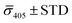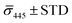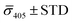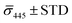Open Access Article
Open Access ArticleTwo-color interferometric scattering (iSCAT) microscopy reveals structural dynamics in discrete plasmonic molecules†
Leslie
Velasco
 ,
Aniqa N.
Islam
,
Koustav
Kundu
,
Aniqa N.
Islam
,
Koustav
Kundu
 ,
Aidan
Oi
,
Aidan
Oi
 and
Björn M.
Reinhard
and
Björn M.
Reinhard

Department of Chemistry and The Photonics Center, Boston University, Boston, MA 02459, USA. E-mail: bmr@bu.edu
First published on 3rd June 2024
Abstract
Plasmonic molecules are discrete assemblies of noble metal nanoparticles (NPs) that are of interest as transducers in optical nanosensors. So far, NPs with diameters of ∼40 nm have been the preferred building blocks for plasmonic molecules intended as optical single molecule sensors due to difficulties associated with detecting smaller NPs through elastic scattering in conventional darkfield microscopy. Here, we apply 405 nm, 445 nm two-color interferometric scattering (iSCAT) microscopy to characterize polyethylene glycol (PEG) tethered dimers of 10 nm and 20 nm Ag NPs and their monomers. Dimers of both NP sizes can be discerned from their respective monomers through changes in the average iSCAT contrast. In the case of 20 nm Ag NPs, dimer formation induces a change in the sign of the iSCAT contrast, providing a characteristic signal for detecting binding events. 20 nm Ag NP dimers with 0.4 kDa and 3.4 kDa polyethylene glycol (PEG) spacers show iSCAT contrast distributions with significantly different averages on both wavelength channels. The iSCAT contrast measured for individual PEG-tethered 10 nm or 20 nm NP dimers as a function of time shows contrast fluctuations indicative of a rich structural dynamics in the assembled plasmonic molecules, which provides an additional metric to discern dimers from monomers and paves the path to a new class of interferometric plasmon rulers.
Detecting and monitoring nanoscale contacts is crucial in many areas of chemistry and biology for diverse applications such as detecting ligand – receptor binding events, unravelling the complex working principles of molecular machines, or elucidating spatial regulation mechanisms of cellular processes, to name only a few. There has consequently been significant interest in developing optical assays and strategies for detecting and quantifying sub-diffraction limit contacts under physiological conditions. The developed strategies include fluorescence resonance energy transfer (FRET),1,2 dual color optical colocalization,3,4 and fluorescence superresolution microscopy.5–7 The fluorescent probes that are commonly applied in these studies have limitations regarding their photophysical stability,8 and blinking and bleaching can disrupt sensitive measurements especially at the single molecule level. Noble metal nanoparticles sustain localized surface plasmon resonances and provide strong scattering signals and are, thus, potentially alternative probes for applications that require long continuous observation times and/or high acquisition rates.9 The scattering cross-sections and resonance wavelengths of NPs in discrete plasmonic molecules are modulated through distance-dependent plasmon coupling between the NPs, and this effect has been utilized to generate dynamic plasmonic rulers for nanoscale distance measurements.10,11 Previous plasmon ruler12–14 and related plasmon coupling microscopy (PCM)15,16 strategies primarily relied on darkfield microscopy17,18 for the optical detection and spectral analysis of NP dimers and discrete clusters. The scattering cross-section of NPs is strongly size-dependent, and one limitation of conventional darkfield microscopy, which relies on an oblique excitation and detection of scattered light by a low numerical aperture objective, is that it requires at least one NP with a diameter of approximately 40 nm or larger for a reliable detection. This size requirement is an impediment for some applications. Fortunately, the sensitivity of darkfield detection can be improved through enhanced discrimination between scattered and excitation light beams,19 and alternative interferometric detection approaches offer greatly enhanced sensitivity for the optical detection of small scatterers.20,21 Interferometric scattering (iSCAT) microscopy, in particular, has demonstrated impressive detection sensitivities for immobilized individual biomolecules and NPs as small as 2 nm.22–28 iSCAT has also been demonstrated to allow distinguishing between different sizes of NPs,20,29,30 molecules,31–33 and molecular assemblies34,35 based on their iSCAT contrast. iSCAT in combination with a phase-sensitive detection has also shown promise to resolve sub-diffraction limit contacts between NPs with separations down to about 150 nm.36 All of these properties make iSCAT a promising strategy to investigate the assembly of sub-40 nm NP into plasmonic molecules and monitor separations within these assemblies on chemical length scales.
In this study, we characterize 10 nm and 20 nm Ag NP monomers and in situ assembled dimers using a two-color iSCAT microscope37 and evaluate the effect of dimerization on the iSCAT signals. We demonstrate that iSCAT imaging does not only facilitate distinction between monomers and dimers but also provides information about the structural dynamics of the assembled dimers.
Results and discussion
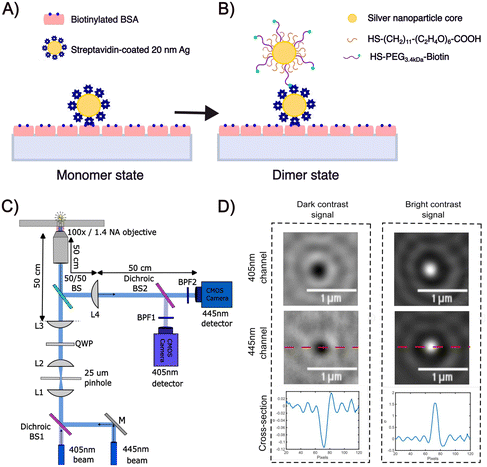
The experimental strategy for assembling dimers of Ag NPs using a biotin/streptavidin chemistry is illustrated in Fig. 1A and B.10 After immobilizing 10 nm or 20 nm diameter streptavidin-coated NPs (NP-STV) on a BSA–biotin functionalized glass surface (Fig. 1A), a second “flavor” of 10 nm or 20 nm NPs functionalized with biotinylated polyethylene glycols (PEGs) is added to bind to the immobilized NP–STV (Fig. 1B). We used Ag NPs that were functionalized with HS–PEG3.4 kDa–biotin and HS–PEG0.4 kDa–COOH in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000. We refer to these NPs in the following as NP–PEG3.4 kDa–biotin. In some experiments HS–PEG3.4 kDa–biotin was replaced with a shorter HS–PEG0.4 kDa–biotin. The contour lengths of the 0.4 kDa, 3.4 kDa PEGs are 2.43 nm and 21.50 nm, respectively, and the predicted worm-like-chain38 end-to-end distances are 1.36 nm and 4.04 nm.39 The length of the PEG tethers together with the diameter of the streptavidin (approx. 5 nm)40 defines the average interparticle separation between the NPs.
1000. We refer to these NPs in the following as NP–PEG3.4 kDa–biotin. In some experiments HS–PEG3.4 kDa–biotin was replaced with a shorter HS–PEG0.4 kDa–biotin. The contour lengths of the 0.4 kDa, 3.4 kDa PEGs are 2.43 nm and 21.50 nm, respectively, and the predicted worm-like-chain38 end-to-end distances are 1.36 nm and 4.04 nm.39 The length of the PEG tethers together with the diameter of the streptavidin (approx. 5 nm)40 defines the average interparticle separation between the NPs.
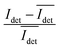
We determined the iSCAT contrast of surface-tethered NP–STV before and after addition of the biotin-functionalized NPs assembled simultaneously on two wavelength channels (405 nm, 445 nm) in a homebuilt iSCAT microscope (Fig. 1C). In iSCAT the signal of a scatterer immobilized on a substrate is generated through interference of the light scattered from the NP with the light reflected at the interface between the substrate and the ambient medium. The acquired iSCAT signal at the detector has contributions from reference beam, NP scattering, and the interference of both:23
Idet ∝ |Einc|2 (r2 + |s|2 + 2r|s|cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ϕ) ϕ) | (1) |
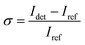 | (2) |
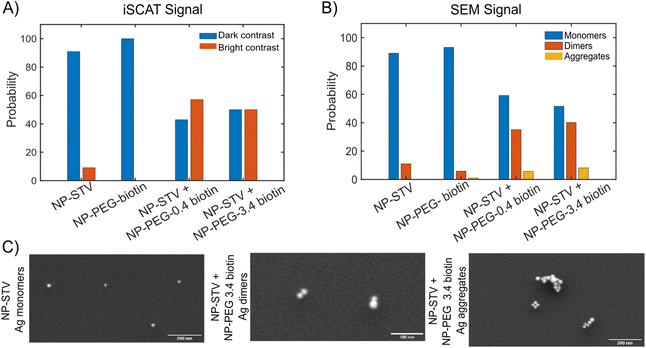
We utilized a two-color iSCAT microscope in this work as it provides some basic spectral information by simultaneously monitoring the iSCAT contrast at 405 nm and 445 nm (σ405, σ445).37 20 nm Ag NPs immobilized on a glass substrate yield negative σ405, σ445 values.37 However, after incubating 20 nm NP–PEG3.4 kDa–biotin with immobilized 20 nm NP–STV we observed two different types of scatterers with either a “dark” (i.e. σ405, σ445 are negative) or “bright” (i.e. σ405, σ445 are positive) iSCAT contrast. Fig. 1D shows representative iSCAT images for both types of scatterers on the two monitored wavelength channels. The peak contrast of the interferograms were determined by 2D Gaussian fits (Fig. S1†). Under the chosen experimental conditions (see Methods for details), the ratio of dark: bright scatterers was approximately 50%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50% (Fig. 2A). Incubation of 20 nm NP–PEG0.4 kDa–biotin with immobilized 20 nm NP–STV resulted in a ratio closer to 40%
50% (Fig. 2A). Incubation of 20 nm NP–PEG0.4 kDa–biotin with immobilized 20 nm NP–STV resulted in a ratio closer to 40%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60%. We independently imaged 20 nm NP–STV and NP–PEG3.4 kDa–biotin (immobilized on BSA–biotin functionalized substrate with free STV) controls (Fig. 2A). The biotinylated NPs exclusively had a dark contrast, and the NP–STV histogram indicates only a small fraction of scatterers with bright contrast. Furthermore, a close inspection of the recorded movies revealed that these bright signals were not immobilized on the surface but quickly diffused out of the field of view after imaging. Nevertheless, they were included in our statistics. Average and standard deviation for contrast values on both wavelength channels for experimental controls were:
60%. We independently imaged 20 nm NP–STV and NP–PEG3.4 kDa–biotin (immobilized on BSA–biotin functionalized substrate with free STV) controls (Fig. 2A). The biotinylated NPs exclusively had a dark contrast, and the NP–STV histogram indicates only a small fraction of scatterers with bright contrast. Furthermore, a close inspection of the recorded movies revealed that these bright signals were not immobilized on the surface but quickly diffused out of the field of view after imaging. Nevertheless, they were included in our statistics. Average and standard deviation for contrast values on both wavelength channels for experimental controls were: 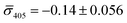 ,
, 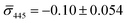 for NP–PEG3.4 kDa–biotin and
for NP–PEG3.4 kDa–biotin and 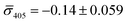 ,
, 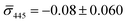 for NP–STV (Fig. S2†). Overall, the much higher probability for bright scatterers after incubation of NP–PEG–biotin with surface-immobilized NP–STV confirm binding between the two flavors of NPs as the cause for the observed contrast change. The change in contrast signal between a monomer and dimer is illustrated in Fig. S3.†
for NP–STV (Fig. S2†). Overall, the much higher probability for bright scatterers after incubation of NP–PEG–biotin with surface-immobilized NP–STV confirm binding between the two flavors of NPs as the cause for the observed contrast change. The change in contrast signal between a monomer and dimer is illustrated in Fig. S3.†
To further characterize the assembled NP structures, we imaged 20 nm NP–STV before and after incubation with 20 nm NP–PEG–biotin in the scanning electron microscope (SEM) (Fig. S4 and S5†). These experiments were performed with NP concentrations that were a factor of 100 higher than in the optical experiments to facilitate sufficient NP counts at the much higher magnification of the SEM. The size histograms in Fig. 2B confirm that NP–STV and NP–PEG3.4 kDa–biotin samples are primarily made up of monomers. NP–STV samples contained 89.0% monomers vs. 11.0% dimers in a population of 100 particles and NP–PEG3.4 kDa–biotin samples contained 93.1% monomers, 5.9% dimers, and 1% aggregates in a population of 102 particles. In comparison, the co-incubated sample NP–PEG3.4 kDa–biotin + NP-STV contains monomers (51.6%) and an increased fraction of dimers (40.2%), in addition to some aggregates (8.2%) in a total population of 122 particles. For the NP–PEG0.4 kDa–biotin + NP–STV samples the fractions of monomer, dimer, and aggregates were 59.2%, 35.1%, 5.7% for a total of 174 particles (Fig. S6†). The ratio of monomers to plasmonic molecules of approximately 50%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50% and 60%
50% and 60%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40% for the co-incubation experiments with 3.4 kDa and 0.4 kDa PEG spacers confirms that the contrast change in the iSCAT experiments is the result of the formation of plasmonic molecules. Fig. 2C shows characteristic SEM images of monomers, dimers and clusters. The SEM data confirm dimers as primary assembly product. Given the higher NP concentrations used to generate the sample for the SEM studies, it is likely that the relative contribution of aggregates is even lower under the conditions of the optical iSCAT experiments. SEM images of the NP–PEG–biotin particles used for iSCAT experiments and for validation experiments are shown in Fig. S7.† The successful assembly of dimers under the outlined experimental conditions was further validated by formation of heterodimers as seen in Fig. S8.†
40% for the co-incubation experiments with 3.4 kDa and 0.4 kDa PEG spacers confirms that the contrast change in the iSCAT experiments is the result of the formation of plasmonic molecules. Fig. 2C shows characteristic SEM images of monomers, dimers and clusters. The SEM data confirm dimers as primary assembly product. Given the higher NP concentrations used to generate the sample for the SEM studies, it is likely that the relative contribution of aggregates is even lower under the conditions of the optical iSCAT experiments. SEM images of the NP–PEG–biotin particles used for iSCAT experiments and for validation experiments are shown in Fig. S7.† The successful assembly of dimers under the outlined experimental conditions was further validated by formation of heterodimers as seen in Fig. S8.†
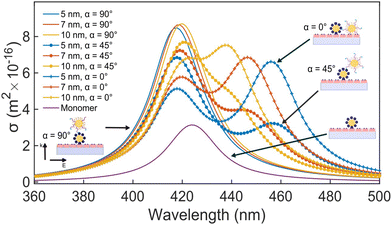
For an individual 20 nm Ag NP monomer, the iSCAT contrast σ as defined by eqn (2) is determined by the interference term 2r|s|cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ϕ as the scattering amplitude for 20 nm monomers is small. The phase ϕ has contributions from the wavelength-dependent phase shift associated with the plasmon (ϕP) as well as a Gouy phase (ϕG) shift, which corresponds to a phase lag of π/2 for light scattered off individual NPs.42 If the NP is modelled as a damped driven harmonic oscillator, the electron oscillation is in phase with the incident light at low frequencies but develops a phase lag of π as the wavelength is scanned across the resonance with a phase shift of π/2 at the resonance frequency. The wavelengths chosen in our two-color iSCAT microscope, 405 nm and 445 nm, overlap with the low and high energy tail of the monomer resonance (Fig. 3). This simple model predicts that ϕP and ϕG give rise to a combined phase lag Δϕ with
ϕ as the scattering amplitude for 20 nm monomers is small. The phase ϕ has contributions from the wavelength-dependent phase shift associated with the plasmon (ϕP) as well as a Gouy phase (ϕG) shift, which corresponds to a phase lag of π/2 for light scattered off individual NPs.42 If the NP is modelled as a damped driven harmonic oscillator, the electron oscillation is in phase with the incident light at low frequencies but develops a phase lag of π as the wavelength is scanned across the resonance with a phase shift of π/2 at the resonance frequency. The wavelengths chosen in our two-color iSCAT microscope, 405 nm and 445 nm, overlap with the low and high energy tail of the monomer resonance (Fig. 3). This simple model predicts that ϕP and ϕG give rise to a combined phase lag Δϕ with 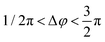 at 405 nm and 445 nm and accounts for negative σ405, σ445 values for the 20 nm monomers.43 Dimer formation by binding of NP–PEG3.4 kDa–biotin to an immobilized Ag–STV increases the scattering amplitude and the contribution from |s|2 to σ in eqn (1). This is illustrated by finite difference time (FDTD) simulations of the scattering spectra of a Ag NP monomer and selected dimer geometries in Fig. 3. We attribute the experimentally observed inversion of the sign of the σ405, σ445 contrast for Ag NP dimers to a sufficiently strong increase in the positive scattering term |s|2 so that it exceeds the absolute value of the negative interference term 2r|s|cos
at 405 nm and 445 nm and accounts for negative σ405, σ445 values for the 20 nm monomers.43 Dimer formation by binding of NP–PEG3.4 kDa–biotin to an immobilized Ag–STV increases the scattering amplitude and the contribution from |s|2 to σ in eqn (1). This is illustrated by finite difference time (FDTD) simulations of the scattering spectra of a Ag NP monomer and selected dimer geometries in Fig. 3. We attribute the experimentally observed inversion of the sign of the σ405, σ445 contrast for Ag NP dimers to a sufficiently strong increase in the positive scattering term |s|2 so that it exceeds the absolute value of the negative interference term 2r|s|cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ϕ. This conclusion is supported by iSCAT experiments with 30 nm diameter Ag NPs whose volume is 1.4 × 104 nm3 compared to 8.4 × 103 nm3 for the 20 nm Ag NP dimers. The slightly larger NPs serve as a model for two coupled 20 nm Ag NPs. The 30 nm Ag NPs have positive iSCAT contrast on both wavelength channels, and the σ405, σ445 distribution overlaps with that of the 20 nm Ag NP dimers (Fig. S9†).
ϕ. This conclusion is supported by iSCAT experiments with 30 nm diameter Ag NPs whose volume is 1.4 × 104 nm3 compared to 8.4 × 103 nm3 for the 20 nm Ag NP dimers. The slightly larger NPs serve as a model for two coupled 20 nm Ag NPs. The 30 nm Ag NPs have positive iSCAT contrast on both wavelength channels, and the σ405, σ445 distribution overlaps with that of the 20 nm Ag NP dimers (Fig. S9†).
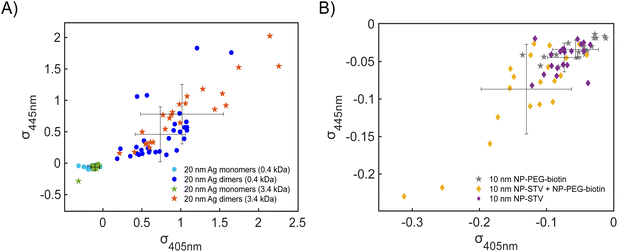
Fig. 4A contains a scatterplot of σ445vs. σ405 measured for 114 individual scatterers after incubation of 20 nm NP–STV with 20 nm NP–PEG3.4 kDa–biotin collected in three experiments. The plot also contains data obtained in binding experiments in which the HS–PEG3.4 kDa–biotin tether was replaced with the shorter HS–PEG0.4 kDa–biotin molecule. The measured data for both spacer lengths cluster in two clearly separated regions of the σ405, σ445 plane with either (i) σ405 and σ445 < 0 or (ii) σ405 and σ445 > 0, consistent with their assignments as monomers and dimers, respectively. Table 1 contains the average 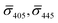 values (±STD) for the different conditions observed for 20 nm NP–STV and NP–PEG–biotin tethered particles. The σ405, σ445 distributions of the monomer subpopulations (σ < 0) with HS–PEG0.4 kDa–biotin and HS–PEG3.4 kDa–biotin ligands show no significant differences, but for the dimer subpopulations (σ > 0) a Student's t-test indicates distributions with different averages for both
values (±STD) for the different conditions observed for 20 nm NP–STV and NP–PEG–biotin tethered particles. The σ405, σ445 distributions of the monomer subpopulations (σ < 0) with HS–PEG0.4 kDa–biotin and HS–PEG3.4 kDa–biotin ligands show no significant differences, but for the dimer subpopulations (σ > 0) a Student's t-test indicates distributions with different averages for both 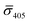 and
and 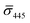 at the p < 0.05 level. The differences in the σ405, σ445 distribution for dimers assembled with different PEG spacers underlines the potential of iSCAT for distinguishing plasmonic assemblies with different tether molecules. For both PEG spacers the σ405, σ445 distributions of the dimer sub-populations are much broader than their counterparts for the monomers. This broadening can have different causes. For one, the assembly strategy does not exclusively form dimers. Contributions from trimers and larger clusters will broaden the σ405, σ445 distributions (Fig. S10†). However, as outlined above in our discussion of Fig. 2A and B the number of clusters formed under our experimental conditions is low, so that cluster formation is not sufficient to account for the observed spread. Another factor that contributes to the spread of the measured σ405, σ445 values is related to the conformational variability of the formed dimers. The spectral response of a dimer depends both on the orientation of the dimer axis relative to the incident E-field and the separation between the NPs (Fig. 3), and these factors can differ between individual dimers. The orientation of the dimer axis also affects the detected scattering signal by determining the direction of the emitted light. These structural degrees of freedom are unique to dimers and can contribute to a broader spread of the iSCAT signal detected for dimers when compared to that of the monomers. The two-color iSCAT detection is useful to detect these differences between dimers. For instance, the 400 Da PEG tethered dimers show in general a much larger variability along σ405 for σ405 < 1 only to increase along σ445 for σ405 ≥ 1. These details would not be resolved with a single-color detection.
at the p < 0.05 level. The differences in the σ405, σ445 distribution for dimers assembled with different PEG spacers underlines the potential of iSCAT for distinguishing plasmonic assemblies with different tether molecules. For both PEG spacers the σ405, σ445 distributions of the dimer sub-populations are much broader than their counterparts for the monomers. This broadening can have different causes. For one, the assembly strategy does not exclusively form dimers. Contributions from trimers and larger clusters will broaden the σ405, σ445 distributions (Fig. S10†). However, as outlined above in our discussion of Fig. 2A and B the number of clusters formed under our experimental conditions is low, so that cluster formation is not sufficient to account for the observed spread. Another factor that contributes to the spread of the measured σ405, σ445 values is related to the conformational variability of the formed dimers. The spectral response of a dimer depends both on the orientation of the dimer axis relative to the incident E-field and the separation between the NPs (Fig. 3), and these factors can differ between individual dimers. The orientation of the dimer axis also affects the detected scattering signal by determining the direction of the emitted light. These structural degrees of freedom are unique to dimers and can contribute to a broader spread of the iSCAT signal detected for dimers when compared to that of the monomers. The two-color iSCAT detection is useful to detect these differences between dimers. For instance, the 400 Da PEG tethered dimers show in general a much larger variability along σ405 for σ405 < 1 only to increase along σ445 for σ405 ≥ 1. These details would not be resolved with a single-color detection.
| Condition | ||
|---|---|---|
| 20 nm NP–STV | −0.14 ± 0.059 | −0.08 ± 0.060 |
| 20 nm NP–PEG3.4 kDa–biotin | −0.14 ± 0.056 | −0.10 ± 0.054 |
| 20 nm NP–STV + NP–PEG0.4 kDa–biotin monomers | −0.13 ± 0.063 | −0.05 ± 0.019 |
| 20 nm NP–STV + NP–PEG0.4 kDa–biotin dimers | 0.74 ± 0.322 | 0.46 ± 0.437 |
| 20 nm NP–STV + NP–PEG3.4 kDa–biotin monomers | −0.10 ± 0.053 | −0.06 ± 0.051 |
| 20 nm NP–STV + NP–PEG3.4 kDa–biotin dimers | 1.02 ± 0.533 | 0.78 ± 0.473 |
So far, our analysis has been focused on dimers and monomers of 20 nm Ag NPs. We performed similar iSCAT imaging experiments with 10 nm NP–STV before and after incubation with 10 nm NP–PEG3.4 kDa–biotin. The σ405, σ445 values are summarized in Fig. 4B. The average 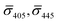 values (±STD) for the 10 nm NP–STV imaged before and after addition of NP–PEG3.4 kDa–biotin as well as the average contrast values for bare NP–PEG3.4 kDa–biotin are summarized in Table 2. Unlike for the 20 nm NPs, dimer formation does not result in a change of the sign of the iSCAT contrast (Fig. 4B and S11†), as the interference term in eqn (1) remains the dominating factor for the smaller NPs. Instead, the distribution of the iSCAT contrast values increases in magnitude and becomes more negative after incubation with NP–PEG3.4 kDa–biotin. A Student's t-test reveals significant differences at the p < 0.007 level for both
values (±STD) for the 10 nm NP–STV imaged before and after addition of NP–PEG3.4 kDa–biotin as well as the average contrast values for bare NP–PEG3.4 kDa–biotin are summarized in Table 2. Unlike for the 20 nm NPs, dimer formation does not result in a change of the sign of the iSCAT contrast (Fig. 4B and S11†), as the interference term in eqn (1) remains the dominating factor for the smaller NPs. Instead, the distribution of the iSCAT contrast values increases in magnitude and becomes more negative after incubation with NP–PEG3.4 kDa–biotin. A Student's t-test reveals significant differences at the p < 0.007 level for both 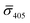 and
and 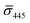 when comparing NP–STV and NP–PEG–biotin to NP–STV + NP–PEG3.4 kDa–biotin. Although the 10 nm Ag NP lack the characteristic change in sign of the iSCAT contrast observed for the 20 nm Ag NPs, the significant differences in the iSCAT contrast distributions indicates that it is still possible to discern monomers from dimers and larger plasmonic molecules at the ensemble level.
when comparing NP–STV and NP–PEG–biotin to NP–STV + NP–PEG3.4 kDa–biotin. Although the 10 nm Ag NP lack the characteristic change in sign of the iSCAT contrast observed for the 20 nm Ag NPs, the significant differences in the iSCAT contrast distributions indicates that it is still possible to discern monomers from dimers and larger plasmonic molecules at the ensemble level.
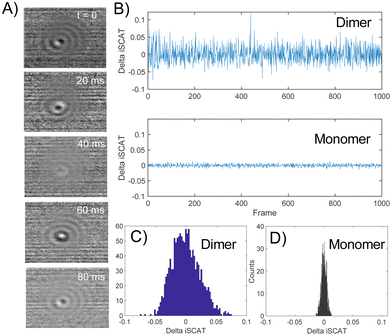
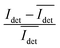
The spread of the iSCAT signals observed for tethered Ag NPs in Fig. 4 has been attributed to variations in the orientation and interparticle separation of the dimer and the orientation of the dimer axis relative to the incident E-field. The structural dependence of the iSCAT signal provides opportunities for monitoring conformational fluctuations in individual polymer tethered NP dimers as function of time. This is demonstrated in Fig. 5A where we plot the normalized iSCAT signal difference, 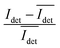 , recorded on the 405 nm channel for one dimer assembled by binding of NP–PEG3.4 kDa–biotin (20 nm) to surface immobilized NP–STV (20 nm). The iSCAT images were recorded with a frame rate of 500 frames per second (fps). By subtracting the moving average,
, recorded on the 405 nm channel for one dimer assembled by binding of NP–PEG3.4 kDa–biotin (20 nm) to surface immobilized NP–STV (20 nm). The iSCAT images were recorded with a frame rate of 500 frames per second (fps). By subtracting the moving average, 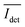 , calculated over an interval of ±10 frames around each time point from the detected iSCAT signal for each pixel in each frame, Idet, the calculated difference images highlight strong changes in the iSCAT signal even between subsequent frames. Fig. 5B contains trajectories of contrast dimer (top) and a monomer (bottom) measured in the center of the respective interferogram over a total of 1000 frames. Fig. 5C and D contain the corresponding histograms of the normalized intensity difference for dimer and monomer. While the iSCAT signal difference of the monomer is narrowly distributed around
, calculated over an interval of ±10 frames around each time point from the detected iSCAT signal for each pixel in each frame, Idet, the calculated difference images highlight strong changes in the iSCAT signal even between subsequent frames. Fig. 5B contains trajectories of contrast dimer (top) and a monomer (bottom) measured in the center of the respective interferogram over a total of 1000 frames. Fig. 5C and D contain the corresponding histograms of the normalized intensity difference for dimer and monomer. While the iSCAT signal difference of the monomer is narrowly distributed around 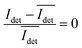 , the iSCAT signal for the dimer shows a much broader distribution as result of both positive and negative fluctuations of the iSCAT signal. The NP dimers investigated in this work are immobilized on the surface with only one NP, while the second can undergo a tethered particle motion (Fig. 1B). This motion continuously reconfigures the separation and relative orientation of the tethered NPs and results in fluctuations of the detected iSCAT signal.
, the iSCAT signal for the dimer shows a much broader distribution as result of both positive and negative fluctuations of the iSCAT signal. The NP dimers investigated in this work are immobilized on the surface with only one NP, while the second can undergo a tethered particle motion (Fig. 1B). This motion continuously reconfigures the separation and relative orientation of the tethered NPs and results in fluctuations of the detected iSCAT signal.
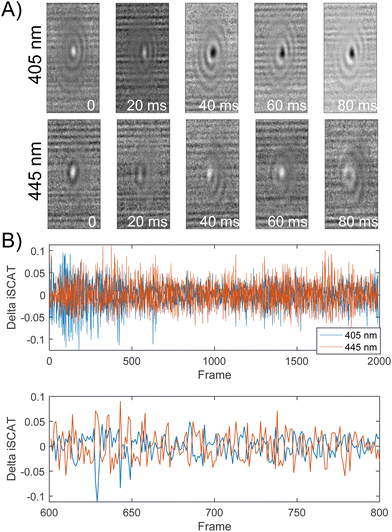
Importantly, the ability to detect conformational dynamics of PEG-tethered NPs is not limited to 20 nm probes. Fig. 6A and B contains selected normalized iSCAT signal difference images and the signal difference trajectories recorded simultaneously on σ405, σ445 channels for a 10 nm NP dimer. Similarly, as for the 20 nm Ag NP dimers, the iSCAT signal of the 10 nm Ag NP dimer shows a high degree of fluctuations indicative of a rich structural dynamics. Intriguingly, the 10 nm dimer signal on the two channels shows noticeable levels of anticorrelation. This is demonstrated in the magnified section of the dimer trajectory included in Fig. 6B. The correlation coefficient for this section is −0.6140. FDTD simulations (Fig. S12†) equivalent to the ones shown above for 20 nm Ag NP dimers indicate that the anticorrelated contrast changes at 405 nm and 445 nm could, for example arise from spectral shifts associated with a change in the elevation (polar orientation) of the 10 nm Ag NP dimer axis or from changes in the interparticle separation of a dimer with an inclined dimer axis.
In conclusion, we have characterized the iSCAT contrast of plasmonic molecules (in particular dimers) assembled from 10 nm and 20 nm Ag NPs in comparison to their respective monomer building blocks. Shifts in the average iSCAT contrast on the two monitored detection channels and differences in the magnitude of the iSCAT signal fluctuations of individual scatterers as function of time were shown to facilitate a distinction between monomers and dimers. For 10 nm Ag NPs, dimer formation resulted in a shift and broadening of the contrast distribution. In the case of 20 nm Ag NPs dimer formation also resulted in a drastic change in the sign of the iSCAT contrast. The differences in iSCAT contrast between 20 nm Ag NP dimers containing molecular tethers with different lengths (PEG0.4 kDavs. PEG3.4 kDa) were more subtle but still detectable. Importantly, the time-dependent fluctuation of the iSCAT signal detected here for dimers of 10 nm or 20 nm Ag NPs tethered by a PEG3.4 kDa molecule with a contour length of 21.50 nm underlines the potential of plasmon rulers in combination with iSCAT detection (interferometric plasmon rulers) to characterize structural dynamics in polymer tethers on nm length scales using NPs with much smaller diameters than required in conventional plasmon ruler or tethered bead assays. The decrease in probe size enables to minimize excluded volume effects in tethered bead assays with short tether molecules whose mechanochemical properties still raise many questions.44
Methods
NP–STV preparation
Commercial citrated-stabilized Ag colloidal solution (Nanocomposix) of concentration 1011 nanoparticles per mL was used for streptavidin coating protocols as described in literature.10 For preparation of NP–STV, 20 mL colloidal solution was combined with 500 μL of streptavidin (1 mg mL−1). Following this, 200 μL of sodium bicarbonate (1 M) was added to the solution. This combined mixture was stirred for 10 minutes and cleaned two times by centrifugation with 100 kDa centrifugal filters (5 krpm, 1 minute) and washed with 20 mM NaCl/5 mM Tris pH 7 buffer. NP–STV pellets were collected by centrifugation and stored in 20 mM NaCl/5 mM Tris pH 7 buffer for stabilization.NP–PEG–biotin preparation
20 nm citrate-stabilized Ag NPs (Nanocomposix) of concentration 1011 nanoparticles per mL were functionalized using two thiolated polyethylene glycol (PEG) molecules. A short carboxylic acid terminated PEG, HS-(CH2)11–(C2H4O)6–COOH (ProChimiaSurfaces), was added to 4 mL of 20 nm Ag NP colloidal solution to a final concentration of 5 uM. The second PEG added to this mixture was a HS–PEG–biotin molecule with a final concentration of 5 nM. The PEG had a molecular weight of either 3.4 kDa, for the long tether structure, or 0.4 kDa, for the short tether. Once combined, the mixture was incubated for 5 hours in the dark and cleaned two times by centrifugation (10.5 krpm, 30 min) and washed with distilled water. Once thoroughly cleaned and recovered a clear supernatant, the pellets were collected and stored in water.Monitoring NP dimer assembly with two-color iSCAT microscopy
The two-color wide-field iSCAT setup shown in Fig. 1C has previously been described in ref. 37. The NPs were illuminated with circularly polarized light using a total power density of 6 kW cm−2 in the sample plane. The assembly of NP dimers was monitored in flow chambers assembled using a rectangular fused silica capillary (VitroTubes) and polyethylene microtubing (Scientific Commodities). The chambers used for this work were treated with 1 mg mL−1 BSA–biotin in T50 buffer (10 mM Tris pH 8, 50 mM NaCl, Millipore water) for 10 minutes followed by a washing step with T50. Then NP–STV were flushed in at a concentration of 109 NPs mL−1. Subsequently, the chamber was washed with T50 buffer and then incubated with biotin–PEG–Ag NPs at a concentration of 109 NPs mL−1 for 10 additional minutes. At the end of this incubation, T50 buffer was flushed through the chamber by gravity flow for the duration of the experiment. For control experiments, a similar protocol was followed except for the imaging of NP–PEG3.4 kDa–biotin. In this case, the chamber was treated with BSA–biotin for 10 minutes, washed with T50, and then treated for an additional 10 minutes of 0.01 mg mL−1, filtered streptavidin solution. NP–PEG3.4 kDa–biotin were introduced into the system by gravity flow, and after 10 minutes, T50 buffer was flown through the chamber until the end of the experiment. Images were recorded at 500 frames per s at a set exposure time of 0.800 ms. The chamber was mounted on a nanopositioner stage (PiezosystemJena) where a frequency of 10 Hz was applied at 1.5 μm during data acquisition (except when iSCAT intensity fluctuations were measured). Unless otherwise noted, data analysis follows procedures described in ref. 37. In a typical experiment, the NPs were illuminated for no longer than 10–30 s. SEM images collected before/after illumination did not indicate any systematic size changes due to illumination (Fig. S13†).NP characterization
NP concentrations were determined using known extinction coefficient by UV-Vis. Scanning electron microscope (SEM) samples were prepared similarly as described for optical iSCAT experiments but on a 0.5 × 0.5 cm2 silicon wafer at a concentration of 1011 particles per mL for both anchor and tethered particles In the SEM, the stage was tilted by 30-degree to observe the overhead stacking of particles on top of each other on the flat silicon-chip surface.Author contributions
BMR and LV conceived the idea and designed the experiments. LV and AI performed the measurements and data analysis. KK and AO contributed analysis technologies and reagents. BMR and LV wrote the manuscript with input from all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
BMR acknowledges support from the National Institutes of Health through grants R21GM143672 and R01CA138509.References
- R. Roy, S. Hohng and T. Ha, A Practical Guide to Single-Molecule FRET, Nat. Methods, 2008, 5(6), 507–516, DOI:10.1038/nmeth.1208.
- E. Lerner, A. Barth, J. Hendrix, B. Ambrose, V. Birkedal, S. C. Blanchard, R. Börner, H. Sung Chung, T. Cordes, T. D. Craggs, A. A. Deniz, J. Diao, J. Fei, R. L. Gonzalez, I. V. Gopich, T. Ha, C. A. Hanke, G. Haran, N. S. Hatzakis, S. Hohng, S.-C. Hong, T. Hugel, A. Ingargiola, C. Joo, A. N. Kapanidis, H. D. Kim, T. Laurence, N. K. Lee, T.-H. Lee, E. A. Lemke, E. Margeat, J. Michaelis, X. Michalet, S. Myong, D. Nettels, T.-O. Peulen, E. Ploetz, Y. Razvag, N. C. Robb, B. Schuler, H. Soleimaninejad, C. Tang, R. Vafabakhsh, D. C. Lamb, C. A. Seidel and S. Weiss, FRET-Based Dynamic Structural Biology: Challenges, Perspectives and an Appeal for Open-Science Practices, eLife, 2021, 10, e60416, DOI:10.7554/eLife.60416.
- H. Tan, S. Bungert-Plümke, D. Kortzak, C. Fahlke and G. Stölting, Dual-Color Colocalization in Single-Molecule Localization Microscopy to Determine the Oligomeric State of Proteins in the Plasma Membrane, Bio-Protoc., 2023, 13(13), e4749, DOI:10.21769/BioProtoc.4749.
- L. S. Churchman, Z. Ökten, R. S. Rock, J. F. Dawson and J. A. Spudich, Single Molecule High-Resolution Colocalization of Cy3 and Cy5 Attached to Macromolecules Measures Intramolecular Distances through Time, Proc. Natl. Acad. Sci. U. S. A., 2005, 102(5), 1419–1423, DOI:10.1073/pnas.0409487102.
- B. Huang, M. Bates and X. Zhuang, Super-Resolution Fluorescence Microscopy, Annu. Rev. Biochem., 2009, 78(1), 993–1016, DOI:10.1146/annurev.biochem.77.061906.092014.
- A. F. Koenderink, R. Tsukanov, J. Enderlein, I. Izeddin and V. Krachmalnicoff, Super-Resolution Imaging: When Biophysics Meets Nanophotonics, Nanophotonics, 2022, 11(2), 169–202, DOI:10.1515/nanoph-2021-0551.
- S. W. Hell and J. Wichmann, Breaking the Diffraction Resolution Limit by Stimulated Emission: Stimulated-Emission-Depletion Fluorescence Microscopy, Opt. Lett., 1994, 19(11), 780, DOI:10.1364/OL.19.000780.
- Q. Zhao, I. T. Young and J. G. S. De Jong, Photon, Budget Analysis for Fluorescence Lifetime Imaging Microscopy, J. Biomed. Opt., 2011, 16(8), 086007, DOI:10.1117/1.3608997.
- J. Yguerabide and E. E. Yguerabide, Light-Scattering Submicroscopic Particles as Highly Fluorescent Analogs and Their Use as Tracer Labels in Clinical and Biological Applications, Anal. Biochem., 1998, 262(2), 137–156, DOI:10.1006/abio.1998.2759.
- C. Sönnichsen, B. M. Reinhard, J. Liphardt and A. P. Alivisatos, A Molecular Ruler Based on Plasmon Coupling of Single Gold and Silver Nanoparticles, Nat. Biotechnol., 2005, 23(6), 741–745, DOI:10.1038/nbt1100.
- B. M. Reinhard, M. Siu, H. Agarwal, A. P. Alivisatos and J. Liphardt, Calibration of Dynamic Molecular Rulers Based on Plasmon Coupling between Gold Nanoparticles, Nano Lett., 2005, 5(11), 2246–2252, DOI:10.1021/nl051592s.
- E. W. A. Visser, M. Horáček and P. Zijlstra, Plasmon Rulers as a Probe for Real-Time Microsecond Conformational Dynamics of Single Molecules, Nano Lett., 2018, 18(12), 7927–7934, DOI:10.1021/acs.nanolett.8b03860.
- R. T. Hill, J. J. Mock, A. Hucknall, S. D. Wolter, N. M. Jokerst, D. R. Smith and A. Chilkoti, Plasmon Ruler with Angstrom Length Resolution, ACS Nano, 2012, 6(10), 9237–9246, DOI:10.1021/nn3035809.
- T. Chen, Y. Hong and B. M. Reinhard, Probing DNA Stiffness through Optical Fluctuation Analysis of Plasmon Rulers, Nano Lett., 2015, 15(8), 5349–5357, DOI:10.1021/acs.nanolett.5b01725.
- L. Wu and B. M. Reinhard, Probing Subdiffraction Limit Separations with Plasmon Coupling Microscopy: Concepts and Applications, Chem. Soc. Rev., 2014, 43(11), 3884–3897, 10.1039/C3CS60340G.
- S. Zhang and B. M. Reinhard, Characterizing Large-Scale Receptor Clustering on the Single Cell Level: A Comparative Plasmon Coupling and Fluorescence Superresolution Microscopy Study, J. Phys. Chem. B, 2019, 123(26), 5494–5505, DOI:10.1021/acs.jpcb.9b05176.
- C. Sönnichsen, S. Geier, N. E. Hecker, G. von Plessen, J. Feldmann, H. Ditlbacher, B. Lamprecht, J. R. Krenn, F. R. Aussenegg, V. Z.-H. Chan, J. P. Spatz and M. Möller, Spectroscopy of Single Metallic Nanoparticles Using Total Internal Reflection Microscopy, Appl. Phys. Lett., 2000, 77(19), 2949–2951, DOI:10.1063/1.1323553.
- P. F. Gao, G. Lei and C. Z. Huang, Dark-Field Microscopy: Recent Advances in Accurate Analysis and Emerging Applications, Anal. Chem., 2021, 93(11), 4707–4726, DOI:10.1021/acs.analchem.0c04390.
- A. Weigel, A. Sebesta and P. Kukura, Dark Field Microspectroscopy with Single Molecule Fluorescence Sensitivity, ACS Photonics, 2014, 1(9), 848–856, DOI:10.1021/ph500138u.
- C.-Y. Cheng, Y.-H. Liao and C.-L. Hsieh, High-Speed Imaging and Tracking of Very Small Single Nanoparticles by Contrast Enhanced Microscopy, Nanoscale, 2019, 11(2), 568–577, 10.1039/C8NR06789A.
- O. Avci, N. Ünlü, A. Özkumur and M. Ünlü, Interferometric Reflectance Imaging Sensor (IRIS)—A Platform Technology for Multiplexed Diagnostics and Digital Detection, Sensors, 2015, 15(7), 17649–17665, DOI:10.3390/s150717649.
- V. Jacobsen, P. Stoller, C. Brunner, V. Vogel and V. Sandoghdar, Interferometric Optical Detection and Tracking of Very Small Gold Nanoparticles at a Water-Glass Interface, Opt. Express, 2006, 14(1), 405, DOI:10.1364/OPEX.14.000405.
- D. Cole, G. Young, A. Weigel, A. Sebesta and P. Kukura, Label-Free Single-Molecule Imaging with Numerical-Aperture-Shaped Interferometric Scattering Microscopy, ACS Photonics, 2017, 4(2), 211–216, DOI:10.1021/acsphotonics.6b00912.
- J. Ortega Arroyo, J. Andrecka, K. M. Spillane, N. Billington, Y. Takagi, J. R. Sellers and P. Kukura, Label-Free, All-Optical Detection, Imaging, and Tracking of a Single Protein, Nano Lett., 2014, 14(4), 2065–2070, DOI:10.1021/nl500234t.
- M. P. McDonald, A. Gemeinhardt, K. König, M. Piliarik, S. Schaffer, S. Völkl, M. Aigner, A. Mackensen and V. Sandoghdar, Visualizing Single-Cell Secretion Dynamics with Single-Protein Sensitivity, Nano Lett., 2018, 18(1), 513–519, DOI:10.1021/acs.nanolett.7b04494.
- Y.-H. Liao, C.-H. Lin, C.-Y. Cheng, W. C. Wong, J.-Y. Juo and C.-L. Hsieh, Monovalent and Oriented Labeling of Gold Nanoprobes for the High-Resolution Tracking of a Single-Membrane Molecule, ACS Nano, 2019, 13(10), 10918–10928, DOI:10.1021/acsnano.9b01176.
- H. Lee, H. J. Park, G. J. Yeon and Z. H. Kim, Amplitude and Phase Spectra of Light Scattered from a Single Nanoparticle, ACS Photonics, 2022, 9(9), 3052–3059, DOI:10.1021/acsphotonics.2c00803.
- J. Sülzle, W. Yang, Y. Shimoda, N. Ronceray, E. Mayner, S. Manley and A. Radenovic, Label-Free Imaging of DNA Interactions with 2D Materials, ACS Photonics, 2024, 11(2), 737–744, DOI:10.1021/acsphotonics.3c01604.
- A. D. Kashkanova, M. Blessing, A. Gemeinhardt, D. Soulat and V. Sandoghdar, Precision Size and Refractive Index Analysis of Weakly Scattering Nanoparticles in Polydispersions, Nat. Methods, 2022, 19(5), 586–593, DOI:10.1038/s41592-022-01460-z.
- K. Holanová, M. Vala and M. Piliarik, [INVITED] Optical Imaging and Localization of Prospective Scattering Labels Smaller than a Single Protein, Opt. Laser Technol., 2019, 109, 323–327, DOI:10.1016/j.optlastec.2018.08.014.
- E. D. B. Foley, M. S. Kushwah, G. Young and P. Kukura, Mass Photometry Enables Label-Free Tracking and Mass Measurement of Single Proteins on Lipid Bilayers, Nat. Methods, 2021, 18(10), 1247–1252, DOI:10.1038/s41592-021-01261-w.
- M. Dahmardeh, H. Mirzaalian Dastjerdi, H. Mazal, H. Köstler and V. Sandoghdar, Self-Supervised Machine Learning Pushes the Sensitivity Limit in Label-Free Detection of Single Proteins below 10 kDa, Nat. Methods, 2023, 20(3), 442–447, DOI:10.1038/s41592-023-01778-2.
- G. Young, N. Hundt, D. Cole, A. Fineberg, J. Andrecka, A. Tyler, A. Olerinyova, A. Ansari, E. G. Marklund, M. P. Collier, S. A. Chandler, O. Tkachenko, J. Allen, M. Crispin, N. Billington, Y. Takagi, J. R. Sellers, C. Eichmann, P. Selenko, L. Frey, R. Riek, M. R. Galpin, W. B. Struwe, J. L. P. Benesch and P. Kukura, Quantitative Mass Imaging of Single Biological Macromolecules, Science, 2018, 360(6387), 423–427, DOI:10.1126/science.aar5839.
- A. Sonn-Segev, K. Belacic, T. Bodrug, G. Young, R. T. VanderLinden, B. A. Schulman, J. Schimpf, T. Friedrich, P. V. Dip, T. U. Schwartz, B. Bauer, J.-M. Peters, W. B. Struwe, J. L. P. Benesch, N. G. Brown, D. Haselbach and P. Kukura, Quantifying the Heterogeneity of Macromolecular Machines by Mass Photometry, Nat. Commun., 2020, 11(1), 1772, DOI:10.1038/s41467-020-15642-w.
- T. Heermann, F. Steiert, B. Ramm, N. Hundt and P. Schwille, Mass-Sensitive Particle Tracking to Elucidate the Membrane-Associated MinDE Reaction Cycle, Nat. Methods, 2021, 18(10), 1239–1246, DOI:10.1038/s41592-021-01260-x.
- G. Cui, D. Y. Kim, D. Zu, G. Chai and S. E. Lee, Interferometric Phase Intensity Nanoscopy (iPINE), Revealing Nanostructural Features, Resolves Proximal Nanometer Objects below the Diffraction Limit: Implications in Long-Time Super-Resolution of Biological Dynamics, ACS Appl. Nano Mater., 2024 DOI:10.1021/acsanm.3c04361.
- L. Velasco, T. Ouyang and B. M. Reinhard, Two-Color iSCAT Imaging of Ag Nanoparticles Resolves Size and Ambient Refractive Index Changes, Nano Lett., 2023, 23(10), 4642–4647, DOI:10.1021/acs.nanolett.3c01306.
- J. N. Milstein and J.-C. Meiners, Worm-Like Chain (WLC) Model, in Encyclopedia of Biophysics, ed. G. C. K. Roberts, Springer Berlin Heidelberg, Berlin, Heidelberg, 2013, pp. 2757–2760. DOI:10.1007/978-3-642-16712-6_502.
- F. Oesterhelt, M. Rief and H. E. Gaub, Single Molecule Force Spectroscopy by AFM Indicates Helical Structure of Poly(Ethylene-Glycol) in Water, New J. Phys., 1999, 1, 6–6, DOI:10.1088/1367-2630/1/1/006.
- A. Kuzuya, K. Numajiri, M. Kimura and M. Komiyama, Single-Molecule Accommodation of Streptavidin in Nanometer-Scale Wells Formed in DNA Nanostructures, Nucleic Acids Symp. Ser., 2008, 52(1), 681–682, DOI:10.1093/nass/nrn344.
- M. Piliarik and V. Sandoghdar, Direct Optical Sensing of Single Unlabelled Proteins and Super-Resolution Imaging of Their Binding Sites, Nat. Commun., 2014, 5(1), 4495, DOI:10.1038/ncomms5495.
- J. Hwang and W. E. Moerner, Interferometry of a Single Nanoparticle Using the Gouy Phase of a Focused Laser Beam, Opt. Commun., 2007, 280(2), 487–491, DOI:10.1016/j.optcom.2007.08.032.
- L. Saemisch, N. F. Van Hulst and M. Liebel, One-Shot Phase Image Distinction of Plasmonic and Dielectric Nanoparticles, Nano Lett., 2021, 21(9), 4021–4028, DOI:10.1021/acs.nanolett.1c00866.
- D. E. Segall, P. C. Nelson and R. Phillips, Volume-Exclusion Effects in Tethered-Particle Experiments: Bead Size Matters, Phys. Rev. Lett., 2006, 96(8), 088306, DOI:10.1103/PhysRevLett.96.088306.
Footnote |
| † Electronic supplementary information (ESI) available: Additional iSCAT images and fits of 20 nm and 10 nm NPs and dimers, σ405, σ445 plot for experimental control, SEM characterization data for 10 nm and 20 nm data before and after assembly. iSCAT images for 20 nm Ag NP dimers formed with 0.4 kDa and 3.4 kDa PEG spacers, SEM microcraphs before/after dimer formation, histogram of 20 nm Ag NP aggregation state obtained with 0.4 Da PEG spacer, SEM images of NP–PEG–biotin controls, SEM images of 20 nm NP–STV + 40 nm NP–PEG3.4 kDa–biotin heterodimers, σ405, σ445 plot for 30 nm Ag NP monomers, σ405, σ445 plot for aggregates of 20 nm Ag NPs, iSCAT image and fit of 10 nm NP monomer and dimer, FDTD simulation for 10 Ag NP dimers, SEM size histograms of Ag NPs before and after illumination. See DOI: https://doi.org/10.1039/d4nr01288g |
| This journal is © The Royal Society of Chemistry 2024 |