Open Access Article
Open Access ArticleStar-polymer unimolecular micelle nanoparticles to deliver a payload across the blood–brain barrier†
Mehak
Malhotra
a,
Meenakshi
Pardasani
b,
Shahidkhan
Pathan
a,
Priyadharshini
Srikanth
b,
Karishma
Shaw
b,
Nixon M.
Abraham
 *b and
Manickam
Jayakannan
*b and
Manickam
Jayakannan
 *a
*a
aDepartment of Chemistry, Indian Institute of Science Education and Research (IISER Pune), Dr Homi Bhabha Road, Pune 411008, Maharashtra, India. E-mail: jayakannan@iiserpune.ac.in
bLaboratory of Neural Circuits and Behaviour (LNCB), Department of Biology, Indian Institute of Science Education and Research (IISER Pune), Dr Homi Bhabha Road, Pune 411008, Maharashtra, India. E-mail: nabraham@iiserpune.ac.in
First published on 29th October 2024
Abstract
Nanocarrier-mediated therapeutic delivery to brain tissue is impeded by tightly controlled transportation across the blood–brain barrier (BBB). Herein, we report a well-defined core–shell star-shaped unimolecular micelle (star-UMM; a single polymer entity) as an efficient BBB-breaching nanoparticle for brain-specific administration of the fluorescent anticancer drug doxorubicin and in vivo mapping of brain tissues by the near-infrared biomarker IR780 in mice. The star-UMM was engineered by precisely programming the polymer topology having hydrophobic and hydrophilic polycaprolactone blocks and in-built with lysosomal enzyme-biodegradation stimuli to deliver the payloads at intracellular compartments. In vivo imaging in mice revealed prolonged circulation of star-UMM in blood for >72 h, and whole-organ image-quantification substantiated its efficient ability to breach the BBB. Star UMM exhibited excellent stability in blood circulation and reduced cardiotoxicity, was non-hemolytic, had substantial uptake in the cortical neurons of the mouse brain, had lysosomal enzymatic-biodegradation, and exhibited negligible immunogenicity or necrosis. This newly designed star-UMM could have long-term applications in brain-specific drug delivery.
Introduction
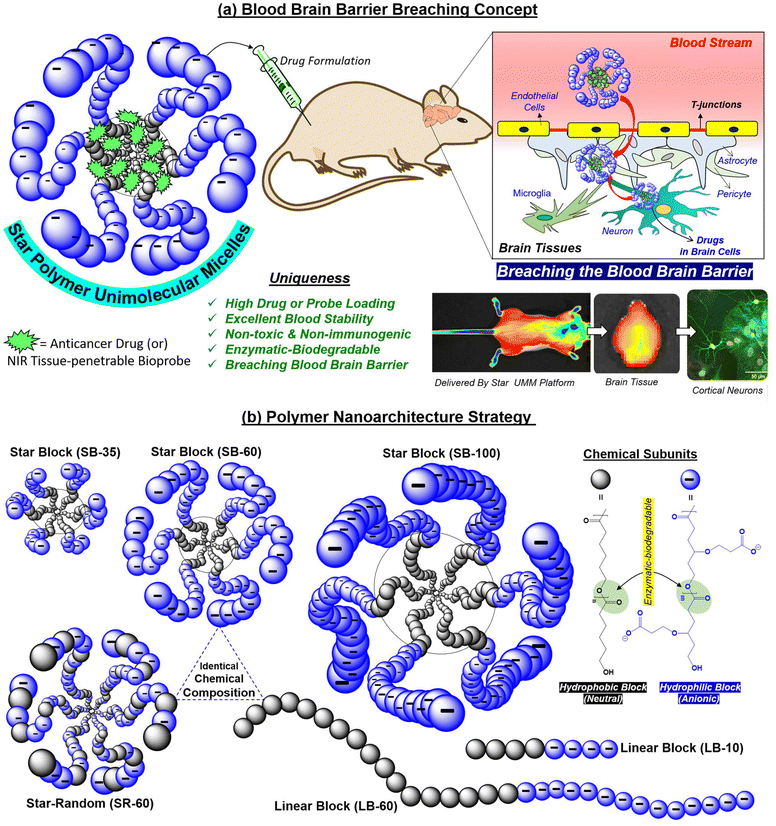
Drug delivery across the tightly regulated vasculature of the blood–brain barrier (BBB) in treating tumors and neurodegenerative diseases has been a major bottleneck.1–3 The BBB maintains the homeostatic balance by regulating the transport of small-molecule nutrients and ions through the vasculature into the brain; thus, restricting the transportation of larger size and extraneous species across this biological barrier.4 Nano-prodrug development has clearly identified that carriers <50 nm in size can penetrate the BBB and enable drug accumulation in the brain.5–9 Polypeptide nanoparticles (NPs), tumor-derived extracellular vesicles, synthetic protein NPs, nano-antioxidants, liposomes, dendrimers, worm-like micelles, and T cell-mediated polymer NPs are some of the important examples reported for BBB research.10–23 Intranasal24 and gut-to-brain oral drug-delivery models25 have also been developed to overcome BBB challenges encountered by intravenous delivery pathways. Near infrared (NIR)-assisted fluorescent NPs have been utilized for image-guided therapy for Parkinson's disease26 and glioblastoma.27 Uncontrollable glomerular renal filtration of smaller (<15 nm) nanocarriers, and splenic and hepatic filtration of bigger NPs (>200 nm) in the body limits the drug concentration of the NP formulation upon intravenous administration which, in turn, reduces the bioavailability of NPs for BBB crossing.28,29 Thus, the next generation of BBB-crossing nanocarriers is mandatorily designed to be substantially stable to evade disassembly in body fluidics in vivo, tiny-size sub-nanometer objects (<50 nm) for penetrating tightly regulated t-junctions, high drug-loading content, reduced cardiotoxicity and, most importantly, biodegradable for safe use in BBB research.30–34 Among the many synthetic NPs, branched macromolecular architectures such as “star block” copolymers exhibit resistance against renal filtration and retain the drug NP formulation in the blood for prolonged periods.29 Star-block copolymers provide excellent structural control to build well-defined core–shell NPs.35–40 Persistent to their three-dimensional globular core–shell geometry, star polymers often exist as unimolecular micelles41,42 which is highly desirable for in vivo drug administration to maintain the drug NP against the concentration gradient in the bloodstream. Herein, for the first time, these unique features of the star-polymer unimolecular micelles (star-UMM) were explored for BBB research based on biodegradable polymer “nanovectors”, and the proof-of-concept was demonstrated in vivo for the clinically important anticancer drug doxorubicin (DOX) and brain tissue-penetrable NIR biomarker IR-780. This new strategy is shown in Fig. 1.“Tweaking” the topology of the macromolecular architectures was found to be a crucial factor in designing the star-UMM. For this purpose, hydrophobic polycaprolactone (PCL) and carboxylic-substituted hydrophilic PCL segments were chosen based on our efforts.43–55 Systematically, several structures, such as linear di-blocks, star di-blocks, and star random copolymers, were “tailor-made” by ring-opening polymerization (ROP) to get the correct polymer geometry with high encapsulation capabilities. All these structures (including their linear polymer counterparts) were traced to obtain the “ideal” star-UMM platform with the ability to breach the BBB. Furthermore, the periphery of the star-UMM was decorated with an anionic charge. This is crucial for efficient penetration into brain tissue,56,57 long circulation against renal filtration in the blood stream58 and, most importantly, to elicit appropriate amphiphilicity for loading and delivering cargoes. The in vivo biodistribution data established the supremacy of the star-UMM platform for crossing the BBB with reduced side-effect of cardiotoxicity. Furthermore, microtubule-assisted protein 2 (MAP2), neuronal nuclei (NeuN) and glial fibrillary acidic protein (GFAP) immunostaining were employed to mark the different cell types across brain tissue to ascertain the neural uptake of star-UMM. Our approach opens up new research opportunities based on biodegradable star polymer macromolecules as potential futuristic single molecular-like star-UMM NPs to breach the BBB and be useful for long-term brain-specific drug delivery.
Results and discussion
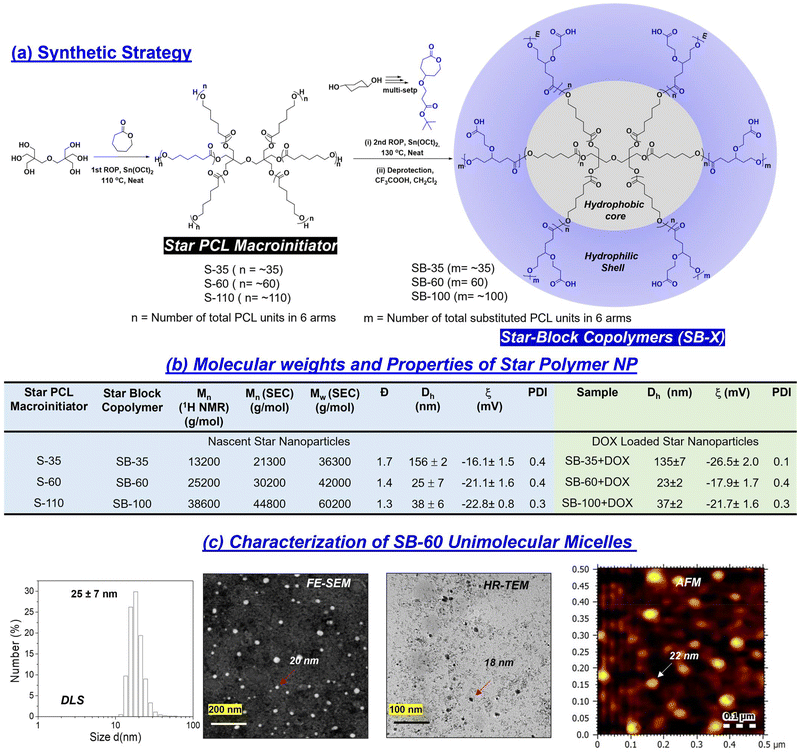
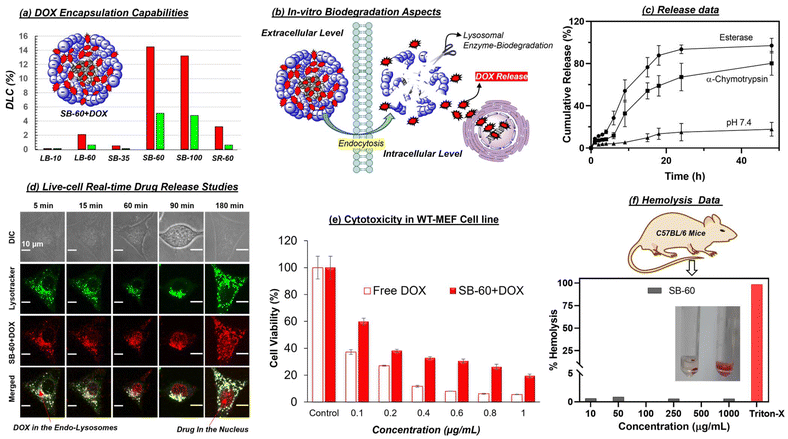
Star-block copolymers were constituted with a PCL core and γ-tButyl ester-substituted PCL segments at the periphery (as shown in Fig. 2a) employing an in-house built melt reactor (Fig. S1a†) to perform solvent-free ring-opening polymerization (ROP). Three star-polymers were synthesized in a sequential ROP process in which initial polymerization of ε-caprolactone (CL) yielded a six-arm PCL macroinitiator (MI) having statistically 5, 10 and 20 units per arm. These PCL MI were subsequently employed for the ROP of γ-tButyl ester-substituted caprolactone monomer48 (t-BECL) (Fig. S1b†), (Fig. 2a and Fig. S1c†) to yield three star-di-block polymers having 35/35, 60/60 and 120/120 units. For instance, the 60/60 di-block had 10 PCL and 10 t-BECL units per arm. The star block copolymers (SB) were referred to as “SB-35”, “SB-60”, and “SB-100” where the number represents the total content of carboxylic ester-substituted PCL segments at the periphery. To determine the structure and degree of polymerization (number of units), the peak intensities in 1H-NMR were analyzed in detail (Fig. S2 and S3a†). For feed [M]/[PCL MI] = 60, the actual incorporation was determined to be 64 ± 3 repeating units, confirming the statistical distribution of 10 units per arm in the second block (ESI Fig. S2a†). Deprotection of t-butyl ester in these star block copolymers yielded their carboxylic acid-substituted PCL block copolymers (1H-NMR) (Fig. S2 and S3a†). Size-exclusion chromatography (SEC) was employed to determine the Mn, Mw, and polydispersity index (PDI) and the values are summarized in the table in Fig. 2b (SEC plots) (Fig. S3b†). Star-block copolymers were produced in very high molecular weights of 40 kDa to 60 kDa, which are sustainably high enough to fold or self-assemble into a single polymer entity. The thermal properties of these new star polymers are described in Fig. S3c.† Hydrophilic carboxylic PCL units in the periphery and hydrophobic PCL units in the core provided perfect molecular geometry for the star polymers to acquire the required amphiphilicity. Star block copolymers were self-assembled by a dialysis method (details in the Experimental section). Dynamic light scattering (DLS) revealed the hydrodynamic diameter (Dh) of the star-block copolymer SB-60 to be Dh = 25 ± 7 nm (Fig. 2c). In Fig. 2c, FESEM and AFM images revealed the formation of spherical NPs with average sizes of 23 ± 3 nm and 22 ± 5 nm, respectively. HR-TEM images were in coherence, and the histogram generated (not shown) from ∼50 particles gave an average value of 24 ± 4 nm. From these data, the hydrodynamic radius (Rh) of the SB-60 NP was estimated to be 12 ± 3 nm. Small-angle X-ray scattering (SAXS) was employed to determine the radius of gyration (Rg) of SB-60 and, based on the Guinier approximation, the Rg was estimated to be 7.1 ± 2.3 nm (Fig. S4a†). The ratio of Rg/Rh was estimated to be 0.77 with respect to the existence of unimolecular micellar formulations.59 A pyrene-encapsulation study showed no change in the ratio of I1/I3 at different polymer concentrations, and depicted the unimolecular micelle self-assembly by the star-block copolymer SB-60 and SB-100 (Fig. S4b†). SB-35 was not readily dispersible in water and produced a turbid solution, rendering it unusable. The molecular weights of the polymers increased with an increase in the number of PCL and carboxylic PCL units in SB-35, SB-60 and SB-100. Interestingly, the increase in molecular weights in star block copolymers varied substantially in their aqueous self-assembly. For instance, the shorter-arm star polymer SB-35 self-assembled into aggregated micelles and produced larger size 150 nm NPs, whereas the higher star analogues SB-60 and SB-100 exhibited the formation of unimolecular micelles. The formation of unimolecular micelle seemed to be driven by high molecular weights and the star-polymer design. This was confirmed by comparing the self-assembly of the linear diblock copolymer LB-60, which had an identical mass as that of star SB-60; however, it differed largely in its aqueous self-assembly into aggregated micelles rather than unimolecular micelles. The above analysis denoted the existence of core–shell <30 nm-sized star-UMMs by both SB-60 and SB-100. The pH-dependent zeta potential and size analysis confirmed negatively charged stable NPs across a pH range from 4 to 11 (Fig. S5†).The encapsulation capabilities of star-UMM NPs were studied for doxorubicin (DOX) and the NIR dye IR780 by the dialysis method (Experimental section). The DLC for SB-60, SB-100, SB-35 was found to be 14.2%, 13.1% and <1% for DOX, respectively (Fig. 3a). This suggested that SB-60 exhibited the most optimized core–shell structure to attain the highest DLC for star-UMM. SB-35 did not have sufficient compartmentalization for DOX loading. The core–shell geometry probably attained maximum packing at SB-60; thus, no significant DLC increase was observed in SB-100. Encapsulation of NIR dye IR780 in SB-60 showed a very good DLC of 5%, which is excellent for deep-tissue bioimaging analysis. The sizes of nascent and DOX-loaded star block copolymer formulations are tabulated in Fig. 2b (DLS plots in Fig. S4c†). All the nano-formulations showed monomodal size distribution with narrow PDI values. The IR780-loaded star block copolymer SB-60 analogue had a size in the range of 120 ± 10 nm. The zeta potential for SB60 and SB100 and their DOX-loaded samples were recorded in Britton–Robinson buffer (10 mM), physiological pH 7.4, and the values are reported in the table in Fig. 2b. The zeta potential of SB-35 could not be measured in Britton–Robinson buffer (10 mM) due to uncontrolled precipitation and, therefore, the values were reported in water. The zeta potential of SB-60 and SB-100 polymers was found to be −21.1 and −22.8 mV, respectively. As anticipated, SB-100 exhibited a slightly higher zeta potential due to the increase in the number of carboxylic acids. Similar trends were observed for their DOX-loaded samples as well. The zeta potential did not show a drastic change when increasing the number of carboxylic units from 60 in SB-60 to 100 in SB-100. This could be attributed to the fact that the charges were saturated on the periphery of the polymer nano-assemblies at 60 carboxylic units. The collapsing of a polymer chain in each-arm is typically a chain length-dependent process, and it seems 60 carboxylic units was the optimum length in the present star-polymer design. We wished to rationalize the role of topology of the star-block copolymer architectures towards its ability to self-assemble into unimolecular micelles in aqueous medium. Hence, two controlled molecules having linear di-block (LB-60) and star-random copolymer (SR-60, no segregation of core and shell) architectures were made. The chemical compositions and molecular weights of SB-60, LB-60 and SR-60 were identical, and they differed only by the arrangements of repeating units (Fig. S6a†). SR-60 self-assembled as a NP of size Dh = 30 ± 5 nm, like that of SB-60 (Fig. S6b†). However, the linear di-block LB-60 exhibited a Dh of 170 ± 10 nm with respect to the formation of large-sized aggregated micelles (FESEM and HR-TEM images in Fig. S6c†). The pyrene-encapsulation experiment for LB-60 showed a breakpoint with respect to a critical micellar concentration of 1 μg mL−1 (Fig. S6c†), as typically reported for aggregated micelles. SR-60 and LB-60 exhibited DLC = 3% and 2% for DOX encapsulation which was almost 7-fold lower than that of SB-60 star-UMM (Fig. 3a). Furthermore, the linear di-block copolymer LB-10 was synthesized with 10-PCL units and 10-carboxyl PCL units to mimic the 1-arm of the SB-60 star di-block copolymer (Fig. S7†) and it demonstrated inferior DOX encapsulation (0.2%). These findings reiterated the importance of polymer topology for producing UMM with a high degree of drug loading and, hence, offers an excellent nanocarrier for drug delivery. All the details of % DLC and drug-loading efficiency (% DLE) for star and linear NPs are summarized in tabular form in Fig. S8.†
An aliphatic polyester backbone in star-UMM makes them fully lysosomal enzymatic-biodegradable, as shown pictorially (Fig. 3b). In the presence of horse-liver esterase enzyme (10 U), >95% drug release was observed within 24 h (Fig. 3c). Enzymatic cleavage by α-Chymotrypsin (8 U) resulted in release of only 60% of DOX molecules. In the control, percentage release was substantially lower (∼15%), indicating that the degradation of the UMM occurred only in the presence of lysosomal enzymes. The horse-liver esterase enzyme seemed to be the most suitable for complete degradation of the nano-assemblies (Fig. 3b gives a pictorial representation of the intracellular enzymatic biodegradation). The time-scan CLSM images in Fig. 3d captured cells incubated with SB-60 + DOX unimolecular micelle for the 5–180 min time points (live cell). LysoTracker™ staining helped to visualize the uptake of NPs by the cells and co-localization of the DOX signal at lysosomal compartments. The signals from LysoTracker (green) and DOX (red) can be seen as yellow in the merged image. This trend was because the star-nanocarrier was taken up readily by the cells via endocytosis and internalized at the lysosomal compartment for biodegradation. A control experiment with free DOX (60 and 90 min time points in Fig. S9†) exhibited no colocalization with LysoTracker, indicating that DOX, being a small molecule, was taken up by the cells via diffusion. To determine the cyto-compatibility of these nanocarriers, the MTT assay was employed in WT-MEF cell lines. Various concentrations of the nascent star block copolymer scaffold SB-60 were incubated with WT-MEF cells for 72 h (Fig. S9†). As evident from the histogram, the polymer scaffold displayed 100% biocompatibility up to 100 μg mL−1, and about 70–80% of cells were viable up to 200–500 μg mL−1. Furthermore, the cytotoxicity of the DOX-loaded scaffold SB-60 + DOX showed that the free DOX was more toxic to cells compared with their delivery from the polymer platform (Fig. 3e). The IC50 values for free DOX and SB60 + DOX in the WT-MEF cell line was 0.09 ± 0.014 and 0.11 ± 0.02 μg mL−1, respectively. Furthermore, the compatibility and efficacy were evaluated for SB-60 and SB-60 + DOX NPs in a neuroblastoma (SH-SY5Y) cell line and data are shown in Fig. S10.† The SB-60 NPs exhibited excellent compatibility, with 100% cell viability up to 100 μg mL−1. On the other hand SB-60 + DOX (IC50 = 1.39 ± 0.11 μg mL−1) showed slightly better killing compared with nascent DOX (IC50 = 1.69 ± 0.12 μg mL−1), as shown in Fig. S10.† Cellular-uptake studies of SB-60 + DOX in SH-SY5Y cells revealed enhanced fluorescence signals with an increase in incubation time, suggesting an effective delivery ability of the star platform in neuroblastoma, as shown in Fig. S10.† Further hemolysis assays clearly exhibited polymer biocompatibility with negligible hemolysis values at concentrations as high as 1000 μg mL−1 (Fig. 3f).
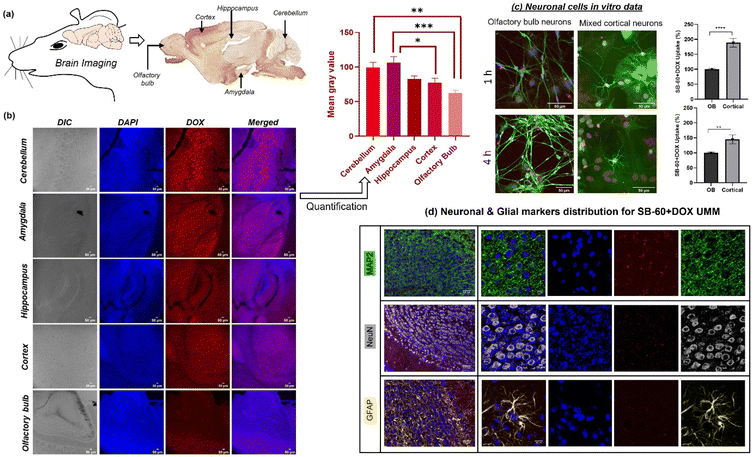
The biodistribution and biochemical analysis of the DOX-loaded star-UMM was investigated. Ten female mice (8–12 weeks, balb/c strain, ∼25 g) were split into two experimental groups: ‘free DOX’ (group 1) and DOX-loaded star-block copolymer ‘SB-60 + DOX’ (group 2). Each group comprised three mice that were used for confocal imaging analysis of drug uptake by organs and for histology. To understand the biocompatibility of the polymers alone, an additional group (n = 3) was constituted as the control group (SB-60; group 3), to confirm that the nascent polymer itself did not alter the physiology of the animal. Histology images of various organs upon SB-60 + DOX uptake via H&E staining are shown in Fig. 4a. The high-magnification images (×40) obtained using a bright field microscope (Carl Zeiss) showed no signs of necrosis, blood clotting, or morphological alterations in any of the tissue samples for mice in the SB-60 + DOX group. However, in the panel corresponding to the free DOX group, some blood clotting was observed in heart tissue and, according to the literature,60 this is a sign of cardiotoxicity augmented by free DOX (Fig. S11†). Hence, the star block copolymer DOX-loaded assemblies did not result in any damage to tissues, unlike free DOX. To study the immune response in mice, plasma samples collected from all 10 mice (three groups plus one mouse injected with 1× PBS as control) at 24 h were employed to determine the concentration of five cytokines (IL-2, IL-4, IL-17A, IFN-γ and TNF-α) using cytokine standards. The mouse injected with PBS acted as the negative control, wherein no immune response was expected because PBS is non-immunogenic. Across all groups, cytokine levels were very similar to that observed using PBS, and values were <5 pg mL−1 (Fig. 4b), thereby suggesting that the nano-formulations were not immunogenic. As can be seen from Fig. 4a, for each mouse, the organs collected 24 h-post-injection were brain, heart, kidneys, liver, and spleen. The DAPI-stained 50 μm sections were imaged to measure DOX uptake in brain and heart tissues across different groups, as shown in Fig. 4c (63× magnification) and Fig. S12† (63× magnification). Images from the kidney, liver, spleen, brain, and heart (10× magnification) are shown in Fig. S12.† In Fig. 4c, in the brain images, only the SB-60 + DOX nano-formulation exhibited a strong red fluorescence signal (DOX) whereas, a substantially low signal was observed in the free DOX group. This finding was attributed to the SB-60 + DOX UMM being 18–30 nm, making them ideal nano-formulations to cross the BBB. These SB-60 + DOX micelles passively and selectively cross the BBB, accumulate in brain tissue and, being stable at infinite dilution, would not result in premature release of the drug. Another positive aspect for these SB-60 + DOX micelles was their ability to reduce uptake in heart tissue, as can be seen from Fig. 4c. The signal of the SB-60 + DOX micelles was extremely weak compared with the strong red signal corresponding to free DOX. This was one of the highlights of our study because the major side effect of chemotherapy with DOX is cardiotoxicity. Hence, reducing uptake of DOX in the heart by means of the nano-formulations would greatly overcome the toxic effects of DOX. Thus, the current design of SB-60 UMM can overcome the limitation, and facilitates the use of even higher doses of DOX for chemotherapy in the long-term. The DOX uptake from confocal images was quantified by determining the normalized mean gray values, and shown in the form of bar plots in Fig. 4c. These values for the SB-60 + DOX micelle exhibited enhanced uptake in the brain and drastically reduced uptake by the heart compared with an opposing trend in the free DOX group. Renal clearance was higher in case of free DOX compared with that in the other group. SB-60 + DOX exhibited reduced RES uptake, as can be seen from the low mean gray values in the liver and spleen tissues. An important observation was the significantly higher uptake of SB-60 + DOX UMM in brain tissue compared with free DOX. This was further closely investigated by measuring DOX biodistribution across brain regions under different conditions.
Five areas of the brain (cerebellum, amygdala, hippocampus, cortex, and olfactory bulb (OB)) were chosen for further analysis (posterior to anterior; see the labelling on the sagittal section of the mouse brain in Fig. 5a, adapted from the Gene Expression Nervous System Atlas).61 Quantifying the DOX intensities from the confocal images in Fig. 5b led to interrogation of whether the nano-formulations were taken up equally by different parts of the brain. The representative confocal images (Fig. 5b) exhibited that the DOX intensities visibly decreased upon going from the posterior part (cerebellum) to the most anterior part (OB). This was corroborated by the quantification of DOX intensities as normalized mean gray values plotted against the corresponding region of the brain (Fig. 5b). This trend could be attributed to the variation in BBB heterogeneity and permeability across different brain regions, which depends on differential astrocyte and pericyte coverage, differences in tight-junction proteins such as zonula occludens (ZO)-1 and ZO-2, variation in cellular interactions between white matter and gray matter in different regions, and changes in vascular density.62 To validate the ability of star block copolymer UMM to enter neurons upon crossing the BBB, an in vitro time-dependent experiment was envisaged. Herein, SB-60 + DOX UMM were incubated with the OB and a mixed cortical primary neuronal culture for 1 h and 4 h followed by immunocytochemistry, and the panels are shown in Fig. 5c. Staining (DAPI and MAP2 antibody) was employed for imaging the differentiated mature neurons, represented via the blue (λexc = 405 nm) and green channels (λexc = 633 nm), respectively. DOX emission (red channel, λexc = 488 nm) from the UMMs had substantial co-localization with neuronal markers. The SB-60 + DOX uptake in OB and cortical neurons was quantified. The plot of uptake (%) vs. time revealed significantly higher uptake of SB-60 + DOX UMM in cortical neurons as compared with that in OB neurons across both time points (Fig. 5c). The in vitro neuronal culture data were in alignment with the in vivo brain biodistribution data, wherein UMM uptake was higher in the cortex as opposed to the OB (Fig. 5b). To examine the cell specificity of these micelles in brain tissue, immunostaining was carried out marking different brain tissue cell types (i.e., MAP2 for staining mature neurons, NeuN as the nuclei marker and GFAP for labelling glial cells and astrocytes). The intent was to investigate the specificity of DOX-loaded unimolecular micelles across brain tissue (if any), and the results can be seen in Fig. 5d. The emission of DOX coming from within mature neurons appeared to co-localize with markers, as can be seen from the merged images (10×-first column and 63×-second column) in Fig. 5d. This affirmed intracellular uptake of the cargo and that uptake was similar across the brain tissue with no specificity for any cell type.
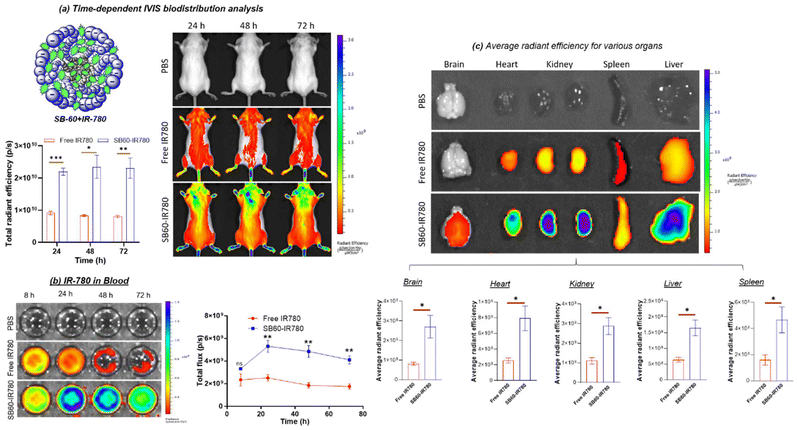
NIR dye IR780, having excitation in the NIR region, overcomes the limitation of tissue auto-fluorescence while offering the advantage of deeper penetration into tissue.63,64 Two groups of mice (n = 3) were used. Mice injected with free IR780 iodide dye (group 1) and SB-60 + IR780 (group 2) were utilized along with a control mouse (injected with 1× PBS) for in vivo biodistribution using the In Vivo Imaging System (IVIS) from PerkinElmer. The IVIS imaging for tracking the biodistribution was carried out at 24 h, 48 h and 72 h, as can be seen in Fig. 6a (dorsal view). Using the dorsal view images of mice, an attempt was made to quantify the IR780 distribution in the most rostral part of the body with time across the two groups by selecting a region of interest (ROI). The plot of total radiant efficiency vs. time in Fig. 6a clearly demonstrated that UMM SB-60 + IR780 exhibited a superior potential to penetrate the BBB as opposed to the free dye, and that this distribution did not change with time up to 72 h. Plasma samples were also subjected to IVIS imaging to quantify the amount of IR780, and the representative photographic image of the wells can be seen in Fig. 6b. The SB60 + IR780 nano-scaffolds demonstrated their ascendancy in their ability to be in circulation for longer than the free dye (see plot of total flux vs. time). The whole-organ representative IVIS image for the major organs captured at the 72 h time point can be seen in Fig. 6c. As can be seen from the average radiant efficiency plot, the SB-60 + IR780 unimolecular micelle had higher uptake in all the tissues compared with free IR780 dye. The whole-organ imaging reiterated the ability of the SB-60 + IR780 star UMM to breach the BBB.
A mechanism illustrating the ability of the nano-scaffold to penetrate the BBB compared with that of the free drug is outlined in Fig. 7a. Transcytosis across the barrier is dictated by factors such as lipophilicity, size, charge, molecular weight and, even with the requisite parameters, molecules undergo efflux out into the blood stream via the P-glycoprotein (Pgp) pump.65–67 The crucial parameters for NPs intended to penetrate the BBB are size <50 nm, appropriate lipophilicity, and the surface charge should be near neutral.65–67 In the current investigation, a plausible mechanism of BBB breach can be ascribed to caveolae-mediated transcytosis, which transports albumin-like macromolecules from the luminal side to brain parenchyma.65–67 Crossing of the BBB by the star block copolymer UMM in the present study could be attributed to three main factors: (i) prolonged circulation in blood making it more bioavailable; (ii) appropriate lipophilicity favorable for transcytosis; (iii) the presence of carboxylate groups on the periphery that circumvent Pgp efflux pumps. These factors, combined with the size range of the UMM (<30 nm), could aid in breaching the BBB. Having established the ability of star UMM to cross the BBB, another crucial investigation was to determine whether the integrity of the BBB was compromised in this process. The Evans Blue (EB) extravasation assay68,69 and IR780 breaching ability across the BBB were employed as tools. EB, being a BBB-impermeable dye, was chosen to evaluate BBB disruption by detecting its presence in brain tissue. As can be seen in Fig. 7b and Fig. S13,† blue coloration was not observed in the brain tissues of control- and SB60-treated groups, suggesting that the integrity of BBB was maintained upon administration of star-UMM polymer (SB60). Furthermore, the amounts of dye leaked into brain tissue were quantified, and it was found to be similar in the SB60 group and control group. In another experiment, mice were initially treated with SB60 nascent NPs for 24 h followed by administration of free IR780 for 24 h, as can be seen in Fig. 7c. In the scenario that the BBB integrity was disrupted by the star UMM, there would be significant uptake of IR780 dye. The IVIS image of brain tissues in Fig. 7c clearly demonstrates that IR780 was taken up significantly when delivered via the star-UMM platform. On the other hand, treatment with nascent SB60 (without IR780) for 24 h followed by treatment with free IR780 did not show significant uptake. These investigations suggested that the star-UMM displayed the unique ability to cross the BBB without compromising its integrity. Taking cognizance of in vivo and in vitro analyses, star-UMM demonstrated an excellent capability to breach the most tightly regulated biological barrier: the BBB. These findings call for further experiments employing potential delivery systems combined with targeted delivery70 and precise behavioral paradigms controlling sensory experiences.71–75 Our results provide a potential drug-delivery method for brain tumors, and add significantly to the emerging field of “cancer neuroscience” research.
Conclusions
Tracing through the plethora of star and linear polymer architectures, the unique star block UMM evinced the crucial factors necessary for a BBB nanocarrier. The factors were the star macromolecular topology, size <50 nm, UMM assembly, as well as the ability to carry and deliver a high amount of desired payload across the BBB. The data for in vitro cellular uptake suggested an internalization mechanism of DOX-loaded UMM via endocytosis and their enzymatic-biodegradation at lysosomal compartments. A myriad of experiments were carefully designed to explore the potential of the star UMM in vivo. The pharmacokinetics evaluation established the high bioavailability of star UMM nanocarriers owing to their stability against dilution. The biodistribution of the star-UMM was visualized in real-time by employing IVIS, which reiterated the potential of the UMM to penetrate the BBB to deliver dyes/drugs. The star polymer nano-formulations were non-immunogenic and biocompatible. UMM uptake in brain tissue was influenced by BBB heterogeneity; in vitro data for primary neuronal cultures also demonstrated enhanced uptake of the UMM by mixed cortical neurons. Hence, the current star-polymer design opens up opportunities for UMMs and demonstrates their ability to penetrate the BBB in vivo to deliver drugs which could be useful for the treatment of brain-related malignancies.Notes
The authors declare no competing financial interest.All animal procedures were carried out at National Facility for Gene Function in Health and Diseases (Indian Institute of Science Education and Research (IISER), Pune, India) with compliance of Institutional Animal Ethics Committee (IAEC) at IISER Pune and the Committee for the Control and Supervision of Experiments on Animals (CCSEA), Government of India guidelines. Animal ethics committee approval number is IISER/IAEC/2018-02/07.
Statistics
All analyses were done using Prism 8.0 (GraphPad). One-way ANOVA, Tukey's test, and unpaired t-test were performed, and values are represented as the mean ± SEM.Author contributions
Mehak Malhotra contributed to polymer synthesis, characterization, nano-formulation, in vitro and in vivo analysis. Meenakshi Pardasani contributed to in vivo and brain-tissue analysis. Shahidkhan Pathan contributed to polymer synthesis and in vivo brain studies, Priya Srikanth conducted analysis of uptake by brain cells. Karishma Shaw conducted the study using the neuroblastoma cell line. Nixon Abraham and Manickam Jayakannan conceptualized the study, drafted and edited the final version of the manuscript.Data availability
The authors declare that the data supporting the findings of this study are available within this document and its ESI† files.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors obtained research grants from Science and Engineering Research Board (CRG/2023/000894; New Delhi, India), DBT/Wellcome Trust India Alliance intermediate (IA/I/14/1/501306 to N.A.), DST-Cognitive Science Research Initiative (DST/CSRI/2017/271 to N.A.), and DBT/Wellcome Trust India Alliance senior grant (IA/S/22/2/506517 to N.A.). We thank IISER Pune Microscopy Facility for cellular imaging and PerkinElmer Centre for Excellence IISER Pune. Part of the work was carried at the National Facility for Gene Function in Health and Disease at IISER Pune, supported by a grant from the Department of Biotechnology, Government of India (BT/INF/22/SP17358/2016).References
- W. A. Banks, Nat. Rev. Drug Discovery, 2016, 15, 275–292 CrossRef PubMed.
- I. U. Ali and X. Chen, ACS Nano, 2015, 9, 9470–9474 CrossRef PubMed.
- J. Kreuter, Adv. Drug Delivery Rev., 2001, 47, 65–71 CrossRef PubMed.
- L. Gastaldi, L. Battaglia, E. Peira, D. Chirio, E. Muntoni, I. Solazzi, M. Gallarate and F. Dosio, Eur. J. Pharm. Biopharm., 2014, 87, 433–444 CrossRef.
- W. Tang, W. Fan, J. Lau, L. Deng, Z. Shen and X. Chen, Chem. Soc. Rev., 2019, 48, 2967–3014 RSC.
- Y. Chen and L. Lu, Adv. Drug Delivery Rev., 2012, 64, 640–665 CrossRef PubMed.
- Y. Zhaou, Z. Peng, E. S. Seven and R. M. Leblanc, J. Controlled Release, 2018, 270, 290–303 CrossRef PubMed.
- J. Kreuter, Adv. Drug Delivery Rev., 2014, 71, 2–14 CrossRef CAS PubMed.
- S. Wohlfart, S. Gelperina and J. Kreuter, J. Controlled Release, 2012, 161, 264–273 CrossRef CAS.
- Y. Liu, Y. Zou, C. Feng, A. Lee, J. Yin, R. Chung, J. B. Park, H. Rizos, W. Tao, M. Zheng, O. C. Farokhzad and B. Shi, Nano Lett., 2020, 20, 1637–1646 CrossRef CAS.
- W. Lv, J. Xu, X. Li, Q. Xu and H. Xin, ACS Nano, 2018, 12, 5417–5426 CrossRef CAS PubMed.
- A. Singh, W. Kim, K. Jeong, C. H. Kang, Y. S. Kim, J. Koh, S. D. Mahajan, P. N. Prasad and S. Kim, Adv. Healthcare Mater., 2016, 26, 7057 CAS.
- G. Morad, C. V. Carman, E. J. Hagedorn, J. R. Perlin, L. I. Zon, N. Mustafaoglu, T.-E. Park, D. E. Ingber, C. C. Daisy and M. A. Moses, ACS Nano, 2019, 13, 13853–13865 CrossRef CAS.
- S. Singh, N. Drude, L. Blank, P. B. Desai, H. Konigs, S. Rutten, K. Langen, M. Moller, F. M. Mattaghy and A. Morgenroth, Adv. Healthcare Mater., 2021, 10, 2100812 CrossRef CAS PubMed.
- R. Prades, B. Oller-Salvia, S. M. Schwarzmaier, J. Selva, M. Moros, M. Balbi, V. Grazu, J. M. de La Fuente, G. Egea, N. Plesnila, M. Teixido and E. Giralt, Angew. Chem., Int. Ed., 2015, 54, 3967–3972 CrossRef CAS.
- D. Gaurnieri, A. Falanga, O. Muscetti, R. Torallo, S. Fusco, M. Galdiero, S. Galdiero and P. A. Netti, Small, 2013, 9, 853–862 CrossRef.
- T. Lin, P. Zhao, Y. Jiange, Y. Tang, H. Jin, Z. Pan, H. He, V. C. Yang and Y. Huang, ACS Nano, 2016, 10, 9999–10012 CrossRef PubMed.
- C. Martinelli, C. Pucci, M. Battaglini, A. Marino and G. Ciofani, Adv. Healthcare Mater., 2020, 9, 1901589 CrossRef.
- B. Surnar, A. S. Shah, M. Park, A. A. Kalathil, M. Z. Kamran, R. J. Jaime, M. Toborel, M. Nair, N. Kolishetti and S. Dhar, ACS Nano, 2021, 15, 15741–15753 CrossRef.
- V. Leiro, S. D. Santos, C. D. F. Lopes and A. P. Pego, Adv. Funct. Mater., 2018, 28, 1700313 CrossRef.
- G. K. Babanyinah, A. Bhadran, H. Polara, H. Wang, T. Shah, M. C. Biewer and M. C. Stefan, Chem. Sci., 2024, 15, 9987–10001 RSC.
- M. Ayer, M. Schuster, I. Gruber, C. Blatti, E. Kaba, G. Enzmann, O. Burri, R. Guiet, A. Seitz, B. Engelhardt and H.-A. Kloak, Adv. Healthcare Mater., 2021, 10, 2001375 CrossRef.
- A. K. Sarkar, G. Kura, P. Seth, N. R. Jana and N. R. Jana, ACS Appl. Nano Mater., 2024, 7, 3468–3478 CrossRef.
- S. Zha, K.-L. Wong and A. H. All, Adv. Healthcare Mater., 2022, 11, 2102610 CrossRef CAS.
- Y.-B. Miao, K.-H. Chen, C.-T. Chen, F.-L. Mi, Y.-J. Lin, Y. Chang, C.-S. Chiang, J.-T. Wang, K.-J. Lin and H.-W. Sung, Adv. Mater., 2021, 33, 2100701 CrossRef CAS PubMed.
- Y. Gao, Y. Cheng, J. Chen, D. Lin, C. Liu, L.-K. Zhang, L. Yin, R. Yang and Y.-Q. Guan, Adv. Healthcare Mater., 2022, 11, 2201655 CrossRef CAS.
- D. Reichel, B. Sagong, J. Teh, Y. Zhang, S. Wagner, H. Wang, L. W. K. Chung, P. Butte, K. L. Black, J. S. Yu and J. Manual Perez, ACS Nano, 2020, 14, 8392–8408 CrossRef CAS PubMed.
- H. S. Choi, W. Liu, P. Misra, E. Tanaka, J. P. Zimmer, B. I. Ipe, M. G. Bawendi and J. V. Frangioni, Nat. Nanotechnol., 2007, 25, 1165 CAS.
- M. E. Fox, F. C. Szoka and M. J. Frechet, Acc. Chem. Res., 2009, 42, 1141–1151 CrossRef CAS PubMed.
- L. Zhang, K. Yao, Y. Wang, Y. L. Zhou, Z. Fu, G. Li, J. Ling and Y. Yang, Nano Lett., 2021, 21, 3007–3015 CrossRef CAS PubMed.
- R. Pahuja, K. Seth, A. Shukla, R. K. Shukla, P. Bhatnagar, L. K. Singh Chauhan, P. N. Saxena, J. Arun, B. P. Chaudhari, D. K. Patel, S. P. Singh, R. Shukla, V. K. Khanna, P. Kumar, R. K. Chaturvedi and K. C. Gupta, ACS Nano, 2015, 9, 4850–4871 CrossRef CAS.
- H. Guerrero-Cazares, S. Y. Tzeng, N. P. Young, A. O. Abutaleb, A. Quinones-Hinojisa and J. J. Green, ACS Nano, 2014, 8, 5141–5153 CrossRef CAS PubMed.
- A. J. Clark and M. E. Davis, Proc. Natl. Acad. Sci. U. S. A., 2015, 6, 12486–12491 CrossRef.
- D.-P. Yang, M. N. N. Linn Oo, G. R. Deen, Z. Li and X. J. Loh, Macromol. Rapid. Commun., 2017, 38, 1700410 CrossRef PubMed.
- J. M. Ren, T. G. McKenzie, Q. Fu, E. H. H. Wong, J. Xu, Z. An, S. Shanmugam, T. P. Davis, C. Boyer and G. G. Qiao, Chem. Rev., 2016, 116, 6743–6836 CrossRef CAS PubMed.
- W. Wu, W. Wang and J. Li, Prog. Polym. Sci., 2015, 46, 55–85 CrossRef CAS.
- M. Trollsas and J. L. Hedrick, J. Am. Chem. Soc., 1998, 120, 4644–4651 CrossRef.
- K. M. Fichter, L. Zhang, K. L. Kiick and T. M. Reineke, Bioconjugate Chem., 2008, 19, 76–88 CrossRef CAS PubMed.
- S. P. Chali, S. Azhadri, A. Galstyan, A. H. Groschel and B. J. Ravoo, Chem. Commun., 2021, 57, 9446–9449 RSC.
- V. Karmegam, S. S. Kuruppu, C. M. Udamulle Gedara, M. C. Biewer and M. C. Stefan, J. Polym. Sci., 2021, 59, 3040–3052 CrossRef CAS.
- G. Chen, Y. Wang, R. Xie and S. Gong, Adv. Drug Delivery Rev., 2018, 130, 58–72 CrossRef CAS.
- H. Liu, S. Farrell and K. Uhrich, J. Controlled Release, 2000, 68, 167–174 CrossRef CAS.
- B. Kulkarni, M. Malhotra and M. Jayakannan, Chem. - Asian J., 2022, e202101337 CrossRef.
- R. Ghosh, M. Malhotra, R. R. Madhuri Sathe and M. Jayakannan, Biomacromolecules, 2020, 21, 2896–2912 CrossRef PubMed.
- B. Kulkarni, M. Malhotra and M. Jayakannan, ACS Appl. Polym. Mater., 2019, 1, 3375–3388 CrossRef.
- B. Kulkarni and M. Jayakannan, ACS Biomater. Sci. Eng., 2017, 3, 2185–2197 CrossRef PubMed.
- B. Surnar and M. Jayakannan, Biomacromolecules, 2016, 17, 4075–4085 CrossRef PubMed.
- M. Malhotra, B. Surnar and M. Jayakannan, Macromolecules, 2016, 49, 8098–8112 CrossRef.
- B. Surnar and M. Jayakannan, ACS Biomater. Sci. Eng., 2016, 2, 1926–1941 CrossRef PubMed.
- B. Kulkarni, B. Surnar and M. Jayakannan, Biomacromolecules, 2016, 17, 1004–1016 CrossRef CAS PubMed.
- B. Surnar, K. Sharma and M. Jayakannan, Nanoscale, 2015, 7, 17964–17979 RSC.
- B. Surnar and M. Jayakannan, Biomacromolecules, 2013, 14, 4377–4387 CrossRef CAS PubMed.
- U. Pranav, M. Malhotra, S. Pathan and M. Jayakannan, ACS Biomater. Sci. Eng., 2023, 9, 743–759 CrossRef CAS PubMed.
- R. Ghosh and M. Jayakannan, Biomacromolecules, 2023, 24, 739–755 CrossRef CAS PubMed.
- S. Pathan and M. Jayakannan, Adv. Healthcare Mater., 2024, 13, 2304599 CrossRef CAS PubMed.
- P. R. Lockman, J. M. Koziara, R. J. Mumper and D. D. Allen, J. Drug Targeting, 2004, 12, 635–641 CrossRef CAS PubMed.
- E. A. Nance, G. F. Woodworth, K. A. Sailor, T.-Y. Shih, Q. Xu, G. Swaminathan, D. Xiang, C. Eberhart and J. Hanes, Sci. Transl. Med., 2012, 4, 149ra119 Search PubMed.
- J. H. Miner, Kidney Int., 2008, 74, 559–561 CrossRef.
- L. J. M. Vagberg, K. A. Cogan and A. P. Gast, Macromolecules, 1991, 24, 1670–1677 CrossRef CAS.
- H. Tang, J. Zhang, J. Tang, Y. Shen, W. Guo, M. Zhou, R. Wang, N. Jiang, Z. Gan and Q. Yu, Biomacromolecules, 2018, 19, 2849–2862 CrossRef CAS.
- S. Gong, C. Zheng, M. L. Doughty, K. Losos, N. Didkovsky, U. B. Schambra, N. J. Nowak, A. Joyner, G. Leblanc, M. E. Hatten and N. Heintz, Nature, 2003, 425, 917 CrossRef CAS PubMed.
- M. Suciu, A. Hermenean and I. Wilhelm, Tissue Barriers, 2016, 4, e1143544 CrossRef.
- L. Wang and C. Niu, J. Mater. Chem. B, 2021, 9, 4079–4097 RSC.
- U. A. Gavhane, D. C. Joshi and M. Jayakannan, Biomacromolecules, 2024, 25, 3756–3774 CrossRef CAS.
- K. K. Pulicherla and M. Kumar Verma, AAPS PharmSciTech, 2015, 16, 223 CrossRef CAS PubMed.
- M. Zhou, S. X. Shi, N. Liu, Y. Jiang, M. S. Karim, S. J. Vodovoz, X. Wang, B. Zhang and A. S. Dumont, J. Clin. Med., 2021, 10, 3795 CrossRef CAS PubMed.
- S. Ayloo and C. Gu, Curr. Opin. Neurobiol., 2019, 57, 32–38 CrossRef CAS PubMed.
- M. P. de Souza Goldim, A. D. Giustina and F. Petronilho, Curr. Protoc. Immunol., 2019, 126, e83 CrossRef PubMed.
- J.-H. Gu, et al. , PLoS One, 2013, 8, e61641 CrossRef CAS PubMed.
- N. M. Abraham, V. Egger, D. R. Shimshek, R. Renden, I. Fukunaga, R. Sprengel, P. H. Seeburg, M. Klugmann, T. W. Margrie, A. T. Schaefer and T. Kuner, Neuron, 2010, 65, 399–411 CrossRef PubMed.
- A. S. Bhattacharjee, S. Konakamchi, D. Turaev, R. Vincis, D. Nunes, A. A. Dingankar, H. Spors, A. Carleton, T. Kuner and N. M. Abraham, Cell Rep., 2019, 28, 2966–2978 CrossRef PubMed.
- N. M. Abraham, D. Guerin, K. Bhaukaurally and A. Carleton, PLoS One, 2012, 7, 112 Search PubMed.
- S. Mahajan, D. Sen, A. Sunil, P. Srikanth, S. D. Marathe, K. Shaw, M. Sahare, S. Galande and N. M. Abraham, Front. Neurosci., 2023, 17, 1180868 CrossRef PubMed.
- M. Pardasani, S. D. Marathe, M. M. Purnapatre, U. Dalvi and N. M. Abraham, FASEB J., 2021, 9, 35 Search PubMed.
- M. Pardasani, A. M. Ramakrishnan, S. Mahajan, M. Kantroo, E. McGowan, S. Das, P. Srikanth, S. Pandey and N. M. Abraham, Mol. Psychiatry, 2023, 28, 4693–4706 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Synthesis, structural characterization and additional biological data are provided. See DOI: https://doi.org/10.1039/d4nr02636e |
| This journal is © The Royal Society of Chemistry 2024 |