Open Access Article
Open Access ArticleExploring the conformational dynamics of the SARS-CoV-2 SL4 hairpin by combining optical tweezers and base analogues†
Vinoth
Sundar Rajan‡
 ab,
Anna
Wypijewska del Nogal‡
ab,
Anna
Wypijewska del Nogal‡
 b,
Sune
Levin
a,
L. Marcus
Wilhelmsson
b,
Sune
Levin
a,
L. Marcus
Wilhelmsson
 *b and
Fredrik
Westerlund
*b and
Fredrik
Westerlund
 *a
*a
aDivision of Chemical Biology, Department of Life Sciences, Chalmers University of Technology, Gothenburg, SE-412 96, Sweden. E-mail: fredrik.westerlund@chalmers.se
bDivision of Chemistry and Biochemistry, Department of Chemistry and Chemical Engineering, Chalmers University of Technology, Gothenburg, SE-412 96, Sweden. E-mail: marcus.wilhelmsson@chalmers.se
First published on 7th December 2023
Abstract
The parasitic nature of the SARS-CoV-2 virus demands selective packaging of its RNA genome (gRNA) from the abundance of other nucleic acids present in infected cells. Despite increasing evidence that stem-loop 4 (SL4) of the gRNA 5′ UTR is involved in the initiation of this process by binding the nucleocapsid (N) protein, little is known about its conformational dynamics. Here, we unravel the stability, dynamics and (un)folding pathways of SL4 using optical tweezers and a base analogue, tCO, that provides a local and subtle increase in base stacking without perturbing hydrogen bonding. We find that SL4 (un)folds mainly in a single step or through an intermediate, encompassing nucleotides from the central U bulge to the hairpin loop. Due to an upper-stem CU mismatch, SL4 is prone to misfold, the extent of which can be tuned by incorporating tCO at different positions. Our study contributes to a better understanding of SARS-CoV-2 packaging and the design of drugs targeting SL4. We also highlight the generalizability of using base analogues in optical tweezers experiments for probing intramolecular states and conformational transitions of various nucleic acids at the level of single molecules and with base-pair resolution.
Introduction
Coronavirus infectious disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), leading to from mild to lethal respiratory infections.1 Like other coronaviruses belonging to the Betacoronavirus genera of the Coronaviridae family, e.g., SARS-CoV (2002 outbreak) or MERS-CoV (Middle East respiratory syndrome-CoV; 2013 outbreak), SARS-CoV-2 contains a large (∼30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nt) and enveloped single-stranded RNA genome (gRNA; Fig. 1A).1 A major challenge for the virus is to distinguish its own gRNA from the pool of cellular RNAs and its own subgenomic mRNAs (sgmRNAs) in virion assembly.2 The nucleocapsid (N) protein is the protein largely responsible for binding, compacting, and packaging of the SARS-CoV-2 genome into the nascent virion.3–6 Growing evidence indicates that the packaging occurs through liquid–liquid phase separation (LLPS; Fig. 1B), i.e., macromolecular self-assembly, leading to the formation of functional micron-scale droplets (condensates) through multivalent interactions.3,4,7,8In vitro work has demonstrated that the formation of gRNA-N protein condensates (Fig. 1B) is initiated at specific regions of the gRNA sequence that have a high affinity for the N protein (termed principal N protein-binding sites; Fig. 1A), while many other regions are bound at higher N protein concentrations.3,4 One of the principal binding sites is flanked by stem-loop 4 (SL4) – the object of this study.3,9 SL4 is a 44 nt long RNA hairpin in the 5′ untranslated region (5′ UTR) of the SARS-CoV-2 gRNA (nts 84–127; Fig. 1A and 2A).3,9 Interestingly, it is the double-stranded RNA (dsRNA) structure of SL4, and not its sequence nor the single-stranded RNA (ssRNA) sequence of the principal binding site, that promotes gRNA condensation with the N protein,4 making conformational studies of SL4 particularly important. Apart from playing a role in LLPS, SL4 is considered to function as a spacer required for sgmRNA synthesis, mediating a proper relative orientation between SL1–3 and the open reading frame 1a (ORF1a) encoding non-structural proteins (NSPs).10 Moreover, SL4 carries a short upstream ORF (uORF; its AUG start codon is positioned in the SL4 hairpin loop – Fig. 2A, arrow) that could reduce translation initiation at the ORF1a start codon.10 Furthermore, a recent study demonstrated that RNA-binding protein 24 (RBM24) binds to SL4 and inhibits the translation of viral polyproteins.11 The SL4 hairpin is structurally conserved among Betacoronavirus genera and does not change in SARS-CoV-2 variants.10,12–14 This suggests that it is important for viral propagation and makes it interesting not only for deciphering the infection process, but also as a promising therapeutic target for novel drugs insensitive to virus evolution.10,12
000 nt) and enveloped single-stranded RNA genome (gRNA; Fig. 1A).1 A major challenge for the virus is to distinguish its own gRNA from the pool of cellular RNAs and its own subgenomic mRNAs (sgmRNAs) in virion assembly.2 The nucleocapsid (N) protein is the protein largely responsible for binding, compacting, and packaging of the SARS-CoV-2 genome into the nascent virion.3–6 Growing evidence indicates that the packaging occurs through liquid–liquid phase separation (LLPS; Fig. 1B), i.e., macromolecular self-assembly, leading to the formation of functional micron-scale droplets (condensates) through multivalent interactions.3,4,7,8In vitro work has demonstrated that the formation of gRNA-N protein condensates (Fig. 1B) is initiated at specific regions of the gRNA sequence that have a high affinity for the N protein (termed principal N protein-binding sites; Fig. 1A), while many other regions are bound at higher N protein concentrations.3,4 One of the principal binding sites is flanked by stem-loop 4 (SL4) – the object of this study.3,9 SL4 is a 44 nt long RNA hairpin in the 5′ untranslated region (5′ UTR) of the SARS-CoV-2 gRNA (nts 84–127; Fig. 1A and 2A).3,9 Interestingly, it is the double-stranded RNA (dsRNA) structure of SL4, and not its sequence nor the single-stranded RNA (ssRNA) sequence of the principal binding site, that promotes gRNA condensation with the N protein,4 making conformational studies of SL4 particularly important. Apart from playing a role in LLPS, SL4 is considered to function as a spacer required for sgmRNA synthesis, mediating a proper relative orientation between SL1–3 and the open reading frame 1a (ORF1a) encoding non-structural proteins (NSPs).10 Moreover, SL4 carries a short upstream ORF (uORF; its AUG start codon is positioned in the SL4 hairpin loop – Fig. 2A, arrow) that could reduce translation initiation at the ORF1a start codon.10 Furthermore, a recent study demonstrated that RNA-binding protein 24 (RBM24) binds to SL4 and inhibits the translation of viral polyproteins.11 The SL4 hairpin is structurally conserved among Betacoronavirus genera and does not change in SARS-CoV-2 variants.10,12–14 This suggests that it is important for viral propagation and makes it interesting not only for deciphering the infection process, but also as a promising therapeutic target for novel drugs insensitive to virus evolution.10,12
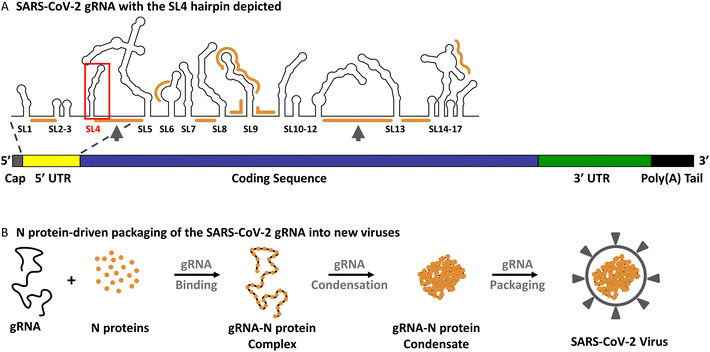 | ||
| Fig. 1 (A) SARS-CoV-2 gRNA with the secondary structure of its 5′ end containing the SL4 hairpin (red frame) depicted. Orange lines – N protein-binding sites and arrows – principal N protein-binding sites. The secondary structure was retrieved from Iserman et al.3 (B) Schematic representation of the N protein-driven packaging of the SARS-CoV-2 gRNA into new virions through liquid–liquid phase separation (LLPS). Adapted from Jack et al.7 | ||
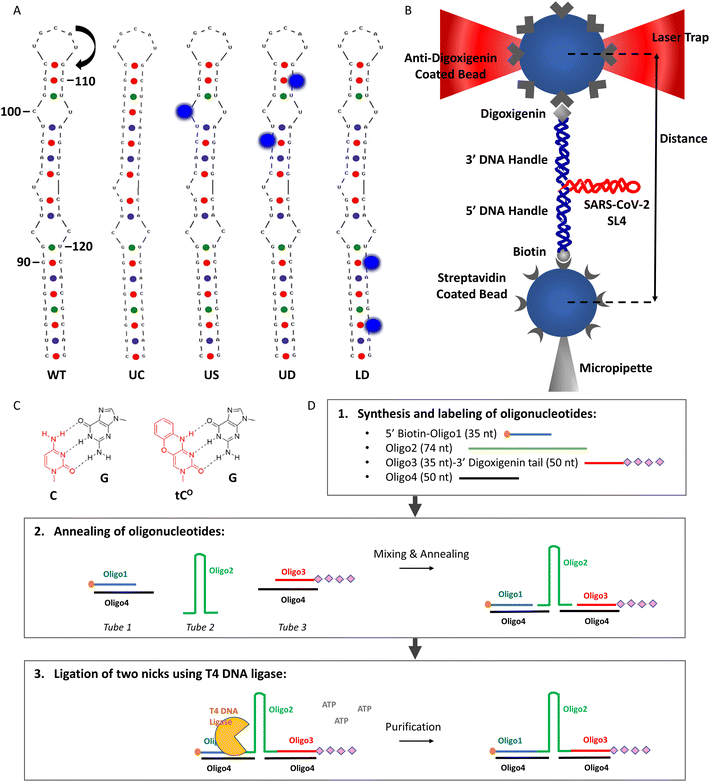 | ||
| Fig. 2 (A) The SARS-CoV-2 SL4 constructs studied. The wild type (WT; genomic numbering) and upper complementary (UC) hairpins are the lowest free energy secondary structures predicted in UNAFold. The upper-single tCO (US), upper-double tCO (UD) and lower-double tCO (LD) structures are drawn schematically (not predicted). GC, AU, and GU base pairs are marked by red, blue, and green dots, respectively. tCO modifications are depicted as cobalt spheres. The start (AUG start codon) of the uORF is indicated with an arrow. (B) The optical tweezers’ setup used for studying SARS-CoV-2 SL4 (adapted from Wypijewska del Nogal et al.15). Drawing is not to scale. (C) The structure of the tCO base analogue paired with guanine, G, in comparison to cytosine, C, paired with G. (D) Method for preparing RNA/DNA constructs for optical tweezers studies. The sequence of interest (here WT or modified SL4) is contained in Oligo2 and is, in the final construct, flanked by a short (50 bp) dsDNA handle at each end. | ||
Optical tweezers (Fig. 2B) is a powerful tool to investigate structure and dynamics of RNAs,15–23 but very few studies have been reported so far, where this single-molecule force spectroscopy technique was employed to examine the SARS-CoV-2 gRNA. The studies that exist focus on the programmed ribosomal frameshifting (PRF) pseudoknot.24–26 Here, we use optical tweezers to study the SARS-CoV-2 SL4 hairpin and in that way define its dynamics and structural rearrangements, as they may be useful to understand packaging of single SARS-CoV-2 genomes into developing virions.
For the first time, we here expand the conformational readout of RNA structures by optical tweezers by incorporating base analogues (BAs) into the studied structure. BAs are synthetic mimics of natural nucleobases designed, in overall, to enhance and/or probe specific physicochemical properties of nucleic acids without significantly perturbing their structure. Until now, they were conventionally studied using ensemble techniques, such as spectroscopy or thermal melting.27–31 Here we demonstrate that using BAs in optical tweezers provides a way to locally modify the RNA of interest and characterize its stability and conformational dynamics with base-pair resolution. In particular, it allows identification of structural elements within the RNA that significantly influence these properties. The BA which we use here is a tricyclic cytosine, tCO (Fig. 2C),27 yet our approach is general for any BA. tCO resembles the natural cytosine (C) in that it forms three hydrogen bonds with guanine (Fig. 2C) and preserves A-form for RNA duplexes,27,32 and B-form for DNA duplexes.33 However, due to the extended aromatic system, it slightly stabilizes RNA/DNA duplexes in most sequence contexts via increased base stacking.27,32,33 In a previous study, we used tC (differing in structure to tCO only in that the oxygen in the middle ring is replaced by a sulfur)34 in place of cytosine in a fully complementary 44 nt-long DNA hairpin and observed an increase in its stability in unfolding experiments with optical tweezers with the number of tC incorporations.35 The added value of using BAs compared to standard mutagenesis, where an intrinsic nucleobase is replaced by another canonical base15,21,22,36 or where it is omitted,16 is that more subtle, close-to-native effects can be observed and that specific intramolecular interactions can be investigated.
In this work, we demonstrate how BA-modified RNA studied using optical tweezers provides a detailed understanding of key features of the SARS-CoV-2 SL4 hairpin, including its spatiotemporal dynamics and conformational ensembles. We identify a large heterogeneity of the hairpin (un)folding that, to a large extent, is explained by a CU mismatch in the upper stem. Our observations may help to elucidate SL4's interactions with, for example, the N protein and how these interactions affect virus propagation. Furthermore, our new method using BAs in optical tweezers is general to any nucleic acid and complements existing techniques37 in determining intramolecular states and conformational transitions at the single-molecule level.
Results
Design of the SARS-CoV-2 SL4 constructs
The lowest free energy (ΔG = −24.3 kcal mol−1) secondary structure of SL4 (encompassing nts 84–127; Fig. 2A, WT – wild type), predicted using UNAFold (Integrated DNA Technologies. https://eu.idtdna.com/UNAFold (accessed on September 23, 2021)), is a hairpin with a non-perfect stem (containing a C92:C119 mismatch, a C100:U112 mismatch and a U95 bulge) and a non-canonical 5 nt hairpin loop (5′-UGCAU-3′). This agrees perfectly with the secondary structure of SL4 determined by selective 2′-hydroxyl acylation analysed by primer extension (SHAPE) probing3 and nuclear magnetic resonance (NMR) spectroscopy.9,38For the optical tweezers’ experiments, we designed five constructs containing either wild-type or modified SL4 sequences (Fig. 2A and ESI Table S1†). In the upper complementary (UC) construct (Fig. 2A and ESI Table S1†), uracil 112 was replaced by guanine (U112G), providing Watson–Crick base pairing in place of the C100:U112 mismatch. Its lowest free energy secondary structure (ΔG = −29.0 kcal mol−1; predicted in UNAFold) was identical to the WT SL4 structure, except for the complementary C100:G112 base pair instead of the C100:U112 mismatch (Fig. 2A). The difference in ΔG between WT and UC (4.7 kcal mol−1) indicates that removing the C100:U112 mismatch stabilizes the hairpin significantly. In the other three modified constructs (Fig. 2A and ESI Table S1†), the tCO cytosine analogue27 (Fig. 2C) replaced an intrinsic cytosine either in the upper or lower region of the SL4 hairpin, in positions C100 (upper-single tCO construct, US), C98 and C110 (upper-double tCO construct, UD) or C121 and C125 (lower-double tCO construct, LD). The tCO BA has the same hydrogen bonding as native cytosine (Fig. 2C), but slightly increased base stacking.27 This makes it possible to modify different sites within SL4 with minimal perturbation to its native structure and probe the importance of specific structural elements within the hairpin (lower stem in LD, CU mismatch in US, and upper-stem base-paired regions flanking the mismatch in UD).
To allow optical tweezers nanomanipulation, the RNA sequences were flanked at each end by a short double-stranded DNA (dsDNA) handle (50 bp; Fig. 2B and ESI Table S1†) for the attachment to beads functionalized with streptavidin and digoxigenin, respectively, enabling formation of a tether between the beads (Fig. 2B). A material- and cost-effective design of the oligonucleotides was developed (Fig. 2D), where each RNA hairpin was initially flanked only by 15 DNA nucleotides at each end (Oligo2). In the subsequent steps (Fig. 2D), the hairpins were annealed and ligated to two dsDNA handles with sticky ends (Oligo1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Oligo4 and Oligo3
Oligo4 and Oligo3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Oligo4), universal for all RNA sequences used in the study due to the sequence complementarity between the sticky ends and the DNA flanks of Oligo2.
Oligo4), universal for all RNA sequences used in the study due to the sequence complementarity between the sticky ends and the DNA flanks of Oligo2.
(Un)folding pathways of the WT, UC, and tCO-modified SL4 hairpins
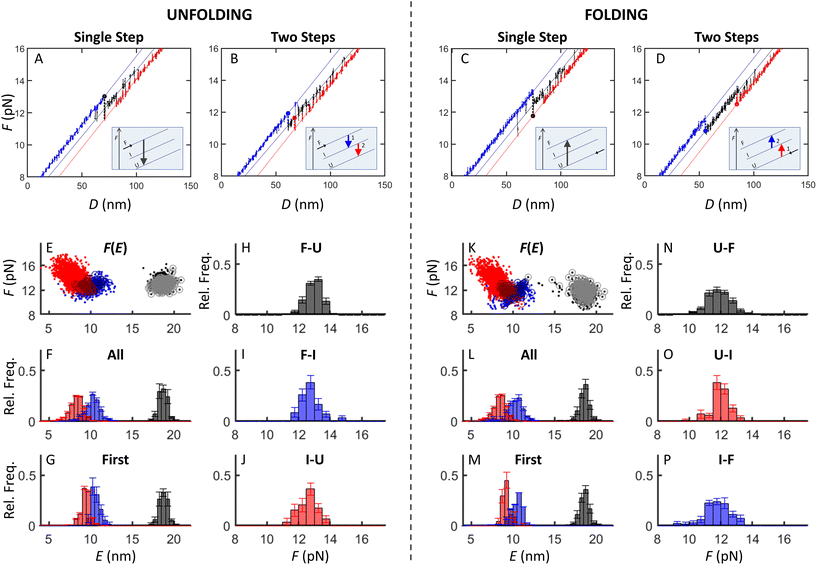
Several types of (un)folding pathways were observed in our optical tweezers’ measurements (Fig. 3A). They were classified based on identifying unfolding (rips) and folding (zips) transitions in the pull and relax force–distance curves (FDCs), respectively,15,16,21,23 considering each relax curve with the succeeding pull curve23,39 (see ESI section 2† for classification details). The (un)folding forces (F) and extensions (E) were determined as described in Materials and methods.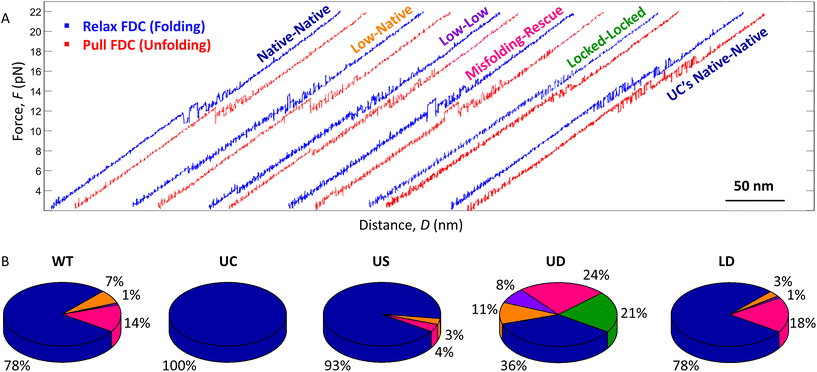 | ||
| Fig. 3 (Un)folding pathways of the WT, UC and tCO-modified SL4 hairpins. (A) Representative relax FDCs (blue) with their consecutive pull FDCs (red) illustrating different types of (un)folding: native–native, low-native, low–low and misfolding-rescue – observed for WT, US, UD, and LD (WT FDCs are depicted), locked–locked for UD, and UC's native–native for UC. (B) Fractions of each (un)folding pathway for the different constructs (statistical analysis in ESI Tables S2–S5†). Colour code as in (A). | ||
For WT SL4, a majority of the FDCs (78%, Fig. 3B) showed multiple unfolding and folding transitions between ∼10–14 pN (referred to as native–native, i.e., displaying a complete folding from the single strand to the native hairpin and vice versa in unfolding; Fig. 3A). The molecule extension during a complete transition in unfolding (EU = 18.8 ± 0.4 nm) and folding (EF = 18.6 ± 0.5 nm) corresponded to 44.0 ± 1.1 nt and 45.0 ± 1.5 nt, respectively, which agrees with the number of nucleotides in the SL4 hairpin (44 nt). The remaining 22% of the FDCs revealed other types of unfolding and folding pathways (referred to as non-native). Among them, 7% demonstrated folding to the native hairpin conformation at a low force (<8 pN), as apparent from the signature of the subsequent pull FDC being the same as for the native–native process (referred to as low-native; Fig. 3A and B). In rare cases (1%), when the hairpin folded at a low force, it also unfolded at a low force (<8 pN, referred to as low-low; Fig. 3A and B). Furthermore, a significant portion of the relax FDCs (14%) indicated that WT SL4 can misfold. The consecutive pull FDC always had a ‘rescue’ pattern, previously described in the literature,16,26 where the hairpin folded to the native conformation prior to unfolding. A pair of such curves is referred to as misfolding-rescue (Fig. 3A and B). The number of nucleotides released in the rescue curves is in a vast majority of the curves lower than for the full hairpin, indicating that it is indeed a misfolded hairpin (ESI Fig. S1†).
In contrast to the heterogeneity of folding and unfolding seen for WT SL4, UC, where the upper mismatch is replaced by a complementary base pair, showed only one relax-pull pattern. This pattern was termed as UC's native–native (Fig. 3A and B), as it resembled the native–native FDCs of WT except for two features: the range of transitions was shifted to higher forces (∼13–17 pN) and both folding and unfolding always occurred in two consecutive steps with multiple transitions between the states. This behaviour of the UC hairpin indicates that the heterogeneous dynamics observed for WT is due to the unpaired bases in the upper stem of the hairpin.
For the tCO-modified constructs, the same (un)folding pathways as for WT SL4 were observed, albeit in different proportions (Fig. 3B). For US, the occurrence of native–native (un)folding was significantly increased compared to WT and observed in 93% of the FDCs (Fig. 3B and ESI Table S2†). In addition, the occurrence of the low-native pathway (3%) decreased (non-significantly) and the misfolding-rescue pathway (4%) showed a significant decrease (Fig. 3B and ESI Tables S3 and S5†). UD, in turn, had a (non-significant) increase in low-native (11%) and low-low (8%) pathways and a significant increase in misfolding-rescue (24%) compared to WT (Fig. 3B and ESI Tables S3–S5†). Native–native (un)folding was significantly decreased and observed in 36% of the FDCs for UD (Fig. 3B and ESI Table S2†). Moreover, UD was the only construct displaying a ‘locked–locked’ pattern (21% of FDCs; Fig. 3A and B), a type of a misfolding without rescue, where we observed only two states in folding and unfolding FDCs (see ESI section 5† for more information). (Un)folding of LD was very similar to WT SL4 (Fig. 3B and ESI Tables S2–S5†), indicating that the two tCO incorporations in the lower stem did not significantly affect the dynamics of the hairpin.
Single vs. two steps in native (un)folding
The FDCs of the WT SL4 hairpin revealed dynamics in unfolding and folding, with multiple transitions (hopping) between three conformations: hairpin (F), intermediate (I) and single strand (U) (Fig. 4A–D). These different conformations were determined from the FDCs and analysed using a custom-made MATLAB code (described in Materials and methods and ESI†), resulting in (un)folding force versus extension (F(E)) plots (Fig. 4E and K). The presence of three clusters of data points in these plots suggests that unfolding proceeded either in a single-step (F–U) or through an intermediate state (in two steps, F–I, and I–U), as discussed further below. For folding, the reverse steps were identified (U–F, U–I, and I–F, respectively). The approach applied for the identification of single vs. two-step (un)folding transitions, and hence for determining the presence of the intermediate state, is described in detail in ESI section 3 and Fig. S2†. To confirm that there are indeed two different pathways both in folding and unfolding we investigated the distribution of the lifetime of the intermediate state in (un)folding, using only the first transition (see further below). Approximately 90% of the intermediates existed for less than 5 ms (ESI Fig. S3†), while lifetimes up to 244 ms (folding) and 326 ms (unfolding) were also observed. This is far more than expected from an exponential distribution of all lifetimes and supports the existence of two different (un)folding pathways. On the other hand, the lifetime distribution of UC SL4 showed an exponential distribution with ∼10% of intermediates with a lifetime of less than 5 ms, suggesting the existence of only one native pathway (ESI Fig. S4†). The lifetime distributions of the tCO-modified SL4 constructs were similar to WT, confirming the existence of two different native pathways (ESI Fig. S5–S7†) also for these constructs.For the WT and tCO-modified constructs, there were from 4.6 (for US) to 7.0 (for WT) times more transitions between I and U (red dots) than between F and I (blue dots) in unfolding and from 4.5 (for US) to 9.6 (for WT) in folding (Fig. 4A–E, K, ESI Fig. S8B–D, and Table S6†). In addition, the U–I transitions occurred in a much broader force and extension range than the I–F transitions and were enriched in transitions at short extension and high force (Fig. 4E and K), indicating transient base-pairing of short hairpin fragments due to the CU mismatch in the upper stem. Accordingly, for UC SL4, the U–I and F–I clusters in (un)folding showed a narrow extension range (ESI Fig. S8A†) as compared to other constructs (Fig. 4E and K, and ESI Fig. S8B–D†), because the complementary upper stem prevents the formation of transient short hairpin fragments. Also, the ratio of the I–U to F–I transitions in unfolding (and U–I to I–F in folding) was close to 1 for UC (Table S6†), meaning that the dynamics of this hairpin before and after the intermediate state is almost the same.
While all transitions inform on the complete dynamics of the molecule between states, we next focused on the first complete transition, i.e., between fully folded hairpin and the single stranded state, and vice versa. The first complete unfolding transition happened either via a single-step (F–U) or in two consecutive steps (F–I followed by I–U), and the reverse steps were observed in folding. First transitions are denoted as circles over dots (which represent all transitions) in the F(E) plots in Fig. 4E and K, where they also can be seen in three clusters. The mean extension of the first transitions overlapped with the mean of all transitions for F–U and F–I (un)folding (compare centres of the extension histograms; Fig. 4F vs. G and L vs. M). For I–U (un)folding, the mean of all transitions was lower than for the first transitions (extension histograms), in line with the previous finding that single-stranded SL4 can spontaneously hybridize on short fragments. A similar observation was made for the force: the mean force of all transitions was similar to the mean force of the first transitions for F–U and F–I (un)folding, and it was higher for I–U (un)folding (compare the range of red dots vs. red circles in (un)folding F(E) plots; Fig. 4E and K).
The (un)folding forces and nucleotide numbers were determined for the first transitions as has been proposed before15,40 and are summarized in Table 1. In single step unfolding (Fig. 4A), WT SL4 unzipped to the single strand at FU of 13.0 ± 0.4 pN (Table 1; FU distribution in Fig. 4H), which corresponded to unfolding of 44.0 ± 1.1 nt. In two-step unfolding (Fig. 4B), the hairpin unzipped partially at 12.7 ± 0.5 pN, releasing 25.0 ± 0.9 nt (corresponding to the lower stem, from the 5′/3′ ends until the U bulge; Fig. 2A), before full unzipping in which 22.0 ± 0.9 nt were released (Table 1 and Fig. 4I, J). This indicates a transient formation of an intermediate structure containing about a half of the hairpin before the complete unfolding as also evident from the free energy landscape (FEL) profile at zero force, calculated as a sum of ΔG per bp using UNAFold (ESI Fig. S9A†). On average, the second step occurred within the same force range as the first step (Table 1). This further supports the conclusion that SL4 is a bipartite hairpin with the central U bulge serving as a separator between the two duplex regions.
| SARS-CoV-2 SL4 | No of molecules | No of FDCs | Unfolding | Folding | ||||
|---|---|---|---|---|---|---|---|---|
| F–U | F–I | I–U | U–F | U–I | I–F | |||
| Unfolding force, FU (pN) | Folding force, FF (pN) | |||||||
| a The reported values are the median of measurements on five or more molecules ± standard error of the median. Measured in 50 mM Tris-HCl, 140 mM NaCl, 5.0 mM KCl, 1.0 mM EDTA and 5.0 mM MgCl2, pH 7.4, at room temperature. NA – not applicable. | ||||||||
| WT | 9 | 1093 | 13.0 ± 0.4 | 12.7 ± 0.5 | 12.7 ± 0.5 | 11.9 ± 0.6 | 12.0 ± 0.4 | 11.8 ± 0.7 |
| UC | 5 | 548 | NA | 15.1 ± 0.5 | 14.9 ± 0.6 | NA | 14.5 ± 0.5 | 14.7 ± 0.5 |
| US | 5 | 481 | 12.5 ± 0.8 | 12.1 ± 0.4 | 11.8 ± 0.4 | 12.5 ± 0.9 | 11.6 ± 0.5 | 11.6 ± 0.8 |
| UD | 5 | 228 | 13.3 ± 0.8 | 12.9 ± 0.8 | 13.2 ± 0.8 | 12.0 ± 0.8 | 12.3 ± 0.4 | 12.1 ± 0.5 |
| LD | 6 | 686 | 13.1 ± 0.5 | 12.8 ± 0.5 | 12.7 ± 0.4 | 12.1 ± 0.6 | 12.2 ± 0.5 | 12.0 ± 0.8 |
| Number of unfolded nucleotides | Number of folded nucleotides | |||||||
| WT | 9 | 1093 | 44.0 ± 1.1 | 25.0 ± 0.9 | 22.0 ± 0.9 | 45.0 ± 1.5 | 22.0 ± 1.1 | 25.0 ± 1.0 |
| UC | 5 | 548 | NA | 23.0 ± 0.6 | 23.0 ± 1.1 | NA | 23.0 ± 0.8 | 23.0 ± 0.6 |
| US | 5 | 481 | 43.0 ± 1.9 | 26.0 ± 1.2 | 22.0 ± 1.3 | 43.0 ± 2.1 | 22.0 ± 1.8 | 27.0 ± 0.8 |
| UD | 5 | 228 | 44.0 ± 1.2 | 25.0 ± 0.8 | 22.0 ± 1.1 | 44.0 ± 1.9 | 22.0 ± 1.4 | 25.0 ± 1.0 |
| LD | 6 | 686 | 43.0 ± 1.0 | 24.0 ± 1.2 | 21.0 ± 1.0 | 43.0 ± 1.5 | 21.0 ± 1.3 | 24.0 ± 1.2 |
The number of nucleotides folded in the single-step process corresponded to the full length of WT SL4 (Table 1). The measured length of the folding intermediate was the same as in unfolding (Table 1) and corresponded to the upper part of the SL4 hairpin from the U bulge to the hairpin loop. This suggests that the two-step unfolding of the WT SL4 hairpin proceeds in unzipping of its lower stem until the U bulge followed by unzipping of the upper stem, and that folding is a reverse process involving the same steps. Most (89%) of the pull FDCs showed the single-step unfolding as the first transition and 11% showed two-step unfolding (Fig. 5A). Similarly, single-step transitions were dominant in folding (86% of FDCs; Fig. 5B).
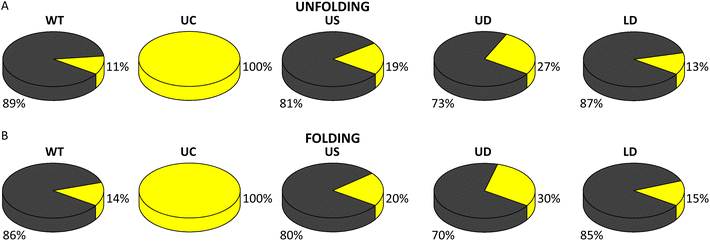 | ||
| Fig. 5 Fraction of single- (grey) and two-step (yellow) unfolding (A) and folding (B) for each SARS-CoV-2 SL4 construct (statistical analysis in ESI Tables S7 and 8†). | ||
Making the upper part of the stem fully base-paired, as in UC, eliminated (un)zipping of the molecule in a single step (Fig. 5A and B) and shifted both unfolding and folding of the two-step transitions to significantly higher forces (Table 1 and ESI Fig. S10†; force distribution for UC can be seen in ESI Fig. S8A†). As for the WT SL4, the number of nucleotides (un)folded in the first step corresponded to the (un)zipping of half of the hairpin (Table 1), in agreement with the FEL analysis (ESI Fig. S9B†). This difference between WT and UC agrees with the analysis of the lifetime distributions in Fig S3–S7.†
The values of (un)folding force and number of nucleotides were the same (within the error) for WT and the three tCO-modified constructs (Table 1 and ESI Fig. S10;† force distribution for US, UD, and LD in ESI Fig. S8B–D†). However, the proportions between single- and two-step (un)folding varied between the constructs (Fig. 5A and B). In unfolding, the percentage of two-step transitions showed a (non-significant) increase from 11% for WT to 19% for US and a further (significant) increase to 27% for UD (ESI Table S7†). A similar trend was observed in folding, where the two-step transitions increased from 14% for WT to 20% for US and 30% for UD (ESI Table S8†). The fraction of two-step transitions for LD (Fig. 5A and B) was similar to WT, both in unfolding (13%) and folding (15%) (ESI Tables S7 and S8†).
Occurrence of hairpin, intermediate, and single-strand conformations in native (un)folding
The above analysis revealed characteristics for the first complete unfolding and folding of the SL4 hairpins via a single and two steps. However, multiple transitions occur between the three conformational states and force-dependent state probability analysis of native FDCs (Fig. 6 and ESI Fig. S11, and section 4†) provides additional information on how the occurrence of folded, intermediate, and unfolded states changes with force. UC SL4 unfolded and formed an intermediate at a significantly higher force than WT and all tCO-containing constructs (Fig. 6A–C). The WT and LD SL4s showed similar hairpin, intermediate, and single-strand populations (Fig. 6A–C). The other two tCO-containing SL4s, US and UD, initiated unfolding and formation of the intermediate at lower forces compared to WT, as well as formed the intermediate over a broader force range (Fig. 6B–C). Interestingly, while the population of the US intermediate shifted to lower forces in unfolding, it overlapped with the WT intermediate population during folding (Fig. 6Bvs. ESI Fig. S11B,† respectively). This highlights that the tCO in the CU mismatch promotes folding to the native SL4 hairpin, which agrees with the previous observation that native folding outcompetes other folding pathways for US (Fig. 3B). We next compared the fraction of a given state for folding and unfolding (Fig. 6D–F). For WT, the intermediate state was more populated during folding than unfolding, at the expense of the folded state, and, again, the behaviour was very similar for LD (Fig. 6D–F). This could be attributed to SL4 visiting the intermediate state while sampling the conformational space prior to finding its native folded conformation. This yields slower folding kinetics compared to unfolding kinetics. For UC, this difference between folding and unfolding is absent (Fig. 6D–F), which indicates that folding of UC is faster than the corresponding kinetics of WT. As determined also in the analyses presented above, US displays features in between WT and UC, the difference in the population of the intermediate state between folding and unfolding was significantly reduced compared to WT, but not negligible as for UC (Fig. 6D–F). The tCO in the mismatch position in US thus speeds up the folding of the hairpin, but not to the same extent as for UC. UD resembled, to a large extent, WT SL4, although additional features, like broadening of the intermediate population and smaller difference between folding and unfolding for the unfolded state compared to WT, were observed (Fig. 6D–F).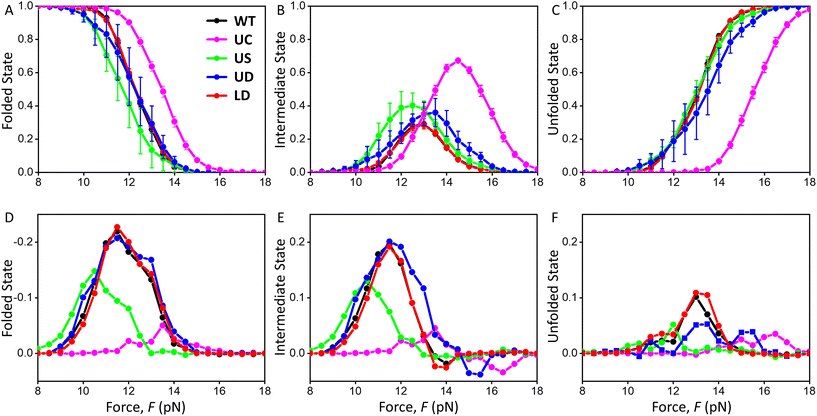 | ||
| Fig. 6 Force-dependent probability of folded (A), intermediate (B), and unfolded (C) states during unfolding for each SARS-CoV-2 SL4 construct. Note the overlap of the WT (in black) and LD (in red) results in (A–C). Difference in the force-dependent probability of folded (D), intermediate (E), and unfolded (F) states between folding and unfolding obtained by subtracting probability values of A–C from ESI Fig. S11A–C,† respectively. Note the flipped y-axis in (D). Colour legend given in (A) also applies to (B)–(F). | ||
Discussion
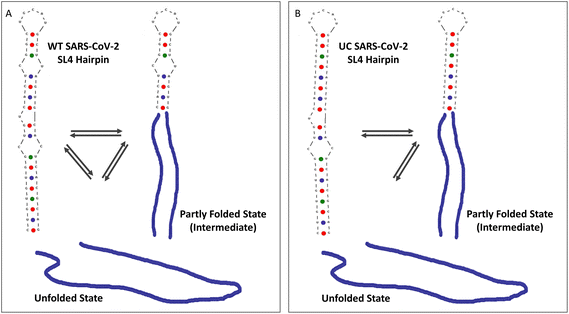
Increasing attention regarding the infection cycle of SARS-CoV-2 is drawn to the intracellular liquid–liquid phase separation of the gRNA driven by the N protein.3,7,41,42 This genome packaging mechanism enables highly specific recognition and selection of the SARS-CoV-2 genetic material from an abundance of viral and cellular RNAs present in infected cells. The SL4 hairpin of the SARS-CoV-2 gRNA 5′-end examined herein has been proposed as one of the N protein-binding sites in the initiation phase of gRNA condensation3,4 and a promising therapeutic target, structurally conserved among SARS-CoV-2 variants and other Betacoronaviruses.10,12 We now discuss how the specific regions within the SL4 structure contribute to its stability and conformational dynamics using RNA BAs in combination with optical tweezers.WT SL4
The experimentally determined structure3,9 and our theoretical prediction of the SARS-CoV-2 SL4 structure is a hairpin containing two mismatches, a bulge, and a 5 nt hairpin loop. Our study shows that both the folding and unfolding of SL4 are highly dynamic. The hairpin explores different pathways and frequently hops between three conformational states: the single-stranded form, an intermediate, and the full-length hairpin (Fig. 7A). Given that SL4 contains the uORF,10 such a conformational dynamic behaviour may facilitate access to its AUG start codon (Fig. 2A, arrow). While most FDCs showed native folding at ∼12 pN in a single or two steps, we also observed folding at much lower force (∼2–8 pN). In addition, a significant portion of the FDCs revealed misfolding that was rescued in the consecutive pull curve. This suggests that the WT SL4 explores various folding pathways, although the energetically most favoured intermediate is the one encompassing approximately half of the molecule, with the middle determined by the U bulge. The experimental findings are well in line with theoretical predictions from other groups, which suggest that SL4 is a bipartite hairpin consisting of two base-paired regions separated by the central U bulge.13,43,44Misfolding of viral hairpins, followed by a rescue mechanism in unfolding, has been previously reported for the HIV transactivation response region (TAR) RNA hairpin and was attributed to its 3 nt bulge (UCU).16 Apart from the misfolding-rescue pathway, the TAR RNA hairpin always unfolded in a single step and without hopping.16 Similarly the human pre-miRNA-377 hairpin from our previous study had only one unfolding pathway via a single step, without hopping, while the folding was heterogenous.15 Altogether, SARS-CoV-2 SL4 has outstanding dynamics with diverse (un)folding pathways and numerous conformational transitions between three states. High dynamics and conformational heterogeneity of the SARS-CoV-2 SL4 hairpin may be needed for gRNA condensation with the N protein upon LLPS and to facilitate access to its uORF starting in the hairpin loop.
LD SL4
The LD SL4 was designed to examine the stability and dynamics of the lower stem by exchanging two Cs with tCOs. LD turned out to have a very similar behaviour to the WT hairpin. Not only were the (un)folding forces the same as for WT, but also there were no significant differences in the dynamics between LD and WT. In our previous optical tweezers study using the tC BA in a fully complementary DNA hairpin, we observed an increase in the hairpin's mechanical stability with tC incorporation.35 tC is chemically similar to tCO (ref. 34) and in UV melting studies, both tCO (ref. 27, 32 and 33) and tC34 have been shown to have, on average, a stabilizing effect on DNA/RNA duplexes. However, for certain nearest neighbours of tCO/tC, no change or a decrease in thermal stability (Tm) of duplexes has been observed,33,34 and not all sixteen nearest neighbours have been investigated.27,32,34 In particular, the Tm has not been examined for the RNA sequence contexts equivalent to the surrounding of the tCO in our SL4 constructs. Therefore, the observation that two tCO incorporations did not affect the stability and dynamics of SL4 is not surprising.UC SL4
The UC SL4 was designed to investigate the effect of the CU mismatch in the upper part of the hairpin (bases at positions 100 and 112). UC always (un)folded in two steps (Fig. 7B) and there were no other folding or unfolding pathways. The fact that the mutation eliminated the non-native (un)folding, strongly suggests that the upper mismatch is sufficient to promote the heterogeneity in the conformational dynamics of SL4. Moreover, transiently paired short fragments of the hairpin (of low extension values) were not observed in UC's (un)folding, highlighting that the upper mismatch contributes to their formation, fostering the spatiotemporal dynamics of SL4 even further. Instead, both transition analysis and force-dependent state probability analysis pointed out that UC SL4 is in a quasi-equilibrium between unfolding and folding at the pulling rate used with faster folding kinetics compared to WT. Finally, the unfolded and intermediate states occurred at a higher force for UC than for WT SL4. This means that the upper mismatch, as expected from our UNAFold prediction, lowers the stability of the hairpin and enhances SL4's dynamics.US SL4
The US SL4 was designed to alter the C100:U112 mismatch, albeit to a lesser extent than in UC, given that the 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 112 base-pair remained mismatched in US. The fraction of misfolding and other non-native pathways was reduced for US compared to WT, making this hairpin more similar to UC. This further confirms that misfolding and other non-native types of SL4 (un)folding can be attributed to the upper mismatch. Moreover, the fraction of two-step transitions increased for US compared to WT, but to a much smaller extent than for UC. Hence, in contrast to LD, replacing C with tCO in the mismatch does affect the dynamics of the hairpin, but to a smaller extent than replacing the mismatch with a complementary CG base pair. The comparison of the population of the different states during folding and unfolding revealed that also the kinetics of US folding was in between that of WT and UC.
112 base-pair remained mismatched in US. The fraction of misfolding and other non-native pathways was reduced for US compared to WT, making this hairpin more similar to UC. This further confirms that misfolding and other non-native types of SL4 (un)folding can be attributed to the upper mismatch. Moreover, the fraction of two-step transitions increased for US compared to WT, but to a much smaller extent than for UC. Hence, in contrast to LD, replacing C with tCO in the mismatch does affect the dynamics of the hairpin, but to a smaller extent than replacing the mismatch with a complementary CG base pair. The comparison of the population of the different states during folding and unfolding revealed that also the kinetics of US folding was in between that of WT and UC.
UD SL4
The rationale behind the design of the UD SL4 was to alter the stability/dynamics of the base-paired regions in the upper stem by a double C-to-tCO exchange. The tCOs in UD are opposing complementary guanines, yet one of them is embedded between a GU wobble base pair and the hairpin loop, which makes the surrounding of the C100:U112 mismatch interesting from a structural stability and dynamics point of view. The UD construct behaved significantly different not only from WT SL4, but also from US and UC. Native (un)folding was significantly reduced and a majority of the FDCs corresponded to non-native events. A unique and frequently explored ‘locked’ pathway of UD, not observed for any other construct, is presumably due to locking of the molecule in a non-native conformation. One possibility is that it corresponds to the thermodynamically second most stable structure of SL4 predicted by UNAFold (ESI Fig. S12B & S13 and section 5†), although other approaches, especially molecular modelling, may provide a greater insight into this matter. Similar locked states and recovery to the native conformation after a single-few folding–unfolding cycles (ESI Fig. S13C and S14†) have been reported for the mechanical (un)folding of the cytochrome C protein.39 Given our previous optical tweezers study on the human pre-miRNA-377,15 which revealed that a large hairpin loop increases folding heterogeneity, we hypothesize that the increased (un)folding heterogeneity of UD compared to WT may be due to the upper tCO affecting the behaviour of the hairpin loop. When it comes to the native folding and unfolding pathways, the stability and dynamics of UD were similar to WT SL4, although with a significantly higher fraction of two-step transitions via the intermediate and broadening of the intermediate population over the examined force range. Overall, the results obtained for UD further emphasize that the complex dynamics of the SARS-CoV-2 SL4 hairpin is due to the upper stem.Base analogues in optical tweezers as a powerful method to study conformational dynamics of nucleic acids
In addition to unveiling important structural and dynamic features of the SARS-CoV-2 SL4 hairpin, our study also demonstrates how BAs can be used to locally vary the base stacking of nucleic acids to investigate their conformational dynamics. In a recent study, we showed that the tC BA can stabilize a fully complementary DNA hairpin.35 Here we demonstrate how BAs can be used in optical tweezers experiments to explore conformational states of RNA structures and transitions between them. Since BAs are similar in size to natural bases and typically designed to (1) maintain specificity for complementary bases; (2) restore hydrogen bonds with their complements; and/or (3) preserve the intrinsic secondary structure of nucleic acids, their incorporation into an RNA/DNA of interest enables studies of a selected region within the molecule with base-pair resolution. Insight into the stability and dynamic properties of individual structural elements within nucleic acids is important for understanding their function, identifying new drug targets, and developing novel therapeutics. Such properties can be distinguished by using optical tweezers together with substituting a natural nucleobase with its close mimic – a BA – in the investigated structural motif. In this way, stability and conformational dynamics of the WT RNA/DNA molecule can be defined and compared with its BA-modified counterpart, expanding the information from the behaviour of the entire molecule to specific regions. A selection of BAs exists27,28,34,45,46 and depending on the choice of a BA as well as its surrounding in the RNA/DNA, the effect of the BA on the stability and conformational transitions of the nucleic acid can be prominent (see US and UD results) or negligible (see LD results), making our new method suitable to a wide range of applications. Our finding of LD being a faithful mimic of WT SL4 is very interesting and useful for future applications as it showcases that depending on the BA and the position of its insertion into the RNA/DNA sequence, the native stability and dynamics of the studied nucleic acid may even be unperturbed.Novel method for synthesis of RNA/DNA constructs for optical tweezers
Our study also provides a technical advancement for optical tweezers. Instead of using the common few kbp-long handles,20–22,47–49 we here used short DNA handles, only 50 bp in length (Fig. 2B and ESI Table S1†). Short handles are advantageous for data analysis, since as an effect of using them the force changes approximately linearly with extension and the signal-to-noise ratio is improved. In comparison to the short DNA handles presented so far in a few DNA folding studies,35,50 our protocol has the advantage that it does not involve ligation in the studied nucleic acid sequence, which is particularly important for stems containing secondary-structure elements, which are common in, for example, pre-miRNAs and viral RNA hairpins. In addition, it involves a purification step (Fig. 2D) which has not been reported before for short handles yet is crucial to obtain data unbiased by interactions between the RNA/DNA of interest and the components of the ligation reaction mixture. Short handles have been used for RNA only in our previous study on pre-miRNA-377, where they were synthesized together with the studied sequence.15 The separate synthesis of handles from the sequence of interest presented herein facilitates material-, time-, and cost-effective preparation of a larger library of constructs.Conclusions
In conclusion, our study employing single-molecule optical tweezers together with base analogue substitutions in the sequence of the SARS-CoV-2 SL4 is the first, thorough study on the stability, (un)folding dynamics, and conformational states of this hairpin. We found that the hairpin dynamics is surprisingly complex with frequent hopping between conformations, as well as folding into non-native structures. Using the tCO base analogue, we demonstrate that a vast majority of this dynamic behaviour is due to the presence of the centrally situated bulge and, in particular, a mismatch in the upper stem of the hairpin. Modifications to the lower stem, on the other hand, do not affect the dynamics. These results are highly relevant to the understanding how viral gRNA motifs control N protein binding in the first steps of virion assembly using a limited pool of proteins. They also aid in the design of drugs targeting SL4 to alter liquid–liquid phase separation, suggesting the upper stem as an attractive drug–target site for impeding SL4's dynamics.Furthermore, our strategy of using base analogues to locally modify the properties of RNA (and DNA) is general and can be expanded to other nucleic acids as well as various base analogues. It can, in combination with our new protocol for synthesizing and purifying optical tweezer's constructs with short handles, be broadly used for studying the structural stability and dynamics of nucleic acids, as well as their interactions with proteins, small-molecule ligands, and other nucleic acids, achieving base-pair resolution.
Materials and methods
Oligonucleotides
The oligonucleotides used in this study (ESI Table S1†) were purchased from Eurogentec, Liège, Belgium (0.2 μmol scale synthesis, RP-HPLC purification for DNA oligos and PAGE purification for DNA–RNA–DNA hybrids, and MS QC). The protocol for the synthesis of constructs and determining the concentration is detailed in ESI section 1†.Optical tweezers measurements
Optical tweezers experiments were carried out in an in-house built instrument equipped with two counter-propagating laser diodes (150 mW; 845 nm), forming an optical trap.15 Data were acquired using force-ramp method at 100 nm s−1 velocity of the optical trap and a frequency of 1.0 kHz. The mechanical force was applied on the anti-digoxigenin coated bead in the optical trap, ramped up until the construct was completely unfolded, and the ramped down to near zero to allow the RNA to refold.To avoid RNase contamination, the custom-made microfluidic chamber,15 together with tubing and syringes, was cleaned in a series of washing steps with RNase-free water, RNase ZAP (Sigma, ref. #: R2020), RNase-free water, ethanol 99.7%, RNase-free water and finally, with the buffer. Polystyrene beads coated with streptavidin were purchased from Kisker Biotech (ref. #: PC-S-2.0; 2.0 to 2.9 mm diameter). Anti-digoxigenin beads were prepared in-house from protein G-coated polystyrene beads from Kisker Biotech (ref. #: PC-PG-3.0; 3.0 to 3.4 mm diameter).15 Prior to measurements, an SL4 construct was incubated with anti-digoxigenin beads for 30 min at room temperature, diluted in 1.0 mL of buffer and injected into the upper channel of the microfluidic chamber. The streptavidin-coated beads were diluted in buffer and injected into the lower channel. The construct was then tethered between the optical trap and the micropipette in the central channel. All measurements were performed in 50 mM Tris-HCl, 140 mM NaCl, 5.0 mM KCl, 1.0 mM EDTA and 5.0 mM MgCl2, pH 7.4, at room temperature (ca. 23 ± 1 °C).
Optical tweezers data analysis
The data were analysed using custom-written MATLAB programs and are presented as the median ± standard error of the median, with the size of the sample, N, being the number of molecules. For each construct, several molecules, and hundreds of force–distance curves (FDCs) were recorded (Table 1). For details regarding the analysis and classification of the FDCs, identification of all and first transitions in FDCs, and force–dependent state probability analysis, see ESI.†Author contributions
Vinoth Sundar Rajan: conceptualization, methodology, investigation, validation, formal analysis, software, visualization, writing – original draft, review & editing. Anna Wypijewska del Nogal: conceptualization, methodology, investigation, validation, formal analysis, software, visualization, writing – original draft, review & editing. Sune Levin: software, visualization. L. Marcus Wilhelmsson: conceptualization, funding acquisition, supervision, writing – review & editing. Fredrik Westerlund: conceptualization, funding acquisition, supervision, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by funding from the Area of Advance Nano at Chalmers University of Technology [to L. M. W. and F. W.]; the Swedish Foundation for Strategic Research [SSF, grant no. IS14-0041 to L. M. W.]; the European Research Council [Consolidator grant no. 866238 to F. W.]; and the Swedish Research Council [VR, grant no. 2020-03400 to F. W.].References
- S. Sood, V. Aggarwal, D. Aggarwal, S. K. Upadhyay, K. Sak, H. S. Tuli, M. Kumar, J. Kumar and S. Talwar, Curr. Pharmacol. Rep., 2020, 6, 212–227 CrossRef CAS PubMed.
- P. S. Masters, Virology, 2019, 537, 198–207 CrossRef CAS PubMed.
- C. Iserman, C. A. Roden, M. A. Boerneke, R. S. G. Sealfon, G. A. McLaughlin, I. Jungreis, E. J. Fritch, Y. J. Hou, J. Ekena, C. A. Weidmann, C. L. Theesfeld, M. Kellis, O. G. Troyanskaya, R. S. Baric, T. P. Sheahan, K. M. Weeks and A. S. Gladfelter, Mol. Cell, 2020, 80, 1078–1091 CrossRef CAS PubMed.
- C. A. Roden, Y. Dai, C. A. Giannetti, I. Seim, M. Lee, R. Sealfon, G. A. McLaughlin, M. A. Boerneke, C. Iserman, S. A. Wey, J. L. Ekena, O. G. Troyanskaya, K. M. Weeks, L. You, A. Chilkoti and A. S. Gladfelter, Nucleic Acids Res., 2022, 50, 8168–8192 CrossRef CAS PubMed.
- M. Morse, J. Sefcikova, I. Rouzina, P. J. Beuning and M. C. Williams, Nucleic Acids Res., 2023, 51, 290–303 CrossRef CAS PubMed.
- S. Lu, Q. Ye, D. Singh, Y. Cao, J. K. Diedrich, J. R. Yates 3rd, E. Villa, D. W. Cleveland and K. D. Corbett, Nat. Commun., 2021, 12, 502 CrossRef CAS PubMed.
- A. Jack, L. S. Ferro, M. J. Trnka, E. Wehri, A. Nadgir, X. Nguyenla, D. Fox, K. Costa, S. Stanley, J. Schaletzky and A. Yildiz, PLoS Biol., 2021, 19, e3001425 CrossRef CAS PubMed.
- C. R. Carlson, J. B. Asfaha, C. M. Ghent, C. J. Howard, N. Hartooni, M. Safari, A. D. Frankel and D. O. Morgan, Mol. Cell, 2020, 80, 1092–1103 CrossRef CAS PubMed.
- A. Wacker, J. E. Weigand, S. R. Akabayov, N. Altincekic, J. K. Bains, E. Banijamali, O. Binas, J. Castillo-Martinez, E. Cetiner, B. Ceylan, L. Y. Chiu, J. Davila-Calderon, K. Dhamotharan, E. Duchardt-Ferner, J. Ferner, L. Frydman, B. Furtig, J. Gallego, J. T. Grun, C. Hacker, C. Haddad, M. Hahnke, M. Hengesbach, F. Hiller, K. F. Hohmann, D. Hymon, V. de Jesus, H. Jonker, H. Keller, B. Knezic, T. Landgraf, F. Lohr, L. Luo, K. R. Mertinkus, C. Muhs, M. Novakovic, A. Oxenfarth, M. Palomino-Schatzlein, K. Petzold, S. A. Peter, D. J. Pyper, N. S. Qureshi, M. Riad, C. Richter, K. Saxena, T. Schamber, T. Scherf, J. Schlagnitweit, A. Schlundt, R. Schnieders, H. Schwalbe, A. Simba-Lahuasi, S. Sreeramulu, E. Stirnal, A. Sudakov, J. N. Tants, B. S. Tolbert, J. Vogele, L. Weiss, J. Wirmer-Bartoschek, M. A. Wirtz Martin, J. Wohnert and H. Zetzsche, Nucleic Acids Res., 2020, 48, 12415–12435 CrossRef CAS PubMed.
- S. P. Ryder, B. R. Morgan, P. Coskun, K. Antkowiak and F. Massi, Evol. Bioinf., 2021, 17 DOI:10.1177/11769343211014167.
- Y. Yao, H. Sun, Y. Chen, L. Tian, D. Huang, C. Liu, Y. Zhou, Y. Wang, Z. Wen, B. Yang, X. Chen and R. Pei, Antiviral Res., 2023, 209, 105478 CrossRef CAS PubMed.
- L. Sun, P. Li, X. Ju, J. Rao, W. Huang, L. Ren, S. Zhang, T. Xiong, K. Xu, X. Zhou, M. Gong, E. Miska, Q. Ding, J. Wang and Q. C. Zhang, Cell, 2021, 184, 1865–1883 CrossRef CAS PubMed.
- R. Rangan, A. M. Watkins, J. Chacon, R. Kretsch, W. Kladwang, I. N. Zheludev, J. Townley, M. Rynge, G. Thain and R. Das, Nucleic Acids Res., 2021, 49, 3092–3108 CrossRef CAS PubMed.
- Z. Miao, A. Tidu, G. Eriani and F. Martin, RNA Biol., 2021, 18, 447–456 CrossRef CAS PubMed.
- A. Wypijewska del Nogal, V. Sundar Rajan, F. Westerlund and L. M. Wilhelmsson, Int. J. Mol. Sci., 2021, 22, 9008 CrossRef CAS PubMed.
- P. T. Li, C. Bustamante and I. Tinoco Jr., Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 7039–7044 CrossRef CAS PubMed.
- D. B. Ritchie, T. R. Cappellano, C. Tittle, N. Rezajooei, L. Rouleau, W. K. A. Sikkema and M. T. Woodside, RNA, 2017, 23, 1376–1384 CrossRef CAS PubMed.
- X. Hu, X. Li, L. Yang, Y. Zhu, Y. Shi, Y. Li, H. Wang and Q. Gong, PLoS One, 2019, 14, e0222938 CrossRef CAS PubMed.
- J. D. Wen, M. Manosas, P. T. Li, S. B. Smith, C. Bustamante, F. Ritort and I. Tinoco Jr., Biophys. J., 2007, 92, 2996–3009 CrossRef CAS PubMed.
- C. V. Bizarro, A. Alemany and F. Ritort, Nucleic Acids Res., 2012, 40, 6922–6935 CrossRef CAS PubMed.
- G. Chen, K. Y. Chang, M. Y. Chou, C. Bustamante and I. Tinoco Jr., Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 12706–12711 CrossRef CAS PubMed.
- L. Yang, Z. Zhong, C. Tong, H. Jia, Y. Liu and G. Chen, J. Am. Chem. Soc., 2018, 140, 8172–8184 CrossRef CAS PubMed.
- Y. J. Wu, C. H. Wu, A. Y. Yeh and J. D. Wen, Nucleic Acids Res., 2014, 42, 4505–4515 CrossRef CAS PubMed.
- K. Neupane, M. Zhao, A. Lyons, S. Munshi, S. M. Ileperuma, D. B. Ritchie, N. Q. Hoffer, A. Narayan and M. T. Woodside, Nat. Commun., 2021, 12, 4749 CrossRef CAS PubMed.
- M. M. Zimmer, A. Kibe, U. Rand, L. Pekarek, L. Ye, S. Buck, R. P. Smyth, L. Cicin-Sain and N. Caliskan, Nat. Commun., 2021, 12, 7193 CrossRef CAS PubMed.
- L. Pekarek, M. M. Zimmer, A. S. Gribling-Burrer, S. Buck, R. Smyth and N. Caliskan, Nucleic Acids Res., 2023, 51, 728–743 CrossRef CAS PubMed.
- A. F. Fuchtbauer, S. Preus, K. Borjesson, S. A. McPhee, D. M. J. Lilley and L. M. Wilhelmsson, Sci. Rep., 2017, 7, 2393 CrossRef PubMed.
- A. Wypijewska del Nogal, A. F. Fuchtbauer, M. Bood, J. R. Nilsson, M. S. Wranne, S. Sarangamath, P. Pfeiffer, V. S. Rajan, A. H. El-Sagheer, A. Dahlen, T. Brown, M. Grotli and L. M. Wilhelmsson, Nucleic Acids Res., 2020, 48, 7640–7652 CrossRef PubMed.
- A. Wypijewska, E. Bojarska, J. Stepinski, M. Jankowska-Anyszka, J. Jemielity, R. E. Davis and E. Darzynkiewicz, FEBS J., 2010, 277, 3003–3013 CrossRef CAS PubMed.
- A. Wypijewska del Nogal, M. D. Surleac, J. Kowalska, M. Lukaszewicz, J. Jemielity, M. Bisaillon, E. Darzynkiewicz, A. L. Milac and E. Bojarska, FEBS J., 2013, 280, 6508–6527 CrossRef CAS PubMed.
- M. Bood, A. Wypijewska del Nogal, J. R. Nilsson, F. Edfeldt, A. Dahlen, M. Lemurell, L. M. Wilhelmsson and M. Grotli, Sci. Rep., 2021, 11, 9396 CrossRef CAS PubMed.
- A. F. Fuchtbauer, M. S. Wranne, M. Bood, E. Weis, P. Pfeiffer, J. R. Nilsson, A. Dahlen, M. Grotli and L. M. Wilhelmsson, Nucleic Acids Res., 2019, 47, 9990–9997 CrossRef CAS PubMed.
- P. Sandin, K. Borjesson, H. Li, J. Martensson, T. Brown, L. M. Wilhelmsson and B. Albinsson, Nucleic Acids Res., 2008, 36, 157–167 CrossRef CAS PubMed.
- K. C. Engman, P. Sandin, S. Osborne, T. Brown, M. Billeter, P. Lincoln, B. Norden, B. Albinsson and L. M. Wilhelmsson, Nucleic Acids Res., 2004, 32, 5087–5095 CrossRef CAS PubMed.
- V. Sundar Rajan, X. Viader-Godoy, Y. L. Lin, U. Dutta, F. Ritort, F. Westerlund and L. M. Wilhelmsson, Phys. Chem. Chem. Phys., 2021, 23, 14151–14155 RSC.
- F. Landuzzi, X. Viader-Godoy, F. Cleri, I. Pastor and F. Ritort, J. Chem. Phys., 2020, 152, 074204 CrossRef CAS PubMed.
- E. Marklund, Y. Ke and W. J. Greenleaf, Nat. Rev. Genet., 2023, 24, 401–414 CrossRef CAS PubMed.
- J. Vogele, D. Hymon, J. Martins, J. Ferner, H. R. A. Jonker, A. E. Hargrove, J. E. Weigand, A. Wacker, H. Schwalbe, J. Wohnert and E. Duchardt-Ferner, Nucleic Acids Res., 2023, 51, 11318–11331 CrossRef PubMed.
- J. Li and H. Li, Chem. Sci., 2022, 13, 7498–7508 RSC.
- M. Manosas, D. Collin and F. Ritort, Phys. Rev. Lett., 2006, 96, 218301 CrossRef CAS PubMed.
- J. Svetlova, E. Knizhnik, V. Manuvera, V. Severov, D. Shirokov, E. Grafskaia, P. Bobrovsky, E. Matyugina, A. Khandazhinskaya, L. Kozlovskaya, N. Miropolskaya, A. Aralov, Y. Khodarovich, V. Tsvetkov, S. Kochetkov, V. Lazarev and A. Varizhuk, Int. J. Mol. Sci., 2022, 23, 15281 CrossRef CAS PubMed.
- S. Wang, T. Dai, Z. Qin, T. Pan, F. Chu, L. Lou, L. Zhang, B. Yang, H. Huang, H. Lu and F. Zhou, Nat. Cell Biol., 2021, 23, 718–732 CrossRef CAS PubMed.
- R. Rangan, I. N. Zheludev, R. J. Hagey, E. A. Pham, H. K. Wayment-Steele, J. S. Glenn and R. Das, RNA, 2020, 26, 937–959 CrossRef CAS PubMed.
- I. Manfredonia and D. Incarnato, Biochem. Soc. Trans., 2021, 49, 341–352 CrossRef CAS PubMed.
- R. W. Sinkeldam, N. J. Greco and Y. Tor, Chem. Rev., 2010, 110, 2579–2619 CrossRef CAS PubMed.
- D. Dziuba, P. Didier, S. Ciaco, A. Barth, C. A. M. Seidel and Y. Mely, Chem. Soc. Rev., 2021, 50, 7062–7107 RSC.
- J. Liphardt, B. Onoa, S. B. Smith, I. Tinoco Jr. and C. Bustamante, Science, 2001, 292, 733–737 CrossRef CAS PubMed.
- G. Chen, J. D. Wen and I. Tinoco Jr., RNA, 2007, 13, 2175–2188 CrossRef CAS PubMed.
- P. T. Li, D. Collin, S. B. Smith, C. Bustamante and I. Tinoco Jr., Biophys. J., 2006, 90, 250–260 CrossRef CAS PubMed.
- N. Forns, S. de Lorenzo, M. Manosas, K. Hayashi, J. M. Huguet and F. Ritort, Biophys. J., 2011, 100, 1765–1774 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nr04110g |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |