 Open Access Article
Open Access ArticleIntracellular delivery strategies using membrane-interacting peptides and proteins
Linh D.
Mai†
a,
Sydney C.
Wimberley†
ab and
Julie A.
Champion
 *ab
*ab
aSchool of Chemical and Biomolecular Engineering, Georgia Institute of Technology, 950 Atlantic Dr NW, Atlanta, GA 30332-2000, USA. E-mail: julie.champion@chbe.gatech.edu
bBioEngineering Program, Georgia Institute of Technology, USA
First published on 25th July 2024
Abstract
While the cellular cytosol and organelles contain attractive targets for disease treatments, it remains a challenge to deliver therapeutic biomacromolecules to these sites. This is due to the selective permeability of the plasma and endosomal membranes, especially for large and hydrophilic therapeutic cargos such as proteins and nucleic acids. In response, many different delivery systems and molecules have been devised to help therapeutics cross these barriers to reach cytosolic targets. Among them are peptide and protein-based systems, which have several advantages over other natural and synthetic materials including their ability to interact with cell membranes. In this review, we will describe recent advances and current challenges of peptide and protein strategies that leverage cell membrane association and modulation to enable cytosolic delivery of biomacromolecule cargo. The approaches covered here include peptides and proteins derived from or inspired by natural sequences as well as those designed de novo for delivery function.
1. Introduction
While biomacromolecules, such as proteins and nucleic acids, have significant potential as intracellular therapeutics, they require delivery to the cytosol to be effective. As a result, most biomacromolecule therapeutics require a carrier, due to their inability to cross the cell membrane directly or be effectively internalized by cells. Therapeutics must be transported past the plasma membrane and, often, endosomal membrane as well. These membranes comprise amphiphilic phospholipids containing a negatively charged polar head and a hydrophobic tail,1,2 making them selective to direct translocation of a limited subset of small and hydrophobic molecules.3 If direct translocation is impossible, cells can internalize macromolecules through the energy-dependent processes of endocytosis and receptor-mediated endocytosis.1 During endocytosis, the cell membrane forms a vesicle called an endosome, surrounding the material to uptake.4–6 After the internalization of the cargo, the endosome begins to acidify due to the proton pumps on the endosomal membrane. Due to charge and ion concentration differences between the cytosol and the endosome, osmosis occurs. The increased pressure within the endosome can cause rupture and delivery of its contents into the cytosol. However, even if the endosome is damaged, the release of endosomal contents is not guaranteed. Endosomes will be trafficked out of the cell or lysosomes containing enzymes will combine with endosomes and degrade the contents.7–9 This is a significant challenge to cytosolic drug delivery and has spurred the engineering of new generations of delivery systems.Different delivery systems have been developed from various synthetic and natural materials, including polymers, lipids, and proteins or peptides. Synthetic polymers have advantages in cost and scale up, but biocompatibility and biodegradability can be challenges.10 Lipid nanoparticles have been approved to deliver different nucleic acids effectively, but their complex formulations and stability can be limiting.11 Proteins and peptides have advantages as carriers due to their biocompatibility and ability to be precisely tuned by altering the amino acid sequence, which dictates structure and function via hydrophobic, charged, and hydrogen bonding interactions.3,10 Proteins and peptides can be mixed with or fused to therapeutic cargos or can form nanocarriers that encapsulate cargos to deliver intracellular therapeutics, including small molecules, nucleic acids, and proteins or peptides.3 However, these delivery systems, along with the others listed above, do not guarantee cytosolic delivery. One way to overcome this delivery hurdle is by using peptides and proteins that interact directly with cellular membranes.
Peptides and proteins can interact with membranes in different ways. Some peptides, particularly those containing positive and hydrophobic amino acids, can induce direct translocation, often in a concentration dependent manner.12,13 The positive and hydrophobic amino acids interacting with the negative amphiphilic cellular membrane cause the peptide to temporarily or permanently disrupt the plasma membrane, allowing direct entry into the cytosol. Other peptides and proteins can interact with an amphiphilic membrane similarly, but require endocytosis to enter the cell before interacting with the endosomal membrane.14,15 The stiffness of materials also affects their uptake. In the case of protein delivery systems, endocytosis increases with increasing stiffness in an α-lactalbumin nanotube system and phagocytosis/micropinocytosis increases with increasing stiffness of lysozyme microcapsules.16,17 These examples are not membrane-active, though. Alpha-helices, discussed in section 3.3, do have variation in stiffness,18 though the effect of their stiffness on membrane interaction and cytosolic delivery has not been evaluated.
This review focuses primarily on literature from the last five years that describes peptides and proteins designed to interact with plasma or endocytic membranes to promote cytosolic delivery of biomacromolecule cargos. Many examples are derived from or inspired by nature, where membrane association and disruption proteins are common, especially in pathogens. Others are the product of de novo design. Multivalent receptor targeting systems are advantageous for their ability to increase endocytosis of the carrier to the target cell type over other cells. However, this review will focus specifically on membrane-active peptides and proteins, including some receptor-specific proteins and others that are not. Multivalent receptor targeting of drug carriers has been extensively reviewed and we refer the reader to the cited references to learn more.19,20 Small peptides will be covered first, followed by larger proteins.
2. Peptide-based strategies for intracellular delivery through membrane association
Peptides are defined as short chains of amino acids that, in the interest of this review, will be limited to those ∼30-amino acids long or less.12 Due to their size, they are typically simpler and cheaper to produce using solid state synthesis compared to a large recombinant protein21 and they do not tend to greatly increase the size of the carrier-cargo complex. Peptides are also often less immunogenic, which contributes to their advantages as therapeutics or as carrier molecules.22 Like proteins, peptides are diverse, and their sequences can be modified to obtain different chemical properties such as hydrophobicity and charge.23,24 The modulation of these traits is especially beneficial in facilitating interactions with cell membranes and initiating responses to different biological stimuli to aid intracellular delivery of chemicals and macromolecules.24 Since the discovery of cell penetrating peptide by Frankel and Pabo in 1988,25 the delivery abilities of peptides have been widely investigated, resulting in other newly discovered classes of peptides with intracellular delivery capacity.24,26,27 This section will cover recent advances aimed at improving existing peptide-based delivery systems, as well as new peptide strategies created to improve cytosolic delivery through association with plasma and organelle membranes.2.1. Advanced cell-penetrating peptide (CPPs)
Cell penetrating peptides, or CPPs, are peptides, often cationic, that can translocate across the cellular membrane.26,27 Because of this ability, they have been extensively studied as a tool for intracellular delivery of biomacromolecules and other hard-to-deliver cargos, with thousands of discovered entities in the CPPSite 2.0 database.28 CPPs are often linked to cargo covalently, using techniques such as chemical conjugation or genetic fusion.29 Some cargos will, in turn, leverage the CPP mechanism and cross the membrane through direct translocation and others will be internalized through an energy-dependent endocytic mechanism in which the CPP may help cargo escape from the endosome.12 Despite promising in vitro delivery results, CPPs face an array of challenges in vivo.30 For example, their positive charges subject them to significant interactions with primarily negatively charged proteins in serum, resulting in serum instability.31,32 Furthermore, CPPs without any targeting or specificity features risk association with other cells before reaching their intended target, leading to inefficient delivery and undesired off-target toxicity.31 Additionally, it should be noted that, contrary to popular belief, CPPs bound to cargo do not often directly translocate through the membrane to deliver that cargo.33,34 Especially at low concentration or with larger biomacromolecule cargos, CPPs are more likely to enter cells through endocytosis or macropinocytosis and as the result, end up trapped in the endosome.8,35–37 However, more recently, there have been alternative strategies to address these challenges, which include different methods of delivery, engineered stimuli responsiveness and alternative assembly that allow for safer and more effective CPP-mediated intracellular delivery (Table 1). This section will focus on these advanced CPP strategies and will not cover simple conjugation of non-specific, monomeric CPPs to cargo, as there are several other reviews that fastidiously covered this topic.12,26,27| Category | Approach | Ref. |
|---|---|---|
| Additive use | Adding excess linear CPPs for plasma membrane permeation | 33 |
| Adding dimerized TAT for endosomal escape | 35 | |
| Stimuli-responsive | pH-responsive helical polypeptide | 36 |
| pH low insertion peptide (pHLIP) | 37 | |
| Fusion peptide with pH-sensitive charge shielding sequence | 38 | |
| UV-Vis-triggered stapled peptide | 41 | |
| Alternative assembly and fusion with functional moieties | TAT trimeric cluster | 44 |
| TAT-based multifunctional chimeric peptide with PAMP | 31 | |
| Fusion of CPP with mitochondrial-penetrating peptide | 48 | |
| Fusion with nucleus-transporting peptide for nucleus delivery | 31, 33 and 50 |
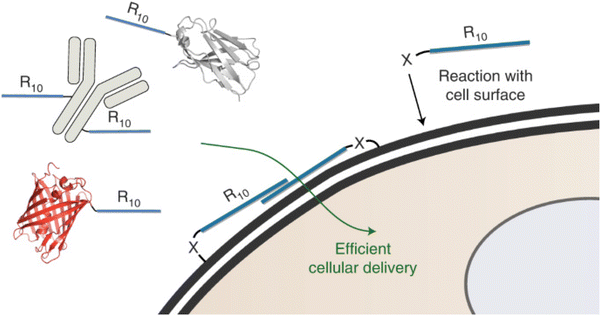 | ||
| Fig. 1 Hypothesized mechanism of action for additive use of a modified CPP. “Cellular uptake of large biomolecules enabled by cell-surface-reactive cell-penetrating peptide additives.” A. F. L. Schneider, M. Kithil, M. C. Cardoso, M. Lehmann and C. P. R. Hackenberger, Nature Chemistry, 13, 2021 reproduced with permission from SNCSC.38 | ||
In addition to direct translocation, additive use of CPPs can also help with endosomal escape in energy-dependent uptake pathways. Allen et al. designed a dimeric peptide based on dfTAT (dimeric version of TAT, a CPP derived from human immunodeficiency virus (HIV) trans-activator of transcription), d(X)TAT, that can be co-delivered with an enzyme payload, without the need for pre-complexion or coincubation prior to injection.40 It was observed that, with this method, d(X)TAT associates with the membrane of late endosomes to allow the protein payload to escape and diffuse in the cytosol, where some cargo reaches the nucleus. In vitro and in vivo data showed that this delivery method was able to effectively deliver Cre recombinase intracellularly to neurons and astrocytes in the central nervous system through stereotactic injection, with comparable efficiency to the gold standard adeno-associated viruses (AAV).40 Remarkably, the addition of d(X)TAT did not change the amount of Cre that entered the cells, but significantly increased the amount of protein that reached the nucleus, as examined through Cre-activated expression of a reporter gene. Additionally, sequential delivery of Cre and d(X)TAT also improved cytosolic delivery, albeit to a lesser degree compared to co-delivery.40 Altogether, the additive approaches to CPP delivery have the advantage of being quite simple, in that they do not require complex chemical conjugation or genetic fusions. Nevertheless, the toxicity of soluble CPPs at off-target sites needs to be carefully evaluated, especially at the dose relevant to their end-point application.
One such system was created by Lee et al., where poly-L-lysine was engineered to have pH-dependent helical propensity.41 At physiological pH, the electrostatic attraction between the side chains compromises the helical structure of the peptide. At lower pH, such as in tumor environments (∼pH 6), the carboxylate group is protonated and induces rearrangement of the side chains to promote a cationic helical structure, which increases the membrane interaction ability of these peptides. Through a set of in vitro experiments, the authors found that the cell penetrating ability of this peptide is inactive at pH 7.4 and cellular uptake is a result of endocytosis. At pH 6, the peptide increased uptake efficiency two-fold, and demonstrated direct translocation across cell membrane instead of endocytosis, as demonstrated by cold incubation and chlorpromazine inhibition studies. They also reported unusual selectivity towards cancer cells compared to normal lung fibroblast, even at lower pH, though more study is needed to verify and explain this observation.41 Another example of this phenomena is pH low insertion peptide, or pHLIP, derived from a helical peptide of bacteriorhodopsin. pHLIPs fold in the acidic tumor environment, directly penetrate the cellular membrane and deliver their cargo to the cytosol. When covalently bound to a STING agonist cargo with a self-immolating moiety and administered intraperitoneally, pHLIPs demonstrated reduction of murine colorectal (CT26) tumor size and development of immune memory, which was not seen with the cargo alone.42
Another system utilizing pH as a stimulus is a multi-component carrier engineered by Yang et al. to only expose the membrane active supercharged polypeptide sequence in a low pH environment in order to deliver proteins, peptides, small molecules, and nucleic acid therapeutics in vitro and in vivo.43 The fusion peptide contains the supercharged polypeptide (SCP) made “activatable” (ASCP) by a pH-sensitive charge shielding sequence (CSS). The supercharged polypeptide (SCP) composed of 40 amino acids, 20 of which are arginine, increases endocytic uptake and cytosolic delivery of attached protein cargo with minimal toxicity.44 Due to the positive charge of the SCP, in vivo use was not explored because of potential negative off-target effects associated with positive charge. With addition of the CSS sequence, comprising glutamic acid and histidine, the authors hypothesized that at neutral pH the negatively charged glutamic acid binds arginine in the SCP and shields it, but at low pH, histidine is protonated and disrupts the electrostatic interaction between the CSS and the SCP.14,43 However, no experiments confirmed the change of charge under different pH conditions. While the route of SCP uptake was still endocytosis, the mechanism of cytosolic delivery was not presented. ASCP delivered cytotoxic beclin-1 protein to breast cancer cells (MDA-MB-231) in vitro at pH 6, detected by reduced metabolic activity. In a xenograft tumor model of MD-MB-231 cells in nude mice, ASCP delivered cytotoxic peptide KLA to tumors and caused a decrease of tumor volume compared to the peptide delivered with the non-activable SCP. There was also more accumulation of ASCP-KLA in the tumor and less accumulation in kidneys and liver compared to SCP-KLA.
In addition to pH as a stimulus, the abundance of certain enzymes in specific extracellular environments is also another attractive trigger for delivery systems. One such class of enzymes is matrix metalloproteinases (MMPs), which are over expressed in the extracellular matrix of tumors and wounds. As MMPs were found to be abundant at the site of traumatic brain injury due to intracellular calcium imbalance after impact, Chen et al. created a lipoprotein nanoparticle displaying MMP9-activatable CPP that includes a MMP9 substrate peptide that shields the positively-charged CPP.45 At the injury site, local MMP9 cleaves the peptides and exposes CPPs, allowing for membrane interaction. This system was able to deliver cyclosporin A (CsA) peptide in vivo, resulting in lesion site accumulation and intracellular delivery confirmed through mitochondrial localization and protective effect of delivered CsA on mitochrondrial function.45 The authors also showed that this system was able to conserve neuropathological integrity and memory function in injured mice.
While biological stimuli, such as pH or enzymes, are convenient for triggering delivery because they are present in the body, in order to gain more precise control of the delivery, external stimuli are sometimes preferred.46 Kim and colleagues devised an amphipathic peptide with an azobenzene staple for photo-switching ability through cis–trans isomerization.47 Under UV-Vis light, azobenzene reversibly switched from its trans to cis conformation, stabilizing the alpha helix structure of the stapled peptide and, thus, enabling cell penetrating effect at the desired site of action. The mechanism was demonstrated in vitro in various cell lines such as HeLa and HEK-293 at sub-micromolar peptide concentration, where after 5 minutes of exposure to light, cellular uptake of the stapled peptides was significantly improved compared to the non-stapled version. However, it was unclear from the confocal images if the peptides ended up in the cytosol or the endosome, raising the need for additional studies to investigate mechanism of uptake and fate of the peptides. Furthermore, UV-triggered uptake was minimal in certain cell lines, like CHO-K1, so there is a need identify appropriate targets and usage for this technology. Potential drawbacks of UV delivery systems, such as phototoxicity or poor tissue penetration, should be considered to evaluate their application practicality.48
While stimuli-responsive CPPs present an elegant solution for selective delivery and limiting off-target toxicity of the cargo and delivery system, there is still room for improvement. For instance, some of these peptides behave differently with each cell line, which begs the question of the reproducibility and versatility of these systems. Selectivity and sensitivity of stimuli responsiveness should also be examined in the presence of other physiological factors, such as serum or off-target stimuli, which could be done in more representative in vitro models or in vivo. Lastly, care must be taken to elucidate the mechanism and efficiency of these systems for cytosolic delivery to enable further optimization. For external stimuli, feasibility of treatment should also be considered, such as accessibility of equipment for stimuli application, or duration and frequency of the treatment.
As previously mentioned, even when CPPs induce cellular uptake of their cargo, they are often trapped in the endosome and are unable to escape to the cytosol where the cargo exerts its function. To circumvent this challenge, multiple moieties have been conjugated to CPPs to improve escape from the endosome. Among these is a class of peptides called pH-dependent membrane active peptides (PMAP), which activate at the low pH condition in the late endosome and lyse the endosomal membrane to release cargo to the cytosol.51,52 To further improve this system, Yu et al. introduced a TAT-based multifunctional chimeric peptide (eTAT) consisting of three additional modules: a PMAP, a dimeric leucine zipper scaffold and a proteolytic cleavage site between the CPP-PAMP system and the cargo.36 They hypothesized that the multimeric display enabled by dimerization of the leucine zipper will help with serum stability and improved endocytosis, while the proteolytic cleavage site will help separate the TAT-PMAP from its cargo, allowing for better escape. Indeed, the authors sequentially proved the effectiveness of each component and demonstrated cytosolic protein delivery both in vivo and in vitro.36In vitro, they saw 70% retention of fluorescence in the nucleus when delivering GFP1–10-NLS in HEK-293T cells in 100% FBS compared to serum-free control. In vivo, eTAT-phosphatase 1B was administered to mice intravenously, which completely prevented death from necroptosis induced by tumor necrosis factor alpha. One drawback to this method, however, is that there is reported evidence of cytotoxicity of CPP-PMAP fusions,51,53 so care must be taken in selecting combinations of CPP, PMAP and cargo, as well as conjugation strategies for the drug delivery complex.51
Once the drug reaches cytosol, intracellular targeting peptides can enable targeted delivery and localization towards a desired organelle for better therapeutic effect. A difficult target is mitochondria, which possess a bilayer membrane that therapeutics need to cross. To overcome this challenge, Cerrato and colleagues covalently fused two penetrating peptides: PepFect14 to help with cytosolic delivery and mtCPP1, a mitochondrial-penetrating peptide. They demonstrated that this fusion peptide was able to form nanoparticles with antisense oligonucleotides through simple mixing and deliver them to silence mitochondrial mRNA in HeLa cells.54 Another attractive intracellular target is the nucleus since it contains the genetic information of the cell.55 One common strategy to gain nucleus-targeting function for CPP systems is by fusion with a nuclear localization sequence (NLS), which allows cargo to leverage nuclear transport to gain direct access to the nucleus, as has been demonstrated in several papers mentioned in this review.36,38 Another peptide, NrTP, derived from rattlesnake toxin, has been genetically fused to a truncated peptide, sC18 with antimicrobial origin, to demonstrate subnuclear localization in the nucleolus of HeLa and MCF7 cell lines.56
Overall, there have been many innovative approaches to tackle the challenges in using CPPs as effective intracellular delivery carriers. That said, there are still questions that remain unanswered. For instance, serum proteins significantly influence the performance of engineered peptides in some studies, without inclusion of ideas for resolution. Furthermore, the relative lack of in vivo studies raises the question of how translatable these technologies are to be used as therapeutics in complex biological systems. Lastly, it is still unclear as to how some peptides associate with the membrane to promote cytosolic delivery, which is crucial knowledge for future improvement of these systems.
2.2. Peptide coacervates
Liquid–liquid phase separation (LLPS) is a multi-step supramolecule formation process driven simultaneously by entropy and enthalpy, where a homogenous mixture is separated into two liquid phases with different compositions.57–59 Often leveraged in the natural formation process of membrane-less organelles, this mechanism is also utilized in peptide solutions to form highly concentrated peptide coacervates or droplets.60,61 These peptide droplets are liquid-like and enable rapid recruitment of small and macromolecules to be loaded into the droplet during phase separation.62 Remarkably, while it is commonly held that nano-sized dimensions are required for molecules to cross the cellular membrane, it was found that micron-sized LLPS peptide droplets were able to cross the cellular membrane in an energy-independent fashion, making them a promising intracellular delivery vessel.63 One challenge with this method, however, is that once in the cytosol, the droplets may remain in an organelle-like state for up to 7 days without releasing their cargo.60 To circumvent this, Sun et al. included a redox-sensitive self-immolative moiety to a droplet-forming, histidine-rich peptide from squid beak.64 In the cytosol, the presence of glutathione triggered reduction and disassembly of the droplet (Fig. 2). Through experiments with endocytosis inhibitors, the coacervate was shown to bypass energy-dependent uptake pathways and efficiently deliver a range of functional biomolecules, including proteins, small peptides and mRNAs, intracellularly to liver cancer cells (HEPG2) in vitro. Delivery was hypothesized to be the result of cholesterol-dependent lipid rafting due to the liquid-like state of the droplets.64,65 Importantly, this complexion method can protect more unstable cargos, such as mRNA, from premature degradation by RNAses.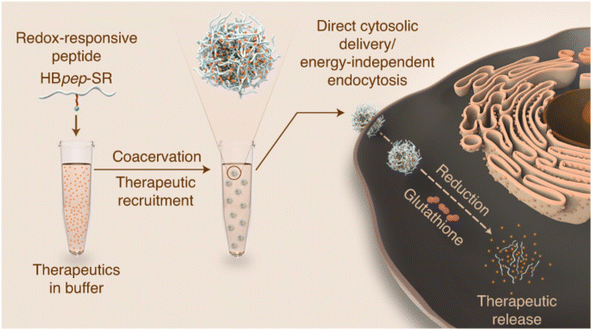 | ||
| Fig. 2 Redox-sensitive LLPS micro-coacervate for intracellular delivery of macromolecules. “Phase-separating peptides for direct cytosolic delivery and redox-activated release of macromolecular therapeutics.” Y. Sun, S. Y. Lau, Z. W. Lim, S. C. Chang, F. Ghadessy, A. Partridge and A. Miserez, Nature Chemistry, 14, 2022 reproduced with permission from SNCSC.64 | ||
In another LLPS delivery study, Futaki et al. created a liquid droplet by attaching a Fc-binding domain on the trimeric version of a lytic peptide, L17E, to enable complexation of IgG. However, they found that the liquid droplets were only formed when IgG is labeled with negatively charged dyes, such as AF488.66 These droplets were able to effectively deliver IgG intracellularly to 60% of treated HeLa cells, proposed to be cytosolic by the diffuse, non-punctate, signal of the fluorescent dyes. Experiments with HeLa cells with labeled actin showed rapid actin alterations at the cellular membrane, followed by rapid increase in cellular fluorescent signal of labeled IgG inside. This observation led to the hypothesized mechanism of action for this delivery system, in which, upon contact with cellular membrane, L17E quickly induced actin polymerization and membrane ruffling, leading to transient perturbation of the cellular membrane, allowing for rapid direct influx of IgG into the cytosol.
These recent developments and positive results of LLPS peptide coacervates suggest the diversity and feasibility of using these droplets as intracellular delivery platforms, whose potential should be further explored.67 For instance, other features could be added to these LLPS coacervates for better intracellular delivery, as they have been previously engineered for extracellular delivery. This includes adding stimuli-responsive moieties into peptide droplets for selective release of cargo, such as in the case of magnetic-responsive peptide coacervates designed by Lim et al.,63 or attaching targeting ligands that enables site specific delivery.62
2.3. Selective lytic peptides for improved endosomal escape
Often derived from pathogenic bacteria, viruses and venomous animals, lytic peptides are able to target and destabilize cellular membranes. Compared to CPPs in the previous section, these peptides are more likely to lyse and damage phospholipid plasma membranes, which might compromise cellular viability.68 However, they are also considered to be more efficient at enabling endosomal escape compared to other membrane-associating peptides, such as CPPs, through complete lysis of endosomal membranes.69 This section will discuss recent efforts in engineering lytic peptides to harness their potential for intracellular delivery, while mitigating their toxicity to cells.One approach to help mask the lytic activity of these peptides until they reach the endosome is through amphiphilicity reduction, to reduce association with the plasma membrane.70 Akishiba et al. substituted leucine with glutamic acid on the hydrophobic region of the amphiphilic spider venom lytic peptide, M-lycotoxin (L17E), which reduced plasma-membrane perturbation activity and cytotoxicity 30-fold (EC50 = 40 μM).69 They then demonstrated that the lytic function is active in the endosome, allowing cytosolic delivery of functional proteins and antibodies in HeLa cells in the presence of serum. This phenomenon was hypothesized to be the result of stronger association of net-positively charged L17E with more negatively charged endosomal membrane, compared to plasma membrane, which is composed of more neutral lipid (Fig. 3).69 Interestingly, this charge difference seems to be the main driving force for the selective lytic activity of this peptide, as protonation of Glu in acidic conditions does not significantly affect the perturbation degree, as often seen otherwise in other peptide-based endosomal escape strategies.71,72 Similarly, Chen et al. demonstrated that modulating selectivity of membrane perturbation could also be achieved through iterative balancing of hydrophobicity between the two termini of an engineered lytic peptide, KL1, that was derived from the apoptotic peptide KLA.70,73,74 Through this process, LP6, a moderately hydrophobic variant of KL1 was identified to have efficient delivery and reduced toxicity (EC50 = 31.1 μM). Once in contact with anionic lipids, such as those in endosomal membranes, LP6 switched to a defined alpha-helix conformation.74 This alpha helix conformation provides stronger membrane association with the endosomal membrane, compared to its non-helical state when arriving at the plasma membrane.74,75 It was demonstrated that LP6 delivered polysaccharide (dextran) and protein (saporin, IgG) cargos intracellularly to the cytosol, through clathrin-dependent endocytosis and subsequent LP6-faciliated endosomolysis.
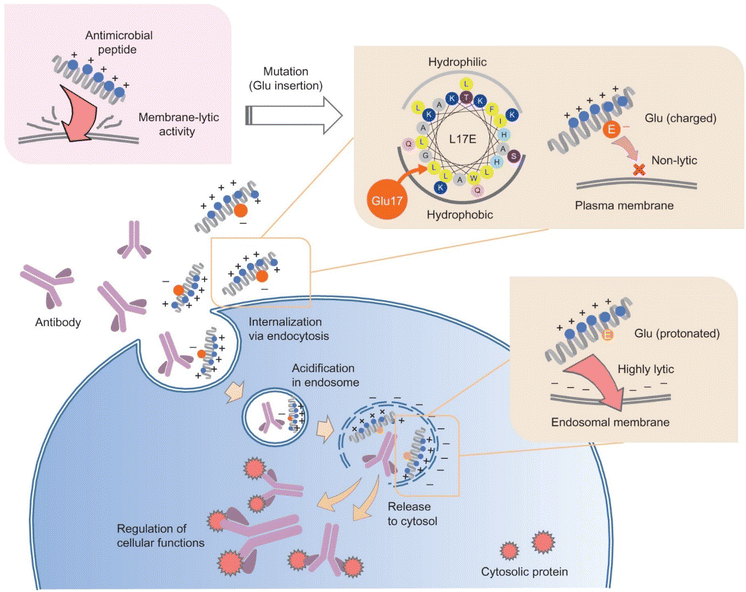 | ||
| Fig. 3 Selective endosomal membrane lytic activity through attenuation of cationic spider venom lytic peptide. “Cytosolic antibody delivery by lipid-sensitive endosomolytic peptide.” M. Akishiba, T. Takeuchi, Y. Kawaguchi, K. Sakamoto, H. H. Yu, I. Nakase, T. Takatani-Nakase, F. Madani, A. Graslund and S. Futaki, Nature Chemistry, 9, 2017 reproduced with permission from SNCSC.69 | ||
To further improve delivery efficiency of lytic peptides, especially for bigger cargoes such as antibodies, different assembly configurations of lytic peptides have been designed. For instance, multimeric assemblies of attenuated lytic peptides were found to enhance cytosolic delivery of macromolecules, such as in the case of L17E/Q21E homodimer.76,77 L17E/Q21E is a less hydrophobic and cytotoxic, though less effective, variant of L17E that was dimerized using a disulfide bridge at the N-terminal. Dimeric L17E/Q21E was able to better deliver functional anti-nuclear pore complex (NPC) antibodies in HeLa cells, compared to using 8X higher dose of monomeric L17E.77 Another approach to alter lytic peptide assembly for better intracellular delivery is through fusion with other functional peptides to grant them additional features. One such attempt attached a macropinocytosis-inducing peptide (SN21) to a highly toxic membrane-lytic peptide, LK15.78 This assembly configuration facilitated macropinocytosis in HeLa cells by triggering dynamic actin reorganization at the cellular membrane and subsequent release of functional antibody cargo from macropinosomes, and also reduced the cytotoxicity of LK15.
Altogether, lytic peptides have been demonstrated to possess promising intracellular delivery capabilities. However, the feasibility of their application for therapeutics could only be realized once their lytic activity and thus, toxicity, be thoroughly evaluated and controlled. Notably, safety concerns still remain after attenuation of the lytic activity, since dimeric assembly of the attenuated L17E is still highly toxic.77 Thus, any attempt, either in using a new lytic peptide or in engineering attenuated lytic peptides must address cytotoxicity or potential immunogenicity when tested in vivo.79
3. Membrane interacting proteins for cytosolic delivery of therapeutic cargo
Protein sequence and structural diversity can be leveraged to solve specific delivery challenges.80,81 This section will focus on proteins engineered to deliver therapeutics to the cytosol. Proteins are larger than peptides and display secondary and tertiary structures that dictate their specific functions. They are typically produced recombinantly in bacterial or eukaryotic cells, depending on the origin of the protein and the need for post-translational modifications. Both whole proteins and selected domains of proteins, frequently derived from those found in nature, have been used to deliver cytosolically active drugs.3.1 Bacterial toxins
Some bacterial toxins can access the host cytosol, primarily for infection. Use of these for delivery has focused on AB bacterial toxins, named for their toxic enzymatic A-domain and a B-domain with membrane penetrating ability.82 AB toxins are endocytosed via receptor-mediated endocytosis and are unique because they traffic directly to the Golgi and avoid degradation in the late endosome. After the toxins are transported to the Golgi, they are trafficked to the endoplasmic reticulum, then directly into the cytoplasm.83 Current research is focused on utilizing the B-domains of these toxic proteins to aid in the delivery of drugs. To use these proteins, AB toxins are recombinantly truncated to only the B-domain or point mutations are made to the active site of the A-domain to diminish toxicity. Then, the modified proteins are conjugated to drug carriers or fusion proteins to deliver the drug of interest.Cholera toxin B (CtxB) can aid in delivering proteins and other attached molecules to the cytosol while avoiding entrapment in the endo-lysosomal system. Once bound to ganglioside GM1 on the surface of neural cells, CtxB gains entry via receptor-mediated endocytosis.84 Jia et al. used the B-subunit from Cholera toxin to deliver GFP and toxic protein apoptin as a proof of concept for intracellular and cytosolic delivery, respectively.85 GFP and CTxB cannot be expressed as fusion proteins due to the requirement for different purification methods to yield and maintain functionality. To address this problem, authors utilized the intein subunits of Cfa on the termini of the respective proteins to conjugate post-translation. As seen in Fig. 4, intein CfaN is fused to the C-terminus of CTxB, and intein CfaC was attached to the N-terminus of GFP or apoptin. When proteins with CfaN and CfaC subunits were mixed in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, the intein excises itself and the proteins GFP or saponin are linked to CTxB via peptide bonds. GFP delivered with CtxB via intein splicing induced higher uptake in vitro in mouse glioblastoma cells (Neuro2A), compared to soluble GFP in media with and without serum. Researchers also deleted a key cysteine from CfaC fused to GFP and showed there is still uptake in Neruo2A cells, indicating there may be a noncovalent interaction between CfaN and CfaC. Lower cell viability was reported when human and mouse glioblastoma cells (U87, and Neuro2A, respectively) were treated with the CTxB intein spliced apoptin, a toxic protein, compared to soluble apoptin indicating functional cytosolic delivery. Toxicity did not occur when CTxB intein spliced GFP was delivered. Lower cell viability and uptake were not observed in HepG2 (human liver cancer line) or HeLa (human cervical cancer line) cells, confirming the specificity of CTxB.85 Another bacterial toxin specific to GM1 is the Escherichia coli heat-labile enterotoxin (LT).86 Its B-domain is a pentamer containing a protease cleavage site. Liu et al. formed a self-assembling pentameric protein delivery vehicle, LEB5, a self-releasing protein carrier capable of trafficking via the trans-Golgi network. LEB5 comprises 5 repeating subunits of the LT B-domain attached to GFP as a model cargo protein. A loop portion of the LT protein was included to provide proteolytic cleavage sites, allowing for the release of GFP once in the cytosol via cytosolic proteases. This study found that proteins transported via the LEB5 carrier were transported to the Golgi apparatus in vitro using A549 (human alveolar epithelial cells). Also, there was no significant change in cellular viability associated with this carrier. However, no evidence was given as to whether this carrier finally results in cytosolic delivery.
1 ratio, the intein excises itself and the proteins GFP or saponin are linked to CTxB via peptide bonds. GFP delivered with CtxB via intein splicing induced higher uptake in vitro in mouse glioblastoma cells (Neuro2A), compared to soluble GFP in media with and without serum. Researchers also deleted a key cysteine from CfaC fused to GFP and showed there is still uptake in Neruo2A cells, indicating there may be a noncovalent interaction between CfaN and CfaC. Lower cell viability was reported when human and mouse glioblastoma cells (U87, and Neuro2A, respectively) were treated with the CTxB intein spliced apoptin, a toxic protein, compared to soluble apoptin indicating functional cytosolic delivery. Toxicity did not occur when CTxB intein spliced GFP was delivered. Lower cell viability and uptake were not observed in HepG2 (human liver cancer line) or HeLa (human cervical cancer line) cells, confirming the specificity of CTxB.85 Another bacterial toxin specific to GM1 is the Escherichia coli heat-labile enterotoxin (LT).86 Its B-domain is a pentamer containing a protease cleavage site. Liu et al. formed a self-assembling pentameric protein delivery vehicle, LEB5, a self-releasing protein carrier capable of trafficking via the trans-Golgi network. LEB5 comprises 5 repeating subunits of the LT B-domain attached to GFP as a model cargo protein. A loop portion of the LT protein was included to provide proteolytic cleavage sites, allowing for the release of GFP once in the cytosol via cytosolic proteases. This study found that proteins transported via the LEB5 carrier were transported to the Golgi apparatus in vitro using A549 (human alveolar epithelial cells). Also, there was no significant change in cellular viability associated with this carrier. However, no evidence was given as to whether this carrier finally results in cytosolic delivery.
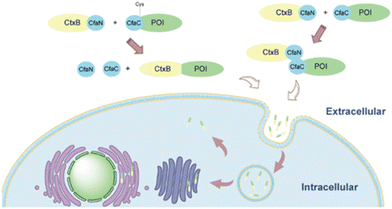 | ||
| Fig. 4 Cholera toxin subunit B attached to protein of interest (POI) via intein splicing or non-covalent interactions. Reprinted with permission from Jia, X. F., et al. “Highly efficient method for intracellular delivery of proteins mediated by cholera toxin-induced protein internalization.” Molecular Pharmaceutics, 18(11): 4067–4078. Copyright 2021 American Chemical Society.85 | ||
Another bacterial toxin that promotes intracellular delivery is the Shiga toxin B-subunit (STxB). This protein is endocytosed via receptor-mediated endocytosis by glycosphingolipid globotriaosylceramide (Gb3), commonly found on many cancer cell surfaces.87 Gb3-mediated endocytosis allows STxB to be trafficked to the cytosol via the trans-Golgi network to avoid endosomal entrapment. Hadjerci et al. identified and replaced residues on STxB to incorporate a non-natural amino acid, allowing the conjugation of synthetic hydrophobic moieties to increase cytosolic delivery in vitro without affecting the key binding site of the protein.88 Also, a biotin-based reporter moiety was conjugated to STxB to interact with a modified HeLa (Gb3 positive) cell line to prove cytosolic delivery.89 Incorporating a hydrophobic moiety increased cytosolic delivery in HeLa cells while maintaining specificity for Gb3. Similar results were reported by Danielewicz et al. for a modified STxB toxin in which an anti-neoplastic cancer drug was conjugated to STxB by incorporating an azide via an unnatural amino acid.90 The protein conjugate showed specificity for highly Gb3-expressing HT-29 cells in vitro and no significant delivery to a low Gb3-expressing colon cancer cell line (LS-174), when evaluated via a cytotoxicity assay. Overall, STxB protein drug carriers open an opportunity for a Gb3-targeted protein delivery system with efficient delivery to the cytosol via the trans-Golgi network, though in vivo efficacy remains to be assessed.
Both diphtheria toxin (DT) and botulinum toxin (BoNT) have been used as carriers for large Cas proteins. Tian et al. created a fusion protein comprised of the receptor binding domain of DT (DTR) and BoNT/X components.91 BoNT/X is a homolog of BoNT that does not induce residual toxicity due to modification of its A domain.92 The fusion protein used to deliver the Cas13a protein is called Cas13a-XLCHN-DTR. To reduce the toxicity of the BoNT/X point mutations were added to the light chain of BoNT/X, called BoNT/X-LC (XLCHN in the fusion protein). The translocation domain of BoNT/X undergoes conformational changes in the acidic late endosome, allowing it to transverse the endosomal membrane with the therapeutic protein still attached. The mechanism of this process is still unknown.91 Cas13a delivered by the fusion protein demonstrated function by degrading mRNA associated with GFP fluorescence in GFP producing HeLa cells in vitro. A limitation of this system is that it cannot deliver ribonucleoprotein (RNP) complex comprising Cas9 protein and single-guide RNA, meaning the guiding crRNA needs to be delivered separately, via lentivirus in this case. The targeting ability of the fusion protein, Cas13a-XLCHN-DTR, is due to DTR specificity to heparin-binding epidermal growth factor precursor, which is commonly found on the surface of many cell types including lung, muscle, heart, and brain.91,93 This system combines the targeting ability of one protein (DT) and membrane disruption ability of another (BoNT). However, there are some efficacy concerns with respect to this system, since many people are vaccinated against diphtheria.
3.2 Viral proteins
Just as bacterial pathogens have evolved proteins to aid in intracellular entry, so have viruses. Hemagglutinin (HA) is a trimeric protein on the surface of influenza viruses that exhibits significant sequence diversity to evade host immune responses.94 HA facilitates intracellular entry of influenza via binding to the terminal sialic acid residue of cell surface glycoproteins and endocytosis. After endocytosis, a change in pH alters the conformation of HA, which then initiates the HA terminus to engage with the vesicle membrane, causing fusion between the viral and endosomal membranes. This fusion event can force viral contents to be released into the cytosol.Park et al. used virus-mimicking cell membrane-coated nanoparticles to deliver messenger RNA (mRNA).95 They employed an engineered cellular membrane displaying HA subtype 7 (HA7) as the outer layer of the nanoparticle, with a poly(lactic-co-glycolic acid) (PGLA) core containing mRNA cargo and lipid-like G0-C14 to help with loading, as seen in Fig. 5. HA7 on the nanoparticle surface was reported to be a driver of membrane fusion. A lysosomal dye was used to determine a decrease in colocalization between the HA7 coated nanoparticles and lysosomes, indicating less endosomal entrapment of the nanoparticles. HA7-coated nanoparticles delivered more mRNA coding for GFP than uncoated nanoparticles to murine melanoma cells (B16F10) in vitro, with increased fluorescence indicating cytosolic delivery of mRNA. HA7 coated nanoparticles containing luciferase coding mRNA were delivered in vivo to CD-1 mice via intranasal and intravenous routes. Increased luminescence in the nasal cavity and serum were evidence of successful delivery and mRNA translation by intranasal and intravenous routes, respectively.
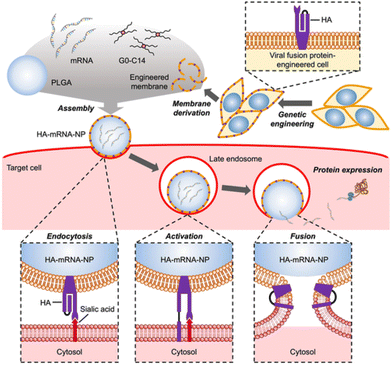 | ||
| Fig. 5 Fabrication of HA coated nanoparticles comprised of an engineered cellular membrane encasing a PGLA core containing cationic G0-C14 and mRNA (top). The process in which HA associates with sialic acid on glycoproteins on cells and fuses with the endosomal membrane in mammalian cells (bottom). Used with permission of John Wiley & Sons-Books from Virus-mimicking cell membrane-coated nanoparticles for cytosolic delivery of mRNA, J. H. Park et al., volume 6, issue 2, 2022; permission conveyed through Copyright Clearance Center, Inc.95 | ||
HA can also be fused directly to cargo for intracellular delivery by recombinant or chemical conjugation, like the bacterial toxins. HA is a trimeric protein and each monomer contains glycoprotein subunits HA1 and HA2. The HA2 domain contains the region that associates with membranes.96 More specifically, the N-terminus of the HA2 domain is pH sensitive and is has been proven to interact with lipid membranes at low pH (pH ∼ 5) due to a change in its conformation.97 Tang et al. conjugated the HA2 N-terminus peptide to a DNA origami carrier via click chemistry to deliver sgRNA/Cas9 complexes to edit the tumor-associated gene PLK1 causing death of MCF7 tumor cells in vitro and in vivo.98 A DNA aptamer specific for glycoprotein Mucin 1, abundant in breast cancer cells, was used as a targeting motif. HA decreased colocalization of the DNA complexes with lysosomes in MCF7 cells as compared to the carrier without HA, though statistical analysis was lacking. In a mouse tumor model, tail vein injection of the HA-modified DNA complexes showed an increase in gene editing within the tumor cells compared to the sgRNA/Cas9 complex alone. However, there was no comparison to DNA complex without HA.
Many other viruses, besides influenza, have membrane interactive proteins. While the bulk of SARS-CoV-2 research has focused on disease treatment and prevention, Zheng et al. were the first to examine if SARS-CoV-2 non-structural protein 2 (NSP2) could be used as a drug delivery tool.99 NSP2 is a secretory protein associated with multiple coronaviruses and membrane translocation activity; however, the mechanism is unknown.100 When NSP2 was genetically fused to a cytosolically active toxic protein cargo Birkholderia Lethal Factor 1 (BLF1) and delivered to Huh7 cells, it caused significantly more cell death than soluble BLF1, proving cytosolic delivery. NSP2 genetically fused to GFP was also delivered with no significant toxicity. Because these experiments were performed in serum-free media, it is not clear if NSP2 fusions will exhibit effective delivery in complex, protein-rich environments more representative of in vivo conditions. A limitation of the described viral delivery systems is the potential for an immune response or neutralization by existing antibodies, depending on the protein selected. HA is an antigen that elicits strong antibody responses from infection or vaccination, but significant NSP2 titers have not been detected in SARS-CoV-2 infected patients, and most COVID-19 vaccines do not include this antigen.101–103 Unlike bacterial toxins, many viral proteins must be expressed in eukaryotic cells for proper folding or glycosylation, which could challenge cost-effective scale-up.
3.3 Coiled coils and other alpha-helical structures
Coiled coils are characterized by two or more α-helices wound together in a super-helix.104 Coiled coils comprise seven repeating subunits that create a knob-and-hole structure with alternating hydrophobic and charged residues in a parallel or anti-parallel orientation. Coiled coils have been shown to associate and, in some cases, fuse with membranes.104–106 They have been used in diverse applications, including forming self-assembled materials for drug delivery.104Our lab has used hexameric coiled coils (Hex) to deliver fused cargo in vitro. When Hex was affinity bound to IgG or recombinantly fused to GFP, it induced higher endocytic uptake and cytosolic localization of cargo in SKBr3 and HeLa cells.107,108 While Hex did not interact with the plasma membrane, it did induce greater endosomal disruption events and this effect was enhanced when the cargo location was moved to one end of the coiled coil to make Hex more solvent exposed.108 There was also a benefit of 6x-histidine tags, used for purification of Hex, on endosomal disruption. While the mechanism of Hex intracellular delivery was not clear, a glutamic acid-rich weakly dimeric leucine zipper coiled-coil fused to GFP did not induce delivery, suggesting the number of helices in the coiled coil does impact delivery.
Li et al. fused cationic arginine-rich alpha helix K5 to promote the delivery of Cas9 ribonuclease protein.109 The K5 alpha helix forms a coiled coil with glutamic acid-rich E5, though in this work K5 was used alone and the membrane activity is due to the alternating positive charge and hydrophobic residues that also drive coiled coil formation.110,111 Mediated by K5 interaction with cell membranes, the fusion protein Cas9-K5 increased cytosolic delivery of Cas9 and template to HEK293 cells in vitro in reduced serum media, proven by using a DNA template to delete the stop codon before TdTomato that results in TdTomato expression (Cre-dependent system).109 Also, in an Ai9 mouse model, researchers delivered Cas9-K5 with DNA coding for the Cre-dependent tdTomato expression via stereotaxic injection. Researchers reported that tdTomato protein was expressed in the hippocampus and striatum of treated mice. No data was presented on Cas9 delivery without K5, so it is difficult to determine the specific effect of K5 on intracellular brain delivery.
Dimeric coiled coils can also make drug carriers that are more analogous to nanoparticles. Takahashi et al. employed anti-parallel heterodimeric coiled coils to fuse to a portion of the hydrophobic protein oleosin to create a bilayer drug carrier.112 As seen in Fig. 6, the bilayer nanocapsule is formed by a water-in-oil emulsion and can hold cargo such as the cytotoxic protein, RNAse. Thioredoxin A (Trx) was added to increase the hydrophobicity of the protein, and the Fc domain binding protein Streptococcal protein G (ProG) was added to enable targeting antibody attachment to the capsules. Cetuximab-coated capsules delivered RNAse to the cytosol of human fibroblasts (NHDF) in vitro. The release of RNAse in the cytosol is thought to be a result of first the degradation of the capsule driven by the low pH environment of the late endosome and then the coiled-coil associating with the endosomal membrane. Both theories are supported via a decrease in ellipticity of circular dichroism at lower pH, and a rhodamine quenching experiment performed on liposomes at pH 7.4 and pH 5.5. To evaluate alpha-helix interactions with cellular membranes, synthetic phospholipid liposomes containing self-quenching rhodamine dye are often used to model the cellular membrane.112,113 When the membrane is perturbed by a drug carrier, it will release the dye, increasing the fluorescence. Typically, this experiment is carried out at different pH values to simulate the lowering of endosomal pH and verify the conformational change of membrane active pH-sensitive alpha-helices. Overall, coiled coils can deliver drugs to the cytosol with varying degrees and have been applied in a number of different formats. In all cases, pH appears to play some role in membrane perturbation activity of these proteins.
 | ||
| Fig. 6 Combination of oleosin protein (Ole), thioredoxin (Trx) and coiled coils (NZ and CZ) facilitate the self-assembly of cargo-loaded nanocapsules. Used with permission of Royal Society of Chemistry, from Delivery of external proteins into the cytoplasm using protein capsules modified with IgG on the surface, created from the amphiphilic two helix-bundle protein OLE-ZIP, K. Takahashi et al., volume 60, issue 8, 2024; permission conveyed through Copyright Clearance Center, Inc.112 | ||
4. Conclusions and outlook
The cytosol and organelles of mammalian cells are attractive targets for the delivery of molecules, both for fundamental studies and therapeutic purposes. However, since knowledge of the factors that influence the degree of uptake and fate of the cargos once they enter cells is still limited, it remains a challenge to ensure that these molecules can reach their intracellular targets.114 Arguably, the biggest challenge in intracellular delivery is the bilayer phospholipid membrane that surrounds the cell and endosomes. These membranes prevent passive diffusion of most macromolecules, stopping them from entering the cell or escaping the endosome to reach the cytosol.115 To circumvent these barriers, biomacromolecular cargoes are often delivered by materials or molecules that can modulate the membranes in ways that allow their cargo to pass. Among these systems, proteins and peptides have the advantages of relatively compact size, good biocompatibility and being easily modifiable, enabling fusion or conjugation with macromolecular cargos or other materials like polysaccharides and lipids for added functions.114,116,117This review paper summarized recent advances in using proteins and peptides to improve intracellular delivery of therapeutics through membrane interaction (Table 2). We noted the ever-expanding library of newly discovered protein and peptide entities with some degree of membrane association or disruption abilities. The proteins and peptides reported were derived from diverse sources, from spiders to microorganisms, then improved through directed evolution or rational design. We also covered recent efforts spent on advancing and finding different ways to utilize the currently available proteins and peptides. The efficacy of these systems was improved by employing different modes of delivery or rational assembly configurations. Specificity and enhanced safety were achieved through engineered stimuli-responsiveness, as well as attenuation of off-target activities. Aided with increasingly better fundamental understanding of the membrane association processes utilized by these macromolecules,8 we expect that future efforts in improving these systems will be more focused on fine-tuning these molecules in order to harness their unique characteristics for more efficient and precise intracellular delivery.
| Delivery approach | Advantages | Disadvantages | Potential applications |
|---|---|---|---|
| Additive usage of CPPs | Ease of use | Potential toxicity due to added soluble CPPs; serum interference | Target that is immediately accessible from the site of drug administration; intratumoral drug delivery |
| Stimuli-responsive CPPs | Specificity and limit off-target toxicity | Stimuli might not be specific to target | Toxic cargos with targets that provide specific and distinct stimuli |
| Functional assembly of CPPs | Added functional features for targeting or gaining entry to intracellular organelles | Some assemblies demonstrate increased toxicity | Therapeutic cargos with specific intracellular targets |
| LLPS peptide coacervates | Micron-sized drug depot | Might need modification of drug cargo for coacervate formation; coacervate might not release drug upon cellular entry | Cargos with suitable properties for forming coacervate through LLPS, which could be identified through prediction tools or LLPS database61 |
| Lytic peptides | Potent, high membrane lytic activity | Cellular membrane lysis and cytotoxicity | Larger cargos that might need endosomal membrane lysis to escape |
| Bacterial toxins | Targeting ability, access to alternative endosome trafficking paths | Limitations on cell types due to necessity of specific surface receptor | Specific targeting for various cancers, and delivery of protein and nucleic acid cargo |
| Viral proteins | Direct interaction with membrane, variety of means to disrupt the membrane | May require expression in eukaryotic host; potential for neutralization by antibodies from prior vaccination/infection | General use to deliver proteins and nucleic acid cargo |
| Coiled coils | Variety of coiled coils able to interact with membranes, pH responsive | Not all coiled coils are membrane active | General use to deliver proteins and nucleic acid cargo |
One major challenge in the field is how to reliably, easily and quantitatively measure and differentiate between cellular uptake and actual cytosolic delivery. Flow cytometry data with fluorescent-tagged cargos is not sufficient, since one cannot differentiate between the molecules entrapped in endosomes or the plasma membrane, from those that functionally delivered to the cytosol. Confocal microscopy provides spatial information, where diffuse signal within the cell boundary is interpreted as cytosolic delivery, and punctate are associated with endosomal entrapment. However, when both punctate and diffuse signal are present, it is quite difficult to evaluate the effectiveness of the delivery given the difference in intensity of the two features. Moreover, it has been observed that even mild fixation can allow membrane-associated peptides to artifactually get into the cytosol.118 Microscopy is frequently used qualitatively, not quantitatively, and thus, moderate improvements are not often captured. Another approach is to use lysosome specific dyes and track co-localization of cargo and lysosomes. However, there can be a large effect of imaging settings like oversaturation or slide thickness on correlation calculation, leading to post-processing bias, and the absence of co-localization does not prove cytosolic location – it only proves the cargo is not in the lysosomes.119 Even more convincing methods to assess cytosolic delivery, such as delivering functional proteins such as split GFP or split luciferase, or antibodies with cytosolic targets and measuring the output of the cargo function, still have drawbacks.120,121 These methods are indirect, heavily relying on downstream processes and reactions to happen properly for accurate interpretation of the data. A more recent analytical method that allows direct measurement of cytosolic concentration of delivered cargos is fluorescent correlation spectroscopy (FCS). This method captures fluctuations in fluorescent intensity of single molecule and uses them to identify its cellular location and concentration.122 Although FCS is direct and quantitative of the molecule of interest, it is quite complicated and might not be suitable for high-throughput experiments or available to all researchers. Thus, the need remains for better, easier and faster assays to analyze and quantify cytosolic delivery.
Prior to uptake, it is also important to characterize the initial steps of membrane interaction and binding. Adhesion of peptides and proteins primarily depends on their amino composition, which dictates charge, hydrophobicity, and structure, though the process of adhesion is not fully understood.123 Experiments to gain insight into the initial steps of membrane adhesion between peptides or proteins and membranes were established mainly using CPPs and viral peptides. Methods include circular dichroism, Fourier transform infrared spectroscopy, and nuclear magnetic resonance to show the structural changes between peptides/proteins and cell or model membranes throughout binding and membrane interaction.123,124 To identify disruption or fusion with cellular membranes, synthetic phospholipid liposomes containing self-quenching dye are used to model the cellular membrane.112,113 Similarly, hemolysis of red blood cells incubated with peptides/proteins can be used as a measure of membrane interaction since the cells are not endocytic.125 The methods presented, while extensive, can be limited by equipment and a broad area of expertise needed to perform a complete analysis of adhesion and membrane perturbation. Further, these methods do not reveal if or which specific membrane proteins are involved in binding events.
Another limitation in the field is the lack of in vivo experiments in the majority of studies mentioned. In some cases, experiments are done strictly in serum-free media, which is not reflective of the complex environments that these peptides would be used. Eventually, it may be revealed that promising candidates in vitro are poor delivery carriers in animal models and, ultimately, humans. Studies from the lipid nanoparticle delivery field have identified the lack of correlation between in vitro and in vivo results,126,127 which could similarly exist for protein and peptide delivery systems. Additionally, many studies that utilized animal or bacterial-derived proteins and peptides lack toxicity and immunogenicity evaluation, which is crucial in assessing the feasibility of their therapeutic application and can only be reliably evaluated in vivo. Therefore, going forward, future studies with delivery peptide and proteins should be tested in conditions that best mimic the in vivo environment and interrogate in vivo function as much as possible, to identify the most translatable candidates.
Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge funding from the National Institutes of Health, 1 R01 HL158159 – 01. We acknowledge the contributions of named and unnamed people whose health, lives, livelihoods, legacy, and privacy were extorted, often without compensation, consent, or regard to their safety, in the name of biomedical research. Adults and children were stripped of their humanity, and often their identity. We commit to educating ourselves on the history and ethical failures of biomedical research and encourage others to do the same.References
- S. Behzadi, V. Serpooshan, W. Tao, M. A. Hamaly, M. Y. Alkawareek, E. C. Dreaden, D. Brown, A. M. Alkilany, O. C. Farokhzad and M. Mahmoudi, Chem. Soc. Rev., 2017, 46, 4218–4244 RSC.
- M. D. Hartmann, Subcell. Biochem., 2017, 82, 63–93 CAS.
- H. Hillaireau and P. Couvreur, Cell. Mol. Life Sci., 2009, 66, 2873–2896 CrossRef CAS PubMed.
- G. Sahay, D. Y. Alakhova and A. V. Kabanov, J. Controlled Release, 2010, 145, 182–195 CrossRef CAS PubMed.
- T. Kanaseki and K. Kadota, J. Cell Biol., 1969, 42, 202–220 CrossRef CAS PubMed.
- S. Mayor and R. E. Pagano, Nat. Rev. Mol. Cell Biol., 2007, 8, 603–612 CrossRef CAS PubMed.
- C. de Duve, Med. Sci., 2005, 21, 8–11 Search PubMed.
- D. Pei and M. Buyanova, Bioconjugate Chem., 2019, 30, 273–283 CrossRef CAS PubMed.
- J. P. Luzio, P. R. Pryor and N. A. Bright, Nat. Rev. Mol. Cell Biol., 2007, 8, 622–632 CrossRef CAS PubMed.
- A. H. Faraji and P. Wipf, Bioorg. Med. Chem., 2009, 17, 2950–2962 CrossRef CAS PubMed.
- R. Tenchov, R. Bird, A. E. Curtze and Q. Zhou, ACS Nano, 2021, 15, 16982–17015 CrossRef CAS PubMed.
- I. Ruseska and A. Zimmer, Beilstein J. Nanotechnol., 2020, 11, 101–123 CrossRef CAS PubMed.
- N. Chen, Y. He, M. Zang, Y. Zhang, H. Lu, Q. Zhao, S. Wang and Y. Gao, Biomaterials, 2022, 286, 121567 CrossRef CAS PubMed.
- J. Yin, Q. Wang, S. Hou, L. Bao, W. Yao and X. Gao, J. Am. Chem. Soc., 2018, 140, 17234–17240 CrossRef CAS PubMed.
- M. Rafalski, A. Ortiz, A. Rockwell, L. C. V. Ginkel, J. D. Lear, W. F. DeGrado and J. Wilschut, Biochemistry, 2002, 30, 10211–10220 CrossRef PubMed.
- C. Bao, B. Liu, B. Li, J. Chai, L. Zhang, L. Jiao, D. Li, Z. Yu, F. Ren, X. Shi and Y. Li, Nano Lett., 2020, 20, 1352–1361 CrossRef CAS PubMed.
- P. Shi, J. Qin, S. Luo, P. Hao, N. Li and X. Zan, Biomater. Sci., 2021, 10, 178–188 RSC.
- O. N. Yogurtcu, C. W. Wolgemuth and S. X. Sun, Biophys. J., 2010, 99, 3895–3904 CrossRef CAS PubMed.
- N. Porebska, K. Ciura, A. Chorazewska, M. Zakrzewska, J. Otlewski and L. Opalinski, Biotechnol. Adv., 2023, 67, 108213 CrossRef CAS PubMed.
- S. Sachdev, C. Cabalteja and R. Cheloha, Methods Cell Biol., 2021, 166, 205–222 CAS.
- K. Fosgerau and T. Hoffmann, Drug Discovery Today, 2015, 20, 122–128 CrossRef CAS PubMed.
- L. Wang, N. Wang, W. Zhang, X. Cheng, Z. Yan, G. Shao, X. Wang, R. Wang and C. Fu, Signal Transduction Targeted Ther., 2022, 7, 48 CrossRef CAS PubMed.
- T. Samec, J. Boulos, S. Gilmore, A. Hazelton and A. Alexander-Bryant, Mater. Today Bio, 2022, 14, 100248 CrossRef CAS PubMed.
- F. Heitz, M. C. Morris and G. Divita, Br. J. Pharmacol., 2009, 157, 195–206 CrossRef CAS PubMed.
- A. D. Frankel and C. O. Pabo, Cell, 1988, 55, 1189–1193 CrossRef CAS PubMed.
- Z. Sun, J. Huang, Z. Fishelson, C. Wang and S. Zhang, Biomedicines, 2023, 11, 1971 CrossRef CAS PubMed.
- R. A. Bottens and T. Yamada, Cancers, 2022, 14, 5546 CrossRef CAS PubMed.
- P. Agrawal, S. Bhalla, S. S. Usmani, S. Singh, K. Chaudhary, G. P. Raghava and A. Gautam, Nucleic Acids Res., 2016, 44, D1098–D1103 CrossRef CAS PubMed.
- K. M. Wagstaff and D. A. Jans, Curr. Med. Chem., 2006, 13, 1371–1387 CrossRef CAS PubMed.
- T. Skotland, T. G. Iversen, M. L. Torgersen and K. Sandvig, Molecules, 2015, 20, 13313–13323 CrossRef CAS PubMed.
- M. Kosuge, T. Takeuchi, I. Nakase, A. T. Jones and S. Futaki, Bioconjugate Chem., 2008, 19, 656–664 CrossRef CAS PubMed.
- J. V. V. Arafiles, J. Franke, L. Franz, J. Gomez-Gonzalez, K. Kemnitz-Hassanin and C. P. R. Hackenberger, J. Am. Chem. Soc., 2023, 145, 24535–24548 CAS.
- M. Lundberg, S. Wikstrom and M. Johansson, Mol. Ther., 2003, 8, 143–150 CrossRef CAS PubMed.
- C. Palm-Apergi, P. Lonn and S. F. Dowdy, Mol. Ther., 2012, 20, 695–697 CrossRef CAS PubMed.
- F. Zakany, I. M. Mandity, Z. Varga, G. Panyi, P. Nagy and T. Kovacs, Cells, 2023, 12, 1700 CrossRef CAS PubMed.
- S. Yu, H. Yang, T. Li, H. Pan, S. Ren, G. Luo, J. Jiang, L. Yu, B. Chen, Y. Zhang, S. Wang, R. Tian, T. Zhang, S. Zhang, Y. Chen, Q. Yuan, S. Ge, J. Zhang and N. Xia, Nat. Commun., 2021, 12, 5131 CrossRef CAS PubMed.
- Y. Huang, Y. Jiang, H. Wang, J. Wang, M. C. Shin, Y. Byun, H. He, Y. Liang and V. C. Yang, Adv. Drug Delivery Rev., 2013, 65, 1299–1315 CrossRef CAS PubMed.
- A. F. L. Schneider, M. Kithil, M. C. Cardoso, M. Lehmann and C. P. R. Hackenberger, Nat. Chem., 2021, 13, 530–539 CrossRef CAS PubMed.
- S. Reissmann and M. P. Filatova, J. Pept. Sci., 2021, 27, e3300 CrossRef CAS PubMed.
- J. K. Allen, T. C. Sutherland, A. R. Prater, C. G. Geoffroy and J. P. Pellois, Sci. Adv., 2022, 8, eabo2954 CrossRef CAS PubMed.
- D. Lee, I. Noh, J. Yoo, N. S. Rejinold and Y. C. Kim, Acta Biomater., 2017, 57, 187–196 CrossRef CAS PubMed.
- A. Moshnikova, M. DuPont, M. Iraca, C. Klumpp, H. Visca, D. Allababidi, P. Pelzer, D. M. Engelman, O. A. Andreev and Y. K. Reshetnyak, Front. Pharmacol., 2024, 15, 1346756 CrossRef CAS PubMed.
- Y. F. Yang, H. C. Zhu, D. K. Liu, H. Luo, R. L. Chang, Y. Ji, W. B. Yao, J. Yin and X. D. Gao, Adv. Funct. Mater., 2023, 33, 2301011 CrossRef CAS.
- Q. Wang, Y. F. Yang, D. K. Liu, Y. Ji, X. D. Gao, J. Yin and W. B. Yao, Nano Lett., 2021, 21, 6022–6030 CrossRef CAS PubMed.
- L. Chen, Q. Song, Y. Chen, S. Meng, M. Zheng, J. Huang, Q. Zhang, J. Jiang, J. Feng, H. Chen, G. Jiang and X. Gao, ACS Nano, 2020, 14, 6636–6648 CrossRef CAS PubMed.
- M. B. Hansen, E. van Gaal, I. Minten, G. Storm, J. C. van Hest and D. W. Lowik, J. Controlled Release, 2012, 164, 87–94 CrossRef CAS PubMed.
- G. C. Kim, J. H. Ahn, J. H. Oh, S. Nam, S. Hyun, J. Yu and Y. Lee, Biomacromolecules, 2018, 19, 2863–2869 CrossRef CAS PubMed.
- V. W. Karisma, W. Wu, M. Lei, H. Liu, M. F. Nisar, M. D. Lloyd, C. Pourzand and J. L. Zhong, Front. Cell Dev. Biol., 2021, 9, 598717 CrossRef PubMed.
- J. Geng, X. Guo, L. Wang, R. Q. Nguyen, F. Wang, C. Liu and H. Wang, Biomolecules, 2020, 10, 217 CrossRef CAS PubMed.
- O. Tietz, F. Cortezon-Tamarit, R. Chalk, S. Able and K. A. Vallis, Nat. Chem., 2022, 14, 284–293 CrossRef CAS PubMed.
- I. Neundorf, R. Rennert, J. Hoyer, F. Schramm, K. Lobner, I. Kitanovic and S. Wolfl, Pharmaceuticals, 2009, 2, 49–65 CrossRef CAS PubMed.
- I. Szabo, M. Yousef, D. Soltesz, C. Bato, G. Mezo and Z. Banoczi, Pharmaceutics, 2022, 14, 907 CrossRef CAS PubMed.
- M. Zhou, X. Zou, K. Cheng, S. Zhong, Y. Su, T. Wu, Y. Tao, L. Cong, B. Yan and Y. Jiang, Clin. Transl. Med., 2022, 12, e822 CrossRef CAS PubMed.
- C. P. Cerrato, T. Kivijarvi, R. Tozzi, T. Lehto, M. Gestin and U. Langel, J. Mater. Chem. B, 2020, 8, 10825–10836 RSC.
- C. P. Cerrato and U. Langel, Expert Opin. Drug Delivery, 2022, 19, 133–146 CrossRef CAS PubMed.
- A. Gronewold, M. Horn and I. Neundorf, Beilstein J. Org. Chem., 2018, 14, 1378–1388 CrossRef CAS PubMed.
- M. J. Harrington, R. Mezzenga and A. Miserez, Nat. Rev. Bioeng., 2024, 2, 260–278 CrossRef.
- S. Mukherjee and L. V. Schafer, Nat. Commun., 2023, 14, 5892 CrossRef CAS PubMed.
- C. Yuan, Q. Li, R. Xing, J. Li and X. Yan, Chem, 2023, 9, 2425–2445 CAS.
- Z. W. Lim, Y. Ping and A. Miserez, Bioconjugate Chem., 2018, 29, 2176–2180 CrossRef CAS PubMed.
- B. Wang, L. Zhang, T. Dai, Z. Qin, H. Lu, L. Zhang and F. Zhou, Signal Transduction Targeted Ther., 2021, 6, 290 CrossRef PubMed.
- J. Wang, M. Abbas, Y. Huang, J. Wang and Y. Li, Commun. Chem., 2023, 6, 243 CrossRef CAS PubMed.
- Z. W. Lim, V. B. Varma, R. V. Ramanujan and A. Miserez, Acta Biomater., 2020, 110, 221–230 CrossRef CAS PubMed.
- Y. Sun, S. Y. Lau, Z. W. Lim, S. C. Chang, F. Ghadessy, A. Partridge and A. Miserez, Nat. Chem., 2022, 14, 274–283 CrossRef CAS PubMed.
- Y. Sun, X. Xu, L. Chen, W. L. Chew, Y. Ping and A. Miserez, ACS Nano, 2023, 17, 16597–16606 CrossRef CAS PubMed.
- T. Iwata, H. Hirose, K. Sakamoto, Y. Hirai, J. V. V. Arafiles, M. Akishiba, M. Imanishi and S. Futaki, Angew. Chem., Int. Ed., 2021, 60, 19804–19812 CrossRef CAS PubMed.
- K. Kamagata, A. Hando, M. Ariefai, N. Iwaki, S. Kanbayashi, R. Koike and K. Ikeda, Sci. Rep., 2023, 13, 5648 CrossRef CAS PubMed.
- L. Chen, Q. Zhang, X. Yuan, Y. Cao, Y. Yuan, H. Yin, X. Ding, Z. Zhu and S.-Z. Luo, Int. J. Biochem. Cell Biol., 2017, 83, 71–75 CrossRef CAS PubMed.
- M. Akishiba, T. Takeuchi, Y. Kawaguchi, K. Sakamoto, H. H. Yu, I. Nakase, T. Takatani-Nakase, F. Madani, A. Graslund and S. Futaki, Nat. Chem., 2017, 9, 751–761 CrossRef CAS PubMed.
- X. Liu, R. Cao, S. Wang, J. Jia and H. Fei, J. Med. Chem., 2016, 59, 5238–5247 CrossRef CAS PubMed.
- W. Li, F. Nicol and F. C. Szoka Jr., Adv. Drug Delivery Rev., 2004, 56, 967–985 CrossRef CAS PubMed.
- S. Kobayashi, I. Nakase, N. Kawabata, H. H. Yu, S. Pujals, M. Imanishi, E. Giralt and S. Futaki, Bioconjugate Chem., 2009, 20, 953–959 CrossRef CAS PubMed.
- X. Chen, S. Ji, A. Li, H. Liu and H. Fei, J. Med. Chem., 2020, 63, 5012 CrossRef CAS PubMed.
- X. Chen, H. Liu, A. Li, S. Ji and H. Fei, J. Biol. Chem., 2021, 297, 101364 CrossRef CAS PubMed.
- D. Kalafatovic and E. Giralt, Molecules, 2017, 22, 1929 CrossRef PubMed.
- E. N. Lorenzon, J. P. Piccoli, N. A. Santos-Filho and E. M. Cilli, Protein Pept. Lett., 2019, 26, 98–107 CrossRef CAS PubMed.
- Y. Nomura, K. Sakamoto, M. Akishiba, T. Iwata, H. Hirose and S. Futaki, Bioorg. Med. Chem. Lett., 2020, 30, 127362 CrossRef CAS PubMed.
- J. V. V. Arafiles, H. Hirose, M. Akishiba, S. Tsuji, M. Imanishi and S. Futaki, Bioconjugate Chem., 2020, 31, 547–553 CrossRef CAS PubMed.
- J. Uusna, K. Langel and U. Langel, Methods Mol. Biol., 2015, 1324, 133–148 CrossRef PubMed.
- W. Lohcharoenkal, L. Wang, Y. C. Chen and Y. Rojanasakul, BioMed. Res. Int., 2014, 2014, 180549 CrossRef PubMed.
- A. Maham, Z. Tang, H. Wu, J. Wang and Y. Lin, Small, 2009, 5, 1706–1721 CrossRef CAS PubMed.
- P. O. Falnes and K. Sandvig, Curr. Opin. Cell Biol., 2000, 20, 407–413 CrossRef PubMed.
- J. M. Williams and B. Tsai, Curr. Opin. Cell Biol., 2016, 41, 51–56 CrossRef CAS PubMed.
- N. L. B. Wernick, D. J.-F. Chinnapen, J. A. Cho, W. I. Lencer, N. L. B. Wernick, D. J.-F. Chinnapen, J. A. Cho and W. I. Lencer, Toxins, 2010, 2, 310–325 CrossRef CAS PubMed.
- X. F. Jia, Y. Zhang, T. Wang and Y. Fu, Mol. Pharm., 2021, 18, 4067–4078 CrossRef CAS PubMed.
- D. Liu, Y. F. Zhan, X. Y. Wu, H. P. Qiao, Y. L. Zhang and B. Li, Int. J. Biol. Macromol., 2023, 237, 124172 CrossRef CAS PubMed.
- L. Johannes, W. Römer, L. Johannes and W. Römer, Nat. Rev. Microbiol., 2009, 8, 105–116 CrossRef PubMed.
- J. Hadjerci, A. Billet, P. Kessler, G. Mourier, M. Ghazarian, A. Gonzalez, C. Wunder, N. Mabrouk, E. Tartour, D. Servent and L. Johannes, Cells, 2023, 12, 1291 CrossRef CAS PubMed.
- N. Danielewicz, F. Rosato, J. Tomisch, J. Gräber, B. Wiltschi, G. Striedner, W. Römer and J. Mairhofer, ACS Omega, 2023, 8, 15406–15421 CrossRef CAS PubMed.
- N. Danielewicz, F. Rosato, J. Tomisch, J. Gräber, B. Wiltschi, G. Striedner, W. Römer and J. Mairhofer, ACS Omega, 2023, 8, 15406–15421 CrossRef CAS PubMed.
- S. H. Tian, Y. Liu, E. Appleton, H. Wang, G. M. Church and M. Dong, Cell Rep., 2022, 38, 110476 CrossRef CAS PubMed.
- S.-I. Miyashita, J. Zhang, S. Zhang, C. B. Shoemaker and M. Dong, Sci. Transl. Med., 2021, 13, 575 Search PubMed.
- G. Huang, G. E. Besner, D. R. Brigstock, G. Huang, G. E. Besner and D. R. Brigstock, Lab. Invest., 2012, 92, 703–712 CrossRef CAS PubMed.
- R. G. Webster, W. J. Bean, O. T. Gorman, T. M. Chambers and Y. Kawaoka, Microbiol. Rev., 1992, 56, 152–179 CrossRef CAS PubMed.
- J. H. Park, A. Mohapatra, J. R. Zhou, M. Holay, N. Krishnan, W. W. Gao, R. H. Fang and L. F. Zhang, Angew. Chem., Int. Ed., 2022, 61, e202113671 CrossRef CAS PubMed.
- S. J. Gamblin, S. G. Vachieri, X. Xiong, J. Zhang, S. R. Martin and J. J. Skehel, Cold Spring Harbor Perspect. Med., 2021, 11, a038638 CrossRef CAS PubMed.
- X. Han, J. H. Bushweller, D. S. Cafiso, L. K. Tamm, X. Han, J. H. Bushweller, D. S. Cafiso and L. K. Tamm, Nat. Struct. Biol., 2001, 8, 8 CrossRef PubMed.
- W. T. Tang, T. Tong, H. Wang, X. H. Lu, C. P. Yang, Y. S. Wu, Y. Wang, J. B. Liu and B. Q. Ding, Angew. Chem., Int. Ed., 2023, 62, 51 Search PubMed.
- N. Z. Zheng, S. R. Liu, J. H. Chen, Y. Xu, W. Y. Cao, J. Y. Lin, G. Lu and G. G. Zhang, Mol. Pharm., 2024, 21, 1149–1159 CrossRef CAS PubMed.
- M. Gupta, C. M. Azumaya, M. Moritz, S. Pourmal, A. Diallo, G. E. Merz, G. Jang, M. Bouhaddou, A. Fossati, A. F. Brilot, D. Diwanji, E. Hernandez, N. Herrera, H. T. Kratochvil, V. L. Lam, F. Li, Y. Li, H. C. Nguyen, C. Nowotny, T. W. Owens, J. K. Peters, A. N. Rizo, U. Schulze-Gahmen, A. M. Smith, I. D. Young, Z. Yu, D. Asarnow, C. Billesbølle, M. G. Campbell, J. Chen, K.-H. Chen, U. S. Chio, M. S. Dickinson, L. Doan, M. Jin, K. Kim, J. Li, Y.-L. Li, E. Linossi, Y. Liu, M. Lo, J. Lopez, K. E. Lopez, A. Mancino, I. Frank, R. Moss, M. D. Paul, K. I. Pawar, A. Pelin, J. Thomas, H. Pospiech, C. Puchades, S. G. Remesh, M. Safari, K. Schaefer, M. Sun, M. C. Tabios, A. C. Thwin, E. W. Titus, R. Trenker, E. Tse, T. K. M. Tsui, F. Wang, K. Zhang, Y. Zhang, J. Zhao, F. Zhou, Y. Zhou, L. Zuliani-Alvarez, Q. S. B. Consortium, D. A. Agard, Y. Cheng, J. S. Fraser, N. Jura, T. Kortemme, A. Manglik, D. R. Southworth, R. M. Stroud, D. L. Swaney, N. J. Krogan, A. Frost, O. S. Rosenberg and K. A. Verba, Res. Square, 2021 DOI:10.21203/rs.3.rs-515215/v1.
- F. Krammer, G. J. D. Smith, R. A. M. Fouchier, M. Peiris, K. Kedzierska, P. C. Doherty, P. Palese, M. L. Shaw, J. Treanor, R. G. Webster and A. García-Sastre, Nat. Rev. Dis. Primers, 2018, 4, 3 CrossRef PubMed.
- Y. Li, Z. Xu, Q. Lei, D. Y. Lai, H. Hou, H. W. Jiang, Y. X. Zheng, X. N. Wang, J. Wu, M. L. Ma, B. Zhang, H. Chen, C. Yu, J. B. Xue, H. N. Zhang, H. Qi, S. J. Guo, Y. Zhang, X. Lin, Z. Yao, H. Sheng, Z. Sun, F. Wang, X. Fan and S. C. Tao, Cell Rep., 2021, 36, 2 Search PubMed.
- Q. Lei, C. Z. Yu, Y. Li, H. Y. Hou, Z. W. Xu, Z. J. Yao, Y. D. Zhang, D. Y. Lai, J. B. Ndzouboukou, B. Zhang, H. Chen, Z. Q. Ouyang, J. B. Xue, X. S. Lin, Y. X. Zheng, X. N. Wang, H. W. Jiang, H. N. Zhang, H. Qi, S. J. Guo, M. A. He, Z. Y. Sun, F. Wang, S. C. Tao and X. L. Fan, J. Adv. Res., 2022, 36, 133–145 CrossRef CAS PubMed.
- J. Utterström, S. Naeimipour, R. Selegård and D. Aili, Adv. Drug Delivery Rev., 2021, 170, 26–43 CrossRef PubMed.
- H. R. Marsden, N. A. Elbers, P. H. H. Bomans, N. A. J. M. Sommerdijk and A. Kros, Angew. Chem., Int. Ed., 2009, 48, 2330–2333 CrossRef PubMed.
- J. Yang, J. Tu, G. E. M. Lamers, R. C. L. Olsthoorn and A. Kros, Adv. Healthcare Mater., 2017, 6, 20 Search PubMed.
- W. Lv and J. A. Champion, Nanomedicine, 2021, 32, 102315 CrossRef CAS PubMed.
- A. Dhankher, W. Lv, W. T. Studstill and J. A. Champion, J. Controlled Release, 2021, 339, 248–258 CrossRef CAS PubMed.
- J. Li, J. Tuma, H. S. Han, H. Kim, R. C. Wilson, H. Y. Lee and N. Murthy, Adv. Healthcare Mater., 2022, 11, 9 Search PubMed.
- J. Seelig, Biochim. Biophys. Acta, Biomembr., 2004, 1666, 40–50 CrossRef CAS PubMed.
- N. Ben-Tal, A. Ben-Shaul, A. Nicholls and B. Honig, Biophys. J., 1996, 70, 1803–1812 CrossRef CAS PubMed.
- K. Takahashi, T. Nishiyama, N. Umezawa, Y. Inoue, I. Akiba, T. Dewa, A. Ikeda and T. Mizuno, Chem. Commun., 2024, 60, 968–971 RSC.
- D. Hoekstra, T. de Boer, K. Klappe and J. Wilschut, Biochemistry, 1984, 23, 5675–5681 CrossRef CAS PubMed.
- M. P. Stewart, A. Lorenz, J. Dahlman and G. Sahay, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2016, 8, 465–478 Search PubMed.
- J. Mueller, I. Kretzschmar, R. Volkmer and P. Boisguerin, Bioconjugate Chem., 2008, 19, 2363–2374 CrossRef CAS PubMed.
- C. Nie, Y. Zou, S. Liao, Q. Gao and Q. Li, Curr. Res. Food Sci., 2023, 7, 100592 CrossRef CAS PubMed.
- A. Egorova, S. Shtykalova, M. Maretina, A. Selutin, N. Shved, D. Deviatkin, S. Selkov, V. Baranov and A. Kiselev, Int. J. Mol. Sci., 2022, 23, 1164 CrossRef CAS PubMed.
- J. P. Richard, K. Melikov, E. Vives, C. Ramos, B. Verbeure, M. J. Gait, L. V. Chernomordik and B. Lebleu, J. Biol. Chem., 2003, 278, 585–590 CrossRef CAS PubMed.
- A. M. Moreno-Echeverri, E. Susnik, D. Vanhecke, P. Taladriz-Blanco, S. Balog, A. Petri-Fink and B. Rothen-Rutishauser, J. Nanobiotechnol., 2022, 20, 464 CrossRef CAS PubMed.
- S. L. Y. Teo, J. J. Rennick, D. Yuen, H. Al-Wassiti, A. P. R. Johnston and C. W. Pouton, Nat. Commun., 2021, 12, 3721 CrossRef CAS PubMed.
- J. S. Kim, D. K. Choi, S. W. Park, S. M. Shin, J. Bae, D. M. Kim, T. H. Yoo and Y. S. Kim, Biochem. Biophys. Res. Commun., 2015, 467, 771–777 CrossRef CAS PubMed.
- R. F. Wissner, A. Steinauer, S. L. Knox, A. D. Thompson and A. Schepartz, ACS Cent. Sci., 2018, 4, 1379–1393 CrossRef CAS PubMed.
- C. Lozada, T. M. A. Barlow, S. Gonzalez, N. Lubin-Germain and S. Ballet, Front. Chem., 2021, 9, 689006 CrossRef CAS PubMed.
- M. S. Freitas, L. P. Gaspar, M. Lorenzoni, F. C. L. Almeida, L. W. Tinoco, M. S. Almeida, L. F. Maia, L. Degreve, A. P. Valente and J. L. Silva, J. Biol. Chem., 2007, 282, 27306–27314 CrossRef CAS PubMed.
- Y. J. Lee, G. Johnson and J. P. Pellois, Biochemistry, 2010, 49, 7854–7866 CrossRef CAS PubMed.
- K. A. Whitehead, J. Matthews, P. H. Chang, F. Niroui, J. R. Dorkin, M. Severgnini and D. G. Anderson, ACS Nano, 2012, 6, 6922–6929 CrossRef CAS PubMed.
- K. Paunovska, C. D. Sago, C. M. Monaco, W. H. Hudson, M. G. Castro, T. G. Rudoltz, S. Kalathoor, D. A. Vanover, P. J. Santangelo, R. Ahmed, A. V. Bryksin and J. E. Dahlman, Nano Lett., 2018, 18, 2148–2157 CrossRef CAS PubMed.
Footnote |
| † These two authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |



