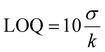Open Access Article
Open Access ArticleBODIPY(aryl)iodonium salts in the efficient synthesis of diversely functionalized BODIPYs and selective detection of serum albumin†
Bintu
Kumar
a,
Anindita
Bhatta
b,
Prakriti
Saraf
a,
Taur Prakash
Pandurang
 a,
Krishnan
Rangan
a,
Krishnan
Rangan
 c,
Madhushree
Sarkar
c,
Madhushree
Sarkar
 a,
Sivaprasad
Mitra
a,
Sivaprasad
Mitra
 *b and
Dalip
Kumar
*b and
Dalip
Kumar
 *a
*a
aDepartment of Chemistry, Birla Institute of Technology and Science, Pilani-333 031, India. E-mail: dalipk@pilani.bits-pilani.ac.in
bDepartment of Chemistry, North-Eastern Hill University, Shillong-793022, India. E-mail: smitra@nehu.ac.in
cDepartment of Chemistry, Birla Institute of Technology and Science, Pilani, Hyderabad Campus-500 078, India
First published on 2nd April 2024
Abstract
BODIPY(aryl)iodonium salts were readily accessible from the high-yielding reaction of BODIPY with iodoarenes or hydroxyl(tosyloxy)iodoarenes in the presence of m-CPBA. The prepared BODIPY(aryl)iodonium salts bearing substituents of varied electronic nature were utilized for the direct syntheses of thiocyanate, azide, amine and acrylate functionalized BODIPYs and β,β′-bis-BODIPYs. The regioselective syntheses of α-piperidinyl and β-piperidinyl substituted BODIPYs were achieved through the reaction of BODIPY(aryl)iodonium salts with piperidine in the absence and presence of copper(I). Expeditious and high yielding (79–82%) synthesis of β,β′-bis-BODIPYs was also developed through the palladium-catalyzed reductive coupling of the easily accessible BODIPY(aryl)iodonium salts. Some of the indole-appended BODIPYs and bis-BODIPYs displayed strong absorption in the visible region (∼610 nm). The BODIPY(aryl)iodonium salts also showed significant binding with serum albumin and were observed to be selective serum protein sensors with estimated limits of detection as low as 7 μg mL−1 in some cases.
Introduction
BODIPY and related compounds are most frequently used as heterocyclic fluorophores offering advantageous features such as narrow absorption and emission bands with high peak intensity, excellent fluorescence quantum yield, good photostability and biocompatibility.1 The structural modification of the BODIPY core is anticipated to tune its absorption and fluorescence profiles to generate useful probes in bio-imaging and photodynamic therapy (PDT). The π-conjugation at the α- and β-positions along with the modification of meso-substituents on the BODIPY core are among the effective ways to achieve dyes with a red-shift in absorption spectra and also large Stokes shift.2–4 Furthermore, modified BODIPYs are increasingly explored due to their wide range of interesting applications in chemiluminescence, organic solar cells, energy transfer systems, fluorescence imaging etc.5–7In the recent past, C–H functionalization of the BODIPY scaffold has become an attractive alternative to prepare BODIPY-based compounds.1,8–10 Although, traditional approaches to prepare BODIPYs are also available, they generally require multistep processes starting from pyrrole or related unstable intermediates.11,12
Direct C–H bond functionalization of BODIPY chromophores is a highly attractive way of exploring the diversity of the BODIPY moieties. You and Wu described a late-stage regioselective decarboxylative direct C–H arylation using substituted benzoic acids as coupling partners of BODIPY in the presence of palladium acetate, PCy3·HBF4 as a ligand and silver carbonate at 140 °C for 24 h.13 Jiao, Hao and co-workers developed a direct β-selective styrylation of BODIPYs involving the reaction of BODIPY with styrene in the presence of palladium acetate and silver acetate as an oxidant in PivOH at 80 °C.14 Recently, gold(I)-catalysed direct C–H alkynylation reaction of BODIPY with ethynylbenziodoxolone was utilized by Furuta, Ishida and co-workers to prepare β-alkynylated BODIPYs in low yields.15 While the present work was in progress, the Ortyl group reported panchromatic pentamethylated BODIPY iodonium salts and successfully used them in 3D printing applications.16 The BODIPY core being an electron-deficient moiety and the co-existence of multiple equivalent C–H bonds on pyrrolic unsubstituted BODIPY leads to regioselective functionalization at the β-position being a challenging task. Limited examples of this involved formylation,17 borylation18 and halogenations19,20 at the β-position of BODIPYs. However, to the best of our knowledge, BODIPY(aryl)iodonium salts are yet to be explored for functionalization of the BODIPY core. The copper-catalyzed reaction of β-iodo BODIPY with primary and secondary amines was reported to furnish α-amino BODIPYs.21 Recently, Xiang and co-workers22 prepared β-arylated and β-vinylated BODIPYs involving Cu-catalyzed reaction of tetrasubstituted BODIPYs with diaryliodonium salts.
Diaryliodonium salts referred to as diaryl-λ3-iodanes are widely considered to be an important and realistic class of hypervalent iodine reagents.23,24 Additionally, diaryliodonium salts have been successfully utilized by chemists as mild and environmentally benign arylating agents owing to advantageous features such as high reactivity, stability and low toxicity.24,25 Leveraging the special features, diaryliodonium salts have been utilized for a wide range of applications in synthetic chemistry involving reactions with various organic or inorganic electron-rich nucleophilic substrates resulting in the formation of new aryl–carbon and aryl–heteroatom bonds.26,27 The diverse arylating reactivity of diaryliodonium salts has been demonstrated in different types of coupling reactions under metal-free and metal-catalyzed conditions.28–31 Remarkable approaches for the direct arylation of electron-rich (hetero)arenes under metal-free conditions were recently accomplished through the use of diaryliodonium salts as arylation reagents.32,33 A vast majority of the existing procedures to prepare diaryliodonium salts usually involve the reaction of iodine(III) compounds with an appropriate arene in the presence of an oxidant in an acidic medium.34–36 Ochiai and co-workers reported vinyl(phenyl)iodonium tetrafluoroborates using the reaction of vinylboronic acids and esters with iodine(III) reagents.37 In 2007, the Olofsson group reported35 the straightforward and facile synthesis of symmetrical or unsymmetrical diaryliodonium salts by employing arenes and aryl iodides or iodine in the presence of m-CPBA and triflic acid. Circumventing the limitations of existing approaches, Widdowson's group prepared various diaryliodonium salts from the reaction of pre-formed iodobenzene diacetate (IBD) and arylboronic acids in the presence of trifluoromethanesulfonic acid.38
The extremely versatile BODIPY derivatives exhibit excellent photophysical properties like red-shifted UV-Visible absorption maxima, high molar absorption coefficients (ε), broad emission bandwidths, large Stokes shifts and high fluorescence quantum yields as well as high photo-stability.39 Due to these advantageous features, BODIPY and derived compounds are used for biochemical labelling,40,41 light-emitting devices,42 and light harvesting systems,43 and as sensitizers in solar cells.44 Since serum albumin is the most abundantly present plasma protein and binds to diverse groups of drugs, hormones, enzymes etc., it is frequently used as a model protein to study the interaction with small molecules using fluorescence spectroscopy.33 Moreover, serum protein binding capacity can majorly influence the distribution and metabolism of drugs and hence these studies can help in better understanding the pharmacokinetics of the synthesized systems and designing more effective drug-like molecules.45 The present work aims to develop a reliable and convenient protocol to access diverse BODIPY(aryl)iodonium salts and investigate their utility in the preparation of structurally diverse BODIPY dyes. Further, detailed photophysical characterization of one of the synthesized BODIPY(aryl)iodonium salts (4e) along with molecular insights into their interaction with bovine serum albumin (BSA) is also reported towards the application of the synthesized systems as potential serum protein sensors.
Results and discussion
In our efforts to prepare BODIPY(aryl)iodonium salt 4, iodoarene 2 was allowed to react with m-CPBA and PTSA 3 in dry dichloromethane (DCM) followed by the addition of BODIPY 1 as illustrated in Scheme 1. The reaction conditions were further optimized to develop a high-yielding protocol for the synthesis of 4a. First, BODIPY 1 and 2a were treated with m-CPBA in the presence of anhydrous PTSA (3) in dichloromethane to afford 4a in 74% yield (entry 1, Table 1). With the use of acetonitrile (MeCN) as a reaction solvent, 4a was formed in 32% yield (entry 2). The yield was increased to 61% with the use of trifluoroethanol (TFE) as a reaction solvent (entry 3).Gratifyingly, the yield of 4a was significantly increased with the use of 1.2 equivalents of anhydrous PTSA 3 (entry 4). When the reaction was performed in a mixture of dichloromethane and trifluoroethanol, 4a was isolated in 78% yield (entry 5). Notably, at 25 °C, the yield of product 4a was diminished to 19% (entry 6). After a series of experiments, 1 (1.1 equiv.), m-CPBA (1.2 equiv.), anhydrous PTSA 3 (1.2 equiv.) and 2a (1 equiv.) in dichloromethane at 0 °C for 2 h, were found to be the optimal reaction conditions to produce BODIPY(aryl)iodonium salt 4a in 93% yield (see Table 1). The use of MesI(OAc)2 instead of iodomesitylene 2a resulted 4a in lower yield (62%, entry 7). The use of MesI(OH)OTs instead of MesI(OAc)2 led to 4a in better yield (80%). The substituted BODIPYs and iodoarenes with various substitution patterns (mesityl 2a and triisopropyl substituted 2e) were well tolerated under the optimized conditions to afford BODIPY(aryl)iodonium salts 4a–g in 65–93% yields (Scheme 2). The structure of 4e was confirmed by single-crystal XRD analysis. The present reaction conditions were utilized for the gram scale synthesis of 4e (2.5 g, 87%) involving readily available precursors BODIPY 1 (1.0 g) and 1-iodo-2,4,6-triisopropylbenzene 2e (Scheme 3).
Crystal structure analysis of 4e
According to the X-ray crystallography data, compound 4e (CCDC 2294690†) is crystallized in the P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) space group of the triclinic system and the asymmetric unit shows the presence of one molecule of 4e and one CHCl3 (Fig. 1a and Fig. S1†). The geometry around the iodine atom is bent or T-shaped due to the presence of lone pairs of electrons on the iodine center and iodine is bonded to the β-C of BODIPY and a carbon of the arene ring. The interplanar angle between the meso-phenyl plane and the BODIPY ring is 53.67°, while the plane of the iodoarene ring forms an angle of about 174° with respect to the BODIPY plane. The positively charged iodine center is involved in non-covalent interactions with the tosylate (OTs) ion (I⋯O: 2.658(3) Å, 2.740(3) Å) (Fig. 1b). The CHCl3 molecule is in close contact with the fluorine of the BODIPY unit and arene ring of tosylate. The iodoarene and meso-phenyl of the BODIPY are involved in C–H⋯π interactions with the phenyl ring of tosylate. The centroid (phenyl of tosylate) to H distances are 2.432 Å (arene ring) and 3.547 Å (meso-phenyl) as shown in Fig. 1c. The crystallographic and selected bonding parameters of compound 4e are listed in the ESI (Tables S1–S6†).
space group of the triclinic system and the asymmetric unit shows the presence of one molecule of 4e and one CHCl3 (Fig. 1a and Fig. S1†). The geometry around the iodine atom is bent or T-shaped due to the presence of lone pairs of electrons on the iodine center and iodine is bonded to the β-C of BODIPY and a carbon of the arene ring. The interplanar angle between the meso-phenyl plane and the BODIPY ring is 53.67°, while the plane of the iodoarene ring forms an angle of about 174° with respect to the BODIPY plane. The positively charged iodine center is involved in non-covalent interactions with the tosylate (OTs) ion (I⋯O: 2.658(3) Å, 2.740(3) Å) (Fig. 1b). The CHCl3 molecule is in close contact with the fluorine of the BODIPY unit and arene ring of tosylate. The iodoarene and meso-phenyl of the BODIPY are involved in C–H⋯π interactions with the phenyl ring of tosylate. The centroid (phenyl of tosylate) to H distances are 2.432 Å (arene ring) and 3.547 Å (meso-phenyl) as shown in Fig. 1c. The crystallographic and selected bonding parameters of compound 4e are listed in the ESI (Tables S1–S6†).
Scope of BODIPY(aryl)iodonium salts
The diaryliodonium salts have been used in the formation of carbon–carbon and carbon–heteroatom bonds by releasing aryl moieties to carbon and heteroatom nucleophiles.46,47 In the recent past, Gaunt48 and Sanford49 independently developed the copper and palladium-catalyzed arylation of indoles by utilizing diaryliodonium salts. Unsymmetrical diaryliodonium salts are reported to selectively transfer aryl moieties to nucleophiles by concordantly releasing the sterically hindered 1-iodo-2,4,6-trimethylbenzene. Therefore, BODIPY(trimethylphenyl)iodonium salt 4a was prepared in an effort to selectively transfer a BODIPY unit by the generation of 1-iodo-2,4,6-trimethylbenzene as a byproduct.33 Given the usefulness of diaryliodonium salts, herein we investigated applications of the prepared BODIPY(trimethylphenyl)iodonium salt 4a as exemplified by the preparation of functionalized BODIPYs 5a–j (Scheme 4). For example, the reaction of 4a with sodium azide or potassium thiocyanate in the presence of cesium carbonate as a base afforded the corresponding products 5a–c in 70–81% yields.The BODIPY azide 5c could be a useful synthon in click chemistry to prepare 1,2,3-triazolyl appended BODIPYs.50
The copper(I) promoted reaction of 4a with phthalimide led to β-phthalimido appended BODIPY 5d in 56% yield. Under metal free conditions, the reaction of 4a with piperidine afforded β-piperidinyl BODIPY 5e in 58% yield. Presumably, initial exchange of the tosylate in 4a with the piperidine moiety followed by reductive elimination of the unstable λ3-iodane intermediate may generate 5e. When the same reaction was performed in the presence of cuprous iodide, a regioisomeric α-piperidinyl BODIPY 5f was selectively formed in 77% yield. Formation of these α-amine derivatives possibly involves a direct oxidative nucleophilic substitution of the α-hydrogen in BODIPY.11 Next, with the use of copper-catalyzed reaction of 4a with toluidine, α-tolyl BODIPY 5g was achieved in 80% yield. Next, the reaction of 4a with indole and 3-methylindole under palladium-catalyzed conditions produced indolyl BODIPYs 5h (57%) and 5i (77%) in good to high yields, respectively.51 Next, Pd-catalyzed reaction of 4a with ethyl acrylate led to Heck product 5j with 50% yield.
The scope of the protocol was further extended by preparing BODIPY dimers which are known for their unique charge localization, high extinction coefficient, and moderate fluorescence quantum yield as well as redox properties.18,52,53 Some of the BODIPY dimers have also been used as photosensitizers via intersystem crossing from the excited singlet to triplet state in generating singlet oxygen.54,55 Inspired by our recent reductive coupling of diaryliodonium salts,56 the palladium-catalyzed reaction of 4a was utilized to prepare β,β′-bis-BODIPY 5k in 71% yield. When the same reaction was performed under MW irradiation, the yield of 5k was increased to 79%. Under similar conditions using BODIPY iodonium salt 4g with a meso-anisoyl moiety, bis-BODIPY 5l was prepared in 82% yield. However, palladium catalyzed coupling of BODIPY iodonium salt 4a with core BODIPY failed to produce the expected unsymmetrical bis-BODIPY. Further, using copper salts and 4a, bis-BODIPY 5k was obtained in only 30% yield (Scheme 5).
The photophysical properties of indole appended-BODIPYs were investigated by absorption and emission spectroscopy in UV-grade dichloromethane. The absorption spectra of 4e, 5h and 5i displayed characteristic absorption bands ranging within 493–600 nm (Table S7 and Fig. S2†). The indole-appended BODIPYs 5h and 5i showed a red shift of about 100 nm when compared to the parent BODIPY (501 nm). The emission intensity of 4e is quenched possibly due to the combined heavy atom and vibrational rotational motion effects.57 On the other hand, indole-appended BODIPYs 5h and 5i also displayed relatively weak emission intensity that may be attributed to the presence of a donor (indole)–acceptor (BODIPY) system.58
Solvatochromism could be considered as experimental evidence for the intramolecular charge transfer (ICT) effect. A bathochromic shift of 13 nm was observed for the longer-wavelength peak of 5h from 590 nm in DCM to 603 nm in DMSO (Fig. S3 and Table S8†), illustrating a remarkable ICT effect. To get insight into the electronic structures, DFT calculations were performed at the B3LYP/6-31G(d,p) level (Fig. S4†). The LUMO lobes are largely localized on the BODIPY moieties in all the compounds, while the HOMO and HOMO−1 lobes are distributed more on the indole moieties in 5h, which explains the more significant ICT effect that was observed in the solvatochromism experiment. However, an almost similar magnitude of red-shift in fluorescence spectra in some of the other weakly polar and/or protic solvents indicates the presence of additional processes, like specific solute–solvent interaction, contributing to this solvatochromic effect. The prepared bis-BODIPY 5k showed an absorption band at 607 nm and 5l showed two absorption bands at 540 and 726 nm, respectively. The prepared 5k and 5l showed emission bands at 672 and 697 nm, respectively, with corresponding red shifts of about 154 and 179 nm compared to 4e (see Fig. S5†). This may be due to the excitonic coupling between the adjacent BODIPY units.59
Plausible mechanism
Based on our results and previous literature reports, a plausible reaction mechanism for the transfer of various nucleophiles on the arene ring is illustrated in Scheme 6. Initially, the oxidative addition of the iodonium adduct to Cu(I) species is likely to release a highly electrophilic species A along with iodoarenes.48 Further, the ligand exchange reaction of species A with electron-rich nucleophiles may furnish an intermediate species B. Finally, the reductive elimination of B is anticipated to generate 5d, 5f and 5gvia the release of Cu(I) to complete the catalytic cycle.60 In the case of Pd-catalyzed C–H activation, the nucleophile is believed to be initially coordinated with the Pd(II) catalyst followed by activation of the C–H bond of the nucleophile to produce an intermediate species A′ accompanied by removal of acetic acid.61 The oxidative addition of iodonium adduct 4a takes place with the release of a highly electrophilic species B′ along with iodoarenes. Finally, the reductive elimination of B′ would result in the formation of 5h, 5i, and 5j emancipating the active Pd(II) species to complete the catalytic cycle.Solvatochromic effect and photophysical properties of BODIPY(aryl)iodonium salts
The photophysical properties of the five BODIPY derivatives 4a–e are checked in four different solvents, namely methyl alcohol, dimethyl sulfoxide, dichloromethane, and water. Broad and unstructured absorption spectra of the investigated systems appear within 480–510 nm range depending on the solvent. On excitation at their corresponding absorption maxima, the emission peaks are seen to be in the range of 510–530 nm (Fig. 2). In all the cases, the excitation spectra measured at the respective emission maximum produce unstructured profiles and match with the corresponding absorption spectra. The observed differences in absorption profiles could be due to variation in the size of aryl ring (phenyl, mesityl, 2,4,6-triisopropyl-phenyl) in the BODIPY(aryl)iodonium salts which may influence the degree of conjugation and the distribution of electron density in the molecules. The fluorescence lifetime values of these compounds are estimated using the time-resolved fluorescence decay data. In most of the cases, the fluorescence decay spectra were fitted with the bi-exponential model and the average fluorescence lifetime is reported. However, a single exponential decay model was enough to fit the data for the investigated systems in DMSO. Representative fluorescence decay spectra of 4b in water with the simulated fitting curve are shown in Fig. 2(d). Different photophysical parameters like absorption maximum (λabs), emission maximum (λem), Stokes shift (Δυss), fluorescence quantum yield (Φf), average lifetime (τav), radiative rate constant (κr) and total non-radiative rate constant (∑knr) are calculated for 4a–e and listed in Table S8.† Interestingly, all the compounds show the highest fluorescence quantum yield in dichloromethane. To identify the optimum dye concentration for the serum protein sensing (discussed later) and to identify the possible intermolecular interactions, the photophysical behavior of these compounds is also examined in an aqueous medium by varying the concentrations of the probe from 0.5 to 50 μM. The maximum absorbance shows a distinct deviation from linearity with increasing concentration of the dye beyond 2.6 μM (Fig. 3(a)). A similar break is also observed in the maximum fluorescence intensity as well as in the average fluorescence lifetime at 1.5–2 μM dye concentration for all the systems. A representative example is shown for compound 4e in Fig. 3. Aggregation of the dye molecules in water at higher probe concentrations could be the reason behind this observation.62 | ||
| Fig. 3 Break point (indicated by the arrow mark) in the variation in absorbance (a), fluorescence intensity (b) and average lifetime (c) with change in concentration of 4e in an aqueous medium. | ||
To ensure the presence of monomeric dye compounds, the concentration of the probe was kept fixed at 1 μM in the protein binding experiments discussed below.
Interaction of BODIPY with serum albumin
Serum albumin can be employed as a very good model target for studying interaction with biomolecules as it is the most profusely present protein in blood plasma and plays a very important role in biological processes. For this experiment, bovine serum albumin (BSA) concentration is varied from 0 to 40 μM keeping the BODIPY concentration fixed at 1 μM in a pH 7.4 buffer medium. With an increase in the BSA concentration, gradual enhancement of emission intensity is observed till 30–35 μM and it saturates beyond this. In Fig. 4(a) the emission spectra of 4e with the addition of increased concentration of BSA at 298 K is shown as a representative example and the inset shows the binding isotherm. Restricted motion of the BODIPY molecule in the binding cavity of BSA may be the reason for the enhancement of fluorescence intensity. Similar observations for BODIPY–BSA interaction have also been reported earlier.50 Interestingly, the fluorescence intensity of 4b is relatively insensitive towards the addition of BSA (Fig. 4(b)) implying that this compound does not bind with BSA. On comparing the structure of 4b with other BODIPY derivatives, it is apparently clear that there is no hydrophobic alkyl substitution on both the BODIPY rings for 4b as well as on the benzene ring attached to I+, which may significantly affect its association with BSA. To get quantitative details about the complexation process, the binding constants in BODIPY–BSA systems for all the four compounds 4a, 4c, 4d and 4e are calculated using the modified Benesi–Hildebrand (B–H) equation (eqn (1)) for 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 interaction at five different temperatures, viz. 298 K, 303 K, 308 K, 313 K and 318 K.
1 interaction at five different temperatures, viz. 298 K, 303 K, 308 K, 313 K and 318 K.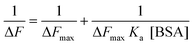 | (1) |
Further, to get a better idea about the nature of the interaction in BODIPY–BSA systems, from the obtained values of binding constant, the thermodynamic parameters, change in enthalpy (ΔH) and change in entropy (ΔS), were calculated using the slope and intercept of the van't Hoff relation (eqn (2)).
 | (2) |
Moreover, the change in Gibbs free energy (ΔG) was calculated using eqn (3).
| ΔG = ΔH − TΔS | (3) |
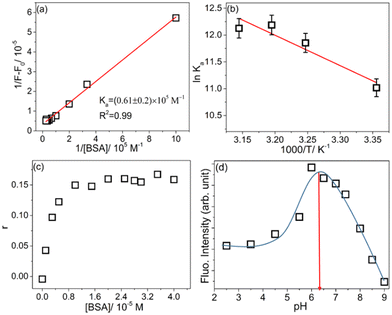
All the thermodynamic parameters are shown in Table S10.† Representative figures of B–H and van't Hoff analyses for compound 4e are shown in Fig. 5(a and b). B–H and van't Hoff plots for the other three compounds are presented in Fig. S6 and S7,† respectively. From the calculated values of ΔH and TΔS, it is found that the association of 4e to BSA is an entropy-driven process, whereas the bindings of 4a, 4c and 4d are enthalpy driven processes. A possible explanation for this observation may involve significant structural deformation in BSA due to bulkier substitution in 4e. Comparing the structural makeup of 4e with the other three, it is clear that 4e contains more hydrophobic substitutions than the other three. The interaction between these hydrophobic groups and BSA can result in positive ΔS, which causes the binding process to be entropy-driven.
The lifetime decay spectra of these systems are also collected at different concentrations of BSA. Like free BODIPY derivatives in an aqueous medium, a bi-exponential decay model is needed to fit the time-resolved fluorescence data in the presence of BSA with a major contribution from a fast component and a minor contribution from a notably long component (about 0.2 ns and 3 ns, respectively, in the case of 4a). The detailed data of lifetime for all the four compounds are presented in Table S11.† It is found that the average lifetime values of 4a and 4e increase significantly with an increase in the concentration of BSA. Because of the structural confinement and restriction enforced by the less polar and rigid BSA cavity, the non-radiative decay processes are significantly reduced, increasing excited state lifetime values, and making the BODIPY dyes more emissive in nature when BSA is present. But in the case of 4c and 4d, the average lifetime decreases with the addition of BSA. Comparing the structures of the four compounds, substitution with electron-releasing methyl groups on both the pyrrole rings of 4c and 4d may induce a different interaction with BSA, which is not present in the case of 4a and 4e.
To obtain more insight about the binding mechanism, steady state fluorescence anisotropy (r) was also checked for the BODIPY–BSA system. When a fluorophore binds with protein, the normal movement or the molecular rotation of the fluorophore gets hindered because of the constrained conditions in the binding environment and this leads to an increase in the fluorescence anisotropy value. For all four BODIPY derivatives 4a, 4c, 4d and 4e, the anisotropy value increases with an increase in the BSA concentration and gets saturated around 20–25 μM, which correlates nicely with the steady state. A representative plot showing the change in fluorescence anisotropy value with increasing BSA concentration for compound 4e is given in Fig. 5(c). The anisotropy value changes from 0.004 in the aqueous buffer medium to 0.167 in the presence of BSA in this case. Thus, the anisotropy study also supports the confined motion of the dye molecule inside the BSA cavity as the reason for the enhancement of fluorescence intensity.
Isothermal titration calorimetry (ITC) measurements
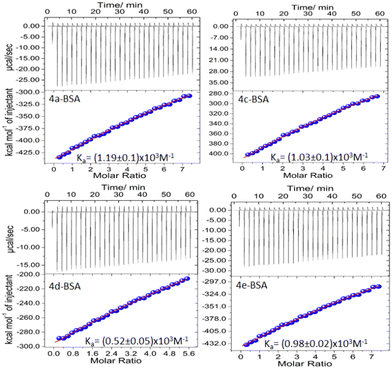
The thermodynamic parameters associated with the interaction of BODIPY derivatives with BSA are also evaluated through ITC measurement. A one site binding model is used to analyze the data obtained from the ITC titration experiment. The estimated binding constants are found to be (1.19 ± 0.1) × 103 M−1, (1.03 ± 0.1) × 103 M−1, (0.52 ± 0.05) × 103 M−1 and (0.98 ± 0.02) × 103 M−1 for 4a, 4c, 4d and 4e, respectively. The integrated binding isotherm as well as the raw heat data obtained from consecutive addition of BODIPY is shown in Fig. 6. From the values obtained it is found that the binding process is exothermic and has a negative entropy change value. The enthalpy and entropy change for binding, ΔH and ΔS, as well as the calculated negative ΔG values, are presented in Table S12.† Due to the intrinsic technical difference in the measurement of thermodynamic parameters through fluorescence and ITC (for example, the fluorescence technique uses the steady state approximation and application of indirect methods like van't Hoff plots in contrast to the “transient effect” and direct techniques in the latter), it is not expected to obtain the same values of the measured quantities in both these experiments.63Additionally, whereas spectroscopic (non-calorimetric) measurements like fluorescence titration experiments only consider the local change in the area close to the fluorescent tryptophan residue, a calorimetric measurement like ITC considers the overall change in protein conformation. This assertion is supported by numerous literature reports, which demonstrate similar discrepancies in results between ITC and fluorescence measurements.64,65 However, the trend in the variation of binding constant Ka and change in Gibbs free energy values for the different systems are in good agreement with those obtained from the fluorescence titration experiment.
Selectivity for BSA
To use the BODIPY derivatives as efficient albumin detectors, it is imperative to check their selectivity towards albumin over other biomolecules. For this comparative experiment, the effect of the fluorescence emission response of BODIPY on the addition of different proteins and amino acids was checked.To a 5 μM solution of 4a, 4c, 4d and 4e, 10 μM solutions of BSA, collagen (Col), N-acetyl-L-cystine (Cys), fibrinogen (Fib), glutathione (Glu) and lysozyme (Lys) were added and fluorescence emission spectra were collected. It is observed (Fig. 7) that the fluorescence intensity in the case of the other proteins and amino acids shows negligible change compared to the BODIPY derivatives (4a, 4c, 4d and 4e) in an aqueous buffer medium. But with the addition of BSA, there is around a 1.5–3 fold increase in fluorescence intensity indicating that bovine serum albumin possesses a suitable hydrophobic cavity for binding with these dye molecules. This result provides good evidence for these BODIPY derivatives’ higher selectivity for serum albumin.
pH optimization for the interaction of BODIPY with BSA
The significant increase in fluorescence intensity of the BODIPY systems in the presence of BSA prompts us to investigate further the use of these compounds as potential serum protein sensors. To optimize the experimental conditions, the variation in fluorescence intensity of BODIPY–BSA interaction was checked in a wide range of pH ranging from 2.5 to 9 for compound 4e. No emission peak position change was observed but there was a significant increase in the fluorescence intensity with an increase in pH up to pH 6 and after that the intensity started to decrease gradually. Fig. 5(d) shows the intensity change of 4e in the presence of BSA with the change of pH. Therefore, the pH range of 6–7 is believed to be the ideal condition for monitoring the serum protein sensing ability of 4e.Estimation of LOD and LOQ
The lowest concentration or quantity of a specific compound that can be detected accurately is called the limit of detection (LOD) and the limit of quantification (LOQ) is the smallest quantity of analyte that can be calculated with ‘acceptable’ accuracy and precision. LOD and LOQ can be calculated using the following equations.Where σ stands for the standard deviation (SD) and k is the slope obtained from the I/I0vs. [analyte] plot. The calculated values of LOD and LOQ are presented in Table S13.† The normal concentration of albumin in blood is 35–55 mg ml−1 and in urine is 0.03 mg ml−1. Albumin levels higher or lower than these values can be an indicator of different health risks like hypoalbuminemia, microalbuminuria, macroalbuminuria, etc. which can lead to cardiovascular and renal damage.66,67 The LOD values obtained for the four BODIPY derivatives show their efficacy as good serum protein indicators.
Conclusion
In summary, we have developed an efficient and convenient protocol for the synthesis of novel BODIPY(aryl)iodonium salts 4a–g by coupling the BODIPY core with iodoarenes. The utility of BODIPY(aryl)iodonium salts for further diversification of the BODIPYs under metal-free and metal-catalyzed reactions was also demonstrated. Prepared BODIPYs 5h, 5i, 5k and 5l with red-shifted absorption and emission bands, and low fluorescence quantum yields could be useful as photosensitizers in photodynamic therapy. BODIPYs 4a–g were used as fluorescent probes to detect serum albumin and compounds 4a, 4c, 4d and 4e were found to show excellent selectivity for bovine serum albumin with substantially low values of LOD compared to the normal concentration of serum albumins present in healthy physiological fluids. Structural comparison shows the need for hydrophobic substitution in the BODIPY structure for better development of serum protein sensors. The strong fluorescence enhancement on interaction with serum albumin shows that these dyes can be applied for the detection of a very little amount of hydrophobic protein. As a result, this study offers a foundation for the production of future amphiphilic dyes that can be utilized to more accurately and sensitively detect protein surface hydrophobicity. This will enable us to comprehend interactions between proteins and ligands, molecular recognition, and their biological roles better.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank DST (CRG/2022/007389) New Delhi for providing financial support. A research fellowship received from BITS Pilani (P. S.) is gratefully acknowledged. We are also thankful for HRMS (DST FIST, New Delhi) and NMR (BITS Pilani) instrumentation facilities.References
- W. Sheng, F. Lv, B. Tang, E. Hao and L. Jiao, Chin. Chem. Lett., 2019, 30, 1825–1833 CrossRef CAS.
- V. Lakshmi, M. R. Rao and M. Ravikanth, Org. Biomol. Chem., 2015, 13, 2501–2517 RSC.
- B. F. Hohlfeld, B. Gitter, K. J. Flanagan, C. J. Kingsbury, N. Kulak, M. O. Senge and A. Wiehe, Org. Biomol. Chem., 2020, 18, 2416–2431 RSC.
- T. Sarma, P. K. Panda and J.-I. Setsune, Chem. Commun., 2013, 49, 9806–9808 RSC.
- J. Banuelos, Chem. Rec., 2016, 16, 335–348 CrossRef CAS PubMed.
- R. Sola-Llano and J. Bañuelos, in BODIPY Dyes-A Privilege Molecular Scaffold with Tunable Properties, IntechOpen, 2018 Search PubMed.
- W. Asahi, R. Kurihara, K. Takeyama, Y. Umehara, Y. Kimura, T. Kondo and K. Tanabe, ACS Appl. Bio Mater., 2019, 2, 4456–4463 CrossRef CAS PubMed.
- B. Verbelen, L. C. D. Rezende, S. Boodts, J. Jacobs, L. V. Meervelt, J. Hofkens and W. Dehaen, Chem. – Eur. J., 2015, 21, 12667–12675 CrossRef CAS PubMed.
- B. Verbelen, V. Leen, L. Wang, N. Boens and W. Dehaen, Chem. Commun., 2012, 48, 9129–9131 RSC.
- X. Yang, L. Jiang, M. Yang, H. Zhang, J. Lan, F. Zhou, X. Chen, D. Wu and J. You, J. Org. Chem., 2018, 83, 9538–9546 CrossRef CAS PubMed.
- H. Chong, H.-A. Lin, M.-Y. Shen, C.-Y. Liu, H. Zhao and H.-H. Yu, Org. Lett., 2015, 17, 3198–3201 CrossRef CAS PubMed.
- K. C. Behera, P. Pushpanandan and M. Ravikanth, Eur. J. Inorg. Chem., 2023, 26, e202300039 CrossRef CAS.
- L. Luo, D. Wu, W. Li, S. Zhang, Y. Ma, S. Yan and J. You, Org. Lett., 2014, 16, 6080–6083 CrossRef CAS PubMed.
- F. Lv, Y. Yu, E. Hao, C. Yu, H. Wang, N. Boens and L. Jiao, Org. Biomol. Chem., 2019, 17, 5121–5128 RSC.
- T. Shimada, S. Mori, M. Ishida and H. Furuta, Beilstein J. Org. Chem., 2020, 16, 587–595 CrossRef CAS PubMed.
- M. Topa-Skwarczyńska, M. Galek, M. Jankowska, F. Morlet-Savary, B. Graff, J. Lalevée, R. Popielarz and J. Ortyl, Polym. Chem., 2021, 12, 6873–6893 RSC.
- C. Yu, L. Jiao, H. Yin, J. Zhou, W. Pang and Y. Wu, Eur. J. Org. Chem., 2011, 5460–5468 CrossRef CAS.
- Y. Hayashi, S. Yamaguchi, W. Y. Cha, D. Kim and H. Shinokubo, Org. Lett., 2011, 13, 2992–2995 CrossRef CAS PubMed.
- L. Jiao, W. Pang, J. Zhou, Y. Wei, X. Mu, G. Bai and E. Hao, J. Org. Chem., 2011, 76, 9988–9996 CrossRef CAS PubMed.
- V. Lakshmi, M. R. Rao and M. Ravikanth, Org. Biomol. Chem., 2015, 13, 2501–2517 RSC.
- J. G. Knight, R. B. Alnoman and P. G. Waddell, Org. Biomol. Chem., 2015, 13, 3819–3829 RSC.
- W. Ren, H. Xiang, C. Peng, Z. Musha, J. Chen, X. Li, R. Huang and Y. Hu, RSC Adv., 2018, 8, 5542–5549 RSC.
- V. V. Zhdankin and J. D. Protasiewicz, Coord. Chem. Rev., 2014, 275, 54–62 CrossRef CAS.
- A. Yoshimura and V. V. Zhdankin, Chem. Rev., 2016, 116, 3328–3435 CrossRef CAS PubMed.
- X. Zhou, Q. Wu, Y. Yu, C. Yu, E. Hao, Y. Wei, X. Mu and L. Jiao, Org. Lett., 2016, 18, 736–739 CrossRef CAS PubMed.
- M. Fananas-Mastral, Synthesis, 2017, 49, 1905–1930 CrossRef CAS.
- K. Aradi, B. Tóth, G. L. Tolnai and Z. Novák, Synlett, 2016, 27, 1456–1485 CrossRef CAS.
- F. Tinnis, E. Stridfeldt, H. Lundberg, H. Adolfsson and B. Olofsson, Org. Lett., 2015, 17, 2688–2691 CrossRef CAS PubMed.
- C. Dey, E. Lindstedt and B. Olofsson, Org. Lett., 2015, 17, 4554–4557 CrossRef CAS PubMed.
- T. L. Seidl and D. R. Stuart, J. Org. Chem., 2017, 82, 11765–11771 CrossRef CAS PubMed.
- K. Yin and R. Zhang, Org. Lett., 2017, 19, 1530–1533 CrossRef CAS PubMed.
- T. Besson and C. Fruit, Pharmaceuticals, 2021, 14, 661 CrossRef CAS PubMed.
- A. Pacheco-Benichou, T. Besson and C. Fruit, Catalysts, 2020, 10, 483 CrossRef CAS.
- M. Zhu, N. Jalalian and B. Olofsson, Synlett, 2008, 592–596 CAS.
- M. Bielawski, M. Zhu and B. Olofsson, Adv. Synth. Catal., 2007, 349, 2610–2618 CrossRef CAS.
- T. Dohi, M. Ito, K. Morimoto, Y. Minamitsuji, N. Takenaga and Y. Kita, Chem. Commun., 2007, 4152–4154 RSC.
- M. Ochiai, M. Toyonari, T. Nagaoka, D.-W. Chen and M. Kida, Tetrahedron Lett., 1997, 38, 6709–6712 CrossRef CAS.
- M. A. Carroll, V. W. Pike and D. A. Widdowson, Tetrahedron Lett., 2000, 41, 5393–5396 CrossRef CAS.
- N. Boens, B. Verbelen, M. J. Ortiz, L. Jiao and W. Dehaen, Coord. Chem. Rev., 2019, 399, 213024 CrossRef CAS.
- K. V. Ksenofontova, A. A. Kerner, A. A. Ksenofontov, A. Y. Shagurin, P. S. Bocharov, M. M. Lukanov, A. R. Kayumov, D. E. Zhuravleva, Z. I. Iskhakova and E. E. Molchanov, Molecules, 2022, 27, 7911 CrossRef CAS PubMed.
- N. Gupta, S. I. Reja, V. Bhalla, M. Gupta, G. Kaur and M. Kumar, Chem. Commun., 2015, 51, 10875–10878 RSC.
- Y. H. Jung, D. Karthik, H. Lee, J. H. Maeng, K. J. Yang, S. Hwang and J. H. Kwon, ACS Appl. Mater. Interfaces, 2021, 13, 17882–17891 CrossRef CAS PubMed.
- V. Engelhardt, S. Kuhri, J. Fleischhauer, M. García-Iglesias, D. González-Rodríguez, G. Bottari, T. Torres, D. M. Guldi and R. Faust, Chem. Sci., 2013, 4, 3888–3893 RSC.
- H. Klfout, A. Stewart, M. Elkhalifa and H. He, ACS Appl. Mater. Interfaces, 2017, 9, 39873–39889 CrossRef CAS PubMed.
- N. Dorh, S. Zhu, K. B. Dhungana, R. Pati, F.-T. Luo, H. Liu and A. Tiwari, Sci. Rep., 2015, 5, 18337 CrossRef CAS PubMed.
- G. L. Tolnai, U. J. Nilsson and B. Olofsson, Angew. Chem., 2016, 128, 11392–11396 CrossRef.
- D. Lubriks, I. Sokolovs and E. Suna, J. Am. Chem. Soc., 2012, 134, 15436–15442 CrossRef CAS PubMed.
- R. J. Phipps, N. P. Grimster and M. J. Gaunt, J. Am. Chem. Soc., 2008, 130, 8172–8174 CrossRef CAS PubMed.
- N. R. Deprez, D. Kalyani, A. Krause and M. S. Sanford, J. Am. Chem. Soc., 2006, 128, 4972–4973 CrossRef CAS PubMed.
- B. Kumar, A. Bhatta, P. Saraf, K. Rangan, M. Sarkar, S. Mitra and D. Kumar, Dalton Trans., 2022, 51, 8169–8176 RSC.
- N. R. Deprez, D. Kalyani, A. Krause and M. S. Sanford, J. Am. Chem. Soc., 2006, 128, 4972–4973 CrossRef CAS PubMed.
- K. X. Teng, W. K. Chen, L. Y. Niu, W. H. Fang, G. Cui and Q. Z. Yang, Angew. Chem., Int. Ed., 2021, 60, 19912–19920 CrossRef CAS PubMed.
- Q. Gong, K. Cheng, Q. Wu, W. Li, C. Yu, L. Jiao and E. Hao, J. Org. Chem., 2021, 86, 15761–15767 CrossRef CAS PubMed.
- B. Köksoy, E. N. Kaya, F. Hacıvelioğlu, S. Yeşilot and M. Durmuş, Dyes Pigm., 2017, 140, 384–391 CrossRef.
- G. Mazzone, A. D. Quartarolo and N. Russo, Dyes Pigm., 2016, 130, 9–15 CrossRef CAS.
- V. Arun, P. V. Reddy, M. Pilania and D. Kumar, Eur. J. Org. Chem., 2016, 2096–2100 CrossRef CAS.
- J. Zou, Z. Yin, K. Ding, Q. Tang, J. Li, W. Si, J. Shao, Q. Zhang, W. Huang and X. Dong, ACS Appl. Mater. Interfaces, 2017, 9, 32475–32481 CrossRef CAS PubMed.
- M. Liu, C. Wang and Y. Qian, New J. Chem., 2021, 45, 18082–18089 RSC.
- Y. Liu, J. Zhao, A. Iagatti, L. Bussotti, P. Foggi, E. Castellucci, M. Di Donato and K.-L. Han, J. Phys. Chem. C, 2018, 122, 2502–2511 CrossRef CAS.
- B. Chen, X.-L. Hou, Y.-X. Li and Y.-D. Wu, J. Am. Chem. Soc., 2011, 133, 7668–7671 CrossRef CAS PubMed.
- A. M. Prendergast, R. Shanahan, A. Hickey, F. Harrington, D. Schönbauer, P. A. Byrne, M. Schnürch and G. P. McGlacken, J. Chem. Educ., 2019, 97, 200–206 CrossRef.
- P. Rybczynski and A. Kaczmarek-KÈ©dziera, Struct. Chem., 2021, 32, 953–965 CrossRef CAS.
- N. C. Garbett and J. B. Chaires, Expert Opin. Drug Discovery, 2012, 7, 299–314 CrossRef CAS PubMed.
- T. S. Banipal, A. Kaur and P. K. Banipal, Spectrochim. Acta, Part A, 2017, 170, 214–225 CrossRef CAS PubMed.
- M. A. Rohman, P. Baruah, A. Bhatta and S. Mitra, J. Mol. Liq., 2019, 290, 111210 CrossRef CAS.
- M. Sasmal, A. S. M. Islam, D. Moni, D. Maiti, A. Dutta and M. Ali, ACS Appl. Bio Mater., 2022, 5, 5854–5864 CrossRef CAS PubMed.
- D. de Zeeuw, H.-H. Parving and R. H. Henning, J. Am. Soc. Nephrol., 2006, 17, 2100–2105 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2294690. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d4ob00336e |
| This journal is © The Royal Society of Chemistry 2024 |