 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The impact of sequence periodicity on DNA mechanics: investigating the origin of A-tract's curvature†
Tania
Gardasevic
 and
Agnes
Noy
and
Agnes
Noy
 *
*
School of Physics, Engineering and Technology, University of York, York, YO10 5DD, UK. E-mail: agnes.noy@york.ac.uk
First published on 4th September 2024
Abstract
Periodic sequences in phase with DNA helical shape are prevalent in genomes due to their capacity to modulate DNA elasticity on a global scale. However, how this occurs is not well understood. We use all-atom molecular dynamics simulations on 40 bp DNA fragments to assess the effect of periodicity on bending, twisting, and stretch elasticity. We observe that DNA static curvature is the mechanical parameter most influenced by periodicity, with A-tract sequences having the greatest effect. A-tracts generate global curvature by bending in distinct directions (minor groove and backbones) that complement the bending of the rest of DNA, which predominantly is towards the major groove. Even if A-tracts are rigid at the local scale, these small bends integrate with the greater bends from the sequences between, producing an amplifying effect. As a result, our findings support a ‘delocalized bend’ model in which the A-tract operates as an ‘adaptable mechanical part’. By understanding how global curvature emerges from local fluctuations, we reconcile previous contradictory theories and open an avenue for manipulating DNA mechanics through sequence design.
Introduction
The mechanical properties of DNA are fundamental to many biological processes, for which decoding the complex rules of flexibility is vital for understanding. The bendability of DNA molecules is responsible for DNA packing within cells,1 gene regulation,2–4 and protein interactions.5,6 DNA looping is involved in regulatory regions by bringing together far away sites along the chain,7–9 and some processes such as DNA repair and methylation require change of flexibility at the individual nucleotide level.10–13A variety of methods have been employed to probe the sequence features responsible for mechanical variation in DNA. Structural imaging methods such as X-ray crystallography give atomistic information but lack dynamics.14,15 There has been success with experiments involving techniques such as atomic force microscopy (AFM),16,17 cyclization,4,18–20 and optical/magnetic tweezers17,21,22 which reveal dynamic behaviour but only at the global scale. This leaves a need for techniques able to study the local dynamics at the atomistic level, which is filled by computational simulations.23
Based on the findings of all-atom simulations and X-ray database analysis, it has been determined that YR (pyrimidine–purine) base-pair (bp) steps are the most flexible, often exhibiting hinge-like behaviour.14,24–26 In contrast, RR (purine–purine) steps function as intermediaries, while RY (purine–pyrimidine) steps are the most rigid. However, this principle is an oversimplified description due to the influence of the particular attributes of individual bases. Through the analysis of all possible tetramers from the ABC simulation database,27 we found A/T rich sequences tend to exhibit more extreme mechanical properties, including the most rigid (AAAA, AATT) and the most flexible (TATA, CACA), compared to sequences abundant in G/C, which are more average.26
An important factor for controlling DNA bendability at the longer scale is the repetitive placement of particular sequences in phase with the helical pitch. A-tracts sequences have been studied extensively due to their periodic distribution in genomes as well as their key role in nucleosome positioning and DNA looping.28,29 It is widely recognised that multiple short phased A-tracts introduce a directional bend in the DNA helical axis, although this is counter-intuitive given that these sequences possess a straight structure and rigid mechanical properties.17,29,30 This has been explained by the fact that DNA in solution tends to bend toward the major groove at the base-pair level, which makes the local molecular contour be writhed or crooked31–33 (see Fig. 1). Then, the action of phased A-tracts would be to provide proper helical phasing and not counteract the net curvature produced by the sequences in between, according to the non-A-tract model.29,34,35 However, the mechanism that determines the strong curvature caused by A-tracts is still not clear. Bending at the edges of A-tracts, as proposed in the junction model,29,36 or at the A-tracts themselves towards the minor groove, as suggested in the wedge model,29,37 might also be the cause.
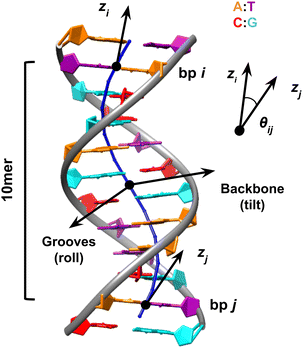 | ||
| Fig. 1 Sequence-averaged DNA presents a crooked shape due to its propensity to bend towards the major groove at the bp step level. This structure is built using average helical parameters27 with A represented in orange, T in purple, C in red, and G in cyan. The local molecular contour (in blue) is defined by the midpoint between bp steps.32 This scheme also illustrates the expansion of 3DNA's algorithm by the SerraNA program to consider pairs of bp that are not adjacent, using a 10mer as an example. The bending angle (θi,j) between two bp i and j is determined by the directional vectors (ẑi and ẑj) originating from them. The direction in which bending occurs is decided at the midpoint, according to the orientation of grooves or backbones. The bending angle is partitioned into two angles, which are equivalent to roll and tilt, as defined at the level of two 2 bp. | ||
Further periodic signals have been detected by the utilisation of high-throughput cyclization assays, which are capable of ascertaining the inherent bendability of thousands of DNA sequences.20 Sequences that have the greatest tendency for cyclization are distinguished by the repeating of in-phase A/T dinucleotides on one hand and in-phase G/C dinucleotides on the other. These two motifs are interconnected by virtue of being separated by half of the helical repeat (5, 15, 25 bp).20 However, the exact influence of these correlations on the curvature and dynamics of DNA is still unknown.
The field of DNA nanotechnology has just started to exploit the growing understanding of DNA elasticity. To construct complex shapes, for instance, twisting and curvature have been altered by removing and adding bp.38 Still, the application of sequence-dependent flexibility in the manipulation of DNA nanostructures is infrequent. Recently, a Brownian ratchet employing sequence-dependent flexibility gradients has been suggested as a novel approach to facilitate the transportation of positively charged nanoparticles.39 However, A-tracts have been the sole motif specifically used in the construction of sequence-specific designs so far, including DNA minicircles40 and lipid bilayers encircled by DNA.41
Although the origins of flexibility at the bp level are well-established,14,42 there have been limited efforts to understand how these bends combine to produce global bending. These studies have primarily relied on simple mesoscopic models using the nearest-neighbour approximation (only interactions between two bp),30,43,44 despite the well-established impact of the sequence context.36,45 A new bp-level model incorporating long-range effects through the implementation of machine learning techniques has been recently developed, although it has not yet been employed to calculate global elastic properties.46 All these limitations hinder our capacity to precisely predict the mechanical characteristics of genomic sequences or design sequences for DNA nanotechnology.
We previously developed the Length-Dependent Elastic Model (LDEM) to characterize the evolution of DNA mechanical properties as length scale progressively increases from a single bp, through the analysis of all-atom molecular dynamics simulations.31 Using this model, we observed that the transition from local to global elasticity typically occurs within the length of one DNA turn, resulting in long-range values that are consistent with DNAs at the kbp scale.26,31 The elastic constants undergo significant changes at small lengths, until they reach approximately 10 bp or a full helix turn.26,31 As the length continues to increase, the elastic constants hit a plateau, indicating that they remain moderately constant without any major alterations (see Fig. S1†). This implies that the global elastic properties of a sequence motif can be determined through all-atom simulations of relatively short DNA fragments (30–50 bp). The implementation of the LDEM in the SerraNA program allows for the execution of all these calculations in a highly efficient manner (https://github.com/agnesnoy/SerraNA)26 and has been employed in the present study.
Here, we conduct a systematic analysis of how sequence periodicity affects DNA mechanical characteristics in terms of bending, twisting and stretching, using all-atom molecular dynamics. The goal is to understand why the repetition of certain sequences results in DNA fragments with exceptional mechanical properties. For these reasons, we designed and modelled a series of 40 bp-long sequences with different periodic patterns, taking into account the general YR/RR/RY principle of DNA mechanics while integrating the most malleable (TATA, CACA) and rigid (AAAA) tetramer motifs26 (see Fig. 2). The 40 bp length was selected as a compromise between capturing the plateau behaviour while optimizing computational resources, as they are among the longest linear DNA molecules simulated with an atomic description of the entire system, including the solvation box.26,47
Materials and methods
Design of DNA sequences
A total of 24 linear DNA fragments containing 40 bp were built using the NAB module from Amber20.48 Sequences were designed under the assumption that helical pitch was effectively 10 bp at this limited scale. They were classified primarily into three categories: (i) uniform, including (R)40, (YR)20; (ii) periodic, including (R5YRYRY)4, (R5Y5)4, (R9Y)4; and (iii) non-periodic, including variants of the motifs listed above. Sequences were also grouped by base composition being A/T, C/G or mixture (see Fig. 2).Simulations
A CG segment was placed at each end of the 40 bp sequences to prevent end melting during the simulation. The resulting 44 bp sequences were built using the nucleic acid builder utility in Amber20 with bsc1 force-field.48,49 The structures were solvated in TIP3P octahedral boxes50 and Na+/Cl− pairs, using Smith-Dang parameters,51 to achieve a physiological salt concentration of 200 mM. Simulations were carried out with the pmemd.cuda code from Amber2048 using periodic boundary conditions and Particle Mesh Ewald.52 Movements of hydrogen atoms were annihilated using SHAKE,53 which allowed us the use of a 2 fs integration step. The systems were subjected to a minimisation/equilibration process according our standard protocols25,26 and followed by 500 ns of productive simulations, which provides an adequate sampling for elastic constant calculations.26 Simulations were performed at constant temperature (300 K) and pressure (1 atm) using the Berendsen thermostat and barostat.54Structural parameters and elastic constants
The elastic constants for bending, twisting, and stretching were estimated for each DNA molecule using SerraNA, as detailed before.26,31 This application employs the helical coordinate system, as defined by the 3DNA program,55 to analyse all the frames from simulations at the bp level. SerraNA expands the 3DNA's algorithm to characterize the geometry of not only two consecutive bp, but also those separated by an increasing number of nucleotides (see Fig. 1).The program utilises the worm-like chain model (WLC) to compute persistence lengths (A) by fitting the directional decay between the tangent vectors (ẑi and ẑj) of two bp (i and j) that are separated by a growing number of bp steps N with a distance rise b of 0.34 nm. By assuming a sufficiently weakly bending molecule, SerraNA simplifies WLC's formula (〈cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θi,j〉 = e−Nb/A), considering only the quadratic approximation:
θi,j〉 = e−Nb/A), considering only the quadratic approximation:
 | (1) |
The net linear increase in 〈θi,j2〉 with length comes from the distribution of static bends along a molecule (〈θs2〉) and the dynamic bends induced by thermal fluctuations (〈θd2〉).56,57 SerraNA obtains the former from the average bp step parameters, which determine the local static bends, and the latter though 〈θ2〉 = 〈θs2〉 + 〈θd2〉.26,31 Then, the static and dynamic contributions of the persistence lengths (As, Ad) are determined using the fitting of the linear directional decays, following the same approximations as before:26,31
 | (2) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 56,57 when combined.
56,57 when combined.
The direction of bending is established by partitioning it into two components: roll (towards the grooves) and tilt (towards the backbone). The decomposition of bending is defined according to the orientation of DNA's grooves and backbones at the mid-point between the two bp considered, as defined in the 3DNA algorithm (see Fig. 1).
The bulk torsional modulus for each sequence (C) is determined by taking the overall average of Ci,j values obtained for all sub-fragments longer than 10 bp (after the transition from short- to long-range behaviour).31 Each individual Ci,j is obtained from the elastic matrix Fi,j = kBTbNVi,j−1, where Vi,j−1 represents the inverse covariance matrix extracted from simulations using the bending, twisting and stretching parameters for that particular sub-fragment comprised between bp i and j.26,31 Covariance matrices (Vi,j) are assumed to have a multi-Gaussian distribution, and while this is not always the case for short sub-fragments due to their bimodality,27,45,58 this behaviour rapidly dissipates as the DNA reaches the length of a complete helical turn.26
Similarly, stretch moduli for each sub-fragment (Bi,j) are calculated using the diagonal term of F associated with the end-to-end distance. Because the stretching response of relatively short DNA molecules is affected by long end-effects, the bulk stretch modulus (B) is determined through the linear fitting of its length dependency, using only the central 18mer and sub-fragments exceeding 10 bp26,31 (see Fig. S1†). This limited fitting has been demonstrated to be enough for capturing long-range behaviour, resulting in values that are comparable to those obtained through single-molecule techniques.
Results and discussion
In order to discern the consequences of sequence periodicity on DNA deformability, we investigated a series of linear DNA fragments of 40 bp, combining different sequence motifs: (i) uniform sequences as (R)40 and (YR)20, expected to be rigid and flexible, respectively; (ii) periodic in-phase sequences of alternating rigid and flexible pentamers ((R5YRYRY)4); (iii) periodic in-phase sequences with the flexible part reduced to minimum ((R9Y)4); (iv) periodic sequences of alternating rigid pentamers (R5), placing hinge (YR) and anti-hinge (RY) steps half-turn from each other and a full turn from themselves (R5Y5)4; (v) out-of-phase variations of (R5YRYRY)4, such as (R4YRYR)5 and (R3YRY)6 R3Y; and (vi) out-of-phase variations of (R5Y5)4, such as (R4Y4)5 and (R6Y6)3R4.Persistence length correlates with the amount of flexible bp steps
Persistence length values A obtained from our simulations are larger than the general experimental measurements of 45–55 nm16,21,22 but in agreement with other simulation data26,30,47,59 (see Fig. 2 and Table S1†). This is common for MD simulations, possibly due to an overestimation of stacking interactions60 or the simulations being conducted in much more controlled salt conditions of monovalent (Na+, Cl−) ions only, compared to experimental conditions where there are other species such as Mg2+, Hepes and Tris16,21,22 which significantly affect DNA flexibility.21,61 Nevertheless, our simulations can reproduce the degree of change observed in single-molecule experiments17 when comparing sequences with phased A-tracts (yielding an average ± standard deviation of 63 ± 6 and 44 ± 3 nm, respectively) and those without (67 ± 8 and 47 ± 4 nm), resulting in an identical decrease of 6%.The variability of persistence lengths observed in Fig. 2 underscores the sequence-dependent nature of DNA flexibility at this global length scale, as documented in prior research.19,20,26,30,62 One can easily see that the nature of bp steps plays a significant role when comparing sequences made up of identical bases. At the A/T block, (A)40 presents a larger A than (TA)20 due to the accumulation of flexible TA steps. However, for the G/C group, this order is reversed and (G)40 is more flexible than the (GC)20. A similar observation was previously made at the tetramer level (GGGG vs. GCGC).26 This can be explained by the fact that the GG step has a considerably lower stacking energy than CG and GC,63,64 which leads to weaker interactions between adjacent bps, thereby permitting bending and deformation. Overall, the remaining sequences, including those in the mixed group, fall within the range of values derived from their respective polydinucleotide sequences, regardless of whether they exhibit periodicity in phase or not (see Fig. 2). Nevertheless, some RNYM sequences are exceptionally stiff as a result of the accumulation of poly(R) and RY steps, both being similarly rigid and not compensated by the flexible YR steps.
All these results suggest that the total number of flexible bp steps determines the persistence lengths of individual DNA fragments. We verified this idea by finding a measurable correlation between the global value of A for each individual DNA fragment and the mean value obtained from all the constituent tetramers in that specific sequence (with a correlation coefficient of 0.52, see Fig. S2†). When we only consider the bendability caused by thermal fluctuations (i.e. Ad), the correlation coefficient increases to 0.57, but it decreases to 0.51 when we only consider the static curvature (i.e. As) (see Fig. S2†). This suggests that the flexible or rigid character of the constituent bp steps dictate Ad. On the contrary, our results indicate that a large number of short curved motifs is not sufficient to predict a strong global curvature; rather, they must be arranged in a particular manner (see below).
In-phase periodicity increases overall DNA curvature
The As data is significantly more varied than the A and Ad data, which agrees with Mitchell et al.30 and Velasco-Berrelleza et al.,26 who found sequence-dependent variation in A to be primarily due to changes in intrinsic shape (i.e. As), rather than to thermal fluctuations (Ad). Additionally, we noted that the A/T group has the greatest degree of variance, while the G/C group presents the least, consistent with our findings at shorter length scales.26By employing the non-curved features of A-tracts, we observe that curvature rises with the in-phase alternation of curved and non-curved blocks (Fig. 2). Our simulations show that (R5YRYRY)4 in the T/A and C/A groups have lower As (i.e., stronger curvature) than the corresponding (RY)20. This impact is decreased when the flexible YRYRY section is reduced to a single RY and YR step in (R9Y)4 sequences. However, curvature rises in (R5Y5)4 sequences, indicating that the separation of alternating RY and YR steps by 5 bps is a very efficient way for causing curvature. Finally, we notice that deviations from in-phase periodicity reduce or maintain DNA curvature, but never increase it (Fig. 2).
In summary, our simulations suggest that alternating curved and non-curved sections on DNA, aligned with its helical structure, are essential for creating a significant overall curvature. This is especially true when A-tracts are present, regardless of the length or composition of sequences between (T/A, C/A, or G/C motifs), as seen in previous experiments.17,29 Our findings are consistent with high-throughput cyclization data, which identified A/T and G/C dinucleotides separated by half a DNA turn.20 Although the same study found a weak spatial association between AA and CA, gel electrophoresis experiments have demonstrated that the presence of CA steps helps to generate a particularly large curvature with A-tracts,65 which is in agreement with our simulations. Lastly, our measurement of the degree of bending per decamer containing an A-tract is 17.7 ± 0.5 degrees, which agrees really well with the value of 18 degrees found by gel electrophoresis66 and NMR structures67 (see Fig. 3).
A-tract curvature is induced via the delocalized-bend model
We then investigated the connection between local and global bending to determine which are the structural factors that contribute to create a significant curvature when A-tracts are present. A clear pattern at the dinucleotide level confirms the basic notion that YR steps exhibit the most pronounced local bends (similar to hinges), RY steps display the least bending (anti-hinges), and RR steps exhibit an intermediate behaviour (Fig. 3). Because the strongest local bends always point towards the major groove (giving positive values for roll; see Fig. S3†), they cancel each other out in uniform or out-of-phase sequences, due to their all-around orientation (Fig. S4†). Then, the bending profiles for these sequences become flatter as we extend the length of the sub-fragments, indicating that there is no prevailing curvature direction and that bending magnitude increases due to thermal fluctuations (Fig. 3). In contrast, in most in-phase periodic sequences, local bends couple to build up a large curvature, due to the consistent placement of their major grooves on the same side of the DNA. This is reflected by the presence of peaks in the bending profiles, showing regardless of the length of the sub-fragments (Fig. 3).Therefore, our findings would initially suggest that the source of bending is outside the A-tracts and that their primary function is simply to be stiff and not counteract the net curvature caused by the sequences between, as per the non-A-tract model. Nevertheless, upon closer inspection of the global bending profiles, we observe that the largest curvature is not centred around YR steps, but rather is displaced in most cases (Fig. 3).
To establish the orientation of the global DNA curvature presented for each fragment, we decomposed the bending angles into the roll and tilt components according to the midpoint of the selected sub-fragments (see Methods and Fig. 1). We chose to examine the 10 possible 30 bp sub-fragments from the 40 bp sequences because their midpoints cover the whole central DNA turn, thus accounting for every conceivable orientation of the global curvature (see Fig. 4). We observe that the largest curvature arises from a combination of bending towards the grooves and backbones, with the latter being equally important as the former (Fig. 4A and S4†). A-tract sequences bend towards the minor groove and backbone, while sequences in between bend towards the major groove, as shown in representative structures matching the two-component bending profiles (Fig. 4B). In contrast the equivalent non-curved sequences, like (G5CGCGC)4 and (G5C5)4, have a flatter profile without bending towards the minor groove and backbones, demonstrating the significance of bending in A-tracts (Fig. 4C, S3 and S4†).
This preferred curved orientation is commonly observed in our simulations for various sequences (such as (A5CACAC)4, (A5CGCGC)4, (A5C5)4 and (A9C)4) and is consistent with previous high-resolution structures67,68 and simulations.10,59 Specifically, A-tracts shift from a positive tilt at the 5′ junction to negative at the 3′ junction, displaying a bending toward the minor groove (negative roll) in the middle closer to the 3′ junction.
Because curvature emerges from bends inside and outside A-tracts, our results support the delocalized-bend model proposed by Crothers and co-workers.68,69 This model amalgamates aspects from the alternative theories: curvature arises from phased combinations of bending towards the major groove (as in the non-A-tract model) and towards the minor groove and backbone (A-tract model). The equivalence in bending between (A5CACAC)4 and (A5C5)4 sequences suggests that the bend at the 5′ junction is critical, hence providing support for the junction model. Nevertheless, our simulations show that YR steps at the 5′ junction are not particularly flexible, contrary to what was initially proposed by this hypothesis.29,36 Rather, their significance is derived from the positioning of A-tracts to bend towards the minor groove and backbone, as well as non-A-tracts to bend towards the major groove. Collectively, the combination of these local effects produces a global curvature that is directed into the minor groove at the A-tracts, slightly shifted towards the 3′-end. This correlates well with the overall bending orientation deduced from gel electrophoresis and hydroxyl-radical cutting.36,66,70
A-tracts are versatile segments that enhance the curvature of adjacent sequences
Based on our calculations, the mechanical property that makes A-tracts essential for inducing large curvature in DNA is the ability to bend in any direction, albeit only marginally, owing to their rigid nature.71,72 This differentiates them from the remaining DNA sequences, which are restricted to bending in the direction of the major groove. As a result, A-tracts contribute to the global curvature through their distinctive bends toward the minor groove and backbones, which complement the common DNA bends toward the major groove. This explains why the nature of the sequences between them has such a negligible effect on this curvature.29 The (A9C)4 sequence demonstrates the adaptability of A-tracts, as they bend towards the minor groove and two backbones, allowing the isolated YR steps to bend in the direction of the major groove (Fig. 4).Stretch and torsion moduli
The average stretch (B) and torsion moduli (C) for all our modelled DNA fragments are 1882 ± 455 and 104 ± 18 (±standard deviation), respectively (see Table S1†). These values are in reasonably good agreement with the results obtained from single-molecule experiments, which are typically around 1500 pN and 100 nm.73–76 Recent experiments have shown that DNA fragments containing A-tracts have higher stretching rigidity (2400 ± 220 pN),17 which could explain the comparatively high value of B in our proposed sequences. By analyzing all simulations without A-tracts, we obtain a lower average (1608 ± 582), which is more consistent with the experiments conducted on random DNA.76 The remaining sequences (with A-tracts) provide a higher average (2018 ± 336), which, although not identical to the experimental result, replicates the observed trend.Our simulations reveal that periodicity has no major effect on the stretch modulus of a certain sequence (Fig. 5A). Instead, the mechanical parameter appears to be influenced by the flexible character of the constituent steps, as evidenced by the correlation between global and local values with a coefficient of 0.48, in a similar manner to the dynamic persistence length (Ad) (Fig. S2†). Contrary to the persistence length, G/C sequences exhibit the largest variability, whereas the A/T group presents the lowest (Fig. 5A). This agrees with the fact that the stretch modulus is determined by the crookedness of the molecule, i.e. the oscillations of bp centres around the molecular axis.33 Then, the change in DNA contour length is mostly due to aligning the base pair centers with the helical axis, rather than separating successive base pairs, which is prevented by the strong stacking interactions.31,33,77 Consequently, GG is the most crooked polydinucleotide sequence33 and presents the lowest stretch modulus, whereas GC is among the least crooked sequences33 with a high modulus. Sequences abundant in A/T are comparatively rigid despite their weak stacking interactions,64 due to their comparatively low crookedness.33
The global torsional modulus of individual DNA segments is not influenced by periodicity and shows a weak correlation with the constituent oligomers (with a coefficient of 0.09), suggesting a complex transition between the short and long-range behaviour (Fig. 5B and S2†). Typically, the torsional modulus shows a crossover from relative soft values of around 30–60 nm at the dinucleotide level to a large-scale asymptotic value between 90–120 nm. However, our results indicate that this transition does not occur in a uniform fashion. Instead, the different sequences exhibit varying capacities to connect local oscillations within neighboring sequences in order to produce global fluctuations. When examining sequences with similar torsion modulus at the dinucleotide level, we notice that they exhibit a distinct rate of change as the DNA length increases (see Fig. 5C). In some sequences, the local fluctuations exhibit a high anti-correlation as length increases, generating a rigid torsional modulus, whereas, in others, the local fluctuations are more independent resulting in a softer global elastic constant (Fig. 5C).
The varying capacity to connect local and global variations for the different sequences could be caused by the distinct levels of crookedness, as suggested by the remarkable correlation between twist and stretch elastic constants (with a coefficient of 0.73, see Fig. S5†). It is worth noting that a crooked DNA fragment can alternatively be described as locally supercoiled or writhed. Therefore, alterations in this structural parameter can be counterbalanced by fluctuations in twist, as per the DNA topology relation. In this, the total number of times one strand revolves around the other (i.e. the linking number) is equal to the sum of twist and writhe. We anticipate that the potential correlation between twist elasticity and crookedness will be the focus of subsequent research.
Conclusions
In this study, we performed all-atom molecular dynamics simulations for numerous 40 bp DNA sequences with varying levels of periodicity and analysed their elasticity and static curvature using our software SerraNA. We found that the dynamic persistence length is primarily influenced by the quantity of flexible bp steps, whereas curvature is influenced by sequence periodicity aligning with the helical shape of DNA. This is due to the fact that, when position periodically, the structural features of a specific sequence (major groove, minor groove, backbones) always face the same side of the DNA, multiplying the effect of its inherent curvature.Our simulations indicate that A-tract is the sequence motif that generates the largest curvature when positioned at in-phase regular intervals, in agreement with previous experiments.29 By examining how global curvature emerges from local bends, we provide a comprehensive understanding of the origin of this curvature, tying together previous theories (A-tract, non-A-tract, and junction), which focused on the separate structural factors. We show that curvature can be attributed to the coordination of bends, both inside and outside A-tracts, supporting the delocalized-bend model proposed by Crothers and co-workers.68 On one hand, DNA bending occurs predominantly at the flexible YR steps and, on average, is directed toward the major groove. On the other hand, A-tracts possess the distinct capacity of flexing in the other directions (minor groove and backbones), even if it is only marginally due to their rigid nature. Major curvature therefore emerges from the phased combination of strong bending toward the major groove in non-A-tract sequences and weak bending toward the minor groove and backbone in A-tracts. The combination of these local effects produces a global curvature that is directed into the major groove at the 5′ junction of A-tracts and into the minor groove at the 3′ junction, explaining the junction model derived from biochemical studies.
A-tracts's fundamental property for inducing large curvature is their mechanical adaptability, which enables them to bend in various directions and couple with the directions of the surrounding sequences, thereby producing an amplifying effect. Thus, A-tracts operate as mechanically versatile elements. The composition of the sequences between A-tracts hardly affects this curvature because the vast majority of DNA sequences predominantly bend in the direction of the major groove.
Finally, our simulations reveal that the stretch and twist elastic constants are independent of DNA periodicity. While the number of flexible bp steps influences stretch modulus in the same way as dynamic persistence length does, torsional modulus has a more complex link between local and global elasticity that needs additional investigation. Overall, the findings presented in this work not only bring new insights into the flexible features of periodic sequences in genomes, but also facilitate the development of sequences with ad hoc mechanical characteristics that can be utilized in DNA nanotechnology.
Author contributions
A. N. conceived the study. T. G. performed the simulations. T. G. and A. N. analysed them. A. N. and T. G. wrote the manuscript.Data availability
Simulations are available at the MDposit database (https://mdposit-dev.mddbr.eu/#/browse), and data are available in the ESI† and the University of York Data Repository (https://doi.org/10.15124/e33eb7cf-b8d8-49bd-9833-ee47d5b56eb3).Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Elliot W. Chan and Victor Velasco-Berrelleza for technical guidance. This work was supported by the Engineering and Physical Sciences Research Council (EPSRC) EP/N027639/1 to AN; EP/T022205/1, EP/T022167/1 and EP/X035603/1 for computational resources]; and the Physics of Life Network UK [EP/T022000/1 to T. G.]References
- A. Marin-Gonzalez, J. G. Vilhena, R. Perez and F. Moreno-Herrero, Q. Rev. Biophys., 2021, 54, e8 CrossRef CAS PubMed.
- J. R. Goñi, A. Pérez, D. Torrents and M. Orozco, Genome Biol., 2007, 8, R263 CrossRef PubMed.
- S. H. Kim, M. Ganji, E. Kim, J. van der Torre, E. Abbondanzieri and C. Dekker, eLife, 2018, 7, e36557 CrossRef.
- A. Basu, D. G. Bobrovnikov, Z. Qureshi, T. Kayikcioglu, T. T. M. Ngo, A. Ranjan, S. Eustermann, B. Cieza, M. T. Morgan, M. Hejna, H. T. Rube, K.-P. Hopfner, C. Wolberger, J. S. Song and T. Ha, Nature, 2021, 589, 462–467 CrossRef CAS PubMed.
- J. Li, J. M. Sagendorf, T.-P. Chiu, M. Pasi, A. Pérez and R. Rohs, Nucleic Acids Res., 2017, 45, 12877–12887 CrossRef CAS PubMed.
- F. Battistini, A. Hospital, D. Buitrago, D. Gallego, P. D. Dans, J. L. Gelpí and M. Orozco, J. Mol. Biol., 2019, 431, 3845–3859 CrossRef CAS PubMed.
- V. Lioy, A. Cournac, M. Marbouty, S. Duigou, J. Mozziconacci, O. Espéli, F. Boccard and R. Koszul, Cell, 2018, 172, 771–783 CrossRef CAS PubMed.
- T. Hsieh, C. Cattoglio, E. Slobodyanyuk, A. Hansen, O. Rando, R. Tjian and X. Darzacq, Mol. Cell, 2020, 78, 539–553 CrossRef CAS PubMed.
- N. Krietenstein, S. Abraham, S. Venev, N. Abdennur, J. Gibcus, T. Hsieh, K. Parsi, L. Yang, R. Maehr, L. Mirny, J. Dekker and O. Rando, Mol. Cell, 2020, 78, 554–565 CrossRef CAS.
- J. Curuksu, K. Zakrzewska and M. Zacharias, Nucleic Acids Res., 2008, 36, 2268–2283 CrossRef CAS PubMed.
- G. Rossetti, P. D. Dans, I. Gomez-Pinto, I. Ivani, C. Gonzalez and M. Orozco, Nucleic Acids Res., 2015, 43, 4309–4321 CrossRef CAS PubMed.
- A. Afek, H. Shi, A. Rangadurai, H. Sahay, A. Senitzki, S. Xhani, M. Fang, R. Salinas, Z. Mielko, M. Pufall, G. Poon, T. Haran, M. Schumacher, H. Al-Hashimi and R. Gordan, Nature, 2020, 587, 291–296 CrossRef CAS PubMed.
- A. Pérez, C. Castellazzi, F. Battistini, K. Collinet, O. Flores, O. Deniz, M. Ruiz, D. Torrents, R. Eritja, M. Soler-López and M. Orozco, Biophys. J., 2012, 102, 2140–2148 CrossRef.
- W. K. Olson, A. A. Gorin, X.-J. Lu, L. M. Hock and V. B. Zhurkin, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 11163–11168 CrossRef CAS PubMed.
- A. A. Kornyshev, D. J. Lee, A. Wynveen and S. Leikin, Nucleic Acids Res., 2011, 39, 7289–7299 CrossRef CAS PubMed.
- A. K. Mazur and M. Maaloum, Nucleic Acids Res., 2014, 42, 14006–14012 CrossRef CAS PubMed.
- A. Marin-Gonzalez, C. L. Pastrana, R. Bocanegra, A. Martín-González, J. G. Vilhena, R. Pérez, B. Ibarra, C. Aicart-Ramos and F. Moreno-Herrero, Nucleic Acids Res., 2020, 48, 5024–5026 CrossRef CAS.
- Y. Zhang and D. M. Crothers, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 3161–3166 CrossRef CAS.
- S. Geggier and A. Vologodskii, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 15421–15426 CrossRef CAS PubMed.
- A. Basu, D. Bobrovnikov, B. Cieza, J. Arcon, Z. Qureshi, M. Orozco and T. Ha, Nat. Struct. Mol. Biol., 2022, 29, 1178–1187 CrossRef CAS.
- C. G. Baumann, S. B. Smith, V. A. Bloomfield and C. Bustamante, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 6185–6190 CrossRef CAS.
- J. Lipfert, G. M. Skinner, J. M. Keegstra, T. Hensgens, T. Jager, D. Dulin, M. Köber, Z. Yu, S. P. Donkers, F.-C. Chou, R. Das and N. H. Dekker, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 15408–15413 CrossRef CAS PubMed.
- G. da Rosa, L. Grille, V. Calzada, K. Ahmad, J. P. Arcon, F. Battistini, G. Bayarri, T. Bishop, P. Carloni, T. Cheatham III, R. Collepardo-Guevara, J. Czub, J. R. Espinosa, R. Galindo-Murillo, S. A. Harris, A. Hospital, C. Laughton, J. H. Maddocks, A. Noy, M. Orozco, M. Pasi, A. Pérez, D. PetkevičiūGerlach, R. Sharma, R. Sun and P. D. Dans, Biophys. Rev., 2021, 13, 995–1005 CrossRef CAS.
- F. Lankaš, J. Šponer, J. Langowski and T. E. Cheatham, Biophys. J., 2003, 85, 2872–2883 CrossRef PubMed.
- A. Noy and R. Golestanian, J. Phys. Chem. B, 2010, 114, 8022–8031 CrossRef CAS.
- V. Velasco-Berrelleza, M. Burman, J. W. Shepherd, M. C. Leake, R. Golestanian and A. Noy, Phys. Chem. Chem. Phys., 2020, 22, 19254–19266 RSC.
- M. Pasi, J. H. Maddocks, D. Beveridge, T. C. Bishop, D. A. Case, T. Cheatham, P. D. Dans, B. Jayaram, F. Lankaš, C. Laughton, J. Mitchell, R. Osman, M. Orozco, A. Pérez, D. Petkevičiūtė, N. Spackova, J. Šponer, K. Zakrzewska and R. Lavery, Nucleic Acids Res., 2014, 42, 12272–12283 CrossRef CAS.
- E. Segal and J. Widom, Curr. Opin. Struct. Biol., 2009, 19, 65–71 CrossRef CAS.
- T. E. Haran and U. Mohanty, Q. Rev. Biophys., 2009, 42, 41–81 CrossRef CAS PubMed.
- J. S. Mitchell, J. Glowacki, A. E. Grandchamp, R. S. Manning and J. H. Maddocks, J. Chem. Theory Comput., 2017, 13, 1539–1555 CrossRef CAS.
- A. Noy and R. Golestanian, Phys. Rev. Lett., 2012, 109, 228101 CrossRef.
- T. Sutthibutpong, S. A. Harris and A. Noy, J. Chem. Theory Comput., 2015, 11, 2768–2775 CrossRef CAS.
- A. Marin-Gonzalez, J. G. Vilhena, F. Moreno-Herrero and R. Perez, Phys. Rev. Lett., 2019, 122, 048102 CrossRef PubMed.
- D. S. Goodsell and R. E. Dickerson, Nucleic Acids Res., 1994, 22, 5497–5503 CrossRef CAS PubMed.
- S. B. Dixit, F. Pitici and D. L. Beveridge, Biopolymers, 2004, 75, 468–479 CrossRef CAS.
- H.-S. Koo, H.-M. Wu and D. M. Crothers, Nature, 1986, 320, 501–506 CrossRef CAS PubMed.
- A. Bolshoy, P. McNamara, R. E. Harrington and E. N. Trifonov, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 2312–2316 CrossRef CAS.
- H. Dietz, S. M. Douglas and W. M. Shih, Science, 2009, 325, 725–730 CrossRef CAS PubMed.
- S. Park, J. Song and J. S. Kim, Sci. Adv., 2019, 5, eaav4943 CrossRef CAS PubMed.
- T. L. Schmidt and A. Heckel, Nano Lett., 2011, 11, 1739–1742 CrossRef CAS PubMed.
- K. Iric, M. Subramanian, J. Oertel, N. P. Agarwal, M. Matthies, X. Periole, T. P. Sakmar, T. Huber, K. Fahmy and T. L. Schmidt, Nanoscale, 2018, 10, 18463–18467 RSC.
- A. Pérez, F. Lankaš, F. J. Luque and M. Orozco, Nucleic Acids Res., 2008, 36, 2379–2374 CrossRef PubMed.
- W. K. Olson, N. L. Marky, R. L. Jernigan and V. B. Zhurkin, J. Mol. Biol., 1993, 232, 530–554 CrossRef CAS PubMed.
- V. B. Zhurkin, M. Y. Tolstorukov, F. Xu, A. V. Colasanti and W. K. Olson, in Sequence-Dependent Variability of B-DNA, Springer US, 2005, pp. 18–34 Search PubMed.
- A. Balaceanu, D. Buitrago, J. Walther, A. Hospital, P. D. Dans and M. Orozco, Nucleic Acids Res., 2019, 47, 4418–4430 CrossRef CAS.
- D. Farré-Gil, J. P. Arcon, C. A. Laughton and M. Orozco, Nucleic Acids Res., 2024, gkae444 Search PubMed.
- H. Dohnalová and F. Lankaš, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2021, 12, e1575 Search PubMed.
- D. Case, K. Belfon, I. Ben-Shalom, S. Brozell, D. Cerutti, T. Cheatham III, V. Cruzeiro, T. Darden, R. Duke, G. Giambasu, M. Gilson, H. Gohlke, A. Goetz, R. Harris, S. Izadi, S. Izmailov, K. Kasavajhala, A. Kovalenko, R. Krasny, T. Kurtzman, T. Lee, S. LeGrand, P. Li, C. Lin, J. Liu, T. Luchko, R. Luo, V. Man, K. Merz, Y. Miao, O. Mikhailovskii, G. Monard, H. Nguyen, A. Onufriev, F. Pan, S. Pantano, R. Qi, D. Roe, A. Roitberg, C. Sagui, S. Schott-Verdugo, J. Shen, C. Simmerling, N. Skrynnikov, J. Smith, J. Swails, R. Walker, J. Wang, L. Wilson, R. Wolf, X. Wu, Y. Xiong, Y. Xue, D. York and K. P. A. Kollman, University of California, San Fransisco, 2020.
- I. Ivani, P. D. Dans, A. Noy, A. Pérez, I. Faustino, A. Hospital, J. Walther, P. Andrio, R. Goñi, A. Balaceanu, G. Portella, F. Battistini, J. L. Gelpí, C. González, M. Vendruscolo, C. A. Laughton, S. A. Harris, D. A. Case and M. Orozco, Nat. Methods, 2015, 13, 55–58 CrossRef PubMed.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
- D. E. Smith and L. X. Dang, J. Chem. Phys., 1994, 100, 3757–3766 CrossRef CAS.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- J. P. Ryckaert, G. Ciccotti and H. J. C. Berendsen, J. Comput. Phys., 1977, 23, 327–341 CrossRef CAS.
- H. J. C. Berendsen, J. P. M. Postma, W. F. van Gunsteren, A. DiNola and J. R. Haak, J. Chem. Phys., 1984, 81, 3684–3690 CrossRef CAS.
- X.-J. Lu and W. Olson, Nucleic Acids Res., 2003, 31, 5108–5121 CrossRef CAS PubMed.
- E. N. Trifonov, R. K. Tan and S. C. T. Harvey, DNA Bending & Curvature, 1988 Search PubMed.
- J. A. Schellman and S. C. Harvey, Biophys. Chem., 1995, 55, 95–114 CrossRef CAS.
- P. D. Dans, A. Balaceanu, M. Pasi, A. S. Patelli, D. Petkevičiūtė, J. Walther, A. Hospital, G. Bayarri, R. Lavery, J. H. Maddocks and M. Orozco, Nucleic Acids Res., 2019, 47, 11090–11102 CrossRef CAS PubMed.
- T. Dršata, N. Špačková, P. Jurečka, M. Zgarbová, J. Šponer and F. Lankaš, Nucleic Acids Res., 2014, 42, 7383–7394 CrossRef PubMed.
- K. Liebl and M. Zacharias, J. Chem. Theory Comput., 2023, 19, 1529–1536 CrossRef CAS PubMed.
- S. Guilbaud, L. Salomé, N. Destainville, M. Manghi and C. Tardin, Phys. Rev. Lett., 2019, 122, 028102 CrossRef CAS.
- H.-M. Chuang, J. G. Reifenberger, H. Cao and K. D. Dorfman, Phys. Rev. Lett., 2017, 119, 227802 CrossRef.
- D. Svozil, P. Hobza and J. Sponer, J. Phys. Chem. B, 2010, 114, 1191–1203 CrossRef CAS PubMed.
- F. Kilchherr, C. Wachauf, B. Pelz, M. Rief, M. Zacharias and H. Dietz, Science, 2016, 353, aaf5508 CrossRef.
- B. A. Beutel and L. Gold, J. Mol. Biol., 1992, 228, 803–812 CrossRef CAS PubMed.
- H. S. Koo and D. M. Crothers, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 1763–1767 CrossRef CAS.
- D. MacDonald, K. Herbert, X. Zhang, T. Polgruto and P. Lu, J. Mol. Biol., 2001, 306, 1081–1098 CrossRef CAS PubMed.
- A. Barbič, D. P. Zimmer and D. M. Crothers, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 2369–2373 CrossRef.
- D. Strahs and T. Schlick, J. Mol. Biol., 2000, 301, 643–663 CrossRef CAS PubMed.
- A. M. Burkhoff and T. D. Tullius, Nature, 1988, 331, 455–457 CrossRef CAS PubMed.
- A. D. DiGabriele, M. R. Sanderson and T. A. Steitz, Proc. Natl. Acad. Sci. U. S. A., 1989, 86, 1816–1820 CrossRef CAS.
- A. D. DiGabriele and T. A. Steitz, J. Mol. Biol., 1993, 231, 1024–1039 CrossRef CAS PubMed.
- Z. Bryant, M. D. Stone, J. Gore, S. B. Smith, N. R. Cozzarelli and C. Bustamante, Nature, 2003, 424, 338–341 CrossRef CAS PubMed.
- F. Mosconi, J. F. M. C. Allemand, D. Bensimon and V. Croquette, Phys. Rev. Lett., 2009, 102, 078301 CrossRef.
- J. Lipfert, J. W. Kerssemakers, T. Jager and N. H. Dekker, Nat. Methods, 2010, 7, 977–980 CrossRef CAS.
- P. Gross, N. Laurens, L. B. Oddershede, U. Bockelmann, E. J. G. Peterman and G. J. L. Wuite, Nat. Phys., 2011, 7, 731–736 Search PubMed.
- J. Luengo-Márquez, J. Zalvide-Pombo, R. Pérez and S. Assenza, Nanoscale, 2023, 15, 6738–6744 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr02571g |
| This journal is © The Royal Society of Chemistry 2024 |




