Open Access Article
Open Access ArticleInvestigating the effects of ultrafine bubbles on bacterial growth†
Mai Phuong Vu ,
Nguyen Le Hanh Tran,
Thien Quang Lam,
Anh Thi Quynh Tran
,
Nguyen Le Hanh Tran,
Thien Quang Lam,
Anh Thi Quynh Tran ,
Thu Phan Anh Le and
Khoi Tan Nguyen
,
Thu Phan Anh Le and
Khoi Tan Nguyen *
*
School of Biotechnology, International University, Vietnam National University, Ho Chi Minh City, 700000, Vietnam. E-mail: ntkhoi@hcmiu.edu.vn; Fax: +84 28 3724 4271; Tel: +84 28 3724 4270
First published on 10th January 2024
Abstract
Several previous studies have considered ultrafine bubbles as a potential research target because their properties can be applied in many different research areas. In particular, the interaction between UFBs and microorganisms has always been one of the aspects that receives much attention due to the high difficulty in controlling a living system. The properties of UFBs, as mobile air–water interfaces, are greatly determined by their gas cores which play a critical role in regulating microbial growth. This study aims to investigate the effects of ultrafine bubbles on bacterial growth. Two well-studied organisms were chosen as models – Escherichia coli and Staphylococcus aureus. Their growing behavior was examined based on the growth rate, phenotype and biomass. Three types of Luria-Bertani cultures were tested, including a standard culture containing distilled water, an air ultrafine bubble culture, and a hydrogen ultrafine bubble culture. The UFBs were generated via ultrasonic cavitation and stabilized by 50 μM SDS, which was proven to have negligible effects on bacterial growth. By comparing among the three cultivation conditions, the bacterial growth rates were observed to be the highest in exposure to HUFBs. The results also signified that UFBs had an enhancement on cell proliferation. On the other hand, while proposing an increase in cell density, bacteria cultured in HUFB media have their sizes decreased uniformly and significantly (p-value < 0.05). This study confirmed that bacterial growth was promoted by UFBs; and better effects recorded were due to the HUFB present in the culture media. However, the average morphological size of bacteria was in negative correlation with their population size.
1. Introduction
Bubbles in a bulk solution are apparent to the eyes when their sizes are large enough, as can be observed in carbonated drinks or from an air diffuser in a water tank. This applies for the bubbles with diameters of a few millimeters, also the presence of microscopic bubbles with diameters of microns can be confirmed with turbidity.1,2 In contrast, the existence of bubbles with a diameter of roughly 100 to 200 nm can only be detected using the dynamic light scattering method.3 Moreover, these bubbles in this size range could remain stable for more than a month.4,5 Among these size ranges, gas bubbles with diameters less than 1 micrometer are defined as ultrafine bubbles (UFBs).6 They are sometimes referred to as bulk nanobubbles. UFBs are commonly formed through hydrodynamic cavitation using a Venturi tube, swirling flow, or the injection of pressured water containing gas.7In many circumstances, UFBs form with the presence of microbubbles.7 Since bubble buoyancy is affected by their size, gas bubbles with diameters greater than 100 micrometers rise quickly to the liquid surface, whereas ultrafine bubbles rise more slowly due to low buoyancy, allowing them to remain in the liquid for a long time.8,9 For UFBs generation, the ultrasonic application method to induce cavitation is widely used nowadays. When water containing a relatively poorly soluble gas, such as ambient air, is exposed to the ultrasonic field, bubble formation owing to gas cavitation occurs. Once the pressure of the liquid falls below the saturated vapor pressure, cavitation bubbles develop and suspend in the liquid by trapping the dissolved gas in solution.10 This approach is appropriate for producing from small amount to a moderately high concentration of UFBs in water.11,12
UFBs have been utilized in a wide range of applications, functioning as both antibacterial and sterilizing agents while also promoting the growth of plants, animals, microorganisms, and marine life.13–17 In addition, bubbles contribute to the interaction of species within natural or artificial ecosystems during their life cycles.18 As mobile air–liquid interfaces, they play an important role in the detachment and transport of microorganisms, for example, bacteria.19 According to current research, the presence of UFBs alone influences the shape of specific bacteria and thickens their membranes, resulting in reduced bacterial cell proliferation.20 The observation of microbial growth is crucial for the understanding of the behavior of living cells. Cell development as well as metabolism monitoring are critical to study. Several essential approaches, either direct or indirect, used for the prediction of cell growth trend includes the measurements of dry cell weight (biomass), optical density, turbidity, respiration, metabolic rate, and metabolite.21
Nowadays, bacterial expression systems are frequently used to produce recombinant proteins as well as other functional materials.22,23 Numerous variables, including the concentration of nutrients and other environmental conditions, have an impact on bacterial development. The microbial growth rate is shown to be considerably influenced by the composition of the culture media. Some of these reported physical and chemical parameters include temperature, moisture, pH levels, and oxygen levels.22,23 However, another important factor as the interaction of bubbles and bacteria has not yet been examined at its most fundamental level, at which a single bubble–bacterium pair is considered.24 Because gas bubbles are negatively charged in water, the electrostatic repulsion between bubbles and cells is often believed to be stronger than the hydrophobic force. Nevertheless, this ionic interaction is not always dominant. It is hypothesized that the attractions observed between bubbles and hydrophobic surfaces are caused by apolar interactions.25 The bubbles could come into close contact with cells, while the water film between them remained stable without rupturing.25 It is also proposed that any gas utilized to form ultrafine bubbles can affect bacterial growth and metabolic rate.26
Escherichia coli (E. coli), a Gram-negative gamma proteobacterium, is currently the most well-studied organism.27 The bacterium lives primarily in the lower intestinal tracts of warm-blooded animals, including humans. It is frequently released into the environment via feces or wastewater effluent. E. coli in environmental waterways has long been thought to be a sign of recent faecal pollution.28 The bacterium E. coli was chosen as an experimental organism because they grow quickly in chemically defined growth media while remaining dispersed from each other.27 This Gram-negative bacterium is not strictly rod-shaped. Based on examination of numerous published electron micrographs, it lacks an area of consistent diameter. The shape and maximal girth of cells appear to be inconsistent throughout the cell division cycle. Furthermore, the typical width and length of the cells vary based on their nutritional environment.29 According to prior studies, for E. coli cultured in bubble-rich media, the exponential phase's growth rate was higher, and the lag phase's duration was shorter. Significantly, the length of the bacteria increased when they were exposed to bubbles with diameters around 200–300 nm.20
It was also reported that ultrasonic-generated microbubbles (5–20 μm in diameter) were stable for possible use in topical treatments of difficult-to-cure wound infections. Particularly, they are efficient against methicillin-resistant Staphylococcus aureus (S. aureus) skin infections.30 Gram-positive S. aureus bacteria is responsible for a wide range of clinical illnesses. This species is harmful toward the skin and soft tissues, as well as endovascular sites and internal organs. Among both community and hospitals, cases of infections brought on by this bacterium have been observed to be rather frequent.31 Compared to the microbubbles, air UFBs and CO2-UFBs (approximately 100 nm in diameter) obtained growth promotion effects on S. aureus, indicating that variation in bubble sizes can differently influence bacterial growth.32
The types of gas filled in bubbles also play a vital role in affecting bacterial growth since different gases influence differently on the life spans of bacteria. For example, ozone ultrafine bubbles were shown to exhibit potential bactericidal activity against multidrug-resistant bacteria and periodontopathic bacteria in vitro.33 In an investigation on bactericidal effects of CO2, N2 and O2-UFBs, it was demonstrated that only carbon dioxide gas suppressed E. coli growth, whilst the rest (N2 and O2) showed a negligible reduction in E. coli survival rate.34 Other research on UFBs had been performed to specify the effect of internal gases on bacterial growth, however, the experimental results lead to inconsistent conclusions about bacterial enhancement and suppression.35,36 More specifically, the action of UFBs and their encapsulated gas are greatly influenced by various factors such as solution pH or bubble concentration.35 This study focused on the investigation of the growing behavior of bacteria in culture media with and without the presence of ultrafine bubbles. The effects of hydrogen UFBs on bacterial growth including growth rates, biomass, morphology and size will be studied and then can be compared with those of air UFBs. The hydrogen UFBs are hypothesized to exhibit certain effects on the growth of Escherichia coli (E. coli) and Staphylococcus aureus (S. aureus). During the experiments, an appropriate amount of sodium dodecyl sulfate (SDS, 50 μM) was introduced to the culture media in order to stabilize UFBs without causing cell death.
2. Materials and methods
2.1. Materials
The bacterial strains used for experiments were E. coli and S. aureus. LB broth was used as standard culture media (HiMedia). The hydrogen bubble-rich water used for hydrogen UFB culture media preparation was produced by a hydrogen bubble generator (Matsushita TK-HS92). A table-top Ultrasonic cleaner (Derui, DR-MS13) was responsible for UFBs formation in culture media after autoclaving. Bacterial growth curves were constructed using UV-Vis spectrophotometer (JASCO, model V-730), their morphology was observed by an optical microscope. The ultrapure deionized (DI) water used for cleaning and preparing chemicals was obtained by a multi-cartridge purification system (Millipore-Burlington, MA, USA) with a resistivity of at least 18 MΩ cm. All of the culturing techniques were performed under strictly sterilized conditions in a safety cabinet.2.2. Methods
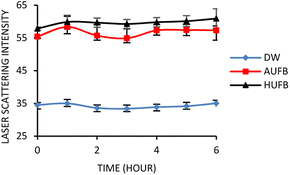
Afterwards, to stabilize the UFBs in bubble-rich culture media, 40 μl of 50 mM sodium dodecyl sulfate (SDS) was added to 40 ml of UFBs-containing media and mixed thoroughly to reach the final SDS concentration of 50 μM. To avoid foam formation, the surfactant was added after the bubble generation stage. The presence of UFBs in bubble-rich media was later confirmed by laser scattering measurements. A CW laser provides a monochromatic light source (beam size 0.5 mm, 5 mW at 532 nm) through a fixed test tube containing the samples. The images of scattered bubbles were captured hourly using a CCD (Venus Engine III) in a darkroom for 6 hours.37
The bacterial growth curves of different treatments were constructed by measuring their optical density at 600 nm using a UV-Vis spectrometer. The growth curves were built hourly for the first 7 hours, then incubated overnight, after 24 hours of incubation, the OD600 value of each species were monitored at the end of their stationary phases.
Additionally, SDS can cause cell death, which reduces the reliability of the OD600 measurement since the dead cells may alter the OD values. To verify that the addition of 50 μM SDS did not negatively impact cell proliferation, the cell counting method was performed. This aimed to confirm that the live cell concentration in UFBs media with SDS did not change significantly in comparison to those without SDS.
Five cultured samples (1 DW, 2 AUFB, 2 HUFB) were prepared following the previously described procedure. Specifically, for the bubble-rich media, two conditions were established: one with SDS and one without SDS. Following a 7 hour incubation period, the bacterial samples were ready for analysis. Subsequently, 20 μl of cell suspension from each sample was diluted with 180 μl of 0.1% methylene blue, creating a 10-fold dilution. After allowing the mixtures to stand for 5 minutes, they were drawn into the hemocytometer's chamber for microscopic cell counting. The live cell concentration was calculated and compared properly to prove that bacterial cells were not damaged by SDS.
3. Results and discussion
3.1. UFBs were generated sufficiently by ultrasonication and well stabilized by 50 μM SDS
Cavitation occurs when the hydrodynamic pressure becomes greater than the vapor pressure of the liquid as they are placed in the ultrasonic field. The liquid creates air bubbles, creating a two-phase system.38 To test for the presence and stability of UFBs in culture media, laser light scattering was employed. The standard errors were calculated based on the average scattering value versus time of three treatments (i.e., normal (DW), air UFB-rich (AUFB) and hydrogen UFB-rich (HUFB) culture media) to indicate the difference in laser intensity measured among them. Between the normal medium and the bubble-rich media, the average value of triplicated light scattering samples was significantly different from each other with no overlap of standard error ranges. Since the AUFB and HUFB were generated over the same amount of time, their obtained scattering values were relatively comparable with overlapping standard error ranges.Fig. 1 illustrates that the scattered light's intensity of bubble-rich samples did not undergo any substantial change over 6 hours of observation. The scattering intensity was shown to be stable with deviations of less than 4% (DW media) and 6% (bubble-rich media) (Table S1†). These results confirm that the presence and stability of UFBs was sufficient to have some influences on bacterial growth during the incubation.
The DW culture media, which did not undergo UFBs generation process, was used as the control sample. In comparison with the normal DW, the AUFB and HUFB culture media showed significantly greater scattering intensity with little change during the observation period, confirming the existence and stability of UFBs.
The term “light scattering” refers to processes occurring in an interaction between light and matters. During light scattering, the incident light is partially absorbed by matters contained in the medium. Then the light is emitted back and deflected in various directions simultaneously with lower intensity.39 In comparison with solid particles, the light scattering effect by bubbles in liquid appears to be more significant due to the influence of their relative refractive index. The gas core of bubbles, either the air or hydrogen gas, which does not have a refractive index deviating too far from unity, exhibits a more crucial scattering effect than diffraction on the incident light.40 In addition, the correlation between particle sizes and the wavelength of the light source is also explained by the Mie scattering theory of homogeneous spherical matters. Mie theory proposes a scattering phenomenon of a particle with a similar or bigger size in comparison to the incident wavelengths,39 which can explain the scattering effect of UFBs (<1 μm in diameter) in exposure to a laser beam (532 nm).
During the autoclave process, the air and hydrogen bubbles that originated in the culture media tend to grow both in size and population. Due to the increasing temperature and pressure, the molecules at the air–water interface become more active and less likely to adhere to the others, which results in possible surface tension increase and bubble burst.41 As the temperature drops, the gas in the presenting bubbles tends to be dissolved into the bulk solution, leading to the decrease in bubble population.42 Therefore, ultrasonication was applied after autoclaving to homogenize the UFBs by means of cavitation. Given that the stability of UFBs in culture media is influenced by temperature, the UFBs during incubation at 37 °C dissolve more slowly than those used for laser scattering measurement at room temperature. Therefore, a small addition of SDS into culture media is doubtlessly critical to ensure the presence of UFBs during bacterial incubation.
3.2. UFBs exhibited certain effects on bacterial populations
From a previous research, the concentration of 50 μM SDS was demonstrated to have no significant effect on S. aureus growth in bubble-rich environment.43 Therefore, the influence of SDS on E. coli growth needs eliminating in order to avoid errors in results due to the co-effect of UFBs and SDS on bacterial growth. Fig. 2 demonstrated that 50 μM SDS had insignificant effects on E. coli growth curves. From Fig. 2, the OD600 value showed that the E. coli growing in the culture media containing 50 μM SDS maintained the normal growth compared to the sample without SDS.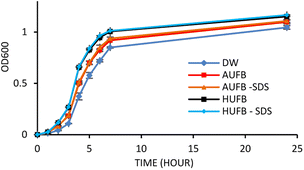 | ||
| Fig. 2 E. coli growth curve with and without the addition of 50 μM SDS into bubble-rich culture media. (Error bars represent the standard errors of samples) | ||
Together with growth curve construction, to mitigate the potential that dead cells caused by SDS may alter OD600 values, a cell counting experiment was conducted to confirm the unsubstantial effect of SDS on bacterial growth. To do that, the cell concentration from culture media (AUFB and HUFB) containing surfactant and those without were tested against one another using the Mann–Whitney U test. The test results are reported in Table S2,† with all tests returning non-significant differences. Therefore, it could be assured that the impact of 50 μM SDS on bacteria is negligible.
The bacterial growth curves of the two species were constructed using optical density measurement. The collected data of each sample was normalized using the data point at 0 hour to calculate the average value and standard error of each measurement point. The growth of S. aureus and E. coli in three culture media (DW, AUFB and HUFB) showed distinct growth rate with no overlapping error bars from the exponential phase. Fig. 3 and 4 indicated that the HUFB culture media best enhanced bacterial growth, followed by the AUFB media, and lastly, normal DW one.
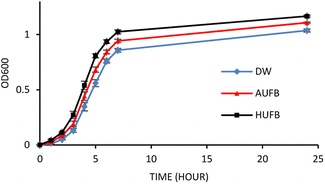 | ||
| Fig. 3 E. coli growth curves in different culture media. (Error bars represent the standard errors of samples) | ||
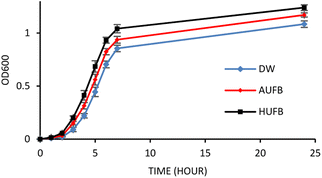 | ||
| Fig. 4 S. aureus growth curves in different culture media. (Error bars represent the standard errors of samples) | ||
The 50 μM-SDS media exhibited no significant effect on E. coli growth, in agreement with the tolerance range of E. coli in exposure to SDS.44 Thus, the growth curves of E. coli and S. aureus were constructed with the contribution of SDS to the bubble-rich culture media. The growth curves of both species shared a similar trend among the three treatments. In particular, E. coli and S. aureus cultured in standard LB media (DW) had identical growth rates to previously reported studies.45,46 However, since their size, shape and division rate are different from each other, the OD600 value of these two strains cannot be compared. Particularly, 1 OD600 of E. coli usually contain 8 × 108 cells per ml while that of S. aureus has 1.5 × 108 cells per ml.47,48 In Fig. 3 and 4, the bacteria underwent the lag phase for the first 3 hours, then entered their exponential phase and reached the stationary phase after 7 hours of cultivation. E. coli and S. aureus exposed to UFBs had significantly higher growth rates than those of DW media. Especially, the HUFB media promoted bacterial growth most efficiently. The difference in growth rates only occurred after the exponential phase was reached since bacteria need adaptation time before multiplying.49 At that time, the UFBs start affecting cell proliferation.
Although having identical growing conditions, the differences in the growing pattern between E. coli and S. aureus are also contributed by the differences in their characteristics. The most important characteristic that distinguishes them is that E. coli is Gram-negative and S. aureus is Gram-positive. Gram-positive bacteria are different from the Gram-negative ones in their cell wall structures, which directly participate in the interaction between bacteria and UFBs. Gram-negative bacteria possess an outer membrane containing lipopolysaccharides, which the Gram-positive bacteria lack. Their peptidoglycan layers are much thicker than that of the Gram-negative bacteria.50 However, the identical growth trend suggested that the influence of UFBs on bacterial cells is likely driven more by their physical interactions rather than biological interactions caused by several bacteria-specific characteristics.
During the UFBs generation, the cavitation UFBs were likely formed evenly throughout the culture media immersed in the ultrasonic field. The UFBs were then coated and stabilized by the surrounding nutrients, which consisted of mostly proteins and their derivatives. Via hydrophobic interaction, the lipophilic residues of proteins concentrated in the gas core while their hydrophilic parts exposed to the liquid media, creating a shield encapsulating UFBs.51 Based on the suspension properties of UFBs, the nutrients were dispersed more thoroughly in the culture media, thus increased solute mass transfer to the bacteria. By releasing digestive enzymes to hydrolyze proteins attached on UFBs, the microbes can better access, absorb the nutrients in bubble-rich and grow better compared to those in the DW media.52
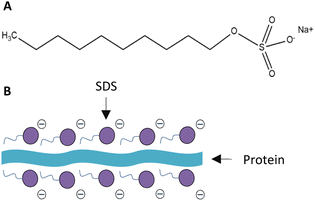
With the contribution of SDS as a surface stabilizer, UFBs were less likely to coalesce; therefore, reducing the risk of bacterial cells being damaged by UFBs bursting. Specifically, the surfactant assembling at the air–water interface can lower the drainage rate of the liquid film and enhance the interfacial characteristics (surface elasticity and surface viscosity) which limit bubble coalescence.53 SDS is a low molecular weight surfactant possesses hydrophilic heads and hydrophobic tails that can form a micelle structure on a particle. Theoretically, when the SDS molecules come into contact with an UFB, their hydrophobic alkyl chains accumulate inside the gas core, and the hydrophilic heads are placed on the UFB's surface, forming a thin film surrounding the UFB and preventing it from coalescence. However, in this study, the surfactant was added after the sonication process, where the protein-UFB adhesion had already developed. Therefore, another mechanism should be proposed to explain the competitive interaction between protein and SDS with UFBs.
As SDS is anionic, it is capable of unfolding proteins with its negatively charged head and forming a surfactant–protein complex. In this study, due to the low SDS concentration (50 μM), this surfactant would mostly adsorb at the air–water interface rather than be solubilized in the bulk. Therefore, only a portion of the protein nutrients accumulating on the UFB's surfaces would interact and form complexes with SDS (Fig. 5). The electrostatic interactions between the anionic surfactant SDS and the counter charged amino acids chain lead to the neutralization of proteins' charge.54 This, in turn, causes the proteins to linearize and lose their three-dimensional structure.55 Owing to this phenomenon, the proteins coupled with SDS become more hydrophobic, thus adsorb more on air–water interfaces.51 However, this effect only occurs at low SDS concentration (10 μM). When the SDS concentration increases up to 70 μM (the SDS concentration used in this study was 50 μM), the hydrophobic interaction will alter the electrostatic force and make the complexes become more hydrophilic.54
Another mechanism could be established regarding the adherence of bacteria to the air–water interface. E. coli and S. aureus, with hydrophobic cell membranes, tend to attach to low-surface energy particles via hydrophobic interaction.56 In aqueous environment, the bacterial cells thus would preferably adhere to the air–water interface of UFBs. By interacting with suspended UFBs, the bacteria could avoid settling at the bottom of their containers, which can potentially prevent them from reaching the soluble nutrients in the culture media.
Between the two types of gas used, the HUFBs promoted a better bacterial growth than the AUFBs for both E. coli and S. aureus. With relatively identical scattering results, bubble concentration was not the determinant of the difference in bacterial growth between AUFB and HUFB culture media. In this case, the internal gas used for UFB generation should be considered. In the HUFB culture media, most of the UFBs were filled with hydrogen gas, which created a uniform bubble-rich environment. In contrast, the air-formed UFBs contain various gas types, each of which may have a unique effect on bacteria. Although the UFB-cell biological interaction was hypothesized to be less significant than their physical interactions, they should still be considered. It is noteworthy that the different types of gas inside the UFBs can affect the biological activities of bacterial cells in different manners. Therefore, bacterial growth would strongly depend on the gas with which they interact. The air can either enhance or suppress bacterial growth inconsistently, while hydrogen has been proven to be a crucial factor in supporting bacterial growth.
Molecular hydrogen (H2) is an indispensable element for aerobic bacteria, which aids their cellular respiration and carbon fixation.57 It was previously stated that hydrogen molecules can be utilized by E. coli as an electron donor for the electron transport chain to facilitate cellular respiration.58 Unfortunately, hydrogen gas only appears in a trace amount in the air although it is important to the long-term survival of bacteria. When bacteria attached to the hydrogen gas–water interface of HUFBs, they would experience similar growth conditions regardless of their binding position on the bubble's surface. As a result, these two facultative anaerobic bacteria in the environment containing HUFBs could grow evenly and optimally. As mentioned earlier, once the coating protein–surfactant complex became less surface-active, the UFBs could not be well stabilized. At this stage, the UFBs might burst and release hydrogen molecules into the surrounding environment, supplying bacteria suspended in the liquid with hydrogen. On the other hand, bubble implosion may also injure bacterial cell membranes within a certain range. In this situation, S. aureus, with its thick peptidoglycan membrane, can withstand physical damage better than E. coli.
In addition, the influence of air on bacterial growth may vary due to the effects of several gas molecules. Particularly, the CO2-UFBs were reported to have a significant bactericidal effect on E. coli.34 It was proposed that carbon dioxide can affect cellular respiration by displacing the oxygen required for bacterial metabolism, hence reducing bacterial growth rates.59 However, the presence of oxygen molecules might activate cell proliferation and metabolism since both E. coli and S. aureus are facultative anaerobes. The energy conservation mechanism of cellular respiration uses oxygen as the terminal electron acceptor to generate the energy required to metabolize nutrients and promote bacterial growth.60 Furthermore, the most abundant gas in the air is nitrogen, which can damage cell growth at high concentration. When the nitrogen concentration is excessive, the oxygen supply may be displaced by nitrogen molecules, causing cell death.61 Based on these impacts, bacteria grown in AUFB culture media cannot maximize their growth rate in comparison to those exposed to HUFBs. Although the growth trends of E. coli and S. aureus were similar, this cannot be inferred that all other species of bacteria would share the same growing behaviors. The interaction between bacterial cells and UFBs is specific for each species; therefore, modifying the concentration, stability, and gas type of UFB or changing bacterial strain would definitely vary the effects of UFBs on bacterial growth.
3.3. Biomass weight verified the reliability of growth curve measurements
E. coli and S. aureus biomass taken from different culture media were obtained and presented in Fig. 6 and 7. The growth curves data and the ascending trend of bacterial biomass going from DW, AUFB to HUFB culture media share the same pattern, which further confirmed the reliability of the growth curves representing the growth rates.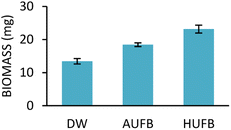 | ||
| Fig. 6 Biomass of E. coli cultured in different culture media. (Error bars represent the standard errors of samples) | ||
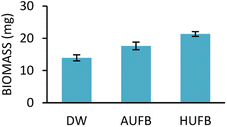 | ||
| Fig. 7 Biomass of S. aureus cultured in different culture media. (Error bars represent the standard errors of samples) | ||
Although the number of viable cells in bubble-rich media was proven to be higher than that of DW media, the OD600 measurement was not sufficiently reliable to confirm that the bacterial populations in the three culture media were substantially different. The insoluble bacterial cells in culture media provided the OD600 value by scattering the incident light beams and preventing them from reaching the detector. Unfortunately, the abundant existence of UFBs in samples might also reduce the detectable light intensity and change the outcome values of UV-Vis measurement by scattering.
To signify the difference in bacterial populations among the culture media, biomass measurement was conducted to eliminate the influence of UFBs on UV-Vis spectroscopy. At the end of the stationary phase, bacterial biomass was collected and weighed. As depicted in Fig. 6 and 7, the biomass from the HUFB medium was the highest for both E. coli and S. aureus, followed by the AUFB and DW media, respectively. These results help verify that bacterial populations follow a descending order as grown in HUFB, AUFB and DW LB media.
3.4. Bacterial morphology changed in exposure to UFBs
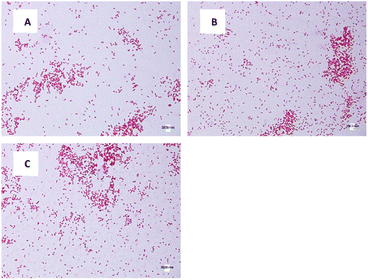
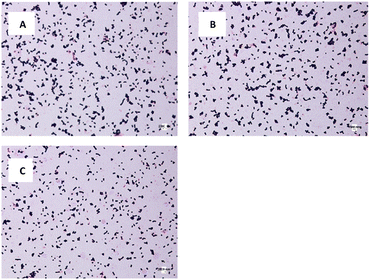
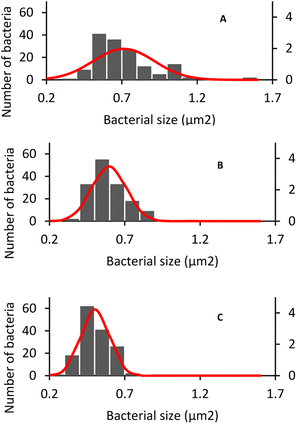
The morphology and size of E. coli and S. aureus were captured using an optical microscope. For E. coli in DW culture media (Fig. 8A), the bacteria possessed a normal morphology, which aligns well with other published data. The bacteria were observed to have a cylindrical body with flat caps, which formed a rod-shaped structure. The average size of DW E. coli was measured to be 0.48 ± 0.15 μm2, consistent with the typical size of E. coli (1.0–2.0 μm long, 0.5 μm radius).62 In AUFB culture media (Fig. 8B), the size average of E. coli decreased to 0.42 ± 0.14 μm2. Their morphology showed no abnormal changes, yet the bacterial size changed due to the reduction in length, according to Fig. 8B.In Fig. 8C, the HUFB culture media affected E. coli's size and shape most significantly in comparison with DW and AUFB culture media. The size area average of E. coli exposed to HUFB was calculated to be 0.35 ± 0.10 μm2. In addition, although still maintaining the rod-shaped morphology, their shape tended to decrease in length and increase in width. Consequently, these E. coli bacteria tends to be shorter but thicker compared with those cultured in DW media.
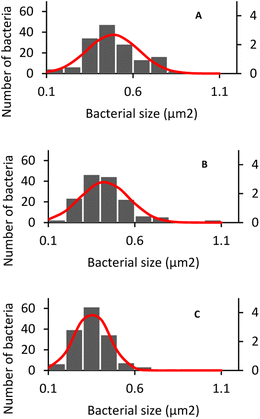
Additionally, the size of HUFB-exposed E. coli was estimated with the highest precision due to the lowest standard deviation (±0.10 μm2) among the three treatments. Fig. 9 showed the histogram of E. coli's average size distribution of the three samples with normal bell curves. It can be seen that the E. coli bacteria from HUFB media have their sizes most concentrated around the mean compared to the other two samples.
The same phenomenon of size change and variation occurred with S. aureus. The average size of S. aureus in DW, AUFB and HUFB was 0.70 ± 0.20 μm2; 0.60 ± 0.11 μm2 and 0.50 ± 0.09 μm2, respectively. In particular, the average size of S. aureus grown in DW media was consistent with their normal radius range.63 S. aureus has a spherical shape, different from the rod shape observed in E. coli. As in Fig. 10, S. aureus from the bubble-rich environments showed no difference in shape compared to those from the DW media. Therefore, the size variation of them only came from the reduction in cell radius. In addition, based on the sample standard deviation, S. aureus in HUFB media also grew in a narrower size range compared to the others (Fig. 11).
To confirm that the E. coli and S. aureus cell sizes of different treatments are statistically different, the t-test was performed to compare the mean bacterial size from each pair of culture media. The test yielded all p-values lower than the significance level (α = 0.05) (Table S3†), implying that the sizes of bacteria in DW, AUFB and HUFB culture media were significantly different. Given that the bacterial biomass increased from DW, AUFB to HUFB media, the growth trend of bacterial size was opposite, from HUFB, AUFB to DW. Therefore, it can be concluded that the difference in bacterial biomass was not because of larger bacteria, but because of their population increase.
The differences in bacterial size and shape in the three culture media for both E. coli and S. aureus were shown in Fig. 8 and 10. The sizes of both species in bubble-rich culture media were observed to be smaller than the originial DW media. According to Fig. 9 and 11, the HUFB media nourished bacterial populations with the smallest size and the most uniform shape. There is no clear mechanism that can explain the varying bacterial size among the treatments, however, their morphology can be affected by several critical factors.
It has been reported that, with an increasing growth rate, an E. coli strain becomes rounder compared to their original rod shape,64 which correlates well to the case of E. coli exposed to UFBs. In addition, with the even distribution of UFBs all around the culture media, it was not necessary for bacteria to move frequently to exploit new sources of nutrients. As bacteria motility decreases, their demand for flagella generation (for E. coli) is reduced.65 Since the resolution of the images from the light microscope was not high enough to distinguish between the bacteria's flagella and their main body, the growth of flagella would contribute to the enlargement of the bacteria; leading to an overall larger size of bacteria cultured in DW media compared to those grown in bubble-rich media. Furthermore, nonmotile bacteria are less probable of aggregating and forming biofilm, which could lower their cell division rate.66,67 Additionally, the motility of bacteria near a surface is observed to be different compared to that of bacteria moving freely in the culture media. Particularly, E. coli would change from forward into circulation movement as they approach a surface.68 If this effect happens frequently, the morphology of bacteria interacting with UFBs may change over time due to their motility alterations.
In the special case where bacteria are exposed to HUFBs, they grew more uniformly in size in comparison with those that are grown in the DW and AUFB culture media. It can be hypothesized that in exposure to hydrogen-dominant UFBs, the bacteria exhibit a consistent growth trend together with a precise morphology among the cells in the population. In contrast, the AUFB and DW culture media possess different gases in the gas core or in the soluble form. These gases can either enlarge or reduce bacterial size depending on their specific properties. The air in AUFBs is composed of several gases; therefore, bacterial growth and size will vary depending on the gas they interact with. It is worth noting that the variation of UFBs size is not a significant factor contributing to the bacterial size change since the AUFBs and HUFBs were generated and stabilized under the same conditions.
Additionally, having a uniform bacterial size can have several benefits. Maintaining a precise range of bacterial size could control the consistency of chromosomes and keep their growth cycles in synchronization.69 For E. coli growth, this trait is crucial because of their common use as a host strain to produce recombinant DNA and proteins.
3.5. Bacterial growth decreased pH values of culture media
The pH values of three media samples were recorded before inoculation and after the incubation terminated. As shown in Fig. 12, the pH value of three samples stayed in the neutral range prior to the inoculation, and the bubble-rich culture media seemed to have no effect on the solutions' pH. However, the pH values of all samples were significantly reduced after a complete bacterial cultivation process. There were no significant differences observed between the pH values of three culture media, hence, it can be considered that the culturing pH changed independently of the influence of UFBs. Therefore, the pH of culture media might have been decreased due to bacterial growth.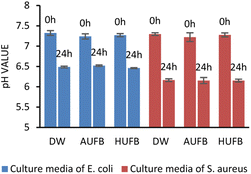 | ||
| Fig. 12 pH values of different culture media before (0 h) and after (24 h) culturing E. coli and S. aureus. (Error bars represent the standard errors of samples.) | ||
In both E. coli and S. aureus growth, the pH values were recorded to be similar after the incubation completed. Thus, this phenomenon should be explained by the influence of bacteria on the pH of the culture media. A study on E. coli and other bacterial growth reported that, based on the bacterial strain, the pH of the surrounding environment can decrease in order to fit the specific optimal pH of each species. With the presence of UFBs, it can be considered that the effects of UFBs on bacterial growth are strongly dependent on the culture media pH because the UFBs do not dominantly control the pH but the bacteria.70 Therefore, the impacts of UFBs on bacteria may vary among different bacterial types.
As shown in Fig. 12, the pH values after 24 hours of cultivation under different treatments tended to concentrate around the theoretical optimal pH range of E. coli (pH 6.5–7.5) and S. aureus (pH 6–7) at room temperature.71,72 In addition, the AUFBs can also modify the pH of the surrounding environment through the dissolution of carbon dioxide (CO2), which could react with water to form the unstable carbonic acid (H2CO3). This weak acid dissociates and releases H+ into the environment, leading to a drop in the pH values. Similarly, the pH of HUFB-rich culture media can be decreased in relation to the H+ concentration in the solution.73
Another mechanism of UFBs promoting bacterial growth in relation to pH was proposed to show the dependence of UFBs behaviors on the surrounding pH. At neutral pH solutions, the UFBs have higher negative zeta potential than in acidic media since they have more OH− concentrating at the air–water interface.74 Therefore, the supplied nutrients with positive counterions can interact and adhere to the UFBs surface, enhancing their distribution inside the culture media and facilitating the bacterial food uptake process.36
4. Conclusion
The experimental data confirmed that ultrasound-induced UFBs were able to intensify bacterial growth rates of both E. coli and S. aureus. The physical interaction between UFBs and bacterial cells is believed to hold the major responsibility for this enhancement. The UFBs were hypothesized to serve as vehicle for transporting nutrients and bacterial cells around the culture media, thus facilitating mass transfer. Furthermore, the adsorption of SDS at the air–water interface is also a crucial factor promoting bacterial growth. This surfactant could form complexes with proteins and compete for adhesion on UFB surfaces, leading to protein unfolding and detachment, thus enhancing bacterial nutrient absorption.Upon comparing AUFB media to the HUFB media, the bacterial growth rates in HUFB cultures were observed to be higher. Due to the contribution of multiple gases in the AUFB media, its promoting effect was not as fully operative as that of HUFBs, which predominantly composed of hydrogen molecules (H2). Additionally, an inverse correlation was observed between the bacterial population and their cell sizes. The smallest but most uniform cell size was obtained in the HUFB media (0.35 ± 0.10 μm2 for E. coli, 0.50 ± 0.09 μm2 for S. aureus). Therefore, this may imply that the HUFBs and AUFBs behaviors were distinct from each other.
However, although the effect of UFBs on bacterial growth has been elucidated, more studies as well as further investigations need to be conducted to shed more light into the interaction at the bubble-cell level and to optimize the impact of UFBs on bacterial growth.
Author contributions
M. V. (investigation, methodology, writing-OD, formal analysis, data curation). N. T. (investigation, data curation, validation, writing-R&E). T. L. (investigation, validation, writing-R&E). A. T. (investigation, writing-R&E). T. L. (investigation, writing-R&E). K. N. (investigation, methodology, formal analysis, data curation, funding acquisition).Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research is funded by Vietnam National Foundation for Science and Technology Development (NAFOSTED) under grant number 106.02.2018.315.References
- S. Maeda, H. Kobayashi, K. Ida, M. Kashiwa, I. Nishihara and T. Fujita, International Conference on Optical Particle Characterization (OPC, 2014), 2014, vol. 9232, pp. 161–165 Search PubMed.
- S. H. Oh, J. G. Han and J.-M. Kim, Fuel, 2015, 158, 399–404 CrossRef CAS.
- K. Yasui, T. Tuziuti and W. Kanematsu, Ultrason. Sonochem., 2018, 48, 259–266 CrossRef CAS PubMed.
- F. Y. Ushikubo, T. Furukawa, R. Nakagawa, M. Enari, Y. Makino, Y. Kawagoe, T. Shiina and S. Oshita, Colloids Surf., A, 2010, 361, 31–37 CrossRef CAS.
- H. Kobayashi, S. Maeda, M. Kashiwa and T. Fujita, Presented in Part at the International Conference on Optical Particle Characterization (OPC 2014), 2014 Search PubMed.
- B. Park, S. Yoon, Y. Choi, J. Jang, S. Park and J. Choi, Pharmaceutics, 2020, 12, 1089 CrossRef CAS PubMed.
- K. Terasaka, K. Yasui, W. Kanematsu and N. Aya, Ultrafine Bubbles, CRC Press, New York, 2021 Search PubMed.
- M. Iijima, K. Yamashita, Y. Hirooka, Y. Ueda, K. Yamane and C. Kamimura, Plant Prod. Sci., 2020, 23, 366–373 CrossRef CAS.
- J. R. Seddon, D. Lohse, W. A. Ducker and V. S. Craig, ChemPhysChem, 2012, 13, 2179–2187 CrossRef CAS PubMed.
- P. Sarkar, G. Ghigliotti, J.-P. Franc and M. Fivel, J. Fluid Struct., 2021, 105, 103327 CrossRef.
- N. Nirmalkar, A. Pacek and M. Barigou, Langmuir, 2019, 35, 2188–2195 CrossRef CAS PubMed.
- K. Yasuda, H. Matsushima and Y. Asakura, Chem. Eng. Sci., 2019, 195, 455–461 CrossRef CAS.
- J.-S. Park and K. Kurata, HortTechnology, 2009, 19, 212–215 Search PubMed.
- K. Ebina, K. Shi, M. Hirao, J. Hashimoto, Y. Kawato, S. Kaneshiro, T. Morimoto, K. Koizumi and H. Yoshikawa, PLoS One, 2013, 8, e65339 CrossRef CAS PubMed.
- F. Kawara, J. Inoue, M. Takenaka, N. Hoshi, A. Masuda, S. Nishiumi, H. Kutsumi, T. Azuma and T. Ohdaira, Digestion, 1958, 90, 10–17 Search PubMed.
- W. Mahakarnchanakul, P. Klintham, S. Tongchitpakdee and W. Chinsirikul, FFTC-KU International Workshop on Risk Management on Ahrochemicals through Novel Technologies for Food Safety in Asia, 2015, pp. 1–19 Search PubMed.
- J. A. Domingos, Q. Huang, H. Liu, H. T. Dong, N. Khongcharoen, P. T. Van, N. H. Nghia, P. T. Giang, P. The Viet and S. St-Hilaire, bioRxiv, 2021, DOI:10.1101/2021.08.27.457885.
- P. L. Walls, J. C. Bird and L. Bourouiba, Am. Zool., 2014, 54, 1014–1025 Search PubMed.
- S. Khodaparast, M. K. Kim, J. E. Silpe and H. A. Stone, Environ. Sci., 2017, 51, 1340–1347 CrossRef CAS PubMed.
- T. Q. Luu, P. N. Hong Truong, K. Zitzmann and K. T. Nguyen, Langmuir, 2019, 35, 13761–13768 CrossRef CAS PubMed.
- G. Najafpour, Biochemical Engineering and Biotechnology, Elsevier, Amsterdam, 2015 Search PubMed.
- M. Winnacker, Soft Matter, 2017, 13, 6672–6677 RSC.
- I. A. Purwasena, D. I. Astuti, M. Syukron, M. Amaniyah and Y. Sugai, J. Petrol. Sci. Eng., 2019, 183, 106383 CrossRef CAS.
- J. Zevnik and M. Dular, Ultrason. Sonochem., 2022, 87, 106053 CrossRef CAS PubMed.
- I. Demir, I. Lüchtefeld, C. Lemen, E. Dague, P. Guiraud, T. Zambelli and C. Formosa-Dague, J. Colloid Interface Sci., 2021, 604, 785–797 CrossRef CAS PubMed.
- P. Sobieszuk, A. Strzyżewska and K. Ulatowski, Chem. Eng. Process, 2021, 159, 108247 CrossRef CAS.
- J. E. Cronan, Escherichia Coli as an Experimental Organism, eLS, USA, 2014 Search PubMed.
- J. Jang, H. G. Hur, M. J. Sadowsky, M. Byappanahalli, T. Yan and S. Ishii, J. Appl. Microbiol., 2017, 123, 570–581 CrossRef CAS PubMed.
- A. L. Koch, Microbiology, 1982, 128, 2527–2539 CAS.
- E. Ronan, N. Edjiu, O. Kroukamp, G. Wolfaardt and R. Karshafian, Ultrasonics, 2016, 69, 182–190 CrossRef CAS PubMed.
- T. J. Foster, Mol. Med. Microbiol., 2002, 839–888 CAS.
- H. Yamada, K. Konishi, K. Shimada, M. Mizutani and T. Kuriyagawa, Int. J. Autom. Technol., 2021, 15, 99–108 CrossRef.
- S. Hayakumo, S. Arakawa, M. Takahashi, K. Kondo, Y. Mano and Y. Izumi, Sci. Technol. Adv. Mater., 2014, 15, 055003 CrossRef CAS PubMed.
- M. Yamaguchi, T. Ma, D. Tadaki, A. Hirano-Iwata, Y. Watanabe, H. Kanetaka, H. Fujimori, E. Takemoto and M. Niwano, Langmuir, 2021, 37, 9883–9891 CrossRef CAS PubMed.
- R. Morishita, S. Itoh and M. Takeda-Morishita, Biocontrol Sci., 2022, 27, 139–142 CrossRef CAS PubMed.
- S. Ogata and Y. Murata, Fluids, 2022, 7, 383 CrossRef CAS.
- R. J. Delahaije, F. J. Lech and P. A. Wierenga, Food Hydrocolloids, 2019, 91, 263–274 CrossRef CAS.
- C. Vargel, Corrosion of Aluminium, Elsevier, Amsterdam, 2020 Search PubMed.
- B. Kruppa and G. Strube, Opt. Meas., 1994, 159–177 Search PubMed.
- X. Han, J. Shen, P. Yin, S. Hu and D. Bi, Opt. Commun., 2014, 316, 198–205 CrossRef CAS.
- T. J. Lin, K. Tsuchiya and L. S. Fan, AIChE J., 1998, 44, 545–560 CrossRef CAS.
- L. K. Wang, N. K. Shammas, W. A. Selke and D. B. Aulenbach, Flotation Technology, 2010, vol. 1249–83 Search PubMed.
- A. Jetten and G. Vogels, Antimicrob. Agents Chemother., 1973, 4, 49–57 CrossRef CAS PubMed.
- L. Dong, H. Liu, L. Meng, M. Xing, T. Lan, M. Gu, N. Zheng, C. Wang, H. Chen and J. Wang, J. Dairy Sci., 2019, 102, 6914–6919 CrossRef CAS PubMed.
- L. E. Amabilis-Sosa, M. Vázquez-López, J. L. G. Rojas, A. Roé-Sosa and G. Moeller-Chávez, Environments, 2018, 5, 47 CrossRef.
- A. A. Tahi, S. Sousa, K. Madani, C. L. Silva and F. A. Miller, Ultrason. Sonochem., 2021, 78, 105743 CrossRef CAS PubMed.
- Y.-C. Chang, C.-Y. Yang, R.-L. Sun, Y.-F. Cheng, W.-C. Kao and P.-C. Yang, Sci. Rep., 2013, 3, 1863 CrossRef PubMed.
- R. A. S. Couto, L. Chen, S. Kuss and R. G. Compton, Analyst, 2018, 143, 4840–4843 RSC.
- T. D. Brock, M. T. Madigan, J. M. Martinko and J. Parker, Brock Biology of Microorganisms, Prentice-Hall, Upper Saddle River (NJ), 2003 Search PubMed.
- T. J. Silhavy, D. Kahne and S. Walker, Cold Spring Harbor Perspect. Biol., 2010, 2, a000414 Search PubMed.
- B. S. Murray, Curr. Opin. Colloid Interface Sci., 2007, 12, 232–241 CrossRef CAS.
- N. E. Diether and B. P. Willing, Microorganisms, 2019, 7, 19 CrossRef CAS PubMed.
- J. Zhou, P. Ranjith and W. Wanniarachchi, Adv. Colloid Interface Sci., 2020, 276, 102104 CrossRef CAS PubMed.
- V. Alahverdjieva, V. Fainerman, E. Aksenenko, M. Leser and R. Miller, Colloids Surf., A, 2008, 317, 610–617 CrossRef CAS.
- D. Winogradoff, S. John and A. Aksimentiev, Nanoscale, 2020, 12, 5422–5434 RSC.
- D. R. Absolom, F. V. Lamberti, Z. Policova, W. Zingg, C. J. van Oss and A. W. Neumann, Appl. Environ. Microbiol., 1983, 46, 90–97 CrossRef CAS PubMed.
- C. Greening, Z. F. Islam and S. K. Bay, Trends Microbiol., 2022, 30, 330–337 CrossRef CAS PubMed.
- M. J. Lukey, A. Parkin, M. M. Roessler, B. J. Murphy, J. Harmer, T. Palmer, F. Sargent and F. A. Armstrong, J. Biol. Chem., 2010, 285, 3928–3938 CrossRef CAS PubMed.
- J. A. Daniels, R. Krishnamurthi and S. S. Rizvi, J. Food Protect., 1985, 48, 532–537 CrossRef CAS PubMed.
- Y. Shan, Y. Lai and A. Yan, Reprogramming Microbial Metabolic Pathways, 2012, pp. 159–179 Search PubMed.
- P. Munsch-Alatossava and T. Alatossava, Front. Microbiol., 2014, 5, 619 Search PubMed.
- S. Baron, Medical Microbiology, University of Texas Medical Branch at Galveston, Galveston (TX), 1996 Search PubMed.
- J. Hudson, in Encyclopedia of Meat Sciences, ed. C. Devine and M. Dikeman, Academic Press, USA, 2004, pp. 820–825 Search PubMed.
- N. Nanninga, Can. J. Microbiol., 1988, 34, 381–389 CrossRef CAS PubMed.
- V. Palma, M. S. Gutiérrez, O. Vargas, R. Parthasarathy and P. Navarrete, Microorganisms, 2022, 10, 563 CrossRef CAS PubMed.
- M. K. Porter, A. P. Steinberg and R. F. Ismagilov, Soft Matter, 2019, 15, 7071–7079 RSC.
- R. M. Donlan and J. W. Costerton, Clin. Microbiol. Rev., 2002, 15, 167–193 CrossRef CAS PubMed.
- K. D. Young, Microbiol. Mol. Biol. Rev., 2006, 70, 660–703 CrossRef PubMed.
- J. Errington, R. A. Daniel and D.-J. Scheffers, Microbiol. Mol. Biol. Rev., 2003, 67, 52–65 CrossRef CAS PubMed.
- R. Sánchez-Clemente, M. I. Igeño, A. G. Población, M. I. Guijo, F. Merchán and R. Blasco, 2018, 2, p. 1297.
- K. Davey, Int. J. Food Microbiol., 1994, 23, 295–303 CrossRef CAS PubMed.
- C. Stewart, Foodborne Microorganisms of Public Health Significance, 2003, pp. 359–379 Search PubMed.
- A. J. Garcia III and J.-M. Ramirez, Elife, 2017, 6, e27563 CrossRef PubMed.
- J. N. Meegoda, S. Aluthgun Hewage and J. H. Batagoda, Environ. Eng. Sci., 2018, 35, 1216–1227 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra07454d |
| This journal is © The Royal Society of Chemistry 2024 |