Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Non-natural sialic acid derivatives with o-nitrobenzyl alcohol substituents for light-mediated protein conjugation and cell imaging†
Guo-Biao
Zhu‡
 a,
Chen
Guo‡
a,
Chen
Guo‡
 a,
Xue-Lian
Ren
a,
Xue-Lian
Ren
 fg,
Ming-Zhe
Li
c,
Di-Ya
Lu
c,
Xi-Le
Hu
fg,
Ming-Zhe
Li
c,
Di-Ya
Lu
c,
Xi-Le
Hu
 *a,
He
Huang
*a,
He
Huang
 *fg,
Tony D.
James
*fg,
Tony D.
James
 *de and
Xiao-Peng
He
*de and
Xiao-Peng
He
 *abc
*abc
aKey Laboratory for Advanced Materials and Joint International Research Laboratory of Precision Chemistry and Molecular Engineering, Feringa Nobel Prize Scientist Joint Research Center, Frontiers Center for Materiobiology and Dynamic Chemistry, School of Chemistry and Molecular Engineering, East China University of Science and Technology, 130 Meilong RD, Shanghai 200237, China. E-mail: xlhu@ecust.edu.cn; xphe@ecust.edu.cn
bThe International Cooperation Laboratory on Signal Transduction, Eastern Hepatobiliary Surgery Hospital, National Center for Liver Cancer, Shanghai 200438, China
cShanghai World Foreign Language Academy, No. 400 Baihua Street, Shanghai 200233, China
dDepartment of Chemistry, University of Bath, Bath, BA2 7AY, UK. E-mail: t.d.james@bath.ac.uk
eSchool of Chemistry and Chemical Engineering, Henan Normal University, Xinxiang 453007, China
fState Key Laboratory of Chemical Biology, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China. E-mail: hhuang@simm.ac.cn
gSchool of Pharmaceutical Science and Technology, Hangzhou Institute for Advanced Study, University of Chinese Academy of Sciences, Hangzhou 310024, China
First published on 4th November 2024
Abstract
We have synthesized two sialic acid derivatives substituted with an ortho-nitrobenzyl alcohol (o-NBA) group that can undergo light-mediated conjugation with primary amines at their 5- or 9-carbon position. The o-NBA derivatives were shown to react with multiple lysine residues of human serum albumin (HSA) when exposed to 365 nm light irradiation within 10 min. The resulting sugar conjugates were characterized by mass spectroscopy and used for fluorescence-based cell imaging.
Glycosylation is an important post-translational modification (PTM) of proteins.1,2 Protein glycosylation plays an essential role in cell physiology mediating immune responses, cell adhesion, cell–cell interactions and signal transduction.3 Abnormal glycosylation levels is a hallmark for health related conditions including inflammation, autoimmune disorders, neurodegenerative diseases and cancer.4–7 Sialic acids are a common terminal sugar of N- and O-linked glycans. Sialylation is abnormally upregulated in a diverse range of cancers including breast,8 pancreatic,9 ovarian,10 brain,11 lung,12 prostate,13 colorectal,14 renal,15 skin16 and liver cancer.17
In order to detect glycosylation in live cells, non-natural sugar probes have been developed. A general strategy is to install biorthogonal handles (such as an azide) to the side chain of a monosaccharide without perturbing its natural metabolic activity.18–20 Then after incubation with live cells, a biorthogonal unit (such as an alkyne) linked with a fluorophore is added to react with the metabolically processed sugar probe, thus enabling the spatiotemporal monitoring of biomolecular glycosylation.21,22 This elegant strategy has not only significantly advanced glycobiology, but provided insight into carbohydrate-based drug discovery.23,24 However, while sugar probes for metabolic labelling of glycans have been extensively reported, those capable of capturing sugar-binding proteins remain much less explored.
To achieve the in situ capture of proteins, a reactive handle needs to be introduced into the biomolecules for biorthogonal conjugation. The prototypical reactive units commonly used include benzophenone, diazirine, aryl azide, tetrazole, and thienyl-substituted α-ketoamide.25 Recently developed photocatalytic biorthogonal reactions are particularly advantageous for live cell imaging because of their biocompatibility and the ability to achieve biomolecular labelling in an on-demand fashion.26 Recently, Chen et al. developed a strategy using ortho-nitrobenzyl alcohol (o-NBA) as a light-activatable reactive handle to modify nucleic acids, and the resulting non-natural biomolecular probes were shown to sensitively capture nucleic acid-binding proteins.27 This is because the nitroso benzaldehyde intermediate generated by UV light irradiation can undergo selective cyclization with primary amines of proteins.28–31 This prompted us to explore the use of o-NBA as a reactive handle to modify sugars for light-mediated protein conjugation.
Here, we designed and synthesized two sialic acid derivatives modified with an o-NBA group attached to the 5- or 9-postion capable of light-mediated coupling with primary amines (Fig. 1a). As a proof-of-concept, the resulting sugar probes were used for light-catalyzed conjugation with human serum albumin (HSA) in which lysine residues (with amino side chain) are abundant (Fig. 1b). The resulting sugar–HSA conjugates were characterized by mass spectroscopy and were used for cell imaging using an encapsulated fluorescent imaging agent.
 | ||
| Fig. 1 (a) Structure of the o-NBA-modified sialic acid derivatives for (b) light-mediated covalent conjugation with human serum albumin (HSA). | ||
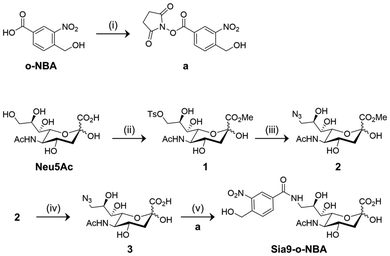
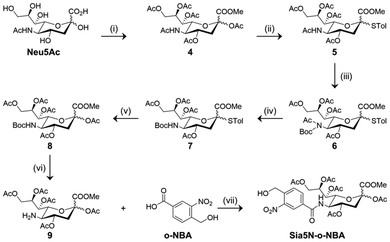
C-2 substituted peracetylated N-acetyl mannosamine (ManNAc) has been used as a non-natural sugar probe to detect cell sialylation after undergoing a series of biochemical transformations.23 However, introducing bulky functional groups to the C-2 position of ManNAc may compromise substrate recognition by endogenous enzymes that mediate sialylation.32 To address this issue, non-natural sialic acid derivatives are being developed to minimize perturbation of the subsequent sialylation biochemical pathways.33,34 According to previous literature reports, we synthesized the key sialic acid intermediates 3 and 9,35–37 and the subsequent introduction of an o-NBA group by an amidation reaction allowed for the 9- (Sia9-o-NBA) and 5-substituted (Sia5N-o-NBA) non-natural sialic acid derivatives to be successfully prepared, respectively (Schemes 1 and 2).
Sialic acid intermediate 3 was obtained according to previous literature methods.35,36 Briefly, the commercially available N-acetylneuric acid (Neu5Ac) was transformed into its methyl ester in the presence of Dowex 50WX2 H+ resin to protect the carboxyl acid group. The hydroxyl group at the C9 position of this intermediate was then treated with tosyl chloride in pyridine to obtain the tosylated derivative 1. Azide 3 was obtained through the reaction of sodium azide with 1 in methanol to afford intermediate 2, followed by saponification with LiOH. Finally, 3 was reduced with Pd/C in water to yield an amine intermediate, followed by amidation with N-hydroxysuccinimide-activated o-NBA (a) to give Sia9-o-NBA in 15% yield (Scheme 1).
Likewise, intermediate 9 was obtained according to a previous literature report.37 Starting from Neu5Ac, esterification and then per-acetylation afforded a fully protected sugar product 4. Treatment of 4 with p-thiocresol (TolSH) in the presence of BF3·Et2O as the Lewis acid catalyst for 24 h afforded thioglycoside 5. In order to selectively remove the C5 N-acetyl group, an additional t-butyloxycarbonyl (Boc) group was introduced to protect the secondary amine, affording intermediate 6. Subsequently, deacetylation in a mixture of MeONa/MeOH followed by acetylation with Ac2O and 4-dimethylaminopyridine (DMAP) gave the C5 Boc-protected sugar 7. A short (2 h) treatment of 7 with N-bromosuccinimide (NBS) in a mixture of acetone/water set free the C1 alcohol, which was immediately acetylated with Ac2O/DMAP to give 8. The Boc group of 8 was selectively removed with trifluoroacetic acid to obtain amine 9, which underwent amidation with o-NBA in the presence of 2-(7-azabenzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HATU) and diisopropylethylamine (DIPEA) to produce Sia5N-o-NBA in 18% yield (Scheme 2).
With the non-natural sialic acid derivatives in hand, we examined their capacity for light-mediated protein conjugation using HSA as a model. The protecting group-free Sia9-o-NBA was used for the study. To a phosphate buffered saline (PBS, 0.01 M, pH 7.4) solution of HSA, an excess of Sia9-o-NBA was added. The resulting mixture was exposed to 365 nm light irradiation for 10 min, and then incubated at 37 °C for another 30 min. Subsequently, the unreacted sialic acid derivatives were removed through dialysis to afford the Sia9-o-NBA-HSA conjugates, which were then digested with trypsin for LC-MS/MS analysis. The results showed that 20 lysine sites of HSA were modified with Sia9-o-NBA after light irradiation (Fig. 2). These results indicate the effectiveness of the sialic acid derivative for light-mediated protein conjugation.
Given that sialic acid-binding proteins are commonly detected in a variety of mammalian cells, we attempted to use the sialic acid–HSA conjugates for fluorescence-based cell imaging. Chlorin e6 (Ce6), which is known to bind HSA in its IB domain, was used as the fluorescent imaging agent.38 HSA without and with conjugation with Sia9-o-NBA were incubated with Ce6 for 6 h, followed by dialysis to obtain Ce6-encapsulated HSA derivatives Ce6@HSA and Ce6@Sia9-o-NBA-HSA. Fluorescence spectroscopy indicated the presence of the typical Ce6 emission band for all protein conjugates when dissolved in PBS solution (Fig. S1†).
RAW264.7 (mouse macrophage) and HeLa (human cervical cancer) cells were used for the imaging experiments. We observed that the HSA-based imaging agents were internalized by both cell lines after incubation for 1 hour at a Ce6 concentration of 5 μM (Fig. 3a). The fluorescence intensity of the acquired images was then quantified using a Columbus™ Image Data Storage and Analysis System (Fig. 3b). Quantitative analysis revealed a higher cellular uptake of Ce6@Sia9-o-NBA-HSA by RAW264.7 than HeLa cells. This difference is probably caused by a higher Siglec (a family of sialic acid-binding proteins) expression level in immune cells.39 Indeed, a subsequent analysis by quantitative real-time polymerase chain reaction corroborated the significantly higher expression level of several Siglec subtypes in RAW264.7 than HeLa cells (Fig. 3c). As a result, our sialic acid-conjugated HSA could be used to target macrophages for a diverse range of biomedical applications.
Conclusions
We synthesized two sialic acid derivatives bearing the o-NBA group at their 5- or 9-positions. The use of 365 nm light catalyzed the conjugation between both non-natural sugar derivatives with HSA. LC-MS/MS analysis indicated that the sugar-protein conjugates were successfully produced after light activation for 10 min, and a subsequent cell imaging assay was suggestive of different uptake efficiency by macrophages that highly express sialic acid-binding proteins. We anticipate that these derivatives will be useful for the light-mediated capture of sialic-acid binding proteins,40,41 as well as for imaging applications in biological samples and live cells.42–45 Relevant chemical biological studies are currently underway in our laboratory.Data availability
All data associated with this study have been included in either the manuscript or the supplementary file associated with the manuscript.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
The authors thank the Natural National Science Foundation of China (NSFC) (No. 92253306, 82130099 and 22108077), the Shanghai Municipal Science and Technology Major Project (No. 2018SHZDZX03), the International Cooperation Program of Shanghai Science and Technology (No. 23490711600), the Fundamental Research Funds for the Central Universities (222201717003), the Programme of Introducing Talents of Discipline to Universities (B16017), the Open Funding Project of the State Key Laboratory of Bioreactor Engineering, State Key Laboratory of Drug Research (SKLDR-2023-KF-10) and Ministry of Education Key Laboratory on Signaling Regulation and Targeting Therapy of Liver Cancer (Naval Medical University) (Grant. 2023-MEKLLC-MS/ZD-00*) for financial support. TDJ wishes to thank the University of Bath and the Open Research Fund of the School of Chemistry and Chemical Engineering, Henan Normal University (2020ZD01) for support. The Research Center of Analysis and Test of East China University of Science and Technology is gratefully acknowledged for assistance in analytical experiments.References
- K. T. Schjoldager, Y. Narimatsu, H. J. Joshi and H. Clausen, Nat. Rev. Mol. Cell Biol., 2020, 21, 729–749 CrossRef CAS PubMed.
- C. Reily, T. J. Stewart, M. B. Renfrow and J. Novak, Nat. Rev. Nephrol., 2019, 15, 346–366 CrossRef.
- A. Varki, Glycobiology, 2017, 27, 3–49 CrossRef PubMed.
- N. Berois, A. Pittini and E. Osinaga, Cancers, 2022, 14, 645 CrossRef PubMed.
- D. Thomas, A. K. Rathinavel and P. Radhakrishnan, Biochim. Biophys. Acta, Rev. Cancer, 2021, 1875, 188464 CrossRef PubMed.
- S. S. Pinho and C. A. Reis, Nat. Rev. Cancer, 2015, 15, 540–555 CrossRef PubMed.
- Y. Wang and H. Chen, Oncogene, 2023, 42, 1970–1979 CrossRef PubMed.
- A. A. Barkal, R. E. Brewer, M. Markovic, M. Kowarsky, S. A. Barkal, B. W. Zaro, V. Krishnan, J. Hatakeyama, O. Dorigo, L. J. Barkal and I. L. Weissman, Nature, 2019, 572, 392–396 CrossRef.
- C. Gao, L. Wisniewski, Y. Liu, B. Staal, I. Beddows, D. Plenker, M. Aldakkak, J. Hall, D. Barnett, M. K. Gouda, P. Allen, R. Drake, A. Zureikat, Y. Huang, D. Evans, A. Singhi, R. E. Brand, D. A. Tuveson, S. Tsai and B. B. Haab, Clin. Cancer Res., 2021, 27, 226–236 CrossRef.
- R. B. Jones, A. D. Silva, K. E. Ankenbauer, C. M. Britain, A. Chakraborty, J. A. Brown, S. W. Ballinger and S. L. Bellis, Glycobiology, 2023, 33, 626–636 CrossRef.
- P. Schildhauer, P. Selke, M. S. Staege, A. Harder, C. Scheller, C. Strauss, R. Horstkorte, M. Scheer and S. Leisz, Cells, 2023, 12, 2758 CrossRef.
- P. Singh, A. Joon, M. Kumari, T. Singh, A. Bal, P. Maan and S. Ghosh, Cell Biochem. Biophys., 2022, 80, 781–793 CrossRef.
- E. Scott, E. A. Goode, R. Garnham, K. Hodgson, M. Orozco-Moreno, H. Turner, K. Livermore, K. P. Nangkana, F. M. Frame, A. Bermudez, F. J. G. Marques, U. L. McClurg, L. Wilson, H. Thomas, A. Buskin, A. Hepburn, A. Duxfield, K. Bastian, H. Pye, H. M. Arredondo, G. Hysenaj, S. Heavey, U. Stopka-Farooqui, A. Haider, A. Freeman, S. Singh, E. W. Johnston, S. Punwani, B. Knight, P. McCullagh, J. McGrath, M. Crundwell, L. Harries, R. Heer, N. J. Maitland, H. Whitaker, S. Pitteri, D. A. Troyer, N. Wang, D. J. Elliott, R. R. Drake and J. Munkley, J. Pathol., 2023, 261, 71–84 CrossRef PubMed.
- M. Zhou, S. Lv, Y. Hou, R. Zhang, W. Wang, Z. Yan, T. Li, W. Gan, Z. Zeng, F. Zhang and M. Yang, Front. Immunol., 2022, 13, 994874 CrossRef PubMed.
- A. Gong, X. Zhao, Y. Pan, Y. Qi, S. Li, Y. Huang, Y. Guo, X. Qi, W. Zheng and L. Jia, J. Cell Sci., 2020, 133, jcs244020 CrossRef PubMed.
- R. Hatanaka, E. Araki, M. Hane, S. Go, D. Wu, K. Kitajima and C. Sato, Biochem. Biophys. Res. Commun., 2022, 608, 52–58 CrossRef PubMed.
- X. Zou, J. Lu, Y. Deng, Q. Liu, X. Yan, Y. Cui, X. Xiao, M. Fang, F. Yang, H. Sawaki, T. Sato, B. Tan, X. Lu, B. Feng, A. Kuno, H. Narimatsu, C. Gao and Y. Zhang, Oncogene, 2023, 42, 516–529 CrossRef PubMed.
- J. Scache, V. Rigolot, C. Lion, M. Mortuaire, T. Lefebvre, C. Biot and A. S. Vercoutter-Edouart, Sci. Rep., 2022, 12, 22129 CrossRef PubMed.
- H. Wang, M. C. Sobral, D. K. Y. Zhang, A. N. Cartwright, A. W. Li, M. O. Dellacherie, C. M. Tringides, S. T. Koshy, K. W. Wucherpfennig and D. J. Mooney, Nat. Mater., 2020, 19, 1244–1252 CrossRef CAS.
- X. Fan, Q. Song, D. E. Sun, Y. Hao, J. Wang, C. Wang and X. Chen, Nat. Chem. Biol., 2022, 18, 625–633 CrossRef PubMed.
- J. Han, R. Bhatta, Y. Liu, Y. Bo, A. Elosegui-Artola and H. Wang, Nat. Commun., 2023, 14, 5049 CrossRef PubMed.
- Y. Zhong, L. Xu, C. Yang, L. Xu, G. Wang, Y. Guo, S. Cheng, X. Tian, C. Wang, R. Xie, X. Wang, L. Ding and H. Ju, Nat. Commun., 2023, 14, 7285 CrossRef.
- H. Wang and D. J. Mooney, Nat. Chem., 2020, 12, 1102–1114 CrossRef PubMed.
- W. Yi, P. Xiao, X. Liu, Z. Zhao, X. Sun, J. Wang, L. Zhou, G. Wang, H. Cao, D. Wang and Y. Li, Signal Transduction Targeted Ther., 2022, 7, 386 CrossRef PubMed.
- H. Wu and J. Kohler, Curr. Opin. Chem. Biol., 2019, 53, 173–182 CrossRef PubMed.
- K. Kozoriz, O. Shkel, K. T. Hong, D. H. Kim, Y. K. Kim and J.-S. Lee, Acc. Chem. Res., 2023, 56, 25–36 CrossRef PubMed.
- A. D. Guo, K. N. Yan, H. Hu, L. Zhai, T. F. Hu, H. Su, Y. Chi, J. Zha, Y. Xu, D. Zhao, X. Lu, Y. J. Xu, J. Zhang, M. Tan and X. H. Chen, Nat. Chem., 2023, 15, 803–814 CrossRef CAS PubMed.
- A. D. Guo, D. Wei, H. J. Nie, H. Hu, C. Peng, S. T. Li, K. N. Yan, B. S. Zhou, L. Feng, C. Fang, M. Tan, R. Huang and X. H. Chen, Nat. Commun., 2020, 11, 5472 CrossRef CAS.
- A. D. Guo, K. H. Wu and X. H. Chen, RSC Adv., 2021, 11, 2235–2241 RSC.
- H. Hu, W. Hu, A. D. Guo, L. Zhai, S. Ma, H. J. Nie, B. S. Zhou, T. Liu, X. Jia, X. Liu, X. Yao, M. Tan and X. H. Chen, Nat. Commun., 2024, 15, 1465 CrossRef CAS PubMed.
- W. Hu, Y. Yuan, C.-H. Wang, H.-T. Tian, A.-D. Guo, H.-J. Nie, H. Hu, M. Tan, Z. Tang and X.-H. Chen, Chem, 2019, 5, 2955–2968 CAS.
- H. Yang, L. Lu and X. Chen, Biotechnol. Adv., 2021, 46, 107678 CrossRef CAS.
- S. J. Moons, G. J. Adema, M. T. Derks, T. J. Boltje and C. Bull, Glycobiology, 2019, 29, 433–445 CAS.
- C. Bull, T. Heise, D. M. Beurskens, M. Riemersma, A. Ashikov, F. P. Rutjes, T. H. van Kuppevelt, D. J. Lefeber, M. H. den Brok, G. J. Adema and T. J. Boltje, ACS Chem. Biol., 2015, 10, 2353–2363 CrossRef.
- Z. S. Chinoy, C. Bodineau, C. Favre, K. W. Moremen, R. V. Duran and F. Friscourt, Angew. Chem., Int. Ed., 2019, 58, 4281–4285 CrossRef CAS.
- X. Wang, S. Lang, Y. Tian, J. Zhang, X. Yan, Z. Fang, J. Weng, N. Lu, X. Wu, T. Li, H. Cao, Z. Li and X. Huang, ACS Cent. Sci., 2020, 6, 382–389 CrossRef CAS PubMed.
- X. Zheng, K. Sun, Y. Liu, X. Yin, H. Zhu, F. Yu and W. Zhao, J. Controlled Release, 2023, 353, 675–684 CrossRef CAS PubMed.
- D. Hu, Z. Sheng, G. Gao, F. Siu, C. Liu, Q. Wan, P. Gong, H. Zheng, Y. Ma and L. Cai, Biomaterials, 2016, 93, 10–19 CrossRef CAS PubMed.
- C. Reily, T. J. Stewart, M. B. Renfrow and J. Novak, Nat. Rev. Nephrol., 2019, 15, 346–366 CrossRef.
- W.-T. Dou, Z.-Y. Qin, J. Li, D.-M. Zhou and X.-P. He, Sci. Bull., 2019, 64, 1902–1909 CrossRef.
- W.-T. Dou, X. Wang, T. Liu, S. Zhao, J. Liu, Y. Yan, J. Li, C. Zhang, A. C. Sedgwick, H. Tian, J. L. Sessler, D. M. Zhou and X.-P. He, Chem, 2022, 8, 1750–1761 Search PubMed.
- W.-T. Dou, P. Qiu, Y.-Y. Shi, L. Zhu, C. Guo, N. Li, Y. Zang, T.-T. Liu, S.-W. Zhao, Y.-F. Pan, L.-W. Dong, J. L. Sessler, Y.-X. Tan, J. Li, H.-Y. Wang, H. Tian and X.-P. He, J. Am. Chem. Soc., 2023, 145, 17377–17388 CrossRef PubMed.
- W.-T. Dou, H.-H. Han, A. C. Sedgwick, G.-B. Zhu, Y. Zang, X.-R. Yang, J. Yoon, T. D. James, J. Li and X.-P. He, Sci. Bull., 2022, 67, 853–878 CrossRef PubMed.
- X. Chai, H.-H. Han, A. C. Sedgwick, N. Li, Y. Zang, T. D. James, J. Zhang, X.-L. Hu, Y. Yu, Y. Li, Y. Wang, J. Li, X.-P. He and H. Tian, J. Am. Chem. Soc., 2020, 142, 18005–18013 CrossRef PubMed.
- X.-P. He and H. Tian, Chem, 2018, 4, 246–268 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental section, additional figures and original spectral copies of new compounds. See DOI: https://doi.org/10.1039/d4ob01563k |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2024 |