Open Access Article
Open Access ArticleCoPolDB: a copolymerization database for radical polymerization
Kei-ichiro
Takahashi
a,
Hiroshi
Mamitsuka
 a,
Masatoshi
Tosaka
a,
Masatoshi
Tosaka
 b,
Nanyi
Zhu
b and
Shigeru
Yamago
b,
Nanyi
Zhu
b and
Shigeru
Yamago
 *b
*b
aBioinformatics Center, Institute for Chemical Research, Kyoto University, Gokasho Uji, 6110011, Japan
bInstitute for Chemical Research, Kyoto University, Gokasho Uji, 6110011, Japan
First published on 29th January 2024
Abstract
Although a large amount of knowledge on copolymers and copolymerization has been already accumulated in the literature, there are no freely available databases with a proper graphical user interface (GUI) on copolymerization. Focusing on copolymerization with only two types of monomers (due to the focus on fundamentals) and copolymerization in radical polymerization, i.e. the most major polymerization, we present CoPolDB, a database of copolymerization with numerous helpful GUI functions, including: (1) graphically showing multiple connections (with different reactivity ratios) between two monomers and the corresponding copolymer, assisting users’ intuitive understanding and; (2) providing a list of alternative monomers for each monomer pair, according to the similarity to the original monomer. We believe that CoPolDB is a useful resource to understand the current copolymerization status entirely and comprehensively as well as an inspiring tool to promote polymer chemistry research. CoPolDB is available at https://www.copoldb.jp/.
1. Introduction
Radical polymerization is one of the most important polymerization methods in the synthesis of polymers in industry.1–4 Its characteristic features include high monomer versatility and functional group tolerance, enabling the fabrication of polymer materials from vinyl monomers with different reactivity and different functional groups. In particular, the copolymerization of two (or more) monomers, 1 and 2 (Fig. 1a), has been extensively used to tailor polymer products by imparting various functions, such as polarity and reacting groups, to polymer materials derived from monomers. One of the authors recently developed the synthesis of structurally controlled hyperbranched polymers via radical copolymerization of conventional monomers with a branch-inducing monomer, and the copolymerization ratio is important for controlling the branch structure.5–9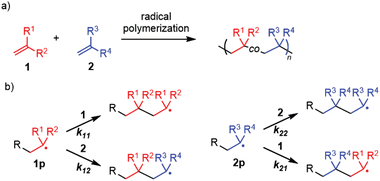 | ||
| Fig. 1 Copolymerization: (a) the general reaction scheme and (b) the four propagation pathways involving the copolymerization. | ||
The structure of copolymers and the properties of the resulting polymer materials are determined by the reactivity of each monomer, defined using the monomer reactivity ratios, r1 = k11/k12 and r2 = k22/k21 (k11 and k12 refer to the rate constant of 1p reacting with 1 and 2, respectively, and k22, and k21 refer to the rate constant of 2p reacting with 2 and 1, respectively) (Fig. 1b). Thus, understanding and predicting the monomer reactivity is a fundamental and important research theme.1–3 Due to the long history of radical polymerization, a large amount of knowledge on copolymers and copolymerization has already been accumulated in the literature. Organizing this knowledge and presenting the organized information in a visually understandable manner would be useful for advancing research in polymer chemistry and relevant fields, not only scientifically but also from an engineering perspective. However, to the best of our knowledge, currently there are no freely available online data resources relevant to copolymerization, except for the following two cases.
(1) PolyInfo10 (https://polymer.nims.go.jp/): a database (DB) of polymers, including homopolymers, copolymers, polymer blends, composites, etc., where 105 properties are given for each polymer entry. PolyInfo is a useful resource, providing the properties of each polymer, but it gives no information on copolymerization kinetics and monomer reactivities.
(2) Recently, an online DB for rate coefficients in radical polymerization (https://sql.polymatter.net) was created.11 This is also a proper resource of monomers, copolymers and even copolymerization. However, this DB consists solely of the rate constant of homopolymerization. In addition, this DB has no graphical user interface (GUI), and this makes it hard for users to intuitively understand the reaction of copolymerization.
With the above considerations in mind, we propose CoPolDB (https://www.copoldb.jp/) equipped with a user-friendly GUI, which allows users to visually see the connection between monomers, copolymers, and copolymerization kinetics. CoPolDB (although it is currently limited to the copolymerization of two monomers) has a number of unique features, particularly the following four: (1) CoPolDB consists primarily of two parts – monomers and copolymers – which are connected to each other, since monomers are building blocks of copolymerization (Fig. 2 and 3). (2) CoPolDB includes kinetic parameters r1 and r2, from which the so-called copolymer composition curve, F1/f1 plot, can be calculated and visualized (F1 and f1 refer to the molar fraction of monomer unit derived from monomer 1 in a copolymer and the feed of monomer 1, respectively) (Fig. 4). This visualization allows users to intuitively understand the effect of the monomer reactivity ratio on the composition of synthesized polymers. Note that all data are included when multiple combinations of r1 and r2 were reported with scatter plots. (3) All copolymerization data are extracted from a polymer handbook,12 and each entry is hyperlinked to the original reference through a digital object identifier (DOI), if the DOI is available. (4) Only the minimum information is kept in CoPolDB, so that the size of the required storage is very small. Relevant information can be provided from internet resources, such as PubChem,13 through the identifiers generated by CoPolDB.
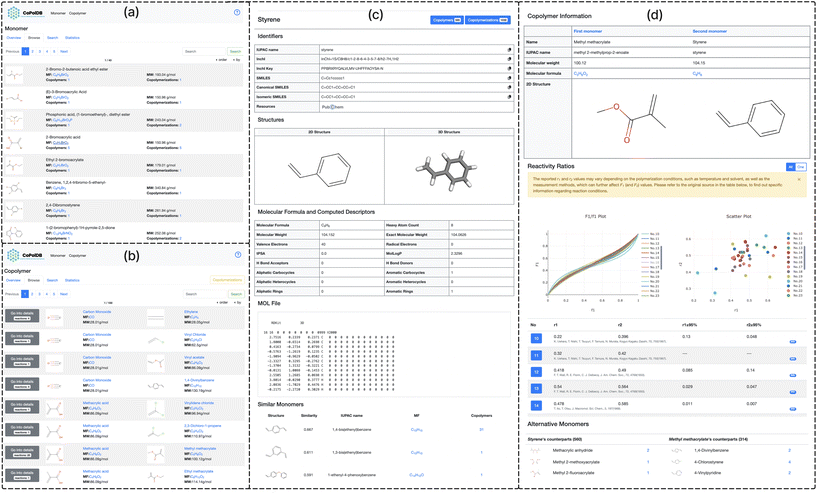 | ||
| Fig. 3 Interface examples: (a) Monomers “Browse”: for each monomer, the chemical structure, the monomer names, the molecular formula, the molecular weight, how many times used for generating copolymers, and how many times used in copolymerization reactions, are shown. The last two items have hyperlinks to the corresponding entries in the copolymers “Browse” page. (b) Copolymers “Browse”: a list of monomer pairs (each corresponding to a copolymer and linking to the detail page of (d)) is shown. This page can be switched to the corresponding Copolymerizations “Browse” page by clicking ‘copolymerizations’ in the upper right corner. (c) Monomers detail page: for a monomer (styrene), the identifiers, the molecular structure, the computed descriptors, the MOL file, and any similar monomers, are shown. (d) Copolymers detail page: the F1/f1 plots, the scatter plots and a reactivity ratio table with DOIs (see also Fig. 4) are shown. The alternative monomers usable in copolymerization reactions (Fig. 5) are suggested. | ||
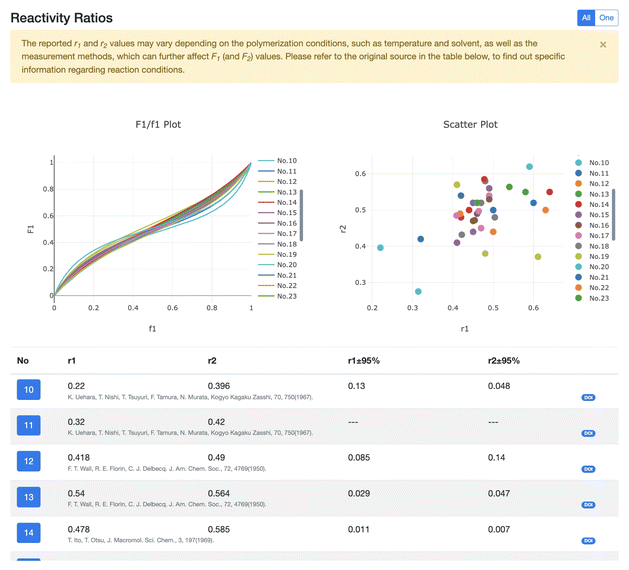 | ||
| Fig. 4 Enlarged Fig. 3d. The reactivity ratios for the same monomer pair are shown in the F1/f1 (upper left) and scatter plots (upper right). The original reference of each ratio can be accessed by clicking the corresponding DOI. We can choose an arbitrary subset of the reactivity ratios, and the plots of only the selected ratios are shown. | ||
We emphasize that CoPolDB is the first freely available DB with a sophisticated GUI on copolymerization in polymer chemistry. Even across chemistry as a whole, it would be hard to find a free DB with an advanced GUI like CoPolDB. On the other hand, in biology, a lot of data and information have been stored in a variety of DBs and presented with proper GUIs, such as GenBank14 for DNA sequences and GEO (Gene Expression Omnibus)15 for gene expression data. These DBs are useful not only for understanding the current state of biology but also for promoting research in biology. This data-based aspect might be a point lacking in current chemistry. We hope that CoPolDB will be one of the first steps to add more data-driven viewpoints to the present chemistry.
2. Database
2.1. Database structure
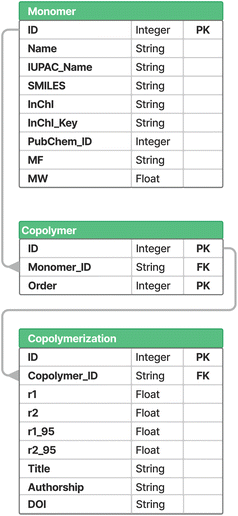
CoPolDB is a relational DB, for which Fig. 2 shows the entity relationship (ER) diagram, with the following three tables: (1) Monomer, (2) Copolymer, and (3) Copolymerization.(1) Monomer: each entry has the entry ID (Monomer_ID), the trivial and IUPAC names, the SMILES (Simplified Molecular Input Line Entry System)16 string, the InChI (International Chemical Identifier), the InChI key,17 the molecular weight (MW) and the molecular formula (MF).
Using the monomer identifiers, i.e. the SMILES and InChI key, the physical or chemical information can be retrieved from external resources and presented.
(2) Copolymer: each entry has the entry ID (Copolymer_ID), a pair of monomers (specified by two Monomer_IDs) and the “order” with a numerical value to specify the first and second monomers.
(3) Copolymerization: each entry has the entry ID, Copolymer_ID, the reactivity ratios r1 and r2, and the reference information (the title, the authorship and the DOI). In addition, each entry has the 95% confidence limits of the ratios if they are provided in the polymer handbook.12
These three tables are connected through Monomer_ID and Polymer_ID (Fig. 2), so that relevant information can be retrieved using these IDs. For instance, given a monomer, CoPolDB can provide the number of copolymers, which are synthesized from this monomer, and reversely, given a reaction of copolymerization, users can obtain the molecular formula of the two monomers used in the reaction.
2.2. Data sources
All entries (of copolymerization) are obtained from a polymer handbook.12 As the primary data source, CoPolDB extracts the monomer names and reactivity ratios from published literature. Regarding monomers, the information other than the monomer names is generated using two software tools, RDKit18 and OpenBabel.19 Below we explain the sources of the three tables in CoPolDB.Monomer: the SMILES code is created with reference to the original source and assigned to each monomer. Using the SMILES code, the molecular formula and molecular weight of a molecule are generated by RDKit.18 Similarly, using the SMILES code, the InChI and InChI key are generated using OpenBabel.19 The trivial and IUPAC names are obtained from PubChem by searching the SMILES code.
Copolymer and copolymerization: the information is generated from the literature based on manual curation (not by any software). The Copolymer table stores the two Monomer_IDs corresponding to the two monomers found in the corresponding literature. The Copolymerization table stores the corresponding Copolymer_ID, reactivity ratio and reference (title, author and DOI) information. The DOIs of all references are stored, unless they are neither accessible nor publicly available.
As of the time of writing this paper, CoPolDB has 864 monomers, 1954 copolymers, and 2991 reactions of copolymerization in the Monomer, Copolymer, and Copolymerization tables, respectively. For the DOIs, we identified 725 unique identifiers, which can be assigned to 2449 out of the 2991 reactions of copolymerization.
3. Graphical user interface (GUI)
The web interface of CoPolDB is composed of two parts – Monomers and Copolymers – which correspond to the Monomer and Copolymer tables (the Copolymerization table is switchable with the Copolymer table), respectively. Both parts have four sub-pages: (i) Overview, (ii) Browse, (iii) Search and (iv) Statistics.3.1. Monomers
This part presents the information on monomers, i.e. the materials for producing copolymers by polymerization.(i) Overview: monomers for radical polymerization, e.g. fundamental concepts of radical polymerization, monomer types, and structures of representative monomers, are described. Examples of representative monomers are also introduced.
(ii) Browse: a comprehensive list of all monomer entries with pagination and a sort function is presented, where each monomer is with six key pieces of information (Fig. 3a): (1) the 2-dimensional structural formula displayed as a PNG image generated by RDKit based on the SMILES notation, (2) the monomer name (the conventional name or the IUPAC name if the conventional name is unavailable), (3) the molecular formula (generated by RDKit), (4) the molecular weight (calculated by RDkit), (5) the number of copolymers linked to the monomer, and (6) the number of reactions of copolymerization linked to the monomer.
Users can sort monomer entries, using any items other than the structural formulas, in ascending or descending order. For example, by sorting the number of copolymers (5) in descending order, users can see the most frequently used monomers in copolymers in CoPolDB. In each page, hyperlinks are given to all items, except for the monomer name and the molecular weight. For example, by clicking the molecular formula, a list of monomers with the same molecular formula is shown. By clicking the number of copolymers (or copolymerizations), a list of copolymers (copolymerizations) synthesized from the same monomer is shown. That is, these hyperlinks allow users to access a variety of information on reactions of copolymerization. Also, by clicking the structural formula, the corresponding monomer detail page (Fig. 3c) is presented.
The monomer detail page has five sections: (1) Identifiers (ID), (2) Structures (ST), (3) Molecular Formula and Computed Descriptors (MD), (4) MOL File (MO) and (5) Similar Monomers (SM), giving users helpful insight into copolymerization reactions, since the properties and behavior of the resulting copolymer often depend on the arrangement of monomers within the polymer chain. These five sections are described further: (1) ID: if a monomer is registered in PubChem, the corresponding IUPAC name, canonical SMILES, and isomeric SMILES are kept linked to the monomer in PubChem. These can be used as the keywords to search external resources and tools to calculate the physical or chemical properties. (2) ST: the 2D and 3D structures obtained by RDKit18 and 3Dmol.js,20 respectively, are shown. To generate the conformation for the 3D structure, CoPolDB uses a distance geometry (DG) method called Experimental–Torsion–Knowledge Distance Geometry (ETKDG).21 (3) MD: the molecular formula and fifteen molecular descriptors, such as the molecular weight, calculated by RDKit are presented. (4) MO: a de facto standard for the data formats in chemoinformatics, MOL, is shown in a text format. This is the result of a conformational calculation to generate the 3D structure in ST. (5) SM: the Tanimoto similarity22 of two molecules (fingerprints) is given as two fingerprints (say A and B), defined as the ratio of the intersection (A∩B) to the union (A∪B). A list of monomers with the Tanimoto similarity of 0.25 or higher, computed using Morgan fingerprints,23 is presented in descending order of the Tanimoto similarity.
(iii) Search: users can search monomers using the following items: (a) the canonical or IUPAC name, (b) the molecular formula, (c) the SMILES code, (d) the InChI Key, (e) the minimum/maximum molecular weight, (f) the minimum/maximum occurrences in copolymers. From (a) to (d), partial text matching, i.e. checking whether the query text is included or not in the target text, is available. Users can search a molecular formula in the text, without thinking about the order of atom symbols. For example, C8H8 and H8C8 are treated as the same molecular formula. Users can input a query, being assisted by an auto-complete function. That is, if a user inputs two or more characters in a text field, such as a part of a name, a SMILES or a molecular formula, up to 10 candidate strings will be displayed for the user to choose from.
(iv) Statistics: the number of monomers in CoPolDB is shown, with the distribution of molecular weights and the distribution of the number of occurrences in copolymers.
3.2. Copolymers
This part allows you to search and compare the reactivity ratios of copolymerization. In order to avoid counting the same monomer pair twice or more, for each monomer pair, the monomer with a smaller molecular weight† is referred to as the first monomer M1, and that with a larger molecular weight is the second monomer M2, similarly the reactivity ratios being r1 and r2, respectively.(i) Overview: a chemical overview of radical polymerization, such as the principles of copolymerization and the structure of copolymers, is explained.
(ii) Browse: unlike in Monomers, there are two browse pages: Copolymers and Copolymerizations, which are switchable. The former shows a list of monomer pairs (Fig. 3b), each with a hyperlink to the detail page (Fig. 3d) of the corresponding copolymer, while the latter shows a list of copolymerizations, each also with a hyperlink to a detail page of the corresponding copolymer. The detail page has an interactive GUI (Fig. 4) to show and compare, for each monomer pair, various different reactivity ratios (from different references) by F1/f1 plots and scatter plots (upper left and right of Fig. 4, respectively), with the reported r1 and r2 pairs (lower half of Fig. 4) accessible to the original reference by clicking the corresponding DOI.
The reactivity ratios of monomers are crucial for the rational design and synthesis of copolymers with specific properties, since the reactivity ratios can vary due to a variety of factors, especially temperature. First, the different functional groups on the same monomer can have different reactivity ratios, due to the different reactivity towards the other monomer. Second, the environment of a reaction, such as temperature, pressure, solvent, and additives, all affects the reactivity ratios. In addition, the reactivity ratios can vary depending on the experimental method. Thus, even for the same monomer pair, various reactivity ratios can be obtained.
A F1/f1 plot is constructed by plotting the mole fraction of monomer 1, F1, in the copolymer against the mole fraction of that same monomer in the feed, f1. This is a well-used graphical tool for understanding copolymerization, more specifically, predicting the composition of the copolymer using the feed composition and then designing the process of copolymerization to have particular desired properties. This plot can also be used to compare the reactivity ratios of different monomer pairs, being useful for selecting particular monomers for copolymerization through the compatibility and expected copolymer properties. CoPolDB presents F1/f1 plots for all copolymerizations in the detail page to help users to predict copolymer properties and access experimental conditions of interest. Particularly, users can select one reactivity ratio among the various available, and check the information on the selected ratio, such as r1, r2 and the original paper, which provides this ratio, by clicking the corresponding DOI.
The detail page further presents – for a monomer of a copolymer – a list of monomers, each being able to be a replacement of the original monomer. Fig. 5 is an example detail page, showing the copolymer of styrene and methyl methacrylate: the replaceable monomers of styrene which are found in the copolymers of styrene, are sorted by the structural similarity to methyl methacrylate, and this is done in the same way for the methyl methacrylate side. Each monomer in this page has a hyperlink (at the number of reactions of the corresponding monomer), and by clicking the link, the detail page on the alternative copolymer appears.
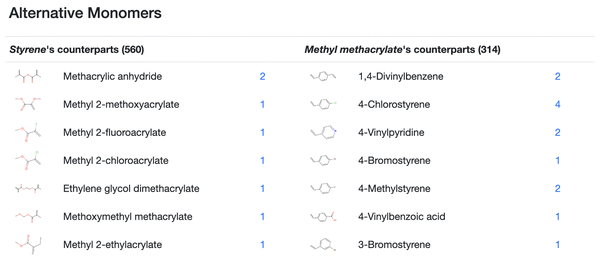 | ||
| Fig. 5 Enlarged Fig. 3d: for the styrene and methyl methacrylate pair, the alternative monomers for styrene and methyl methacrylate are listed on the left and right columns, respectively. The number of registered reactions of copolymerization, including the corresponding molecule is shown, and the number is linked to the corresponding copolymers detail page. For a pair of monomers (A and B), CoPolDB generates a list of the alternative monomers for A in two steps: (1) in the registered monomer pairs, searching for monomers, which can be a pair with B, and (2) sorting the found monomers, according to the structural similarity to A. | ||
(iii) Search: an interface to search for the copolymerization reactions through the input queries of monomer information, such as the molecular formula, the SMILES code, and the InChI key. The interface accepts a range of monomer reactivity ratios as an input query.
(iv) Statistics: the number of polymers, copolymerization reactions and references with DOIs are shown, with the distributions of the number of copolymerizations per copolymer and the r1 and r2 reactivity ratios.
3.3. User guide
The top right-hand side corner of all pages has a clickable question mark, through which users can visit a user guide, which carefully explains the usage of each page in CoPolDB.4. Conclusions
We have presented CoPolDB, the first DB with a proper, user-friendly GUI for copolymerization. All entries on copolymerization reactions in CoPolDB are retrieved from the published literature. CoPolDB has two parts – monomers and copolymers – where these two parts are connected through the copolymerization that a copolymer is synthesized from its building blocks, i.e. monomers. These three parts – i.e. two monomers, copolymers and copolymerizations – are graphically presented, and have a lot of functions to visually present the stored information, such as the F1/f1 plots for the multiple (different) reactivity ratios for the same monomer pair. Also, for each monomer of a monomer pair used for synthesizing the corresponding copolymer, alternative monomers, i.e. the similar monomers in terms of chemical structures, are presented. We believe that CoPolDB is useful for understanding copolymerization, particularly different reactivity ratios for the same monomer pair, and promoting the research in the relevant fields.As mentioned in the Introduction, there are already a variety of DBs in biology. Currently these DBs are important resources as input for data-driven approaches, such as statistics, data science and machine learning (ML), for biology. For example, a ML model is trained from the gene expression data of patients with a certain disorder, and the trained model is used for finding future patients or biomarkers relevant to the disorder. The data stored in CoPolDB also would be able to be used for the same direction of the research in polymer chemistry.
Currently, CoPolDB stores only data published in the literature, and there are no functions for users to input the data into the DB. In biology, experimentally identified gene sequences must be registered in a DB, such as GenBank, when published. This type of rule has promoted the increasing size of DBs in biology. Similarly, in the future, we can modify CoPolDB to allow users to register copolymerization results. Also, we will provide CoPolDB with a function to allow users to read access to the data of CoPolDB. This extension will allow users to use the CoPolDB data for further studies on their own computers, such as building a ML model for predicting reactivity ratios. In fact, we think that applying the built ML models to the data in CoPolDB would be useful to find and fix incorrect data hidden in the database.
Author contributions
The following roles were conducted by the authors: K. T., conceptualization, data curation, formal analysis, investigation, methodology, resources, software, validation, visualization, writing – original draft and writing – review & editing; H. M., conceptualization, investigation, methodology, project administration, software, supervision, validation, visualization, writing – original draft and writing – review & editing; M. T., data curation, formal analysis, investigation, resources, validation and writing – review & editing; N. Z., formal analysis, investigation, resources; S. Y., conceptualization, formal analysis, funding acquisition, investigation, project administration, resources, supervision, validation and writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors were in part supported by MEXT KAKENHI #21H05027.References
- G. Odian, Principles of Polymerization, John Wiley & Sons, Ltd, NJ, USA, 2004 Search PubMed.
- G. Moad and D. H. Solomon, The Chemistry of Radical Polymerization, Second Edition, Elsevier, Amsterdam, Netherlands, 2005 Search PubMed.
- P. Hiemenz and T. Lodge, Polymer Chemistry, Second Edition, Taylor & Francis, 2007 Search PubMed.
- P. Nesvadba, in Radical Polymerization in Industry, John Wiley & Sons, Ltd, 2012 Search PubMed.
- S. Yamago, Polym. J., 2021, 53, 1–18 CrossRef.
- Y. Lu, T. Nemoto, M. Tosaka and S. Yamago, Nat. Commun., 2017, 8, 1863 CrossRef PubMed.
- Y. Lu and S. Yamago, Angew. Chem., Int. Ed., 2019, 58, 3952–3956 CrossRef CAS PubMed.
- Y. Lu and S. Yamago, Macromolecules, 2020, 53, 3209–3216 CrossRef CAS.
- H. Kojima, Y. Imamura, Y. Lu, S. Yamago and T. Koga, Macromolecules, 2022, 55, 7932–7944 CrossRef CAS.
- S. Otsuka, I. Kuwajima, J. Hosoya, Y. Xu and M. Yamazaki, 2011 International Conference on Emerging Intelligent Data and Web Technologies, 2011, 22–29.
- J. Van Herck, S. Harrisson, R. A. Hutchinson, G. T. Russell and T. Junkers, Polym. Chem., 2021, 12, 3688–3692 RSC.
- Polymer handbook, ed. J. Brandrup, E. Immergut and E. Grulke, Wiley, New York, 4th edn, 1999 Search PubMed.
- Y. Wang, J. Xiao, T. O. Suzek, J. Zhang, J. Wang and S. H. Bryant, Nucleic Acids Res., 2009, 37, W623–W633 CrossRef CAS PubMed.
- K. Clark, I. Karsch-Mizrachi, D. J. Lipman, J. Ostell and E. W. Sayers, Nucleic Acids Res., 2016, 44, D67–D72 CrossRef CAS.
- T. Barrett, S. E. Wilhite, P. Ledoux, C. Evangelista, I. F. Kim, M. Tomashevsky, K. A. Marshall, K. H. Phillippy, P. M. Sherman, M. Holko, A. Yefanov, H. Lee, N. Zhang, C. L. Robertson, N. Serova, S. Davis and A. Soboleva, Nucleic Acids Res., 2012, 41, D991–D995 CrossRef.
- D. Weininger, J. Chem. Inf. Comput. Sci., 1988, 28, 31–36 CrossRef CAS.
- S. R. Heller, A. McNaught, I. Pletnev, S. Stein and D. Tchekhovskoi, J. Cheminf., 2015, 7, 23 Search PubMed.
- https://www.rdkit.org/ .
- N. M. O'Boyle, M. Banck, C. A. James, C. Morley, T. Vandermeersch and G. R. Hutchison, J. Cheminf., 2011, 3, 33 Search PubMed.
- N. Rego and D. Koes, Bioinformatics, 2015, 31, 1322–1324 CrossRef PubMed.
- S. Wang, J. Witek, G. A. Landrum and S. Riniker, J. Chem. Inf. Model., 2020, 60, 2044–2058 CrossRef CAS.
- D. Bajusz, A. Rácz and K. Héberger, J. Cheminf., 2015, 7, 20 Search PubMed.
- H. L. Morgan, J. Chem. Doc., 1965, 5, 107–113 CrossRef CAS.
Footnote |
| † If the molecular weights are the same, the order is determined based on molecular descriptor values, such as HeavyAtomCount and NumValenceElectrons, which are calculated by RDKit. |
| This journal is © The Royal Society of Chemistry 2024 |