Open Access Article
Open Access ArticleNon-enzymatic synthesis of C-methylated fluostatins: discovery and reaction mechanism†
Bidhan Chandra
De
ab,
Chunfang
Yang
 ab,
Chunshuai
Huang
a,
Changsheng
Zhang
ab,
Chunshuai
Huang
a,
Changsheng
Zhang
 ab and
Wenjun
Zhang
ab and
Wenjun
Zhang
 *ab
*ab
aKey Laboratory of Tropical Marine Bioresources and Ecology, Guangdong Key Laboratory of Marine Materia Medica, South China Sea Institute of Oceanology Chinese Academy of Sciences, Guangzhou 510301, China. E-mail: wzhang@scsio.ac.cn
bUniversity of Chinese Academy of Sciences, 19 Yuquan Road, Beijing 100049, China
First published on 3rd January 2024
Abstract
Two C-methylated fluostatins (FSTs) B3 (1) and B4 (2) were synthesized from flavin-mediated nonenzymatic epoxide ring-opening reactions of FST C. The structures of 1 and 2 were elucidated by HRESIMS, NMR, and ECD spectroscopic analyses. A subsequent 13C labeling study demonstrated that the C-methyl groups of 1 and 2 were derived from DMSO and enabled the mechanistic proposal of a nonenzymatic C-methylation.
The methyl group is the simplest alkyl fragment appearing in small molecule drugs. The addition of methyl groups to pharmaceutical compounds often leads to notable enhancements in their lipophilicity, membrane solubility, bioavailability, and protection from enzymatic degradation in vivo.1–3 In biological systems, methyl transfer is typically catalyzed by S-adenosylmethionine (SAM) dependent methyltransferases.2,4 The molecular mechanism of SAM-dependent methylation generally entails a nucleophilic substitution taking place at the sulfonium methyl carbon of SAM, resulting in a variety of C-, O-, N- and S-methylated products,4,5 or involves radical chemistry to yield C(sp3)-methyl bonds (Fig. 1A).6,7 In synthetic chemistry, considerable strategies have been developed to selectively and efficiently incorporate methyl groups into pharmaceutical scaffolds at both sp2 and sp3 carbon centres.1 Traditionally, the C–H methylation of both sp2 and sp3 centres has primarily involved the deprotonation of acidic C–H bonds followed by alkylation using electrophilic methyl sources like methyl iodide (Fig. 1B).8 Very recently, a bioinspired reaction via a hydrogen atom transfer (HAT)-SH2 dual catalytic strategy has been developed for the direct C(sp3)–H methylation of drug compounds.9 Despite their sophisticated designs, these synthetic methods face challenges, including multi-step synthesis with low efficiency or the requirement of complex starting materials.1
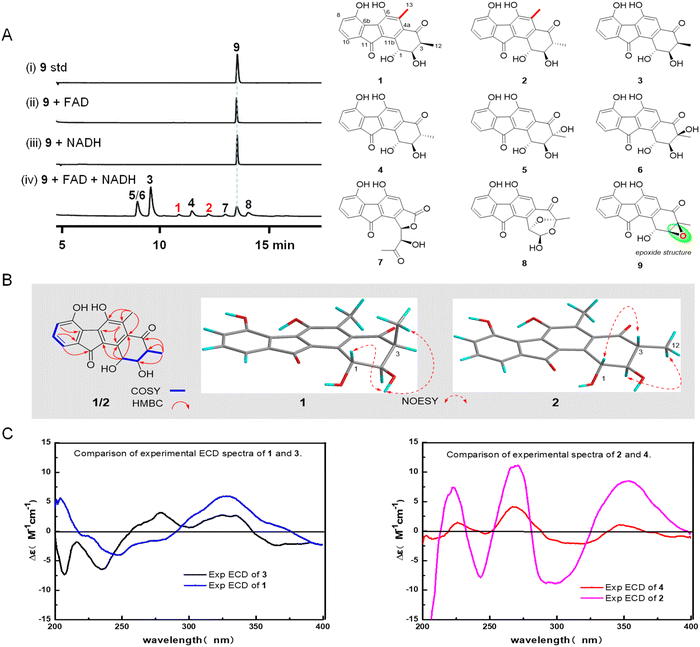
Fluostatins (FSTs) are distinguished by their characteristic tetracyclic benzo[a]fluorene skeleton.10,11,22 To date, approximately fifty analogues of FSTs have been identified, encompassing FST monomers,11–15 the benzo[cd]indeno[2,1-f]indazole skeleton,10 racemic aminobenzo[b]fluorenes,13 a pentacyclic skeleton fused with both benzo[b]fluorine and a six-membered lactone ring,16 and C–C or C–N coupled homo- or heterodimers.15,17 Notably, all these FSTs feature a single C-methyl group at C-3. FSTs are frequently characterized by the presence of an epoxide group at C-2/C-3. Recently, the epoxide group in FST C (9) has been demonstrated to undergo flavin-mediated, nonenzymatic reductive and oxidative ring-opening reactions, leading to diverse products, including FSTs B1 (3), B2 (4), C2 (5), C3 (6), C4 (7) and C5 (8) (Fig. 2A).18 In this work, we identified two additional C5-methylated FSTs, FSTs B3 and B4 (1 and 2), from the epoxide ring opening reactions. Subsequent experiments indicated that the C5-methyl group was nonenzymatically derived from dimethyl sulfoxide (DMSO) (Fig. 1C). Herein, we report the isolation and structure elucidation of 1 and 2, and propose a C-methylation mechanism.
In a previous study, we demonstrated that the α/β hydroxylase Alp1U can hydrolyze the epoxide of FST C (9), resulting in the chiral vicinal diol FST C1 and FST C2 (5).19 More recently, we have described nonenzymatic epoxide ring opening reactions: the incubation of FST C (9) with flavin adenine dinucleotide (FAD) and nicotinamide adenine dinucleotide (NADH) resulted in the production of various redox products, including 3–8 (Fig. 2A and Fig. S1†), while the control assays of 9, lacking either FAD or NADH, showed no conversions (Fig. 2A).18 Furthermore, we provided evidence for the spontaneous tautomerization of 3 to 4 in buffers with neutral pH.18 This phenomenon can be further amplified under alkaline pH conditions, yielding FST A.18 Interestingly, in our further studies of the nonenzymatic epoxide ring opening reactions, we occasionally encountered two very minor products, FST B3 (1, yield 1%) and FST B4 (2, yield 1.2%; Fig. 2A).
To structurally characterize these minor products, 1 and 2 were isolated from a large-scale reaction of FST C (9), carried out in the presence of FAD and NADH, in a PBS buffer at pH 7.0. The molecular formula of 1 was established to be C19H16O6 by HRESIMS (m/z 339.0874 [M − H]−, calcd for 339.0881, Fig. S2†). The 1H and 13C NMR data of 1 were similar to those of FST B1 (3); the difference is that the H-5 in 3 was replaced by a methyl group. This assignment was supported by the HMBC correlations from H3-13 to C-4a/C-5/C-6. Further detailed analysis of the 2D NMR data of 1 confirmed its planar structure (Fig. 2B). The relative configuration of 1 was assigned by NOESY correlations. The trans-configuration of H-1 and H-2 in 1 was deduced by the NOE correlations of H-1/OH-2, and a cis-configuration of H-2/H-3 in 1 was suggested by the NOESY correlations of 2-OH/H3-12 (Fig. 2B). Based on comparison of the experimental ECD spectra of 1 and 3 (Fig. 2C), the absolute configuration of 1 was assigned as 1R,2R,3S. The molecular formula of 2 was established as C19H16O6 by HRESIMS (m/z 339.0877 [M − H]−, calcd for 339.0881, Fig. S3†), the same as that of 1. The 1D and 2D NMR data of 2 (Table S1 and Fig. S3†) and 1 were highly similar, suggesting that 2 was a stereoisomer of 1. The planar structure of 2 was confirmed to be identical to that of 1 by comparing their 2D NMR data. The observed NOESY correlations of H-1/H-3 and H-2/H3-12 (Fig. 2B) indicated a trans configuration of H-1/H-2 and H-2/H-3 in 2. Moreover, the absolute configuration of 2 was decided by comparison of the ECD spectra of 2 and 4 (Fig. 2C). Their almost identical cotton effects suggested that 2 should have the 1R,2R,3R configuration.
Since the compound 9 we used for the nonenzymatic epoxide ring opening reactions was dissolved in DMSO, we inferred that DMSO might play an essential role in the formation of the C5-methylated products 1 and 2. Additionally, the structural similarity between 1/2 and 3/4 suggested that ring-opening products 3/4 likely served as precursors for the formation of 1/2. To verify this hypothesis, we conducted an experiment in which compound 3 was dissolved in DMSO and then incubated in PBS buffer (pH 7.0) for half an hour at 30 °C. As a result, traces of 1 were observed (Fig. 3A). A subsequent LC-MS analysis of the reaction products detected the presence of a molecular ion peak at m/z 339.7 ([M − H]−) (Fig. 3B). Furthermore, 3 was dissolved in DMSO-d6 and incubated in PBS buffer (pH 7.0). An LC-MS analysis then identified a +3 Da-shifted molecular ion peak at m/z 342.7 ([M − H]−) for 1 (Fig. 3B), denoting the incorporation of three deuterons into the C5-methylated 1. This evidence strongly suggests that DMSO acts as a methyl donor in this methylation process, and 3 serves as a precursor. A similar reaction was observed between 4 and 2. Given the previous research demonstrating the spontaneous conversion of 3 to 4,18 we also postulated that 1 might similarly convert to 2 under the same conditions. This proposal was subsequently validated by observing the facile transformation of 1 into 2 at pH 9 (Fig. 3C and Fig. S4†).
FSTs possess a unique tetracyclic benzo[a]fluorene skeleton.11 The distinctive structural features of FSTs provide them with the ability to generate a reactive para-quinone methide (p-QM) intermediate through an autocatalytic process that involves the 1,6-elimination reaction in FSTs.15 In a previous report, we described that a reactive p-QM-like intermediate undergoes coupling with a nucleophilic donor, leading to a diverse spectrum of C–C/C–N dimer FSTs.15 Here, we propose that the formation of 1/2 follows a mechanism similar to the one previously described. Specifically, the reaction involves the following key steps (Fig. 3D): (i) DMSO undergoes protonation, forming an activated electrophilic species M;20 (ii) 3 undergoes a 1,6-elimination reaction, yielding a transient p-QM intermediate M1;15 (iii) OH− participates in a nucleophilic reaction at C-1 of M1, facilitating electron transfer to C5 of M1; (iv) M1 then undergoes a subsequent nucleophilic reaction with the methyl group of M, culminating in the formation of M2; and (v) M2 undergoes tautomerization, leading to the formation of 1.
Conclusions
In conclusion, C5-methylated FSTs, 1 and 2, were identified in the ring-opening reaction. Subsequent experimentation confirmed the chemical synthesis of 1 and 2 from precursors 3 and 4 in the presence of DMSO. The unique structural composition of p-QMs involves reactive carbonyl and olefinic moieties, facilitating resonance between neutral and zwitterionic structures.21 This characteristic enables p-QMs to engage frequently in 1,6-conjugate addition reactions. Nonetheless, it is hypothesized that the acquisition of 1 and 2 is a result of the 1,4-addition of p-QM intermediates. This scenario presents a specific instance demonstrating the reaction pattern of p-QMs. Moreover, the study highlights the extension of methylation of C(sp2)–H bonds under mild conditions, eliminating the need for metal and non-metal catalysts.Author contributions
Bidhan Chandra De, C. Huang, and W. Zhang performed compound isolation and structure determination. Bidhan Chandra De and C. Yang carried out the reaction. Bidhan Chandra De, W. Zhang and C. Zhang wrote the manuscript. C. Zhang directed the research.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research has been partially funded by the National Natural Science Foundation of China (31820103003, 41676165, and 22377131), the Key Science and Technology Project of Hainan Province (ZDKJ202018), and the Youth Innovation Promotion Association CAS (2022349). B. C. D. acknowledges the support of CAS-TWAS President's PhD Fellowship. We extend our gratitude to Prof. Hung-wen Liu from the University of Texas at Austin for his valuable discussions on the mechanism. We also appreciate the analytical facilities provided by SCSIO.References
- D. Aynetdinova, M. C. Callens, H. B. Hicks, C. Y. X. Poh, B. D. A. Shennan, A. M. Boyd, Z. H. Lim, J. A. Leitch and D. J. Dixon, Chem. Soc. Rev., 2021, 50, 5517–5563 RSC.
- E. J. Barreiro, A. E. Kümmerle and C. A. M. Fraga, Chem. Rev., 2011, 111, 5215–5246 CrossRef CAS PubMed.
- H. Schönherr and T. Cernak, Angew. Chem., Int. Ed., 2013, 52, 12256–12267 CrossRef PubMed.
- A.-W. Struck, M. L. Thompson, L. S. Wong and J. Micklefield, ChemBioChem, 2012, 13, 2642–2655 CrossRef CAS PubMed.
- Y.-H. Lee, D. Ren, B. Jeon and H.-W. Liu, Nat. Prod. Rep., 2023, 40, 1521–1549 RSC.
- Q. Zhang, W. A. van der Donk and W. Liu, Acc. Chem. Res., 2012, 45, 555–564 CrossRef CAS PubMed.
- S. Ma, D. Mandalapu, S. Wang and Q. Zhang, Nat. Prod. Rep., 2022, 39, 926–945 RSC.
- Y. Chen, Chem. – Eur. J., 2019, 25, 3405–3439 CrossRef CAS PubMed.
- E. Mao and D. W. C. MacMillan, J. Am. Chem. Soc., 2023, 145, 2787–2793 CrossRef CAS PubMed.
- W. Zhang, C. Yang, C. Huang, L. Zhang, H. Zhang, Q. Zhang, C.-S. Yuan, Y. Zhu and C. Zhang, Org. Lett., 2017, 19, 592–595 CrossRef CAS PubMed.
- W. Zhang, Z. Liu, S. Li, Y. Lu, Y. Chen, H. Zhang, G. Zhang, Y. Zhu, G. Zhang, W. Zhang, J. Liu and C. Zhang, J. Nat. Prod., 2012, 75, 1937–1943 CrossRef CAS PubMed.
- J. Jin, X. Yang, T. Liu, H. Xiao, G. Wang, M. Zhou, F. Liu, Y. Zhang, D. Liu, M. Chen, W. Cheng, D. Yang and M. Ma, Mar. Drugs, 2018, 16, 87 CrossRef PubMed.
- C. Huang, C. Yang, Z. Fang, L. Zhang, W. Zhang, Y. Zhu and C. Zhang, Mar. Drugs, 2019, 17, 150 CrossRef CAS PubMed.
- C. Huang, C. Yang, Y. Zhu, W. Zhang, C. Yuan and C. Zhang, Front. Chem., 2018, 6, 528 CrossRef CAS PubMed.
- C. Huang, C. Yang, W. Zhang, L. Zhang, B. C. De, Y. Zhu, X. Jiang, C. Fang, Q. Zhang, C.-S. Yuan, H.-W. Liu and C. Zhang, Nat. Commun., 2018, 9, 2088 CrossRef PubMed.
- C. Yang, C. Huang, C. Fang, L. Zhang, S. Chen, Q. Zhang, C. Zhang and W. Zhang, J. Org. Chem., 2021, 86, 11019–11028 CrossRef CAS PubMed.
- C. Yang, C. Huang, W. Zhang, Y. Zhu and C. Zhang, Org. Lett., 2015, 17, 5324–5327 CrossRef CAS PubMed.
- B. C. De, W. Zhang, C. Yang, A. Mándi, C. Huang, L. Zhang, W. Liu, M. W. Ruszczycky, Y. Zhu, M. Ma, G. Bashiri, T. Kurtán, H.-W. Liu and C. Zhang, Nat. Commun., 2022, 13, 4896 CrossRef CAS PubMed.
- L. Zhang, B. C. De, W. Zhang, A. Mándi, Z. Fang, C. Yang, Y. Zhu, T. Kurtán and C. Zhang, J. Biol. Chem., 2020, 295, 16987–16997 CrossRef CAS PubMed.
- G. Rasul, G. K. S. Prakash and G. A. Olah, J. Org. Chem., 2000, 65, 8786–8789 CrossRef CAS PubMed.
- J.-Y. Wang, W.-J. Hao, S.-J. Tu and B. Jiang, Org. Chem. Front., 2020, 7, 1743–1778 RSC.
- C. Yang, L. Zhang, W. Zhang, C. Huang, Y. Zhu, X. Jiang, W. Liu, M. Zhao, B. C. De and C. Zhang, Nat. Commun., 2022, 13, 5386 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ob01920a |
| This journal is © The Royal Society of Chemistry 2024 |