Open Access Article
Open Access ArticleUnderstanding the nanoscale adhesion forces between the fungal pathogen Candida albicans and antimicrobial zinc-based layered double hydroxides using single-cell and single-particle force spectroscopy
Jazia
Awassa
a,
Samantha
Soulé
 a,
Damien
Cornu
a,
Damien
Cornu
 a,
Christian
Ruby
a and
Sofiane
El-Kirat-Chatel
a,
Christian
Ruby
a and
Sofiane
El-Kirat-Chatel
 *ab
*ab
aUniversité de Lorraine, CNRS, LCPME, F-54000 Nancy, France. E-mail: sofiane.elkirat@u-bordeaux.fr
bUniv. Bordeaux, CNRS, Bordeaux INP, CBMN, UMR 5248, F-33600 Pessac, France
First published on 2nd February 2024
Abstract
Antifungal resistance has become a very serious concern, and Candida albicans is considered one of the most opportunistic fungal pathogens responsible for several human infections. In this context, the use of new antifungal agents such as zinc-based layered double hydroxides to fight such fungal pathogens is considered one possible means to help limit the problem of antifungal resistance. In this study, we show that ZnAl LDH nanoparticles exhibit remarkable antifungal properties against C. albicans and cause serious cell wall damage, as revealed by growth tests and atomic force microscopy (AFM) imaging. To further link the antifungal activity of ZnAl LDHs to their adhesive behaviors toward C. albicans cells, AFM-based single-cell spectroscopy and single-particle force spectroscopy were used to probe the nanoscale adhesive interactions. The force spectroscopy analysis revealed that antimicrobial ZnAl LDHs exhibit specific surface interactions with C. albicans cells, demonstrating remarkable force magnitudes and adhesion frequencies in comparison with non-antifungal negative controls, e.g., Al-coated substrates and MgAl LDHs, which showed limited interactions with C. albicans cells. Force signatures suggest that such adhesive interactions may be attributed to the presence of agglutinin-like sequence (Als) adhesive proteins at the cell wall surface of C. albicans cells. Our findings propose the presence of a strong correlation between the antifungal effect provided by ZnAl LDHs and their nanoscale adhesive interactions with C. albicans cells at both the single-cell and single-particle levels. Therefore, ZnAl LDHs could interact with C. albicans fungal pathogens by specific adhesive interactions through which they adhere to fungal cells, leading to their damage and subsequent growth inhibition.
1. Introduction
Fungal infections lead to various human diseases, ranging from superficial cutaneous to serious invasive fungal diseases (IFDs), which together are estimated to affect more than a billion people worldwide.1,2 The number of antifungal agents used in clinics is very limited compared to that of antibiotics.3 Historically, fungal infection treatment relied on four classes of clinical antifungal drugs: polyenes, azoles, echinocandins, and the pyrimidine analogue 5-flucytosine.4 However, the incidence of invasive fungal infections has always been on the rise, and the problem of antifungal resistance has become an emerging concern.5,6 For example, Candida albicans is a major opportunistic fungal pathogen responsible for several human infections in different body parts such as the oral cavity, skin, bloodstream, gastrointestinal tract, and vagina.7 Infections by C. albicans could further lead to severe morbidity and mortality, especially in immunocompromised patients.8 Given the significant concern of fungal infections and the limited arsenal of existing antifungal drugs, inorganic-based nanomaterials were considered good candidates for developing new antifungal agents.6,9 In this respect, the use of pristine layered double hydroxides (LDHs) as inorganic antifungal agents could play a very significant role. LDHs are anionic clays comprising structures based on the stacking of brucite-like sheets of divalent and trivalent metal hydroxides. The net positive charge of these sheets is compensated by the presence of negatively charged intercalating anions and water molecules in the interlaminar region.10–12Several recent studies have shown that zinc-based pristine LDHs could act as efficient antibacterial agents against various Gram-positive and Gram-negative bacterial strains.13–20 To the best of our knowledge, the only study claiming the antifungal activity of LDHs against fungal pathogens is that of Moaty et al.,13 in which the antibacterial and antifungal activities of ZnFe LDHs were tested against a broad range of bacterial and fungal pathogens. However, the authors admitted that the antifungal mechanism of LDHs was far from being understood, and therefore real experimental setups explaining the antifungal effect of LDHs are still lacking.
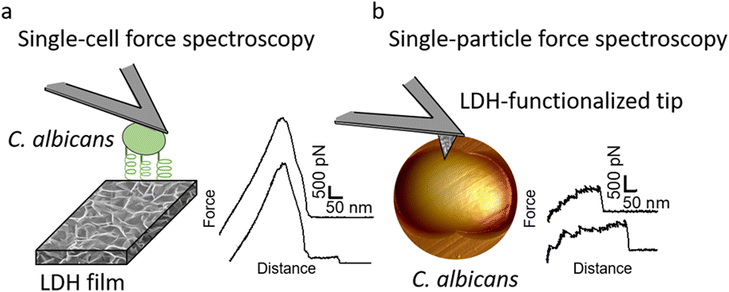
One possible explanation of the antifungal activity of zinc-based LDHs is direct surface interactions between the LDH surface and the cell wall of yeast cells. In our recent previous work, we have linked the importance of the direct surface interactions mechanism between the Gram-positive bacteria Staphylococcus aureus and ZnAl LDH nanoparticles (NPs) to their antibacterial mode of action.14 Here, our interest was to determine whether ZnAl LDH NPs could interact with C. albicans fungal pathogens to provide an efficient antifungal effect. Recently, remarkable advances were employed in the use of atomic force microscopy (AFM)-based single-cell and single-particle force spectroscopy (SCFS and SPFS, respectively) to probe the forces driving cell–cell and cell–solid interactions.21–26 Yet, these methods were rarely implemented to study the interactions between fungal pathogens and antifungal agents, e.g., ZnAl LDHs. In fact, SCFS and SPFS are powerful tools for probing the forces that drive cell–surface interactions.26,27 The general principle of SCFS is to immobilize a single living cell on AFM tips and to record the force–distance curves between the cell probe and a substrate, whereas SPFS mainly relies on functionalizing AFM probes with particles/molecules to probe interactions with cells at the single particle/molecule level.
In this work, the antifungal activity of ZnAl LDH NPs is evaluated against the fungal pathogen C. albicans using an agar disc diffusion assay and a broth microdilution test to determine their minimum inhibitory concentration (MIC). AFM imaging is employed to show the damage imposed by ZnAl LDH NPs on the cell wall of C. albicans cells. C. albicans cells were further attached to AFM tipless cantilevers to probe the nanoscale adhesion forces between these pathogenic yeasts and ZnAl LDHs using SCFS (Fig. 1a). Moreover, we took advantage of our original approach to functionalize AFM tips with LDH films to probe their interactions at the single-particle level using AFM-based SPFS (Fig. 1b). Compared to the negative controls, e.g., Al-coated surfaces and MgAl LDH films, ZnAl LDHs exerted specific adhesion forces on the cell wall of C. albicans at both the single-particle and single-cell levels. Such findings strongly support the correlation between the efficient antifungal effect of ZnAl LDH NPs and their interactions with C. albicans cells mediated by Als protein adhesion molecules.
2. Results and discussion
2.1. ZnAl LDH NPs exhibit significant antifungal activity against C. albicans
Before testing the antifungal activity of LDH NPs, ZnAl LDH NPs were prepared by a fast coprecipitation method. MgAl LDH NPs were also prepared using a similar synthesis method which served as a negative control.Nitrate-intercalated MgAl and ZnAl LDH NPs having a ZnII![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AlIII molar ratio of ∼2 were prepared by coprecipitation–hydrothermal method.14 Both LDH NPs presented a hexagonal lamellar structure as revealed by their SEM and AFM images (Fig. 2). The NPs also exhibited similar average particle sizes extracted from SEM imaging (169 ± 18 nm and 167 ± 40 nm, for MgAl and ZnAl NPs, respectively). Similar average particle sizes were also obtained from the AFM height images (170 ± 30 nm and 167 ± 33 nm, for MgAl and ZnAl LDH NPs, respectively). Both LDH NPs were well crystallized presenting neither crystalline nor amorphous impurities as suggested by our previously performed XRD, XPS, and IR analyses.14 The detailed structural and morphological characterization of these LDH NPs can be found in our recent previous study.14
AlIII molar ratio of ∼2 were prepared by coprecipitation–hydrothermal method.14 Both LDH NPs presented a hexagonal lamellar structure as revealed by their SEM and AFM images (Fig. 2). The NPs also exhibited similar average particle sizes extracted from SEM imaging (169 ± 18 nm and 167 ± 40 nm, for MgAl and ZnAl NPs, respectively). Similar average particle sizes were also obtained from the AFM height images (170 ± 30 nm and 167 ± 33 nm, for MgAl and ZnAl LDH NPs, respectively). Both LDH NPs were well crystallized presenting neither crystalline nor amorphous impurities as suggested by our previously performed XRD, XPS, and IR analyses.14 The detailed structural and morphological characterization of these LDH NPs can be found in our recent previous study.14
The antifungal activity of MgAl and ZnAl LDH NPs was then tested against the fungal pathogen C. albicans using the agar disc diffusion method. The generated inhibition zone diameters and their mean values obtained from three repetitive experiments are presented in Fig. 3. Moreover, the minimum inhibitory concentration (MIC) required to inhibit the growth of C. albicans by the two LDH NPs was determined using broth microdilution turbidimetric tests and three repetitive trials were performed as well (Fig. 3).
MgAl LDH NPs did not provide any antifungal effect, as revealed by the two antifungal activity tests, and thus played the role of negative control. On the other hand, ZnAl LDH NPs showed efficient antifungal activity against C. albicans with an average inhibition zone diameter of 15.3 mm and MIC value of 375 μg mL−1. The antifungal activity against the fungal pathogen C. albicans was weaker than the antibacterial effect achieved by ZnAl LDH NPs against S. aureus (inhibition zone diameter of 20.6 mm and MIC value of 93.8 μg mL−1).14 Such a difference between the antifungal and antibacterial activities of ZnAl LDH NPs may be explained by the different microorganisms tested, e.g., C. albicans and S. aureus cells, where each type of microorganism responds differently to the same biocide.13,28,29 Similarly, the antifungal activity of our ZnAl LDH NPs against C. albicans fungal pathogens was weaker than that of ZnO NPs synthesized by Yassin et al. (inhibition zone diameter = 24.18 mm and MIC value of 10 μg mL−1),30 but was still greater than the antifungal activity achieved by MgO and SiO2 NPs prepared by Karimiyan et al. (MIC > 3200 μg mL−1).31 Moreover, Jalal et al. also reported weaker antifungal activity of ZnO NPs against the same fungal strain, which showed an MIC value of 250 μg mL−1.32 The difference in the antifungal activity of our ZnAl LDH NPs, in this case, is attributed to the nature of the antifungal agent, or to the differences between followed protocols used to perform the antifungal activity tests.15,16
Compared to MgAl LDH NPs, the superior antifungal activity of ZnAl NPs could be attributed to the nature of their constituent divalent metal, e.g., Zn2+. We have previously shown that the antimicrobial activity of pristine LDHs is strongly dependent on the nature of the constituent divalent metal,14,15 and it was previously reported that zinc-based LDHs have remarkable antibacterial properties towards a broad range of bacteria.13–20 Moreover, Moaty et al., who performed the only antifungal study of ZnFe LDHs, also claimed that the antifungal effect of the tested LDH sample was attributed to the antifungal nature of the constituent divalent zinc metal.13 The antimicrobial properties of zinc-based LDHs are generally attributed to their ability to adhere to microorganisms and change their charge distribution, thus blocking the transport of nutrients in and out of the membrane, or else by binding to the intracellular DNA leading to its denaturation.20,33,34 Such an antimicrobial action mode requires the presence of direct surface interactions between the LDH and the tested microorganism, here C. albicans.
2.2. ZnAl LDH NPs impose serious cell wall damage on C. albicans cells
AFM imaging was used to visualize the surface topography of C. albicans cells before and after treatment with MgAl and ZnAl LDH NPs at a concentration of 375 μg mL−1, corresponding to the MIC value of ZnAl LDH NPs. Additionally, the roughness parameter Rq (root mean square of the heights) was obtained from high maginifcation AFM images acquired by scanning areas of 1 × 1 μm2 on the top of yeast cells.As shown in Fig. 4a and b, untreated yeast cells as well as those treated with non-antimicrobial MgAl LDH NPs presented well-defined, intact morphology with smooth surfaces (Rq = 17.9 ± 0.5 nm, and 18.2 ± 0.4 nm, for untreated and MgAl NP-treated cells, respectively).
On the other hand, cells treated with antimicrobial ZnAl LDH NPs were distorted with highly corrugated surfaces presenting hollows and crests (Fig. 4c). Similar cell damage observations were previously reported on C. albicans cells treated with different biocides under air and liquid conditions.35–38 Moreover, some cells were even completely damaged presenting irregular morphologies and dissociated cell walls leaving behind some cell debris within the polycarbonate membrane pores. Additionally, cells treated with ZnAl LDH NPs were found to have ∼2.5 times rougher surfaces (Rq = 50.4 ± 8.6 nm) than untreated and MgAl LDH NP-treated ones. The increase in surface roughness of yeast cells suggests the existence of serious cell wall damage induced by ZnAl LDH NPs. The structural integrity of the cell wall of C. albicans is an essential element for its survival. Therefore, damage to the cell wall might cause osmotic disorders in the fungal cell, rupture of the cell membrane, and subsequently the outflow of cellular and cytoplasmic components leading to cell death.39,40
All these observations suggest that ZnAl LDH NPs induce an efficient antifungal effect on C. albicans cells by targeting their cell walls leading to their distortion, damage, and subsequently the death of the microorganism.
2.3. Nanoscale adhesion forces between C. albicans and ZnAl LDHs
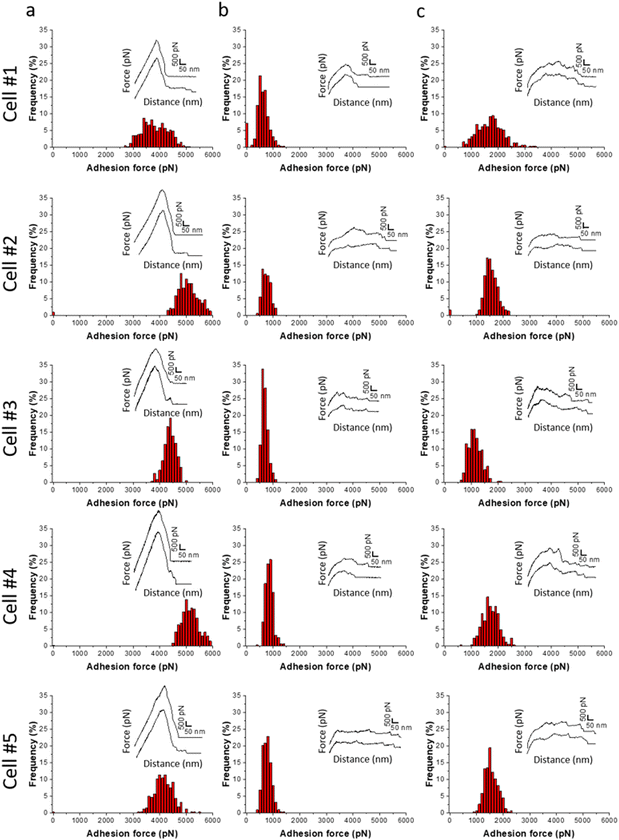
To determine whether the antifungal activity of ZnAl LDHs is related to their ability to interact with yeast cells leading to their damage, the nanoscale adhesion forces between C. albicans and ZnAl LDHs were probed at the single-cell level. For this purpose, living C. albicans cells were attached to tipless cantilevers coated with polydopamine acting as a bio-adhesive (Fig. 5a and b).21,26,27,41,42 The obtained fungal cell probes were brought into contact with ZnAl LDH films grown on silicon substrates using a wet-chemistry method adapted from Zhang et al.43 (Fig. 5c and d). As controls, Al-coated silicon substrates and MgAl LDH films were also probed using the same cell probes. Force–distance curves were recorded during constant approach and retraction of the yeast cell to and from the different substrates.Fig. 6a shows adhesion force histograms as well as representative force–distance (FD) curves obtained from probing ZnAl LDH films with different fungal probes. The adhesion frequency was ∼100% as all recorded retraction FD curves showed an adhesion signature. The slight variation (<2%) in adhesion frequencies among the probed ZnAl LDH films may be attributed to the heterogeneity in the attached cell microarchitecture and cell wall composition. The measured adhesion forces ranged from 3000 to 5000 pN with rupture lengths ranging between 140 and 650 nm (data not shown). Such findings suggest strong surface interactions (thousands of pN) occurring at the interface between the attached C. albicans cells and ZnAl LDH films.
Upon probing Al-coated silicon substrates using the same cell probes, adhesion force magnitudes did not exceed 1200 pN, which represents much weaker adhesion behavior than that observed with ZnAl LDH films (Fig. 6b).
MgAl LDH films probed with the same fungal probes showed slightly higher adhesion force magnitudes (500–2500 pN) in comparison with Al-coated surfaces. However, their adhesion behavior in terms of force magnitudes (Fig. 6c) was remarkably weaker than those of ZnAl LDH films. Considering that MgAl and ZnAl LDH films possess similar morphologies (Fig. 5c and d), one can deduce that the higher adhesion force magnitudes on ZnAl LDH films are not directly linked to their morphology but rather to their constituent divalent metal ions, e.g., Zn2+, playing a major role in the previously observed antifungal effect of Zn-based LDHs.13
Adhesion interactions between C. albicans cells and ZnAl LDHs were further probed at the single LDH particle level using SPFS. For this purpose, AFM tips were functionalized with ZnAl and MgAl LDH films in a similar procedure to that of the synthesis of LDH (Fig. 5d and e).14 Living C. albicans cells were immobilized in porous membranes and further probed using different functionalized tips, e.g., ZnAl-, MgAl- and Al-functionalized AFM tips. Due to their very small size, the physico-chemical characterizations on ZnAl- and MgAl-functionalized tips were performed on LDH films grown on silicon wafers using the exact same protocol used to synthesize LDH-functionalized tips. The detailed characterizations of these films were reported in our previous work.14 It is important here to mention that the morphology and the thickness of ZnAl and MgAl LDH films were very similar. The thickness values obtained by SEM cross-sectional imaging were 312 ± 24 nm and 315 ± 25 nm for MgAl LDH films and ZnAl LDH films, respectively. Moreover, the scratch method was also used to determine the thickness of the obtained films by AFM imaging and the obtained values (314 ± 10 nm and 316 ± 12 nm for MgAl film and ZnAl LDH films, respectively) were in good agreement with those found by SEM imaging.14
Fig. 7b and e shows adhesion force histograms associated with adhesion force maps and representative FD curves, respectively, obtained from different C. albicans cells probed with ZnAl tips. Adhesive white–grey pixels were randomly distributed on the maps suggesting the homogeneous adhesion of ZnAl LDH to the fungal cell surface. Most recorded FD curves showed an adhesion signature with a frequency ranging from 44 to 61% (Fig. 7b). Adhesion curves showed strong interactions (Fig. 7e), with multiple large force peaks (300–3000 pN, Fig. 7b) and long ruptures ranging from 200 to 500 nm (data not shown). These sawtooth force signatures are characteristic of Als proteins sequentially unfolding in the C. albicans cell wall.21,38,44,45
Upon probing the same cells (Fig. 7c) using Al-functionalized tips, no considerable adhesion between aluminum and C. albicans was noticed, where the adhesion frequency did not exceed 14%. Additionally, most rupture lengths are below 200 nm with force magnitudes not exceeding 1200 pN. These few adhesive events are considered weak non-specific interactions (FD curves did not show sawtooth sequential unfolding, Fig. 7f) as compared to signatures obtained with of ZnAl-functionalized tips.
MgAl-functionalized tips showed slightly higher adhesion frequencies (17 to 27%, Fig. 7d) and force magnitudes (300 to 1500 pN, Fig. 7d) with FD curves showing slightly longer sawtooth sequential unfolding that we attribute to Als adhesive proteins (Fig. 7g). However, the interactions observed for non-antifungal MgAl LDHs with C. albicans cells were much weaker than those obtained by ZnAl-functionalized tips. Such an observation comes into line with that of the results obtained from SCFS (Fig. 6), indicating that the strong adhesive interactions observed between ZnAl LDHs and C. albicans cells are related to their antifungal nature. In fact, we have previously observed such differences in adhesion behavior between antimicrobial ZnAl LDHs and non-antimicrobial MgAl LDHs toward S. aureus bacterial cells.14
We had previously established in this work that ZnAl LDH NPs possessed specific and efficient antifungal properties in comparison with non-antifungal MgAl LDH NPs, which were validated by C. albicans growth inhibition (Fig. 3) and imposing serious morphological damage on their cell walls (Fig. 4). Therefore, the specific adhesion interactions observed for antifungal ZnAl LDHs, which may be attributed to the stretching of Als adhesive proteins, provide strong evidence for the contribution of surface adhesive interactions to the antifungal activity of ZnAl LDHs.
Indeed, some researchers have previously reported the antimicrobial activity of zinc-based LDHs through direct surface interactions,13,20 however, none of them provided any evidence on the presence of these interactions. The researchers claimed that zinc present in the LDH structure can bind and then interact with the membranes of microorganisms causing damage to the cell wall, which could further lead to cell death. Zinc ions present on the surface of LDHs may bind to the membranes of microorganisms, and it may then be transported by the uptake system for essential metal ions to the cell where they could accumulate and exert toxic effects. Therefore, the interactions translated in the form of forces measured between ZnAl LDHs and C. albicans provided evidence on the presence of surface interactions that may contribute one key element to the antifungal behavior of ZnAl LDHs.
In this context, ZnAl LDH NPs were able to specifically interact and adhere to C. albicans cells with high adhesion frequency and great force magnitudes, thus imposing serious damage on their cell walls, which consequently leads to an efficient antifungal effect against these yeast pathogens.
3. Conclusion
In the present study, the role of adhesive interactions in the antifungal activity of ZnAl LDH NPs against C. albicans was studied. ZnAl LDH NPs exhibited an efficient antifungal effect against C. albicans in comparison with non-antifungal MgAl LDH NPs, which was evidenced by greater inhibition zone diameters generated in agar disc diffusion assay and MIC determination by broth microdilution tests. Additionally, serious morphological impacts including roughened surfaces, surface distortion, and dissociation into cell debris were imposed on C. albicans cells by ZnAl LDH NPs treatment at the MIC.Moreover, AFM-based SCFS and SPFS were employed to probe the nanoscale adhesive interactions between C. albicans cells and the prepared ZnAl LDH NPs to further correlate them to the observed antifungal activity of the latter. For SCFS, living C. albicans cells were attached to AFM tipless cantilevers and used to probe the surface of the prepared ZnAl, MgAl and Al films at the single-cell level, whereas for SPFS, Al-coated and LDH-functionalized AFM tips were prepared and used to probe surface adhesion interactions with the cell surface at the single LDH particle level. SCFS showed that C. albicans cells interacted with ZnAl LDHs with much higher force magnitudes than Al-coated surfaces and non-antifungal MgAl LDHs. Such strong adhesive interactions were attributed to the antifungal nature of Zn-based LDHs arising from their constituent divalent metal ions, e.g., Zn2+. The comparison of the adhesion behavior of Al-coated, MgAl-functionalized, and ZnAl-functionalized tips at the single-particle level further indicated that specific surface interactions occur at high adhesion frequencies and remarkable force magnitudes between ZnAl LDHs and C. albicans cells, which may be attributed to the presence of Als adhesive proteins in the cells. Such findings suggested a strong correlation between the specific ability of zinc-based LDHs to interact with and damage C. albicans cell walls, thus correlating the antifungal properties of ZnAl LDHs to the nanoscale adhesive forces occurring at the single-cell and single-particle levels with C. albicans fungal pathogens.
4. Experimental part
4.1. LDH NPs synthesis
Nitrate-intercalated MgAl and ZnAl LDH NPs were synthesized similarly to the method of our previous work.14 Briefly, a mixture of 0.2 mol L−1 M(NO3)2·6H2O (M = Zn, Mg; SIGMA-ALDRICH) and 0.1 mol L−1 Al(NO3)3·9H2O (SIGMA-ALDRICH) was added quickly to 0.15 M NaOH solution (VWR Chemicals), under a nitrogen atmosphere and vigorous stirring for 20 min. An LDH slurry was obtained by centrifugation, washed twice, and dispersed again in decarbonated water. The resultant suspension was then transferred to a stainless-steel autoclave with a Teflon lining and heated for 24 h at 80 °C. LDH powders were then collected by centrifugation and air drying at 80 °C.4.2. Synthesis of LDH films and LDH-functionalized AFM tips
Nitrate-intercalated MgAl and ZnAl LDH films were grown on aluminum-coated silicon wafers using a wet-chemistry protocol, as previously described.14 Briefly, a thin layer of aluminum (∼10 nm) was deposited on silicon wafers previously cleaned with piranha solution by plasma sputtering (Q15OT S, Quorum). Aluminum-coated silicon substrates were then transferred to a stainless-steel autoclave with a Teflon lining containing a solution of 50 mL of M(NO3)2·6H2O (M = Mg, Zn; 2.5 mmol) and NH4NO3 (30 mmol, SIGMA-ALDRICH). The autoclave was heated at 80 °C for 24 h. The obtained LDH films were then washed with deionized water and ethanol and then dried using a nitrogen flush. LDH-functionalized AFM silicon nitride probes (MLCT-C Bruker Corporation) were synthesized in a similar method to that of LDH films grown on silicon wafers.4.3. Scanning electron microscopy imaging
The morphology of LDH NPs, films, LDH-functionalized AFM tips, and cell probes was examined by scanning electron microscopy (SEM) using a JEOL JSM-IT500HR, with a field emission gun (FEG) and a voltage of 2 to 10 kV. The analysis was performed under high vacuum, and settled 60 μm diaphragm aperture. A secondary electron detector (SED) was used for imaging.4.4. Antimicrobial activity tests
The antimicrobial activity of MgAl and ZnAl LDH NPs was evaluated using agar disc diffusion and broth microdilution turbidity methods against the fungal pathogen C. albicans (SC5314).Inhibition zone diameters generated by MgAl and ZnAl LDH powders were determined by an agar disc diffusion test. C. albicans cells were precultured in 5 mL of sterile yeast extract peptone dextrose (YPD, SIGMA ALDRICH) overnight at 30 °C and then diluted to 1/1500 in sterile 3-(N-morpholino)propane sulfonic acid (MOPS, Sigma). Then, 250 μL of diluted fungal suspension was spread on sterilized YPD agar Petri dishes using sterile cotton-tipped swabs (COPAN) to prepare a homogeneous fungal layer. Filter discs (diameter = 6 mm, Dominique Dutscher) were placed on the dishes and 20 μL LDH dispersions (10 mg mL−1) were loaded onto the respective discs. Petri dishes were then incubated at 30 °C for 24 h. Afterwards, inhibition zones were observed around the discs. The diameters of the circular zones were measured using a centimeter ruler and the mean values of three repeated trials were recorded and represented in millimeters.
The minimum inhibitory concentration (MIC) of LDH NPs was evaluated using the broth microdilution turbidimetric method. C. albicans cells were precultured in 5 mL of YPD overnight at 30 °C. An inoculum of C. albicans was then prepared and diluted to obtain an optical density of 0.1 at λ = 600 nm (OD600 = 0.1). MgAl and ZnAl LDH NPs were dispersed in the YPD broth, and two-fold dilution series were then prepared in a 96-well microtiter plate. Each well included 25 μL of diluted LDH powder and 175 μL of yeast inoculum. Un-inoculated broth wells containing the different LDH dispersion concentrations were used as a negative standard growth control. As a positive control, 12 wells were prepared with 175 μL of yeast inoculum, and 25 μL of YPD. The micro-titer plate was then sealed with parafilm and incubated at 30 °C under agitation for 24 h. The MIC value was recognized as the lowest concentration that had no visible turbidity due to C. albicans growth.
4.5. Atomic force microscopy (AFM) imaging and force spectroscopy
All AFM experiments were performed at room temperature using a Bioscope Resolve AFM (Bruker Corporation, Santa Barbara, CA), and the spring constants of the used cantilevers were determined using the thermal noise method.46For topography images of C. albicans, cells were cultivated alone (untreated) or with (treated at MIC) ZnAl and MgAl LDH NPs for 24 h in YPD at 30 °C. C. albicans cells were then centrifuged, washed twice in MOPS, and filtered into porous polycarbonate membranes (pore size = 3 μm, Millipore). After filtration and rough rinsing, a 1 × 1 cm2 piece of the membrane was cut and attached using a double-sided adhesive tape to the bottom of a Petri dish and immersed in MOPS. The roughness parameter Rq (root mean square of the heights) of the cells (6 cells for each condition, e.g., untreated cells, ZnAl LDH NP- and MgAl LDH NP-treated cells) was calculated using the commercial Nanoscope analysis software (Bruker Corporation) to analyse high resolution AFM images acquired by scanning areas of 1 × 1 μm2 (512 samples/line; 262![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 144 data points) on the top of yeast cells. AFM images were acquired in peak-force tapping mode using SNL-C tips.
144 data points) on the top of yeast cells. AFM images were acquired in peak-force tapping mode using SNL-C tips.
For force measurements on cells using AFM-based single-cell force spectroscopy, yeast cell probes were prepared using triangular-shaped tipless cantilevers (NP-O10, Microlevers, Bruker Corporation) coated with polydopamine wet adhesives. The cantilevers were immersed for 1 h in a 10 mmol L−1 Tris buffer solution (pH = 8.5) containing 4 mg mL−1 dopamine hydrochloride (99%, Sigma), and dried under an N2 flow. Single yeast cells deposited on a Petri dish were then attached to the polydopamine-coated cantilevers by bringing the cantilever into contact with an isolated cell for 1 min. Proper attachment and positioning of yeast cells on the cantilever were achieved using a Bioscope Resolve instrument (Bruker Corporation, Santa Barbara, CA) equipped with a Leica inverted microscope. When the proper attachment of the yeast cell was achieved, the probe was positioned over the Al, MgAl and ZnAl LDH films. Multiple force–distance curves were recorded across 5 mm × 5 mm areas of the films using a maximum applied force of 500 pN, an interaction time of 0.1 s, and constant approach and retraction speed of 1 μm s−1. Adhesion force histograms were obtained by calculating for each force curve the maximum adhesion peak. Data were processed using commercial Nanoscope analysis (Bruker Corporation) and MATLAB (The MathWorks, Natick, MA) software.
For force measurements on cells using AFM-based single-particle force spectroscopy, stationary phase cultures of C. albicans were trapped in porous membranes in a similar procedure used for cell imaging in MOPS. First, bare MLCT-C tips were used to localize and image individual cells. Then, the tips were replaced by aluminum-coated tips, MgAl, and ZnAl LDH-functionalized tips. Adhesion force maps and histograms were obtained by recording 32 × 32 force–distance curves on cell surface areas of 1 × 1 μm2, calculating the adhesion force of the last adhesion peak for each force curve, and displaying the value as a grey pixel. All curves were recorded using a maximum applied force of 500 pN, a contact time of 0.1 s, and constant approach and retraction speed of 1 μm s−1. Data were processed using commercial Nanoscope analysis (Bruker Corporation) and MATLAB (the MathWorks, Natick, MA) software.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support was received from the French Ministry of Higher Education (MESR), and the French National Scientific Centre (CNRS). We would like to acknowledge the spectroscopy and microscopy service facility of SMI LCPME (Université de Lorraine-CNRS – https://www.lcpme.ul.cnrs.fr). Jazia Awassa acknowledges the French Ministry of Higher Education for her Ph.D. grant.References
- F. Bongomin, S. Gago, R. Oladele and D. Denning, J. Fungi, 2017, 3, 57 CrossRef PubMed.
- G. D. Brown, D. W. Denning, N. A. R. Gow, S. M. Levitz, M. G. Netea and T. C. White, Sci. Transl. Med., 2012, 4(165), 165rv13 CrossRef PubMed.
- M. C. Fisher, A. Alastruey-Izquierdo, J. Berman, T. Bicanic, E. M. Bignell, P. Bowyer, M. Bromley, R. Brüggemann, G. Garber, O. A. Cornely, S. J. Gurr, T. S. Harrison, E. Kuijper, J. Rhodes, D. C. Sheppard, A. Warris, P. L. White, J. Xu, B. Zwaan and P. E. Verweij, Nat. Rev. Microbiol., 2022, 20, 557–571 CrossRef CAS PubMed.
- N. Robbins, T. Caplan and L. E. Cowen, Annu. Rev. Microbiol., 2017, 71, 753–775 CrossRef CAS PubMed.
- S. Costa-de-Oliveira and A. G. Rodrigues, Microorganisms, 2020, 8, 154 CrossRef CAS PubMed.
- N. Wiederhold, IDR, 2017, vol. 10, pp. 249–259 Search PubMed.
- N. M. Packter, Biochem. Educ., 1995, 23, 115 CrossRef.
- D. W. Denning, Clin. Infect. Dis, 1998, 26, 781–803 CrossRef CAS PubMed.
- J.-L. Xu, Y.-X. Luo, S.-H. Yuan, L.-W. Li and N.-N. Liu, Innov. digit. health diagn. biomark., 2020, 1, 3–7 CrossRef.
- A. Vaccari, Appl. Clay Sci., 2002, 22, 75–76 CrossRef CAS.
- M. Bini and F. Monteforte, J. Anal. Pharm. Res., 2018, 7 DOI:10.15406/japlr.2018.07.00206.
- F. L. Theiss, G. A. Ayoko and R. L. Frost, Appl. Surf. Sci., 2016, 383, 200–213 CrossRef CAS.
- S. A. A. Moaty, A. A. Farghali and R. Khaled, Mater. Sci. Eng., C, 2016, 68, 184–193 CrossRef PubMed.
- J. Awassa, S. Soulé, D. Cornu, C. Ruby and S. El-Kirat-Chatel, Nanoscale, 2022, 14, 10335–10348 RSC.
- J. Awassa, D. Cornu, S. Soulé, C. Carteret, C. Ruby and S. El-Kirat-Chatel, Appl. Clay Sci., 2022, 216, 106369 CrossRef CAS.
- J. Awassa, D. Cornu, C. Ruby and S. El-Kirat-Chatel, Colloids Surf., B, 2022, 217, 112623 CrossRef CAS PubMed.
- S. Dutta, T. K. Jana, S. K. Halder, R. Maiti, A. Dutta, A. Kumar and K. Chatterjee, ChemistrySelect, 2020, 5, 6162–6171 CrossRef CAS.
- M. Li, L. Li and S. Lin, Chin. Chem. Lett., 2020, 31, 1511–1515 CrossRef CAS.
- A. M. León-Vallejo, F. D. Velázquez-Herrera, Á. Sampieri, G. Landeta-Cortés and G. Fetter, Appl. Clay Sci., 2019, 180, 105194 CrossRef.
- F. Peng, D. Wang, D. Zhang, H. Cao and X. Liu, Appl. Clay Sci., 2018, 165, 179–187 CrossRef CAS.
- S. El-Kirat-Chatel and Y. F. Dufrêne, Nanoscale Horiz., 2016, 1, 69–74 RSC.
- M. Benoit, D. Gabriel, G. Gerisch and H. E. Gaub, Nat. Cell Biol., 2000, 2, 313–317 CrossRef CAS PubMed.
- M. Benoit and H. E. Gaub, Cells Tissues Organs, 2002, 172, 174–189 CrossRef CAS PubMed.
- D. J. Müller, J. Helenius, D. Alsteens and Y. F. Dufrêne, Nat. Chem. Biol., 2009, 5, 383–390 CrossRef PubMed.
- J. Friedrichs, J. Helenius and D. J. Muller, Nat. Protoc., 2010, 5, 1353–1361 CrossRef CAS PubMed.
- D. Alsteens, A. Beaussart, S. Derclaye, S. El-Kirat-Chatel, H. R. Park, P. N. Lipke and Y. F. Dufrêne, Anal. Methods, 2013, 5, 3657 RSC.
- A. Beaussart and S. El-Kirat-Chatel, Cell Surf., 2019, 5, 100031 CrossRef CAS PubMed.
- A. D. Russell, J. Antimicrob. Chemother., 2003, 52, 750–763 CrossRef CAS PubMed.
- E. de Souza Araújo, A. S. Pimenta, F. M. C. Feijó, R. V. O. Castro, M. Fasciotti, T. V. C. Monteiro and K. M. G. de Lima, J. Appl. Microbiol., 2018, 124, 85–96 CrossRef PubMed.
- M. T. Yassin, A. A.-F. Mostafa, A. A. Al-Askar and F. O. Al-Otibi, Crystals, 2022, 12, 774 CrossRef CAS.
- A. Karimiyan, H. Najafzadeh, M. Ghorbanpour and S. H. Hekmati-Moghaddam, Zahedan J. Res. Med. Sci., 2015 DOI:10.17795/zjrms-2179.
- M. Jalal, M. A. Ansari, S. G. Ali, H. M. Khan and S. Rehman, Artif. Cells, Nanomed., Biotechnol., 2018, 46, 912–925 CrossRef CAS PubMed.
- M. Sharma and P. Prasher, in Nanotechnology, ed. C. Bhargava and A. Sachdeva, CRC Press, 1st edn, 2020, pp. 261–276 Search PubMed.
- G. V. Vimbela, S. M. Ngo, C. Fraze, L. Yang and D. A. Stout, Int. J. Nanomed., 2018, 12, 3941–3965 CrossRef PubMed.
- S. El-Kirat-Chatel, A. Beaussart, D. Alsteens, D. N. Jackson, P. N. Lipke and Y. F. Dufrêne, Nanoscale, 2013, 5, 1105–1115 RSC.
- F. Quilès, I. Accoceberry, C. Couzigou, G. Francius, T. Noël and S. El-Kirat-Chatel, Nanoscale, 2017, 9, 13731–13738 RSC.
- M. A. Hussain, D. Ahmed, A. Anwar, S. Perveen, S. Ahmed, I. Anis, M. R. Shah and N. A. Khan, Int. Microbiol., 2019, 22, 239–246 CrossRef CAS PubMed.
- C. Formosa, M. Schiavone, H. Martin-Yken, J. M. François, R. E. Duval and E. Dague, Antimicrob. Agents Chemother., 2013, 57, 3498–3506 CrossRef CAS PubMed.
- M. J. Fiołka, S. Mieszawska, P. Czaplewska, A. Szymańska, K. Stępnik, W. Sofińska-Chmiel, T. Buchwald and K. Lewtak, Sci. Rep., 2020, 10, 16352 CrossRef PubMed.
- T. Sultan and S. A. Ali, J. Recept. Signal Transduction, 2011, 31, 39–44 CrossRef CAS PubMed.
- A. Beaussart, S. El-Kirat-Chatel, R. M. A. Sullan, D. Alsteens, P. Herman, S. Derclaye and Y. F. Dufrêne, Nat. Protoc., 2014, 9, 1049–1055 CrossRef CAS PubMed.
- M. Laviale, A. Beaussart, J. Allen, F. Quilès and S. El-Kirat-Chatel, ACS Appl. Mater. Interfaces, 2019, 11, 48574–48582 CrossRef CAS PubMed.
- F. Zhang, L. Zhao, H. Chen, S. Xu, D. G. Evans and X. Duan, Angew. Chem., Int. Ed., 2008, 47, 2466–2469 CrossRef CAS PubMed.
- A. Beaussart, P. Herman, S. El-Kirat-Chatel, P. N. Lipke, S. Kucharíková, P. Van Dijck and Y. F. Dufrêne, Nanoscale, 2013, 5, 10894 RSC.
- A. Beaussart, D. Alsteens, S. El-Kirat-Chatel, P. N. Lipke, S. Kucharíková, P. Van Dijck and Y. F. Dufrêne, ACS Nano, 2012, 6, 10950–10964 CrossRef CAS PubMed.
- J. L. Hutter and J. Bechhoefer, Rev. Sci. Instrum., 1993, 64, 1868–1873 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2024 |