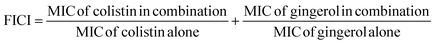Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synergistic action of 6-gingerol as an adjuvant to colistin for susceptibility enhancement in multidrug-resistant Klebsiella pneumoniae isolates
Maheswata Sahoo,
Dibyajyoti Uttameswar Behera,
Rajesh Kumar Sahoo,
Saubhagini Sahoo,
Suchanda Dey and
Enketeswara Subudhi *
*
Centre for Biotechnology, Siksha ‘O’ Anusandhan (Deemed to be University), Kalinga Nagar, Ghatikia, Bhubaneswar 751003, Odisha, India. E-mail: enketeswarasubudhi@soa.ac.in; maheswatasahoo89@gmail.com; dibya01bioinfo@gmail.com; rajeshkumarsahoo@soa.ac.in; saubhagini2015@gmail.com; suchandadey1993@gmail.com; Tel: +91-9861075829
First published on 5th March 2024
Abstract
The growing threat to human health posed by multidrug-resistant Klebsiella pneumoniae (MDR-KP) indicates an urgent need to develop alternative therapeutic options. The emergence of colistin resistance further adds to the complexity. The study aims to explore in silico-screened phytomolecule 6-gingerol, the most potent active constituent of ginger, as an adjuvant to restore sensitivity in MDR-KP isolates to colistin. The screening of phytocompounds of Zingiber officinale were obtained from the spiceRx database, and molecular docking with efflux pump protein AcrB was performed using Schrödinger's Glide program. The synergistic and bactericidal effects of 6-gingerol in combination with colistin against MDR-KP isolates were determined following broth micro-dilution (MIC), checkerboard assay, and time-kill study. 6-Gingerol showed a good binding affinity with AcrB protein (−9.32 kcal mol−1) and followed the Lipinski rule of (RO5), demonstrating favourable drug-like properties. Further, the synergistic interaction of 6-gingerol with colistin observed from checkerboard assays against efflux-mediated colistin resistance MDR-KP isolates reveals it to be a prospectus adjuvant. The time-killing assays showed the effect of 6-gingerol in combination with colistin to be bactericidal against MSK9 and bacteriostatic against MSK4 and MSK7. Overall, the study provides insights into the potential use of 6-gingerol as a safe and easily available natural product to treat multidrug-resistant K. pneumoniae infections combined with colistin but needs in vivo toxicity evaluation before further recommendations can be made.
1. Introduction
The global emergence of multidrug-resistant (MDR) Gram-negative bacteria poses a growing threat to human health. According to the World Health Organization (WHO), 700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 annual deaths are associated with MDR bacteria, which could reach up to 10 million by 2050.1–3 WHO has earmarked some highly resistant Critical Priority pathogens under the acronym ESKAPE, which include Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species.4 Among them, Klebsiella pneumoniae is the most frequently reported healthcare-associated MDR pathogen that causes urinary tract infections, pneumonia, meningitis, and sepsis.5 The frequent use of polymyxin E (colistin) in recent decades as a last resort drug, driven by its efficacy in achieving satisfactory serum levels and low minimum inhibitory concentrations (MIC) against carbapenem-resistant K. pneumoniae, has led to the emergence of resistance.4,6 The over-expression of the efflux pump systems helps extrude antibacterial molecules out of the bacterial cell, thereby reducing their concentrations to an insufficient quantity for proven effectiveness. It is becoming the predominant mechanism behind the emergence of MDR.4,7 Bacterial efflux pumps are essential for drug extrusion and play a role in their virulence and adaptive responses, according to recent clinical and laboratory data.8 Phenotypic profiling has shown that exposure to antimicrobial drugs frequently causes complex bacterial reactions, including altered expressions of several genes encoding the transporters.9 Bacterial efflux pumps are recognized as either primary active transporters that use ATPs as an energy source or secondary active transporters that are obtained as a result of the electrochemical potential difference produced by pumping out Na+ and H+ outside the membrane.10 Hence, it is necessary to select specific inhibitors of efflux pumps, which can be combined with conventional antibiotics to restore their use.
000 annual deaths are associated with MDR bacteria, which could reach up to 10 million by 2050.1–3 WHO has earmarked some highly resistant Critical Priority pathogens under the acronym ESKAPE, which include Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species.4 Among them, Klebsiella pneumoniae is the most frequently reported healthcare-associated MDR pathogen that causes urinary tract infections, pneumonia, meningitis, and sepsis.5 The frequent use of polymyxin E (colistin) in recent decades as a last resort drug, driven by its efficacy in achieving satisfactory serum levels and low minimum inhibitory concentrations (MIC) against carbapenem-resistant K. pneumoniae, has led to the emergence of resistance.4,6 The over-expression of the efflux pump systems helps extrude antibacterial molecules out of the bacterial cell, thereby reducing their concentrations to an insufficient quantity for proven effectiveness. It is becoming the predominant mechanism behind the emergence of MDR.4,7 Bacterial efflux pumps are essential for drug extrusion and play a role in their virulence and adaptive responses, according to recent clinical and laboratory data.8 Phenotypic profiling has shown that exposure to antimicrobial drugs frequently causes complex bacterial reactions, including altered expressions of several genes encoding the transporters.9 Bacterial efflux pumps are recognized as either primary active transporters that use ATPs as an energy source or secondary active transporters that are obtained as a result of the electrochemical potential difference produced by pumping out Na+ and H+ outside the membrane.10 Hence, it is necessary to select specific inhibitors of efflux pumps, which can be combined with conventional antibiotics to restore their use.
The absence of novel antibiotics has led to exploring alternative sources, most preferably the secondary metabolites with therapeutic properties, such as terpenoids, phenolics, and alkaloids from medicinal plants, due to their historical use, low cost, and easy availability.11 Recently, these plants have gained considerable attention as approximately 40% of current medications are derived from phytochemicals.12 Phytochemicals are known to exert a direct antimicrobial effect and improve the efficacy of conventional antibiotics when used in combinations.11,12 Furthermore, these plant-derived compounds can interact with critical stages of the pathogenic process, reducing the potential of bacteria to develop resistance.12 Consequently, combining these compounds with conventional antibiotics seems promising by allowing their reutilization, which has lost effectiveness due to the overactivity of the efflux pump system in Gram-negative bacteria.13,14 Therefore, restoring the sensitivity of bacteria to colistin using potential phytochemicals as an adjuvant could considerably improve therapeutic outcomes and may be a potential approach for treating infections caused by colistin-resistant MDR-KP.12
Previous research has demonstrated that Zingiber officinale phytoconstituents are promising candidates for treating bacterial infections and biofilm inhibition; these have also been used safely in home remedies for a long time.15,16 In silico screening has been a proven method to narrow down a larger library of molecules from the Spice Rx database to a single molecule against the target. In this study, we selected 6-gingerol based on molecular docking and its pharmacological properties for additional in vitro investigation. 6-Gingerol is one of the most abundant natural polyphenols found in ginger rhizomes that exhibits multiple biological activities, including anti-inflammatory, antitumor, antioxidant, and antibacterial. Studies confirmed that the 6-gingerol has anti-biofilm activities against drug-resistant Candida albicans15,17 and quorum-sensing inhibition activity in P. aeruginosa.18 Furthermore, 6-gingerol is highly effective in overcoming the complications of multidrug resistance associated with chemotherapeutic agents.19
Based on the above facts, in this study, we collected 23 MDR-KP from a six-month surveillance study of the Central Laboratory of IMS and SUM hospital, Bhubaneswar, Odisha. These were subjected to a series of in vitro analyses: the broth micro-dilution technique, checkerboard assay, and time-kill kinetics to determine its role as an adjuvant to colistin for enhancing susceptibility in MDR-KP. This study provides valuable insights into using the easily available natural molecule 6-gingerol to inhibit the growth of MDR-KP bacteria and enhance the efficacy of routine antibiotics. These findings represent a significant contribution to the emerging research on the validation of the use of medicinal plant constituents for treating MDR infections as an efflux pump inhibitor.
2. Material and methods
2.1. Chemicals and solvents
Cation adjusted Mueller Hinton broth (CaMHB), Luria Bertani agar (LBA), Luria Bertani broth (LB), Tryptic soy agar (TSA), dimethyl sulfoxide (DMSO), carbonyl cyanide m-chloro phenylhydrazine (CCCP), ethidium bromide (EtBr), colistin sulfate, and triphenyl tetrazolium chloride (TTC) were procured from Himedia, India, and 6-gingerol was purchased from Sigma-Aldrich, India.2.2. In silico assay
2.3. In vitro assay
If the FICI value ≤0.5, ≤1, ≤4, and >4, there would be synergistic, additive, indifferent, and antagonistic interactions between 6-gingerol and colistin.
3. Results and discussion
3.1. In silico analysis
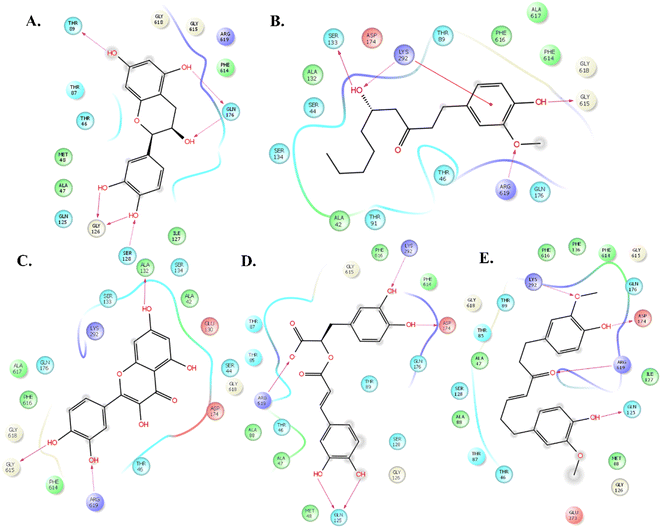
The molecular docking study of 69 phyto compounds with AcrB protein screened out the top five compounds with high XP docking scores. Epicatechin exhibited a binding affinity of −10.78 kcal mol−1, forming four hydrogen bonds with SER128, GLY126, ASP 276, and ARG 619.6-Gingerol displays a binding affinity of −9.32 kcal mol−1, establishing four hydrogen bonds with ASP174, GLN176, LYS292, and ARG619. Quercetin demonstrates a binding affinity of −9.12 kcal mol−1, forming five hydrogen bonds with ASP83, THR87, THR91, SER133, and LYS292. Rosmarinic acid exhibited a binding affinity of −8.52 kcal mol−1, establishing four hydrogen bonds with THR89, GLN125, PHE616, and ARG619. Gingerenone A has a binding affinity of −8.45 kcal mol−1, forming three hydrogen bonds with GLN125, LYS292, and ARG619 (Fig. 1). Additionally, based upon drug-likeness filters, such as Log S (ESOL), molecular weight, number of heavy atoms, number of rotatable bonds, Lipinski rule of (RO5), GI absorption, BBB permeant, and bioavailability score of the five compounds, 6-gingerol demonstrated favourable drug-like properties (Table 1). Thus, 6-gingerol was further considered for the in vitro antimicrobial efficacy study.| Compound name | Docking score | ADMET screening properties | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Phytocompounds | XP score (kcal mol−1) | Log S (ESOL) | Molecular weight | Number of heavy atoms | Number of rotatable bonds | Lipinski (RO5) | GI absorption | BBB permeant | Bioavailability score |
| a RO5: Rule of five; XP: extra precision; ESOL: estimated solubility; ADMET: absorption, distribution, metabolism, excretion, and toxicity; GI absorption: gastrointestinal absorption; BBB permeant: blood–brain barrier permeant. | |||||||||
| Epicatechin | −10.78 | Soluble | 290.27 | 21 | 1 | 0 | High | No | 0.55 |
| 6-Gingerol | −9.32 | Soluble | 294.39 | 21 | 10 | 0 | High | Yes | 0.55 |
| Quercetin | −9.12 | Soluble | 302.24 | 22 | 1 | 0 | High | No | 0.55 |
| Rosmarinic acid | −8.52 | Soluble | 360.31 | 26 | 7 | 0 | Low | No | 0.56 |
| Gingerenone A | −8.45 | Moderately soluble | 356.41 | 26 | 9 | 0 | High | Yes | 0.55 |
3.2. In vitro analysis
| Antimicrobial | MSK4 (MIC) | Interpretation | MSK7 (MIC) | Interpretation | MSK9 (MIC) | Interpretation | |
|---|---|---|---|---|---|---|---|
| a S: susceptible; I: intermediate; R: resistant; MIC: minimum inhibitory concentration. | |||||||
| β-lactam | Ampicillin | ≥32 | R | ≥32 | R | ≥32 | R |
| β-lactam+ β-lactamase inhibitor | Piperacillin/tazobactam | ≥128 | R | ≥128 | R | ≥128 | R |
| Amoxicillin/clavulanic acid | ≥32 | R | ≥32 | R | ≥32 | R | |
| Cephalosporin | Cefepime | ≥64 | R | 32 | R | 32 | R |
| Cefoperazone/sulbactam | ≥64 | R | ≥32 | *R | ≥64 | R | |
| Ceftriaxone | ≥64 | R | ≥64 | R | ≥64 | R | |
| Cefuroxime | ≥64 | R | ≥64 | R | ≥64 | R | |
| Cefuroxime axetil | ≥64 | R | ≥64 | R | ≥64 | R | |
| Carbapenem | Meropenem | ≥16 | R | 8 | R | 8 | R |
| Imipenem | 8 | R | ≤0.25* | *I | ≤0.25* | *I | |
| Ertapenem | ≥8 | R | 4 | R | ≥8 | R | |
| Aminoglycosides | Amikacin | ≥64 | R | ≥64 | R | ≥64 | R |
| Gentamicin | ≥16 | R | ≥16 | R | ≥16 | R | |
| Fluroquinolone | Ciprofloxacin | ≥4 | R | ≥4 | R | ≥8 | R |
| Nalidixic acid | ≥32 | R | ≥32 | R | ≥32 | R | |
| Nitrofuran | Nitrofurantoin | 128 | R | 64 | I | 128 | R |
| Polymyxin | Colistin | ≥16 | R | ≥8 | R | ≥16 | R |
| Folate pathway | Trimethoprim/sulfamethoxazole | ≤20 | S | ≥320 | R | ≥320 | R |
| Glycylcyclines | Tigecycline | 2 | S | ≤0.5 | S | 2 | S |
| Strain name | MIC colistin (μg mL−1) | MIC CCCP (μg mL−1) | MIC 6-gingerol (μg mL−1) | MIC colistin + CCCP (μg mL−1) | Fold reduction (colistin) | MIC colistin+6-gingerol (μg mL−1) | Fold reduction (colistin) | FICI | Outcome |
|---|---|---|---|---|---|---|---|---|---|
| a FICI: fractional inhibitory concentration index; MIC: minimum inhibitory concentration; CCCP: carbonyl cyanide m-chlorophenylhydrazine. | |||||||||
| MSK7 | 512 | 16 | 512 | 32/2 | 16 | 64/32 | 8 | 0.18 | Synergistic |
| MSK4 | 256 | 16 | 128 | 16/4 | 16 | 8/16 | 32 | 0.15 | Synergistic |
| MSK9 | ≥1024 | 16 | 512 | 64/2 | 16 | 128/64 | 8 | 0.25 | Synergistic |
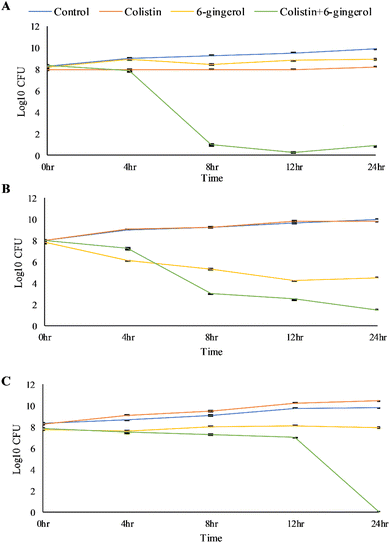
A time-kill assay was performed to validate the ability of 6-gingerol to potentiate the bactericidal effect of colistin. The result demonstrated that MSK4 and MSK7 showed a significant reduction in the CFU from 4 to 12 h with a bacteriostatic effect. In comparison, a complete reduction in CFU count after 12 h was found in MSK9, indicating the bactericidal effect (Fig. 3).
Bacteria develop antibiotic resistance and make the routine treatment critical in the infected patients.28 While increasing the antibiotic dose may overcome resistance but is often not recommended for toxic effects, particularly for antibiotics such as colistin. The development of resistance to colistin, the sole treatment option for infection due to MDR-KP, has made the world reach the antibiotics-era29,30 and prompted the use of adjuvants combined with existing treatment avenues to enhance its efficacy.30 Combinatorial therapy for treating these superbugs has been attempted for several reasons to replace monotherapy; primarily, its synergistic effects make the treatment more effective by preventing the emergence of new resistant strains and reducing antibiotic dose-related toxicity.31 In addition, it may achieve a broad spectrum of antimicrobial activity.29,30 Several edible natural products and food ingredients have been recently found to augment the antibacterial efficacy of nitrofurantoin and clindamycin.32
Herein, we screened phytocompounds of Z. officinale using molecular docking techniques. Based on its pharmacological properties, we identified 6-gingerol as the most promising drug candidate to target the AcrB efflux pump protein. Furthermore, we explored the antimicrobial activity of 6-gingerol against colistin-resistant MDR-KP, which could significantly reduce the MIC of colistin by exhibiting a synergistic effect. The recorded synergistic effect of the 6-gingerol and colistin against efflux-mediated colistin-resistant Gram-negative MDR-KP isolates provides the first direct, immediate evidence for the reduction of MIC of colistin through susceptibility enhancement. 6-Gingerol, the most prevailing constituent among the list of phytochemicals of ginger, has been studied to have antimicrobial activity against methicillin-resistant Staphylococcus aureus and Candida albicans.17,33 The 1/4 MIC of 6-gingerol alone showed bacteriostatic activity but combined with 1/4 MIC of colistin, it could considerably reduce the CFU count when compared to employing colistin alone. While various studies have investigated the biological activities of 6-gingerol and its combinatorial effects with different antibiotics, they remain unexplored. In this study, it was demonstrated that 6-gingerol enhances the antibacterial properties of colistin against MDR-KP isolates.
Currently, the enhancement in susceptibility in colistin resistance mediated by efflux pump MDR-KP isolates was observed, and it has been suggested that 6-gingerol might be inhibiting the efflux pump system responsible for bacterial resistance to colistin. Alternatively, we may hypothesize that the susceptibility enchantment could be for the enhanced influx of 6-gingerol into these bacteria due to increased permeability of the outer leaflet by colistin binding to lipopolysaccharide.34 However, the underlying mechanism of the natural adjuvant, 6-gingerol, which responded differentially to time-killing assay (bactericidal and bacteriostatic) to different MDR-KP strains remains unknown and warrants further investigation. Given its diverse pharmacological properties, 6-gingerol has garnered considerable attention from the scientific community. It has been increasingly incorporated as a food ingredient globally and accredited as GRAS (generally recognized as safe).16,35
However, this study is limited to assaying with fewer colistin-resistant MDR-KP. The in vitro outcome of the above combination needs further in vivo validation before use in clinical practice as a generalized recommendation.
4. Conclusion
Our results demonstrate that the combination of 6-gingerol and colistin exhibits synergistic effects against colistin-resistant MDR-KP isolates. These findings highlight the potential of 6-gingerol, a safe and natural compound, as a valuable resource for finding the links between the structure and function of antibiotic-reversing agents. Our study suggests that this compound has considerable potential for combination therapy against MDR-KP bacterial infections and can enhance the efficacy of colistin in vitro. These findings provide a promising alternative approach for overcoming colistin resistance. Thus, more attempts should be made to elucidate the mode of action of this compound in inciting the antibacterial activity against a larger number of MDR-KP, followed by in vivo toxicity and pharmacokinetics studies before recommending it as a potential adjuvant.Author contributions
Conceptualization: MS, ES, SD; Formal Analysis: MS, DUB, and SS; Investigation: MS, DUB, and SS; Methodology: MS, RKS; Supervision: ES; Writing original draft: MS; Writing-review, and editing: ES, and RKS.Conflicts of interest
The authors report that there are no competing interests to declare.Acknowledgements
The authors are grateful to Prof (Dr) S.C. Si, Dean, Centre for Biotechnology and Prof (Dr) M.R. Nayak, President, Siksha' O′ Anusandhan University, for providing infrastructure and encouragement throughout the work.References
- Z. Fatima, D. Purkait, S. Rehman, S. Rai and S. Hameed, in Biological and Environmental Hazards, Risks, and Disasters, Elsevier, 2023, pp. 197–220 Search PubMed.
- P. Dadgostar, Infect. Drug Resist., 2019, 12, 3903–3910 CrossRef CAS PubMed.
- T. M. Uddin, A. J. Chakraborty, A. Khusro, B. R. M. Zidan, S. Mitra, T. Bin Emran, K. Dhama, M. K. H. Ripon, M. Gajdács, M. U. K. Sahibzada, M. J. Hossain and N. Koirala, J. Infect. Public Health, 2021, 14, 1750–1766 CrossRef PubMed.
- M. S. Mulani, E. E. Kamble, S. N. Kumkar, M. S. Tawre and K. R. Pardesi, Front. Microbiol., 2019, 10, 539 CrossRef PubMed.
- K. L. Wyres, M. M. C. Lam and K. E. Holt, Nat. Rev. Microbiol., 2020, 18, 344–359 CrossRef CAS PubMed.
- K. S. Kaye, J. M. Pogue, T. B. Tran, R. L. Nation and J. Li, Infect. Dis. Clin. N. Am., 2016, 30, 391–414 CrossRef PubMed.
- K. Nishino, S. Yamasaki, R. Nakashima, M. Zwama and M. Hayashi-Nishino, Front. Microbiol., 2021, 12, 737288 CrossRef PubMed.
- W. DU, A. TEPPER and A. VERDELHAN, J. Finance, 2018, 73, 915–957 CrossRef.
- R. J. Nichols, S. Sen, Y. J. Choo, P. Beltrao, M. Zietek, R. Chaba, S. Lee, K. M. Kazmierczak, K. J. Lee, A. Wong, M. Shales, S. Lovett, M. E. Winkler, N. J. Krogan, A. Typas and C. A. Gross, Cell, 2011, 144, 143–156 CrossRef CAS PubMed.
- V. Shriram, T. Khare, R. Bhagwat, R. Shukla and V. Kumar, Front. Microbiol., 2018, 9, 2990 CrossRef PubMed.
- D. Ramírez-Rendon, A. K. Passari, B. Ruiz-Villafán, R. Rodríguez-Sanoja, S. Sánchez and A. L. Demain, Appl. Microbiol. Biotechnol., 2022, 106, 1855–1878 CrossRef PubMed.
- F. N. Idris and M. M. Nadzir, Arch. Microbiol., 2023, 205, 115 CrossRef CAS PubMed.
- J. A. Seukep, L. P. Sandjo, B. T. Ngadjui and V. Kuete, BMC Complementary Altern. Med., 2016, 16, 193 CrossRef PubMed.
- A. J. Seukep, V. Kuete, L. Nahar, S. D. Sarker and M. Guo, J. Pharm. Anal., 2020, 10, 277–290 CrossRef PubMed.
- T. Rathinavel, M. Palanisamy, S. Palanisamy, A. Subramanian and S. Thangaswamy, Int. J. Adv. Sci. Eng., 2020, 06, 1482–1489 CrossRef CAS.
- A. Das, S. Dey, R. K. Sahoo, S. Sahoo and E. Subudhi, J. Essent.Oil Bear. Plants, 2019, 22, 1163–1171 CrossRef CAS.
- J.-H. Lee, Y.-G. Kim, P. Choi, J. Ham, J. G. Park and J. Lee, Front. Cell. Infect. Microbiol., 2018, 8, 299 CrossRef PubMed.
- H.-S. Kim, S.-H. Lee, Y. Byun and H.-D. Park, Sci. Rep., 2015, 5, 8656 CrossRef CAS PubMed.
- F. Al-Abbasi, E. Alghamdi, M. Baghdadi, A. Alamoudi, A. El-Halawany, H. El-Bassossy, A. Aseeri and A. Al-Abd, Molecules, 2016, 21, 886 CrossRef PubMed.
- K. A. Peele, C. Potla Durthi, T. Srihansa, S. Krupanidhi, V. S. Ayyagari, D. J. Babu, M. Indira, A. R. Reddy and T. C. Venkateswarulu, Informatics Med. Unlocked, 2020, 19, 100345 CrossRef PubMed.
- T. A. Halgren, R. B. Murphy, R. A. Friesner, H. S. Beard, L. L. Frye, W. T. Pollard and J. L. Banks, J. Med. Chem., 2004, 47, 1750–1759 CrossRef CAS PubMed.
- E. Mateev, M. Georgieva and A. Zlatkov, Biointerface Res. Appl. Chem., 2022, 13, 159 Search PubMed.
- D. U. Behera, K. Ratnajothy, S. Dey, M. Gaur, R. K. Sahoo, S. Sahoo, B. Rautaraya, M. K. Rout and E. Subudhi, 3 Biotech, 2023, 13, 127 CrossRef PubMed.
- S. Dey, M. Gaur, R. K. Sahoo, A. Das, B. Jain, S. Pati and E. Subudhi, J. Global Antimicrob. Resist., 2020, 22, 54–56 CrossRef PubMed.
- C.-H. Lai, Y.-S. Lin, C.-M. Wang, P.-C. Chang and W.-Y. Shia, Microbiol. Spectrum, 2023, 11(3), e02916 Search PubMed.
- Y. Ding, J. Hao, W. Xiao, C. Ye, X. Xiao, C. Jian, M. Tang, G. Li, J. Liu and Z. Zeng, Front. Microbiol., 2023, 14, 1207441 CrossRef PubMed.
- R. K. Sahoo, E. Subudhi and M. Kumar, Lett. Appl. Microbiol., 2014, 58, 610–616 CrossRef CAS PubMed.
- J. M. Munita and C. A. Arias, Microbiol. Spectrum, 2016, 481–511 CAS.
- L. Yu, J. Zhang, Y. Fu, Y. Zhao, Y. Wang, J. Zhao, Y. Guo, C. Li and X. Zhang, Front. Cell. Infect. Microbiol., 2019, 9, 422 CrossRef CAS PubMed.
- J. Osei Sekyere, U. Govinden, L. A. Bester and S. Y. Essack, J. Appl. Microbiol., 2016, 121, 601–617 CrossRef CAS PubMed.
- T. Tängdén, Upsala J. Med. Sci., 2014, 119, 149–153 CrossRef PubMed.
- A. Malik, S. Bai, L. Seasotiya, P. Bharti and S. Dalal, Am. J. Adv. Drug Delivery, 2014, 2, 522–533 Search PubMed.
- B. O. Oyedemi, E. M. Kotsia, P. D. Stapleton and S. Gibbons, J. Ethnopharmacol., 2019, 245, 111871 CrossRef CAS PubMed.
- N. K. Khadka, C. M. Aryal and J. Pan, ACS Omega, 2018, 3, 17828–17834 CrossRef CAS PubMed.
- I. R. Kubra and L. J. M. Rao, Crit. Rev. Food Sci. Nutr., 2012, 52, 651–688 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |