Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Engineering ssRNA tile filaments for (dis)assembly and membrane binding†
Nicola
De Franceschi
 ,
Baukje
Hoogenberg
,
Allard
Katan
,
Baukje
Hoogenberg
,
Allard
Katan
 and
Cees
Dekker
and
Cees
Dekker
 *
*
Department of Bionanoscience, Kavli Institute of Nanoscience Delft, Delft University of Technology, Delft, The Netherlands. E-mail: c.dekker@tudelft.nl
First published on 7th February 2024
Abstract
Cytoskeletal protein filaments such as actin and microtubules confer mechanical support to cells and facilitate many cellular functions such as motility and division. Recent years have witnessed the development of a variety of molecular scaffolds that mimic such filaments. Indeed, filaments that are programmable and compatible with biological systems may prove useful in studying or substituting such proteins. Here, we explore the use of ssRNA tiles to build and modify filaments in vitro. We engineer a number of functionalities that are crucial to the function of natural proteins filaments into the ssRNA tiles, including the abilities to assemble or disassemble filaments, to tune the filament stiffness, to induce membrane binding, and to bind proteins. This work paves the way for building dynamic cytoskeleton-mimicking systems made out of rationally designed ssRNA tiles that can be transcribed in natural or synthetic cells.
Introduction
Cells feature a variety of cytoskeletal proteins that arrange into filamentous structures that are responsible for conferring mechanical properties to the cell. These systems are key to sustaining cellular functions such as cell shape, cell motility, intracellular transport, and cell division. Examples include actin,1 tubulin,2 ESCRT-III,3 and the bacterial FtsZ4 and archaeal Cdv.5 All of these form filaments that act as recruiting hubs for many other proteins that work in concert with the membrane. A variety of molecular scaffolds have been developed that mimic such cytoskeletal filaments, for example for in vitro studies with reconstituted minimal systems. Notably, in bottom-up synthetic biology, efforts have recently started to build synthetic cells with various cellular components including natural cytoskeletal filaments and mimics thereof.6 In such an approach, the use of natural proteins presents obvious advantages to create a cytoskeleton but it also has drawbacks such as a significant burden on the protein translation system as well as challenges in fine-tuning protein structure and function and encoding new functionalities. It is therefore of interest to study complementary systems that mimic cytoskeletal filaments which may provide advantages as substitutes for mechanical stability and cell division of future synthetic cells. The development of rationally designed tuneable synthetic scaffolding systems is of great interest, in particular if these can be functionalized similarly to biological cytoskeletal systems.Nucleic acids nanotechnology allows the rational design of nanoscale objects that are able to self-assemble into programmable shapes ranging from smileys7 to transmembrane channels,8–10 and rotary motors.11,12 In mimicking cytoskeletal systems, a straightforward approach is to assemble synthetic filaments made of tiles. Such filaments have been obtained using a number of original designs13,14 and the tiles that compose them can be made using either multiple strands of ssDNA15,16 or ssRNA,17 an approach called “RNA tectonics”.13,18 Micrometers-long, stiff filaments13 and lattices19 can be obtained by this approach, which has the further advantage of including the presence of multiple 3′ and 5′ free ends. These “sticky ends” allow easy functionalization of the tiles, for instance with fluorophores and multiple siRNAs, to achieve controlled delivery.20 It is also possible to control the polymerization of such filaments21 also when encapsulated inside water-in-oil droplets.22–24 However, such a design requires both the maintenance of a precise stoichiometry of all the strands composing the tiles and their folding by thermal annealing, which may limit their range of applications. A different approach is based on self-folded ssRNA tiles that are formed from a single ssRNA molecule and that can interact with each other via kissing loops (KLs).25 This presents a number of advantages such as the lack of restraints in terms of stoichiometry and the ability to fold co-transcriptionally at room temperature. Such ssRNA scaffolds have great potential to be expanded for a variety of applications, as can for example be modified to encode a range of curvatures,26 binding to specific proteins27,28 and can be scaled up to yield kilobase-sized filaments.29 Moreover, ssRNA molecules can be directly transcribed from the genome and thus are fully cell biology compatible. So far however, only limited attempts have been pursued to expand their capabilities by functionalizing the ssRNA scaffolds, e.g. by going beyond static structures by engineering dynamic filaments that can assemble as well as disassemble in a controlled way.
Here, we assemble and functionalize ssRNA tile filaments, and we demonstrate that we are able to tune their stiffness, control their polymerization, encode direct membrane binding, and create binding sites for proteins. We introduce additional designing principles, e.g., we design softer tiles that allow filament disassembly by strand displacement. Our work provides a toolbox that paves the way for using ssRNA tile filaments in dynamic cell-like systems.
Results and discussion
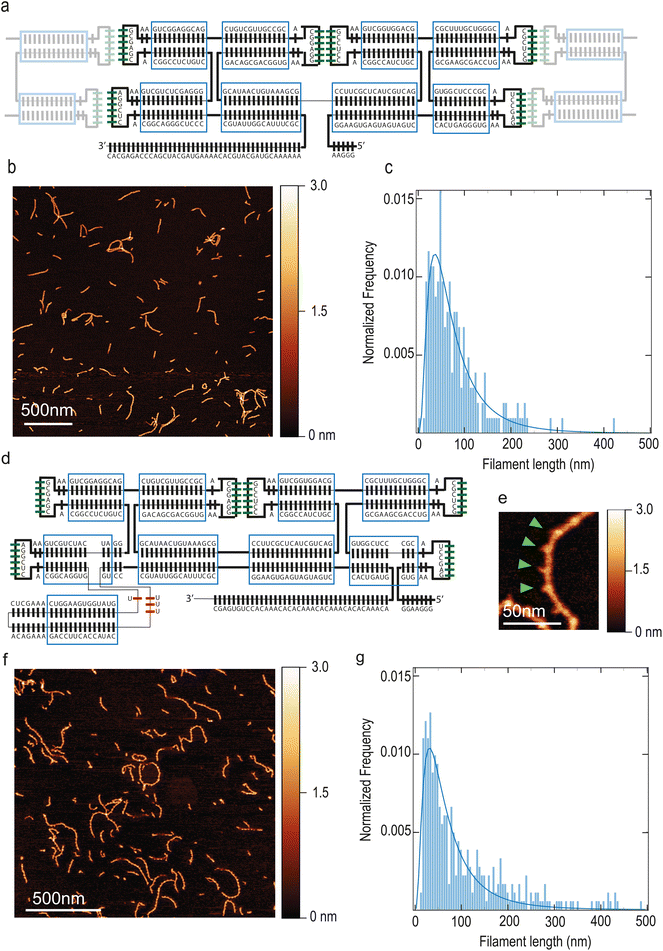
Our starting point was previous work by Geary and co-workers on ssRNA tiles.25 We made a 2-helix ssRNA origami tile design with “Antiparallel Even” (AE) crossovers by substituting the 120° Kissing Loops (KLs) present at the extremities of each helix with 180° KLs, so that tiles can polymerize in a linear, head-to-tail fashion. This initial tile design is hereafter named T1 and it is detailed in Fig. 1A, whereas we will also introduce other tiles variants with different modifications and functionalities. Assuming dsRNA is in A form,30 the designed tile dimensions were 18.2 nm × 4.8 nm × 2.4 nm (length × width × height). Tiles were folded by thermal annealing, deposited on mica and imaged by Atomic Force Microscopy (AFM).The KL title–tile interactions resulted in the formation of linear filaments, see Fig. 1B. These filaments featured a submicron length (Fig. 1C) and the height was 1.7 nm ± 0.3 nm, as measured by using dry AFM which provides a lower limit (ESI Fig. 1A†). T1 filaments exhibited a length of 62 nm (median; 95% CI: 58–67 nm) and a persistence length of 173 nm (95% CI: 152–194 nm). Representative imaged that were quantified are presented in ESI Fig. 2.† Residual monomeric tiles were not observed, indicating that the tile–tile interaction mediated by merely two KLs was very efficient, sequestering single tiles away into the filaments. Supramolecular assembly of tiles were also visible on agarose gel (ESI Fig. 1B†) and required Mg++ to form (ESI Fig. 1C†).
In order to visualize individual tiles within the filaments, we modified the design by introducing a branching point. In a previous report, a branching point creating a 90° bend was demonstrated.31 Here, we introduce a 3-way-junction motif from the bacteriophage Phi29 DNA packaging motor,32 which forms a rigid branching at a ≈ 60° angle within the helix that it originates from.33 We inserted this junction so that it will exit the plane of the tile in a perpendicular fashion. The 3′–5′ junction was also moved along the RNA strand towards the distal stem in order to avoid having two nick close to each other. Furthermore, we modified the 3′-end into an A–C rich sequence, in order to minimize potential aggregation due to self-dimerization. These tiles, named T2 (Fig. 1D and F) can be identified within the filament due to the distinctive spike created by the 3-way junction (Fig. 1E). This allowed to quantify the number of tiles present in each filament and estimate an average tile length of 18.8 ± 1.5 nm (mean ± SD; n = 54), which is in very good agreement with the expected length of 18.2 nm. T2 filaments exhibited a length of 58 nm (median; 95% CI: 53–64 nm) (Fig. 1G) and a persistence length of 38 nm (median; 95% CI: 36–41 nm). Notably, the persistence length of the filaments is greatly reduced, becoming close to that of dsDNA (∼50 nm (ref. 34)). This is likely due to both the insertion of the 3-way junction and the placement of the 3′–5′-end junction closer to the KL.
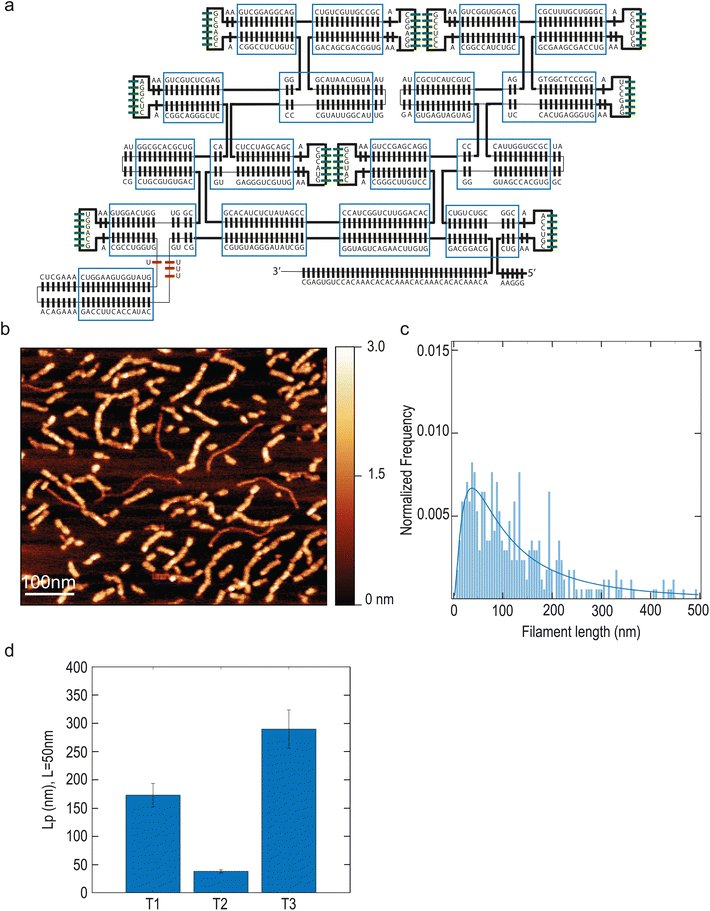
We sought to increase filament stiffness by modifying the design with the addition of 2 helices (named T3, Fig. 2A). The resulting larger tiles formed filaments with a length of 106 nm (median; 95% CI: 89–127 nm) (Fig. 2B) and with a markedly higher persistence length of 290 nm (median; 95% CI: 256–324 nm) (Fig. 2D). Thus, the T3 design allowed to increase filament stiffness compared to T1.
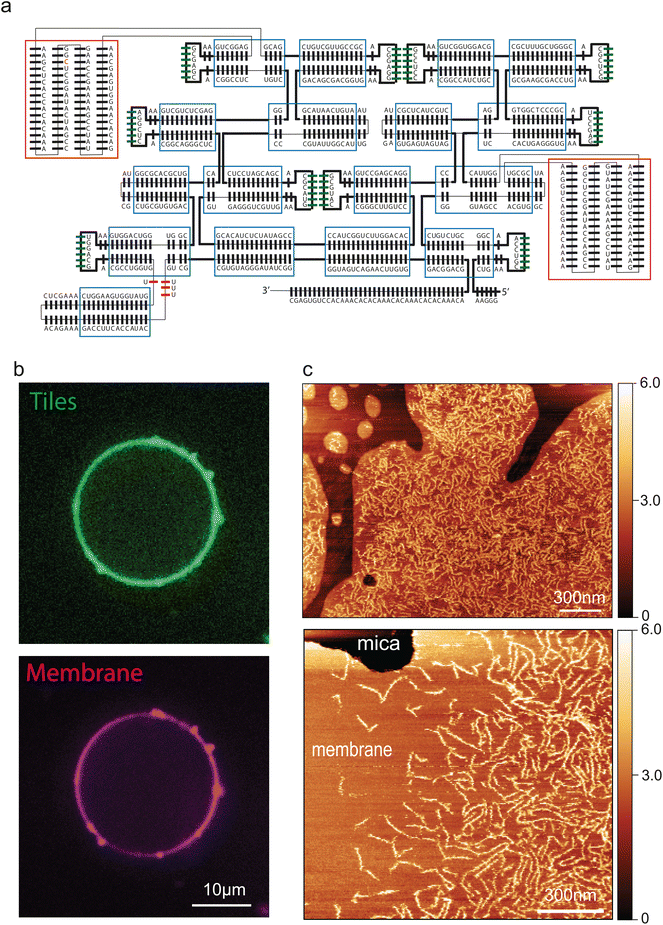
Next, we engineered a membrane-binding site into the tiles to induce attachment of the filaments to a bilayer lipid membrane. A common approach to bind nucleic acid nanostructures to membranes is the use of a short DNA oligomer that hybridizes to the DNA/RNA nanostructure and that is chemically functionalized with cholesterol moieties that anchors to the membrane.35 We initially used this strategy to bind our filaments to the membrane, by indeed hybridizing a cholesterol-functionalized oligo to the 5′ end of each RNA tile. However, while this was partially successful, the filaments appeared to be only weakly attached to the membrane, as multiple passages of the AFM tip during imaging drastically reduced their number (ESI Fig. 3A†). We found that we could significantly improve the stability of filaments-membrane binding by engineering a direct interaction between the tile and the lipid membrane. To this aim, we inserted biotin aptamers36 into the T3 tiles, yielding the T4 design (Fig. 3A). T4 tile filaments exhibited very robust binding to membranes (ESI Fig. 3B†). Thus, they could be imaged extensively both by liquid AFM as well as with confocal microscopy (Fig. 3B and C)upon being fluorescently labelled by 3′-pCp-Cy5. Fig. 3C clearly show an abundance of filaments on the lipid bilayer, whereas no filaments were seen to attach to the bare mica. In addition to simplifying the system, insertion of the biotin aptamer allowed a tight and close contact between filaments and the membrane, which mimics cytoskeletal proteins such as the ESCRT-III complex.37
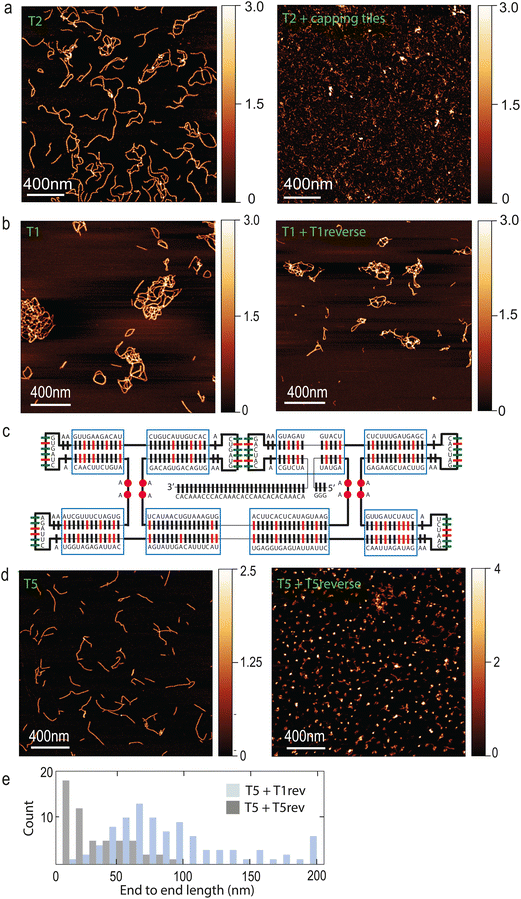
A common feature of cytoskeletal systems is their ability to be regulated by other proteins, which can either promote or inhibit filament polymerization.38 In order to mimic such a functionality, we designed “capping tiles” (hereafter referred to as (“T-cap”) that feature only one pair of KLs (ESI Fig. 4A†). T-caps should effectively inhibit further polymerization of tiles T2. Indeed, we found that addition of T-caps completely abolished filament formation (Fig. 4A). Another common feature of cytoskeletal filamentous systems is the presence of unstructured sequences that act as recruiting hubs for additional proteins that may modify the filament. An example is the linear C-terminal sequence of ESCRT proteins, which recruits the AAA ATPases Vps4 and CdvC37,39 which ultimately leads to filament disassembly. In our tile design, the 3′ extension represents a convenient site for recruiting enzymes such as helicases, that may be able to act on the filaments. Indeed, we observed a robust recruitment of the thermophilic helicase Hel308 to T2 filaments, see ESI Fig. 4B.†
Finally, a crucial aspect of cytoskeletal filaments is their ability to undergo cycles of assembly and disassembly. While tile–tile interactions spontaneously drive filaments towards assembly in our system, we sought to engineer ways to disassemble them as well. Strand displacement is a well-established approach to disassemble DNA and RNA nanostructures.40 In our design, the existing 3′ end can conveniently act as toehold, since it is longer (Fig. 1A) than 7 nucleotides, which were shown to be sufficient to disassemble nanotubes composed of DNA tiles.41 We reasoned that an equimolar amount of reverse strand should be sufficient to disassemble filaments. However, we did not observe T1 filament disassembly upon addition of the T1 reverse strand, even after prolonged incubation (Fig. 4B). Since this may be due to the high GC content of the tiles which leads to a very stable structure, we modified the original design in order to “soften” the tiles. Regions with high GC content (70–83%, indicated in red in Fig. 4C) were lowered to a GC content of only 26–50%. Moreover, we added some unpaired bases between RNA helices (purple circles in Fig. 4C). The resulting tiles (T5) were still able to form filaments, but now the addition of the reverse T5 strand to pre-formed T5 filaments caused complete filament disassembly (Fig. 4D and E) at room temperature.
Conclusions
In this paper, we developed a range of functionalities on ssRNA tiles that facilitates the formation of cytoskeletal-like filaments. By changing the number of helices, we obtained filaments of variable stiffness. By branching the RNA sequence with specific motifs, we could engineer membrane binding via biotin aptamers, resulting in strong and direct binding. Moreover, we showed that binding sites for RNA-binding proteins such as helicases can easily be added with minimal modification of the design. Lastly, we modified the tile sequence to render them softer to achieve filament disassembly by strand displacement.The use of self-folded ssRNA tiles presents a number of advantages over proteins, DNA, or multi-strand RNA tiles. An obvious advantage of self-folding ssRNA tiles is the possibility of directly producing them inside natural or synthetic cells by transcription, thus avoiding the need for a protein translation system. ssRNA tiles are furthermore able to undergo co-transcriptional folding at room temperature.25 In our experiments we found that ssRNA tile can also undergo fast spontaneous isothermal refolding without the need to be coupled with transcription (ESI Fig. 5†). Therefore, if a suitable enzyme-driven pathway for tile disassembly could be implemented, filaments made of tiles could in principle undergo cycles of assembly and disassembly driven by ATP consumption, rather than disassembly driven by strand displacement. Enzymes such as helicases, which we showed to be able to bind ssRNA tiles, and our improved “softer” design that facilitates disassembly, appear to be promising candidates that set the stage to build such system in future research.
In this paper, we showed how the ssRNA tiles design can be significantly expanded with a number of functionalizations, allowing parameters to be fine-tuned and providing a versatile platform for mimicking cytoskeletal filaments. Reconstituting the functionalities of cytoskeletal filaments is an important goal in bottom-up synthetic biology, where one of the most ambitious aims is the reconstitution of cell division.42 The system we describe here paves the way for the development of molecular scaffolds that are programmable, dynamic and compatible with biological systems, on the route to the development of artificial cells.
Methods
Reagents
DPPC (1,2-dipalmitoyl-sn-glycero-3-phosphocholine), DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine), DSPE-PEG(2000) Biotin (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[biotinyl(polyethylene glycol)-2000]) and Egg Liss Rhod PE (L-α-Phosphatidylethanolamine-N-(lissamine rhodamine B sulfonyl)) were purchased from Avanti. The dsDNA templates for tile transcription were ordered from IDT.Tiles design
The original design from ref. 23 was modified manually by inserting the number of base-pairs required based on well-established structural parameters of the RNA duplex. No software was used for optimization. The sequence of the different tiles designs are indicated as the coding strand on the artificial gene. The promoter region in underlined:>T1
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) AAGGAAGTGAGTAGTAGTCCACTGAGGGTGAAGAGCCTACGCCCTCGGTGGCGAAGCGACCTGAAGCTCGCACGGGTCGTTTCGCGCAGGTGGCTGAAGCCTCCACGGCCATCTGCCACTGCTACTCGCTTCCGCGAAATGTCAATACGGACAGCGACGGTGAAGGAGGCACGCCGTTGCTGTCGACGGAGGCTGAAGCGAGCACGGCCTCTGTCGGGAGCTCTGCTGAAAGGCTCACGGCAGGGCTCCCCGTATTGGCATTTCGCAAAAACGTAGCATGCACAAAAGTAGCATCGACCCAGAGCAC
AAGGAAGTGAGTAGTAGTCCACTGAGGGTGAAGAGCCTACGCCCTCGGTGGCGAAGCGACCTGAAGCTCGCACGGGTCGTTTCGCGCAGGTGGCTGAAGCCTCCACGGCCATCTGCCACTGCTACTCGCTTCCGCGAAATGTCAATACGGACAGCGACGGTGAAGGAGGCACGCCGTTGCTGTCGACGGAGGCTGAAGCGAGCACGGCCTCTGTCGGGAGCTCTGCTGAAAGGCTCACGGCAGGGCTCCCCGTATTGGCATTTCGCAAAAACGTAGCATGCACAAAAGTAGCATCGACCCAGAGCAC
>T2
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) AAGGGTGAAGAGCCTACGCCCTCGGTGGCGAAGCGACCTGAAGCTCGCACGGGTCGTTTCGCGCAGGTGGCTGAAGCCTCCACGGCCATCTGCGACTGCTACTCGCTTCCGCGAAATGTCAATACGGACAGCGACGGTGAAGGAGGCACGCCGTTGCTGTCGACGGAGGCTGAAGCGAGCACGGCCTCTGTCGGATCATCTGCTGAAAGGCTCACGGCAGGTGTGTATGGTGAAGGTCAAAGCTCACAGAAAGACCTTCACCATACTTTGTCCCGTATTGGCATTTCGCGGAAGTGAGTAGTAGTCCACTGATGACAAACACACAAACACAAACACACAAACACCTGTGAGC
AAGGGTGAAGAGCCTACGCCCTCGGTGGCGAAGCGACCTGAAGCTCGCACGGGTCGTTTCGCGCAGGTGGCTGAAGCCTCCACGGCCATCTGCGACTGCTACTCGCTTCCGCGAAATGTCAATACGGACAGCGACGGTGAAGGAGGCACGCCGTTGCTGTCGACGGAGGCTGAAGCGAGCACGGCCTCTGTCGGATCATCTGCTGAAAGGCTCACGGCAGGTGTGTATGGTGAAGGTCAAAGCTCACAGAAAGACCTTCACCATACTTTGTCCCGTATTGGCATTTCGCGGAAGTGAGTAGTAGTCCACTGATGACAAACACACAAACACAAACACACAAACACCTGTGAGC
>T3
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) AACTGAACGTCCAACGGCGTCTGTCGGGTAGCCACGTGGCATCGCGTGGTTACTCCACTGAGGGTGAAGAGCCTACGCCCTCGGTGGCGAAGCGACCTGAAGCTCGCACGGGTCGTTTCGCGCAGGTGGCTGAAGCCTCCACGGCCATCTGCGACTGCTACTCGCTAGAGAGTGAGTAGTAGCCGGACGAGCCTGAAGCGTACACGGGCTTGTCCCACAGGTTCTGGCTACCCCGATATCTCTACACGGTGAGGGTCGTTGAAGTACGCACGACGATCCTCCCCGTATTGGCATTGTAATGTCAATACGGACAGCGACGGTGAAGGAGGCACGCCGTTGCTGTCGACGGAGGCTGAAGCGAGCACGGCCTCTGTCGGGAGCTCTGCTGAAAGGCTCACGGCAGGGCTCACGTCGCACGCGGTACGCTGCGTGTGACCGGTGGTCAGGTGAATGGACGACGCCTGGTGTGTATGGTGAAGGTCAAAGCTCACAGAAAGACCTTCACCATACTTTGTCGCGTGTAGGGATATCGGGGTAGTCAGAACTTGTGGACGGACGACAAACACACAAACACAAACACACAAACACCTGTGAGC
AACTGAACGTCCAACGGCGTCTGTCGGGTAGCCACGTGGCATCGCGTGGTTACTCCACTGAGGGTGAAGAGCCTACGCCCTCGGTGGCGAAGCGACCTGAAGCTCGCACGGGTCGTTTCGCGCAGGTGGCTGAAGCCTCCACGGCCATCTGCGACTGCTACTCGCTAGAGAGTGAGTAGTAGCCGGACGAGCCTGAAGCGTACACGGGCTTGTCCCACAGGTTCTGGCTACCCCGATATCTCTACACGGTGAGGGTCGTTGAAGTACGCACGACGATCCTCCCCGTATTGGCATTGTAATGTCAATACGGACAGCGACGGTGAAGGAGGCACGCCGTTGCTGTCGACGGAGGCTGAAGCGAGCACGGCCTCTGTCGGGAGCTCTGCTGAAAGGCTCACGGCAGGGCTCACGTCGCACGCGGTACGCTGCGTGTGACCGGTGGTCAGGTGAATGGACGACGCCTGGTGTGTATGGTGAAGGTCAAAGCTCACAGAAAGACCTTCACCATACTTTGTCGCGTGTAGGGATATCGGGGTAGTCAGAACTTGTGGACGGACGACAAACACACAAACACAAACACACAAACACCTGTGAGC
>T4
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) AACTGAACGTCCAACGGCGTCTGTCGGGTAGCCACGTGGCATCGCGTAAGGTCAGGAACAAAACCGACCATAGGCTCGGGTATCGAAAAGCCTATGAACAAACCTGACCAAGGTTACTCCACTGAGGGTGAAGAGCCTACGCCCTCGGTGGCGAAGCGACCTGAAGCTCGCACGGGTCGTTTCGCGCAGGTGGCTGAAGCCTCCACGGCCATCTGCGACTGCTACTCGCTAGAGAGTGAGTAGTAGCCGGACGAGCCTGAAGCGTACACGGGCTTGTCCCACAGGTTCTGGCTACCCCGATATCTCTACACGGTGAGGGTCGTTGAAGTACGCACGACGATCCTCCCCGTATTGGCATTGTAATGTCAATACGGACAGCGACGGTGAAGGAGGCACGCCGTTGCTGTCGACGAAGCTCACCACACAAACCGATCATAGGCTCGGGAACCGAAAAGCCTATGACACAAGGTGAGCAAGAGGCTGAAGCGAGCACGGCCTCTGTCGGGAGCTCTGCTGAAAGGCTCACGGCAGGGCTCACGTCGCACGCGGTACGCTGCGTGTGACCGGTGGTCAGGTGAATGGACGACGCCTGGTGTGTATGGTGAAGGTCAAAGCTCACAGAAAGACCTTCACCATACTTTGTCGCGTGTAGGGATATCGGGGTAGTCAGAACTTGTGGACGGACGACAAACACACAAACACAAACACACAAACACCTGTGAGC
AACTGAACGTCCAACGGCGTCTGTCGGGTAGCCACGTGGCATCGCGTAAGGTCAGGAACAAAACCGACCATAGGCTCGGGTATCGAAAAGCCTATGAACAAACCTGACCAAGGTTACTCCACTGAGGGTGAAGAGCCTACGCCCTCGGTGGCGAAGCGACCTGAAGCTCGCACGGGTCGTTTCGCGCAGGTGGCTGAAGCCTCCACGGCCATCTGCGACTGCTACTCGCTAGAGAGTGAGTAGTAGCCGGACGAGCCTGAAGCGTACACGGGCTTGTCCCACAGGTTCTGGCTACCCCGATATCTCTACACGGTGAGGGTCGTTGAAGTACGCACGACGATCCTCCCCGTATTGGCATTGTAATGTCAATACGGACAGCGACGGTGAAGGAGGCACGCCGTTGCTGTCGACGAAGCTCACCACACAAACCGATCATAGGCTCGGGAACCGAAAAGCCTATGACACAAGGTGAGCAAGAGGCTGAAGCGAGCACGGCCTCTGTCGGGAGCTCTGCTGAAAGGCTCACGGCAGGGCTCACGTCGCACGCGGTACGCTGCGTGTGACCGGTGGTCAGGTGAATGGACGACGCCTGGTGTGTATGGTGAAGGTCAAAGCTCACAGAAAGACCTTCACCATACTTTGTCGCGTGTAGGGATATCGGGGTAGTCAGAACTTGTGGACGGACGACAAACACACAAACACAAACACACAAACACCTGTGAGC
>T5
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) TATGAAAGAATGATACTCACTTCAGTGAAATGTCAATACTAAGACAGTGACAGTGAAGTAGTCACACTGTTACTGTCTACAGAAGTTGAAGTGATCACAACTTCTGTAAAGTGATCTTTGCTAAAAGATTCATGGTAGAGATTACAGTATTGACATTTCATTGAGGTGAGTATTATTCCAATTAGATAGAAGAATCTACTATCTAGTTGAAGAGAAGCTACTTGAAGATCACACGAGTAGTTTCTCTCATGTAGATGAAGACTACACGTCTAACAAACACACAACCACAAACACCCAAACAC
TATGAAAGAATGATACTCACTTCAGTGAAATGTCAATACTAAGACAGTGACAGTGAAGTAGTCACACTGTTACTGTCTACAGAAGTTGAAGTGATCACAACTTCTGTAAAGTGATCTTTGCTAAAAGATTCATGGTAGAGATTACAGTATTGACATTTCATTGAGGTGAGTATTATTCCAATTAGATAGAAGAATCTACTATCTAGTTGAAGAGAAGCTACTTGAAGATCACACGAGTAGTTTCTCTCATGTAGATGAAGACTACACGTCTAACAAACACACAACCACAAACACCCAAACAC
>T5reverse
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) GTGTTTGGGTGTTTGTGGTTGTGTGTTTGTTAGACGTGTAGTCTTCATCTACATGAGAGAAACTACTCGTGTGATCTTCAAGTAGCTTCTCTTCAACTAGATAGTAGATTCTTCTATCTAATTGGAATAATACTCACCTCAATGAAATGTCAATACTGTAATCTCTACCATGAATCTTTTAGCAAAGATCACTTTACAGAAGTTGTGATCACTTCAACTTCTGTAGACAGTAACAGTGTGACTACTTCACTGTCACTGTCTTAGTATTGACATTTCACTGAAGTGAGTATCATTCTTTCATA
GTGTTTGGGTGTTTGTGGTTGTGTGTTTGTTAGACGTGTAGTCTTCATCTACATGAGAGAAACTACTCGTGTGATCTTCAAGTAGCTTCTCTTCAACTAGATAGTAGATTCTTCTATCTAATTGGAATAATACTCACCTCAATGAAATGTCAATACTGTAATCTCTACCATGAATCTTTTAGCAAAGATCACTTTACAGAAGTTGTGATCACTTCAACTTCTGTAGACAGTAACAGTGTGACTACTTCACTGTCACTGTCTTAGTATTGACATTTCACTGAAGTGAGTATCATTCTTTCATA
>T-cap
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ACACTGAGGGTGAAGAGCCTACGCCCTCGGTGAAAAGCGAAGCGACCTGAAGCTCGCACGGGTCGTTTCGC
ACACTGAGGGTGAAGAGCCTACGCCCTCGGTGAAAAGCGAAGCGACCTGAAGCTCGCACGGGTCGTTTCGC
Tiles transcription and folding
The tile templates were fully double-stranded linear DNA containing the promoter region; they were used directly as a template for RNA transcription using the T7 RiboMAX™ system (Promega) and subsequently purified using the RNeasy Micro Kit (Qiagen). The transcription reactions yielded essentially pure full-length RNA species of the expected molecular weight, and no subsequent purification steps were necessary. Tiles were folded in 40 mM Tris-Acetate (pH 8.0) + 2 mM EDTA + 12.5 mM Mg-Acetate using the following thermal cycle: 5′ at 80 °C; from 80 °C to 70 °C, −1 °C min−1, 10 cycles; from 70 °C to 22 °C, −0.2 °C min−1, 240 cycles. Folding and AFM imaging was performed at a tile concentration of 200 nM in 40 mM Tris-Acetate (pH 8.0) + 2 mM EDTA + 12.5 mM Mg-Acetate.Production of Giant Unilamellar Vesicles, binding of RNA tiles and confocal imaging
GUVs were generated by the PVA-swelling method.43 Briefly, the lipid mix (94.5% DOPC + 5% DSPE-PEG(2000) Biotin + 0.5% Egg Liss Rhod PE) were dissolved in chloroform and spread on the PVA substrate, air-dried and incubated in vacuum at room temperature for 20 minutes. Inner buffer (40 mM Tris-Acetate (pH 8.0) + 2 mM EDTA + 12.5 mM Mg-Acetate + 200 mM sucrose) was added on top, and the GUVs were allow to be generated by swelling for 1 hour. The osmolarity of the outer buffer (40 mM Tris-Acetate (pH 8.0) + 2 mM EDTA + 12.5 mM Mg-Acetate) was adjusted with glucose until reaching iso-osmolarity with the inner buffer. GUVs were collected and added to outer buffer in the presence of 1 μM of tiles directly in the observation chamber. Fluorescence images were acquired using spinning disk confocal laser microscopy (Olympus IXB1/BX61 microscope, 60× objective, iXon camera) with Andor iQ3 software.Preparation of supported lipid bilayer on mica and binding of RNA tiles
Small Unilamellar Vesicles (SUVs) were prepared from a lipid mix composed of 95% DPPC + 5% DSPE-PEG(2000) Biotin by dissolving the DPPC in chloroform, drying the chloroform with N2 stream and re-hydrating the lipid film with water. The sample was then vortexed and sonicated extensively to obtain SUVs. The SUV suspension was deposited on freshly cleaved mica for 20 minutes at 50 °C in a humid chamber to promote vesicle rupture on the mica, obtaining patched of supported lipid bilayer. The mica was washed with buffer (40 mM Tris-Acetate (pH 8.0) + 2 mM EDTA + 12.5 mM Mg-Acetate) and the tiles were added at a concentration of 200 nM. The sample was incubated for 30 minutes at room temperature, washed once with buffer (40 mM Tris-Acetate (pH 8.0) + 2 mM EDTA + 12.5 mM Mg-Acetate) and imaged by liquid AFM.AFM imaging
Filaments from the folding reaction were imaged directly without purification. Samples with high concentrations of filaments were diluted to 10 nM to prevent overcrowding on the surface. The sample was deposited on freshly cleaved mica and incubated at room temperature for 30 seconds to 1 minute. The mica was washed with distilled water and dried using a nitrogen stream. Images were taken with a multimode-8 AFM from Bruker (Bruker Nano GMbH, Berlin, Germany) using Scanasyst-Air-HR probes from Bruker. The AFM was operated using Peak-Force HR mode for imaging in air, with a tapping rate of 8 kHz, at room conditions. Imaging in liquid was performed on the same instrument, using ScanAsyst Fluid + probes and ScanAsyst mode, with a tapping rate of 4 kHz.Quantification of filaments height
The height of the filaments was obtained by applying a threshold mask to the filament image, selecting all contiguous regions of height larger than 0.3 nm above the background and larger than 9 pixels (215 nm2). These regions consist of partial, complete or multiple filaments. The distribution of maximum heights of these regions was fitted with a normal distribution to obtain mean and standard deviation.Quantification of filaments length and persistence length
Image data and processing (plane subtraction and flattening) was done with Gwyddion software. Quantification of filament length and persistence length was done with home-written Matlab software. Briefly, the filaments are selected by thresholding, and the binary image is skeletonized. From the skeleton, a beginning-to-end trajectory is calculated using a Dijkstra algorithm, with optional correction of the path via user guidance. These paths are optimized to a smooth sub-pixel trajectory along the highest points of the filament, via an algorithm that balances the height with the local bending radius. Persistence length is calculated based on fitting the distribution of tangent angles to a Boltzmann distribution, using a spacing of 50 nm. The histograms were normalized as probability densities. All fitting and calculations are done in Matlab R2012b (MathWorks, USA). The code is publicly accessible on TU Delft Gitlab: https://gitlab.tudelft.nl/ajkatan/dnacontour_public.Filament disassembly
Pre-folded filaments (T5) were incubated with equimolar amount (200 nM) of reverse strand (either T1_reverse or T5_reverse) and incubated at room temperature for 1 hour, deposited on mica and imaged.Author contributions
C. D. and N. D. F. designed the study. N. D. F. designed the experiments. N. D. F., B. H. and A. K. carried out the experiments. N. D. F., B. H. and A. K. analysed the data. C. D. supervised the study. C. D. and N. D. F. wrote the manuscript with input from all other authors. All the authors provided critical feedback on the research and the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We thank Chenxiang Lin (Yale University) for discussions and training of N. D. F. in RNA design and handling and Alejandro Martin Gonzalez for assistance in AFM imaging. We acknowledge funding support from the BaSyC program of NWO-OCW and from the ERC Advanced Grant 883684.References
- C. Schwayer, M. Sikora, J. Slováková, R. Kardos and C.-P. Heisenberg, Actin Rings of Power, Dev. Cell, 2016, 37, 493–506 CrossRef CAS PubMed.
- N. B. Gudimchuk and J. R. McIntosh, Regulation of microtubule dynamics, mechanics and function through the growing tip, Nat. Rev. Mol. Cell Biol., 2021, 22, 777–795 CrossRef CAS PubMed.
- Y. Olmos, The ESCRT Machinery: Remodeling, Repairing, and Sealing Membranes, Membranes, 2022, 12, 633 CrossRef CAS PubMed.
- M. Wang, C. Fang, B. Ma, X. Luo and Z. Hou, Regulation of cytokinesis: FtsZ and its accessory proteins, Curr. Genet., 2020, 66, 43–49 CrossRef CAS PubMed.
- Y. Caspi and C. Dekker, Dividing the Archaeal Way: The Ancient Cdv Cell-Division Machinery, Front. Microbiol., 2018, 9, 174 CrossRef PubMed.
- T. Litschel, et al., Reconstitution of contractile actomyosin rings in vesicles, Nat. Commun., 2021, 12, 2254 CrossRef CAS PubMed.
- P. W. K. Rothemund, Folding DNA to create nanoscale shapes and patterns, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- M. Langecker, et al., Synthetic lipid membrane channels formed by designed DNA nanostructures, Science, 2012, 338, 932–936 CrossRef CAS PubMed.
- S. Krishnan, et al., Molecular transport through large-diameter DNA nanopores, Nat. Commun., 2016, 7, 12787 CrossRef CAS PubMed.
- A. Fragasso, et al., Reconstitution of Ultrawide DNA Origami Pores in Liposomes for Transmembrane Transport of Macromolecules, ACS Nano, 2021, 15, 12768–12779 CrossRef CAS PubMed.
- E. Bertosin, et al., A nanoscale reciprocating rotary mechanism with coordinated mobility control, Nat. Commun., 2021, 12, 78 CrossRef PubMed.
- X. Shi, et al., A nanopore-powered DNA turbine. arXiv:2206.06612, 2022.
- L. Nasalean, S. Baudrey, N. B. Leontis and L. Jaeger, Controlling RNA self-assembly to form filaments, Nucleic Acids Res., 2006, 34(5), 1381–1392 CrossRef CAS PubMed.
- L. J. Stenke and B. Saccà, Design, Mechanical Properties, and Dynamics of Synthetic DNA Filaments, Bioconjugate Chem., 2023, 34(1), 37–50 CrossRef CAS PubMed.
- P. W. Rothemund, A. Ekani-Nkodo, N. Papadakis, A. Kumar, D. K. Fygenson and E. Winfree, Design and characterization of programmable DNA nanotubes, J. Am. Chem. Soc., 2004, 126(50), 16344–16352 CrossRef CAS PubMed.
- P. Yin, R. F. Hariadi, S. Sahu, H. M. Choi, S. H. Park, T. H. LaBean and J. H. Reif, Programming DNA tube circumferences, Science, 2008, 321(5890), 824–826 CrossRef CAS PubMed.
- J. M. Stewart, C. Geary and E. Franco, Design and Characterization of RNA Nanotubes, ACS Nano, 2019, 13, 5214–5221 CrossRef CAS PubMed.
- C. Geary, A. Chworos and L. Jaeger, Promoting RNA helical stacking via A-minor junctions, Nucleic Acids Res., 2011, 39(3), 1066–1080 CrossRef CAS PubMed.
- J. M. Stewart, H. K. K. Subramanian and E. Franco, Self-assembly of multi-stranded RNA motifs into lattices and tubular structures, Nucleic Acids Res., 2017, 45(9), 5449–5457 CrossRef CAS PubMed.
- L. Rackley, J. M. Stewart, J. Salotti, A. Krokhotin, A. Shah, J. R. Halman, R. Juneja, J. Smollett, L. Lee, K. Roark, M. Viard, M. Tarannum, J. Vivero-Escoto, P. F. Johnson, M. A. Dobrovolskaia, N. V. Dokholyan, E. Franco and K. A. Afonin, RNA Fibers as Optimized Nanoscaffolds for siRNA Coordination and Reduced Immunological Recognition, Adv. Funct. Mater., 2018, 28(48), 1805959 CrossRef PubMed.
- D. K. Agrawal, R. Jiang, S. Reinhart, A. M. Mohammed, T. D. Jorgenson and R. Schulman, Terminating DNA Tile Assembly with Nanostructured Caps, ACS Nano, 2017, 11(10), 9770–9779 CrossRef CAS PubMed.
- S. Agarwal, M. A. Klocke, P. E. Pungchai and E. Franco, Dynamic self-assembly of compartmentalized DNA nanotubes, Nat. Commun., 2021, 12(1), 3557 CrossRef CAS PubMed.
- P. Zhan, K. Jahnke, N. Liu and K. Göpfrich, Functional DNA-based cytoskeletons for synthetic cells, Nat. Chem., 2022, 14(8), 958–963 CrossRef CAS PubMed.
- N. Arulkumaran, M. Singer, S. Howorka and J. R. Burns, Creating complex protocells and prototissues using simple DNA building blocks, Nat. Commun., 2023, 14(1), 1314 CrossRef CAS PubMed.
- C. Geary, P. W. K. Rothemund and E. S. Andersen, A single-stranded architecture for cotranscriptional folding of RNA nanostructures, Science, 2014, 345, 799–804 CrossRef CAS PubMed.
- D. Liu, et al., Branched kissing loops for the construction of diverse RNA homooligomeric nanostructures, Nat. Chem., 2020, 12, 249–259 CrossRef CAS PubMed.
- A. Krissanaprasit, C. Key, M. Fergione, K. Froehlich, S. Pontula, M. Hart, P. Carriel, J. Kjems, E. S. Andersen and T. H. LaBean, Genetically Encoded, Functional Single-Strand RNA Origami: Anticoagulant, Adv. Mater., 2019, 31(21), e1808262 CrossRef PubMed.
- G. Pothoulakis, M. T. A. Nguyen and E. S. Andersen, Utilizing RNA origami scaffolds in Saccharomyces cerevisiae for dCas9-mediated transcriptional control, Nucleic Acids Res., 2022, 50(12), 7176–7187 CrossRef CAS PubMed.
- C. Geary, G. Grossi, E. K. S. McRae, P. W. K. Rothemund and E. S. Andersen, RNA origami design tools enable cotranscriptional folding of kilobase-sized nanoscaffolds, Nat. Chem., 2021, 13, 549–558 CrossRef CAS PubMed.
- J. Lipfert, et al., Double-stranded RNA under force and torque: similarities to and striking differences from double-stranded DNA, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 15408–15413 CrossRef CAS PubMed.
- A. Chopra, S. Sagredo, G. Grossi, E. S. Andersen and F. C. Simmel, Out-of-Plane Aptamer Functionalization of RNA Three-Helix Tiles, Nanomaterials, 2019, 9(4), 507 CrossRef CAS PubMed.
- D. Shu, Y. Shu, F. Haque, S. Abdelmawla and P. Guo, Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics, Nat. Nanotechnol., 2011, 6, 658–667 CrossRef CAS PubMed.
- E. F. Khisamutdinov, et al., Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square and pentagon nanovehicles, Nucleic Acids Res., 2014, 42, 9996–10004 CrossRef CAS PubMed.
- G. S. Manning, The persistence length of DNA is reached from the persistence length of its null isomer through an internal electrostatic stretching force, Biophys. J., 2006, 91, 3607–3616 CrossRef CAS PubMed.
- A. Khmelinskaia, J. Mücksch, E. P. Petrov, H. G. Franquelim and P. Schwille, Control of Membrane Binding and Diffusion of Cholesteryl-Modified DNA Origami Nanostructures by DNA Spacers, Langmuir, 2018, 34, 14921–14931 CrossRef CAS PubMed.
- J. Nix, D. Sussman and C. Wilson, The 1.3 A crystal structure of a biotin-binding pseudoknot and the basis for RNA molecular recognition, J. Mol. Biol., 2000, 296, 1235–1244 CrossRef CAS PubMed.
- K. Azad, et al., Structural basis of CHMP2A-CHMP3 ESCRT-III polymer assembly and membrane cleavage, Nat. Struct. Mol. Biol., 2023, 30, 81–90 CrossRef CAS PubMed.
- F. Merino, S. Pospich and S. Raunser, Towards a structural understanding of the remodeling of the actin cytoskeleton, Semin. Cell Biol., 2020, 102, 51–64 CrossRef CAS PubMed.
- A. Blanch Jover, N. de Franceschi, D. Fenel, W. Weissenhorn and C. Dekker, The archaeal division protein CdvB1 assembles into polymers that are depolymerized by CdvC, FEBS Lett., 2022, 596, 958–969 CrossRef CAS PubMed.
- F. C. Simmel, B. Yurke and H. R. Singh, Principles and Applications of Nucleic Acid Strand Displacement Reactions, Chem. Rev., 2019, 119, 6326–6369 CrossRef CAS PubMed.
- L. N. Green, H. K. K. Subramanian, V. Mardanlou, J. Kim, R. F. Hariadi and E. Franco, Autonomous dynamic control of DNA nanostructure self-assembly, Nat. Chem., 2019, 11(6), 510–520 CrossRef CAS PubMed.
- L. Olivi, et al., Towards a synthetic cell cycle, Nat. Commun., 2021, 12, 4531 CrossRef CAS PubMed.
- N. de Franceschi, M. Alqabandi, W. Weissenhorn and P. Bassereau, Dynamic and Sequential Protein Reconstitution on Negatively Curved Membranes by Giant Vesicles Fusion, Bio-Protoc., 2019, 9(13), e3294 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nr06423a |
| This journal is © The Royal Society of Chemistry 2024 |