Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The N-biphenyldihydroisoquinolinium scaffold as a novel motif for selective fluorimetric detection of quadruplex DNA†‡
Denisa
Soost
 a,
Gerhard
Bringmann
a,
Gerhard
Bringmann
 b and
Heiko
Ihmels
b and
Heiko
Ihmels
 *a
*a
aDepartment of Chemistry – Biology, and Center of Micro- and Nanochemistry and (Bio-)Technology (Cμ), University of Siegen, Adolf-Reichwein-Str. 2, 57068 Siegen, Germany. E-mail: ihmels@uni-siegen.de
bInstitute of Organic Chemistry, University of Würzburg, Am Hubland, 97074 Würzburg, Germany
First published on 28th August 2024
Abstract
The spectroscopic investigation of the DNA-binding properties of (S)-6,8-dimethoxy-2-(4′-methoxy-[1,1′-biphenyl]-4-yl)-1,3-dimethyl-3,4-dihydroisoquinolin-2-ium revealed the cationic donor–acceptor-substituted biaryl unit as a promising motif for selective fluorimetric detection of particular quadruplex DNA (G4-DNA) forms. The title compound exhibits the characteristic solvent-dependent dual emission of biaryl fluorophores; however, the emission quantum yields are very low (Φfl ≤ 0.01) because of radiationless deactivation of the excited state by conformational changes. In contrast, the emission intensity of the biaryl derivative increases by a factor of 2–10 on association with G4-DNA (KB = 8 × 104 M−1–4 × 105 M−1), because the structural relaxation of the exited ligand is suppressed in the binding site. The signal pattern of the dual emission, specifically the relative contribution of each band, varies with the different G4-quadruplex forms 22AG, c-kit, c-myc, c-kras, and h-ras1, most likely caused by the different dynamic flexbility of the ligand in the distinct binding sites. These effects enable the fluorimetric identification of sterically constrained binding sites, such as in c-kit and h-ras1, even with the naked eye.
Introduction
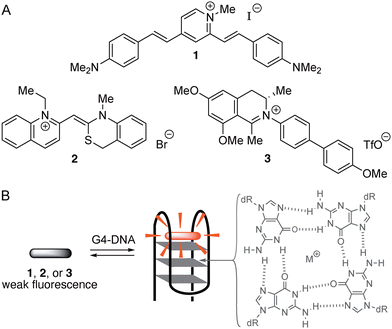
The selective staining and detection of nucleic acids is an important bioanalytical technique in chemistry and biology.1 For this purpose, organic fluorescent probes figure as valuable and versatile tools.2 Specifically, dyes with a very low emission intensity that increases upon association with the target analyte, often referred to as fluorescent “light-up probes”, have proven to be very useful and provide valuable chemosensors for the fluorimetric detection of DNA and similar targets in vitro and in vivo.3 In this context, especially the non-canonical quadruplex DNA (G4-DNA) is an important target.4 Quadruplex DNA is formed upon folding and assembly of guanine-rich DNA strands to a stack of usually three neighboring guanine quartets.5 The structures of quadruplex DNAs vary depending mainly on the particular sequence of the parent single strand(s).6 Most importantly, G4-DNA structures have essential biological functions, for example in the promoter regions of oncogenes, which influence the growth of cancer cells,7 or in the single-stranded overhang of telomeric DNA.8 Therefore, G4-DNA is an attractive target for the development of fluorescent probes that enable the detection and monitoring of this DNA form.9 Indeed, various fluorescent probes have already been developed that may be used to detect G4-DNA structures and to differentiate them from other DNA forms10in vitro11 and in cells.12 Hence, it has been demonstrated that suitable cationic dyes, for example distyrylpyridinium derivatives such as 1, bind selectively to particular target G4-DNA structures and that this complex formation is accompanied by a characteristic, G4-DNA-specific fluorescence light-up effect (Scheme 1).13 As another notable example, the cyanine dye 2 exhibits a strong fluorescence light-up effect upon selective association with the G4-DNA form c-myc (Scheme 1),14 which has been used to identify and distinguish this quadruplex form from other nucleic acid structures.15 But, despite the highly favorable detection properties of these and other2,13 fluorescent probes, there is still demand for novel approaches and for structural features that enable the selective detection of G4-DNA. Thus, there are, so far, only few examples of fluorescent probes available that are able to distinguish different topologies of G4-DNA with sufficient selectivity.16 At the same time, it appears that selectivity is more easily accomplished for some G4-DNA forms, e.g. for c-myc,14,15 whereas fluorescent probes for others, such as c-kit,17 are still rare. Against this background, we report the coincidental observation of the selective fluorimetric response of an N-biphenyldihydroisoquinolinium fluorophore upon binding to G4-DNA, which may contribute to the search for novel functional features of G4-DNA-targeting fluorescent probes. During our studies of the DNA-binding properties of dihydroisoquinolinium-based natural products,18 we have noticed that the very weakly fluorescent cationic biphenyl-substituted derivative 3 showed a distinct fluorescence in the presence of nucleic acids, especially pronounced with G4-DNA. Therefore, we investigated the photophysical and the DNA-binding properties of this particular biphenyl derivative. And we will demonstrate herein that, indeed, this compound is a promising starting point for the development of G4-DNA-selective fluorescent probes.Results and discussion
For a first orientation, the absorption and emission properties of biphenyl 3 were investigated in different solvents (Fig. 1 and Table 1). The absorption spectra of compound 3 are almost independent of the solvent, and the long-wavelength maxima only changed in a small range, from 350 nm in water to 354 nm in EtOH (Fig. 1A). In contrast, the fluorescence properties of compound 3 were found to depend on the solvent (Fig. 1B). While in water, a blue-shifted emission maximum was detected at 439 nm, a red-shifted maximum at 560 nm was found in EtOH. At the same time, dual emission of biphenyl 3 was observed in MeOH, MeCN, and DMSO, namely with two distinct emission bands at 352–353 nm and 556–580 nm (Fig. 1B and Table 1). In all employed solvents the emission quantum yields of 3 are very low (Φfl ≤ 0.01, Table 1).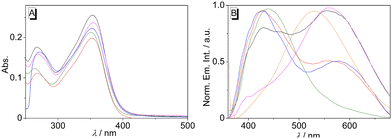 | ||
| Fig. 1 Absorption (c = 10 μM) (A) and normalized emission spectra (B) (λex = 350 nm) of 3 in H2O (green), DMSO (blue), MeOH (black), EtOH (magenta), MeCN (red), and glycerol (orange). | ||
| Solvent | λ abs /nm | ε /L mol−1 cm−1 | λ fl /nm |
Φ
fl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) d d |
|---|---|---|---|---|
| a Long-wavelength absorption maximum; c = 10 μM. b Molar extinction coefficient. c Emission maximum; λex = 350 nm. d Fluorescence quantum yield relative to anthracene in cyclohexane (Φfl = 0.36). The smaller maximum is given in parantheses. | ||||
| H2O | 350 (264) | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 300 |
439 | <0.01 |
| DMSO | 352 (272) | 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 280 |
428 (580) | 0.01 |
| MeOH | 353 (267) | 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 532 532 |
555 (429) | <0.01 |
| EtOH | 354 (269) | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 713 713 |
560 | 0.01 |
| MeCN | 352 (268) | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 849 849 |
426 (552) | <0.01 |
Dual emission has been frequently observed in fluorescent biphenyl derivatives that are integrated in a donor–acceptor system. And it is commonly explained by the dynamic photophysical processes in the excited state and the resulting different emitting excited species.19 Specifically, the excitation induces an intramolecular charge transfer (CT), which is often referred to as charge shift (CS) in the case of ionic chromophores, and to conformational changes in the excited state leading to twisted (TICT) or planar structures (PICT).19b,20 Accordingly, the emission properties of compound 3 are also influenced by the donor–acceptor interplay between the electron-donating methoxy group and the electron-withdrawing iminum functionality, in combination with the conformational changes of the biphenyl unit. Thus, compound 3 showed the expected emission band of a biphenyl chromophore at around 435 nm, which was assigned to the locally excited (LE) state, most likely formed after conformational change to an almost planar structure and a resonance-type excited state (Scheme 2).20f After further rotation around the biaryl axis, the TICT/CS state is formed with an almost perpendicular biphenyl structure, which results in the red-shifted emission band at ca. 550 nm (Scheme 2).20f,21 Apparently, these two states are stabilized or destabilized to a different extent by several solvent properties, because there is no obvious relationship between the formation of the emission bands with the solvent parameters.20c,22 As the fluorescence quantum yields are very low in all tested solvents, an additional non-radiative deactivation of the excited state obviously takes place with high efficiency, which may be caused by torsional relaxation around the biphenyl bond or around the Car–N or the Car–O bond of the donor and acceptor substituents.23 To verify this assumption, the fluorescence spectra were recorded in media with different viscosity, namely in glycerol/water with increasing glycerol content (Fig. S4, cf. ESI‡), because the viscosity of the solvent has a considerable effect on the rate of conformational changes, thus, on the emission quantum yield of excited molecules.24 Indeed, it was observed that in glycerol/water mixtures with higher viscosity, namely with glycerol fraction >40%, the emission intensity of 3 increased significantly with increasing glycerol content (Fig. 1B and Fig. S4 cf. ESI‡). Usually, this observation indicates a non-radiative deactivation of the excited state by conformational changes, such as torsional relaxation,25 because in highly viscous glycerol the conformational changes are slowed down and emission becomes competitive.26 Hence, torsional relaxation from either the LE state or the TICT/CS state of 3 to a non-emitting conformer most likely contributes to a significant extent to the non-radiative deactivation of the excited state.
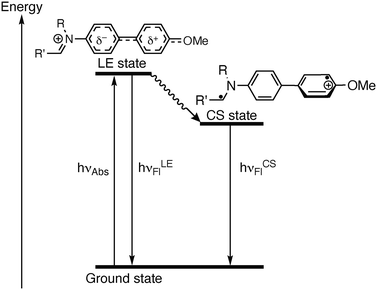 | ||
| Scheme 2 State diagram of the photoexcitation and deactivation pathways of the N-biphenyldihydroisoquinolinium 3. | ||
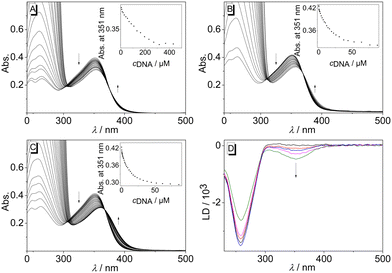
The interactions of biphenyl 3 with calf thymus (ct) DNA, as a typical duplex DNA structure, and with representative quadruplex DNA (G4-DNA) forms 22AG, c-kit, c-kras, c-myc, and h-ras1 were investigated. These G4-DNA forms relate to specific, biologically relevant DNA sequences and have a unique sequence and structure, as defined by the arrangement of the G-quartets and loop regions.27 The ligand-DNA interactions were monitored by photometric and fluorimetric titrations in aqueous buffer solutions. In almost all cases, a continuous decrease of the absorption maxima of compound 3 at 351 nm and a slight red shift (ΔλctDNA = 3 nm, Δλ22AG = 6 nm, Δλckit = 9 nm) were observed upon addition of DNA (Fig. 2A, B, C and Fig. S1, cf. ESI‡). This effect was most pronounced upon addition of G4-DNA c-kit. Moreover, during titrations with G4-DNA, an isosbestic point was formed at 365 nm, whereas with ct DNA an initially formed isosbestic point faded during titration. The binding isotherms derived from the photometric DNA titrations of compound 3 were used to determine the binding constants Kb (Table 2 and Fig. S2, cf. ESI‡),28 which revealed moderate binding affinities towards ct DNA (Kct = 1.3 × 104 M−1) and to G4-DNA (K22AG = 7.6 × 104 M−1; Kckit = 1.6 × 105 M−1; Kckras = 1.8 × 105 M−1, Kcmyc = 4.3 × 105 M−1, Khras1 = 3.1 × 105 M−1). However, the fitting curves of the binding isotherms of ct DNA, c-kras, and h-ras1 showed deviations, presumably because of heterogeneous binding at relatively high or low ligand loading on the DNA. To further assess the binding mode of compound 3 with duplex DNA, the DNA-ligand complexes were analyzed by linear dichroism (LD) spectroscopy in the hydrodynamic field of a rotating cuvette.29 In the presence of ct DNA, a negative LD signal between 300 nm and 400 nm was observed, that is in a range where only the ligand 3 absorbs. And the intensity of these LD bands increased with increasing ligand-to-DNA ratio (LDR) (Fig. 2D). At the same time, the intensity of LD bands of the DNA bases decreased during the titration.
| ct DNA | 22AG /F21Tb | c-kit /FkitTb | c-kras /FkrasTb | c-myc /FmycTb | h-ras1 | |
|---|---|---|---|---|---|---|
| a Binding constant determined from photometric titrations with 22AG, c-kit, c-kras, c-myc, and h-ras1. b Determined from fluorimetric analysis of dye-labeled oligonucleotides; estimated error ±0.5 °C. F21T = fluo-G3(TTAG3)3-tamra, FkitT = fluo-AG3AG3CGCTG3AG2AG3-tamra, FmycT = fluo-TGAG3TG3TAG3TG3TA-tamra, FkrasT = fluo-AG3CG2TG2A2GAG3A-tamra, fluo = fluorescein, tamra = tetramethylrhodamine. c Not determined. | ||||||
| K b /104 M−1 | 1.3 | 7.6 | 16 | 18 | 43 | 31 |
| ΔTmb/°C | 6 | 3 | 6 | 2 | ||
Overall, the photometric analyses indicated the association of the biphenyl 3 with nucleic acids. In particular, the changes in the absorption spectrum, namely hypochromism and the red shift of the absorption bands, are characteristic features of ligands that bind to duplex DNA30 or G4-DNA.31 Moreover, there are no large differences between the binding constants with the employed nucleic acids, which showed that there is no prounounced binding selectivity of the ligand towards a particular DNA form. In the case of duplex DNA, the additional LD-spectroscopic studies indicated an intercalation of the ligand, as confirmed by the negative LD bands of the ligand, which are characteristic of DNA intercalators.32 Accordingly, the observed binding constant of 3 with ct DNA is in a range commonly observed for cationic DNA binders.33 In contrast, the affinity of this ligand towards G4-DNA is relatively low as compared with the ones of known cationic ligands, which usually have binding constants of Kb > 105 M−1.34 In most cases, ligands that intercalate into duplex DNA tend to bind with G4-DNA by terminal π stacking to the quadruplex unit.35 Therefore it is assumed that the ligand 3 binds with essentially the same binding mode to c-kit, c-kras, c-myc, and h-ras1, and that the low binding constants reflect a somewhat hindered fit of the ligand structure to the binding site. The latter may be caused by the three-dimensional steric demand of the ligand as it does not have the required planar structure.
In addition, the stabilization of the dye-labeled G4-DNA forms F21T, FkitT, FkrasT and FmycT towards thermally induced unfolding after binding of ligand 3 was investigated by thermal DNA-denaturation experiments (Table 2 and Fig. S4, cf. ESI‡).36 Analysis of the melting temperature after ligand binding revealed a moderate stabilization of the quadruplex forms F21T (ΔTm = 6 °C) and FkrasT (ΔTm = 6 °C), whereas FkitT (ΔTm = 3 °C) and FmycT (ΔTm = 2 °C) were only slightly stabilized under the same conditions. These results indicated that upon binding the ligand 3 stabilizes the G4-DNA forms to a different extent depending on their topology. Notably, the shifts of the melting temperatures, ΔTm, of the different G4-DNA forms in the presence of ligand 3 do not correlate well with the binding constants. This apparent contradiction has been observed frequently during studies of ligand-quadruplex interactions and may be explained by different dependencies of equilibrium constants and Gibbs free energies on the temperature,6a specifically as the binding constant was determined at temperatures below the respective DNA melting temperature. Furthermore, it was shown exemplarily that the stabilization of the G4-DNA FkitT by ligand 3 did not significantly change in the presence of the double-stranded DNA ds26 [d(CA2TCG2ATCGA2T2CGATC2GAT2G); (ΔΔTm = −0.8 °C)], which indicated that the ligand binds selectively to the G4-DNA in competition with duplex DNA.37,38
To assess the influence of the association of ligand 3 with DNA on its emission properties, fluorimetric titrations with duplex and quadruplex DNA were conducted. In aqueous buffer solution, compound 3 showed a weak, hardly detectable emission band with a maximum at 460 nm. Upon addition of ct DNA, 22AG, c-kit, c-kras, c-myc, and h-ras1, the emission intensity of the bound ligand 3 increased; however, the development of the emission bands varied distinctly with the different DNA forms (Fig. 3A, B, C and Fig. S1, cf. ESI‡). Specifically, the contribution of the two emission maxima resulting from the dual fluorescence of the ligand 3 (see above) depends on the respective DNA form. Upon addition of ct DNA, only one clear emission band developed with a maximum at 530 nm and with an increase of emission intensity by a factor of 2 (at ligand![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA = 2), whereas the band at ca. 460 nm remained very weak. A similar development of the red-shifted fluorescence band at 535–538 with an intensity increased by a factor of 4–5 was observed with G4-DNA 22AG, c-kras, c-myc, respectively. But in these cases, the intensity of the initial blue-shifted band also gradually increased, albeit to a smaller extent than the red-shifted band. In sharp contrast, the addition of c-kit and h-ras1 to compound 3 led to a much stronger increase of the emission intensity of the blue-shifted band at 431 nm (c-kit) and 450 nm (h-ras1) by a factor of 5 and 10 (at ligand
DNA = 2), whereas the band at ca. 460 nm remained very weak. A similar development of the red-shifted fluorescence band at 535–538 with an intensity increased by a factor of 4–5 was observed with G4-DNA 22AG, c-kras, c-myc, respectively. But in these cases, the intensity of the initial blue-shifted band also gradually increased, albeit to a smaller extent than the red-shifted band. In sharp contrast, the addition of c-kit and h-ras1 to compound 3 led to a much stronger increase of the emission intensity of the blue-shifted band at 431 nm (c-kit) and 450 nm (h-ras1) by a factor of 5 and 10 (at ligand![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA = 2), along with the rise of the red-shifted band with smaller intensity. In each case, the fluorescence light-up effect of the ligand 3 upon addition of DNA can be followed by the naked eye (Fig. 3). Moreover, the solutions with different DNA forms could be distinguished by different emission colors, whereas the distinct clear blue emission in the presence of G4-DNA c-kit, and h-ras1 stood out as a special indicative element.
DNA = 2), along with the rise of the red-shifted band with smaller intensity. In each case, the fluorescence light-up effect of the ligand 3 upon addition of DNA can be followed by the naked eye (Fig. 3). Moreover, the solutions with different DNA forms could be distinguished by different emission colors, whereas the distinct clear blue emission in the presence of G4-DNA c-kit, and h-ras1 stood out as a special indicative element.
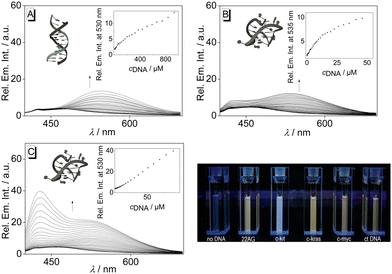 | ||
| Fig. 3 Fluorimetric titration of 3 (c = 20 μM) with ct DNA (A), 22AG (B), and c-kit (C); (A: in BPE buffer, cNa+ = 16 mM, pH = 7.0, B and C: in K-phosphate buffer, cK+ = 95 mM, pH = 7.0). The arrows indicate the development of the absorption or emission bands during the titration. Inset: plot of the emission at λ = 330 nm or λ = 431 nm versus cDNA; of ct DNA (A), 22AG (B), and c-kit (C). Structures in insets taken from the Protein Data Bank (PDB, https://www.rcsb.org/; PDB ID: A, 2k0v; B, 1kf1; C; 6gh0). | ||
The increased emission intensity of compound 3 after addition of DNA is most likely caused by the limited conformational freedom of the ligand within the DNA binding site,3b which in turn suppresses the torsional relaxation of the biphenyl unit as non-radiative deactivation pathway of the excited molecule. Such a fluorescent light-up effect based on this mechanism has been frequently observed for cationic DNA binders that contain a biaryl substructure.21 Moreover, this effect of the binding site on the fluorescence properties of the ligand may also explain the varying development of its emission bands upon association with the different DNA forms (Fig. 3). Specifically, the size and shape of the binding sites of the employed DNA forms vary distinctly,39 so that the bound ligand experiences different degrees of restricted conformational freedom in each case. Such an effect of the binding sites on the emission intensity and energy has already been demonstrated with the structurally resembling compound 9-(4-dimethylamino)benzo[b]quinolizinium, which exhibits a fluorescence light-up effect with different emission maxima in duplex and quadruplex DNA.40 Likewise, this influence of the binding sites on the emission color has been explained by the different conformational flexibility of the ligand within the binding sites.40
In the presence of ct DNA, the emission bands resemble the ones in glycerol and indicate the restricted free volume within the DNA binding site, which, in turn, should hinder conformational changes. Accordingly, the relaxation to the non-emissive conformation is suppressed under these conditions. But obviously, minor structural changes to a relaxed TICT/CS state are still possible, so that the related red-shifted emission band is also formed. In contrast, upon binding to G4-DNA a clear dual emission of the bound ligand was observed, which indicates that both the LE state and the TICT/CS state are populated to a significant extent within the respective binding sites. However, as a striking difference the contribution of each emission band varies with the different G4-DNA forms. And in the case of c-kit and h-ras1, the blue-shifted emission band dominates the overall spectrum, and this combination of bands results in an overall blue emission. For an explanation of this special effect, it is proposed that this effect is caused by the exceptional binding pocket provided by the quadruplex structure.39c Specifically, in the case of c-kit the particular loop structures allow the formation of a cleft at the 3′-end of the quadruplex in which organic ligands may be accommodated.41 Indeed, simple docking studies42 with the ligand 3 and c-kit revealed that the biphenyl 3 may bind in this binding pocket (Fig. 4) and point to a suppression of conformational freedom of the biphenyl unit in this constrained environment. As a result, the emission occurs mainly from the LE state leading to a distinct blue emission.43 Likewise, it is proposed that the conformational flexibility of ligand 3 is similarly restricted in the binding site of h-ras1. In this particular case, the available free volume is probably even more confined because the fluorescence light-up factor is larger. The “regular” terminal π stacking of the ligand at the other G4-DNA forms c-kras, c-myc, 22AG, by contrast, leaves sufficient conformational freedom to give mainly the red-shifted emission of the relaxed state. Notably, these latter G4-DNA forms have different quadruplex structures regarding the DNA-strand orientation, namely parallel (c-kras, c-myc) and hybrid (22AG); and likewise, the quadruplex forms, with which a more pronounced blue shift was observed, have different strand alignment (c-kit: parallel, h-ras1: antiparallel). Therefore, it may be concluded that this particular structural parameter does not influence significantly the emission properties of the bound ligand 3. At the same time, it cannot be excluded that the different polarity and/or availability of hydrogen bonds within the binding sites of the different G4-DNA forms also contributes to the varying emission properties of the bound ligand, because similar effects were observed in different solvents (Fig. 1). In this context, it should be emphasized that the different fluorescence response of the ligand 3 upon association with different DNA forms is not the result of a selective DNA recognition, because the binding studies revealed similar affinities to all tested DNA forms (see above). Instead, the varying emission properties are most likely caused by the different binding modes in combination with the high sensitivity of the dual emission of the biphenyl fluorophore on the surrounding medium.
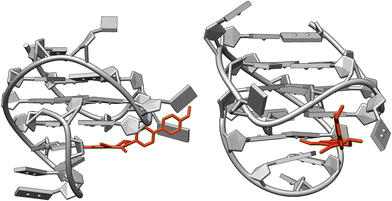 | ||
| Fig. 4 Structure of the complex between ligand 3 and G4-DNA c-kit (PDB ID: 2O3M) as estimated from molecular docking (AutoDock Vina, UCSF Chimera 1.15). | ||
Conclusions
In summary, we have identified the cationic, donor–acceptor-substituted biphenyl structure 3 as a promising complementary motif of quadruplex DNA binders. It has, to the best of our knowledge, not been introduced to this research field so far. However, biaryl-type fluorophores are already known that may be used for fluorimetric DNA detection, with thioflavin T (ThT) being the most prominent example.44 But these fluorescent probes do not exhibit dual emission, so that their response to DNA binding is a fluorescence light-up effect without a color change.In the present case, neither the affinity nor the selectivity of the ligand towards a particular DNA form was especially pronounced. But still, the high sensitivity of the emission properties of the biphenyl unit, specifically the pronounced dual emission, to the surrounding medium may be used to detect and characterize different binding sites of G4-DNA by distinct emission properties of the bound ligand. Specifically, sterically constrained binding sites, such as the one in c-kit, may be identified by the characteristic stronger contribution of the blue-shifted emission band. Therefore, it is proposed that similar cationic biphenyl derivatives with optimized structures for stronger binding affinities will figure as useful and efficient tools for the fluorimetric analysis of quadruplex DNA.
Data availability
The data supporting this article have been included as part of the ESI.‡Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the University of Siegen for financial support. D. S. thanks the House of Young Talents (University of Siegen) for a Ph.D. fellowship. We thank Jennifer Hermann, Hannah Karola Strunk, and Sandra Uebach (University of Siegen) for technical assistance.References
- (a) S. Munan, Y.-T. Chang and A. Samanta, Chronological development of functional fluorophores for bio-imaging, Chem. Commun., 2024, 60, 501–521 RSC; (b) R. Zhai, B. Fang, Y. Lai, B. Peng, H. Bai, X. Liu, L. Li and W. Huang, Small-molecule fluorogenic probes for mitochondrial nanoscale imaging, Chem. Soc. Rev., 2023, 52, 942–972 RSC; (c) D. B. Sung and J. S. Lee, Natural-product-based fluorescent probes: recent advances and applications, RSC Med. Chem., 2023, 14, 412–432 RSC; (d) H. Li, Y. An, J. Gao, M. Yang, J. Luo, X. Li, J. Lv, X. Li, Z. Yuan and H. Ma, Recent Advances of Fluorescence Probes for Imaging of Ferroptosis Process, Chemosensors, 2022, 10, 233–255 CrossRef CAS.
- (a) Y. Li, T. Liu and J. Sun, Recent Advances in N-Heterocyclic Small Molecules for Synthesis and Application in Direct Fluorescence Cell Imaging, Molecules, 2023, 28, 733 CrossRef CAS PubMed; (b) H. Niu, J. Liu, H. M. O'Connor, T. Gunnlaugsson, T. D. James and H. Zhang, Photoinduced electron transfer (PeT) based fluorescent probes for cellular imaging and disease therapy, Chem. Soc. Rev., 2023, 52, 2322–2357 RSC; (c) S. Samanta, K. Lai, F. Wu, Y. Liu, S. Cai, X. Yang, J. Qu and Z. Yang, Xanthene, cyanine, oxazine and BODIPY: the four pillars of the fluorophore empire for super-resolution bioimaging, Chem. Soc. Rev., 2023, 52, 7197–7261 RSC; (d) D. Cheng, W. Xu, X. Gong, L. Yuan and X.-B. Zhang, Design Strategy of Fluorescent Probes for Live Drug-Induced Acute Liver Injury Imaging, Acc. Chem. Res., 2021, 54, 403–415 CrossRef CAS PubMed; (e) C. Guo, A. C. Sedgwick, T. Hirao and J. L. Sessler, Supramolecular fluorescent sensors: An historical overview and update, Coord. Chem. Rev., 2021, 427, 213560 CrossRef CAS PubMed; (f) A. Saini, J. Singh and S. Kumar, Optically superior fluorescent probes for selective imaging of cells, tumors, and reactive chemical species, Org. Biomol. Chem., 2021, 19, 5208 RSC; (g) Y. V. Suseela, N. Narayanaswamy, S. Pratihar and T. Govindaraju, Far-red fluorescent probes for canonical and non-canonical nucleic acid structures: current progress and future implications, Chem. Soc. Rev., 2018, 47, 1098–1131 RSC.
- (a) J. Gebhard, L. Hirsch, C. Schwechheimer and H.-A. Wagenknecht, Hybridization-Sensitive Fluorescent Probes for DNA and RNA by a Modular “Click” Approach, Bioconjugate Chem., 2022, 33, 1634–1642 CrossRef CAS PubMed; (b) S. B. Wagh, V. A. Maslivetc, J. La Chair and A. Kornienko, Lessons in Organic Fluorescent Probe Discovery, ChemBioChem, 2021, 22, 3109–3139 CrossRef CAS PubMed; (c) A. Granzhan, H. Ihmels and M. Tian, The benzo[b]quinolizinium ion as a water-soluble platform for the fluorimetric detection of biologically relevant analytes, ARKIVOC, 2015, 494–523 Search PubMed.
- H. Tateishi-Karimata and N. Sugimoto, Chemical biology of non-canonical structures of nucleic acids for therapeutic applications, Chem. Commun., 2020, 56, 2379–2390 RSC.
- (a) K. Weisz, A world beyond double-helical nucleic acids: the structural diversity of tetra-stranded G-quadruplexes, ChemTexts, 2021, 7, 25 CrossRef; (b) Y. Xu, Chemistry in human telomere biology: structure, function and targeting of telomere DNA/RNA, Chem. Soc. Rev., 2011, 40, 2719–2740 RSC.
- (a) J. Jana and K. Weisz, Thermodynamic Stability of G-Quadruplexes: Impact of Sequence and Environment, ChemBioChem, 2021, 22, 2848–2856 CrossRef CAS PubMed; (b) M. Sanderson and S. Neidle, Principles of Nucleic Acid Structure, Academic Press, 2nd edn, 2021 Search PubMed.
- G. W. Collie and G. N. Parkinson, The application of DNA and RNA G-quadruplex to therapeutic medicines, Chem. Soc. Rev., 2011, 40, 5867–5892 RSC.
- A. Verma, V. K. Yadav, R. Basundra, A. Kumar and S. Chowdhury, Evidence of genome-wide G4 DNA-mediated gene expression in human cancer cells, Nucleic Acids Res., 2009, 37, 4194–4204 CrossRef CAS PubMed.
- X. Xie, M. Zuffo, M.-P. Teulade-Fichou and A. Granzhan, Identification of optimal fluorescent probes for G-quadruplex nucleic acids through systematic exploration of mono- and distyryl dye libraries, Beilstein J. Org. Chem., 2019, 15, 1872–1889 CrossRef CAS PubMed.
- (a) L.-S. Rao, L. Hao, L.-Y. Liu, Y.-L. Zeng, B.-B. Liang, W. Liu and Z.-W. Mao, Detection and tracking of cytoplasmic G-quadruplexes in live cells, Chem. Commun., 2023, 59, 13348–13351 RSC; (b) H. Yang, Y. Zhou and J. Liu, G-quadruplex DNA for construction of biosensors, TrAC, Trends Anal. Chem., 2020, 132, 116060 CrossRef CAS; (c) P. Chilka, N. Desai and B. Datta, Small Molecule Fluorescent Probes for G-Quadruplex Visualization as Potential Cancer Theranostic Agents, Molecules, 2019, 24, 752 CrossRef CAS PubMed.
- M. Zuffo, X. Xie and A. Granzhan, Strength in Numbers: Development of a Fluorescence Sensor Array for Secondary Structures of DNA, Chem. – Eur. J., 2019, 25, 1812–1818 CrossRef CAS PubMed.
- (a) J.-N. Han, C. Zhong, M. Ge, S. Kuang and Z. Nie, Engineering fluorescent protein chromophores with an internal reference for high-fidelity ratiometric G4 imaging in living cells, Chem. Sci., 2023, 14, 4538–4548 RSC; (b) A. S. Oshchepkov, O. Reznichenko, D. Xu, B. S. Morozov, A. Granzhan and E. A. Kataev, Dye-functionalized phosphate-binding macrocycles: from nucleotide to G-quadruplex recognition and “turn-on” fluorescence sensing, Chem. Commun., 2021, 57, 10632–10635 RSC.
- X. Xie, B. Choi, E. Largy, R. Guillot and A. Granzhan, Asymmetric Distyrylpyridinium Dyes as Red-Emitting Fluorescent Probes for Quadruplex DNA, Chem. – Eur. J., 2013, 19, 1214–1226 CrossRef CAS PubMed.
- L. Chen, J. Dickerhoff, S. Sakai and D. Yang, DNA G-Quadruplex in Human Telomeres and Oncogene Promoters: Structures, Functions, and Small Molecule Targeting, Acc. Chem. Res., 2022, 55, 2628–2646 CrossRef CAS PubMed.
- R. Sun, X. Guo, D. Yang, Y. Tang, J. Lu and H. Sun, c-Myc G-quadruplex is sensitively and specifically recognized by a fluorescent probe, Talanta, 2021, 226, 122125 CrossRef CAS PubMed.
- (a) H.-Z. He, K. Li, K.-K. Yu, P.-L. Lu, M.-L. Feng, S.-Y. Chen and X.-Q. Yu, Additive- and column-free synthesis of rigid bis-coumarins as fluorescent dyes for G-quadruplex sensing via disaggregation-induced emission, Chem. Commun., 2020, 56, 6870–6873 RSC; (b) M. Deiana, K. Chand, J. Jamroskovic, I. Obi, E. Chorell and N. Sabouri, A Light-up Logic Platform for Selective Recognition of Parallel G-Quadruplex Structures via Disaggregation-Induced Emission, Angew. Chem., Int. Ed., 2020, 59, 896–902 CrossRef CAS PubMed.
- A. T. Phan, V. Kuryavyi, S. Burge, S. Neidle and D. J. Patel, Structures of an Unprecedented G-Quadruplex Scaffold in the Human c-kit Promoter, J. Am. Chem. Soc., 2007, 129, 4386–4392 CrossRef CAS PubMed.
- G. Bringmann, T. Gulder, B. Hertlein, Y. Hemberger and F. Meyer, Total Synthesis of the N,C-Coupled Naphthylisoquinoline Alkaloids Ancistrocladinium A and B and Related Analogues, J. Am. Chem. Soc., 2010, 132, 1151–1158 CrossRef CAS PubMed.
- (a) J. L. Belmonte-Vazquez, Y. A. Amador-Sanchez, L. A. Rodriguez-Cortes and B. Rodriguez-Molina, Dual-State Emission (DSE) in Organic Fluorophores: Design and Applications, Chem. Mater., 2021, 33, 7160–7184 CrossRef CAS; (b) C. Wang, W. Chi, Q. Qiao, D. Tan, Z. Xu and X. Liu, Twisted intramolecular charge transfer (TICT) and twists beyond TICT: from mechanisms to rational designs of bright and sensitive fluorophores, Chem. Soc. Rev., 2021, 50, 12656–12678 RSC.
- (a) C. Chen and C. Fang, Fluorescence Modulation by Amines: Mechanistic Insights into Twisted Intramolecular Charge Transfer (TICT) and Beyond, Chemosensors, 2023, 11, 87 CrossRef CAS; (b) M. Wang, R.-Z. Wang and C.-H. Zhao, Temperature-Dependent Dual Fluorescence from Small Organic Molecules, Org. Mater., 2022, 4, 204–215 CrossRef CAS; (c) S. K. Behera, S. Y. Park and J. Gierschner, Dual Emission: Classes, Mechanisms, and Conditions, Angew. Chem., Int. Ed., 2021, 60, 22624–22638 CrossRef CAS PubMed; (d) S. Sasaki, G. P. C. Drummen and G.-i. Konishi, Recent advances in twisted intramolecular charge transfer (TICT) fluorescence and related phenomena in materials chemistry, J. Mater. Chem. C, 2016, 4, 2731–2743 RSC; (e) V. V. Volchkov and B. M. Uzhinov, Structural relaxation of excited molecules of heteroaromatic compounds, High Energy Chem., 2008, 42, 153–169 CrossRef CAS; (f) Z. R. Grabowski, K. Rotkiewicz and W. Rettig, Structural Changes Accompanying Intramolecular Electron Transfer: Focus on Twisted Intramolecular Charge-Transfer States and Structures, Chem. Rev., 2003, 103, 3899–4032 CrossRef PubMed.
- M. Tian and H. Ihmels, Synthesis of Fluorescent 9-Aryl-Substituted Benzo[b]quinolizinium Derivatives, Synthesis, 2009, 24, 4226–4234 Search PubMed.
- S. Nigam and S. Rutan, Principles and Applications of Solvatochromism, Appl. Spectrosc., 2001, 55, 362A–370A CrossRef CAS.
- S. I. Druzhinin, S. R. Dubbaka, P. Knochel, S. A. Kovalenko, P. Mayer, T. Senyushkina and K. A. Zachariasse, Ultrafast Intramolecular Charge Transfer with Strongly Twisted Aminobenzonitriles: 4-(Di-tert-butylamino)benzonitrile and 3-(Di-tert-butylamino)benzonitrile, J. Phys. Chem. A, 2008, 112, 2749–2761 CrossRef CAS PubMed.
- (a) M. M. Sreejaya, V. M. Pillai, A. Ayesha, M. Baby, M. Bera and M. Gangopadhyay, Mechanistic analysis of viscosity-sensitive fluorescent probes for applications in diabetes detection, J. Mater. Chem. B, 2024, 12, 2917–2937 RSC; (b) H. Xiao, P. Li and B. Tang, Small Molecular Fluorescent Probes for Imaging of Viscosity in Living Biosystems, Chem. – Eur. J., 2021, 27, 6880–6898 CrossRef CAS PubMed; (c) M. Vogel and W. Rettig, Efficient Intramolecular Fluorescence Quenching in Triphenylmethane-Dyes Involving Excited States with Charge Separation and Twisted Conformations, Ber. Bunsenges. Phys. Chem., 1985, 89, 962–968 CrossRef CAS.
- M. Vogel and W. Rettig, Efficient Intramolecular Fluorescence Quenching in Triphenylmethane-Dyes Involving Excited States with Charge Separation and Twisted Conformations, Ber. Bunsenges. Phys. Chem., 1985, 89, 962–968 CrossRef CAS.
- T. Förster and G. Hoffmann, Die Viskositätsabhängigkeit der Fluoreszenzquantenausbeuten einiger Farbstoffsysteme, Z. Phys. Chem., 1971, 75, 63–76 CrossRef.
- (a) K. B. Wang, Y. Liu, J. Li, C. Xiao, Y. Wang, W. Gu, Y. Li, Y. Z. Xia, T. Yan, M. H. Yang and L. Y. Kong, Structural insight into the bulge-containing KRAS, oncogene promoter G-quadruplex bound to berberine and coptisine, Nat. Commun., 2022, 13, 6016 CrossRef CAS PubMed; (b) M. Sanderson and S. Neidle, Principles of Nucleic Acid Structure, Academic Press, 2nd edn, 2021 Search PubMed; (c) A. T. Phan, V. Kuryavyi, S. Burge, S. Neidle and D. J. Patel, Structure of an Unprecedented G-Quadruplex Scaffold in the Human c-kit Promoter, J. Am. Chem. Soc., 2007, 129, 4386–4392 CrossRef CAS PubMed.
- F. H. Stootman, D. M. Fisher, A. Rodger and J. R. Aldrich-Wright, Improved curve fitting procedures to determine equilibrium binding constants, Analyst, 2006, 131, 1145–1151 RSC.
- B. Nordén, A. Rodger and T. Dafforn, Linear Dichroism and Circular Dichroism: A Textbook on Polarized-Light Spectroscopy, RSC Publishing, Cambridge, 2010 Search PubMed.
- M. M. Aleksić and V. Kapetanović, An Overview of the Optical and Electrochemical Methods for Detection of DNA-Drug Interactions, Acta Chim. Slov., 2014, 61, 555–573 Search PubMed.
- (a) F. Nagatsugi and K. Onizuka, Functional G-Quadruplex Binding Molecules, Chem. Lett., 2020, 49, 771–780 CrossRef CAS; (b) M. P. O'Hagan, J. C. Morales and M. C. Galan, Binding and Beyond: What Else Can G-Quadruplex Ligands Do?, Eur. J. Org. Chem., 2019, 4995–5017 CrossRef.
- T. M. Aliyeu, D. V. Berdnikova, O. A. Fedorova, E. N. Gulakova, C. Stremmel and H. Ihmels, Regiospecific Photocyclization of Mono- and Bis-Styryl-Substituted N-Heterocycles: A Synthesis of DNA-Binding Benzo[c]quinolizinium Derivatives, J. Org. Chem., 2016, 81, 9075–9085 CrossRef CAS PubMed.
- (a) P. Bosch, G. Marcelo, A. Matamoros-Recio, D. Sucunza, F. Mendicuti, A. Domingo and J. J. Vaquero, A new family of fluorescent pyridazinobenzimidazolium cations with DNA binding properties, Dyes Pigm., 2021, 192, 109443 CrossRef CAS; (b) A. K. Das, H. Ihmels and S. Kölsch, Diphenylaminostyryl-substituted quinolizinium derivatives as fluorescent light-up probes for duplex and quadruplex DNA, Photochem. Photobiol. Sci., 2019, 18, 1373–1381 CrossRef CAS PubMed.
- V. Pirota, M. Stasi, A. Benassi and F. Doria, Chapter Six – An overview of quadruplex ligands: Their common features and chemotype diversity, Annu. Rep. Med. Chem., 2020, 54, 163–196 CAS.
- I. Frasson, V. Pirota, S. N. Richter and F. Doria, Multimeric G-quadruplexes: A review on their biological roles and targeting, Int. J. Biol. Macromol., 2022, 204, 89–102 CrossRef CAS PubMed.
- A. De Cian, L. Guittat, M. Kaiser, B. Sacca, S. Amrane, A. Bourdoncle, P. Alberti, M.-P. Teulade-Fichou, L. Lacroix and J.-L. Mergny, Fluorescence-based melting assay for studying quadruplex ligands, Methods, 2007, 42, 183–195 CrossRef CAS PubMed.
- J. Becher, D. V. Berdnikova, H. Ihmels and C. Stremmel, Synthesis and investigation of quadruplex-DNA-binding, 9-O-substituted berberine derivatives, Beilstein J. Org. Chem., 2020, 16, 2795–2806 CrossRef CAS PubMed.
- Y. Zhang, Y. Cheng, Q. Luo, T. Wu, J. Huo, M. Yin, H. Peng, Y. Xiao, Q. Tong and H. You, Distinguishing G-Quadruplexes Stabilizer and Chaperone for c-MYC Promoter G-Quadruplexes through Single-Molecule Manipulation, J. Am. Chem. Soc., 2024, 146, 3689–3699 CrossRef CAS PubMed.
- (a) K.-B. Wang, Y. Liu, J. Li, C. Xiao, Y. Wang, W. Gu, Y. Li, Y.-Z. Xia, T. Yan, M.-H. Yang and L.-Y. Kong, Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine, Nat. Commun., 2022, 13, 6016 CrossRef CAS PubMed; (b) J. Spiegel, S. Adhikari and S. Balasubramanian, The Structure and Function of DNA G-Quadruplexes, Trends Chem., 2020, 2, 123–136 CrossRef CAS PubMed; (c) D. Wei, G. N. Parkinson, A. P. Reszka and S. Neidle, Crystal structure of a c–kit promoter quadruplex reveals the structure role of metal ions and water molecules in maintaining loop conformation, Nucleic Acids Res., 2012, 40, 4691–4700 CrossRef CAS PubMed.
- R. Bortolozzi, H. Ihmels, L. Thomas, M. Tian and G. Viola, 9-(4-Dimethylaminophenyl)benzo[b]quinolizinium: A Near-Infrared Fluorophore for the Multicolor Analysis of Proteins and Nucleic Acids in Living Cells, Chem. – Eur. J., 2013, 19, 8736–8741 CrossRef CAS PubMed.
- (a) K. G. Moghaddam, A. H. de Vries, S. J. Marrink and S. Faraji, Binding of quinazolinones to c-KIT G-quadruplex; an interplay between hydrogen bonding and π-π stacking, Biophys. Chem., 2019, 253, 106220 CrossRef CAS PubMed; (b) A. Głuszyńska, B. Juskowiak, M. Kuta-Siejkowska, M. Hoffmann and S. Haider, Carbazole Derivatives Binding to c-KIT G-Quadruplex DNA, Molecules, 2018, 23, 1134–1154 CrossRef PubMed; (c) D. Wei, J. Husby and S. Neidle, Flexibility and structural conversation in a c-KIT G-quadruplex, Nucleic Acids Res., 2015, 43, 629–644 CrossRef CAS PubMed; (d) A. T. Phan, V. Kuryavyi, S. Burge, S. Neidle and D. J. Patel, Structure of an Unprecedented G-Quadruplex Scaffold in the Human c-kit Promoter, J. Am. Chem. Soc., 2007, 129, 4386–4392 CrossRef CAS PubMed.
- O. Trott and A. J. Olson, AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J. Comput. Chem., 2009, 31, 455–461 CrossRef PubMed.
- (a) A. Pandith, U. Nagarajachari, R. K. G. Siddappa, S. Lee, C.-J. Park, K. Sannathammegowda and Y. J. Seo, Loop-mediated fluorescent probes for selective discrimination of parallel and antiparallel G-Quadruplexes, Bioorg. Med. Chem., 2021, 35, 116077 CrossRef CAS PubMed; (b) V. Dhamodharan and P. I. Pradeepkumar, Specific Recognition of Promoter G-Quadruplex DNAs by Small Molecule Ligand and Light-up Probes, ACS Chem. Biol., 2019, 14, 2102–2114 CAS.
- (a) R.-X. Wang, Y. Ou, Y. Chen, T.-B. Ren, L. Yuan and X.-B. Zhang, Rational Design of NIR-II G-Quadruplex Fluorescent Probes for Accurate In Vivo Tumor Metastasis Imaging, J. Am. Chem. Soc., 2024, 146, 11669–11678 CrossRef CAS PubMed; (b) M. Lv, J. Ren and E. Wang, Topological effect of an intramolecular split G-quadruplex on thioflavin T binding and fluorescence light-up, Chem. Sci., 2024, 15, 4519–4528 RSC; (c) S. Liu, P. Peng, H. Wang, L. Shi and T. Li, Thioflavin T binds dimeric parallel-stranded GA-containing non-G-quadruplex DNAs: a general approach to lighting up double-stranded scaffolds, Nucleic Acids Res., 2017, 45, 12080–12089 CrossRef CAS PubMed; (d) Q. Xu, M. Yang, Y. Chang, S. Peng, D. Wang, X. Zhou and Y. Shao, Switching G-quadruplex to parallel duplex by molecular rotor clustering, Nucleic Acids Res., 2022, 50, 10249–10263 CrossRef CAS PubMed; (e) R. Kumar, K. Chand, S. Bhowmik, R. N. Das, S. Bhattacharjee, M. Hedenström and E. Chorell, Subtle structural alterations in G-quadruplex DNA regulate site specificity of fluorescence light-up probes, Nucleic Acids Res., 2020, 48, 1108–1119 CrossRef CAS PubMed.
Footnotes |
| † Dedicated to Prof. Dr. Frank Würthner on the occasion of his 60th birthday. |
| ‡ Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4qo01193g |
| This journal is © the Partner Organisations 2024 |