Open Access Article
Open Access ArticleShape-dependent cellular uptake of iron oxide nanorods: mechanisms of endocytosis and implications on cell labeling and cellular delivery†
Anbu Mozhi
Thamizhchelvan
 a,
Hedi
Ma
b,
Tianhe
Wu
a,
Darlene
Nguyen
a,
Jonathan
Padelford
b,
Ted J.
Whitworth
c,
Yuancheng
Li
ab,
Lily
Yang
a,
Hedi
Ma
b,
Tianhe
Wu
a,
Darlene
Nguyen
a,
Jonathan
Padelford
b,
Ted J.
Whitworth
c,
Yuancheng
Li
ab,
Lily
Yang
 de and
Hui
Mao
de and
Hui
Mao
 *ae
*ae
aDepartment of Radiology and Imaging Sciences, Emory University School of Medicine, Atlanta, Georgia 30322, USA. E-mail: hmao@emory.edu
b5M Biomed, LLC, Atlanta, Georgia 30303, USA
cRobert P. Apkarian Integrated Electron Microscopy Core, Emory University School of Medicine, Atlanta, Georgia 30322, USA
dDepartment of Surgery Emory University School of Medicine, Atlanta, Georgia 30322, USA
eWinship Cancer Institute, Emory University School of Medicine, Atlanta, Georgia 30322, USA
First published on 18th September 2024
Abstract
The effects of nanoparticle morphology, especially size and shape, on their interactions with cells are of great interest in understanding the fate of nanoparticles in biological systems and designing them for biomedical applications. While size and shape-dependent cell behavior, endocytosis mechanism, and subcellular distribution of nanoparticles have been investigated extensively with gold and other nanoparticles, studies on iron oxide nanoparticles (IONP), one of the most promising and well-thought-of nanomaterials in biomedical applications, were limited. In this study, we synthesized oligosaccharide-coated water-soluble iron oxide nanorods (IONR) with different core sizes (nm) and different aspect ratios (i.e., length/width), such as IONR(L) at 140/6 nm and IONR(S) at 50/7 nm as well as spherical IONP (20 nm). We investigated how their sizes and shapes affect uptake mechanisms, localization, and cell viability in different cell lines. The results of transmission electron microscopy (TEM) and confocal fluorescence microscopic imaging confirmed the internalization of these nanoparticles in different types of cells and subsequent accumulation in the subcellular compartments, such as the endosomes, and into the cytosol. Specifically, IONR(L) exhibited the highest cellular uptake compared to IONR(S) and spherical IONP, 1.36-fold and 1.17-fold higher than that of spherical IONP in macrophages and pediatric brain tumor medulloblastoma cells, respectively. To examine the cellular uptake mechanisms preferred by the different IONR and IONP, we used different endocytosis inhibitors to block specific cellular internalization pathways when cells were treated with different nanoparticles. The results from these blocking experiments showed that IONR(L) enter macrophages and normal kidney cells through clathrin-mediated, dynamin-dependent, and macropinocytosis/phagocytosis pathways, while they are internalized in cancer cells primarily via clathrin/caveolae-mediated and phagocytosis mechanisms. Overall, our findings provide new insights into further development of magnetic IONR-based imaging probes and drug delivery systems for biomedical applications.
1. Introduction
The extensive research and development of engineered nanomaterials for biomedical applications stem from their unique physical and chemical properties, as well as their functions at the nanoscale. As a great number of novel nanomaterials are developed for biomedical applications,1 such as probes for imaging and carriers for drug delivery, there is an increasing need to better understand the mechanisms and controlling factors that regulate the interactions between exogenous nanomaterials and biological systems at the molecular, cellular, and sub-organelle levels.2 Given the complexity of biological systems under different biological, physiological, and disease conditions, and their specific applications for targeted and precision medicine problems, the knowledge of such interactions is critical for the design, preparation, and deployment of engineered nanomaterials, especially inorganic nanomaterials, for optimal biosafety and efficacy of the intended applications. Currently, interactions between nanoparticles and biological systems are considered to be intricately influenced by the physicochemical properties of nanoparticles, which encompass size, shape, and surface chemistry, as well as the types and functions of cells.3,4 Prior investigations on the effects of the physical and structural properties of nanoparticles on nanoparticle–cell interactions are largely based on gold nanoparticles,5–7 since methods for preparing and controlling the structural and morphological properties of gold nanoparticles are well established. It is found that size and shape are critical parameters in the processes and mechanisms of cellular uptake and transport of nanoparticles in the endocytic compartments,8 while surface chemistry can significantly influence how nanoparticles interact within biological systems and their potential toxicity.9–11 However, there are fewer studies on these parameters based on magnetic iron oxide nanoparticles (IONP), which are one of the most attractive inorganic nanomaterials for medical applications owing to their distinctive magnetic properties and biocompatibility. In situ polymerization of small water-soluble glucose molecules onto magnetic IONP enhances their stability and solubility and provides further surface modification for additional applications.12,13 The glucose-coated IONP facilitates targeted interactions with cells expressing glucose transporters (GLUTs) on their membranes, presenting promising opportunities for biomedical interventions.14–16 Various forms of IONP have been developed as contrast agents for magnetic resonance imaging (MRI), carriers for magnetically guided drug delivery, or mediators for heat-based therapies for hyperthermia, with some already used in clinical applications.17–19 Notably, most IONP used in biomedical applications in vivo are made in spherical or isotropic shapes with core sizes of 10–200 nm. However, anisotropic one-dimensional (1D) nanostructures, such as nanorods, have always been of great interest due to their shape-dependent physicochemical or even physiological properties. For example, studies found that anisotropic nanostructures exhibit prolonged blood circulation time and enhanced retention at tumor sites compared to spherical nanostructures.20While most of these early investigations on the shape-dependent properties and applications of nanomaterials were conducted on gold nanomaterials, in which shape-controlled preparation is fairly established, preparations of water-stable and highly uniform iron oxide nanorods (IONR) have been reported in recent years.21,22 The obtained IONR exhibited enhanced magnetic characteristics in contrast to the widely utilized spherical IONP with equivalent iron content. The augmentation in magnetic properties correlates with the aspect ratio (i.e., the length-to-width ratio of the core crystal), as evidenced by the gradual rise in magnetic saturation with the elongation of the IONR.22 Importantly, investigations have revealed that rod-shaped nanostructures do not induce shape-specific toxicity.23 As most research on applications of IONP and NP–cell interactions have focused on spherical IONP, the impact of varied dimensions and aspect ratios of shape-controlled IONR on cellular uptake, subcellular accumulation, and cytotoxicity4,24 is very limited.
The current study re-investigated the effects of size- and shape-dependent cell–nanoparticle interactions and cellular uptake in the context of IONR, given the great interest in applying such a new class of engineered magnetic nanomaterials developed for MRI applications, including labeling cells for imaging-based cell tracking, and targeted drug delivery with imaging-monitoring. For this purpose, we synthesized water-soluble IONR with two different aspect ratios and spherical IONP to investigate the size and shape-dependent cellular uptake and toxicity of these nanoparticles in different cell lines, including macrophages, normal tissue cells, and cancer cells, respectively, using fluorescence confocal microscopy, transmission electron microscopy (TEM), and MTT assay. In addition, different endocytosis inhibitors were utilized to facilitate the investigation of the shape-dependent uptake mechanisms and intracellular distribution of these nanomaterials.
2. Experimental section
Materials
All chemicals and reagents utilized in the experiments were sourced from commercial suppliers and used as received without further purification. Iron(III) chloride hexahydrate ((FeCl3·6H2O), 97%) was ordered from Beantown Chemical (USA). Sodium oleate (NaOA) was supplied by TCI (USA). Oleylamine (70%), polyethyleneimine (PEI, 50%, molecular weight 50–100 kDa), 1-octadecene (purity: 99.8%), D-(+)-glucose, ammonium hydroxide (ACS grade), iron(III) nitrate nonahydrate ((FeN3O9·9H2O); ACS reagent, ≥98%), dimethyl sulfoxide (ACS grade, 99.9%), hexane (reagent grade, ≥98.5%), dimethylformamide (ACS grade; ≥99.8%), and chloroform (HPLC grade; ≥99.8%) were brought from Sigma-Aldrich (Missouri, USA). Ethyl alcohol (200 proof/100%) was obtained from KOPTEC (USA). Feraheme® (ferumoxytol) was supplied by AMAG Pharmaceuticals Inc., Waltham, MA USA. SHP-10 (magnetic IONP with the carboxylic group) was obtained from Ocean Nanotech LLC, San Diego, CA, USA. Bis-benzimide H-33342 trihydrochloride trihydrate (Hoechst-33342, Cat.no. J62134; 98%) was ordered from Alfa Aesar (Ward Hill, MA, USA), LysoTrackerTM red DND-99 (Ref: L7528) was purchased from Life Technology, USA. Cyanine 7 (Cy7) NHS ester was brought from Lumiprobe, Life Science Solutions (USA). μ-Slide 8-well-high glass bottom (Cat. No. 80807) was purchased from Ibidi (Madison, USA). Chlorpromazine hydrochloride (98+%, Cat. no. J63659.09) and cytochalasin D (Ref: PHZ1063) were obtained from Thermo Fisher Scientific (USA), genistein (>99%, Cat. no. RG004) was supplied by TSZ Chem. Methyl-beta-cyclodextrin (Average MW 1310, Code: 377110050) was ordered from Acros Organics (USA). Dynasore (Dynamin inhibitor I, SC-202592) and amiloride HCl (SC-3578A) were purchased from Chem Cruz (USA). 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Ref. M6494) was brought from Invitrogen (Thermo Fisher Scientific, USA). Polyethersulfone (PES) sterile syringe filter membrane (0.2 μm; Product no. 431229) was purchased from Corning®, Life Sciences (New York, USA). Carbon film 300 mesh copper grid was supplied by Electron Microscopy Sciences CF300CU50, USA. Amicon® Ultra – 15 centrifugal filters (30 kDa; Ref: UFC903024) were purchased from Merck Millipore Ltd (USA).Murine macrophage (RAW264.7), human embryo kidney cell line (HEK293), pediatric brain tumor medulloblastoma cell line (D556), and triple-negative breast cancer cell line (MDA-MB-453) were purchased from American Type Culture Collection (ATCC, USA). RPMI 1640 (Rosewell Park Memorial Institute) medium with L-glutamate (Cat. no. 112-025-101) and Dulbecco's phosphate buffered saline, 1× without Ca & Mg (DPBS, Cat. no. 114-057-101) were purchased from Quality Biological (USA). Dulbecco's modified Eagle's medium with 4.5 g L−1 glucose & L-glutamine without sodium pyruvate (DMEM, Ref: 10-017-CV) and Eagle's minimum essential medium with 1.5 g L−1 sodium bicarbonate, nonessential amino acids, L-glutamine and sodium pyruvate (EMEM; Ref; 10-009-CV) were ordered from Corning® (USA). Heat-inactivated fetal bovine serum (FBS) was provided by Avantor® Seradigm (USA). Penicillin/streptomycin (Ref. 15070-063) and TRypLETMSelect (1×) (Ref. 12563-029) were purchased from GibcoTM (USA). DMSO (dimethyl sulfoxide, Bio-Max, cell culture grade, SKU: 40470005-1) was obtained from BioWorld, BioPLUS Chemicals (USA) for cell storage.
Synthesis of IONR and IONP with different sizes and shapes
In a typical procedure of IONR synthesis, β-FeOOH nanorod precursors were first prepared by mixing FeCl3·6H2O (3 g) in 20 mL of deionized (DI) water with PEI for hydrolysis at 120 °C for 4 h using an autoclave. β-FeOOH (akageneite) precursors were precipitated by centrifugation before removing the supernatant. The residual precursors were washed with ethanol twice. The β-FeOOH precursors were then collected and dried at 55 °C overnight to yield a light brownish solid. β-FeOOH precursors with different aspect ratios in their core sizes, i.e., length × width (L/W): 50 × 7 and 145 × 8 (in nm), can be obtained by varying the volume of PEI (0 and 0.1 mL), respectively. Under an argon atmosphere, β-FeOOH precursor (100 mg) and oleylamine (10 mL) were mixed at 120 °C for 15 min, before heating to 220 °C for 4 h. After the reaction had been cooled, the product was washed and centrifuged three times with ethanol/hexane (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and then dispersed in chloroform as a stock solution.
1), and then dispersed in chloroform as a stock solution.
Spherical IONP were prepared using the thermo-decomposition method published in the literature.12,13 In brief, the iron–oleate complex was first prepared by reacting ferric nitride nonahydrate (4.04 g) with sodium oleate (9.13 g) in a mixed solvent consisting of hexane (50 mL), ethanol (10 mL), and water (40 mL) for 4 h at room temperature. Afterward, the mixture was kept overnight to allow the separation of the organic and aqueous phases. The organic phase containing the iron–oleate complex was then collected to synthesize IONP. The solution of the iron–oleate complex (5 mL) was mixed with 1-octadecene (5 mL), which was then degassed with argon for 10 min before heating up to 320 °C with the temperature ramping at 0.6 °C s−1. The reaction was kept at 320 °C for 30 min to synthesize IONP with core sizes of 20 nm. The resulting IONP were cleaned with a hexane/ethanol solvent mixture (v/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and dispersed in chloroform (1 mg IONP per mL) before the following surface functionalization.
1) and dispersed in chloroform (1 mg IONP per mL) before the following surface functionalization.
Preparation of oligosaccharide coated IONR and spherical IONP
We used an oligosaccharide coating to make IONR and IONP water-soluble and stable, based on our previously published method.13 Briefly, 3 mL of chloroform solution of IONR (1 mg mL−1) was slowly added into preheated N,N-dimethylformamide (DMF) solution (50 mL) containing 900 mg of glucose. The mixture was gradually heated to 120 °C at a rate of 5 °C min−1 and maintained for 2.5 h with continuous stirring. The products were washed with pure ethanol three times to remove DMF and redispersed in DI H2O to yield oligosaccharide-coated IONR. The IONR solution was adjusted to pH 10 using 0.5 M sodium hydroxide (NaOH) for storage. The solution of IONR was passed through a 0.2 μm polyethersulfone (PES) filter to remove large aggregates. The synthesis of oligosaccharide coated IONP followed the same procedure as that described for IONR. Iron concentration was quantified by the phenanthroline colorimetric method.Surface functionalization of IONR and spherical IONP
IONR and IONP were partially ammoniated before being labeled with the Cy7 dye to enable fluorescence imaging during cell internalization and subcellular distribution study. Briefly, 1 mL of IONR(L) with a core length and width of 140 nm × 6 nm, IONR(S) with a core length and width of 50 nm × 7 nm, and spherical IONP with a core size of 20 nm were reacted at a concentration of 1 mg Fe per mL with 2 mL of ammonium hydroxide at 37 °C overnight. Then, the ammoniated IONR were purified with Amicon Ultra-4 centrifugal filters (50 kDa) to remove any ammonia hydroxide residues before being dispersed into a coupling buffer (PBS, pH = 7.4) at a concentration of 1 mg Fe per mL. To label nanoparticles for fluorescence-based cell imaging, the near-infrared dye Cy7-NHS ester (50 μL, 1 mg mL−1 in DMSO) was reacted with ammoniated IONR(L), IONR(S), and 20 nm spherical IONP (1 mL of 1 mg Fe per mL) in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 molar ratio [IONP: Cy7] for 3 h at room temperature to prepare Cy7-labeled IONR(L), Cy7-labeled IONR(S), and Cy7-labeled spherical IONP, which were further purified with Amicon Ultra-4 centrifugal filters (30 kDa) in 1× PBS buffer (1 mg Fe per mL) for the future cellular uptake studies.
500 molar ratio [IONP: Cy7] for 3 h at room temperature to prepare Cy7-labeled IONR(L), Cy7-labeled IONR(S), and Cy7-labeled spherical IONP, which were further purified with Amicon Ultra-4 centrifugal filters (30 kDa) in 1× PBS buffer (1 mg Fe per mL) for the future cellular uptake studies.
Characterization of IONR and spherical IONP
Malvern Zetasizer Nano ZS (Malvern, Worcs, U.K.) was used to measure the hydrodynamic size and surface charges of prepared IONR with different aspect ratios (L/W), i.e., IONR(L), IONR(S), and spherical IONP with a core size of 20 nm. Typically, 40 μg of each sample was dissolved in 1 mL of DI H2O and transferred to a 1.5 mL polystyrene cuvette and measured three times at 25 °C temperature. The core sizes and morphology of IONR(L), IONR(S), and IONP were examined using TEM. The samples were prepared by dropping the solution on the carbon film and drying at room temperature before being visualized using JEOL JEM1400 transmission electron microscope (Peabody, MA, USA, accelerating voltage 120 kV). To measure the size distribution of each type of nanoparticles, we randomly selected 50 IONR or IONP in the field of view and measured the size using ImageJ (NIH, Bethesda, MD, USA). The coating surface was characterized by Fourier transform infrared (FTIR) spectroscopy using a Thermo Scientific (NICOLET iS50 FT-IR) spectrometer. The crystalline structure of IONR and IONP was examined by powered X-ray diffraction (XRD) using a Philips X'Pert-Diffractometer with Bragg–Brentano-geometry (Cu Kα-rays, U = 45 kV; I = 40 mA).Preparation of cell culture
We selected four different cell lines, i.e., normal cell lines of RAW264.7 (macrophage) and HEK293 (kidney cells), as well as cancer cell lines of D556 (human medulloblastoma) and MDA-MB-453 (triple negative breast cancer cells), to investigate how nanoparticles of different shapes and sizes interact with different types of cells that have different biological and physiological functions. RAW264.7 cells are murine macrophages that are known to efficiently phagocytose exogenous substances, including nanomaterials. Macrophages also play a crucial role in the immune system, involving in many inflammatory responses that can be targeted for imaging and therapeutic interventions via nanomedicine. HEK293 cells, derived from human kidney tissue and immortalized, are particularly suitable for studying nanoparticles interactions with normal cells and tissue as they have often been used in previous studies.25,26 For comparison with cancer cells, we selected human medulloblastoma D556 cells and breast cancer MDA-MB-453 cells to investigate how cancer cells interact with nanoparticles.RAW264.7 cells were grown as monolayer cultures in Roswell Park Memorial Institute (RPMI) medium, supplemented with 10% (v/v) fetal bovine serum (FBS), 1% (v/v) 50 units per ml penicillin and 50 μg ml−1 streptomycin. FBS was sterilized using a 0.22 μm vacuum filtration system (Cat. No. SCGP00525; EMD Millipore Corp., Burlington, USA) before mixing in the cell culture medium. D556 and HEK293 were grown as monolayer cultures in Eagle's minimal essential medium (EMEM), supplemented with 10% (v/v) fetal bovine serum (FBS), 1% (v/v) 50 units per mL penicillin and 50 μg mL−1 streptomycin. MDA-MB-453 cells were grown as monolayer cultures in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% (v/v) fetal bovine serum (FBS), 1% (v/v) 50 units per mL penicillin and 50 μg mL−1 streptomycin. Cell cultures were performed in an incubator with 5% CO2 plus 95% air at 37 °C. Depending on each experiment's requirements, the cells were seeded in T-75 flasks, 8-well chambered slides, 6-well chambered slides, and 96-well plates. Sub-confluent cell cultures were used for experimentation.
Visualization of cellular uptake and subcellular distribution of IONR and IONP
For qualitative analysis of cellular uptake and endosomal escape, cells were seeded in 8-well chambered glass dishes (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) and supplied with their respective growth medium containing 10% FBS and 1% antibiotics (penicillin–streptomycin, 10
000 cells per well) and supplied with their respective growth medium containing 10% FBS and 1% antibiotics (penicillin–streptomycin, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U mL−1). After 24 h, the cells were incubated with IONR(L), IONR(S), and IONP at a concentration of 50 μg mL−1 for 3 h. The medium was removed, and cells were rinsed with DPBS three times. Soon after being washed, the cells were incubated with LysoTracker, a fluorescent dye commonly used to label acidic organelles, such as lysosomes and endosomes, for 15–20 min in the serum-free medium (1
000 U mL−1). After 24 h, the cells were incubated with IONR(L), IONR(S), and IONP at a concentration of 50 μg mL−1 for 3 h. The medium was removed, and cells were rinsed with DPBS three times. Soon after being washed, the cells were incubated with LysoTracker, a fluorescent dye commonly used to label acidic organelles, such as lysosomes and endosomes, for 15–20 min in the serum-free medium (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 times dilution). LysoTracker works by accumulating in acidic intracellular compartments and typically emitting fluorescence upon binding to the acidic environment of lysosomes and endosomes. After incubation, the cells were washed with DPBS three times and fixed with 4% paraformaldehyde for 15 min, before being washed twice with DPBS, and stained with Hoechst 33342 (1
1000 times dilution). LysoTracker works by accumulating in acidic intracellular compartments and typically emitting fluorescence upon binding to the acidic environment of lysosomes and endosomes. After incubation, the cells were washed with DPBS three times and fixed with 4% paraformaldehyde for 15 min, before being washed twice with DPBS, and stained with Hoechst 33342 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 times dilution in 1× DPBS) for 10 min. The treated cells were gently washed and replaced with 200 μL DPBS, and then were observed using a Leica Stellaris-8 confocal laser scanning microscopy. Cy7 and LysoTracker Red DND-99 showed excitation/emission at 756/779 nm and 577/590 nm, respectively.
400 times dilution in 1× DPBS) for 10 min. The treated cells were gently washed and replaced with 200 μL DPBS, and then were observed using a Leica Stellaris-8 confocal laser scanning microscopy. Cy7 and LysoTracker Red DND-99 showed excitation/emission at 756/779 nm and 577/590 nm, respectively.
Measurement of iron concentrations for quantification of cellular uptake
The Fe concentrations of IONR and IONP taken up by cells were measured by the 1,10-phenanthroline colorimetric assay, as described in our previous report.27,28 Briefly, RAW264.7, HEK293, and D556, cells were treated with IONR(L), IONR(S), and IONP for 4 h and then rinsed with DPBS to eliminate non-internalized NP. RAW264.7 cells were collected by scraping, whereas HEK293 and D556 were collected by TRypLETMSelect and centrifugation. The cells were resuspended in 1 mL of DPBS, counted with a hemocytometer, and normalized to 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells. These cells were digested in nitric acid (400 μL) at 70 °C for 3 h to digest internalized IONR or IONP into Fe ions. The Fe concentration was quantified in a 96-well plate by adding sodium citrate solution (25 g L−1, 95 μL), followed by digested nanoparticle solution (5 μL), hydroquinone (20 mg mL−1 in water, 40 μL), 1,10-phenanthroline (1 g L−1, 40 μL), and acetate buffer (pH 3.5, 20 μL). After mixing for 20 min, the absorbance at 508 nm was measured using a plate reader (Synergy Biotek, USA) with the Fe concentration determined by comparing to the standard curve of the absorbance vs. the Fe(III) concentrations, ranging from 3 to 100 μg mL−1.
000 cells. These cells were digested in nitric acid (400 μL) at 70 °C for 3 h to digest internalized IONR or IONP into Fe ions. The Fe concentration was quantified in a 96-well plate by adding sodium citrate solution (25 g L−1, 95 μL), followed by digested nanoparticle solution (5 μL), hydroquinone (20 mg mL−1 in water, 40 μL), 1,10-phenanthroline (1 g L−1, 40 μL), and acetate buffer (pH 3.5, 20 μL). After mixing for 20 min, the absorbance at 508 nm was measured using a plate reader (Synergy Biotek, USA) with the Fe concentration determined by comparing to the standard curve of the absorbance vs. the Fe(III) concentrations, ranging from 3 to 100 μg mL−1.
Visualization of cellular compartmentation of IONR and IONP
We used TEM to investigate the subcellular compartments of IONR and IONP after being taken up by RAW264.7, HEK293, and D556 cells. Cells were grown in 6-well plates (8 × 105 cells per well) for 24 h, then treated with IONR(L), IONR(S), IONP, SHP-10 or ferumoxytol for 2 and 4 h. Subsequently, the cells were rinsed with DPBS and immobilized with PFGPA fixative (2.5% glutaraldehyde, 0.1% glutaraldehyde, 0.03% picric acid, 0.03% Calcium chloride) for 1 h at room temperature. They were washed with cacodylate buffer and refrigerated at 4 °C until further processing. Cells were then fixed with 1% osmium tetroxide/cacodylate for 1 h, rinsed with cacodylate, and dehydrated in a graded ethanol series (25, 50, 70, 95, and 100%). They were then immobilized in epoxy resin overnight and finally polymerized at 60 °C for 48 h. Using a Leica EM UC6 ultramicrotome, samples were trimmed and sectioned into 70 nm ultrathin sections. These sections were collected onto a formvar/carbon grid before observation under the TEM coupled to a Gatan Camera. In this experiment, post-staining with heavy metals was avoided to eliminate the possibility of confusion of nanoparticles with any metal aggregates on the surface of the trimmed sections.Inhibition assay for investigation of cellular uptake mechanisms
We investigated the mechanisms of shape or size-dependent cellular uptake using a published protocol.29,30 RAW264.7, HEK293, and D556 cells were grown in 6-well plates (5 × 105 cells per well) for 24 h. Cells were pre-incubated with different endocytic selective inhibitors, i.e., chlorpromazine (10 μg ml−1) for clathrin-mediated endocytosis,31 genistein (10 μg ml−1) for caveolae-dependent endocytosis,32 MβCD (5 mg mL−1) for lipid rafts/cholesterol-enriched microdomains,33 amiloride (10 μg ml−1) for micropinocytosis,34 dynasore (20 μg mL−1) for dynamin-GTPase inhibition,35 and CytoD (2.5 μg mL−1) for F-actin polymerization36 in a serum-free medium. The serum-free medium minimizes interference from serum components and enhances inhibitor uptake, ensuring that observed effects are due to inhibited endocytosis pathways. After 1 h, the inhibitors were removed, and cells were co-incubated with nanoparticles (50 μg Fe per mL) and inhibitors for 4 h at 37 °C. Treating cells with inhibitors twice maintains sustained inhibition, minimizing pathway reactivation. Untreated cells served as the negative control, while cells treated only with nanoparticles acted as the positive control. Cells were washed with DPBS and the total amount of internalized iron was quantified by a 1, 10 phenanthroline calorimetric assay.27Evaluation of cytotoxicity of IONR and IONP
We used the MTT assay to evaluate the cell viability and cytotoxicity of prepared IONR and IONP in different cells. This colorimetric assay measures the reduction of MTT into formazan by mitochondrial succinate dehydrogenase, an enzyme active during metabolic processes in viable cells. Cells were seeded for 24 h, then incubated with IONR(L), IONR(S), and IONP at different concentrations (12.5, 25, 50, 100, and 200 μg ml−1) for 24 h and 48 h at 37 °C. After washing with DPBS, the cells were incubated in a medium (90 μL) and 10 μL MTT solution (5 mg mL−1 in DPBS) for 2 h. The medium was then replaced with DMSO (100 μL) for 15 min to dissolve the formazan crystals. The absorbance of the MTT formazan was determined at 540 nm using a plate reader (Synergy Biotek, USA). Viability was expressed as a percentage of the absorbance of treated cells to untreated cells.Estimation of compartmentalization of IONR and IONP in lysosomes
We used LysoTrackers to visualize the lysosomal structures by confocal microscopy. The compartmentalization of nanoparticles in lysosomes can be confirmed when the fluorescence signals of LysoTrackers and Cy7 labeled nanoparticles are co-localized. We then estimated the levels of IONR or IONP in lysosomes by measuring the percentage of co-localized or overlapping fluorescence signals of Cy7 and LysoTrackers after extracting pixels from the confocal images based on their corresponding colors. Image analysis was conducted using Python 3.11.4 with OpenCV 4.6.0, NumPy 1.25.2, and Pandas 2.1.4.Examination of mitochondrial depolarization by the JC-1 assay
D556 and RAW264.7 cells were cultured in μ-slide 8 well high glass bottom dish (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) for 24 h. The cells were incubated for 24 h with IONR(L), IONR(S), and IONP at a dose concentration of 50 μg mL−1 in their respective cell culture medium (200 μL) supplemented with 10% FBS and 1% antibiotics. In the positive control, the cells were treated with 1 μL of 50 mM CCCP in 200 μL of serum-free medium and incubated for 5 min. The cells that are not treated serve as a negative control. The control and treated groups were rinsed with PBS three times and incubated with 200 μL of 2 μM JC-1 dye (10 μL dye from 200 μM JC-1 stock + 1 mL serum-free medium) for 20 min. Then, the cells were rinsed thoroughly with DPBS three times and fixed with 4% paraformaldehyde for 15 min; after being washed twice with DPBS, the cells were collected and then examined using confocal microscopy.
000 cells per well) for 24 h. The cells were incubated for 24 h with IONR(L), IONR(S), and IONP at a dose concentration of 50 μg mL−1 in their respective cell culture medium (200 μL) supplemented with 10% FBS and 1% antibiotics. In the positive control, the cells were treated with 1 μL of 50 mM CCCP in 200 μL of serum-free medium and incubated for 5 min. The cells that are not treated serve as a negative control. The control and treated groups were rinsed with PBS three times and incubated with 200 μL of 2 μM JC-1 dye (10 μL dye from 200 μM JC-1 stock + 1 mL serum-free medium) for 20 min. Then, the cells were rinsed thoroughly with DPBS three times and fixed with 4% paraformaldehyde for 15 min; after being washed twice with DPBS, the cells were collected and then examined using confocal microscopy.
Measurement of intracellular reactive oxygen species (ROS)
We measured intracellular ROS levels using the 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) assay.37 Cells were seeded on μ-slide 8 well-high glass bottom dish for 24 h. After exposure to IONR(L), IONR(S), and IONP (50 μg mL−1) for 24 h. In the positive control, the cells were treated with 30% hydrogen peroxide (H2O2) in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution for 20 min in a complete medium to induce oxidative stress. Cells were washed with DPBS and incubated with DCFH-DA (10 μM) in serum-free culture media for 1 h. The control and treated groups were washed with DPBS thrice, fixed with 4% paraformaldehyde, and counterstained with Hoechst 33342 for 15 min. The cells were washed with DPBS and imaged using confocal microscopy.
1000 dilution for 20 min in a complete medium to induce oxidative stress. Cells were washed with DPBS and incubated with DCFH-DA (10 μM) in serum-free culture media for 1 h. The control and treated groups were washed with DPBS thrice, fixed with 4% paraformaldehyde, and counterstained with Hoechst 33342 for 15 min. The cells were washed with DPBS and imaged using confocal microscopy.
Statistical analysis
In this study, the quantitative data are presented as mean ± standard deviation (SD). The data from three experiments—the effects of endocytosis inhibitors on nanoparticle uptake, the mean fluorescence intensity of cellular uptake and internalization of IONR and IONP, and the uptake of IONR and IONP by different cell lines based on iron concentrations—were analyzed using one-way ANOVA. After establishing the statistical significance with ANOVA, Tukey's HSD test was applied as a post-hoc analysis to find specific pairwise groups. Significance levels were denoted as follows: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. GraphPad Prism software (version 9.5.1) was utilized for all statistical analyses.3. Results and discussion
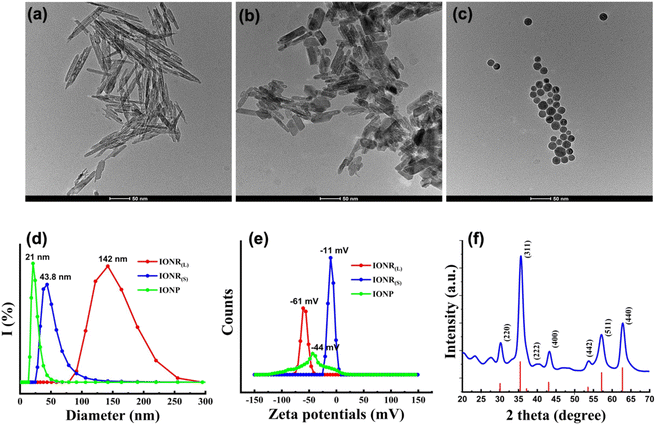
Characterization of oligosaccharide coated IONR and IONP
The β-FeOOH nanorods which serve as precursors for IONR were synthesized initially by hydrolyzing a FeCl3 solution with PEI functioning as a capping agent. PEI controlled the dimensions by adsorbing its protonated form onto the lateral plane (200) of the β-FeOOH nanorods.13 The organic phase IONR was then prepared by reducing the β-FeOOH nanorods in the presence of oleylamine, which acted as a solvent and capping agent. Oleylamine as an electron donor, facilitated the reduction process at an elevated temperature of 220 °C, leading to the formation of IONR. Interestingly, TEM images (Fig. S1a and b, ESI†) revealed hollow structures in the prepared IONR, likely due to the collapse of the β-FeOOH nanorod framework due to the release of water molecules during reduction. These unique hollow structures offer significant potential for drug loading,38 photoacoustic imaging,39 and photocatalysis40 applications. IONP was synthesized through the thermal decomposition of the iron-oleate complex. TEM images (Fig. S1c, ESI†) unveiled that the IONP exhibited high uniformity, with a diameter of 20 nm. Hydrophobic IONR and IONP were then mixed with glucose in a DMF solvent and heated to polymerize glucose on their surfaces, forming a thin oligosaccharide coating layer and rendering them water-soluble. Oligosaccharide-coated IONR(L), IONR(S), and spherical IONP were highly stable and monodispersed in aqueous solvents with good dispersibility as shown in the TEM images in Fig. 1a–c. The core sizes of oligosaccharide coated IONR(L) and IONR(S) were measured from the TEM images by averaging 50 nanoparticles found in the selected field of view. Obtained average lengths and widths were 141 × 6 nm ± 16.4 and 50 × 7 ± 9.9 nm for IONR(L) and IONR(S) with aspect ratios of 28 and 7.14, respectively. The average core size of oligosaccharide coated spherical IONP was measured to be 22 ± 1.8 nm in diameter (Fig. S1d–f, ESI†). The hydrodynamic size, zeta potential, and stability of oligosaccharide coated IONR(L), IONR(S), and spherical IONP were measured using DLS analysis. Oligosaccharide-coated IONR(L), IONR(S), and IONP dispersed in the water had an average hydrodynamic size of 142 nm, 43.8 nm, and 21 nm, respectively, as shown in Fig. 1d. Correspondingly, zeta potential measurements (Fig. 1e) indicate that IONR(L), IONR(S), and IONP have a negative charge of −61, −11, and −44 mV, respectively, due to the deprotonation of the carboxyl groups (-COOH) in the oligosaccharide coating, resulting in negatively charged carboxylate ions (–COO−).41 Additionally, oligosaccharides contain hydroxyl groups (–OH), which can ionize to a lesser extent, also contributing to the negative surface charge. Moreover, during the synthesis or in the dispersion medium, negatively charged ions from the solution might adsorb onto the surface of the IONR and IONP, further enhancing their negative zeta potential. Notably, the negatively charged oligosaccharide coated IONR(S) and IONP are stable in water for several weeks. While IONR(L) exhibited strong magnetism, a small portion tended to precipitate within one or two days but could be resuspended via vortexing. The presence of an oligosaccharide coating on IONR surfaces was confirmed by FTIR analysis (Fig. S2, ESI†). The peak at 670 cm−1 is ascribed to the vibrational mode of the Fe–O bonds in the crystalline lattice of Fe3O4. The small peak at around 963 cm−1 could be attributed to the in-plane wagging of the trans CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH bond.42 The absorbance bands at 1377 and 1456 cm−1 correspond to –CH3 vibrations, and the two peaks at 2852 and 2919 cm−1 represent the C–H stretching vibrations from the alkyl chain of oleylamine,43 indicating successful capping and rendering the particles hydrophobic. A broad intense peak in the 3294 cm−1 region corresponds to the O–H bond stretching vibration, and the appearance of a C
CH bond.42 The absorbance bands at 1377 and 1456 cm−1 correspond to –CH3 vibrations, and the two peaks at 2852 and 2919 cm−1 represent the C–H stretching vibrations from the alkyl chain of oleylamine,43 indicating successful capping and rendering the particles hydrophobic. A broad intense peak in the 3294 cm−1 region corresponds to the O–H bond stretching vibration, and the appearance of a C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C band at 1635 cm−1 represents the presence of aromatic structures from the oligosaccharide coating on nanoparticle surfaces.12 The formation of these aromatic structures can be attributed to intermolecular dehydration and aldol condensation reactions during the glycosylation process. These reactions occur even at a lower temperature of 120 °C, facilitated by the specific conditions around the nanoparticle surfaces.12 Additionally, XRD analysis of the oleylamine coated IONR(L) exhibits good crystallinity, as evidenced by the intensity of the diffraction peaks. All peaks observed in the diffractogram can be assigned to the magnetite spinel structure (JCPDS No. 19-0629).44 The peaks at 32° and 56.16° likely correspond to the crystallographic planes (221) and (510) of the corundum structure of the α-Fe2O3 (hematite) crystalline phase as previously reported.45 Whereas, strong peaks observed at 30.29°, 35.65°, 40.12°, 43.26°, 53.79°, 57.02°, and 62.90° are indicative of the seven distinct diffraction planes (220), (311), (222), (400), (422), (511), and (440) of the crystalline structure of Fe3O4 with magnetite composition (Fig. 1f).12,46 The calculated mesh parameter for stoichiometric magnetite is 8.360 Å, which also suggests that synthesized nanoparticles are predominately composed of magnetite not maghemite (γ-Fe2O3) or hematite. Importantly, both IONR(L) and IONR(S) have the same crystal structure and phase composition.
C band at 1635 cm−1 represents the presence of aromatic structures from the oligosaccharide coating on nanoparticle surfaces.12 The formation of these aromatic structures can be attributed to intermolecular dehydration and aldol condensation reactions during the glycosylation process. These reactions occur even at a lower temperature of 120 °C, facilitated by the specific conditions around the nanoparticle surfaces.12 Additionally, XRD analysis of the oleylamine coated IONR(L) exhibits good crystallinity, as evidenced by the intensity of the diffraction peaks. All peaks observed in the diffractogram can be assigned to the magnetite spinel structure (JCPDS No. 19-0629).44 The peaks at 32° and 56.16° likely correspond to the crystallographic planes (221) and (510) of the corundum structure of the α-Fe2O3 (hematite) crystalline phase as previously reported.45 Whereas, strong peaks observed at 30.29°, 35.65°, 40.12°, 43.26°, 53.79°, 57.02°, and 62.90° are indicative of the seven distinct diffraction planes (220), (311), (222), (400), (422), (511), and (440) of the crystalline structure of Fe3O4 with magnetite composition (Fig. 1f).12,46 The calculated mesh parameter for stoichiometric magnetite is 8.360 Å, which also suggests that synthesized nanoparticles are predominately composed of magnetite not maghemite (γ-Fe2O3) or hematite. Importantly, both IONR(L) and IONR(S) have the same crystal structure and phase composition.
Shape specific cellular uptake of IONR and IONP by different cells
We observed the shape and size dependent cellular uptake of IONR and spherical IONP in three types of cells, i.e., RAW264.7, HEK293, and D556, after 4 h incubation with IONR(L), IONR(S), and spherical IONP at the same Fe concentration of 50 μg mL−1. Fig. 2 shows levels of uptake of IONR with different aspect ratios and IONP by different cells. Specifically, RAW264.7 murine macrophage cells internalized the highest amount of IONR(L) (50.1 ± 2.2 Fe μg per million cells) compared to short IONR(S) (44.7 ± 1.6 Fe μg per million cells) and spherical IONP (36.7 ± 0.8 Fe μg per million cells), respectively. Notably, IONR(L) exhibited a 1.36-fold higher cellular uptake compared to spherical IONP. While cells appeared to take up more IONR(L) than IONR(S), the difference is not statistically significant. The statistically significant higher uptake of both types of IONR over IONP indicates the shape dependent uptake preference by macrophages. In HEK293 embryonic kidney cells, a similar amount of nanoparticle internalization was observed for IONR(L) (i.e., 43.7 ± 2.9 Fe μg per million cells) and IONP (i.e., 43.8 ± 3.3 Fe μg per million cells). However, IONR(S) exhibited a slightly lower uptake (34.9 ± 3.7 Fe μg mL−1), but the difference is not statistically significant. Interestingly, the higher internalization of IONP by HEK293 cells compared to RAW264.7 cells could be attributed to the inherent differences in cell membrane composition, endocytic pathways, and metabolic activity between these cell types. HEK293 cells, being epithelial in origin, might possess a more efficient endocytic machinery for certain nanoparticle sizes, whereas RAW264.7 macrophages preferentially uptake larger and rod-shaped particles due to their phagocytic role. Different from RAW264.7 and HEK293 cells, an approximately 1.83-fold higher uptake of IONR(L) by the D556 medulloblastoma cells was observed (i.e., 39.7 ± 1.56 Fe μg per million cells) than IONR(S) (21.5 ± 1.37 Fe μg per million cells) and 1.17-fold higher than spherical IONP (33.8 ± 0.5 Fe μg per million cells), respectively. In contrast, ferumoxytol, an IONP formulation with a smaller core size (ranging from 7–10 nm and overall size 17–31 nm in diameter) commercially available as an iron supplement for treating anemia but as an off-label used alternative MRI contrast agent in some clinical studies,47–49 exhibited much lower levels of cellular uptake across all three cell lines compared to the rod-shaped IONR and IONP with a 20 nm core size prepared in this study. It is notable that the cellular uptake levels of ferumoxytol are cell-line dependent, with RAW264.7 showing the highest at 7.5 ± 0.3 Fe μg per million cells compared to HEK293 (2.4 ± 0.3 Fe μg per million cells) and D556 (0.43 ± 0.07 Fe μg per million cells). The differences in the cellular uptake of ferumoxytol between these cell lines are statistically significant.Notably the surface charge of nanoparticles can affect the interactions between the cells and nanoparticles, and eventually pathways for internalization and the overall effectiveness of cellular uptake.50 Generally, cell surfaces are negatively charged. Thus, nanoparticles with a positive surface charge can interact with cells non-specifically with the cell membrane via charge–charge interactions, thereby improving internalization efficiency. However, the cell membrane also features specific binding sites with cationic receptors that facilitate interaction with anionic nanoparticles through a process known as “adsorptive endocytosis”. Thus, nanoparticles can cluster on the cell surface due to repulsive interaction with anionic charged receptor domains.50–53 it is possible that the adsorptive endocytosis may play a role in the cell internationalization of reported anionic IONR and IONP. Consequently, IONR(L) with a higher negative charge (−60 mV) can interact with cells more effectively than IONR(S) (−11 mV) and IONP (−44 mV) with a lower negative charge, leading to a higher level of cellular uptake of IONR(L).
Additionally, the observed shape-enhanced cellular uptake of IONR demonstrated a similar trend reported in other studies of anisotropic nanoparticles, such as gold54–56 and silver,57 suggesting that iron oxide-based nanoparticles likely follow similar cellular uptake mechanisms and inherent characteristics of cells, wherein nanorods are preferentially internalized through endocytic pathways. The unique geometry of nanorods results in reduced cell membrane bending energies during their endocytosis, potentially contributing to their preferential uptake. The increased cellular uptake of IONR compared to spherical IONP may benefit magnetism-based theranostic applications in biomedicine. These applications can exploit the unique magnetic properties of IONR for the development of targeted MRI probes for precise imaging,58,59 magnetic field-guided drug delivery systems,38 cell labeling,60 and magnetic separations.22,61
The results from the MTT assay at both 24 and 48 h showed that IONP and IONR with different aspect ratios exhibited robust biocompatibility profiles across diverse cell types tested in this study. Even at the highest concentration of 200 μg Fe per mL, the viability of cells treated with IONR(L), IONR(S), and IONP were greater than 85%, with no significant change in cell death, compared to the cells without nanoparticle treatment (Fig. 3 and S3, ESI†). This is crucial considering that the increased cellular uptake of IONR and IONP is important for enhancing cell-targeted applications but does not directly impact cell viability. However, further studies are warranted to fully investigate their specific mechanisms of action, pharmacokinetics, and long-term side effects to assess their clinical translatability and potential therapeutic benefits.
Shape specific cellular internalization and subcellular localization
To investigate cell interactions with different IONR and possible subcellular localization, we treated cells with Cy7 dye-labeled IONR(L), IONR(S), and spherical IONP at 50 μg Fe per mL, followed by staining the treated cells with organelle-specific fluorescent markers LysoTracker and Hoechst 33342. Using confocal microscopy under the same excitation and acquisition parameters, we observed significantly higher intracellular Cy7 fluorescence signals in all three types of cells with IONR(L) compared to IONR(S) and spherical IONP as shown in Fig. 4. Consistent with the results of intracellular iron quantification shown in Fig. 2, RAW264.7 macrophages appear to exhibit the highest uptake of IONR(L), as evidenced by the most intense Cy7 signals (Fig. 4a), while D556 and HEK293 cells have much less intracellular uptake. Specifically, RAW264.7 cells treated with IONR(L) exhibited the highest mean fluorescence intensity, which was almost 7.5-fold and 4-fold higher than those of HEK293 and D556, respectively (Fig. 4d). In addition, IONR(L) exhibited the highest intracellular signal intensity vs. IONR(S) and IONP in RAW264.7 cells. The differences between these nanoparticles of different shapes and sizes are statistically significant. These results are attributed to the endocytosis of exogenous materials as the major function of macrophages compared to other cells.In addition to the shape-dependent increase in cellular uptake, IONR also exhibited improved subcellular localization, notably efficient distribution within endosomes/lysosomes. Their elongated shape causes more efficient interactions with cell membranes. To further interrogate the intracellular trafficking of the endocytosed IONR and IONP of different shapes and sizes, we examined the co-localization of Cy7-labeled NP and Lysotracker that are typically used visualizing intracellular endosomes and lysosomes using fluorescence microscopic imaging coupled with software-based image analysis. We performed a quantitative assessment and comparison of endo–lysosomal accumulation between different IONR and IONP based on analyzing the pixels exhibiting signals from Lysotracker and Cy7-labeled NP, as well as pixels, in which signals from nanoparticles and lysosomes are overlapped as depicted in Fig. 4e. In HEK293 and D556 cells, the percentage of IONR(L), IONR(S), and IONP co-localized with endo–lysosome was >92%. Whereas in the RAW264.7 cell line, the co-localization percentage was found to be 64.0, 82.9, and 73.2% for IONR(L), IONR(S), and IONP, respectively. Interestingly, while RAW264.7 macrophage cells showed the highest cellular uptake of IONR(L) compared to other cell lines, its endo–lysosomal accumulation under the reported experimental conditions is much lower than others. One possible explanation is that long nanorods may have a greater propensity for escaping the lysosomal degradation pathway in the macrophage cells. This could be due to the rod-shape facilitating their escape from lysosomal compartments into the cytosol. Additionally, RAW264.7 macrophages, known for their phagocytic activity, follow the intracellular trafficking pathways favoring the escape of long nanorods from lysosomes. Conversely, cancer cells and normal kidney cells may employ different mechanisms when taking up nanoparticles, favoring lysosomal retention.
Understanding the shape or size related differences in the accumulation or release of nanoparticles in endosomes and lysosomes is important for nanoparticle-based drug delivery and cell labeling. It is preferred that drug-carrying nanoparticles remain stable, and rapidly escape from endosomes/lysosomes to release the therapeutic cargo into the cytosol, where it can exert its therapeutic effects.62 When nanoparticles are trapped within membrane-bound vesicles, like endosomes and lysosomes, they may not be able to release their cargo into the cytosol. This can lead to the degradation of the biological cargo and possibly cause the cells to expel nanoparticles. In this case, we did not see statistically significant differences in lysosomal accumulation of different IONR and IONP in normal and cancer cells, unlike macrophages.
Lysosomal packing of IONP(L), IONR(S), or IONP in three different cells was further confirmed by TEM examination. Irrespective of variations in size, shape anisotropy, and cell types, all nanoparticles were observed to be located within subcellular vesicular compartments as shown in the selected TEM images in Fig. 5, but not translocated to the nucleus.
Shape dependent endocytic mechanisms of cellular uptake and cellular internalization
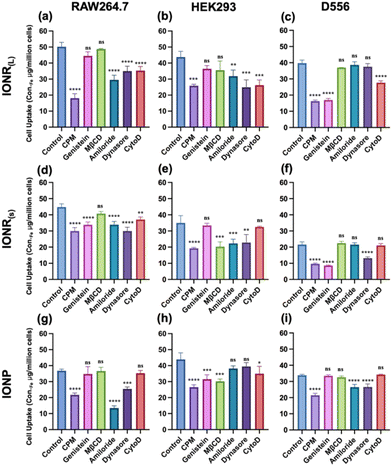
The cellular internalization of foreign substances, such as engineered nanomaterials, typically occurs through two primary mechanisms: pinocytosis and phagocytosis. Pinocytosis, in turn, can be further categorized into macropinocytosis and micropinocytosis. Larger particles and macromolecules with diameters exceeding 200 nm are taken up non-selectively via phagocytosis or macropinocytosis. In comparison, smaller nanoparticles undergo internalization through micropinocytosis pathways, including clathrin-mediated, caveolae/lipid raft-mediated, and clathrin/caveolae-independent mechanisms across all cell types.63 To further elucidate the underlying cellular uptake mechanisms contributing to the observed size and shape-dependent variations in cellular uptake of IONR(L), IONR(S), and spherical IONP, we treated D556, RAW264.7, and HEK293 cells with six different inhibitors, i.e., chlorpromazine (CPM), genistein, methyl-β-cyclodextrin (MβCD), amiloride, dynasore hydrate, or cytochalasin D (CytoD) for 1 h in a serum-free medium followed by 4 h incubation with different nanoparticles in the presence of FBS, antibiotics, and the inhibitor. In this case, CPM disrupts clathrin-mediated endocytosis by relocating clathrin and its adapter proteins from the plasma membrane to intracellular vesicles.31 Genistein, a tyrosine kinase inhibitor, impedes caveolae-dependent endocytosis by blocking actin depolymerization and dynamin 2 recruitment.32 MβCD binds to cholesterol within the cell membrane, inhibiting lipid-raft-mediated mechanisms and enabling assessment of cholesterol impact on the uptake mechanism.33 Amiloride, inhibits macropinocytosis by reducing the sub-membranous pH and disrupting actin polymerization through the inhibition of Rac1 and Cdc42 signaling.34 Dynasore inhibits dynamin, a key protein involved in clathrin-mediated endocytosis and other dynamin-dependent mechanisms35 by facilitating the scission of the cell membrane, which is critical for endosome formation. Lastly, the involvement of the actin cytoskeleton and microtubules was investigated using CytoD, since actin orchestrates the formation of membrane ruffles in macropinocytosis.36Table 1 summarizes the results from the measurement of the intracellular iron concentrations of different cells after being treated with IONR(L), IONR(S), and IONP in the presence of different inhibitors, respectively. Specifically, RAW264.7 macrophage cells (Fig. 6a) exhibited the strongest inhibition or reduction of IONR(L) uptake when exposed to CPM (18.0 ± 2.2 Fe μg per million cells), amiloride (29.5 ± 2.3 Fe μg per million cells), dynasore (34.9 ± 2.4 Fe μg per million cells), and CytoD (35.2 ± 2.1 Fe μg per million cells) compared to cells without inhibitor treatment. However, after being treated with CPM, amiloride, dynasore, or CytoD, RAW264.7 cells showed a 2.7, 1.65, 0.84, and 0.99-fold inhibition of IONR(L) uptake. These results suggest the possible involvement of clathrin-mediated, macropinocytosis, dynamin-dependent, and phagocytosis pathways in macrophage internalization of IONR(L). Analyzing the pattern of RAW264.7 cell internalization of IONR(S) indicates a preference for clathrin-mediated, caveolae-mediated, macropinocytosis, dynamin-dependent, and phagocytosis pathways based on the intracellular Fe concentrations of cells pretreated with CPM (29.9 ± 1.7 Fe μg per million cells), genistein (33.8 ± 1.4 Fe μg per million cells), amiloride (33.8 ± 1.6 Fe μg per million cells), dynasore (29.9 ± 1.9 Fe μg per million cells) and CytoD (37.0 ± 1.3 Fe μg per million cells) as shown in Fig. 6d. In contrast, the uptake of spherical IONP by murine macrophage cells predominantly relied on clathrin-mediated, micropinocytosis, and dynamin-dependent endocytosis. Intracellular Fe concentrations of treated RAW264.7 cells were reduced by 1.6, 2.7, and 0.5-fold, respectively, relative to the inhibitor untreated RAW cells upon exposure to CPM, amiloride, and dynasore. Notably, inhibitors such as genistein, MβCD, and CytoD exhibited a negligible effect on IONP internalization (Fig. 6g), suggesting that caveolae-mediated, lipid raft, and phagocytosis routes may not significantly contribute to the uptake process.
| Cell lines | Nanoparticles | Control | Inhibitors | |||||
|---|---|---|---|---|---|---|---|---|
| CPM | Genistein | MβCD | Amiloride | Dynasoore | CytoD | |||
| Results are presented as the cellular uptake level of Fe concentration in μg per million cells in the presence of inhibitor treatment, compared to the uptake in control (inhibitor untreated) cells under the same conditions. Inhibitors: chlorpromazine (CPM) – clathrin-mediated; genistein – caveolin-mediated; methyl-β-cyclodextrin (MβCD) – lipid rafts/cholesterol depletion; amiloride – macropinocytosis; dynasore – dynamin-dependent; and CytoD – micropinocytosis/phagocytosis. The values are averaged between three independent replicas of three independent experiments. | ||||||||
| RAW264.7 | IONR(L) | 50.1 ± 2.2 | 18.0 ± 2.2 | 44.4 ± 2.0 | 48.7 ± 0.3 | 29.5 ± 2.3 | 34.9 ± 2.4 | 35.2 ± 2.1 |
| IONR(S) | 44.7 ± 1.6 | 29.9 ± 1.7 | 33.8 ± 1.4 | 40.7 ± 1.1 | 33.8 ± 1.6 | 29.9 ± 1.9 | 37.0 ± 1.3 | |
| IONP | 36.7 ± 0.8 | 21.7 ± 0.9 | 34.7 ± 3.7 | 36.5 ± 1.9 | 13.4 ± 1.1 | 25.4 ± 1.0 | 35.2 ± 1.3 | |
| HEK293 | IONR(L) | 43.7 ± 2.9 | 25.8 ± 0.7 | 36.4 ± 1.6 | 35.5 ± 4.7 | 31.8 ± 3.1 | 24.8 ± 3.8 | 26.1 ± 2.6 |
| IONR(S) | 34.9 ± 3.7 | 19.2 ± 0.4 | 33.4 ± 1.0 | 20.2 ± 2.4 | 22.3 ± 2.0 | 22.8 ± 4.0 | 32.4 ± 0.3 | |
| IONP | 43.8 ± 3.3 | 26.5 ± 1.2 | 31.5 ± 2.2 | 30.1 ± 1.2 | 38.1 ± 1.3 | 39.4 ± 1.9 | 34.9 ± 3.7 | |
| D556 | IONR(L) | 39.7 ± 1.5 | 16.2 ± 0.5 | 16.9 ± 0.7 | 36.9 ± 0.1 | 38.6 ± 1.6 | 37.5 ± 1.6 | 27.6 ± 0.9 |
| IONR(S) | 21.5 ± 1.3 | 9.6 ± 0.3 | 8.6 ± 0.2 | 22.4 ± 0.9 | 21.5 ± 0.9 | 13.1 ± 0.6 | 21.0 ± 0.9 | |
| IONP | 33.8 ± 0.5 | 21.4 ± 1.1 | 33.4 ± 0.5 | 32.5 ± 0.66 | 26.4 ± 1.3 | 26.5 ± 1.5 | 34.2 ± 0.1 | |
Compared to macrophages, IONR(L) treated D556 medulloblastoma cells shown in Fig. 6c displayed a significant reduction in uptake indicated by the lower Fe contents in the presence of CPM (16.2 ± 0.55 Fe μg per million cells), genistein (16.9 ± 0.78 Fe μg per million cells), and CytoD (27.6 ± 0.95 μg per million cells) compared to those without inhibitor treatment (39.7 ± 1.56 μg per million cells). However, no statistically significant change in IONR(L) uptake was observed in the presence of MβCD, amiloride, and dynasore. These results suggest that IONR(L) likely undergoes endocytosis by the combination of clathrin-mediated, caveolae-mediated, and macropinocytosis/phagocytosis mechanisms, but not through cholesterol/lipid-raft and dynamin-mediated endocytosis. For IONR(S), D556 cells pretreated with CPM, genistein, and dynasore yielded intracellular iron concentrations of 9.6 ± 0.3, 8.6 ± 0.2, and 13.1 ± 0.6 (Fe μg per million cells), respectively, exhibiting 2.2, 1.1, and 1.6-fold reduction of cellular uptake by these inhibitors (Fig. 6f). In addition, pretreating D556 cells with MβCD, amiloride, and CytoD did not affect the uptake of IONR(S). Therefore, the results suggested that clathrin-mediated, caveolae-mediated, and dynamin-dependent pathways mediate ION uptake. In the case of spherical IONP, the participation of clathrin-mediated, macropinocytosis, and dynamin-dependent pathways was noticed. Compared to 33.9 ± 0.5 Fe μg per million cells in the inhibitor untreated sample, intracellular Fe concentrations were found to be significantly lower in cells treated with CPM (21.4 ± 1.05 Fe μg per million cells), amiloride (26.4 ± 1.3 Fe μg per million cells), and dynasore (26.5 ± 1.5 Fe μg per million cells). Conversely, intracellular Fe concentrations of cells treated with genistein, MβCD, and CytoD did not show a statistically significant difference (Fig. 6i), indicating that there is little involvement of caveolae-mediated, lipid-raft-mediated, and phagocytosis in D556 medulloblastoma cells internalizing spherical IONP.
For HEK293 cells, the uptake of IONR(L) was reduced significantly as indicated by the intracellular Fe concentration due to inhibition of CPM (25.8 ± 0.7 Fe μg per million cells), amiloride (31.8 ± 3.1 Fe μg per million cells), dynasore (24.8 ± 3.8 Fe μg per million cells), and CytoD (26.1 ± 2.6 Fe μg per million cells). However, there was no statistically significant difference in cellular uptake of IONR(L) when treating cells with genistein and MβCD, indicating that clathrin-mediated, dynamin-dependent, and phagocytic mechanisms are involved in HEK293 cellular uptake of IONR(L) (Fig. 6b). For IONR(S), treatment with CPM (19.2 ± 0.4 Fe μg per million cells), MβCD (20.2 ± 2.4 Fe μg per million cells), amiloride (22.3 ± 2.04 Fe μg per million cells), and dynasore (22.8 ± 4.09 Fe μg per million cells) has substantially reduced cellular internalization by approximately 1.8-fold, 1.6-fold, 0.9-fold, and 0.97-fold, respectively, as shown in Fig. 6e. In contrast, genistein and CytoD did not significantly inhibit the internalization of IONR(S). These results suggest that clathrin-mediated, lipid-raft, macropinocytosis, and dynamin-dependent endocytosis mechanisms are key drivers facilitating the potential entry of IONR(S) into HEK293 cells. For spherical IONP, inhibitor-pretreated cells exhibited significant effects on cellular uptake, evidenced by the reduction of intracellular Fe concentrations of cells treated with different inhibitors, i.e., CPM (26.5 ± 1.2 Fe μg per million cells), genistein (31.5 ± 2.2 Fe μg per million cells), MβCD (30.1 ± 1.2 Fe μg per million cells) and CytoD (34.9 ± 3.7 Fe μg per million cells), compared to the control cells without inhibitor treatment (43.8 ± 3.3 Fe μg per million cells). Thus, IONP potentially enters cells via clathrin-mediated, caveolae-mediated, lipid raft-mediated, and phagocytosis pathways (Fig. 6h).
To further validate and substantiate the internalization pathways of different IONR and IONP, we used TEM to examine the cellular structures in correlation with the uptake of IONR or IONP and their subsequent intracellular distribution. We distinguished four distinct architectural configurations indicative of different endocytosis pathways, i.e., clathrin-coated pits for clathrin-mediated endocytosis, flask-shaped formations for caveolae-mediated endocytosis, enlarged macropinocytic vesicles (macropinosomes) for macropinocytosis,64 and cellular pseudopodia formation for phagocytosis initiation.65
In RAW264.7 macrophage cells, the uptake of IONR(L) encompassed the appearance of the clathrin-coated pit, flask-shaped caveolae on the cytosolic side of the plasma membrane, membrane protrusions forming cup-shaped ruffles, and the emergence of cytoskeletal filaments forming pseudopodia, indicating the involvement of clathrin/caveolae-mediated, macropinocytosis and phagocytosis mechanisms (Fig. 7a and S4a–c, ESI†). Conversely, the uptake mechanisms of IONR(S) and IONP were primarily driven by processes associated with clathrin-mediated endocytosis, macropinocytosis, and phagocytosis (Fig. 7b, c, and S4d–f in ESI†). Integration of TEM observations with intracellular Fe concentration measurements after inhibiting cellular uptake using inhibitors compared to inhibitor untreated cells, we found that the internalization of IONR(L) predominantly occurred through clathrin-mediated, macropinocytosis, and phagocytosis mechanisms. In addition to these endocytosis pathways, a small amount of IONR(L) was internalized through caveolae-mediated endocytosis. Therefore, we are possibly able to observe the flask-shaped morphological changes in TEM upon contact of IONR(L) with the macrophage cell surface suggesting that caveolae-mediated endocytosis also contributed to a lesser extent to their uptake process. Similarly, although pseudopodia formation indicative of phagocytosis was observed in TEM, it does not appear to be a major mechanism for the endocytosis of IONP. The Fe concentration measurement for IONR(S) revealed the involvement of clathrin-mediated, caveolae-mediated, macropinocytosis, and phagocytosis pathways. However, TEM examination of the caveolae-mediated endocytosis was less appealing due to its laborious nature and limited ability to capture dynamic cellular processes at the plasma membrane, which requires better resolution of the TEM instrument or implementation of additional cell sample preparation methods.
In HEK293 embryonic kidney cells, localization of IONR(L) in flask-shaped structures and macropinosomes, indicated caveolae-mediated endocytosis and macropinocytosis activation (Fig. 7d and S5a, ESI†). IONR(S) can be seen packed in clathrin-coated pits, flask-shaped structures, and actin-dependent ruffle formations, suggesting clathrin/caveolae-mediated endocytosis and micropinocytosis involvement (Fig. 7e and S5b in ESI†). Additionally, compartmentalization of IONP in clathrin-coated pits and distinct cytoskeleton membrane invaginations confirmed the involvement of clathrin-mediated and macropinocytosis pathways (Fig. 7e and S5c, ESI†). Based on the Fe concentration measurement, we are unable to capture the cell structure changes in the TEM for clathrin-coated pit for clathrin-mediated and pseudopodia formation in phagocytosis for IONR(L) uptake. Conversely, the caveolae-mediated endocytosis pathway did not play a significant role in the internalization of IONR(S). For IONP, caveolae-mediated and phagocytosis morphological changes were not visible due to TEM limitations.
D556 cell interaction with IONR(L) and spherical IONP feature compartmentalization in various endocytic structures, including clathrin-coated pits, flask-shaped structures, and large-sized membrane ruffles ranging from 200 nm to 5 μm in diameter66 (Fig. 7g, i and S5d, e, g, h, ESI†). This observation highlights the prevalence of clathrin-mediated, caveolae-mediated, and macropinocytosis pathways. However, in the case of IONR(S), intracellular locations are within clathrin-coated pits and distinguishable membrane invaginations on the cell membrane, suggesting clathrin-mediated and macropinocytosis pathways as the predominant uptake mechanisms (Fig. 7h and S5f in ESI†). TEM examination of IONR(L) endocytosis mechanisms, corresponding to Fe concentration measurement, was consistent. In contrast, caveolae did not significantly favor IONP internalization. Conversely, in the case of IONR(S), the presence of flask-shaped structures for caveolae-mediated endocytosis was less conclusive to capture through TEM. Additionally, macropinocytosis contributed to a limited degree in the IONR(S) uptake mechanism.
For comparative analysis, we further elucidated the uptake mechanisms and cellular interactions by performing additional control studies using different types of spherical nanoparticles with varying sizes including SHP-10 (a commercial spherical nanoparticle with 10 nm in diameter)67 and ferumoxytol. D556 cells showed a marked preference for clathrin-coated pits and macropinocytosis pathways similar to IONR(S), with SHP-10 demonstrating increased cellular uptake efficiency (Fig. S6a–c, ESI†). This indicates a potentially enhanced receptor-mediated endocytosis facilitated by SHP-10 nanoparticles. Phagocytosis of SHP-10 begins with binding to specialized receptors on phagocyte membranes, triggering actin polymerization that causes plasma membrane deformation into pseudopodia. These pseudopodia completely encase the SHP-10 nanoparticles in a phagocytic cup, which then closes into a phagosome, and then matures through fusion with endosomal compartments and lysosomes (Fig. S6a, b, and d in ESI†). In contrast, ferumoxytol predominantly utilized clathrin-mediated endocytosis and macropinocytosis for internalization (Fig. S7a and c, ESI†). After internalization, ferumoxytol nanoparticles are distributed into various cellular organelles, including endosomes/lysosomes, the nucleus, mitochondria, and the endoplasmic reticulum, offering a broad range of interactions within the cell (Fig. S7b, and c in ESI†). This divergence in uptake mechanisms underscores the significance of nanoparticle physicochemical properties in determining cellular internalization routes.
Our observations reveal that multiple endocytosis pathways were utilized in the cellular uptake and compartmentalization of nanoparticles of different shapes and sizes regardless of the cell type. Long nanorods can undergo quicker invagination and wrapping during the endocytosis process, due to their larger initial contact area and rotation with the membrane. Short nanorods have a lesser extent due to their weaker shape anisotropy.68 Spherical IONP offers much fewer contact points for their interaction and adhesion with the cell membrane due to their curved surface topology, resulting in reduced internalization compared to IONR(L).69 Zhang et al. demonstrated that elongated nanoparticles, such as IONR, align with the curvature of the cell membrane more favorably with more contact compared to spherical particles, promoting faster and more efficient uptake than spherical particles. This is because the nanorods with an elongated shape have more surface area in contact with the cell membrane, providing more points for endocytic initiation.70 Recent theoretical and computational studies have investigated the receptor-mediated membrane wrapping efficiency of nanoparticles during endocytosis based on different sizes, shapes, and elasticity. These studies highlight that receptor recruitment speed and free energy barriers govern the membrane-wrapping efficiency. Specifically, the wrapping efficiency is determined by the receptor diffusion flux and the contact edge length between the nanoparticle and the membrane. Compared to rigid spherical nanoparticles, soft nanoparticles face higher energy barriers due to their ability to deform, requiring more receptors for efficient wrapping. This highlights the role played by nanoparticle elasticity in determining endocytosis efficiency.71 Furthermore, it is noteworthy that the endocytic process is not solely governed by the lipid membrane but also involves key protein components such as clathrin,72 caveolin,73,74 and the cross-linked F-actin network.75,76 These proteins play crucial roles in orchestrating the intricate membrane dynamics and cytoskeleton rearrangements necessary for efficient nanoparticle uptake. Actin filaments support membrane invaginations and provide the necessary force for membrane deformation and vesicle scission. Studies have highlighted the involvement of actin in the dynamic restructuring of the cell membrane during clathrin-mediated endocytosis,77,78 macropinocytosis,65,79 and phagocytosis.80 Fig. S8 in the ESI† demonstrates that IONR(L) cellular uptake induces a mechanical response, resulting in increased cellular rigidity due to cytoskeleton remodeling, which facilitates the formation of actin-rich lamellipodia. This is evidenced by the increased presence of actin filaments in the cell cytoplasm and membrane, suggesting that actin filaments act as a driving force for IONR(L) internalization upon contact with the cell membrane.
Cell membrane tension is crucial for regulating endocytic pathways, presenting an energy barrier during vesicle formation. Zhang et al. have modulated membrane tension by varying substrate stiffness and disrupting actin stress fibers to examine its effect on magnetic force-mediated magnetic IONP uptake. The results showed that increased substrate stiffness decreased IONP uptake under magnetic force, indicating that higher membrane tension inhibits uptake. Conversely, disrupting stress fibers enhanced nanoparticle uptake, suggesting that reduced membrane tension promotes nanoparticle internalization.81 A simulation study by Yue et al. shows how orientation and adhesion strength affect the membrane–mediated interactions between neighboring anisotropic nanoparticles. For rod-like nanoparticles with weak nanoparticle-membrane adhesion, orientation-dependent interactions arise from non-homogeneous membrane curvature, leading to equilibrium arrangements. Strong adhesion induces asymmetrical wrapping, causing short-range repulsion and intermediate-range attraction. In multi-rodlike nanoparticle systems, equilibrium arrangements depend on nanoparticle interaction and dynamic traps, revealing the complex interplay of geometry, adhesion, and membrane forces.82 Overall, the shape anisotropy and initial orientation significantly influence the membrane invagination stage, highlighting the advantage of long rod-like nanoparticles in cellular uptake.
Measurement of intracellular ROS generation
The escalation of ROS beyond the cellular antioxidant threshold induces oxidative stress, a condition marked by the potential for profound cellular injury. This dysregulation risks oxidative damage to crucial cellular components, including proteins, lipids, RNA, and DNA, ultimately fostering apoptosis-driven cell demise.37 In this study, the DCFH-DA probe technique was utilized to elucidate the correlation of IONR with different aspect ratios and IONP in triggering ROS production, subsequently leading to oxidative stress and cellular toxicity in RAW264.7 and D556 cells. The cells treated with H2O2 exhibited a higher ROS signal in the presence of DCFH-DA because H2O2, with the aid of cellular peroxidases, efficiently oxidizes DCFH to highly fluorescent DCF. The fluorescence intensity of DCF serves as a direct indicator of ROS levels. Fig. S9 and S10 in the ESI† illustrate that treatment with IONR(L), IONR(S), and IONP resulted in a low level of ROS production. Low levels of ROS are naturally present within cells under non-stress conditions, maintained by the cell's antioxidant defenses, and are involved in normal cellular functions such as signaling and homeostasis.83 Therefore, IONR and IONP ensure enhanced biocompatibility and reduced cellular toxicity, supporting their safe and effective use in biomedical applications such as drug delivery and imaging.Impact of IONR and IONP on mitochondrial membrane potential
The mitochondrion is a pivotal organelle crucial for cell energy metabolism and ATP synthesis. Disruption of the mitochondrial membrane potential (Δψm) may directly trigger the activation of the mitochondria-mediated intrinsic apoptosis pathway. The impact of IONR and IONP on mitochondrial integrity was evaluated in RAW264.7 and D556 cells through the assessment of mitochondrial membrane potential using a JC-1 assay kit. The JC-1 probe was employed to gauge the changes in Δψm, where it forms aggregates within healthy mitochondria, resulting in red J-aggregates.29 Conversely, the green fluorescence of the JC-1 monomer in the cytosol indicates the loss of Δψm, signifying mitochondrial dysfunction.29 In this context, the untreated negative control exhibits red fluorescence, while the CCCP-treated positive control yields green fluorescence. Compared with the controls, the cells treated with IONR(L), IONR(S), and IONP showed a negligible green signal and a bright red signal suggesting that there was no change in the mitochondrial potential (Fig. S11 and S12 in ESI†). Thus, IONR and IONP did not cause mitochondrial dysfunction under the conditions tested.4. Conclusions
Numerous experimental and theoretical investigations have emphasized the impact of size,84,85 shape,86,87 orientation,88,89 stiffness,90 and surface properties91 of nanomaterials on their cellular uptake. Our re-examination of shape and size dependent cell interactions with nanoparticles revealed that IONR with different aspect ratios are selectively taken up by different cells using different endocytosis pathways. IONR(L) with a higher aspect ratio exhibited a higher cellular uptake in RAW264.7 and D556 cells compared to those with low aspect ratios, such as IONR(S), spherical IONP, and ferumoxytol. Specifically, IONR(L) showed greater uptake in macrophages than in normal kidney cells and cancer cells, likely due to their larger initial contact area and rotation with the membrane. Our further investigation with pathway inhibition and TEM experiments showed that the preferential uptake mechanism of IONR(L) in macrophage cells involved internalization via clathrin-mediated, macropinocytosis, dynamin-dependent, and phagocytosis pathways. Conversely, in normal kidney cells, uptake occurred predominantly through clathrin-mediated, dynamin-dependent, and macropinocytosis/phagocytosis mechanisms. In cancer cells, internalization primarily utilized clathrin/caveolae-mediated and phagocytosis mechanisms. Furthermore, we have found that there is no detectable iron-induced ROS in cells treated with the reported IONR, which does not appear to affect the mitochondrial membrane integrity. The shape/size specific uptake by different cell lines and their cellular responses are informative for further investigation and development of nanoparticles for cell targeted biomedical applications in vivo, including drug delivery, hyperthermia, and MRI.Author contributions
The manuscript was written through the contributions of all authors. All authors have given approval to the final version of the manuscript.Data availability
The data supporting this article have been included as part of the ESI.†TEM images of oleylamine coated and size distribution of oligosaccharide coated IONR(L), IONR(S), and IONP (Fig. S1†). FTIR spectra of oleylamine coated IONR and oligosaccharide coated IONR are shown in Fig. S2†. The results of the MTT assay conducted for 48 h are shown in Fig. S3†). TEM images of 2 h treatment with oligosaccharide coated IONR(L), IONR(S), and IONP on RAW264.7, HEK293, and D556 cell lines (Fig. S4 and S5†). TEM images of D556 cells treated with SHP-10 and ferumoxytol for 2 and 4 h and IONR(L) for 2 h (Fig. S6, S7, and S8†). CLSM was used for the detection of intracellular ROS in RAW264.7 and D556 cells (Fig. S9 and S10†). CLSM images reporting the mitochondrial membrane potential changes in RAW264.7 and D556 cells are shown in Fig. S11 and S12†).
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was partly supported by grants from the National Institutes of Health (R01AG067736 to HM, R01CA261251 to YL and HM, and R43AG078718 to YL and HM). The research reported in this publication was also supported in part by the Emory University Integrated Cellular Imaging Core of the Winship Cancer Institute of Emory University and National Institutes of Health under award number, P30CA138292-04. This study used the instrument at the Emory University Robert P. Apkarian Integrated Electron Microscopy Core (IEMC), which is supported by the grants provided by the Georgia Clinical & Translational Science Alliance of the National Institutes of Health under award number UL1TR000454. Transmission electron micrographs were recorded on the JEOL JEM-1400, 120 kV TEM instrument purchased with the shared instrumentation grant from the National Institutes of Health (S10 RR025679).References
- J. Shi, P. W. Kantoff, R. Wooster and O. C. Farokhzad, Nat. Rev. Cancer, 2017, 17, 20–37 Search PubMed.
- O. C. Farokhzad and R. Langer, ACS Nano, 2009, 3, 16–20 Search PubMed.
- Q. Feng, Y. Liu, J. Huang, K. Chen, J. Huang and K. Xiao, Sci. Rep., 2018, 8, 1–13 Search PubMed.
- J. H. Lee, J. E. Ju, B. I. Kim, P. J. Pak, E. K. Choi, H. S. Lee and N. Chung, Environ. Toxicol. Chem., 2014, 33, 2759–2766 Search PubMed.
- C. Carnovale, G. Bryant, R. Shukla and V. Bansal, ACS Omega, 2019, 4, 242–256 Search PubMed.
- A. Woźniak, A. Malankowska, G. Nowaczyk, B. F. Grześkowiak, K. Tuśnio, R. Słomski, A. Zaleska-Medynska and S. Jurga, J. Mater. Sci.: Mater. Med., 2017, 28, 1–11 Search PubMed.
- S. Sultana, N. Djaker, S. Boca-Farcau, M. Salerno, N. Charnaux, S. Astilean, H. Hlawaty and M. L. de La Chapelle, Nanotechnology, 2015, 26, 055101 Search PubMed.
- Y. Xiao, W. Xu, Y. Komohara, Y. Fujiwara, H. Hirose, S. Futaki and T. Niidome, ACS Omega, 2020, 5, 32744–32752 Search PubMed.
- M. Bhamidipati and L. Fabris, Bioconjugate Chem., 2017, 28, 449–460 Search PubMed.
- X. Ma, J. Sun, L. Zhong, Y. Wang, Q. Huang, X. Liu, S. Jin, J. Zhang and X.-J. Liang, Nano Lett., 2019, 19, 8476–8487 Search PubMed.
- Y. Jiang, S. Huo, T. Mizuhara, R. Das, Y.-W. Lee, S. Hou, D. F. Moyano, B. Duncan, X.-J. Liang and V. M. Rotello, ACS Nano, 2015, 9, 9986–9993 Search PubMed.
- J. Huang, L. Wang, X. Zhong, Y. Li, L. Yang and H. Mao, J. Mater. Chem. B, 2014, 2, 5344–5351 Search PubMed.
- Y. Xu, H. Wu, Q. Xiong, B. Ji, H. Yi, H. Duan and H. Mao, ACS Appl. Bio Mater., 2019, 2, 3362–3371 Search PubMed.
- D. Barbaro, L. Di Bari, V. Gandin, C. Marzano, A. Ciaramella, M. Malventi and C. Evangelisti, PLoS One, 2022, 17, e0269603 Search PubMed.
- K. S. Sharma, A. K. Dubey, A. S. Koijam, C. Kumar, A. Ballal, S. Mukherjee, P. P. Phadnis and R. K. Vatsa, New J. Chem., 2020, 44, 13863–13874 Search PubMed.
- X. H. Shan, H. Hu, F. Xiong, N. Gu, X. D. Geng, W. Zhu, J. Lin and Y. F. Wang, Eur. J. Radiol., 2012, 81, 95–99 Search PubMed.
- L. Zhu, Z. Zhou, H. Mao and L. Yang, Nanomedicine, 2017, 12, 73–87 Search PubMed.
- J. Huang, X. Zhong, L. Wang, L. Yang and H. Mao, Theranostics, 2012, 2, 86 Search PubMed.
- J. Huang, Y. Li, A. Orza, Q. Lu, P. Guo, L. Wang, L. Yang and H. Mao, Adv. Funct. Mater., 2016, 26, 3818–3836 Search PubMed.
- Y. Geng, P. Dalhaimer, S. Cai, R. Tsai, M. Tewari, T. Minko and D. E. Discher, Nat. Nanotechnol., 2007, 2, 249–255 Search PubMed.
- J. Mohapatra, A. Mitra, H. Tyagi, D. Bahadur and M. Aslam, Nanoscale, 2015, 7, 9174–9184 Search PubMed.
- A. Orza, H. Wu, Y. Xu, Q. Lu and H. Mao, ACS Appl. Mater. Interfaces, 2017, 9, 20719–20727 Search PubMed.
- M. Nejabat, A. Samie, M. Ramezani, M. Alibolandi, K. Abnous and S. M. Taghdisi, J. Controlled Release, 2023, 354, 221–242 Search PubMed.
- X. Zhou, Y. Shi, L. Ren, S. Bao, Y. Han, S. Wu, H. Zhang, L. Zhong and Q. Zhang, J. Solid State Chem., 2012, 196, 138–144 Search PubMed.
- R. Agarwal, V. Singh, P. Jurney, L. Shi, S. Sreenivasan and K. Roy, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 17247–17252 Search PubMed.
- V. Reshma and P. Mohanan, Colloids Surf., B, 2017, 157, 182–190 Search PubMed.
- Y. Li, R. Lin, L. Wang, J. Huang, H. Wu, G. Cheng, Z. Zhou, T. MacDonald, L. Yang and H. Mao, J. Mater. Chem. B, 2015, 3, 3591–3603 Search PubMed.
- Y. Li, M. Xie, J. B. Jones, Z. Zhang, Z. Wang, T. Dang, X. Wang, M. Lipowska and H. Mao, Adv. Healthcare Mater., 2022, 11, 2102816 Search PubMed.
- A. Mozhi, I. Ahmad, C. I. Okeke, C. Li and X.-J. Liang, RSC Adv., 2017, 7, 12886–12896 Search PubMed.
- K. Saha, S. T. Kim, B. Yan, O. R. Miranda, F. S. Alfonso, D. Shlosman and V. M. Rotello, Small, 2013, 9, 300–305 Search PubMed.
- L.-H. Wang, K. G. Rothberg and R. Anderson, J. Cell Biol., 1993, 123, 1107–1117 Search PubMed.
- J. Lee, M. Twomey, C. Machado, G. Gomez, M. Doshi, A. J. Gesquiere and J. H. Moon, Macromol. Biosci., 2013, 13, 913–920 Search PubMed.
- O. Zimmer and A. Goepferich, Nanoscale Horiz., 2023, 8, 256–269 Search PubMed.
- M. Al Soraj, L. He, K. Peynshaert, J. Cousaert, D. Vercauteren, K. Braeckmans, S. De Smedt and A. T. Jones, J. Controlled Release, 2012, 161, 132–141 Search PubMed.
- B. G. Park, Y. J. Kim, J. H. Min, T.-C. Cheong, S. H. Nam, N.-H. Cho, Y. K. Kim and K. B. Lee, Nanoscale Res. Lett., 2020, 15, 1–10 Search PubMed.
- L. M. Fujimoto, R. Roth, J. E. Heuser and S. L. Schmid, Traffic, 2000, 1, 161–171 Search PubMed.
- A. Mozhi, V. Sunil, W. Zhan, P. B. Ghode, N. V. Thakor and C.-H. Wang, J. Controlled Release, 2020, 323, 502–518 Search PubMed.
- P. Yu, X.-M. Xia, M. Wu, C. Cui, Y. Zhang, L. Liu, B. Wu, C.-X. Wang, L.-J. Zhang and X. Zhou, Colloids Surf., B, 2014, 120, 142–151 Search PubMed.
- P. J. Kempen, S. Greasley, K. A. Parker, J. L. Campbell, H.-Y. Chang, J. R. Jones, R. Sinclair, S. S. Gambhir and J. V. Jokerst, Theranostics, 2015, 5, 631 Search PubMed.
- X. Meng, Y. Peng, Z. Shao, X. Huang, H. Wei, Z. Wei, Q. Sun and S. Zhao, Chem. Eng. J., 2024, 152282 Search PubMed.
- L. Wang, J. Huang, H. Chen, H. Wu, Y. Xu, Y. Li, H. Yi, Y. A. Wang, L. Yang and H. Mao, ACS Nano, 2017, 11, 4582–4592 Search PubMed.
- H. Sun, B. Chen, X. Jiao, Z. Jiang, Z. Qin and D. Chen, J. Phys. Chem. C, 2012, 116, 5476–5481 Search PubMed.
- X. Sun, K. Sun, S. Wang, S. Zhang, Z. Liu and Y. Wang, Micro Nano Lett., 2016, 11, 118–121 Search PubMed.
- J. Santoyo Salazar, L. Perez, O. De Abril, L. Truong Phuoc, D. Ihiawakrim, M. Vazquez, J.-M. Greneche, S. Begin-Colin and G. Pourroy, Chem. Mater., 2011, 23, 1379–1386 Search PubMed.
- A. A. Ayachi, H. Mechakra, M. M. Silvan, S. Boudjaadar and S. Achour, Ceram. Int., 2015, 41, 2228–2233 Search PubMed.
- J. A. A. Abdullah, L. S. Eddine, B. Abderrhmane, M. Alonso-González, A. Guerrero and A. Romero, Sustainable Chem. Pharm., 2020, 17, 100280 Search PubMed.
- B. Chen, J. Sun, F. Fan, X. Zhang, Z. Qin, P. Wang, Y. Li, X. Zhang, F. Liu and Y. Liu, Nanoscale, 2018, 10, 7369–7376 Search PubMed.
- S. Yan, K. Hu, M. Zhang, J. Sheng, X. Xu, S. Tang, Y. Li, S. Yang, G. Si and Y. Mao, Bioact. Mater., 2023, 19, 418–428 Search PubMed.
- Y. Huang, J. C. Hsu, H. Koo and D. P. Cormode, Theranostics, 2022, 12, 796 Search PubMed.
- M. P. Calatayud, B. Sanz, V. Raffa, C. Riggio, M. R. Ibarra and G. F. Goya, Biomaterials, 2014, 35, 6389–6399 Search PubMed.
- V. S. Kalambur, E. K. Longmire and J. C. Bischof, Langmuir, 2007, 23, 12329–12336 Search PubMed.
- P. Pradhan, J. Giri, R. Banerjee, J. Bellare and D. Bahadur, J. Magn. Magn. Mater., 2007, 311, 282–287 Search PubMed.
- A. Verma and F. Stellacci, Small, 2010, 6, 12–21 Search PubMed.
- Arnida, A. Malugin and H Ghandehari, J. Appl. Toxicol., 2010, 30, 212–217 Search PubMed.
- K. Nambara, K. Niikura, H. Mitomo, T. Ninomiya, C. Takeuchi, J. Wei, Y. Matsuo and K. Ijiro, Langmuir, 2016, 32, 12559–12567 Search PubMed.
- P. Zhang, B. Li, J. Du and Y. Wang, Colloids Surf., B, 2017, 157, 18–25 Search PubMed.
- C. Graf, D. Nordmeyer, C. Sengstock, S. Ahlberg, J. R. Diendorf, J. R. Raabe, M. Epple, M. Köller, J. R. Lademann and A. Vogt, Langmuir, 2018, 34, 1506–1519 Search PubMed.
- S. Nigam, J. Mohapatra, A. V. Makela, H. Hayat, J. M. Rodriguez, A. Sun, E. Kenyon, N. A. Redman, D. Spence, G. Jabin, B. Gu, M. Ashry, L. F. Sempere, A. Mitra, J. Li, J. Chen, G. Wei, S. Bolin, B. Etchebarne, J. P. Liu, C. H. Contag and P. Wang, Small, 2023, 20, 2305300 Search PubMed.
- A. F. Rebolledo, S. Laurent, M. Calero, A. Villanueva, M. Knobel, J. F. Marco and P. Tartaj, ACS Nano, 2010, 4, 2095–2103 Search PubMed.
- S. Gil, C. R. Correia and J. F. Mano, Adv. Healthcare Mater., 2015, 4, 883–891 Search PubMed.
- M. H. Rizvi, R. Wang, J. Schubert, W. D. Crumpler, C. Rossner, A. L. Oldenburg, A. Fery and J. B. Tracy, Adv. Mater., 2022, 34, 2203366 Search PubMed.
- D. Ma, Nanoscale, 2014, 6, 6415–6425 Search PubMed.
- J. Zhao and M. H. Stenzel, Polym. Chem., 2018, 9, 259–272 Search PubMed.
- C. G. Hansen and B. J. Nichols, J. Cell Sci., 2009, 122, 1713–1721 Search PubMed.
- Y. Hui, X. Yi, D. Wibowo, G. Yang, A. P. Middelberg, H. Gao and C.-X. Zhao, Sci. Adv., 2020, 6, eaaz4316 Search PubMed.
- Y.-X. Li and H.-B. Pang, J. Controlled Release, 2021, 329, 1222–1230 Search PubMed.
- E. Ying and H.-M. Hwang, Sci. Total Environ., 2010, 408, 4475–4481 Search PubMed.
- Y. Li, T. Yue, K. Yang and X. Zhang, Biomaterials, 2012, 33, 4965–4973 Search PubMed.
- S. Dasgupta, T. Auth and G. Gompper, Nano Lett., 2014, 14, 687–693 Search PubMed.
- S. Zhang, H. Gao and G. Bao, ACS Nano, 2015, 9, 8655–8671 Search PubMed.
- Z. Shen, H. Ye, X. Yi and Y. Li, ACS Nano, 2018, 13, 215–228 Search PubMed.
- M. Saleem, S. Morlot, A. Hohendahl, J. Manzi, M. Lenz and A. Roux, Nat. Commun., 2015, 6, 6249 Search PubMed.
- M. Tachikawa, N. Morone, Y. Senju, T. Sugiura, K. Hanawa-Suetsugu, A. Mochizuki and S. Suetsugu, Sci. Rep., 2017, 7, 7794 Search PubMed.
- F.-N. Lolo, N. Walani, E. Seemann, D. Zalvidea, D. M. Pavón, G. Cojoc, M. Zamai, C. Viaris de Lesegno, F. Martínez de Benito, M. Sánchez-Álvarez, J. José Uriarte, A. Echarri, D. Jiménez-Carretero, J. Escolano, S. A. Sánchez, V. R. Caiolfa, D. Navajas, X. Trepat, J. Guck, C. Lamaze, P. Roca-Cusachs, M. Kessels, B. Qualmann, M. Arroyo and M. Angel del Pozo, Nat. Cell Biol., 2023, 25, 120–133 Search PubMed.
- P. Rougerie, V. Miskolci and D. Cox, Immunol. Rev., 2013, 256, 222–239 Search PubMed.
- S. Boulant, C. Kural, J.-C. Zeeh, F. Ubelmann and T. Kirchhausen, Nat. Cell Biol., 2011, 13, 1124–1131 Search PubMed.
- M. Kaksonen, C. P. Toret and D. G. Drubin, Nat. Rev. Mol. Cell Biol., 2006, 7, 404–414 Search PubMed.
- M. Kaksonen, Y. Sun and D. G. Drubin, Cell, 2003, 115, 475–487 Search PubMed.
- H. Meng, S. Yang, Z. Li, T. Xia, J. Chen, Z. Ji, H. Zhang, X. Wang, S. Lin and C. Huang, ACS Nano, 2011, 5, 4434–4447 Search PubMed.
- J. A. Champion and S. Mitragotri, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 4930–4934 Search PubMed.
- L. Zhang, S. Hajebrahimi, S. Tong, X. Gao, H. Cheng, Q. Zhang, D. T. Hinojosa, K. Jiang, L. Hong and J. Huard, ACS Appl. Mater. Interfaces, 2023, 15, 50574–50585 Search PubMed.
- T. Yue, X. Wang, F. Huang and X. Zhang, Nanoscale, 2013, 5, 9888–9896 Search PubMed.
- A. J. P. Oliveira de Almeida, J. C. P. Lucio de Oliveira, L. Virgolino da Silva Pontes, J. Frederico de Souza Júnior, T. A. F. Gonçalves, S. H. Dantas, M. Silva de Almeida Feitosa, A. O. Silva and I. Almeida de Medeiros, Oxid. Med. Cell. Longevity, 2022, 2022, 1225578 Search PubMed.
- T. Yue and X. Zhang, Soft Matter, 2011, 7, 9104–9112 Search PubMed.
- S. Zhang, J. Li, G. Lykotrafitis, G. Bao and S. Suresh, Adv. Mater., 2009, 21, 419 Search PubMed.
- S. Barua, J.-W. Yoo, P. Kolhar, A. Wakankar, Y. R. Gokarn and S. Mitragotri, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 3270–3275 Search PubMed.
- Y. Li, M. Kröger and W. K. Liu, Nanoscale, 2015, 7, 16631–16646 Search PubMed.
- X. Shi, A. von Dem Bussche, R. H. Hurt, A. B. Kane and H. Gao, Nat. Nanotechnol., 2011, 6, 714–719 Search PubMed.
- X. Yi, X. Shi and H. Gao, Nano Lett., 2014, 14, 1049–1055 Search PubMed.
- J. Sun, L. Zhang, J. Wang, Q. Feng, D. Liu, Q. Yin, D. Xu, Y. Wei, B. Ding, X. Shi and X. Jiang, Adv. Mater., 2014, 27, 1402–1407 Search PubMed.
- Y. Li, X. Li, Z. Li and H. Gao, Nanoscale, 2012, 4, 3768–3775 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: DETAILS. See DOI: https://doi.org/10.1039/d4nr02408g |
| This journal is © The Royal Society of Chemistry 2024 |