Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Exploiting cyclodextrins as artificial chaperones to enhance enzyme protection through supramolecular engineering†
Ali
Foroutan Kalourazi‡
 ab,
Seyed Amirabbas
Nazemi‡
ab,
Seyed Amirabbas
Nazemi‡
 a,
Ajmal Roshan
Unniram Parambil
a,
Ajmal Roshan
Unniram Parambil
 ac,
Ruben
Muñoz-Tafalla
ac,
Ruben
Muñoz-Tafalla
 de,
Paula
Vidal
f,
S. Shirin
Shahangian
b,
Victor
Guallar
de,
Paula
Vidal
f,
S. Shirin
Shahangian
b,
Victor
Guallar
 dg,
Manuel
Ferrer
dg,
Manuel
Ferrer
 f and
Patrick
Shahgaldian
f and
Patrick
Shahgaldian
 *ac
*ac
aSchool of Life Science, University of Applied Sciences and Arts Northwestern Switzerland, Hofackerstrasse 30, Muttenz CH-4132, Switzerland. E-mail: patrick.shahgaldian@fhnw.ch
bDepartment of Biology, Faculty of Sciences, University of Guilan, Rasht, Iran. E-mail: shahangian@guilan.ac.ir
cSwiss Nanoscience Institute, Klingelbergstrasse 82, Basel CH-4056, Switzerland
dBarcelona Supercomputing Center (BSC), 08034 Barcelona, Spain
eFaculty of Pharmacy and Food Science, Universitat de Barcelona (UB), 08007 Barcelona, Spain
fInstituto de Catalisis y Petroleoquimica (ICP), CSIC, 28049 Madrid, Spain
gInstitution for Research and Advanced Studies (ICREA), 08010 Barcelona, Spain
First published on 13th February 2024
Abstract
We report a method of enzyme stabilisation exploiting the artificial protein chaperone properties of β-cyclodextrin (β-CD) covalently embedded in an ultrathin organosilica layer. Putative interaction points of this artificial chaperone system with the surface of the selected enzyme were studied in silico using a protein energy landscape exploration simulation algorithm. We show that this enzyme shielding method allows for drastic enhancement of enzyme stability under thermal and chemical stress conditions, along with broadening the optimal temperature range of the biocatalyst. The presence of the β-CD macrocycle within the protective layer supports protein refolding after treatment with a surfactant.
Introduction
Molecular chaperones are a family of multidomain proteins involved in the protein production machinery.1–4 They support nascent proteins to achieve functional folding. They are also involved in unfolding misfolded proteins or folded proteins sentenced for proteolysis.5,6 Chaperones are also used to enable or increase yields of recombinant protein expression.7–9 In addition, they substantially increase protein stability under thermal stress conditions.10–12 Inspired by natural chaperones, chemists have created a myriad of strategies to produce artificial chaperone molecules or materials that support protein folding.13,14 For example, mixed-shell polymeric micelles, when carefully designed, assist protein folding.15–17 Among artificial molecular chaperones, cyclodextrins (CDs), cyclic oligomers of glucose, have been shown to act as chaperones and support protein folding.18,19 This effect arises from a combination of hydrogen-bonding of the hydroxyl rims of the CD with the surface of the protein, and inclusion of aromatic amino acids (e.g., Phe, Tyr) in the cavity of the macrocycle.20,21 CDs have also been evaluated as pharmacological chaperones. For example, Garcia-Moreno et al. demonstrated that a fluorinated cyclodextrin acts as a pharmaceutical chaperone and rescues lysosomal glucocerebrosidase enzyme mutants involved in neuropathic forms of Gaucher disease. CDs’ chaperone properties have also been exploited to design materials allowing for recombinant protein refolding from protein aggregates and inclusion bodies; such materials are commercially available.22–24 CDs have also been utilised to protect enzymes. For example, CD-responsive nanogels were demonstrated to provide stabilisation to enzymes such as carbonic anhydrase B25 and horseradish peroxidase.26In our effort to stabilise enzymes and produce robust nanobiocatalysts, we worked on the concept of supramolecular enzyme engineering. It refers to a concept of enzyme supramolecular modification without manipulating the protein sequence. It aims at meticulously controlling the enzyme nano-environment. Applying this approach, we developed a method to protect enzymes immobilised at the surface of silica nanoparticles by growing, in a thickness-controlled fashion (down to nanometre precision), a protective organosilica layer.27 The protection of the enzyme was shown to arise from a large set of supramolecular interactions established between the protein surface and the organosilica layer.28 These interactions may also be optimised in order to engineer the active site of the protected enzyme and to switch a non-enantioselective enzyme into a selective one.29
Herein, we report a method of enzyme immobilisation and supramolecular organosilica shielding. This method exploits the protein chaperone properties of a novel cyclodextrin building block, Fig. 1. The enzyme selected to establish the proof of concept of this method, a lipase from the Bacillus genus, when shielded in CD-containing organosilica, exhibits not only higher thermal stability but also an improved capacity to refold after chaotropic treatment. We expect this method to be versatile in that it can be applied to a range of enzymes for which thermal stability is an issue. This method can eventually contribute to improve the efficiency of industrial biocatalytic processes.
Results
As model enzyme, we used a lipase enzyme (LipMRD9) from the Bacillus genus (WP_034624255.1), identified by sequence-based metagenomic bioprospecting (see ESI† for details). First, we studied the possible CD-binding sites at the surface of this model protein. To that end, we employed the SiteFinder PELE protocol. The simulation encompassed 191![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 sampling steps, comprehensively exploring the protein surface. Fig. 2A illustrates the energy profile (created with Matplotlib library)30 of the LipMRD9-CD interaction, where we monitored the ligand solvent accessible surface area (SASA) versus the protein–CD binding energy. Notably, few binding poses reached energies of −60 kcal mol−1, indicative of a robust interaction between the CD and the protein's surface. Furthermore, the lowest binding energies align with the minimum ligand SASA and minimum total energies of the system, reinforcing the concept of heightened affinity in that specific protein region. A focused analysis was then performed on those poses with the lowest binding energy, based on filtering the CD orientation where secondary alcohols are required to be oriented towards the protein surface.21 This was justified by the fact that the CD building block produced is modified at the primary rim; this is expected to drastically reduce its ability to bind to the protein surface via the primary rim. Out of the 25 steps with a binding energy lower than −50 kcal mol−1, 9 conformed to the correct orientation. Within these 9 steps, we identified three distinct binding sites (Fig. 2B).
000 sampling steps, comprehensively exploring the protein surface. Fig. 2A illustrates the energy profile (created with Matplotlib library)30 of the LipMRD9-CD interaction, where we monitored the ligand solvent accessible surface area (SASA) versus the protein–CD binding energy. Notably, few binding poses reached energies of −60 kcal mol−1, indicative of a robust interaction between the CD and the protein's surface. Furthermore, the lowest binding energies align with the minimum ligand SASA and minimum total energies of the system, reinforcing the concept of heightened affinity in that specific protein region. A focused analysis was then performed on those poses with the lowest binding energy, based on filtering the CD orientation where secondary alcohols are required to be oriented towards the protein surface.21 This was justified by the fact that the CD building block produced is modified at the primary rim; this is expected to drastically reduce its ability to bind to the protein surface via the primary rim. Out of the 25 steps with a binding energy lower than −50 kcal mol−1, 9 conformed to the correct orientation. Within these 9 steps, we identified three distinct binding sites (Fig. 2B).
After identifying the most promising three CD binding sites, a subsequent round of the SiteFinder PELE protocol was initiated. In this phase, the protein bound to the three CDs was considered the receptor, while an additional CD molecule was designated as the ligand. This strategy aimed to comprehensively explore the protein's surface with the three top binding sites already occupied, shedding light on potential tendencies of CDs to bind proteins that were already engaged with other CDs. As depicted in the scatter plot in Fig. 3A, just a few simulation steps managed to surpass the −50 kcal mol−1 barrier. However, upon closer examination of the poses exhibiting binding energy below −40 kcal mol−1, a total of 44 structures, we identified 17 accurately positioned, ultimately leading to the selection of 5 distinct binding sites. Consequently, a total of 8 binding sites were now discernible (Fig. 3B). For each of the identified CD binding sites, a structural analysis was conducted, and the results are presented in Table S1 (ESI†). As depicted in the table, every pose exhibits a minimum of 5 hydrogen bonds between the protein and the ligand, among other interactions. The residues found within the CD cavities comprise Tyr, Leu, Asn and Ile. Three representative poses showcasing these interactions are shown in Fig. 2B. This set of results collectively supported the hypothesis of CDs binding at the protein surface through synergistic hydrogen bonding and hydrophobic inclusion.
Considering that shield formation occurs via organosilane self-sorting at the protein surface followed by polycondensation,27,31 we decided to produce a CD-derivative that can be used as a layer building block, i.e. bearing a tri-ethoxysilane moiety. To that end, native β-CD was reacted with 3-isocyanatopropyltrimethoxysilane in dimethylformamide (80 °C); Fig. 4. The reaction product, CD-TES, was characterised by means of nuclear magnetic resonance (1H and 13C), Fourier transform infrared spectroscopy and mass spectrometry. Collectively, the results confirmed the successful carbamate bond formation with an average of 1.5 moiety per CD macrocycle (Fig. S2–5, ESI†).
 | ||
| Fig. 4 Chemical synthesis of a CD-TES, a β-CD derivative bearing tri-ethoxysilane functions, n ≈ 1.5 as estimated by 1H NMR. | ||
Our model enzyme, LipMRD9 (specific activity 19.3 U mg−1) was immobilised at the surface of amino-modified SPs (average diameter 290 nm ± 20 nm) following a procedure previously published.27 The SPs were further reacted with a mixture of tetra-ethyl-orthosilicate (TEOS), aminopropyltriethoxysilane (APTES) and CD-TES to yield SPs with immobilised LipMRD9 and shielded within a CD-containing organosilica layer, hereafter referred to as SP-LipMRD9-OSCD. Reference particles, SP-LipMRD9-OSREF, were produced by omitting the addition of CD-TES. The particles produced were characterised by scanning electron microscopy (SEM); Fig. 5. The micrographs showed, for SP-LipMRD9-OSCD, a close-to-linear increase of the particles’ diameter at a rate of ca. 6.5 nm h−1; Fig. 5E.
It was accompanied by a moderate increase in surface roughness over the reaction duration, without impact on the polydispersity index which remained of 0.005. The layer growth kinetics for SP-LipMRD9-OSREF followed a similar trend confirming the lack of relevant effect of CD-TES on the layer growth kinetics; Fig. 5F.
Next, we measured the biocatalytic activity of the immobilised enzymes using an established spectrophotometric assay of p-nitrophenyl butyrate hydrolysis; Fig. 6. The specific activity of SP-LipMRD9 was measured to be 14.0 U mg−1. For SP-LipMRD9-OSCD, an activity increase to 127% and 133% (normalized to the specific activity of SP-LipMRD9) was measured for layer thickness values of 4.2 and 8.1 nm, respectively. When the layer was thicker at 10.9 and 14.5 nm, the activity slightly decreased to 118% and 97%. A similar trend was found for SP-LipMRD9-OSREF; this is in good agreement with results obtained with other enzymes such as β-galactosidase or esterase.27–29
Next, we measured the influence of the artificial chaperone building block on the enzyme resistance to thermal stress conditions (Fig. 7A). To that end, the nanobiocatalysts produced were incubated at 50 °C for increasing durations. The soluble enzyme showed a decrease of activity to values of 62.1, 28.8 and 7.8% after 10, 20 and 30 min of reaction. This corresponds to a half-life of ca. 13 min. SP-LipMRD9 and SP-LipMRD9-OSREF displayed similar trends, with a slightly higher thermal stability than the soluble enzyme, with half-life values of 15 and 17 min, respectively. The stability of SP-LipMRD9-OSCD, however, is markedly higher, with values of activity of 93.3, 76.8, 53.6% after 10, 20 and 30 min of incubation. This corresponds to a half-life of ca. 32 min and represent an increase of 246% when compared to the soluble enzyme. This consistent increase in enzyme stability may be directly attributed to the presence of the β-CD macrocycle in the protective layer. A stronger set of interactions between the surface of the protective shell and that of the protein may contribute to limit the detrimental thermodynamics effects of temperature on the protein conformation. Further, the biocatalytic activity the nanobiocatalysts produced was also studied at increasing reaction temperatures (Fig. 7B). The soluble enzyme, LipMRD9, showed an optimum activity at 30 °C and activity values over 80% in the range of 25–35 °C. The engineered counterpart, SP-LipMRD9-OSCD, showed a maximum activity value at 40 °C and maintained more than 80% of activity over a broad range of 25–55 °C. The shift in the optimal temperature value and the relevant broadening of the temperature profile may be attributed to enzyme interactions with the support matrix. Collectively, these results bring compelling evidence of the stabilising effect of β-CD in the engineered organosilica layer.
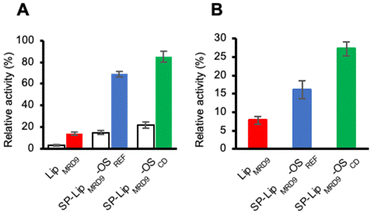
Thermal and chemical protein denaturation are taking place following different molecular mechanisms involving different thermodynamic pathways.32 The encouraging results obtained with thermal stabilisation prompted us to test the stabilisation effect of the chaperone-based protective layer against chemical stress conditions. First, we tested the influence of an anionic surfactant, namely sodium dodecyl sulphate (SDS). LipMRD9, SP-LipMRD9-OSREF, and SP-LipMRD9-OSCD were incubated for 20 min in a buffered solution containing SDS (1%) prior to activity measurements; results are shown in Fig. 8A. The activity of LipMRD9 dropped to 3%; activity values measured for SP-LipMRD9-OSREF and SP-LipMRD9-OSCD were consistently higher, with values of 15% and 21%, respectively. This demonstrated a moderate positive impact of the β-CD on the protein stability. Additionally, the possibility to refold the enzyme after SDS treatment was tested. To that end, LipMRD9-OSREF and SP-LipMRD9-OSCD were submitted to centrifugation and resuspended in Tris–HCl buffer. For the soluble enzyme, the surfactant was removed by dialysis against the same buffer. Activity measurements showed that while LipMRD9 recovered only 20% of activity, SP-LipMRD9-OSREF and SP-LipMRD9-OSCD showed significantly better results with 68 and 84%, respectively. This effect was previously observed for soluble cyclodextrins and a carbonic anhydrase enzyme.18 It was attributed to the capacity of the cyclodextrin macrocycle to strip away surfactant molecule. In the present work, it is more likely that the enzyme recovered its three-dimensional structure owing to the appropriate positioning of the CD moieties in the organosilica matrix. This effect of protein imprinting in organosilica layers was previously demonstrated with viruses.31
Next, we studied the denaturing effect of urea on LipMRD9 and engineered counterparts by measuring the enzymatic activity after 20 minutes incubation with urea solution (6 M); as shown in Fig. 8B, SP-LipMRD9-OSREF, and SP-LipMRD9-OSCD were able to retain 16 and 27% activity after treatment, respectively, while LipMRD9 could only retain 7% of its initial activity. This set of results suggest that the organosilica layer surrounding the enzyme supports its refolding as strongly suggested by the activity recovery values measured. While it is expected that the soluble enzyme may undergo irreversible aggregation and that immobilisation may prevent that, the significantly enhanced recovery for SP-LipMRD9-OSCD brings additional evidence of the stabilising effect of β-CD within the protective layer.
Conclusions
In summary, we have demonstrated that the use of a cyclodextrin tri-alkoxysilane derivative in the formulation of an enzyme via organosilica shielding provides the immobilised biocatalyst with improved stability. In silico modelling using a Monte Carlo-based simulation algorithm allowed the identification of putative binding sites, where the macrocycle interacts with the protein through synergistic hydrogen bonding and hydrophobic inclusion. Once immobilised at the surface of SPs and shielded in a CD-containing organosilica layer, the model enzyme exhibited thermal stability markedly higher than its counterparts. Additionally, the range of temperature at which the enzyme is active was broadened. When subjected to chaotropic stress conditions, supramolecularly engineered enzymes exhibited moderately higher stability. The protein's ability to refold in an active state, however, was markedly enhanced. This increased stability suggests that supramolecularly engineered enzymes with artificial chaperone systems could withstand harsh chemical and physical operational conditions. This is expected to provide a longer shelf life and potential for broader applications in biotechnology.Materials and methods
Chemicals
Tetraethyl orthosilicate (TEOS, ≥99%), (3-aminopropyl)triethoxysilane (APTES, ≥98%), 3-(triethoxysilyl)-propylisocyanate (95%), Trizma® hydrochloride (Tris–HCl, ≥98%), ammonium hydroxide (ACS grade, 28–30%), ethanol (ACS grade, anhydrous), 2-(N-morpholino)ethanesulfonic acid monohydrate (MES, ≥99.0%) and ammonium bicarbonate (≥98%) were purchased from Sigma-Aldrich (Switzerland). β-Cyclodextrin (97%) was purchased from BLD Pharmatech (Germany). Dimethylformamide (DMF, anhydrous, 99.9%) and Pierce™ BCA protein assay kit were purchased from Thermo Fisher Scientific (Switzerland).Buffer solutions
Buffer solutions were prepared with nanopure water. ABC buffer: 10 mM ammonium bicarbonate, 1.5 mM MgCl2, pH 7.8. MES buffer: 10 mM MES, 1.5 mM MgCl2, pH 7.5. Tris buffer: 50 mM Tris-HCl, pH 8.5.Characterisation
Activity assays and bioconjugation reactions were performed and monitored using a Synergy H1 spectrometer (BioTek, Switzerland) into a 96-well plate (Microplate 96 Well Half Area). Nanopure water (resistivity ≥18 MΩ cm) was produced with a Millipore Synergy® purification system (Merck). Particles were imaged using a Zeiss SUPRA® 40VP scanning electron microscope (SEM). 1H and 13C NMR (400 and 100 MHz respectively) were recorded on Bruker Avance-III HD spectrometer in DMSO-d6 or MeOD using residual solvent signals as the internal standard. Infrared (IR) spectra were collected in attenuated total reflection (ATR) using a single reflection diamond ATR and Agilent Cary 630 FTIR Spectrometer. High resolution mass spectra were obtained with Bruker MaXis 4G spectrometer (Electrospray ionisation). For SEM imaging, a solution of bare SPs, SP-LipMRD9-OSCD and SP-LipMRD9-OSREF in nanopure water was prepared and spread on a silicon substrate. The samples were allowed to dry at 20 °C under atmospheric conditions and were subsequently sputter-coated with gold for 15 seconds at 20 mA. Secondary electron micrographs were acquired using the InLens mode with an accelerating voltage of 10 kV at a magnification of 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×. Particle sizes were measured on the acquired micrographs using the ImageJ free software. 100 measurements, at least, were performed for each sample (Tables S3 and S4, ESI†). Statistical analysis of SEM micrographs on SP-LipMRD9-OSREF is shown in Fig. S6 (ESI†).
000×. Particle sizes were measured on the acquired micrographs using the ImageJ free software. 100 measurements, at least, were performed for each sample (Tables S3 and S4, ESI†). Statistical analysis of SEM micrographs on SP-LipMRD9-OSREF is shown in Fig. S6 (ESI†).
Modelling and simulations
PELE (Protein Energy Landscape Exploration) is a Monte Carlo-based simulation algorithm that unfolds in two phases.33 In the initial phase, the ligand undergoes random translation and rotation, while the protein backbone is perturbed using a normal mode method based on an anisotropic network model (ANM).34 Moving to the second phase, optimisation of all rotamers from the ligand and its surrounding side chains occurs through a rotamer library, followed by an overall structural minimisation. Subsequently, acceptance or rejection of the new conformation is determined according to the Metropolis criterion. PELE can be used to study a large number of biological problems, including mapping molecular mechanism,35 enzyme engineering,36,37 or binding site finder.38 This last case is the one of interest in this work. The Site Finder protocol in PELE is a tool that conducts a global exploration across the protein surface to identify an appropriate binding site for a specified ligand. For each processor, a protein–ligand replica simulation is created, where the ligand is placed randomly and explores a defined area of the protein's surface. Note that the random position depends on previously positioned ligands, aiming at exploring novel surface areas; with enough processors (>32), the whole surface will be explored. For the best bound poses, those with lower binding energy and correct positioning (secondary alcohols pointing to the protein surface) were selected and studied. In some cases, a residue was found inside the cavity of the CD macrocycle. The structure of the protein used in this work, LipMRD9, was generated using AlphaFold2,39 and prepared with the protein preparation wizard, Schrödinger (United States).40 Then the structure was set up for the site finder protocol using 192 processors, for a total of 191![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Monte Carlos steps.
000 Monte Carlos steps.
CD-TES synthesis
The cyclodextrin derivative was synthesised according to the procedure of Yilmaz et al. with slight adaptations.41 Cyclodextrin was dried overnight at 70 °C prior to the reaction. 2 mmol of cyclodextrin was dissolved in 20 mL dry DMF. 2 equivalents (4 mmol) of 3-(triethoxysilyl)-propylisocyanate were added and the reaction mixture was stirred for 12 hours at 80 °C. The mixture was cooled to room temperature and precipitated with acetone. The precipitate, CD-TES, was washed several times with acetone, collected and dried under vacuum.LipMRD9 immobilisation and shielding
Extensive details regarding the sequence-based metagenomic bioprospecting and the source and production of LipMRD9 are provided in the ESI section.† Silica nanoparticles (SPs) were produced following a method described by Cumbo et al.31 inspired by the Stöber method.42 The nanoparticles synthesised were diluted in 400 mL H2O to obtain a solution of [SPs] = 3.2 mg mL−1. APTES (260 μL) was added to the previous solution, and the reaction mixture was stirred at 20 °C, 400 rpm for 90 minutes. Subsequently, the suspension was centrifuged at 4000 rpm for 15 minutes, and the supernatant was removed. The particles were resuspended in nanopure H2O (50 mL) and submitted to ultrasonic treatment. This washing step was repeated twice. The particles produced, SP-NH2, were then resuspended in water (50 mL) and stored at 4 °C. To a suspension of SP-NH2 (10 mL, 3.2 mg mL−1) was added glutaraldehyde (25%, 40 μL). The reaction mixture was stirred at 20 °C, 400 rpm for 30 min. Then, the suspension was washed twice with nanopure water, resuspended using an ultrasonic bath and resuspended in 5 mL MES buffer to yield SP-Glu (6.4 mg mL−1). To a suspension of SP-Glu (2 mL, 6.4 mg mL−1) was added 2 mL (in MES buffer) LipMRD9 (final concentration of 73.6 μg mL−1) to reach [SP-Glu] = 3.2 mg mL−1. The resulting mixture was stirred at 20 °C, 400 rpm for 1 h to produce SP-LipMRD9. The nanoparticles were centrifuged at 700 rcf for 5 minutes. A BCA assay was performed on the supernatant collected after enzyme immobilisation, which showed that 81% of the enzyme was immobilised at the surface of SPs; A summary of the results is presented in Table S2 (ESI†).SP-LipMRD9 (1.8 mL, 3.2 mg mL−1) was incubated with TEOS (4 μL) and CD-TES (13 mg) in 10 mM ABC buffer with 1.5 mM MgCl2 at 10 °C, 400 rpm for 1 h. Subsequently, APTES (1.6 μL) was added to the reaction media. The resulting mixture was stirred at 10 °C, 400 rpm for 120 minutes. Aliquots of 400 μL were collected every 30 minutes. The resulting nanoparticles were centrifuged at 700 rcf for 5 minutes, washed and resuspended in 400 μL Tris–HCl buffer ([SP-LipMRD9-OSCD] = 3.2 mg mL−1). A control experiment was conducted by replacing β-CD derivative with a volume of 2 μL of TEOS and following the same shielding procedure as previously described ([SP-LipMRD9-OSREF] = 3.2 mg mL−1). The resulting suspension was allowed to cure at 20 °C for 18 hours for further activity recovery and stored at 4 °C.
LipMRD9 activity assay
LipMRD9 activity was measured spectrophotometrically using p-nitrophenyl butyrate (p-NPB) as substrate at 410 nm. The reaction mixture contained immobilised/soluble LipMRD9 (3 μg mL−1), 1 mM p-NPB in isopropanol and 50 mM Tris–HCl (pH 8.5).Author contributions
AFK, MF, SSS and PS conceptualised the work. AFK carried out enzymatic assays. SAN carried out the SP synthesis and microscopic characterisation. SSS and PS contributed to biocatalytic results’ analyses. ARUP synthesised and characterised CD-TES. RMT and VG were in charge of in silico modelling and interactions studies. PV contributed by enzyme bioprospecting, synthesis, expression, production, and characterisation. PS and SSS supervised the project. The manuscript was written through the contributions of all authors who gave their approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the FuturEnzyme project, which is funded by the European Union's Horizon 2020 program, under grant agreement number 101000327. Funding from the Ministerio de Ciencia e Innovación and Agencia Estatal de Investigación (AEI) and the Fondo Europeo de Desarrollo Regional (ERDF) A way of making Europe and the European Union NextGeneration EU/PRTR are gratefully acknowledged. We also thank David Almendral and Laura Fernandez-Lopez for their support in enzyme analysis.References
- H. Saibil, Nat. Rev. Mol. Cell Biol., 2013, 14, 630–642 CrossRef CAS PubMed.
- P. Koldewey, F. Stull, S. Horowitz, R. Martin and J. C. A. Bardwell, Cell, 2016, 166, 369–379 CrossRef CAS PubMed.
- F. U. Hartl, A. Bracher and M. Hayer-Hartl, Nature, 2011, 475, 324–332 CrossRef CAS PubMed.
- D. Balchin, M. Hayer-Hartl and F. U. Hartl, FEBS Lett., 2020, 594, 2770–2781 CrossRef CAS PubMed.
- B. Margulis, A. Tsimokha, S. Zubova and I. Guzhova, Cells, 2020, 9, 1308 CrossRef CAS PubMed.
- F. den Brave, L. V. Cairo, C. Jagadeesan, C. Ruger-Herreros, A. Mogk, B. Bukau and S. Jentsch, Cell Rep., 2020, 31, 107680 CrossRef CAS PubMed.
- K. Fatima, F. Naqvi and H. Younas, Cell Biochem. Biophys., 2021, 79, 153–174 CrossRef CAS PubMed.
- M. Martinez-Alonso, V. Toledo-Rubio, R. Noad, U. Unzueta, N. Ferrer-Miralles, P. Roy and A. Villaverde, Appl. Environ. Microbiol., 2009, 75, 7850–7854 CrossRef CAS PubMed.
- R. Zhao, M. Davey, Y. C. Hsu, P. Kaplanek, A. Tong, A. B. Parsons, N. Krogan, G. Cagney, D. Mai, J. Greenblatt, C. Boone, A. Emili and W. A. Houry, Cell, 2005, 120, 715–727 CrossRef CAS PubMed.
- M. Shanmugasundaram, N. V. Pavlova, A. R. Pavlov, I. K. Lednev and F. T. Robb, Methods Enzymol., 2021, 659, 145–170 CAS.
- L. Zhao, G. Vecchi, M. Vendruscolo, R. Korner, M. Hayer-Hartl and F. U. Hartl, Cell Rep., 2019, 28, 1335–1345 CrossRef CAS PubMed.
- D. Xu, S. Lim, Y. Cao, A. Abad, A. N. Kang and D. S. Clark, Chem. Commun., 2021, 57, 5511–5513 RSC.
- F. H. Ma, C. Li, Y. Liu and L. Q. Shi, Adv. Mater., 2020, 32, E1805945 CrossRef PubMed.
- O. Hanpanich and A. Maruyama, Biomaterials, 2020, 254, 120150 CrossRef CAS PubMed.
- F. Huang, J. Wang, A. Qu, L. Shen, J. Liu, J. Liu, Z. Zhang, Y. An and L. Shi, Angew. Chem., Int. Ed., 2014, 53, 8985–8990 CrossRef CAS PubMed.
- J. Wang, T. Yin, F. Huang, Y. Song, Y. An, Z. Zhang and L. Shi, ACS Appl. Mater. Interfaces, 2015, 7, 10238–10249 CrossRef CAS PubMed.
- J. Z. Wang, Y. Q. Song, P. C. Sun, Y. L. An, Z. K. Zhang and L. Q. Shi, Langmuir, 2016, 32, 2737–2749 CrossRef CAS PubMed.
- D. Rozema and S. H. Gellman, J. Am. Chem. Soc., 1995, 117, 2373–2374 CrossRef CAS.
- A. Barzegar, A. A. Moosavi-Movahedi, K. Mahnam and S. H. Ashtiani, Carbohydr. Res., 2010, 345, 243–249 CrossRef CAS PubMed.
- D. E. Otzen, B. R. Knudsen, F. Aachmann, K. L. Larsen and R. Wimmer, Protein Sci., 2002, 11, 1779–1787 CrossRef CAS PubMed.
- A. Barzegar, A. A. Moosavi-Movahedi, K. Mahnam and S. H. Ashtiani, Carbohydr. Res., 2010, 345, 243–249 CrossRef CAS PubMed.
- D. Rozema and S. H. Gellman, J. Am. Chem. Soc., 1995, 117, 2373–2374 CrossRef CAS.
- D. Nath and M. Rao, Eur. J. Biochem., 2001, 268, 5471–5478 CrossRef CAS PubMed.
- A. Singh, V. Upadhyay, A. K. Upadhyay, S. M. Singh and A. K. Panda, Microb. Cell Fact., 2015, 14, 41 CrossRef PubMed.
- Y. Nomura, Y. Sasaki, M. Takagi, T. Narita, Y. Aoyama and K. Akiyoshi, Biomacromolecules, 2005, 6, 447–452 CrossRef CAS PubMed.
- S.-i. Sawada, Y. Sasaki, Y. Nomura and K. Akiyoshi, Colloid Polym. Sci., 2011, 289, 685–691 CrossRef CAS.
- M. R. Correro, N. Moridi, H. Schutzinger, S. Sykora, E. M. Ammann, E. H. Peters, Y. Dudal, F. X. Corvini and P. Shahgaldian, Angew. Chem., Int. Ed., 2016, 55, 6285–6289 CrossRef CAS PubMed.
- M. R. Correro, M. Takacs, S. Sykora, P. F. X. Corvini and P. Shahgaldian, RSC Adv., 2016, 6, 89966–89971 RSC.
- C. I. Giunta, I. Cea-Rama, S. Alonso, M. L. Briand, R. Bargiela, C. Coscolin, P. F. X. Corvini, M. Ferrer, J. Sanz-Aparicio and P. Shahgaldian, ACS Nano, 2020, 14, 17652–17664 CrossRef CAS PubMed.
- J. D. Hunter, Comput. Sci. Eng., 2007, 9, 90–95 Search PubMed.
- A. Cumbo, B. Lorber, P. F. X. Corvini, W. Meier and P. Shahgaldian, Nat. Commun., 2013, 4, 1503 CrossRef PubMed.
- A. Narayan, K. Bhattacharjee and A. N. Naganathan, Biochemistry, 2019, 58, 2519–2523 CrossRef CAS PubMed.
- K. W. Borrelli, A. Vitalis, R. Alcantara and V. Guallar, J. Chem. Theory Comput., 2005, 1, 1304–1311 CrossRef CAS PubMed.
- E. Eyal, L. W. Yang and I. Bahar, Bioinformatics, 2006, 22, 2619–2627 CrossRef CAS PubMed.
- S. Roda, G. Santiago and V. Guallar, Adv. Protein Chem. Struct. Biol., 2020, 122, 1–31 CrossRef CAS PubMed.
- L. Fernandez-Lopez, S. Roda, A. Robles-Martín, R. Muñoz-Tafalla, D. Almendral, M. Ferrer and V. Guallar, Int. J. Mol. Sci., 2023, 24, 13768 CrossRef CAS PubMed.
- G. Santiago, M. Martínez-Martínez, S. Alonso, R. Bargiela, C. Coscolín, P. N. Golyshin, V. Guallar and M. Ferrer, Biochemistry, 2018, 57, 2245–2255 CrossRef CAS PubMed.
- L. Díaz, D. Soler, G. Tresadern, C. Buyck, L. Perez-Benito, S. S. Saen-Oon, V. Guallar and R. Soliva, RSC Adv., 2020, 10, 7058–7064 RSC.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Zídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli and D. Hassabis, Nature, 2021, 596, 583–589 CrossRef CAS PubMed.
- G. M. Sastry, M. Adzhigirey, T. Day, R. Annabhimoju and W. Sherman, J. Comput.-Aided Mol. Des., 2013, 27, 221–234 CrossRef PubMed.
- M. Arslan, S. Sayin and M. Yilmaz, Tetrahedron: Asymmetry, 2013, 24, 982–989 CrossRef CAS.
- W. Stober, A. Fink and E. Bohn, J. Colloid Interface Sci., 1968, 26, 62–69 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available: Details of synthetic procedures, enzyme expression, SEM, immobilisation quantification, shielding kinetics, and enzyme-CD docking bioinformatics. See DOI: https://doi.org/10.1039/d3nr06044f |
| ‡ These authors equally contributed to the work. |
| This journal is © The Royal Society of Chemistry 2024 |