Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Development of polyethyleneimine cross-linked fucoidan nanoparticles as delivery systems for improved anticancer efficiency of cytarabine in breast adenocarcinoma cell lines†
Deepa
Geethakumari
a,
Santhini Pulikkal
Veettil
ad,
Sivakumar Krishnankutty
Nair Chandrika
ab,
Anoop Bhaskaran
Sathyabhama
a,
Rojin
Joseph
a,
Shibin Sobhanam
Padmini
a,
Jisha V.
Somasekharan
 c and
Sajeevan
Thavarool Puthiyedathu
c and
Sajeevan
Thavarool Puthiyedathu
 *e
*e
aNational Centre for Aquatic Animal Health, Cochin University of Science and Technology, Cochin, Kerala-16, India
bRajiv Gandhi Centre for Biotechnology, Poojappura, Thiruvananthapuram, Kerala-695014, India
cResearch and Post Graduate Department of Chemistry, MES Keveeyam College, Valanchery-676552, Kerala, India
dDepartment of Applied Chemistry, Cochin University of Science and Technology, Cochin, Kerala-682022, India
eDepartment of Marine Biology, Microbiology and Biochemistry, Cochin University of Science and Technology, Cochin, Kerala-16, India. E-mail: sajeev@cusat.ac.in; Fax: +91484 2381120; Tel: +91-9946099408
First published on 19th March 2024
Abstract
Cytarabine, generally used for the treatment of haematological malignancies, has minimal activity in solid tumours. The present work focuses on the evaluation of the cytotoxic efficiency of cytarabine in MCF-7 cell lines with the aid of fucoidan nanoparticles as drug delivery systems. Polyethyleneimine (PEI) crosslinked fucoidan nanoparticles were synthesised by polyelectrolyte complexation and were characterised by FTIR and 1H NMR. TEM analysis revealed cytarabine-loaded fucoidan nanoparticles (CFNPs) with a size of less than 40 nm. In vitro release kinetics studies showed that the release of cytarabine (82.17 ± 1.24%) from CFNPs was higher at pH 6.4. Molecular simulation studies revealed that fucoidan–cytarabine binding is mainly facilitated by hydrogen bonds. Internalisation of fucoidan nanoparticles by MCF-7 cells was tracked using the fluorophore, SQ 650. Cell viability studies showed improved cytotoxicity in CFNP-treated MCF-7 cell lines compared to free cytarabine. Quantitative real-time PCR showed upregulation of the expression of apoptotic genes, bax, cyt c and cas 9 in cells treated with CFNPs. Flow cytometry using Annexin V/PI displayed an increased percentage of apoptotic cells on treatment with CFNPs compared to cytarabine alone. The result of this study shows that the cytotoxic efficiency of cytarabine in MCF-7 cells can be enhanced using fucoidan nanoparticles as delivery systems.
1. Introduction
Cytarabine (1β-arabinofuranosylcytosine, Ara-C), a synthetic pyrimidine nucleoside analogue approved by the FDA for the treatment of myeloid leukaemia, non-Hodgkin's lymphoma and meningeal leukaemia, was originally isolated from a Caribbean sponge Tectitethya crypta.1 Cytarabine inhibits cell proliferation by competing with deoxycytidine triphosphate and inhibits DNA synthesis by inhibiting DNA polymerase. It inhibits the S phase of the cell cycle. It has been reported that cytarabine shows little cytotoxic activity in solid tumours due to the high expression of the enzyme cytidine deaminase that causes cytarabine degradation.2 However, it showed synergistic effects against solid tumours in combination with other anticancer drugs.3 Cytarabine is rapidly deaminated to a biologically inactive uracil derivative that necessitates continuous infusion for the treatment. The development of drug resistance and toxic side effects are reported due to the overdosing of cytarabine.4 The limitations of cytarabine such as its short plasma half-life, low bioavailability, drug resistance and side effects due to overdosing can be overcome by nanoencapsulation strategies.5Fucoidan is a marine heterogeneous sulphated polysaccharide commonly isolated from brown seaweed and certain marine invertebrates. Fucoidans isolated from marine algae are more complex in structure than marine invertebrates, comprised of repeating units of fucosyl disaccharides substituted with sulphates or uronic acids. The structure and composition of fucoidan may vary with the type of extraction source. The biological properties of fucoidan depend on the molecular weight, degree of sulphation, molecular structure and sugar content, which are related to the source and method of extraction. Fucoidan is reported as a pH-sensitive polymer due to the presence of anionic sulphate groups. Fucoidan has been extensively studied for biomedical applications such as drug delivery, tissue engineering and diagnostic purposes. Fucoidan is reported as a P-selectin binding ligand due to the presence of sulphate moieties. P-selectin is a vascular adhesion molecule expressed on many metastatic tumour cells such as lung, breast and kidney cancers in humans.6 It plays a major role in tumour metastasis through the promotion of angiogenesis by facilitating endothelial cell migration.7 Studies have shown an enhanced expression of P-selectin on the tumour periphery of breast tumours.8 Inhibition of P-selectin mediated platelet adhesion with ligands expressed on tumour cells attenuates cancer metastasis.9
Fucoidan nanoparticles and fucoidan-based nanostructures are reported as ideal carriers for pH-sensitive drug delivery, diagnostics and regenerative medicine.10 These can be synthesised by various methods such as polyelectrolyte complexation, conjugation and functionalization. The most common and simple method is polyelectrolyte complexation in which anionic fucoidan is allowed to complex with positively charged molecules such as chitosan, protamine, polyethyleneimine, etc.11 Polymeric drug delivery vehicles offer controlled drug release, which improves the efficacy of the desired drug.12 The present study focuses on fucoidan nanoparticles as carrier systems for cytarabine to enhance the bioavailability and thereby reduce the toxic side effects, especially on non-targeted sites. The human breast adenocarcinoma cell lines, MCF-7, were chosen as a model system to evaluate the improvement in the anticancer efficiency of cytarabine-loaded PEI cross-linked fucoidan nanoparticles (CFNPs) (Fig. 1).
2. Materials and methods
2.1 Materials
Fucoidan (isolated from Fucus vesiculosus), polyethyleneimine (branched; Mw ∼1300), cytarabine and 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazoliumbromide (MTT) were purchased from Sigma Aldrich. The reagents used in the study were of analytical grade. MCF-7 and Vero cell lines used for the study were purchased from NCCS, Pune, India, and cultured in DMEM/F-12 media supplemented with 10% FBS (Gibco, India).2.2 Synthesis and characterisation of PEI crosslinked fucoidan nanoparticles
Fucoidan nanoparticles (FuNPs) were synthesised by polyelectrolyte complexation. Fucoidan (Fu) and PEI were taken at a concentration ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Branched PEI with a concentration of 1% (w/v) was taken and the pH was adjusted to 7 using 0.1M HCl and was allowed to stir. Fucoidan (1%) was added dropwise to PEI under stirring. The resultant mixture was further allowed to stir for 3 hours and then centrifuged at 15
1. Branched PEI with a concentration of 1% (w/v) was taken and the pH was adjusted to 7 using 0.1M HCl and was allowed to stir. Fucoidan (1%) was added dropwise to PEI under stirring. The resultant mixture was further allowed to stir for 3 hours and then centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min. The precipitate was collected, followed by dialysis against distilled water for 72 hours and freeze-dried. The freeze-dried powder was used for characterisation. Cytarabine was loaded to fucoidan nanoparticles (CFNPs) during the synthesis before the addition of fucoidan solution.
000 rpm for 30 min. The precipitate was collected, followed by dialysis against distilled water for 72 hours and freeze-dried. The freeze-dried powder was used for characterisation. Cytarabine was loaded to fucoidan nanoparticles (CFNPs) during the synthesis before the addition of fucoidan solution.
2.3 Characterisation of fucoidan nanoparticles
Fucoidan nanoparticles synthesised by the polyelectrolyte complexation method were characterised by FTIR and 1H NMR. FTIR-ATR spectroscopy was carried out to characterise the Fu–PEI complex using the FTIR-ATR model IR Prestige-21 Shimadzu spectrophotometer. The spectra of fucoidan, PEI and FuNPs were recorded at 4 cm−1 resolution with 20 scans. The absorbance bands were measured as a function of wave numbers between 400 and 4000 cm−1. Pure fucoidan and the fucoidan–PEI complex were dissolved in methanol-d4, and 1H NMR spectra were recorded using a Bruker AMX500 spectrometer. The chemical shifts δ are given in ppm and referenced to the external standard TMS or an internal solvent standard. Crystallinity and thermal stability of fucoidan nanoparticles were evaluated by XRD and thermogravimetric analysis, respectively.2.4 Encapsulation and loading efficiencies
The encapsulation and loading efficiencies of CFNPs were determined by the indirect method through quantification of the unencapsulated cytarabine in the supernatant after centrifugation of the reaction mixture. The amount of cytarabine in the supernatant was quantified by taking absorbance at 277 nm. The encapsulation and loading efficiencies of CFNPs were calculated using the following formula:| Loading efficiency = W ÷ Wf × 100 | (1) |
| Encapsulation efficiency = W ÷ Wc × 100 | (2) |
where W is the weight of encapsulated cytarabine, Wc is the initial weight of cytarabine, and Wf is the weight of fucoidan nanoparticles.
2.5 Morphology, hydrodynamic diameter and zeta potential
Dynamic light scattering (DLS) based on photon correlation spectroscopy (Horiba Scientific, Nanopartica, Nanoparticle analyser SZ – 100, Japan) was used to study the hydrodynamic diameter, polydispersity index (PDI) and zeta potential of CFNPs. A known concentration of CFNPs was dispersed in Milli-Q water and analyzed at 25° C with a scattering angle of 90°.2.6 In vitro release kinetics
The release kinetics of cytarabine from CFNPs was evaluated at pH 6.4 and 7.4 equivalent to the pH of the tumour and blood. CFNPs (1 mg ml−1) were dispersed in phosphate buffer of pH equivalent to the tumour and blood i.e., 6.4 and 7.4 and dialysed (MW cut off – 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da) at 37 °C for 72 hours. At definite intervals, 1 ml of the sample was withdrawn to determine released cytarabine and replaced with an equal volume of fresh buffer. The released cytarabine from the nanoformulation was quantified using spectrophotometry by taking absorbance at 277 nm.
000 Da) at 37 °C for 72 hours. At definite intervals, 1 ml of the sample was withdrawn to determine released cytarabine and replaced with an equal volume of fresh buffer. The released cytarabine from the nanoformulation was quantified using spectrophotometry by taking absorbance at 277 nm.
2.7 Hemocompatibility
The hemolysis assay was conducted to study the hemocompatibility of the synthesised fucoidan nanoparticles.13 The RBCs were isolated by centrifuging anticoagulated blood at 800 rpm for 5 min and diluted with PBS. The diluted RBCs were treated with fucoidan nanoparticles at concentrations of 50, 100, 250, 500 and 1000 μg at 37 °C for 3 h. Phosphate buffered saline (PBS) and distilled water were taken as the negative and positive control, respectively. The samples were centrifuged at 800 rpm for 5 min. The haemoglobin released due to hemolysis in the supernatant showed absorbance at 541 nm on a UV-visible spectrophotometer. The percentage of haemolysis was determined using eqn (3).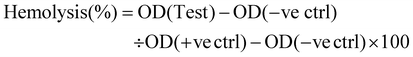 | (3) |
RBC aggregation was studied by incubating 100 μl of diluted RBCs with 100 μl of nanoparticle suspension (1000 μg) for 1 h at 37° C and visualized under a microscope (Leica, Germany) at a magnification of 40×.
2.8 Cell uptake studies
Intracellular uptake of the synthesised nanoparticles in MCF-7 cell lines was studied using the fluorophore, Squaraine 650 (SQ 650).14 SQ 650 was loaded into the fucoidan nanoparticles during the polyelectrolyte complexation. Cells were treated with SQ 650 loaded FuNPs for time intervals of 2, 4, 6 and 12 h and washed with ice-cold PBS thrice and visualised under a fluorescence microscope (Leica, Switzerland).2.9 Cytotoxicity assay
The cytotoxicity of CFNPs was studied in MCF-7 (tumour) and Vero (non-cancerous) cell lines by the MTT assay.15 Cells grown in DMEM with 10% FBS were seeded at a density of 1 × 104 cells per well for the MTT assay. Both cells were treated with different concentrations of cytarabine and CFNPs (0.025–0.4 μM) for 72 h. 20 μl MTT (5 mg ml−1) was added to each well after the treatment period following incubation in the dark for 4 h at 37° C. 200 μl DMSO was added to each well to dissolve formazan crystals. The absorbance was measured at 570 nm using a microplate reader (TECAN Infinite M 200, Austria). The percentage of relative cell viability with respect to the control cells (i.e., cells without treatment) was evaluated.2.10 Acridine orange/ethidium bromide (AO/EB) differential staining
Acridine orange/ethidium bromide (AO/EB) differential staining was used to study the morphological changes associated with apoptosis in CFNP-treated MCF-7 cell lines. Cells were cultured at a density of 1 × 106 cells per well in a 24 well plate and treated with IC50 of CFNPs for 72 h and the same concentration of cytarabine. Cells were washed thrice with PBS after incubation, stained with AO/EB and kept for 1 min. Live and apoptotic cells were visualised under a fluorescence microscope (Leica, Switzerland) with an excitation filter at 480 nm.2.11 Annexin V assay
Annexin V assay with the aid of flow cytometry was performed to quantify the apoptosis induced by cytarabine as well as CFNPs. MCF-7 cells were incubated with the same concentration (0.05 μM) of CFNPs and cytarabine for 72 h. Cells treated with blank nanoparticles i.e. without drug were taken as the control. Precisely 100 μL of the Muse™ Annexin V & dead cell reagent was added to the cell suspension of treatment groups and the control. The tubes were mixed well and incubated for 20 min at room temperature in the dark. After incubation, the cells were analysed in a flow cytometer using Muse flow cytometry software, Muse FCS 3.0 software.2.12 Differential gene expression
The expressions of genes regulating apoptosis, bcl-2, bax, cyt c and cas 9, were evaluated by quantitative real-time PCR. RNA was isolated from the control, cytarabine, and CFNP-treated MCF-7 cells following 72 h incubation. cDNA was synthesised from the isolated RNA and quantitative real-time PCR analysis was performed in a Step OnePlus Real-Time PCR system (Applied Biosystems, USA) using the Power SYBR Green PCR Master Mix (Applied Biosystems, UK) with reported primers.16,17 The ΔCT method was used to study the expression of genes involved in cell proliferation and normalised using an endogenous housekeeping gene, GAPDH. The conditions and sequences of the primers for selected genes were the same as those reported in previous studies18 and the 2−ΔΔ CT method was employed for the quantification of relative gene expression.192.13 In silico studies
The molecular dynamic simulation was used to investigate the encapsulation of cytarabine (CBR) by fucoidan (FDN) oligosaccharide, as well as its release at pH (6.4 and7.4). The fucoidan oligomer, which is made up of six monomer units arranged at random, was chosen. The CBR molecule was able to move freely around inside the FDN oligomer. As a result, the all-atom system consisted of the FDN oligomer while the encapsulation of CBR prior to its release from the FDR inner region at pH 6.4 and 7.4 was investigated. The PubChem database was used to obtain the structures of CBR (CID: 6253) and FDR (CID: 129532628). The PRODRG2 server20 was used to acquire FDR and CBR force field parameters. The United-atom force field GROMOS 53a6 was used to simulate atomic interactions in GROMACS v5.1.4.21 CBR was aligned at the core of 6 FDR repeating units as an initial configuration and then spaced suitably far apart (>3 nm) to minimise the impact of starting orientations. The system was solvated utilising SPC216 water molecules, and the energy was lowered using the steepest descent algorithm. The temperature was kept constant at 323 K using a Nosé–Hoover thermostat throughout the experiment.22 A constant pressure of 1 bar was maintained using the Parrinello–Rahman barostat.23 The Ewald method24 was used to calculate long-range electrostatic interactions of the particle mesh with a cut-off of 1.0 nm, although electrostatic interactions between charged groups below 0.9 nm were computed explicitly. By using Lennard-Jones potential, a smooth 0.9 nm cut-off for van der Waals interactions was computed. In both equilibration and production runs, the LINCS algorithm25 was used to limit all bond lengths, including those to hydrogen atoms, that used a time step of 2 fs for the integration of the motion equation. The water topology, on the other hand, was constrained using SETTLE.26 After energy minimisation to remove proximal contacts where CBR, FDN, and water molecules could relax, all of the atoms in the systems were equilibrated under NPT conditions for 100 ps. The simulations were run for 1.5 ns following reaching equilibrium, with coordinates stored every 100 ps.2.14 Statistical analysis
All experiments were carried out at least in triplicate, and the results are expressed as mean ± SD. One-way ANOVA followed by Tukey's post-hoc analysis using SPSS software (version 20) was used to express the statistical significance, and a p-value <0.05 indicated a statistically significant difference.3. Results and discussion
3.1 Synthesis and characterisation of fucoidan nanoparticles
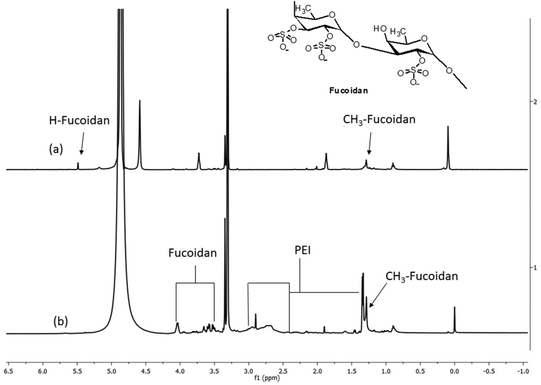
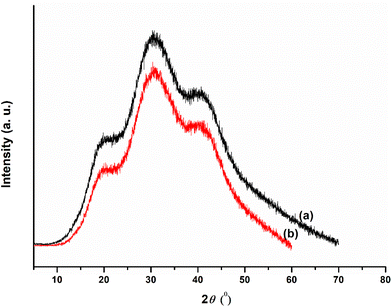
The formation of the fucoidan–PEI nanocomplex was confirmed by IR spectroscopy (Fig. 2). Fucoidan shows characteristic peaks at 823, 1014, 1219, 1382 (stretching frequency of sulphate), 1645 (asymmetrical stretching band of carboxylate ion), 2947 and 3417 cm−1. The PEI shows peaks at 1045, 1471, 1633 (N–H bending vibration of primary amine), 2845, 2956 and 3331 (N–H stretching vibration of primary amine) cm−1. The spectrum of FuNPs shows both peaks of fucoidan and PEI. The band at 1392 cm−1 of FuNPs shows the stretching frequency of sulphate, and the shift from native fucoidan is due to the ionic interaction with PEI. The shift in the peak at 1645 of fucoidan to a shorter wavelength, 1641 cm−1 in FuNPs might be due to hydrogen bonding. The structure of PEI-fucoidan nanoparticle was further confirmed by 1H NMR analysis (Fig. 3). The anomeric proton of fucoidan appeared at 5.51 ppm (H-1 of the fucose ring) and other protons in the range of 4.00–3.00 ppm. The methyl protons of fucoidan appeared at 1.25 ppm. Along with the peaks of fucoidan, characteristic peaks of PEI were observed on the 1H NMR spectra of fucoidan PEI nanoparticles. This indicates that PEI was successfully incorporated into the fucoidan. Crystallinity and thermal stability of fucoidan nanoparticles were studied by XRD and Thermo Gravimetric Analysis (TGA), respectively. The XRD pattern of FCD/PEI NP shows the same spectral pattern of pure fucoidan, indicating the amorphous nature of this material (Fig. 4). The results of XRD show that the synthesised FuNPs can form an amorphous solid dispersion. The TGA curve of the pure fucoidan appears to be with a weight loss of about 3–5% until 110 °C. This could be attributed to dehydration. No major weight loss was observed until 220 °C. Next, a decrease in the weight of 20–25% has occurred slowly between 250 and 500 °C. A similar trend in weight loss of around 10% is observed up to 100 °C in the case of PEI-fucoidan nanoparticle, which is attributed to the removal of moisture or residual solvent in the nanoparticle. The weight loss substantially increases up to 25% at 270 °C and then loses up to 40% between 300 and 600 °C. The results (ESI Fig. 1†) clearly indicate that the nanoparticles can sustain up to higher temperatures. The shift in the final decomposition temperature for weight loss of PEI-fucoidan nanoparticles from pure fucoidan illustrates that the thermal stability enhances when PEI is added to pure fucoidan.3.2 Characterisation of cytarabine loaded fucoidan nanoparticles (CFNPs)
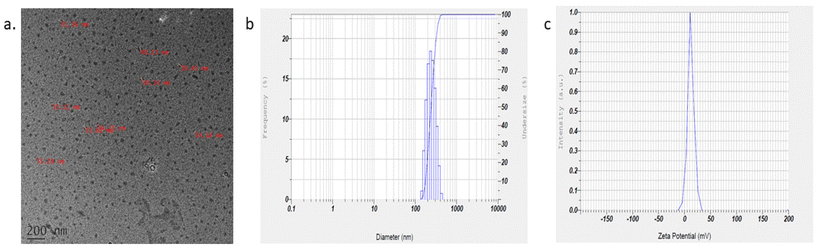
Cytarabine encapsulated fucoidan nanoparticles were synthesised by the polyelectrolyte complexation method. The encapsulation and loading efficiencies of CFNPs were 76.72 ± 1.08 and 48.04 ± 0.867%, respectively. TEM images of CFNPs show roughly spherical particles of size less than 40 nm (Fig. 5a). The hydrodynamic diameter, distribution and surface charge of nanoparticles were studied by DLS based on photon correlation spectroscopy. The mean hydrodynamic diameter obtained from DLS was 283 nm with a polydispersity index of 0.587 and a zeta potential of +12.2 mV (Fig. 5b & c). The zeta potential of unencapsulated fucoidan nanoparticles showed −80 mV (ESI Fig. 3†) The negative charge of the fucoidan nanoparticles are found to decrease after drug loading. The size obtained from DLS is bigger than that obtained from TEM. The nanoparticles observed by TEM analysis are very small compared to the hydrodynamic diameter obtained by DLS. The particle size ranging between 50 and 250 nm is ideal for cancer therapy as it significantly contributes to cell internalisation through the Enhanced Permeability and Retention (EPR) effect.27 It is reported that particles of size around 200 nm were studied more effectively for targeting.283.3 In vitro release studies
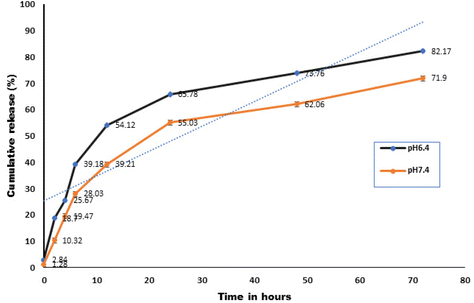
The drug release profile of CFNPs was studied in phosphate buffer of pH 6.4 and 7.4 to mimic the tumour microenvironment and blood, respectively. The release profile of cytarabine from CFNPs at pH 6.4 and 7.4 is shown in Fig. 6. It has been observed that 82.17 ± 1.24% of cytarabine was released at pH 6.4, whereas 71.9 ± 0.89% of the drug was released at pH 7.4. This release shows a biphasic pattern as observed by chitosan nanoparticles.18,29 After 12 h, more than 50% of the drug was released at lower pH. The results implicate that the release of cytarabine was higher at lower pH when compared to the physiological pH, 7.4. The drug release profile of fucoidan nanoparticle formulation showed controlled release of cytarabine. The results showed that around 50% of the drug was in the carrier up to 24 h at pH 7.4, the pH under normal physiological conditions. It indicates the potential of prolonged drug retention time in blood and also reduces the side effects on normal tissues. The nonspecific electrostatic interactions in the polyelectrolyte complex shall be affected by the change in pH.30 It has been reported that the drugs undergo protonation at lower pH and cause faster release into the medium.31 The drug release profile studies were further validated by molecular simulation experiments.3.4 Hemocompatibility studies
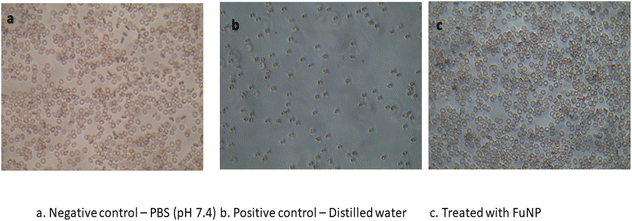
A hemolysis assay was done to evaluate the compatibility of fucoidan nanoparticles towards the blood cells as hemolysis leads to anaemia, jaundice and other pathological conditions. The assay determines the extent of hemolysis caused by the nanoparticles from the concentration of haemoglobin released when the blood is exposed to the nanoformulation. The percentage of hemolysis was calculated after the incubation of RBCs with fucoidan nanoparticles with concentrations of 5, 10, 25, 50 and 100 μg ml−1. The obtained values were less than 2% in all the concentrations. Results of the hemolysis assay show that the synthesised fucoidan nanoparticles are non-hemolytic (Table 1). Fig. 7 illustrates the morphology of RBCs after treatment with the highest concentration of FuNPs (100 μg ml−1) along with the negative (PBS) and positive control (distilled water). Hemolytic activity was assessed according to ASTM E2524-08 (ASTM E2524-08, 2013. The standard test method for analysis of the hemolytic properties of nanoparticles. ASTM Int, West Conshohocken, PA).32| Concentration of FuNPs (μg ml−1) | Hemolysis (%) ± SD |
|---|---|
| 5 | 0.12 ± 0.004 |
| 10 | 0.17 ± 0.0037 |
| 25 | 0.19 ± 0.005 |
| 50 | 0.23 ± 0.009 |
| 100 | 0.45 ± 0.016 |
3.5 Cellular uptake studies
Cell internalisation of fucoidan nanoparticles was studied using the fluorophore, Squarine 650 in MCF-7 cell lines. Cell uptake of the particles was studied at different time intervals (2, 4, 6 and 12 h) and qualitatively visualised by fluorescence microscopy (Fig. 8a). The relative fluorescence intensity was measured at each time interval using a microplate reader (Fig. 8b). The intensity of fluorescence was found to increase with the increase in time. As shown in the figure, the intensity was low in the initial two hours, the nucleus showed little fluorescence, and the intensity was increased after 4 h. The intensity of fluorescence was found to increase with increasing time. The results show that the uptake of dye loaded nanoparticles is time dependent, and increased linearly with the incubation time. Enhanced permeability and retention (EPR) effect is one of the reasons for cell internalization of the dye loaded nano-sized FuNPs (Fig. 8).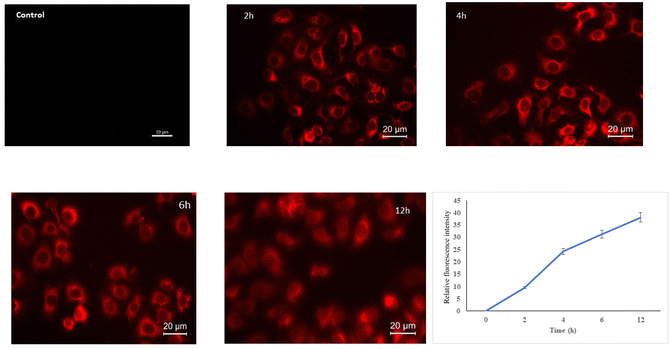 | ||
| Fig. 8 Cell uptake of Squaraine 650 (SQ650) loaded FuNPs in MCF-7 cells by fluorescence microscopy at different time intervals (2nd, 4th, 6th and 12th hours). | ||
3.6 Cytotoxicity studies
Cytotoxicity of CFNPs was studied in MCF-7 (carcinoma) and Vero (non-cancerous) cell lines by the MTT assay. Cells were treated with cytarabine and CFNPs at a concentration of 0.025–0.4 μM for 72 h, and the results of the MTT assay are shown in Fig. 9. The obtained IC50 value for CFNPs was 0.049 μM, whereas 0.329 μM for cytarabine in MCF-7 cell lines. The cell viability was found to decrease in a dose-dependent manner, and cytotoxicity was higher for CFNP-treated cells compared to cells treated with unencapsulated cytarabine. African green monkey kidney epithelial cells (Vero) were used to study the cytotoxicity of CFNPs in normal cells. The IC50 value of CFNPs and cytarabine could not be calculated for Vero cells as the cell viability was above 80% for the studied concentration range (ESI Fig. 4†). The results imply that the nanoformulation and cytarabine of the above concentration do not show significant toxicity in non-cancerous cells.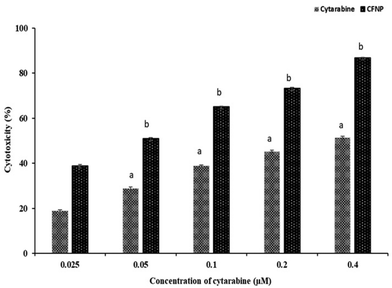 | ||
| Fig. 9 Cell viability studies of cytarabine and CFNPs in MCF-7 cell lines by the MTT assay. Results are expressed as mean + SD (p value ≤ 0.05). | ||
3.7 Acridine orange/ethidium bromide staining for apoptosis
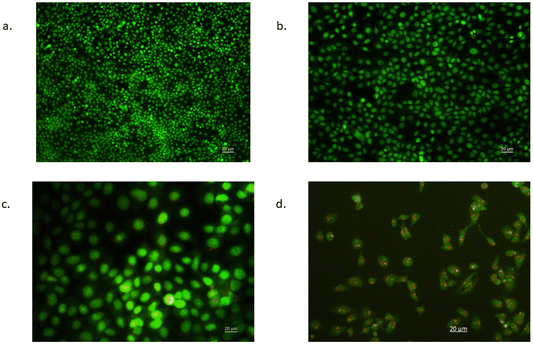
Morphological changes associated with CFNP induced apoptosis were observed in fluorescence microscopy by acridine orange/ethidium bromide dual staining. Fig. 10 shows the differential staining of CFNP and cytarabine treated MCF-7 cells along with control cells and cells treated with nanoparticles without cytarabine. A bright green nucleus was observed in control cells, indicative of the presence of live cells as acridine orange penetrates the intact cell membrane of viable cells. CFNP-treated cells show an orange, and red nucleus indicating apoptotic cells. The membrane blebbing and shrinking during apoptosis lead to the entry of ethidium bromide, producing a red colour. Cells treated with blank nanoparticles also exhibited bright green nuclei as in control cells. Acridine orange enters all cells and stains the nuclei green, whereas ethidium bromide is taken up by apoptotic cells due to loss of integrity of the cell membrane integrity and makes the nuclei of apoptotic cells appear orange-red.183.8 Annexin V assay
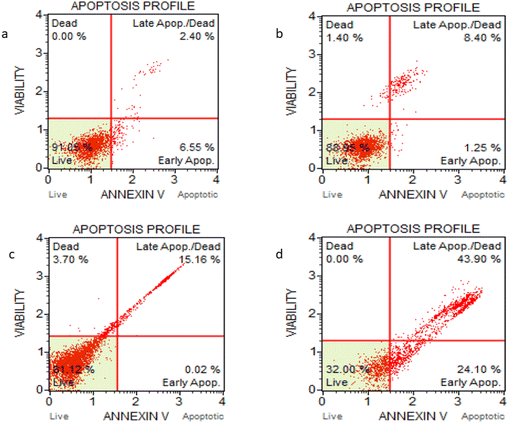
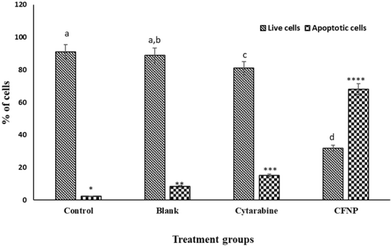
Annexin V assay with the aid of a flow cytometer was used to determine the percentage of viable, apoptotic and dead cells. MCF-7 cells treated with cytarabine and CFNPs of the same concentration (0.05 μM) were subjected to flow cytometry to quantify apoptosis. Cells treated with CFNPs showed 68% apoptotic cells, whereas cells treated with the same concentration of cytarabine showed 18.86% apoptotic cells (Fig. 11). The population profile of the cells obtained from flow cytometry is shown in Fig. 12. Flow cytometry results show that the apoptotic rates were significantly higher in the CFNP-treated cells than in those treated with unencapsulated cytarabine.3.9 Differential gene expression
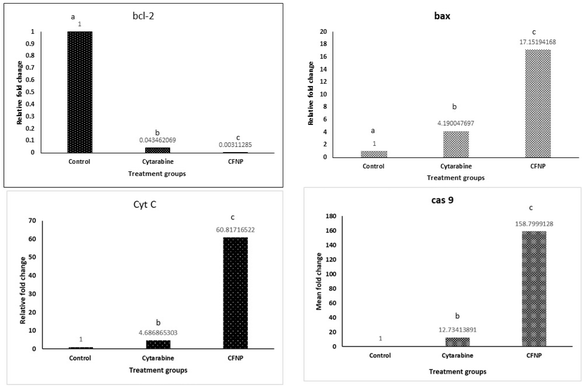
The effect of CFNPs and cytarabine on the mRNA expression of selected genes involved in apoptosis, such as bcl-2, bax, cyt c and cas 9, were analysed by quantitative real-time PCR. Fig. 13 illustrates the expression of bcl-2, bax, cyt c and cas 9 in MCF-7 cells treated with CFNPs as well as free cytarabine. The results of RT-PCR analysis showed that the expression of bcl-2 declined in cells treated with free cytarabine as well as CFNPs compared to control cells, whereas bax expression was significantly increased. The bax/bcl-2 ratio showed a 57-fold increase in CFNP-treated cells compared to cells treated with unencapsulated cytarabine. The bax expression showed a 4-fold enhancement, which in turn enhanced the expression of cyt c as well as cas 9 in cells treated with CFNPs compared to free cytarabine (p < 0.05). CFNP-treated cells showed a significant decrease in the expression of bcl-2 gene, whereas the bax gene expression was enhanced. The downregulation of antiapoptotic bcl-2 expression and the upregulation of proapoptotic bax expression lead to the induction of apoptosis. Similar reports are observed for the drug sulforaphane while encapsulated in the mPEG-PCL copolymer for delivery in MCF-7 cell lines.33 It has been demonstrated that the overexpression of bax leads to the induction of apoptosis through the release of cytochrome c and caspase activation.343.10 Molecular docking and simulation studies
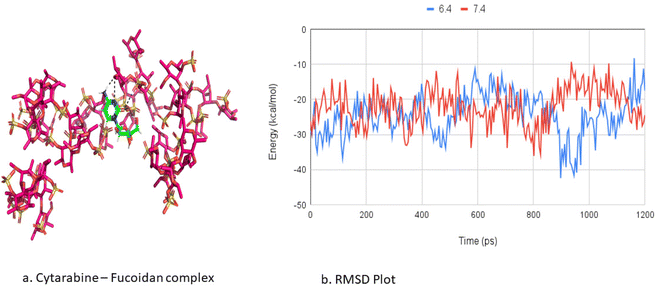
Molecular dynamics simulations were used to investigate the aggregation of cytarabine (CBR) and fucoidan (FDR) and their interactions as well as the pattern of drug release at pH 6.4 and 7.4.The binding free energies of cytarabine with fucoidan complexes were calculated using the MM/PBSA method. Table 2 lists the projected binding free energies (ΔG) of cytarabine at pH 6.4 and 7.4. A robust electrostatic interaction, van der Waals interaction, and solvent-accessible energy interactions favoured a better complex formation at pH 7.4. In contrast, a more positive polar energy contribution at pH 6.4 opposes complex formation and leads to faster cytarabine release, as shown by a significant increase in solvent-accessible energy. Molecular simulation studies support the pH-responsive targeting of the tumour with the aid of the fucoidan polymer.
| Interactions | pH 7.4 | pH 6.4 |
|---|---|---|
| Electrostatic interactions | –1224 ± 0.87 | –1295 ± 0.91 |
| van der Waals interactions | –148 ± 0.02 | –149 ± 0.01 |
| Polar energy | 1224 ± 0.81 | 1321 ± 0.84 |
| Non-polar energy | –17 ± 0.29 | –17 ± 0.31 |
| Solvent accessible energy | –81 ± 1.49 | –96 ± 1.49 |
| Binding energy (ΔG) | –187 ± 1.52 | –145 ± 1.52 |
4. Conclusion
Cytarabine-loaded fucoidan nanoparticles (CFNPs) were synthesised by the polyelectrolyte complexation method. The synthesised particles showed a uniform size distribution, with nanosized particles showing a pH-sensitive drug release profile. The molecular interactions between cytarabine and fucoidan were confirmed by FTIR. Molecular docking studies were used to validate the interactions between fucoidan and cytarabine. The results of the MTT assay showed that CFNPs exhibited dose-dependent cytotoxicity in MCF-7 cell lines, whereas little toxicity was observed in Vero cell lines. The Annexin V assay by flow cytometry showed that apoptosis increased while encapsulating cytarabine in fucoidan nanoparticles. The expression pattern of apoptotic genes from real-time PCR showed that apoptosis was triggered by an intrinsic pathway in MCF-7 cells after treatment with CFNPs. The above study demonstrated that the developed fucoidan nanoparticles can be used to improve the therapeutic effect of cytarabine in MCF-7 cell lines.Author contributions
Deepa Geethakumari: conceptualization, methodology, formal analysis, data curation, investigation, and writing – original draft. Santhini Pulikkal Veettil: methodology, formal analysis, investigation, and data curation. Sivakumar Krishnankutty Nair Chandrika: methodology, formal analysis, investigation, and data curation. Anoop Bhaskaran Sathyabhama: methodology, formal analysis, and data curation. Rojin Joseph: methodology, formal analysis, and investigation. Shibin Sobhanam Padmini: methodology, formal analysis, and investigation. Jisha V Somasekharan: methodology and editing. Sajeevan Thavarool Puthiyedathu: conceptualization, methodology, writing – review & editing, supervision, and project administration.Declaration of originality
The authors declare that the submitted work has not previously been published in full and is not being considered for publication elsewhere. Publication of abstracts and presentations at scientific meetings will not jeopardise full publication.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
The first author is thankful to the University Grants Commission (UGC), Government of India, for the BSR fellowship. Santhini Pulikkal Veettil thanks DST-SERB, India (PDF/2021/003145) for the research fellowship. The authors acknowledge the facilities extended by the Sophisticated Test and Instrumentation Centre (STIC) and the National Centre for Aquatic Animal Health, Cochin University of Science and Technology.References
- X. Thomas, Chemotherapy of acute leukemia in adults, Expert Opin. Pharmacother., 2009, 10(2), 221–237 CrossRef CAS PubMed . Available from: https://www.tandfonline.com/doi/full/10.1517/14656560802618746.
- H. Kojima, M. Iida, H. Miyazaki, T. Koga, H. Moriyama and Y. Manome, Enhancement of cytarabine sensitivity in squamous cell carcinoma cell line transfected with deoxycytidine kinase, Arch. Otolaryngol. Head Neck Surg., 2002, 128(6), 708–713 CrossRef PubMed.
- R. Liu, J. Zhang, D. Zhang, K. Wang and Y. Luan, Self-assembling nanoparticles based on cytarabine prodrug for enhanced leukemia treatment, J. Mol. Liq., 2018, 251, 178–184 CrossRef CAS . Available from: https://linkinghub.elsevier.com/retrieve/pii/S0167732217352893.
- B. S. Chhikara and K. Parang, Development of cytarabine prodrugs and delivery systems for leukemia treatment, Expert Opin. Drug Delivery, 2010, 7(12), 1399–1414 CrossRef CAS PubMed . Available from: https://www.ncbi.nlm.nih.gov/pubmed/20964588.
- J. Liu, D. Zhao, W. He, H. Zhang, Z. Li and Y. Luan, Nanoassemblies from amphiphilic cytarabine prodrug for leukemia targeted therapy, J. Colloid Interface Sci., 2017, 487, 239–249 CrossRef CAS PubMed.
- Y. Shamay, M. Elkabets, H. Li, J. Shah, S. Brook and F. Wang, et al. P-selectin is a nanotherapeutic delivery target in the tumor microenvironment, Sci. Transl. Med., 2016, 8(345), 345ra87 Search PubMed.
- C. L. Qi, B. Wei, J. Ye, L. B. Yang and Q. Q. Zhang, et al. P-selectin-mediated platelet adhesion promotes the metastasis of murine melanoma cells, PLoS One, 2014, 9(3), 1–7 Search PubMed.
- S. P. Gunningham, M. J. Currie, H. R. Morrin, E. Y. Tan, H. Turley, G. U. Dachs AIW, C. Frampton, B. A. Robinson and S. B. Fox, The angiogenic factor thymidine phosphorylase up-regulates the cell adhesion molecule P-selectin in human vascular endothelial cells and is associated with P-selectin expression in breast cancers, J. Pathol., 2007, 212(3), 335–344, DOI:10.1002/path.2174 . Available.
- H. Läubli and L. Borsig, Selectins promote tumor metastasis, Semin. Cancer Biol., 2010, 20(3), 169–177 CrossRef PubMed.
- C. Oliveira, N. M. Neves, R. L. Reis, A. Martins and T. H. Silva, A review on fucoidan antitumor strategies: From a biological active agent to a structural component of fucoidan-based systems, Carbohydrate Polymers. Elsevier Ltd, 2020, vol. 239 Search PubMed.
- P. H. L. Tran, W. Duan and T. T. D. Tran, Fucoidan-based nanostructures: A focus on its combination with chitosan and the surface functionalization of metallic nanoparticles for drug delivery, International Journal of Pharmaceutics. Elsevier B.V., 2020, vol. 575 Search PubMed.
- B. Joseph, A. George, S. Gopi, N. Kalarikkal and S. Thomas, Polymer sutures for simultaneous wound healing and drug delivery – A review, International Journal of Pharmaceutics. Elsevier B.V., 2017, vol. 524, pp. 454–466 Search PubMed.
- R. Augustine, S. K. Nethi, N. Kalarikkal, S. Thomas and C. R. Patra, Electrospun polycaprolactone (PCL) scaffolds embedded with europium hydroxide nanorods (EHNs) with enhanced vascularization and cell proliferation for tissue engineering applications, J. Mater. Chem. B, 2017, 5(24), 4660–4672 RSC.
- L. Li, D. Patil, G. Petruncio, K. K. Harnden, J. v. Somasekharan and M. Paige, et al. Integration of Multitargeted Polymer-Based Contrast Agents with Photoacoustic Computed Tomography: An Imaging Technique to Visualize Breast Cancer Intratumor Heterogeneity, ACS Nano, 2021, 15(2), 2413–2427 CrossRef CAS PubMed.
- M. Dhaneesha, O. Hasin, K. C. Sivakumar, R. Ravinesh, C. B. Naman and S. Carmeli, et al. DNA Binding and Molecular Dynamic Studies of Polycyclic Tetramate Macrolactams (PTM) with Potential Anticancer Activity Isolated from a Sponge-Associated Streptomyces zhaozhouensis subsp. mycale subsp. nov, Mar. Biotechnol., 2018, 21(1), 124–137 CrossRef PubMed.
- Y. Asara, J. A. Marchal, E. Carrasco, H. Boulaiz, G. Solinas and P. Bandiera, et al. Cadmium modifies the cell cycle and apoptotic profiles of human breast cancer cells treated with 5-fluorouracil, Int. J. Mol. Sci., 2013, 14(8), 16600–16616 CrossRef PubMed.
- D. N. Zacks, Q. D. Zheng, Y. Han, R. Bakhru and J. W. Miller, FAS-mediated apoptosis and its relation to intrinsic pathway activation in an experimental model of retinal detachment, Invest. Ophthalmol. Visual Sci., 2004, 45(12), 4563–4569 CrossRef PubMed.
- D. Geethakumari, A. Bhaskaran Sathyabhama, K. Raji Sathyan, D. Mohandas, J. v. Somasekharan and S. Thavarool Puthiyedathu, Folate functionalized chitosan nanoparticles as targeted delivery systems for improved anticancer efficiency of cytarabine in MCF-7 human breast cancer cell lines, Int. J. Biol. Macromol., 2022, 199, 150–161 CrossRef CAS PubMed.
- K. J. Livak and T. D. Schmittgen, Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method, Methods, 2001, 25(4), 402–408 CrossRef CAS PubMed.
- A. W. Schüttelkopf and D. M. F. van Aalten, PRODRG: A tool for high-throughput crystallography of protein-ligand complexes, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60(8), 1355–1363 CrossRef PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith and B. Hess, et al. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers, SoftwareX, 2015, 1–2, 19–25 CrossRef.
- G. J. Martyna, M. L. Klein and M. Tuckerman, Nosé-Hoover chains: The canonical ensemble via continuous dynamics, J. Chem. Phys., 1992, 97(4), 2635–2643 CrossRef.
- M. Parrinello, A. Rahman and M. R. a. Parrinello, Crystal Structure and Pair Potentials: A Molecular_Dynamics Study, Phys. Rev. Lett., 1980, 45(14), 1196–1199 CrossRef CAS . Available from: https://link.aps.org/doi/10.1103/PhysRevLett.45.1196.
- U. Essmann, L. Perera, M. L. Berkowitz, T. Darden, H. Lee and L. G. Pedersen, A smooth particle mesh Ewald method, J. Chem. Phys., 1995, 103(19), 8577–8593 CrossRef CAS.
- B. Hess, H. Bekker, H. J. C. Berendsen and J. G. E. M. Fraaije, LINCS: A Linear Constraint Solver for molecular simulations, J. Comput. Chem., 1997, 18(12), 1463–1472 CrossRef CAS.
- S. Miyamoto and P. A. Kollman, Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models, J. Comput. Chem., 1992, 13(8), 952–962 CrossRef CAS.
- P. Wang, R. K. Kankala, B. Chen, R. Long, D. Cai and Y. Liu, et al. Poly-allylamine hydrochloride and fucoidan-based self-assembled polyelectrolyte complex nanoparticles for cancer therapeutics, J. Biomed. Mater. Res., Part A, 2019, 107(2), 339–347 CrossRef CAS PubMed.
- D. Peer, J. M. Karp, S. Hong, O. C. Farokhzad, R. Margalit and R. Langer, Nanocarriers as an emerging platform for cancer therapy, Nat. Nanotechnol., 2007, 2(12), 751–760 CrossRef CAS PubMed . Available from: https://www.nature.com/naturenanotechnology.
- G. Deepa, K. C. Sivakumar and T. P. Sajeevan, Molecular simulation and in vitro evaluation of chitosan nanoparticles as drug delivery systems for the controlled release of anticancer drug cytarabine against solid tumours, 3 Biotech, 2018, 8(12), 1–11, DOI:10.1007/s13205-018-1510-x . Available.
- E. J. Lee and K. H. Lim, Formation of chitosan-fucoidan nanoparticles and their electrostatic interactions: Quantitative analysis, J. Biosci. Bioeng., 2016, 121(1), 73–83 CrossRef CAS PubMed.
- R. Vivek, V. Nipun Babu, R. Thangam, K. S. Subramanian and S. Kannan, PH-responsive drug delivery of chitosan nanoparticles as Tamoxifen carriers for effective anti-tumor activity in breast cancer cells, Colloids Surf., B, 2013, 111, 117–123 CrossRef CAS PubMed.
- M. Imran, S. Riaz, S. M. H. Shah, T. Batool, H. N. Khan and A. N. Sabri, et al. In vitro hemolytic activity and free radical scavenging by sol-gel synthesized Fe3O4 stabilized ZrO2 nanoparticles, Arabian J. Chem., 2020, 13(11), 7598–7608 CrossRef CAS.
- H. Danafar, A. Sharafi, H. Kheiri Manjili and S. Andalib, Sulforaphane delivery using mPEG–PCL co-polymer nanoparticles to breast cancer cells, Pharm. Dev. Technol., 2017, 22(5), 642–651 CrossRef CAS PubMed.
- S. M. Chiu, L. Y. Xue, J. Usuda, K. Azizuddin and N. L. Oleinick, Bax is essential for mitochondrion-mediated apoptosis but not for cell death caused by photodynamic therapy, Br. J. Cancer, 2003, 89(8), 1590–1597 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3pm00078h |
| This journal is © The Royal Society of Chemistry 2024 |