Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Recent progress in biosensor regeneration techniques
Yizhen
Jia
a,
Shulin
Chen
a,
Qi
Wang
a and
Jinghua
Li
 *ab
*ab
aDepartment of Materials Science and Engineering, The Ohio State University, Columbus, OH 43210, USA
bChronic Brain Injury Program, The Ohio State University, Columbus, OH 43210, USA. E-mail: li.11017@osu.edu
First published on 24th January 2024
Abstract
Biosensors are widely used in various applications, from medical diagnostics to environmental monitoring. Their widespread and continuous use necessitates regeneration methods to ensure cost-effectiveness and sustainability. In the realm of advancing human-centric bioelectronics for continuous monitoring, employing these sensors for real-time, in situ detection of biomarkers presents a considerable challenge. This mini-review examines diverse strategies utilized for the regeneration of biosensors, categorizing them based on their underlying mechanisms and discussing representative works. We explore methods ranging from surface engineering/re-functionalization, chemical treatments, allosteric regulation of bioreceptors, to manipulations of electric/magnetic fields, highlighting their working principles and exemplary studies. The advantages of each method, such as simplicity, high regeneration efficiency, and versatility, are discussed alongside their challenges, including degradation over cycles, limited applicability, and potential damage to sensors. As the demand for continuous and real-time biosensing escalates, the development of efficient and reliable regeneration strategies becomes essential. This mini-review offers an overview of the current landscape of biosensor regeneration, aiming to guide future research and innovations in this area.
Introduction
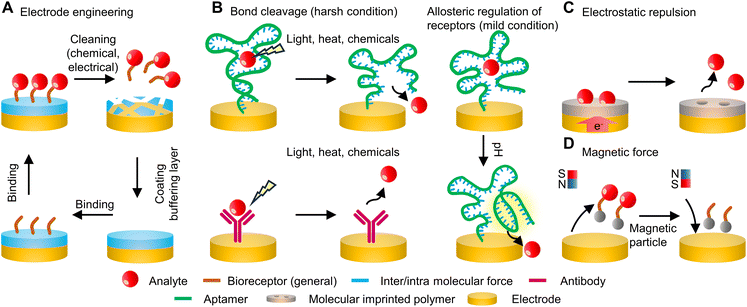
Over the past few decades, biosensing technologies have seen remarkable advancements across a spectrum of applications, encompassing the domains of medicine, food testing, environmental monitoring, and process control in both research and industrial contexts.1–3 A common objective shared by numerous fields is the pursuit of continuous, real-time, and low-cost detection of target analytes in various settings. Conventional biosensing techniques, such as capillary electrophoresis (CE), gas chromatography (GC), high-performance liquid chromatography (HPLC), and nuclear magnetic resonance (NMR) spectroscopy, place demands on centralized equipment, highly skilled personnel for operation, and high-grade analytical reagents and materials, resulting in substantial operational expenses.3–5 To this end, biosensors have emerged as a solution that significantly enhances accessibility, efficiency, and cost-effectiveness in continuous analyte detection across a wide range of applications and settings. Despite the great promise, it is crucial to acknowledge and address the challenges associated with stability and reversibility that require further investigation and resolution: overall, regeneratable sensors are important because they mitigate potential errors arising from chip-to-chip variance during continuous measurements and address the need for highly accurate, cost-intensive transducers, offering a key technique to reduce the overall cost per test in critical applications. Additionally, this significance is particularly pronounced in healthcare and diagnostics, where regeneratable sensors are crucial for establishing time-sequential biometric signature profiles in patients. With the rapid advancement of bioelectronics, especially in bio-integrated forms, the development of regeneratable sensing platforms has become increasingly essential and in demand.6Biosensors are typically characterized as comprising a bioreceptor, a transducer, and a signal-processing unit. In general, the challenge in achieving continuous monitoring with biosensor chips lies in the reusability of the immobilized biorecognition elements, as these functionalized biochemical interfaces are essential for ensuring the necessary sensitivity and selectivity. Recognition elements in biosensors commonly encompass biometric receptors, such as enzymes, antibodies, and nucleic acids, alongside biomimetic components like inorganic nanomaterials and molecularly imprinted polymers (MIPs). The regeneration process typically involves refreshing the receptors, which can be achieved either by cleaning and applying new receptors or by detaching the target analytes from the existing receptors. A common approach for detachment is to overcome the binding affinity between the targets and the receptors. While the underlying mechanism of disrupting this affinity can be similar, the means of achieving it can vary widely, including but not limited to the application of light, heat, or changes in the chemical environment. Additionally, the use of electric potential to induce oxidation or reduction reactions is another effective strategy for removing the target analytes. The development of regeneratable sensors across diverse sensing schemes is still in its early stages, primarily because the interaction modes and requirements differ significantly from one system to another. Furthermore, there is a lack of consensus in the literature regarding the definitions and criteria for regeneration, adding to the complexity and challenges in developing regeneratable sensors. This mini-review aims to consolidate and present recent advances in regeneratable sensing schemes. These strategies encompass a diverse range of approaches (Fig. 1), including thermal and chemical treatments, magnetic manipulation, the creation of novel receptors, engineering enhancements of the sensing interface, and the development of integrated systems combining several of these methods. Finally, the review also addresses the existing challenges and emerging trends in regeneratable biosensors, aiming to provide insights into the future of this evolving field.
Engineering the electrode – re-functionalization after use
A typical biosensor consists of two primary components: a signal transducer and an electrochemical interface with bioreceptors. Modifications to either of these elements can facilitate the regeneration of the sensor. The transducer, in particular, can be designed for easy cleaning and re-functionalization after each use, offering a straightforward approach to regeneration. A recent study introduces a novel method that combines electrochemical affinity-based biosensors with microfluidic chips, which enables real-time, noninvasive measurement of biomarkers directly within organ-on-a-chip setups through in-line microelectrode functionalization, biomarker detection, and sensor regeneration.7 Specifically, this design includes a two-step cleaning and functionalization process, as illustrated in Fig. 2A. This regeneration process is seamlessly automated through an integrated circuitry and microfluidics system controlled by MATLAB code and an automated valve controller.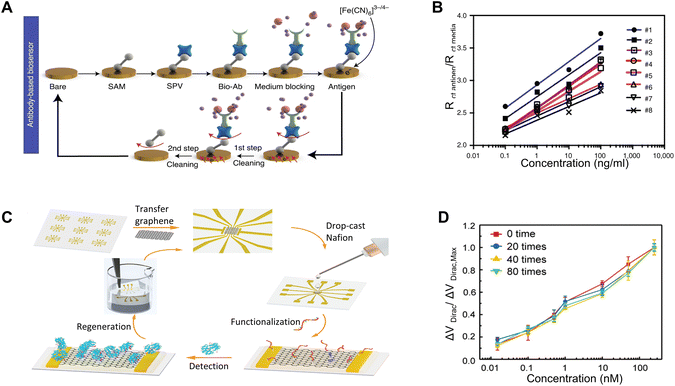 | ||
| Fig. 2 Re-functionalization after use: (A) schematic illustrating the two-step cleaning and functionalization process for a creatine kinase–myocardial band (CK–MB) aptamer biosensor on a microfluidic chip. Reprinted with permission from ref. 7. Copyright 2021 Springer Nature. (B) Calibration curve showcasing the sensitivity of the sensors in Fig. 2A after various regeneration cycles. Reprinted with permission from ref. 7. Copyright 2021 Springer Nature. (C) Schematic depicting the fabrication of a graphene-Nafion based FET biosensor, the detection of interferon-γ, and its regeneration using ethanol. Reprinted with permission from ref. 8. Copyright 2020 Wiley. (D) Consistent sensitivity of the sensors in (C) towards interferon-γ during 80 regeneration cycles. Reprinted with permission from ref. 8. Copyright 2020 Wiley. | ||
The cleaning is first conducted by cyclic voltammetry (CV) scans during continuous flow of H2SO4 and then K3Fe(CN)6, removing all the immobilized molecules on the sensor surface. Then the sensor is refunctionalized by a series of chemical flow and incubation. The versatile strategy enables incorporation of bioreceptor molecules in the form of proteins and nucleic acids such as antibodies and aptamers: for aptamer-based sensors, the procedure involves immobilizing amine group-functionalized creatine kinase–myocardial band (CK–MB) aptamers onto the electrode surface following the formation of a self-assembled monolayer (SAM). This immobilization is achieved using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) and N-hydroxysuccinimide (NHS) coupling. Alternatively, for specific antibody-based sensors, such as the albumin biotinylated antibody or glutathione S-transferase alpha (GST-α) biotinylated antibody, these antibodies are introduced after the formation of the SAM, the EDC/NHS coupling, and the application of streptavidin (SPV). The sensing platforms can be effectively regenerated through a complete refunctionalization process using fresh chemicals and materials. This procedure ensures a high level of consistency and reliability for the devices. As demonstrated in Fig. 2B, the calibration curve generated through electrochemical impedance spectroscopy (EIS) for the resulting electrochemical sensors consistently demonstrates the capacity of the devices to maintain sensitivity throughout five regeneration cycles. The entire process requires approximately four hours for completion.
In view of the potential damage that chemicals may pose to the transducers, the incorporation of a buffering layer can facilitate the straightforward removal of receptors, thereby streamlining the regeneration process. In a recent work, a class of flexible and regenerative biosensors based on aptameric field-effect transistors has been reported to be able to monitor cytokines (e.g., interferon-gamma (IFN-γ)) consistently and sensitively in undiluted human sweat. The functionalization process involves initially applying a Nafion coating onto a graphene surface, subsequently followed by the functionalization of the film with aptamers specifically designed for the targeted biomarkers.8 After each detection cycle, treating the graphene FET biosensor with ethanol removes the Nafion film. This results in a refreshed graphene-based FET ready for reuse in following biomarker detection cycles (Fig. 2C). After up to 80 regeneration cycles, the performance in detecting IFN-γ remains consistent, with a signal variation of less than 8.3% (Fig. 2D). This indicates the structural integrity of graphene after the repetitive treatment with chemicals and the consistent regenerative capability of the sensing platform.
Due to the straightforward nature of these re-functionalization methods, they require only minimal modifications to the sensor design to achieve successful implementation. Nevertheless, some limitations are associated with these methods, including time-consuming processes, the need for manual intervention, and reliance on additional chemicals that may lack biocompatibility. Consequently, while these methods can potentially reduce costs, these limitations make them unsuitable for the development of in vivo biosensors intended for continuous monitoring applications.
Regeneration enabled by light and heat
On the other hand, regeneration can also be achieved by modifying and designing the bioreceptors. By releasing the target analytes, the sensor can restore its functionality. Another practical approach for sensor regeneration involves manipulating inter- and intra-molecular forces through external stimuli, such as heat and light. These external energy sources are applied to break the chemical bonds between the bioreceptor and the analyte, thereby freeing the bioreceptor for subsequent detection. Specifically, this concept has been employed in aptamer-mediated biosensing by leveraging the adaptable characteristics of oligonucleotide sequences: derived from single-stranded DNA or RNA (ssDNA/RNA), aptamers exhibit the unique ability to fold into secondary and intricate three-dimensional configurations.9 This conformational versatility enables them to recognize a broad spectrum of target molecules with exceptional specificity.9 Their targets span from proteins, small molecules, and metal ions to bacterial cells, viruses, and even tissues.10 The underlying binding mechanisms encompass a wide range of interactions, such as hydrogen bonds, van der Waals forces, and electrostatic interactions.11 The inherent reversibility of these noncovalent interactions has provided opportunities to regulate the binding affinity between biomarkers and receptors, allowing for target dissociation through the application of external triggers.Given the versatility of aptamers in recognizing a large variety of targets, and with the dynamic and reversible nature of their binding mechanisms, works have been done to harness these properties to create regeneratable biosensors. A recent study has reported employing electrochemical sensors with ssDNA aptamers for detecting adenosine triphosphate.12 A ferrocene group affixed to the 3′-end of the aptamers serves as the signal-transformation component, while an azobenzene moiety embedded near the 5′-end of the DNA chain functions as the regeneration element. Upon UV-light exposure, these azobenzene units undergo a trans-to-cis isomerization, prompting the hairpin structure of the aptamer to unravel and its base pairs to dissociate (Fig. 3A). This process facilitates the release of the target molecule, effectively regenerating the aptamers. Regeneration can be achieved through an exposure to a 6 W UV lamp for one hour, and after seven cycles, the functionality of the aptamer remains uncompromised (Fig. 3B).
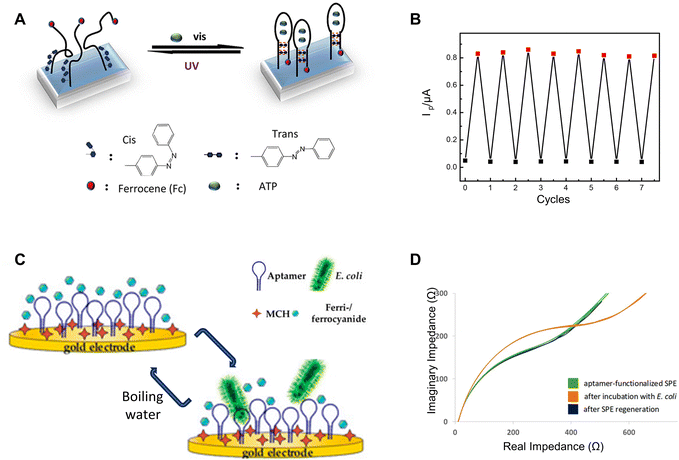 | ||
| Fig. 3 UV-light and heat-based regeneration: (A) schematic illustrating the UV-light-induced regeneration mechanism of the adenosine triphosphate aptamer sensor and (B) its regeneration performance over different cycles. Reprinted with permission from ref. 12. Copyright 2018 American Chemical Society. (C) Schematic showing regeneration using boiling water for the aptamer-based impedimetric sensor targeting the E. coli crooks strain, and (D) EIS characterization of the sensor before and after regeneration. Reprinted with permission from ref. 13. Copyright 2020 MDPI. | ||
In addition to UV light, another study employs the use of boiled water as a regeneration method for aptamer-based sensors designed to detect Escherichia coli (E. coli).13 When exposed to boiling water for 2 minutes, the unfolding of aptamer structures allows for the release of potentially captured targets. Following this, an incubation in a solution containing sodium chloride, tris buffer (tris-hydrogen chloride) and magnesium chloride enables these oligonucleotides to revert to their functional conformation, making it ready for the next detection cycle (Fig. 3C). EIS characterization indicates nearly identical performances before detection and after regeneration, suggesting successful regeneration with minimal to no degradation (Fig. 3D). Another study involving aptamer-based endotoxin detection employs a similar regeneration technique, which involves incubating the sensors at 95 °C for 15 minutes.14 This temperature surpasses the melting point of the DNA aptamer, leading to the disruption of both aptamer-to-aptamer and aptamer-to-endotoxin hydrogen bonds. Consequently, the aptamer unfolds and releases the endotoxin to re-gain its functionality for biorecognition.
Due to the nature of light and heat, these regenerative methods possess the potential for remote-controlled regeneration, offering significant advantages in applications where direct physical access to the biosensor is challenging or impractical. These regenerative methods, while based on simple concepts and requiring minimal setup, can sometimes be too harsh for some in vivo environments. Consequently, while these approaches have shown success in restoring sensor functionality, they are not yet well-suited for the development of in vivo biosensors for continuous monitoring. Moreover, these methods are primarily applicable to aptamer-based biosensors. There are limited reports of regeneration involving other biorecognition elements, such as protein-based ones, due to stability limitations during such treatments.
Regeneration enabled by chemicals
The concept of adjusting the affinity between receptors and analytes is also relevant in the use of chemicals. This approach aids in freeing the analyte by either reducing the binding affinity or facilitating a conformational change that expels the analyte. Using sodium hydroxide (NaOH) represents a common method to elute target molecules from the sensing interface, which has been shown to be effective in removing proteins, nucleic acids, yeast, fungi, bacteria, and viruses from chromatographic systems.15 In a study employing single-walled carbon nanotube (SWCNT)-based biosensors equipped with anti-Yersinia antibodies for Y. enterocolitica detection, the application of a 100 mM NaOH solution enables the biosensor to be effectively reused for up to five times (Fig. 4A).15 Another research study describes a regenerable electrochemical DNA biosensor, which employs a composite of acrylic microspheres and gold nanoparticles coated onto a screen-printed electrode.16 The primary objective of this biosensor is to detect the 35S promoter from the cauliflower mosaic virus (CaMV 35S) gene in soybeans. In this setup, the DNA probe is immobilized onto the acrylic microspheres. The presence of the modified gene is then detected through hybridization, which is monitored by employing differential pulse voltammetry with the anthraquinone-2-sulfonic acid monohydrate sodium salt (AQMS) serving as a redox indicator. Remarkably, the biosensor exhibits an impressively low detection limit, high reproducibility, and the capability for reuse for at least seven cycles following regeneration. The ability to regenerate the genetically modified DNA biosensor lies in the capacity to reinitiate the DNA hybridization reaction between complementary DNA (cDNA) and an immobilized DNA probe. This is achieved by breaking the hydrogen bonds in the double-stranded DNA (dsDNA) using 0.1 M NaOH to revert it back to single-stranded one (Fig. 4B).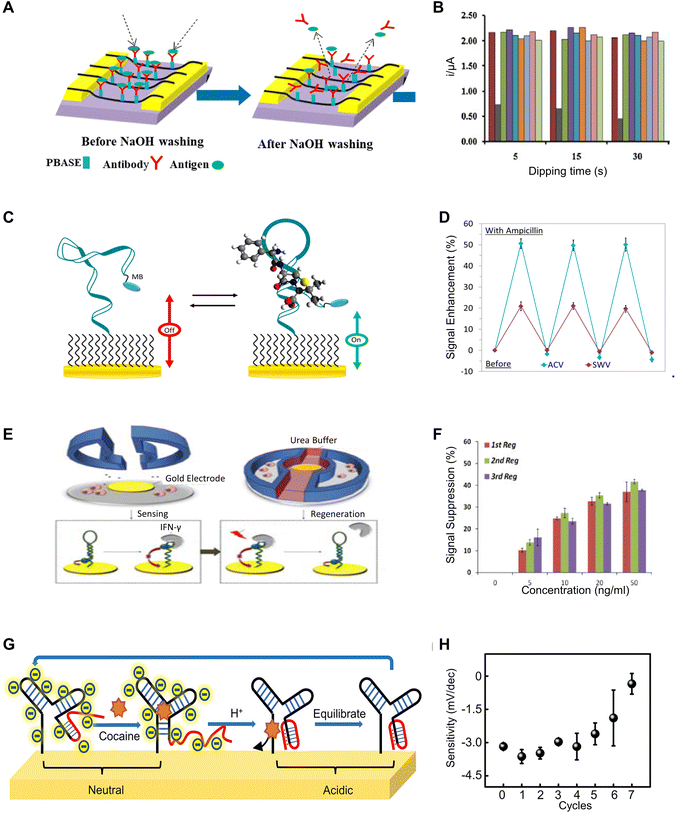 | ||
| Fig. 4 Chemical-based regeneration methods: (A) schematic of regenerating SWCNT-based biosensors for detection of Y. enterocolitica using antibodies through NaOH washing. Reprinted with permission from ref. 15. Copyright 2020 Elsevier. (B) The response of a DNA Biosensor comprising acrylic microsphere–gold nanoparticle composite, displaying its sensitivity towards the cauliflower mosaic virus (CaMV 35S) gene, across different durations of immersion in NaOH solutions over 7 cycles. Reprinted with permission from ref. 16. Copyright 2014 Elsevier. (C) Design and sensing mechanism of a methylene blue-modified aptamer probe for ampicillin sensing and its (D) regenerated sensitivity towards ampicillin with deionized water rinse. Reprinted with permission from ref. 26. Copyright 2018 Elsevier. (E) Schematic illustrating the regeneration of a DNA hairpin aptamer specific to interferon-γ using urea buffers and (F) the corresponding response to varying IFN-γ concentrations over 3 regeneration cycles. Reprinted with permission from ref. 29. Copyright 2014 Royal Society of Chemistry. (G) Mechanism of regeneration enabled by pH changes for a cocaine-sensing DNA sequence, and (H) its changes in sensitivity across different cycles of pH change-induced regeneration. Reprinted with permission from ref. 32. Copyright 2022 American Chemical Society. | ||
Similar to alkaline environments, acidic conditions can also enhance the dissociation of target molecules. Glycine, widely utilized as a regenerative agent in acidic settings, operates effectively within a pH range of 2–7.17 Owing to its zwitterionic nature, possessing both positive and negative charge groups, glycine effectively mitigates charge interactions between the target molecule and the bioreceptor when conjugated to their surfaces, thereby promoting efficient sensor regeneration. Glycine provides a buffering effect, safeguarding bioreceptors against irreversible damage caused by pH fluctuations. In research centered on an immunosensor for the organophosphorus pesticide ethyl parathion (EP), a 0.2 M Gly-HCl buffer at a pH of 2.3 yields a dissociation efficiency of 79.54%.18 Moreover, incorporating 1% DMSO into this buffer significantly improved the dissociation efficiency, elevating it to approximately 97%. The enhanced dissociation is attributed to the high solubility of EP molecules in DMSO, which weakens the interaction between EP molecules and antibodies, thus facilitating their dissociation. Additionally, the immobilized antibodies can maintain adequate activity for conducting 14 consistent assays of EP.
Although acidic environments can facilitate the dissociation of target molecules, they can also cause irreversible damage to substances that specifically bind to these targets, such as antibodies and nucleic acids. Therefore, another effective strategy is to immobilize target molecules with higher stability on the sensor surface and employ competitive detection methods for indirect quantification of target molecule concentrations. In a study focusing on an immunoassay for Microcystin-LR (MC-LR), MC-LR immobilizes onto the surface of an optical fiber.19 Subsequently, a mixture of Cy5.5-labeled anti-MC-LR antibodies (Ab-Cy5.5) and MC-LR comes into contact with the fiber, initiating an indirect competitive immunobinding reaction. Since the fluorescence signal is inversely proportional to the concentration of MC-LR in the sample, this allows for an indirect measurement of MC-LR concentration. Treating the fiber surface with a 0.5% SDS solution at pH 1.9 enables sensor regeneration. Remarkably, even after 100 consecutive measurements, the signal experiences only a 10.7% decrease. Multiple studies have reported similar concepts and achieved comparably impressive regeneration performance.20–22
This remarkable regeneration performance extends to nucleic acid-based biosensors utilizing the specific interaction between dethiobiotin (DTB) and streptavidin (STV). In an aptamer-based evanescent wave biosensor targeting Ochratoxin A, the sensor functionalized with aptamer-modified magnetic beads (MB-Ap) serves for capturing target molecules.23 STV-conjugated aptamer-complementary DNA oligonucleotides serve as signal probes (Sp), and a dethiobiotin-modified fiber incorporates into the biosensor. During operation, the capture of target molecules triggers the release of Sp, which the EWA biosensor detects through DTB-STV recognition. The binding affinity of DTB-STV reduces in acidic environments, and the interaction is effectively blocked by SDS, enabling sensor regeneration using 0.5% SDS at pH 1.9. This method demonstrates the capability for up to 300 regeneration cycles with signal recovery exceeding 96%. Additionally, its versatility allows expansion to other targets by employing different aptamers and signal probes, as similar methods have been reported in other studies.24,25
In contrast to the harsh environment created by chemicals such as concentrated NaOH, there are several alternative strategies reported for analyte dissociation under milder conditions. In a study focused on the development of a class of reagent-less and reusable electrochemical sensors, a methylene blue-modified aptamer probe enables the direct detection of ampicillin in a variety of complex samples, such as serum, saliva, and milk (Fig. 4C) with alternating current voltammetry (ACV) and square wave voltammetry (SWV) used for characterization.26 The sensor can be readily regenerated with a 60-second rinse process in deionized water, and it maintains a consistent sensitivity after the third use (Fig. 4D). An alternative method employs urea to induce reversible denaturation of aptamers by disrupting the hydrogen bonds between the DNA strands.27,28 In a study involving aptasensors designed for continuous monitoring of cell-secreted products, an IFN-γ-binding aptamer with methylene blue as the redox indicator has been employed, and the performance is assessed using SWV scans (Fig. 4E).29 The sensor is regenerated through a urea buffer flush, which disrupts the secondary bonds between the aptamer and IFN-γ. The effectiveness is demonstrated through three regeneration cycles, with consistent limits and ranges of detection, as well as similar sensitivity observed in each cycle (Fig. 4F).
Due to the adaptable nature and responsiveness to external stimuli, allosteric receptors based on aptamers offer an alternative regeneration approach that can be accomplished under relatively mild conditions. This capability opens possibilities for potential applications in biological environments. Recent studies have revealed an increasing enthusiasm for triplex nucleic acids, recognizing them as integral components within the wide range of structures utilized in the development of DNA-centric nanomaterials and constructs.10,30 These DNA triplexes are particularly valuable in constructing sensing platforms as well as molecular toggling mechanisms. Transitions from duplex to triplex structures, triggered by changes in pH, can rapidly disrupt the stem-loop configurations of aptamers in a short time.31 This quick alteration leads to the subsequent release of the ligands they bind to. A recent study utilizes this mechanism for in vivo regeneration of cocaine sensors through mild shifts in pH levels.32 The sensing is facilitated by a modified DNA sequence that includes a pH-sensitive domain at the 5′ end of the oligonucleotide for cocaine detection. As the pH level decreases, the formation of an intramolecular triplex interferes with the stem of the cocaine-aptamer complex that has a three-way junction structure, resulting in the liberation of the bound ligands (i.e., cocaine). Conversely, at elevated pH levels, the disassembly of the triplex structure, due to the absence of hydrogen bonds, restores the capability of the aptamers to bind to cocaine (Fig. 4G). The sensor can restore most of its sensitivity after being treated with a pH of 5.0, and it has little degradation on sensitivity within 5 cycles of regeneration (Fig. 4H).
Beyond regeneratable cocaine sensors, the concept of utilizing allosteric DNA enabled by pH regulation holds promise for the application of this technology to other biomolecules. A study has presented an aptamer sequence that binds to streptavidin and maintains nanomolar affinity at pH 7.4; however, its binding affinity significantly reduces, approximately by a factor of 100, when the pH is adjusted to 5.2.31 This change in affinity causes the release of the target molecules, enabling sensor regeneration, and such pH sensitivity is due to a single-G-A mismatch structure that tends to be more stable in an acidic environment. Another strategy is based on the G-quadruplex conformation changes enabled by A + (anti)-G-(syn) mispairing in an acidic environment. In a recent work, a special human α-thrombin binding aptamer is designed to exist in a G-quadruplex conformation.33 The thrombin aptamer component on G-quadruplex is only exposed in the conformation at pH neutral environment and available for binding to the target. In an acidic environment, the A + (anti)-G-(syn) mispairing will happen and lead to a conformational change in the G-quadruplex which will cause the quadruplex to release the target. Therefore it is able to complete such a “catch and release” process on the target controlled by pH value change.
Regeneration using chemicals encompasses diverse methods for removing the target analyte from the bioreceptors, offering a broader range of applications across different bioreceptors and targets. While some chemical regeneration conditions are still considered harsh, there exist milder approaches that are more suitable for in vivo use. Current efforts are focused on expanding the range of bioreceptors and targets that can be regenerated under these milder conditions. Additionally, significant work is being done on system integration to autonomously manage these environments. However, the time required for regeneration to complete remains a critical issue.
Regeneration enabled by electric field
Although heat-based methods have been demonstrated to be effective, they carry the risk of potentially distorting the structural integrity of sensors. On the other hand, chemical approaches may introduce contaminating residues to the electrodes, thereby introducing additional variability in the transducers during regeneration cycles. As an alternative, recent studies have also documented the regeneration of biosensors through direct electrochemical methods, where the reductive desorption of surface species is accomplished by applying an electrical potential to the sensor surface. This potential, which can be either sweeping or constant, exerts a repulsion force to repel the analyte or facilitates an oxidation reaction, effectively removing the analyte from the sensor surface. It offers the advantage of establishing a highly localized regeneration environment that can be intricately controlled without requiring additional reagents. Several reported examples of this approach are associated with the utilization of molecularly imprinted polymers (MIPs), which are synthetic polymers with selective binding sites or cavities designed to recognize and capture specific target molecules.A recent study reports a regeneration method following this concept, which involves wearable biosensors consisting of laser-engraved graphene (LEG) electrodes, functionalized with polypyrrole (PPy)-based MIPs for detecting five amino acids (Trp, Tyr, Leu, Ile and Val) and Prussian blue nanoparticles (PBNP)-based redox-active reporter (RAR) nanoparticles.34 Given that some target biomarkers are electroactive (e.g., Tyr, Trp), while others are not (e.g., Leu, Val, Ile), the study employs two distinct operative detection strategies: direct and indirect. Each strategy involves its unique set of regeneration protocols (Fig. 5A). In the first category, the biosensors undergo regeneration facilitated by the application of a constant current (∼600 nA mm−2) under an applied voltage (∼0.7 V) over time. Within this electrochemical framework, the electroactive molecules bonded within the LEG-MIP matrix can undergo oxidation at their respective oxidation potentials, and subsequently be removed during the sweat renewal process. This methodology enables the in situ regeneration process without the need for washing steps. As indicated in Fig. 5B, this protocol allows for a robust series of up to 18 successful regenerations for an LEG-MIP Trp sensor in 50 μM Trp solution. Given that multiple electroactive molecules can undergo oxidation at similar potentials, the LEG–MIP approach effectively tackles both challenges in sensitivity and selectivity.
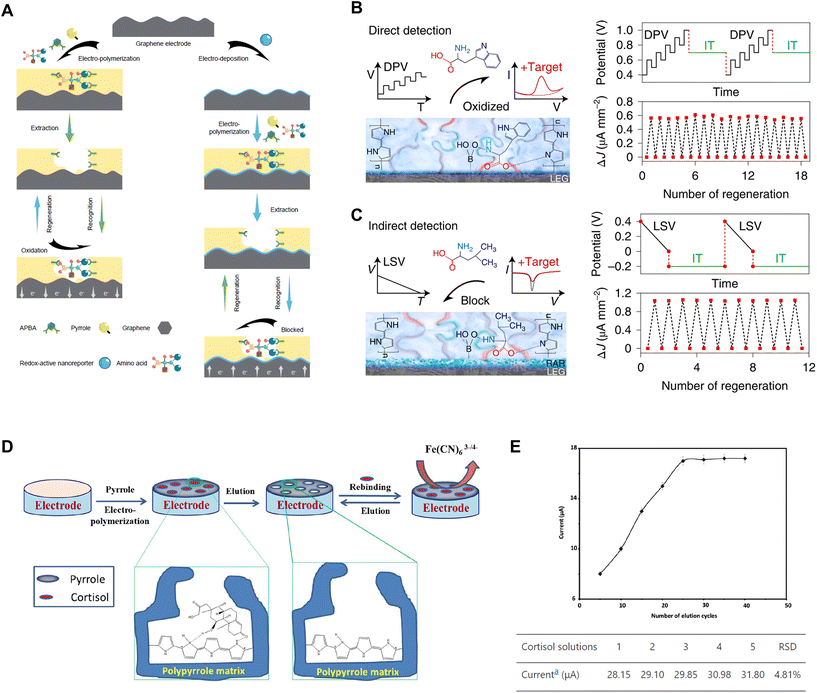 | ||
| Fig. 5 Electric potential-enabled regeneration: (A) schematic illustration of the fabrication, detection and regeneration processes of LEG-MIP sensors for direct detection and LEG-RAR-MIP sensors for indirect detection. Reprinted with permission from ref. 34. Copyright 2022 Springer Nature. (B) Direct detection of electroactive molecules using LEG-MIP biosensors, regenerated by oxidizing bound targets through the application of a constant current (∼600 nA mm−2) under a high voltage (∼0.7 V). Reprinted with permission from ref. 34. Copyright 2022 Springer Nature. (C) Indirect detection using LEG-RAR-MIP sensors, regenerated by repelling bound targets through a constant electrical potential (∼−0.2 V). Reprinted with permission from ref. 34. Copyright 2022 Springer Nature. (D) Schematic illustration of the fabrication and regeneration processes for MIP-based cortisol sensors via an over-oxidation procedure using cyclic voltammetry (0 to 0.8 V) and (E) response of the sensor after several times of electrochemical elution-based regeneration. Reprinted with permission from ref. 35. Copyright 2017 The Electrochemical Society. | ||
On the other hand, the indirect sensing strategy utilizes an extra layer of RAR sandwiched between the LEG and MIP layers as a signalling component that enables the detection of non-electroactive molecules. The adsorption of the target molecules onto the MIP layer decreases the exposure of the RAR, lowering the peak height current density and providing information about the concentration of analytes. To regenerate these LEG-RAR-MIP sensors detecting negatively charged amino acids, a constant electrical potential (∼−0.2 V) is applied to the working electrode. The electric field repels bound target molecules from the MIP layer, thereby restoring the functional baseline of the sensing platform. This process is exemplified by a Leu sensor regenerating 12 times in a 50 μM Leu solution (Fig. 5C).34
Similar findings have been reported in another work, which has demonstrated a regeneratable MIP-based cortisol-sensing interface composed of PPy (Fig. 5D).35 Following the adsorption of cortisol molecules, the sensor regeneration is conducted via electrochemical elution. This process entails an over-oxidation procedure carried out using cyclic voltammetry, covering a voltage range from 0 to 0.8 V over 25 cycles in a PBS solution, to eliminate the adsorbed cortisol from the matrix. However, it is worth noting that this over-oxidation can lead to a compromised adhesion of the PPy polymer to the electrode substrate, which may result in reduced sensor functionality. As illustrated in Fig. 5E, a maximum of seven effective regeneration cycles can be obtained.35
Overall, the utilization of electric field as a regenerative strategy is promising due to its potential for achieving regeneration of sensing capability without the need for additional washing steps and/or chemicals. This method also tends to enable a faster regeneration process. However, its applicability is currently limited, restricting the range of biosensors that can effectively employ this technique. Nonetheless, it remains a relatively underrepresented approach and necessitates a careful design of the sensing interfaces, which may consist of multiple functional layers, such as MIPs and RARs.
Regeneration enabled by magnetic field
Leveraging magnetic fields constitutes another innovative strategy capable of reversibly attaching and detaching receptors from the electrode in a controllable manner. By regulating the magnetic field around the electrode, sensor elements can be autonomously guided into precise alignment at the interface. This includes, but is not limited to, controlling the attachment and removal of the bioreceptors to facilitate the regeneration. An example is depicted in Fig. 6A, which demonstrates the working principle of a reusable magnetic bead-based electrochemiluminescence (ECL) DNA biosensor for the detection of genetically modified organisms (GMOs). Activation of the magnetic field generated by a removable magnet secures a bead-bound biotin probe-connected via a biotin–streptavidin linkage-to the working electrode situated within the electrochemical detection chamber.36 Subsequently, the targeted GMO DNA goes through a hybridization process with the immobilized biotin-probe and the ruthenium(II) tris-bipyridal (TBR)-probe. This leads to the formation of a sandwiched complex, wherein TBR interacts with tripropylamine (TPA), to elicit luminescence emission for ECL-based identification. Upon completion of the detection process, deactivating the magnetic field eliminates the magnetic bead-associated sandwich complex, achieving the regeneration of the entire system. In line with a similar concept, reactivating the magnetic field after the removal of saturated receptors can generate rejuvenated sensing platforms.36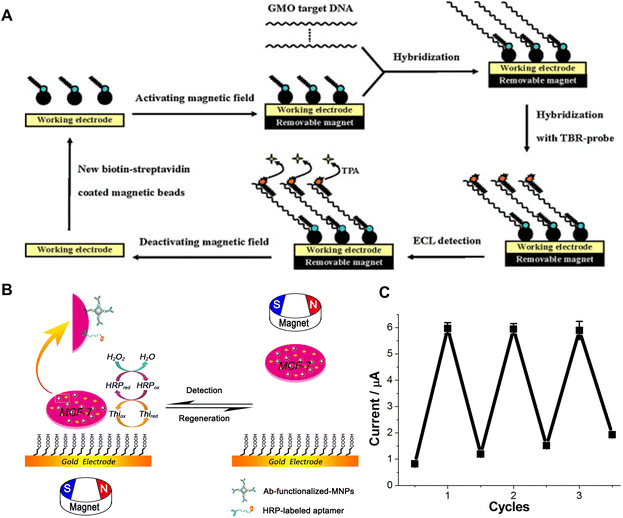 | ||
| Fig. 6 Magnetic field-enabled regeneration: (A) schematic illustration of the basic principle behind reusable magnetic bead-based ECL DNA biosensors for the detection of GMO, consisting of GMO target DNA, TBR-probes, biotin-probes, and streptavidin-coated magnetic beads. Reprinted with permission from ref. 36. Copyright 2010 Elsevier. (B) Schematic illustration of MCMA-based reusable electrochemical biosensors for the detection of human breast cancer cells, and the attraction and removal of the MCMA to and from the gold electrode surface by magnetic field, and (C) the current response for the measurement of 1 × 106 cells over three regeneration cycles. Reprinted with permission from ref. 37. Copyright 2014 Springer Nature. | ||
Another study introduces a magneto-controlled moveable architecture (MCMA), in which, the bio-detecting elements are immobilized on colloidal magnetic nanoparticles (MNPs) that can be attracted to or removed from the gold electrode surface by setting the magnet under or over the electrode.37 This study uses human breast cancer cells (Michigan cancer foundation-7 (MCF-7) cells) as detection targets (Fig. 6B). A sensitive biological element consisting of anti-CEA-antibody-functionalized MNPs (Ab@MNPs) and horseradish-peroxidase-labelled mucin-1 aptamer (HRP-apt) recognizes and binds to the MCF-7 cell. The MNPs enable magnetic manipulation, while the HRP offers electrochemical catalytic activity. When attracted to the electrode surface, the system, comprising hydrogen peroxide, thionine, and HRP, delivers sensitive electrochemical signals. After a measurement is performed, the electrode can be refreshed easily by attracting the MCMA away from the electrode surface using a magnetic field. The whole process can be finished in 2 min. The current response for the measurement of 1 × 106 cells after three cycles appears in Fig. 6C. However, it is worth noting that there is a slight increase in the background current, indicating an inevitable adsorption of the MCMA to the gold electrode.37
Conclusions
In summary, this mini-review discusses a wide range of strategies for the regeneration of biosensors, encompassing every aspect from re-functionalization methods to specialized techniques applied to specific biorecognition interfaces/elements. Table 1 summarizes the important parameters of the representing work discussed in the paper along with their major advantages and disadvantages. Given the growing emphasis on long-term monitoring of biomarkers in biofluids, approaches that permit in situ regeneration of the biosensing interface offer distinct advantages. To this end, while numerous successful regeneration strategies exist in the literature, attention remains particularly focused on those methods that obviate the need for extensive re-engineering and avoid harsh conditions. Recent studies, which are discussed in this mini-review, hold significant promise for achieving the stated goal. These studies include the allosteric regulation of pH-sensitive aptamers, the electrical field-driven regeneration of MIP-based biosensors, and the magnetic field control for the reversible attachment and removal of receptors. However, determining the optimal regeneration conditions is complex due to the variations depending on specific biosensors and target analytes. Achieving the balance of regeneration conditions is essential for maintaining the performance of the sensors/transducers and sensitivity across diverse applications.| Ref. | Sensing target | Sensing mechanism | Regeneration mechanism | Signal recovery percentage | Regeneration times | Major advantages | Disadvantages |
|---|---|---|---|---|---|---|---|
| 12 | Adenosine triphosphate | Aptamer | UV light | >90% for 5 cycles | 7 | Remote regeneration | Requires 1 hour of UV light treatment |
| 13 | E. coli crooks strain | Aptamer | Boiled water | >90% for 5 cycles | More than 5 | Good recovery ability | Harsh condition |
| 16 | CaMV 35S | DNA | 0.1 M NaOH | >90% for 5 cycles | 7 | Fast regeneration | Harsh condition |
| 26 | Ampicillin | Aptamer | Deionized water rinse | Close-to-identical | 3 | Simple, mild condition | Hard to extend to other biomarkers |
| 29 | Interferon-γ | DNA | Urea | >90% for 5 cycles | More than 10 | Good recovery ability | Requires integrated microfluidics |
| 32 | Cocaine | Aptamer | pH change | >90% at 5 cycles | Up to 7 | In vivo regeneration | Long regeneration time (1 h) |
| 34 | Electroactive amino acids | MIP | Oxidation potential | >90% for 5 cycles | Up to 18 | Fast and free of damage | Hard to extend to other biomarkers |
| 34 | Non-electroactive amino acids | RAR-MIP | Negative potential | >90% for 5 cycles | 12 | Fast and free of damage | Hard to extend to other biomarkers |
| 35 | Cortisol | MIP | Cyclic voltammetry | Relative standard deviation (RSD) = 4.81% at 5 cycles | 25 | Fast and free of damage | Hard to extend to other biomarkers |
| 36 | GMOs | DNA | Magnetic field | >90% for 5 cycles | Unlimited | Fast and free of damage | Complex system required |
| 37 | MCF-7 cells | DNA | Magnetic field | RSD < 5.9% at 3 cycles | 3 | Fast and free of damage | Complex system required |
The development of regeneration methods for biosensors is of importance for health monitoring. For short-term wearable sensors and point-of-care systems, the ability to regenerate biosensors plays a crucial role in cost reduction and resource conservation. This aspect gains even greater importance for biosensors dedicated to real-time and continuous monitoring, as they require the capability to conduct a series of chemical and biological biomarker measurements over extended periods. Currently, the predominant biorecognition elements, such as antibodies, aptamers, and molecularly imprinted polymers (MIPs), are designed for single use. This approach limits their application in long-term monitoring scenarios, especially in implantable systems, as these biosensors are generally incapable of being reused after one measurement cycle. Addressing this limitation, the exploration and development of regeneration technologies emerge as a promising direction for future research. These advancements could significantly enhance the sustainability and functionality of biosensors, particularly in applications requiring prolonged and repeated use.
Despite the discovery of various regeneration methods, achieving in vivo regeneration continues to be a significant challenge. This includes dealing with the continuous presence of biomarkers, the difficulty of dissociating reagents such as chemicals on the chip, managing the flow of chemicals to introduce analytes and remove waste, and consistently achieving the conditions necessary for effective regeneration. Alongside addressing these challenges, the sensor also needs to meet critical requirements for in vivo use, including miniaturization and biocompatibility.
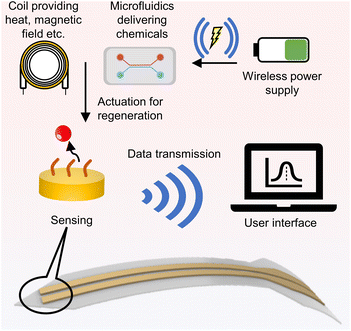
The solution for real-time in vivo detection and regeneration may lie in the development of an integrated system. This platform should incorporate a sensing interface, a regeneration-actuating component, and a wireless data transmission and power supply system. To illustrate this concept, Fig. 7 demonstrates the feedback loop of the system, showcasing how the integrated platform functions in a real-time detection scenario. For effective long-distance control of regeneration, utilizing electrical potential as the primary control factor is optimal due to the mature and simple wireless-control technologies. This integrated platform allows researchers to remotely apply voltage to the platform from outside the body, thereby initiating direct or indirect regeneration processes.
The methodologies presented in Fig. 4G and 5A exemplify effective strategies for in vivo regeneration via wireless potential control. The study showcased in Fig. 4G demonstrates how wireless control of potential through RF energy harvesting can instigate a localized pH alteration, consequently enabling the regeneration of anti-cocaine aptamers. Although the current a lack of in vivo testing, this study offers promising prospects for future in vivo monitoring applications. Furthermore, the research illustrated in Fig. 5A introduces a method of directly using potential for biosensor regeneration, facilitated by Bluetooth and a microcontroller. Despite its primary focus on a wearable sensor, this research effectively validates the regeneration process in an on-body trial, making it a viable option for in vivo monitoring. The integration of these components, particularly the wireless system, plays a vital role in ensuring the efficient functioning and control of the regeneration processes.
The challenge of minimizing performance degradation during the regeneration procedures is a critical one. Consequently, preserving the long-term lifetime of biosensors is a complex task. It is therefore essential for researchers to develop protocols that not only restore sensor performance but also ensure its durability, ultimately enhancing the lifespan of the sensing platform while maintaining its sensitivity and accuracy for reliable, long-lasting applications.
Another remaining challenge in this field is the lack of consistent reporting and benchmarking guidelines for determining the successful regeneration of biosensors. This issue is prevalent in the literature, where various biosensor techniques produce data that is not easily comparable. Consequently, it becomes challenging to evaluate the effectiveness of different regeneration procedures in different studies, hindering progress in this field. Therefore, in evaluating biosensor regeneration, we recommend prioritizing the ability of the sensor to nearly restore its original sensitivity as the primary criterion. Additionally, factors such as the number of regeneration cycles before noticeable degradation occurs, the duration of the sensor remains functional in working condition, and the speed of regeneration should be reported. These parameters collectively paint a picture of the working environment and reveal its potential applications. Based on these applications, the regeneration environment can then be evaluated for its safe applicability.
Integrating regeneration steps into sample handling systems can be technically demanding but is essential for practical deployment. In medical and biological applications, the biocompatibility of regeneration reagents is paramount to prevent damage to biotissues. Furthermore, the cost-efficiency is crucial for widespread adoption, as some regeneration methods can be expensive. Complex data analysis and interpretation are necessary, especially in multi-analyte systems or continuous monitoring processes. Overall, there remains a significant journey ahead in developing universally applicable regeneration strategies that can efficiently accommodate a higher number of regeneration cycles through simplified protocols. Tackling these challenges is imperative to drive progress in the field and unlock the complete potential of regeneratable biosensors across a wide range of applications.
Author contributions
Yizhen Jia and Shulin Chen contributed equally to this work.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by The Ohio State University start-up funds and the Chronic Brain Injury Pilot Award Program at The Ohio State University. This work was also supported by the Ohio State University Materials Research Seed Grant Program, funded by the Center for Emergent Materials; NSF-MRSEC, grant DMR-2011876; the Center for Exploration of Novel Complex Materials; and the Institute for Materials and Manufacturing Research. J. L. acknowledges the support from NSF award ECCS-2223387.References
- V. Naresh and N. Lee, Sensors, 2021, 21, 1109 CrossRef CAS PubMed.
- K. Yang, H. Peretz-Sorok, Y. Liu and F. Lin, Lab Chip, 2016, 16, 943–958 RSC.
- G. Rateni, P. Dario and F. Cavallo, Sensors, 2017, 17, 1453 CrossRef PubMed.
- N. M. Blebea, G. Hancu, R. A. Vlad and A. Pricopie, Molecules, 2023, 28, 638 CrossRef CAS PubMed.
- M. Mortas and H. NourAyvaz, Discov. Food, 2022, 2, 15 CrossRef.
- J. A. Goode, J. V. Rushworth and P. A. Millner, Langmuir, 2015, 31, 6267–6276 CrossRef CAS PubMed.
- J. Aleman, T. Kilic, L. S. Mille, S. R. Shin and Y. S. Zhang, Nat. Protoc., 2021, 16, 2564–2593 CrossRef CAS PubMed.
- Z. Wang, Z. Hao, X. Wang, C. Huang, Q. Lin, X. Zhao and Y. Pan, Adv. Funct. Mater., 2020, 31, 2005958 CrossRef.
- Y. Ning, J. Hu and F. Lu, Biomed. Pharmacother., 2020, 132, 110902 CrossRef CAS PubMed.
- F. Pfeiffer and G. Mayer, Front. Chem., 2016, 4, 25 Search PubMed.
- S. Cai, J. Yan, H. Xiong, Y. Liu, D. Peng and Z. Liu, Analyst, 2018, 143, 5317–5338 RSC.
- X. Zhang, C. Song, K. Yang, W. Hong, Y. Lu, P. Yu and L. Mao, Anal. Chem., 2018, 90, 4968–4971 CrossRef CAS PubMed.
- I. G. Siller, J.-A. Preuss, K. Urmann, M. R. Hoffmann, T. Scheper and J. Bahnemann, Sensors, 2020, 20, 4421 CrossRef CAS PubMed.
- P. Zhu, V. A. Papadimitriou, J. E. Van Dongen, J. Cordeiro, Y. Neeleman, A. Santoso, S. Chen, J. C. T. Eijkel, H. Peng, L. I. Segerink and A. Y. Rwei, Sci. Adv., 2023, 9, eadf5509 CrossRef CAS PubMed.
- A. Sobhan, J.-H. Oh, M.-K. Park and J. Lee, Microelectron. Eng., 2020, 225, 111281 CrossRef CAS.
- A. Ulianas, L. Y. Heng, M. Ahmad, H.-Y. Lau, Z. Ishak and T. L. Ling, Sens. Actuators, B, 2014, 190, 694–701 CrossRef CAS.
- P. P. Dillon, S. J. Daly, B. M. Manning and R. O'Kennedy, Biosens. Bioelectron., 2003, 18(2–3), 217–227 CrossRef CAS PubMed.
- V. B. Kandimalla, N. S. Neeta, N. G. Karanth, M. S. Thakur, K. R. Roshini, B. E. A. Rani, A. Pasha and N. G. K. Karanth, Biosens. Bioelectron., 2004, 20(4), 903–906 CrossRef CAS PubMed.
- J. Liu, Y. Xing, X. Zhou, G. Y. Chen and H. Shi, Biosens. Bioelectron., 2021, 176, 112902 CrossRef CAS PubMed.
- L. Liu, D. Shan, X. Zhou, H. Shi, B. Song, F. Falke, A. Leinse and R. Heideman, Biosens. Bioelectron., 2018, 106, 117–121 CrossRef CAS PubMed.
- L. Liu, X. Zhou, R. Ma, M. He, H. Shi and Q. Yi, Sens. Actuators, B, 2018, 259, 888–893 CrossRef CAS.
- W. Sun, L. Liu, A. G. Memon, X. Zhou and H. Zhao, Biosensors, 2020, 10, 191 CrossRef CAS PubMed.
- R. Wang, Y. Xiang, X. Zhou, L. Liu and H. Shi, Biosens. Bioelectron., 2015, 66, 11–18 CrossRef CAS PubMed.
- R. Wang, X. Zhou, H. Shi and Y. Luo, Biosens. Bioelectron., 2016, 78, 418–422 CrossRef CAS PubMed.
- R. Wang, X. Zhou and H. Shi, Biosens. Bioelectron., 2015, 74, 78–84 CrossRef CAS PubMed.
- Z. G. Yu and R. Y. Lai, Talanta, 2018, 176, 619–624 CrossRef CAS PubMed.
- J.-G. Walter and F. Scheper, Eng. Life Sci., 2012, 12, 496–506 CrossRef CAS.
- K. Rahimizadeh, H. AlShamaileh, M. Fratini, M. Chakravarthy, M. Stephen, S. Shigdar and R. N. Veedu, Molecules, 2017, 22, 2070 CrossRef PubMed.
- Q. Zhou, T. Kwa, Y. Gao, Y. Liu, A. Rahimian and A. Revzin, Lab Chip, 2014, 14, 276–279 RSC.
- L. Sun, B. Cao, Y. Liu, P. Shi, Y. Zheng, B. Wang and Q. Zhang, J. Phys. Chem. B, 2022, 126, 8708–8719 CrossRef CAS PubMed.
- C. K. L. Gordon, M. Eisenstein and H. T. Soh, ACS Sens., 2018, 3, 2574–2580 CrossRef CAS PubMed.
- S. Chen, T.-L. Liu, Y. Dong and J. Li, ACS Nano, 2022, 16, 20922–20936 CrossRef CAS PubMed.
- E. M. McConnell, R. Bolzon, P. Mezin, G. Frahm, M. Johnston and M. C. DeRosa, Bioconjugate Chem., 2016, 27, 1493–1499 CrossRef CAS PubMed.
- M. Wang, Y. Yang, J. Min, Y. Song, J. Tu, D. Mukasa, C. Ye, C. Xu, N. Heflin, J. S. McCune, T. K. Hsiai, Z. Li and W. Gao, Nat. Biomed. Eng., 2022, 6, 1225–1235 CrossRef CAS PubMed.
- P. Manickam, S. K. Pasha, S. A. Snipes and S. Bhansali, J. Electrochem. Soc., 2016, 164, 54–59 CrossRef.
- D. Zhu, J. Liu, Y. Tang and D. Xing, Sens. Actuators, B, 2010, 149, 221–225 CrossRef CAS.
- X. Zhu, C. Feng, Z. Ye, Y. Chen and G. Li, Sci. Rep., 2014, 4, 4169 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2024 |