma, Y. Zhang, G. Gao, J. Liu, X. Zhang, M. Wang and S. Hou, Anal. Chem. Acta, 2018, 1033, 193–198 CrossRef CAS PubMed
.
- X. Zhao, W. Yang, S. Fan, Y. Zhou, H. Sheng, Y. Cao and Y. Hu, J. Luminescence, 2019, 205, 310–317 Search PubMed
.
- X. Zhang, X. Chen, Y. Zhang, K. Liu, H. Shen, E. Zheng, X. Huang, S. Hou and X. Ma, Anal. Bioanal. Chem., 2019, 411, 7957–7966 CrossRef CAS PubMed
.
- X. Chen, X. Zhang, X. Ma, Y. Zhang, G. Gao, J. Liu and S. Hou, Talanta, 2019, 192, 308–313 CrossRef CAS PubMed
.
- C. H. Tung, Q. Zeng, K. Shah, D. E. Kim, D. Schellingerhout and R. Weissleder, Cancer Res., 2004, 64, 1579–1583 CrossRef CAS PubMed
.
- Y. Li, F. Liu, D. Zhu, T. Zhu, Y. Zhang, Y. Li, J. Luo and L. Kong, Talanta, 2022, 237, 122952 CrossRef CAS PubMed
.
- D. Liu, Z. Zhang, A. Chen and P. Zhang, Spectrochim. Acta A, 2022, 265, 120345 CrossRef CAS PubMed
.
- Z. Zhou, X. Pang, C. Fang, H. Li, B. Gu and Y. Zhang, Dyes. Pigments, 2023, 208, 110769 Search PubMed
.
- M. Chen, L. Mu, X. Cao, G. She and W. Shi, Chin. J. Chem., 2019, 37, 330–336 Search PubMed
.
- E.-J. Kim, R. Kumar, A. Sharma, B. Yoon, H. M. Kim, H. Lee, K. S. Hong and J. S. Kim, Biomaterials, 2017, 122, 83–90 CrossRef CAS PubMed
.
- T. S. Wehrman, G. von Degenfeld, P. O. Krutzik, G. P. Nolan and H. M. Blau, Nat. Methods, 2006, 3, 295–301 CrossRef CAS PubMed
.
- Y. Koide, Y. Urano, A. Yatsushige, K. Hanaoka, T. Terai and T. Nagano, J. Am. Chem. Soc., 2009, 131, 6058–6059 CrossRef CAS PubMed
.
- X.-X. Zhang, H. Wu, P. Li, Z.-J. Qu, M.-Q. Tan and K.-L. Han, Chem. Commun., 2016, 52, 8283–8286 RSC
.
- H. W. Lee, C. H. Heo, D. Sen, H.-O. Byun, I. H. Kwak, G. Yoon and H. M. Kim, Anal. Chem., 2014, 86, 10001–10005 CrossRef CAS PubMed
.
- D. Oushiki, H. Kojima, Y. Takahashi, T. Komatsu, T. Terai, K. Hanaoka, M. Nishikawa, Y. Takakura and T. Nagano, Anal. Chem., 2012, 84, 4404–4410 Search PubMed
.
- O. Green, S. Gnaim, R. Blau, A. Eldar-Boock, R. Satchi-Fainaro and D. Shabat, J. Am. Chem. Soc., 2017, 139, 13243–13248 Search PubMed
.
- Z. Li, M. Ren, Y. Zhao, W. Song, J. Cheng and W. Lin, Spectrochim. Acta A, 2021, 251, 119446 Search PubMed
.
- F. Fan, L. Zhang, X. Zhou, F. Mu and G. Shi, J. Mater. Chem. B, 2021, 9, 170–175 Search PubMed
.
- S. Chen, L. Wang, X. Ma, Y. Wu and S. Hou, Sens. Actuators, B, 2022, 367, 132061 Search PubMed
.
- K. Gu, W. Qiu, Z. Guo, C. Yan, S. Zhu, D. Yao, P. Shi, H. Tian and W.-H. Zhu, Chem. Sci., 2019, 10, 398–405 Search PubMed
.
- J.-A. Chen, H. Pan, Z. Wang, J. Gao, J. Tan, Z. Ouyang, W. Guo and X. Gu, Chem. Commun., 2020, 56, 2731–2734 RSC
.
- T. Gao, H. Li, Y. Wu, C. Deng, Y. Xie, J. Wang, Y. Yang, Q. Lv, Q. Jin, Y. Chen, L. Yi, Y. Zhong, X. Li, Q. Zhao, L. Zhang and M. Xie, Talanta, 2021, 235, 122659 CrossRef CAS PubMed
.
- H. B. Bosmann and T. C. Hall, Proc. Natl. Acad. Sci. U. S. A., 1974, 71, 1833–1837 Search PubMed
.
- H. D. Li, D. Kim, Q. C. Yao, H. Y. Ge, J. Chung, J. L. Fan, J. Y. Wang, X. J. Peng and J. Yoon, Angew. Chem., Int. Ed., 2021, 60, 17268–17289 CrossRef CAS PubMed
.
- N.-C. Yang and M.-L. Hu, Anal. Biochem., 2004, 325, 337–343 CrossRef CAS PubMed
.
- F. Debacq-Chainiaux, J. D. Erusalimsky, J. Campisi and O. Toussaint, Nat. Protoc., 2009, 4, 1798–1806 CrossRef CAS PubMed
.
- B. ROTMAN, Proc. Natl. Acad. Sci. U. S. A., 1981, 47, 1981–1991 CrossRef PubMed
.
- H. W. Lee, V. Juvekar, D. J. Lee, S. M. Kim and H. M. Kim, Anal. Chem., 2021, 93, 14778–14783 CrossRef CAS PubMed
.
- Q. Wu, Q.-H. Zhou, W. Li, T.-B. Ren, X.-B. Zhang and L. Yuan, ACS Sens., 2022, 7, 3829–3837 CrossRef CAS PubMed
.
- K. Gu, Y. Xu, H. Li, Z. Guo, S. Zhu, S. Zhu, P. Shi, T. D. James, H. Tian and W.-H. Zhu, J. Am. Chem. Soc., 2016, 138, 5334–5340 CrossRef CAS PubMed
.
- X. Z. Chai, H. H. Han, A. C. Sedgwick, N. Li, Y. Zang, T. D. James, J. Zhang, X. L. Hu, Y. Yu, Y. Li, Y. Wang, J. Li, X. P. He and H. Tian, J. Am. Chem. Soc., 2020, 142, 20270 CrossRef CAS PubMed
.
- J. Zhang, C. Li, C. Dutta, M. Fang, S. Zhang, A. Tiwari, T. Werner, F.-T. Luo and H. Liu, Anal. Chem. Acta, 2017, 968, 97–104 CrossRef CAS PubMed
.
- J. Huang, N. Li, Q. Wang, Y. Gu and P. Wang, Sens. Actuators, B, 2017, 246, 833–839 CrossRef CAS
.
- X. Li, Y. Pan, H. Chen, Y. Duan, S. Zhou, W. Wu, S. Wang and B. Liu, Anal. Chem., 2020, 92, 5772–5779 CrossRef CAS PubMed
.
- D. Asanuma, M. Sakabe, M. Kamiya, K. Yamamoto, J. Hiratake, M. Ogawa, N. Kosaka, P. L. Choyke, T. Nagano, H. Kobayashi and Y. Urano, Nat. Commun., 2015, 6, 7463 CrossRef PubMed
.
- L. Franková and S. C. Fry, Plant J., 2012, 71, 45–60 Search PubMed
.
- Q. Husain, Crit. Rev. Biotechnol., 2010, 30, 41–62 CrossRef CAS PubMed
.
- L. Jia, Y. Li, G. Liu and J. He, Plant Physiol. Biochem., 2023, 194, 383–393 CrossRef CAS PubMed
.
- S. Mohebbi, M. Babalar, Z. Zamani and M. A. Askari, Sci. Hortic., 2020, 259, 108822 CrossRef CAS
.
- M. Kamiya, D. Asanuma, E. Kuranaga, A. Takeishi, M. Sakabe, M. Miura, T. Nagano and Y. Urano, J. Am. Chem. Soc., 2011, 133, 12960–12963 CrossRef CAS PubMed
.
- Y. Ichikawa, M. Kamiya, F. Obata, M. Miura, T. Terai, T. Komatsu, T. Ueno, K. Hanaoka, T. Nagano and Y. Urano, Angew. Chem., Int. Ed., 2014, 126, 6890–6893 CrossRef
.
- S. Chen, K. Niu, L. Wang, Y. Wu, S. Hou and X. Ma, Anal. Chem. Acta, 2022, 1232, 340459 CrossRef CAS PubMed
.
- X. Pang, Y. Li, Z. Zhou, Q. Lu, R. Xie, C. Wu, Y. Zhang and H. Li, Talanta, 2020, 217, 121098 CrossRef CAS PubMed
.
- S. Chen, M. Liu, Y. Zi, J. He, L. Wang, Y. Wu, S. Hou and W. Wu, Spectrochim. Acta A, 2023, 285, 121879 Search PubMed
.
- S. Chen, X. Ma, L. Wang, Y. Wu, Y. Wang, S. Hou and W. Fan, Sens. Actuators, B, 2023, 387, 383–393 CrossRef
.
- Z. Li, P.-Z. Liang, L. Xu, X.-X. Zhang, K. Li, Q. Wu, X.-F. Lou, T.-B. Ren, L. Yuan and X.-B. Zhang, Nat. Commun., 2023, 14, 1843 Search PubMed
.
- H. Chen, K. Shou, S. Chen, C. Qu, Z. Wang, L. Jiang, M. Zhu, B. Ding, K. Qian, A. Ji, H. Lou, L. Tong, A. Hsu, Y. Wang, D. W. Felsher, Z. Hu, J. Tian and Z. Cheng, Adv. Mater., 2021, 33, 2006902 CrossRef CAS PubMed
.
- P. Gao, W. Pan, N. Li and B. Tang, Chem. Sci., 2019, 10, 6035–6071 RSC
.
| This journal is © The Royal Society of Chemistry 2024 |
