Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Turn-on fluorescent nanoprobe for ATP detection based on DNA-templated silver nanoclusters†
Yuxia Li,
Zeting Meng,
Yating Liu and
Baozhu Zhang *
*
Department of Chemistry and Chemical Engineering, Jinzhong University, Yuci 030619, P. R. China. E-mail: zhangbaozhu518@126.com
First published on 13th February 2024
Abstract
A turn-on fluorescence nanoprobe was constructed for the determination of adenosine 5′-triphosphate (ATP) based on DNA-templated silver nanoclusters (DNA-AgNCs). The significant enhancement fluorescence intensity of DNA-AgNCs in the presence of ATP is due to the high special binding affinity between ATP and the aptamer, resulting in the environment of DNA-AgNCs with darkish fluorescence lying at one terminus of DNA slightly altering owing to the change of ATP aptamer conformation. A good linear range runs from 9 to 24 mM with a satisfactory detection limit of 3 μM. Furthermore, the proposed nanoprobe exhibited good performance for ATP detection in diluted fetal bovine serum.
1 Introduction
Fluorescent metallic nanoclusters (e.g. AgNCs, AuNCs, and CuNCs), as ultra-small particles composed of several to about a hundred atoms, since they have sizes of less than 2 nm, the Fermi wavelength of electrons, but have properties like molecules, such as discrete electronic transitions and strong fluorescence,1 have gradually become popular fluorescence probes due to their good water dispersibility, size adjustability and unique optical properties.2–5 In particular, oligonucleotide-templated silver nanoclusters (DNA-AgNCs) possess more advantages, such as great biocompatibility, high photostability, hypotoxicity, ease of synthesis, adjustable light emission wavelength and large Stokes shift, than fluorophores or organic quantum dots.6–8 Therefore, they have been widely utilized to probe various analytes, including metal ions,9,10 enzymes,11–13 DNA,14–16 and biomolecules, in fluorescent biosensors.17,18ATP, as a primary energy storage molecule, plays a key role in regulating biochemical pathways and intra-cellular metabolism in diverse tissues and organs19 and provides energy for various types of biochemical reaction in most living organisms.20 In addition, it has been used as a biomarker for some diseases, such as Alzheimer's disease, hypoxia,21 ischemia,22 Parkinson's disease,23 hypoglycemia,24 and some malignant tumors.25 Thus, in the past few decades, a lot of detection methods for ATP have been reported, such as fluorescence analysis,26 colorimetric methods,27 liquid chromatography,28,29 and electrochemical analysis.30 However, these methods inevitably suffer from time-consuming and laborious modification of electrodes and fluorescent probes and/or preparation of complex samples. In recent years, fluorescence detection methods based on distyrylbenzene (1) with two arms of dipicolylaminomethyl groups at the central benzene ring and its zinc complex (1-Zn),26 rhodamine derivatives,31 and labeled aptamer and magnetic nanoparticles32 have been applied to analyze ATP with good sensitivity and selectivity. However, the first two types of probes require complex organic synthesis, while the latter require 6-carboxyflurrescein labelling on the aptamer. These operations are not only complex but may also affect the detection effect. In addition, the biocompatibility of organic probes is poor. Hence, the development of a simple and sensitive detection strategy for ATP is urgently needed for biochemical studies, food hygiene applications and clinical diagnosis.
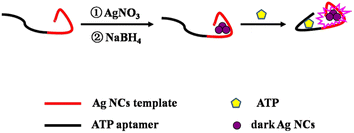
Inspired by the significant increase in fluorescence when darkish DNA-AgNCs are placed together utilizing complementary linkers,33 a series of probes for the detection of ATP were constructed by our team based on this principle. These methods based on DNA-Ag NCs34,35 and DNA-Cu/Ag NCs36 have good sensitivity and selectivity and simple operation. However, LODs are not satisfactory at present. Hence, a novel strategy was constructed for the quantitative detection of ATP based on DNA-Ag NCs with an ATP aptamer, whose DNA template was designed based on a thesis from Zhang's team.34 Namely, two bases (TT or AA) are removed from the bases “TTTTT” or “AAAAA” of the DNA template34 so that they connect nucleation sequences and adapter sequences, respectively. Consequently, the LOD of the proposed probe becomes lower than that obtained without removing connecting base sequences. As shown in Scheme 1, the template contains an ATP aptamer lying on the left and an AgNC-nucleation sequence on the right. Darkish AgNCs on the right become significantly brighter in the presence of ATP owing to the conformational alteration of the ATP aptamer, leading to a change in the environment of AgNCs. Furthermore, a feasibility experiment was performed. As shown in ESI Fig. S1,† the fluorescence was obviously enhanced in the presence of 24 mM ATP. Thus, the proposed nanoprobe is not only simple, convenient and sensitive but also a green ATP detection assay.
2 Experimental section
2.1 Reagents and apparatus
Oligonucleotides, ATP, guanosine 3′-triphosphate (GTP), cytidine 3′-triphosphate (CTP), and uridine 3′-triphosphate (UTP) used in this study were purchased from Sangon Biotechnology Inc. (Shanghai, China), and the names and sequences of the DNA are listed in ESI Table S1.† Fetal bovine serum was provided by Yuanye Biotechnology Co. Ltd (Shanghai, China). Cu(NO3)2, Fe(NO3)2, Fe(NO3)3, Co(NO3)2, Mg(NO3)2, Ca(NO3)2, glucose (Glu), L-histidine (L-His), sodium borohydride (NaBH4, 98%) and silver nitrate (AgNO3, 99.8%) were obtained from Aladdin Bio-chem Technology Co. Ltd (Shanghai, China). All chemical reagents were of analytical grade and used without further purification. Phosphate buffer solution (PBS, 20 mM, pH 7.0) was used in all experiments. All solutions were prepared using Milli-Q water (18.2 MΩ cm).Fluorescence measurements were recorded with an Edinburgh Instruments FS5 fluorescence spectrophotometer at room temperature, and slit widths were 2.0 nm and 4.0 nm for excitation and emission, respectively. The morphologies and average sizes of AgNCs were studied using a JEOL JEM-2100 transmission electron microscope with an acceleration voltage of 200 kV. Time-resolved fluorescence measurements were performed using an FS5 fluorescent lifetime spectrometer (Edinburgh Instruments, Livingston, UK) operating in the time-correlated single photon counting (TCSPC) mode using a semiconductor laser (405 nm) as the excitation source. Commercial software from Edinburgh Instruments was used for data analyses. When 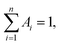 the average excited state lifetime is expressed by the equation
the average excited state lifetime is expressed by the equation 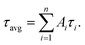 The reported spectrum of each sample represents the average of three scans. X-ray photoelectron spectroscopy (XPS) (ESCALAB 220i-XL, VG Scientific, England) was performed using monochromic Al Kα as the source at 1486.6 eV.
The reported spectrum of each sample represents the average of three scans. X-ray photoelectron spectroscopy (XPS) (ESCALAB 220i-XL, VG Scientific, England) was performed using monochromic Al Kα as the source at 1486.6 eV.
2.2 Synthesis of Ag NCs
DNA-AgNCs were synthesized according to the procedure previously reported in the literature.36 Briefly, Ag NO3 (18 μM) and DNA (3.0 μM) were sequentially added to PBS solution (20 mM, pH 7) under uniform stirring. The mixture was incubated in the dark at 4 °C for 20 min, followed by the addition of fresh NaBH4 (18 μM) and stirred for 1 min; then, the mixture was kept away from light for 1 h at 4 °C. After 1 h, the reduced DNA-AgNC solution was stored at 4 °C for the ATP assay.2.3 Detection of ATP
ATP (0–39 mM) of different concentrations were sequentially added into the abovementioned DNA-AgNC solution. The fluorescence spectrum measurements were performed at room temperature. For the assay of the specificity of the proposed probe, ATP, and ATP analogues, including GTP, UTP, and CTP, were measured under same conditions as ATP. Additionally, other interfering agents, including Cu2+, Fe2+, Fe3+, Co2+, Mg2+, Ca2+, Glu and L-His, were investigated.2.4 Application of the proposed nanoprobe
To further investigate the practicability of the proposed nanoprobe, ATP in fetal bovine serum solution which was diluted 100 times was analyzed. The samples were spiked with ATP of varying different concentrations and measured by employing the same method as for ATP detection.2.5 Method of quantum yield measurement and calculation
The measurement of absolute photoluminescence quantum yield (APLQY) in this study used the SC-30 Integrating Sphere Module of an FS5 Sample Module, diffuse reflectance spectra and absorbance spectra based on diffuse reflectance. The absolute quantum yield, η, is the ratio of the number of photons emitted to the number of photons absorbed: η = Nem/Nabs. It was calculated using “direct excitation” measurements where one records the scatter and emission of a sample being directly excited by the radiation from the excitation monochromator alone.3 Results and discussion
3.1 Optical characterization of DNA-Ag NCs
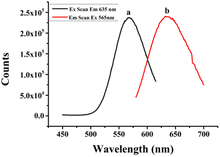
It has been reported that the optical properties of AgNCs are determined by different DNA templates, which depend on the characteristic of their base sequences and secondary structures.37,38 Therefore, templates of DNA named (L)BT3A3, BT3A3 and BT3A3(R) were designed. ESI Table S1† lists their sequences. As shown in ESI Table S1,† the aptamer of ATP is located in the middle part (italic) of the DNA, while the C-rich sequence stabilized AgNCs lie at 5′ and/or 3′ ends (bold). BT3A3(R) and (L)BT3A3 come from BT3A3, from which right (or left) C-rich segments and AAA (or TTT) linkers (underlined) of BT3A3 have been removed. Fig. 1 shows the optical characterization of (L)BT3A3-AgNCs. Excitation (curve a) and emission (curve b) peaks are at 565 and 635 nm, respectively. Similarly, the excitation and emission spectra of BT3A3 and BT3A3(R)-AgNCs were also studied, and it can be seen from ESI Fig. S2A and B† that excitation and emission wavelengths are 560 and 640 nm for BT3A3-Ag NCs and 565 and 650 nm for BT3A53(R)-AgNCs. In addition, the UV-Vis absorption spectra of (L)BT3A3-AgNCs were collected for different concentrations of ATP. Two peaks appear in every curve of ESI Fig. S3† and 430 nm peaks and 565 nm peaks should be attributed to the characteristic plasmon absorption band of Ag nanoparticles and the absorption band of AgNCs, respectively.35 Therefore, Ag nanoparticles exist in (L)BT3A3-AgNCs. The absorption gradually increases with increasing ATP concentration, demonstrating the amount of enhancement of (L)BT3A3-AgNCs.In addition, the images of the probe before and after exposure to UV light (λmax = 365 nm) were taken. As shown in ESI Fig. S4A,† it looks colourless (sample 0 in ESI Fig. S4A†) before exposure to UV light and looks light red (sample 1 in ESI Fig. S4A†) after exposure to UV light. This indicates the emission of red light under irradiation by UV light. Similarly, the images of the probe were measured before and after the addition of 10 mM ATP under illumination by UV light. As shown in ESI Fig. S4B,† the images of the probe upon the addition of 10 mM ATP show light red (sample 1 in ESI Fig. S4B†), while others with 0 mM ATP look colourless (sample 0 in ESI Fig. S4B†). Hence, the color of the probe changed after adding ATP, indicating that ATP specifically binds to its adapter.
The fluorescence intensity of each DNA-AgNC against storage time was detected since the stability of the probe has a great effect on its detection performance. As shown in ESI Fig. S5,† the fluorescence intensity of (L)BT3A3-AgNCs reaches a plateau after 3 h, then remains almost unchanged for 1.5 h. However, the fluorescence intensity of BT3A3-AgNCs declines with storage time. The fluorescence intensity of BT3A3(R)-AgNCs gradually increases, then declines. The changes in fluorescence intensity of (L)BT3A3-AgNCs and BT3A3(R)-AgNCs are similar, as their nucleation sequences are located at one end of the DNA template. The changes in fluorescence intensity of BT3A3-AgNCs are different from them, as its nucleation sequence is located at both ends of the DNA template. It can be seen that the stability of (L)BT3A3-AgNCs is better than those of others.
The quantum yields (QY) of (L)BT3A3-AgNC, BT3A3-AgNC, and BT3A3(R)-AgNC probes were measured to evaluate the ability of these probes to emit fluorescence. As displayed in ESI Fig. S6,† they are 28.28%, 43.07%, and 29.48%, respectively. Apparently, the highest quantum yield of BT3A3-AgNCs is consistent with its two rich-C sequences, and the quantum yields of (L)BT3A3-AgNCs and BT3A3 (R)-AgNCs are similar, owing to them only having a rich-C sequence.
3.2 Characteristics of (L)BT3A3-Ag NCs
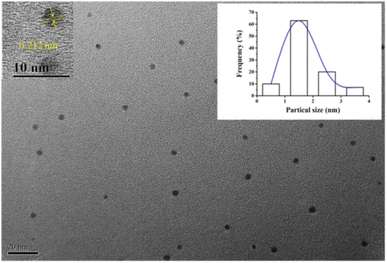
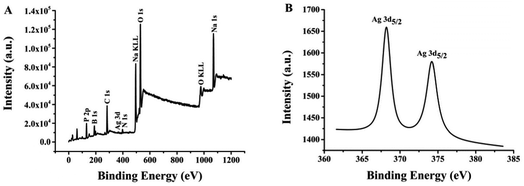
The morphologies and size of (L)BT3A3-AgNCs were studied using transmission electron microscopy. (L)BT3A3-AgNCs are distributed uniformly in Fig. 2. The average diameter of about 2 nm for proposed DNA-AgNCs is in accordance with the size of metal NCs being less than 2 nm (ref. 39) and was obtained from a histogram fitted using the Lorentzian function. Additionally, HRTEM experiments were carried out. The obvious crystal lattice structures of AgNCs are presented in the inset of Fig. 2, which reveals their high crystallinities, where the lattice spacing is 0.212 nm.Ag 3d spectra and XPS wide scan survey spectra were performed to confirm the elemental and valence states of Ag element in (L)BT3A3-AgNCs. Nine peaks in Fig. 3A corroborate the presence of P, B, C, Ag, N, Na and O elements. As shown in Fig. 3B, 2 peaks appear in the expanded spectrum of Ag 3d, and the binding energies of 368.2 eV for Ag 3d5/2 and 374.2 eV for Ag 3d3/2 could be assigned to Ag(1) and Ag(0) in (L)BT3A3-Ag NCs,40,41 respectively. Hence, (L)BT3A3-Ag NCs are compounds of Ag(1) and Ag(0).
3.3 Optimization of experimental conditions
3.4 Assay of ATP
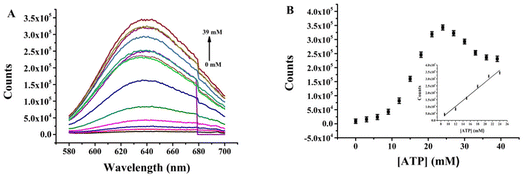
Quantitative determination of ATP exploiting the proposed nanoprobe was performed under optimal conditions. The fluorescence intensity of (L)BT3A3-AgNCs was recorded with different concentrations of ATP. Fig. 4A and B depict that the fluorescence intensity continues to increase when ATP concentration increases, with a good linear relationship within the range of 9–24 mM (R2 = 0.9755, where the linear regression equation is F = −162507.3 + 21888.6CATP) and a limit of detection (LOD) of 3.0 μM, which was calculated according to 3σ0/k (where the value of σ0 is obtained from the standard deviation of the background, and k represents the slope of the calibration line). ESI Table S2† shows that the LOD of the proposed nanoprobe is superior to that of previous sensors for ATP.35,36,42,43 Therefore, the proposed nanoprobe has high sensitivity.To investigate the quenching mechanism of fluorescence in (L)BT3A3-AgNCs, the lifetimes of (L)BT3A3-AgNCs were studied at different concentrations of ATP with an emission wavelength of 635 nm (ESI Fig. S11†). The fluorescence transients of (L)BT3A3-AgNCs exhibit haploid exponential time constants (ESI Table S3†) with an average lifetime of 3.26 ns. The lifetimes of (L)BT3A3-AgNCs hardly change as the ATP concentration increases, therefore, showing that the interaction between (L)BT3A3-AgNCs and ATP is a static process.
3.5 Selectivity of the nanoprobe toward ATP
Selectivity is one of the most important parameters for a fluorescence probe with good properties therefore, ATP and other nucleotide analogues, including CTP, UTP and GTP, were investigated at concentrations of 21 mM. As shown in Fig. 5, relative fluorescences (F/F0) are 51.4, 27.9, 8.73, and 15.9 in the presence of ATP, CTP, GTP, and UTP. Therefore, the relative fluorescence in the presence of ATP increases to a maximum, due to the high specific binding ability of ATP and its aptamers.44 While other nucleotide analogs cause a certain degree of increase, and F/F0 even reaches 27.9 for CTP. Conversely, the fluorescence response of the proposed probe towards ATP was monitored in the presence of interfering analytes, including CTP, UTP, and GTP, as shown in ESI Fig. S12.† The black bars represent the relative fluorescent intensity (F/F0) of (L)BT3A3-Ag NCs in the presence 10 mM ATP, and the red bars represent F/F0 of (L)BT3A3-AgNCs when 10 mM ATP and 5 mM ATP, UTP, CTP, and GTP. It exhibits significant fluorescence enhancement towards ATP. Although the fluorescence also increases a little towards other interference analytes, it is not obvious. CTP increases more than UTP and GTP, which is consistent with above-mentioned experimental results. Therefore, the selectivity of the proposed probe needs future improvement for the assay of ATP. In addition, other interfering agents, including Cu2+, Fe2+, Fe3+, Co2+, Mg2+, Ca2+, Glu and L-His, were investigated for the evaluation of the specificity of the proposed nanoprobe. As shown in ESI Fig. S13,† Cu2+, Fe2+, Fe3+ and Co2+ hardly cause any change in the relative fluorescent intensity (F/F0). F/F0 for Mg2+ and Ca2+ are less than 1 and for Glu and L-His are not more than 0.4. The transitional metallic elements, Cu2+, Fe2+, Fe3+ and Co2+, can quench AgNCs through energy transfer or electron transfer,45 while the influence on AgNCs of Mg2+, Ca2+, Glu and L-His can be neglected.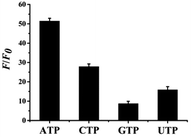 | ||
| Fig. 5 Selectivity of the ATP detection system. The concentrations of ATP, CTP, UTP and GTP are 21 mM, respectively. Error bars represent the standard deviation of three repeated experiments. | ||
3.6 Application of the proposed nanoprobe
To evaluate the practicability of the proposed nanoprobe, (L)BT3A3-AgNCs were exploited to determine ATP spiked in a 100-fold diluted fetal bovine serum. As presented in Table 1, recoveries used this strategy are between 99.2% and 104.3%. Moreover, relative standard deviations are in the range of 1.09 and 1.92, which are comparable with the value previously reported.46,47 Therefore, results demonstrate that the nanoprobe not only has reliability but also good precision for determining ATP in fetal bovine serum.4 Conclusions
In conclusion, a turn-on fluorescence nanoprobe has been developed for ATP detection based on DNA-AgNCs. It has a lot of advantages, including simplicity, sensitivity, specificity, hypotoxicity, low cost, and the avoidance of chemical modification. Furthermore, this proposed nanoprobe succeeded in determining ATP in diluted fetal bovine serum. Therefore, the proposed probe has great potential in clinical diagnosis and biological fields.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Important Research and Development Plan of Shanxi Province (201803D121036), the Startup Foundation of Doctors of Jinzhong University (23E00345), the National Innovation Training Program for College Students of China (202310121017), and the Innovation Team Construction Plan of “1331 Project” of Jinzhong University (jzxycxtd2019010).Notes and references
- L. Liu, Q. Zhang, F. Li, M. Wang, J. Sun and S. Zhu, Spectrochim. Acta, Part A, 2021, 253, 119584–119591 CrossRef CAS PubMed.
- L. M. Peng, Y. C. Bian, X. Q. Shen, H. C. Yao, H. J. Chen and Z. J. Li, Chin. Chem. Lett., 2020, 31, 2871–2875 CrossRef CAS.
- S. Naaz, S. Poddar, S. P. Bayen, M. K. Mondal, D. Roy, S. K. Mondal, P. Chowdhury and S. K. Saha, Sens. Actuators, B, 2018, 255, 332–340 CrossRef CAS.
- S. Naaz and P. Chowdhury, Sens. Actuators, B, 2017, 241, 840–848 CrossRef CAS.
- P. Chowdhury, A. Hazra, M. Kr, B. Mondal, D. Roy, S. P. Roy and S. Pal Bayen, J. Macromol. Sci., Part A: Pure Appl.Chem., 2019, 56, 773–780 CrossRef CAS.
- D. Lyu, J. Li, X. Wang, W. Guo and E. Wang, Anal. Chem., 2019, 91, 2050–2057 CrossRef CAS PubMed.
- N. Enkin, E. Sharon, E. Golub and I. Willner, Nano Lett., 2014, 14, 4918–4922 CrossRef CAS PubMed.
- K. S. Park and H. G. Park, Curr. Opin. Biotechnol., 2014, 28, 17–24 CrossRef CAS PubMed.
- I. M. Khan, S. Zhao, S. Niazi, A. Mohsin, M. Shoaib, N. Duan, S. J. Wu and Z. P. Wang, Sens. Actuators, B, 2018, 277, 328–335 CrossRef CAS.
- J. L. Maclean, K. Morishita and J. W. Liu, Biosens. Bioelectron., 2013, 48, 82–86 CrossRef CAS PubMed.
- J. L. Li, J. H. Ma, Y. C. Zhang, Z. L. Zhang and G. W. He, Microchim. Acta, 2019, 186, 48–54 CrossRef PubMed.
- S. Y. Zhu, X. E. Zhao, W. Zhang, Z. Y. Liu, W. J. Qi, S. Anjuma and G. B. Xu, Anal. Chim. Acta, 2013, 786, 111–115 CrossRef CAS PubMed.
- L. Y. Guo, D. L. Chen and M. H. Yang, Microchim. Acta, 2017, 184, 2165–2170 CrossRef CAS.
- Y. Yuan, Y. Y. Ma, L. Luo, Q. Wang, J. Huang, J. B. Liu, X. H. Yang and K. M. Wang, Microchim. Acta, 2019, 186, 613–618 CrossRef PubMed.
- L. Ge, X. M. Sun, Q. Hong and F. Li, ACS Appl. Mater. Interfaces, 2017, 9, 13102–13110 CrossRef CAS PubMed.
- L. Ge, X. M. Sun, Q. Hong and F. Li, ACS Appl. Mater. Interfaces, 2017, 9, 32089–32096 CrossRef CAS PubMed.
- Y. Qin, M. Daniyal, W. M. Wang, Y. Q. Jian, W. Yang, Y. X. Qin, C. Y. Tong, W. Wang and B. Liu, Sens. Actuators, B, 2019, 291, 485–492 CrossRef CAS.
- J. Y. Chen, X. H. Ji, P. Tinnefeld and Z. K. He, ACS Appl. Mater. Interfaces, 2016, 8, 1786–1794 CrossRef CAS PubMed.
- S.-J. Chen, Y.-F. Huang, C.-C. Huang, K.-H. Lee, Z.-H. Lin and H. T. Chang, Biosens. Bioelectron., 2008, 23, 1749–1753 CrossRef CAS PubMed.
- T. Tatsumi, J. Shiraishi, N. Keira, K. Akashi, A. Mano, S. Yamanaka, S. Matoba, S. Fushiki, H. Fliss and L. Nakagawa, Cardiovasc. Res., 2003, 59, 428–440 CrossRef CAS PubMed.
- D. Janssens, C. Michiels, E. Delaive, F. Eliaers and K. Drieu, J. Remacle, 1995, 50, 991–999 CAS.
- K. K. Arora and P. L. Pedersen, J. Biol. Chem., 1988, 263, 17422–17428 CrossRef CAS PubMed.
- J. D. Weille and M. Lazdunski, Proc. Natl. Acad. Sci., 1988, 85, 1312–1316 CrossRef PubMed.
- I. H. Chaudry, M. G. Clemens, M. Ohkawa, S. Schleck and A. E. Baue, Adv. Shock Res., 1982, 8, 177–186 CAS.
- F. Li, Z. Du, L. Yang and B. Tang, Biosens. Bioelectron., 2013, 41, 907–910 CrossRef CAS PubMed.
- J. Motoyoshiya, J. Y. Wada, K. Itoh, K. Wakabayashi, T. Maruyama, K. Ono, K. Fukasawa, T. Fujimoto, Y. Akaiwa and E. Nonaka, Spectrochim. Acta, Part A, 2018, 195, 223–229 CrossRef CAS PubMed.
- Z. Q. Gao, Z. L. Qiu, M. H. Lu, J. Shu and D. P. Tang, Biosens. Bioelectron., 2016, 89, 1006–1012 CrossRef PubMed.
- J. C. M. Coolen, C. W. Arts, L. R. Swennen, A. Bast, A. C. Stuart and C. Dagnelie, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2008, 864, 43–86451 CrossRef PubMed.
- D. R. Webster, G. D. Boston and D. M. Paton, J. Pharmacol. Methods, 1985, 13, 339–350 CrossRef CAS PubMed.
- X. Li, J. M. Yang, J. Q. Xie, B. Y. Jiang, R. Yuan and Y. Xiang, Biosens. Bioelectron., 2018, 102, 296–300 CrossRef CAS PubMed.
- Y. F. Liu, D. Lee, D. Wu, K. M. K. Swamy and J. Yoon, Sens. Actuators, B, 2018, 265, 429–434 CrossRef CAS.
- X. J. Liu, B. X. Lin, Y. Yu, Y. J. Cao and M. L. Guo, Microchim. Acta, 2018, 185, 243–250 CrossRef PubMed.
- B.-C. Yin, J.-L. Ma, H.-N. Le, S. Wang, Z. Xu and B.-C. Ye, Chem. Commun., 2014, 50, 15991–15994 RSC.
- B. Z. Zhang, Z. Y. Yang, Y. X. Li, L. Ma, F. F. Li, X. Q. Lv and G. M. wen, RSC Adv., 2022, 12, 30024–30029 RSC.
- J. Xu and C. Wei, Biosens. Bioelectron., 2017, 87, 422–427 CrossRef CAS PubMed.
- B. Z. Zhang and C. Y. Wei, Anal. Bioanal. Chem., 2020, 412, 2529–2536 CrossRef CAS PubMed.
- E. G. Gwinn, P. O'Neill, A. J. Guerrero, D. Bouwmeester and D. K. Fygenson, Adv. Mater., 2008, 20, 279–283 CrossRef CAS.
- K. Loo, N. Degtyareva, J. Park, B. Sengupta, M. Reddish, C. C. Rogers, A. Bryant and J. T. Petty, J. Phys. Chem. B, 2010, 114, 4320–4326 CrossRef CAS PubMed.
- X.-R. Song, N. Goswami, H.-H. Yang and J. Xie, Analyst, 2016, 141, 3126–3140 RSC.
- K. R. Krishnadas, A. Ghosh, A. Baksi, I. Chakraborty, G. Natarajan and T. Pradeep, J. Am. Chem. Soc., 2016, 138, 140–148 CrossRef CAS PubMed.
- Q. Zhou, Y. Lin, M. Xu, Z. Gao, H. Yang and D. Tang, Anal. Chem., 2016, 88, 8886–8892 CrossRef CAS PubMed.
- L. Lu, Y. Qian, L. Wang, K. Ma and Y. Zhang, ACS Appl. Mater. Interfaces, 2014, 6, 1944–1950 CrossRef CAS PubMed.
- J.-L. Ma, B.-C. Yin and B.-C. Ye, Analyst, 2016, 141, 1301–1306 RSC.
- C.-H. Lu, F. Wang and I. Willner, Chem. Sci., 2012, 3, 2616–2622 RSC.
- X. Y. Zheng, T. M. Yao, Y. Zhu and S. Shi, Biosens. Bioelectron., 2015, 66, 103–108 CrossRef CAS PubMed.
- Q.-W. Song, R.-H. Wang, F.-F. Sun, H.-K. Chen, Z.-M.-K. Wang, N. Na and O.-Y. Jin, Biosens. Bioelectron., 2017, 87, 760–763 CrossRef CAS PubMed.
- Y. Zhu, X.-C. Hu, S. Shi, R.-R. Gao, H.-L. Huang, Y.-Y. Zhu, X.-Y. Lv and T.-M. Yao, Biosens. Bioelectron., 2016, 79, 205–212 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra07077h |
| This journal is © The Royal Society of Chemistry 2024 |