Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Comparing total chemical synthesis and total biosynthesis routes to fungal specialized metabolites
Dong-Song
Tian
 a,
Xiao
Zhang
a,
Xiao
Zhang
 *a and
Russell J.
Cox
*a and
Russell J.
Cox
 *b
*b
aCollege of Pharmaceutical Sciences, Southwest University, 400715 Chongqing, China. E-mail: zhangxiao023@swu.edu.cn; dstian@swu.edu.cn
bInstitute for Organic Chemistry, Leibniz University of Hannover, Schneiderberg 38, 30167 Hannover, Germany. E-mail: russell.cox@oci.uni-hannover.de
First published on 15th August 2024
Abstract
Covering the period 1965–2024
Total synthesis has been defined as the art and science of making the molecules of living Nature in the laboratory, and by extension, their analogues. At the extremes, specialised metabolites can be created by total chemical synthesis or by total biosynthesis. In this review we explore the advantages and disadvantages of these two approaches using quantitative methodology that combines measures of molecular complexity, molecular weight and fraction of sp3 centres for bioactive fungal metabolites. Total biosynthesis usually involves fewer chemical steps and those steps move more directly to the target than comparable total chemical synthesis. However, total biosynthesis currently lacks the flexibility of chemical synthesis and the ability to easily diversify synthetic routes.
1. Introduction
The production and use of specialized metabolites (SM) has long been important to human society and their value in healthcare, agriculture and other fields is undeniable. At the present time there are three main ways by which such compounds may be obtained. First, they are obtained directly from producing organisms; secondly they may be produced by total chemical synthesis;1 and third, they may be produced by total biosynthesis.2 Combinations of these strategies may also be effective. For example the production of paclitaxel3 and artemisinin4 is achieved by a combination of biosynthesis and chemical synthesis (i.e. semi-synthesis).5Each production method comes with its own advantages and disadvantages. For example, production of many SMs by plants allows the methods of horticulture or agriculture to be deployed, for example in the cases of morphine6 and the cannabinoids.7 However, harvest of wild organisms may be ecologically damaging, as in the case of the pacific yew that produces paclitaxel in low amounts. In terms of domesticated plant production of SMs, variable weather, and pressures on land and water resources can mean that agricultural production competes with food production. Additionally, many compounds obtained directly from host organisms may have imperfect biological properties and require further chemical modification. In the case of microorganisms, production by fermentation can often be optimal, such as the production of beta-lactams in 100 g L−1 titres.8 However, many organisms produce SMs much less well.
In contrast, total chemical synthesis is highly flexible and almost any desired compound can be produced, especially the structurally modified analogues of SMs and natural metabolites that are toxic to the host organisms. Until now, chemical synthesis as a predominate tactic has contributed tremendously to life-saving medicine discovery as well as fundamental chemical manufacturing. But chemical synthesis routes very often feature prohibitively high step counts, and they are highly carbon intensive, especially for structurally complex SMs with fused polycyclic skeletons and multiple stereocenters. The urgent requirement to significantly curtail carbon emissions places additional pressures on the use of total chemical synthesis for SM production.9,10
Total biosynthesis routes can be inherently energy- and carbon-efficient because biological production normally involves a single process, followed by extraction and purification. Biosynthetic routes can be used for production of SMs where the pathways are well understood and where suitable host organisms are available and easily manipulated, but these features are not always available. Furthermore, routes to known SMs can be inflexible, and it is currently difficult to expand the footprint of biosynthetic pathways to encompass the synthesis of congeners that don't exist in nature.
It would therefore be useful to find a systematic way in which chemical and biological production of SMs could be directly compared. Recent advances in informatic methodology have focussed on determining measures of the complexity of individual molecules. Such measures can include the molecular weight of metabolites (MW), the fraction of sp3 hybridised carbon atoms (Fsp3)11,12 and the so-called complexity index (Cm).13,14 No single measure is perfect, but the combination of all three of these measures can be useful for capturing the complexity of individual compounds, or for observing how complexity changes during a synthetic pathway.15 The rapidity by which complexity is gained stands as a proxy of pathway efficiency: efficient pathways should create complex SMs in as few processes as possible. In particular, 3D plots of complexity values allow visualization and comparison of different synthetic routes in a chemical space in which distances represent chemical changes in complexity, molecular weight and hybridisation. This allows a direct comparison of different biological and chemical synthetic strategies.
Fungi produce a very broad range of complex and bioactive SMs, and there is a very long history of biosynthetic16 and chemical synthetic17 investigations of these compounds. Fungi produce all the major classes of SMs including varied fatty acids and polyketides, peptides, terpenes, alkaloids, and compounds of mixed origin such as meroterpenoids.18 Much is now known of their biosynthesis, and recent rapid developments in molecular and genetic methods in fungi mean that many more biosynthetic routes have been fully determined. Fungal SMs thus present a rich group of compounds where comparisons between total chemical and total biosynthetic19 methods can be made directly. In this review we attempt to apply these ideas to a variety of total chemical synthesis and total biosynthesis routes to fungal SMs. For simplicity of discussion we select a range of compounds that reflect the different biosynthetic strategies by which fungi assemble SMs.
1.1 Molecular complexity calculations
Descriptors Fsp3, Cm (mcbit) and MW (Da) can be used to define molecular complexity. In the following sections we calculated and measured these values for the intermediates of both biosynthetic and chemical synthetic pathways. During biosynthesis, CoA thiolesters (and sometimes carrier proteins) are often precursors and intermediates. However, since coenzyme A and carrier proteins are always regenerated and reused in biological systems, we did not include these moieties in molecular complexity calculations. Likewise, in the synthetic routes, we did not include calculations of the complexity changes in reagents and catalysts. So here we use methyl-thiolesters instead of CoA or ACP thiolesters for molecular calculations. However, since protecting groups are essential to most of the chemical routes, we do include these in the calculations. 3D plots parameterized by Fsp3, Cm and MW were drawn using Matplotlib,20 a visualization tool based on the python environment. In all cases discussed below the blue colour indicates the biosynthetic route, while orange represents the chemical synthesis.2. Fatty acid: sporothriolide
Sporothriolide 1 is the major SM produced by the fungi Hypomontagnella monticulosa, H. spongiphila and H. submonticulosa. It possesses a unique alkyl furofurandione architecture and exhibits potent and specific antifungal activity, (e.g. 500 ppm protection against Botrytis cinerea infection of pepper seedlings),21 and it has therefore attracted investigations of its biosynthesis and total synthesis.The genomes of three sporothriolide producers were sequenced and these revealed genes encoding: two fungal fatty acid synthase components (SpofasA & B); an alkyl citrate synthase (SpoE); a methylcitrate dehydratase homolog (SpoL); a decarboxylase (SpoK); a non-heme iron dioxygenase (SpoG); and hydrolases SpoH and SpoJ. The biosynthetic pathway was fully reconstructed in Aspergillus oryzae to produce 1 (Scheme 1A).22 Decanoyl-CoA 2 is synthesised by SpofasA and SpofasB from acetyl- and malonyl-CoA. SpoE (alkyl citrate synthase) then catalyses condensation with oxaloacetate to form alkyl citrate 3. Notably two chiral centres are constructed at this stage by the citrate synthase.
However, the precise stereochemical course of SpoE is still elusive, since presumed intermediate 3 was not observed in the WT host or via heterologous expression in A. oryzae. Dehydration of tertiary alcohol 3 then leads to the formation of alkene 4. Next, decarboxylation catalysed by SpoK gives the alkyl itaconic acid 5. Thereafter, alpha-ketoglutarate dependent oxygenase SpoG mediates two rounds of hydroxylation of the saturated alkyl chain to give 7. The mono- and di-oxygenised itaconic acids are then spontaneously cyclized to lactones 8 and 1. However, in the presence of lactonases SpoH and SpoJ, conversion of 7 to 1 was observed in vivo.
Total synthesis of sporothriolide has been achieved by Kimura and co-workers.23 The route started from a mixed anhydride of 9 reacting with lithium oxazolidinone salt 18 to give N-acyl oxazolidinone 10. Then Michael addition to nitroalkene 19 gave 11 as a single diastereomer in 73% yield. Subsequent Sharpless asymmetric dihydroxylation of 11 allowed spontaneous lactonization (5-exo-trig) and loss of the chiral auxiliary resulting in 12 (75%). The hydroxy group was then protected as a TES ether 13, prior to ruthenium tetroxide oxidation to carboxylic acid 14. This was readily converted to bis-lactone 15 after treatment with aqueous hydrochloric acid. Finally, elimination of HNO2 led to formation of sporothriolide 1 in 71% yield. Summarily, a seven-step enantioselective total synthesis accomplished the construction of sporothriolide 1 in 21% overall yield.
Construction of a 3D plot that represents the pathways in a chemical space shows that they start at similar positions (Fig. 1A). We also calculated the linear distance between each intermediate of the synthesis in the chemical space, in other words, the “chemical distance” of each step using the equation:
Plotting the results vs. step number (e.g.Fig. 1B) shows that the chemical route usually involves longer steps in the chemical space.
Finally, we also measured the distance of each intermediate to the final target and again plotted this vs. step (e.g.Fig. 1C). In the case of sporothriolide it is clear that while the two pathways to the target both share 7 steps, the chemical synthesis pathway is considerably longer in terms of distance in the complexity space. This is most easily visualised when considering the distance of each intermediate from the target. The biosynthetic pathway reaches the target in steps that usually move each intermediate closer to the target. However, in the synthetic pathway the steps from 10 to 11 and from 12 to 13 both dramatically move the intermediates further away from the target. The step from 10 to 11 involves addition of the para-methoxy phenyl unit where all carbons bar one are later removed, while the step from 12 to 13 involves addition of a TES protecting group that is again later removed.
Thus the journey through the 3D chemical complexity space reveals synthetic steps that are atom inefficient because they move the intermediates further away from the target. It is noticeable that the biosynthetic pathway only features two steps where the distance from the target increases: intermediates 3 to 4 to 5. Once again superfluous atoms are shed during this process, but the intermediates do not move significantly away from the final target.
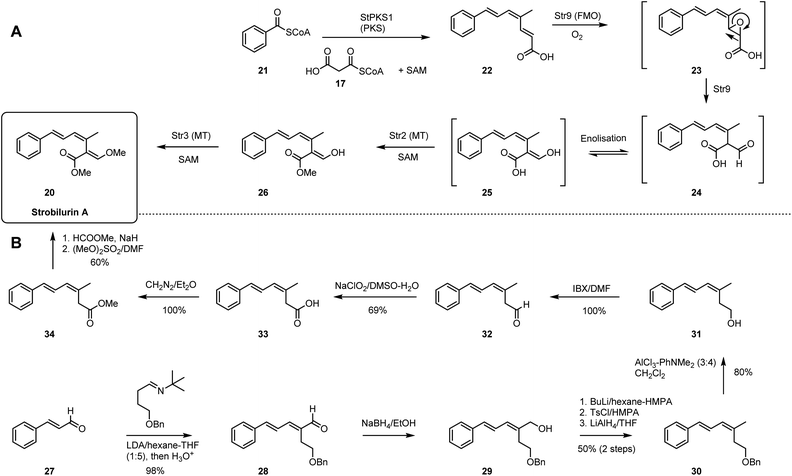
3. Highly-reduced polyketide: strobilurin A
Strobilurins are antifungal agents isolated from basidiomycetes. The unique β-methoxyacrylate moiety is the toxophore and inspiration for the development of the QoI fungicides widely used in crop protection.24,25 Syntheses of strobilurins have been achieved by both total chemical synthesis26,27 as well as by total biosynthesis.28In the biosynthetic pathway (Scheme 2A), benzoyl CoA 21 serves as the precursor to synthesize prestrobilurin 22. An unusual highly-reducing PKS (StPKS1), with hydrolase domain and unique C-terminal methyltransferase domain, is responsible for the assembly of EZE triene 22 using malonyl CoA 17 and S-adenosyl methionine (SAM). Then, a flavin-dependent monooxygenase (FMO) Str9 catalyses the oxidative rearrangement of olefin 22 to give aldehyde 24, presumably via intermediate epoxide 23. Rapid enolization to 25 is followed by two SAM-mediated methylations, firstly by Str2 acting at the carboxyl group, followed by Str3 acting at the enol to obtain strobilurin A 20.29
The chemical synthesis of strobilurin A started from condensation of cinnamaldehyde 27 and benzyloxybutanimine to afford dienal 28 (Scheme 2B). Reduction with NaBH4 in ethanol gave dienol 29, and subsequent O-tosylation and reduction gave 30. Removal of the benzyl protection then gave 31. Oxidation with IBX and Pinnick oxidation gave carboxylic acid 33, which was further methylated to ester 34. Finally, treatment with base and methyl formate followed by dimethyl sulfate gave strobilurin A 20.
In comparison the two routes differ significantly in step-count. The biosynthetic route is particularly short, requiring only a PKS, an oxidative rearrangement and two methylations. In contrast, the chemical synthesis requires eight main steps, although only two of these are carbon–carbon bond-forming steps.
3D Plot of the complexity data shows that the chemical and biological pathways differ significantly. Although the starting points of 21 and 27 are far apart, they are roughly equidistant from the target (Fig. 2C). In the biosynthetic route there are two main processes. First, creation of the polyketide 22 and its oxidative rearrangement move the pathway decisively towards the final Cm value. Then the methylations via26 increases only the molecular weight and Fsp3. Each step only moves the route closer to the target (Fig. 2C). In contrast, the total synthesis route first builds 28 that is close to 20 in all dimensions. But the subsequent seven steps then dramatically move around the complexity space. For example, while intermediate 30 is still close to the final product, step-4 (30 to 31) moves Cm by 90 mcbits away from the target and it takes another four steps to recover. This is clearly a feature of the requirement for a protecting group strategy and redox manipulation in the chemical synthesis route.
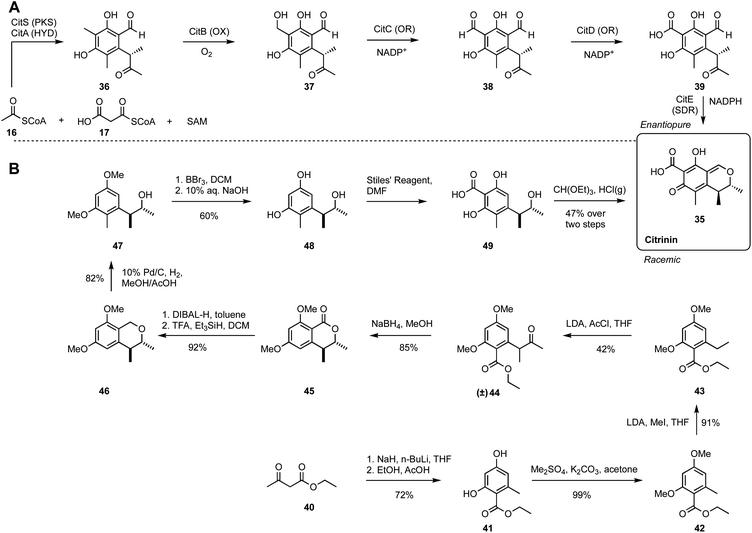
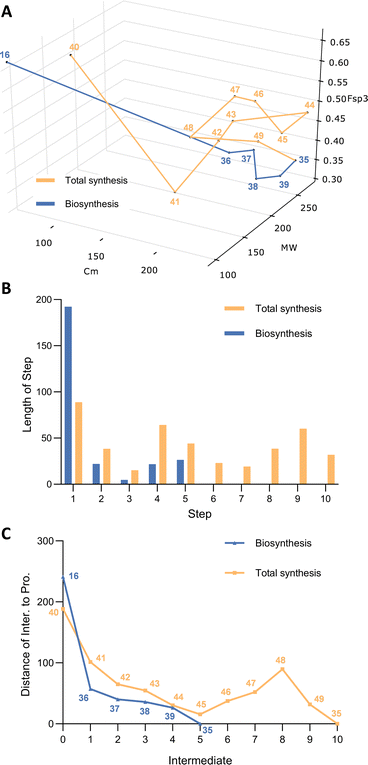
4. Non-reduced polyketide: citrinin
Citrinin 35 was first discovered in the 1930s and is a widely reported mycotoxin causing food contamination and other systemic toxic effects.30 Gene knockout and heterologous expression strategies have fully elucidated the citrinin biosynthetic pathway (Scheme 3A).31,32 CitS, a non-reducing iterative PKS is responsible for the construction of a trimethylated pentaketide aldehyde 36 from acetyl CoA, malonyl CoA and SAM. Then a non-heme iron oxidase CitB catalyses the oxidation of the methyl of 36 to an alcohol 37, followed by two sequential oxidations catalysed by oxidoreductases CitC and CitD, to produce aldehyde 38 and carboxylic acid 39, respectively. The final conversion is performed by CitE, that stereoselectively reduces the 9-ketone and thus facilitates the spontaneous formation of the quinomethide architecture of citrinin 35.The chemical synthesis of racemic citrinin was reported by Humpf and co-workers.33 It commenced with a 2-step preparation of ethyl orsellinate 41 from ethyl acetoacetate 40. After methylation of the hydroxyl groups to 42, a C-methylation is achieved with methyl iodide under base conditions, to give the ethyl congener 43. Acetyl chloride was then used to form ketone 44, and borohydride reduction then allowed spontaneous lactonization to 45. There then followed a series of reductive steps and deprotections to reach the key intermediate 48. The stage is now set for the addition of the final two carbons, one by one, to first carboxylate and then formylate the aromatic ring, followed by final oxa-Pictet–Spengler reaction to form citrinin 35.
Once again, 3D plot of the two pathways reveals interesting differences. The biosynthetic route is short, in terms of step-count with five steps to reach the destination (Fig. 3A). The PKS takes the decisive step in producing 36, but all steps of the pathway move closer to the target. The chemical synthesis pathway is longer in terms of step count despite the fact that the starting point is close to the biosynthetic starting point. Interestingly the first five synthetic steps to intermediate 45 move the pathway monotonically towards the target. But the next three steps to 46, 47 and 48 all move the intermediates away from the target in the complexity space. Especially significant is the requirement for a late-stage introduction of two single carbon atoms to 48 that finally brings the synthetic path back on track to the target. It then takes the final two steps of the chemical synthesis to reach the target 35.
The key differences between the two routes here are that the biosynthetic route installs all the required carbon atoms at the start of the synthesis. This then only leaves redox manipulations and spontaneous cyclisations as requirements to reach the target. In contrast, the synthetic route builds up the carbon-count stepwise. Although additions of carbons in the synthetic route are usually associated with closer approaches to the target, the three reductive steps and the deprotections in the middle of the route required to reach key intermediate 48 contribute to the increased step count overall, while moving the intermediates further away from the target 35.
5. Dual PKS system: squalestatin S1
Squalestatins,34 also known as zaragozic acids,35 are highly oxidized polyketide derivatives which were discovered as lead compounds for the treatment of hypercholesterolemia in 1990s. Extensive investigations of bioactivities suggested that squalestatins could reduce cholesterol production through the inhibition of squalene synthase. Among them, squalestatin S1 50, exhibits nanomolar inhibition of mammalian and fungal squalene synthase, and accordingly displays antifungal properties.36The highly complex caged 4,8-dioxa-bicyclo[3.2.1]octane architecture has attracted constant chemical synthetic investigations.37,38 Meanwhile, biosynthetic studies have revealed the complete pathway.39–42 Early isotope labelling experiments revealed the highly reduced polyketide origin of squalestatins, as well as atmosphere and acetate-derived oxygens,43 while knockout and heterologous expression studies41,42 revealed the order of the steps and the roles of the biosynthetic enzymes.
Genomic and bioinformatic analysis led to the identification of the squalestatin S1 gene cluster that encodes two highly reducing polyketide synthases (hrPKS) and several oxygenases. Benzoyl CoA 21, derived by degradation of phenylalanine, is the starter unit for assembly of intermediate 51 by hexaketide synthase SQHKS. Co-expression with a hydrolase (Mfm8) and the citrate synthase (Mfr3) produces the hexaketide citrate 52. Further introduction of NHI (non-heme iron) oxygenase Mfr1, resulted in the detection of a panel of congeners from LCMS chromatograms, which were consistent with alcohol, ketone, unsaturated ketone and epoxide intermediates on the route to 53. Since these are derived from the stepwise oxidations of 52 by Mfr1 alone, we refer to the transformations induced by Mfr1 as ‘step-2’. Mfr2 then catalyses oxygenations on the oxaloacetate moiety, in which 54 is formed after two rounds of hydroxylation of 53. A subsequent likely Payne rearrangement to 55, precedes epoxide opening and cyclisation to yield the key bicyclo unit 56. We refer to the global conversion from 53 to 56 mediated by Mfr2 as ‘step-3’. Next, a copper-dependent oxygenase Mfm1 catalyses allylic alcohol 58 production through rearrangement of 57, an epoxidized form of 56. Acetylation of 58 gives 59. The final step is the O-acylation of 59 to make squalestatin S1 50.
Assays conducted in vitro40 showed that acyl transferase Mfm4 selectively transfers tetraketide CoA thiolester 60 to the 6-hydroxyl of 59. Mfm4 has a broad substrate scope, catalysing transfer of acyl CoAs from 2 to 10 carbons. Squalestatin tetraketide synthase (SQTKS) is responsible for the construction of the tetraketide moiety in squalestatin S1 biosynthesis based on directed gene knockout and heterologous expression experiments.39
The total chemical synthesis of squalestatin S1 50 has been achieved by Heathcock and co-workers in two overall processes. First, the SM 50 itself was degraded to the relay compound 62 and this was used to develop an 11-step pathway back to 50.44 The relay compound 62 was itself synthesised in 25 steps from an advanced precursor 61,45 giving a formal total chemical synthesis of 36 steps.
The synthesis of 62 started with 1,6-anhydropyranohexose 61 through a sequence that led to the relay compound 62 that contains the 4,8-dioxabicyclo[3.2.1]octane core (Scheme 4B). Treatment of 62 with pyridinium p-toluenesulfonate afforded cyclic methyl acetal 63 as a mixture of diastereomers. The free hydroxyl of 63 was acylated with α,β-unsaturated acid to yield 64, while unsaturated acid 73 was supplied from a multiple-step conversion of the starting material tert-butyl acetate. Acetal 64 was hydrolysed to give hydroxy aldehyde 65, then treated with triethylsilyl chloride to make 66. Addition of aldehyde 66 with stannane achieved the side chain installation, and the newly formed alcohol 67 was then oxidized by Dess–Martin reagent to yield ketone 68. The trial in alkene 69 production by employing Wittig reaction resulted in only low yield, however by using Tebbe reaction, a satisfactory synthesis of 69 was achieved. Deprotection and acetylation were conducted smoothly to obtain advanced intermediate 71. Finally, two-step removal of the protection groups led to a production of the target squalestatin S1 50.
In order to able to sensibly compare the pathways we condensed the synthesis of 62 into a single process. While these steps in combination do move the synthesis closer to the target in the chemical structure space (Fig. 6A), it should be remembered that the overall yield for this process is a remarkably good 4.7%.
Even starting from the late relay compound 62, the chemical synthesis of squalestatin S1 50 requires many more transformations than the biosynthesis (Fig. 4A and B). Once again, the biosynthetic pathway is fairly linear through the chemical space, and each of the seven transformations pushes the intermediates closer to the target. In fact, distance to target decreases almost linearly in the biosynthetic synthesis, whereas the chemical synthesis meanders (Fig. 4C). In particular, chemical steps from 64 to 67 increase mass and complexity through the addition of protecting groups that are later removed in three further steps. These six protection/deprotection steps are required to allow three fairly trivial transformations of the skeleton: oxidation of a secondary alcohol to a ketone and its conversion to the methylene (67 to 69); and O-acetylation (70 to 71) that are the only contributions to the required structure of 50. The very high efficiency of the biosynthetic pathway is achieved because all of the main skeleton carbons are built-in in the first step, and because of the very high inherent selectivity of the key tailoring functionalisations that include multiple oxygenations, cyclisation and regioselective acylations that do not require any protecting steps.
6. Dimeric polyketide: bisorbicillinol
Bisorbicillinoids are biologically active compounds with diverse structures that are isolated from species of Trichoderma and other ascomycetes. They possess numerous interesting bioactivities and their fascinatingly complex skeletons have always been attractive synthetic targets. Extensive biosynthetic investigations have completely unveiled the key construction and dimerization process during the biosynthesis of bisorbicillinol 73.46–48 The bisorbicillinoid BGC encodes: a highly reducing polyketide synthase SorA; a non-reducing polyketide synthase SorB; two FAD-dependent oxygenases SorC and SorD; as well as transcription factors and transporters. Heterologous expression of functional genes in Aspergillus oryzae and bipartite knockout of target genes in Trichoderma reesei led to a full understanding of the roles of these enzymes during the biosynthesis.The first biosynthetic step involves the collaboration of SorA and SorB to assemble the polyketide backbone (Scheme 5). Sorbicillins are often found as mixtures of 2′–3′-saturated and unsaturated forms in the linear side-chains. This is caused by a partially active enoyl reductase (ER) domain of SorA that produces the triketide starter unit 74 for SorB. SorA appears not to be able to fully control the activity of its ER, resulting in mixed products. However, downstream enzymes seem insensitive to this heterogeneity. SorB extends the triketide starter unit 74 three times, methylating the growing chain twice. Finally, reductive release from the PKS gives an aldehyde 75 that undergoes intramolecular Knoevenagel condensation to form the aromatic phenol sorbicillin 76, that is a critical monomer for the biosynthesis of diverse compounds in this family.
SorC then performs the oxidative dearomatisation of sorbicillin 76 to afford sorbicillinol 77. The reactive intermediate 77 is both a diene and a dienophile, and it can undergo intermolecular Diels–Alder (DA) cycloaddition to yield the remarkably complex bisorbicillinol 73. Sorbicillinol 77 is highly reactive, and it can undergo both spontaneous dimerisation (especially during extraction and workup procedures) and enzyme-catalysed Diels–Alder cyclisation, for example by the FAD-dependent SorD. The selectivities of these reactions appears to be affected by solvents in many cases.
Sorbicillin 76 is formed in essentially a single process from acetyl and malonyl CoA and SAM, and thus the journey through chemical space is roughly linear, with each enzyme-bound intermediates such as 74 already on a direct path to the target. The final dimerization covers more than 50% of the total journey to 73. Clearly, bisorbicillinol is biosynthesized by nature with extremely high efficiency.
Total chemical synthesis of bisorbicillinol 73 has been achieved by several research groups, for example Nicolaou49 and Corey et al. have reported a 2-step synthesis of the related trichodimerol,50 but usually as a racemic mixture in low yield. However, Deng et al. have reported a highly enantioselective chemical synthesis route to bisorbicillinol 73 using a cyanosilylation as the stereochemistry-defining step, with an overall yield of 12–19%.51 In this synthesis, acetal ketone 81 was first converted into enantiopure cyanohydrin 82 catalysed by a modified cinchona alkaloid. Next, condensation of nitrile 82 with EtMgBr led to a smooth production of ketone 83. Then a 1-carbon Wittig reaction to afford the corresponding methylene, followed by PMB protection of tertiary alcohol using PMBOC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NH)CCl3 gave PMB-protected ether 84. The acetal of 84 was hydrolysed under acidic conditions, resulting in aldehyde 85, which was subjected to Knoevenagel condensation with a readily accessible ynone ester, to yield 86 with high Z selectivity.
NH)CCl3 gave PMB-protected ether 84. The acetal of 84 was hydrolysed under acidic conditions, resulting in aldehyde 85, which was subjected to Knoevenagel condensation with a readily accessible ynone ester, to yield 86 with high Z selectivity.
Following ozonolysis of the methylene to 87, the C5 side chain was isomerized to dienone 88. After numerous trails, Ph3COK was elected as the suitable base for the enolate generation to prompt the intramolecular Claisen–Vorländer cyclisation to PMB-protected sorbicillinol 89. The final step involves the removal of PMB ether to sorbicillinol 77 with trifluoroacetic acid, and then spontaneous Diels–Alder cycloaddition to the target bisorbicillinol 73 in one pot.
Plotting the biosynthetic and chemical synthetic pathways in 3D gives some insight into the key points of similarity and difference. Once again the biosynthetic pathway reveals its efficiency by its monotonic approach to the target – each intermediate is closer to the target in the chemical space than the last. The chemical route tends to be more meandering and longer in terms of step count, and in particular intermediates 86–88 do not significantly advance the synthetic pathway, reflecting the requirement to remove a single carbon to reveal a ketone and catalyse an isomerisation. However despite these steps not ‘advancing’ the synthesis, the overall yield is very high. Thus despite the need for several protective and deprotective steps the overall synthesis is fairly efficient.
Recently, Gulder's group has taken advantages of the highly active enzyme SorC to significantly shorten the chemical synthesis route.52 SorC offers dual advantages: highly regio-selective oxidative dearomatisation requires no protecting groups; and very high stereoselectivity means that achiral (and thus easier to make) intermediates are required. Here, simple formylation, LiAlH4 reduction and Friedel–Crafts acylation afford sorbicillin 76 in 3 steps. SorC then catalyses the final oxidation, while spontaneous dimerisation then leads to the target 73. This route closely follows the biosynthesis and is highly efficient. However, it offers additional advantages because SorC activates various differently-functionalised substrates, in-turn allowing the synthesis of varied analogs of 73 that would be difficult to reach by the biosynthetic route alone. SorC has also been exploited in the synthesis of the non-symmetrical sorbicillactone, again in a remarkably short and efficient process (Fig. 5).53
7. PKS-NRPS: tenellin
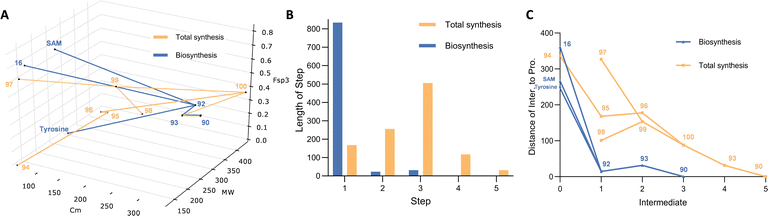
Tenellin 90 is a 2-pyridone with sub-micromolar cytotoxicity vs. human cancer cell lines.54 It belongs to a large family of fungal SMs that derive from the condensation of an amino acid with a polyketide. Related 2-pyridones such as ilicicolin H possess antifungal activity,55 and may be responsible for the function of natural biocontrol agents.56 The 2-pyridones are constructed by oxidative ring-expansion of the corresponding acyl tetramic acids57 that are also very commonly produced by fungi. This family also includes potent bioactive compounds such as the cytochalasans58 and mycotoxins such as the fusarins.59,60 The widespread occurrence, and often potent bioactivity of these compounds has therefore led to numerous total syntheses and detailed investigations of their biosynthesis.Convergent total synthesis of tenellin was achieved by Rigby and Qabar (Scheme 6).61 The strategy involves the combination of a vinyl isocyanate 96 with a β-keto ester enolate 99 to yield the tenellin skeleton of pyridone 100. In order to avoid the side reactions and to increase production yields, removal of MEM protection (2-methoxyethoxymethyl) of 100 to make pretenellin-B 93 was performed first, followed by N-hydroxylation to provide the expected racemic tenellin 90. The requisite vinyl isocyanate 96 was routinely produced from the commercially available 4-hydroxybenzaldehyde 94 in two steps via an intermediate carboxylic acid 95. In parallel, diene keto ester 99 resulted from the Horner–Wittig condensation of hexenal 97 with phosphine oxide 98 in a good yield.
Biosynthesis of tenellin is also a remarkably short process. A hybrid polyketide synthase nonribosomal peptide synthetase (PKS-NRPS), TenS, constructs the tenellin backbone. The PKS is a highly efficient system that builds the required pentaketide from acetate 16, malonate 17 and SAM using a single iterative module.
Programming of this system has been extensively studied,62 and effective execution of the programme requires the presence of a trans-acting enoyl reductase known as TenC. The completed pentaketide constructed by TenS/TenC is then passed to the NRPS where the condensation (C) domain catalyses the condensation of the ACP tethered pentaketide with thiolation (T) domain bound tyrosine thiolester to afford the ketoacyl amino thiolester 91. Prior to the condensation process, the tyrosine moiety is initially selected and activated by the adenylation (A) domain, followed by transfer to the T-domain phosphopantetheine arm. A Dieckmann cyclisation is then catalysed by the C-terminal DKC domain to release the enzyme linked 91 from PKS-NRPS system, which generates the first isolatable intermediate pretenellin-A 92 with a typical tetramic acid core. The subsequent oxidative ring expansion from 92 to 93 is carried out by P450 monooxygenase TenA. Finally, a second cytochrome P450 oxygenase, TenB, catalyses the required N-hydroxylation to give tenellin 90.
On the face of it, the two routes to tenellin are rather similar. Both routes approach the target almost monotonically (Fig. 6C) and the chemical synthesis route is only 2 steps longer than the biosynthesis. The chemical synthesis route benefits from the convergent creation of the 2-pyridone, that requires only deprotection and N-hydroxylation to reach the target. However, the preceding steps to the pyridone nucleus are rather flattering, appearing to give very quick access to the required precursors. It should be remembered, however, that both 97 and 98 must themselves be made by multi-step routes, and the overall 5-step sequence is therefore longer in reality.
The biosynthetic route is also extremely efficient. The initial PKS-NRPS TENS/TENC takes advantage of an iterative highly programmed PKS to effectively reach late-stage intermediate 92 in a single step. And, as found in other pathways, the very high selectivity of the late-stage oxidations obviates the need for protecting and redox steps. The TENS/TENC system has also formed the basis for systematic investigations into the programming mechanisms and it has proven possible to control both the chain-length and methylation pattern of the polyketide component.63–65 Thus, while the biosynthetic pathway already offers very high efficiency, it can also be reengineered to make related compounds such as bassianin.63
8. Alkaloid: communesin F
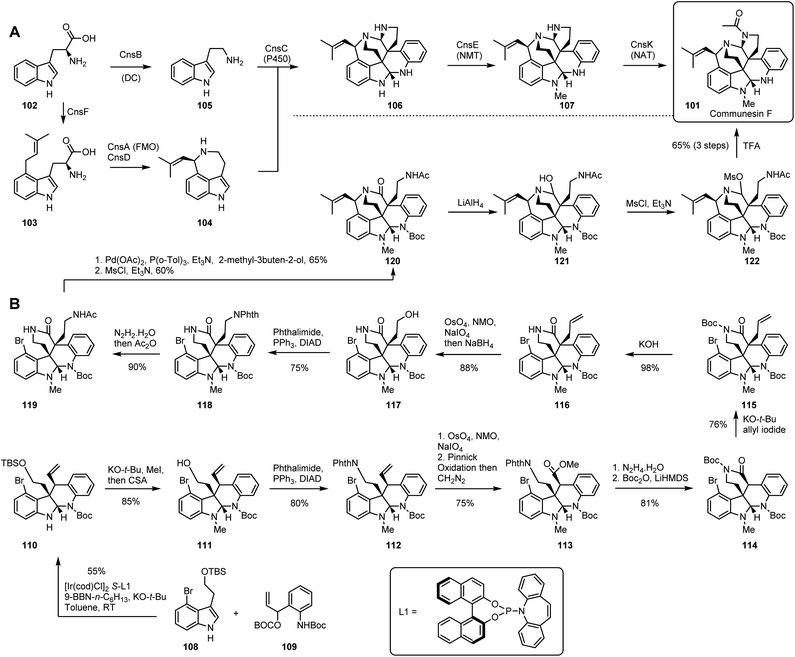
The communesins are a family of indole alkaloids isolated from Penicillium species.66 Structurally, communesins possess seven inter-connected rings, two aminal linkages, and five or six stereocentres. The presence of vicinal quaternary carbon centres makes their chemical synthesis particularly challenging.67 However, biosynthesis of alkaloids differs from the biosynthesis of polyketides, peptides and terpenes because there is no ‘core’ synth(et)ase that sets up the skeleton in a single efficient early step. Thus comparison of the biosynthetic and chemical synthetic routes to the same compound is illuminating in a case where nature might be expected to use multiple steps to form the backbone (Scheme 7).Tryptophan 102 is the precursor for the biosynthetic pathway. Tryptamine 105 is produced by a decarboxylase CnsB. In a parallel pathway, dimethylallylation of tryptophan 102 by the action of 4-dimethylallyl tryptophan synthase (DMATS) CnsF yields 103, and then oxidative cyclisation gives aurantioclavine 104. It is known that an FAD-dependent monooxygenase CnsA is responsible for this intramolecular conversion, aided by the catalase CnsD. Biochemical evidence shows that the tryptophan derived units 104 and 105 are then linked via a heterodimeric coupling to afford the heptacyclic scaffold 106 of the communesins.68 The key catalyst is CnsC, a cytochrome P450 monooxygenase that creates the contiguous quaternary centres.69 Finally, highly selective N-methylation (CnsE) and N-acetylation give the target 101.
Yang and co-workers described the first asymmetric synthesis of communesin F 101, featuring an iridium catalysed intermolecular cyclization in the first step, such that tetracyclic core 110 was produced in gram scale with excellent stereoselectivity from advanced and protected precursors 108 and 109.70
N-methylation of the indole and removal of TBS protection of 110 afforded 111, then nitrogen was introduced via Mitsunobu displacement of hydroxyl with phthalimide to give protected amine 112. The olefin originating in 109 then underwent oxidative cleavage, Pinnick oxidation and methylation to yield ester 113. Standard deprotection of the phthalimide using hydrazine led to a primary amine, then a spontaneous lactamization afforded the pentacyclic scaffold, which was Boc protected to give N-Boc lactam 114. Stereoselective α-allylation and Boc group deprotection afforded alkene 116 which was oxidized to the corresponding aldehyde, then treated with NaBH4 to furnish the primary alcohol 117. Installation of nitrogen was again performed using phthalimide to make protected amine 118, followed by a routine deprotection and acylation to 119.
Next, a Heck reaction installed the butenyl moiety to form an allylic alcohol that was subsequently cyclised into hexacyclic benzazepine 120via a mesylated intermediate. Under the treatment of LiAlH4, amide 120 was reduced to hemiaminal 121. Finally, mesylation of 121 to give 122, which is transformed into a highly reactive iminium intermediate, allowed cyclization to the heptacyclic target 101.
Evaluation of the molecular complexity during the biosynthetic pathway highlights its remarkable efficiency. In particular the dimerisation of 104 and 105 send the route dramatically towards the target, and only two short steps are required to reach the goal. This is remarkable given that it is achieved by a single catalyst. In comparison the chemical synthesis is both very long (14 steps from fairly advanced precursors) and meandering in the chemical space (Fig. 7A). Ultimately this is the result of having to introduce small groups, or even individual nitrogen atoms, one by one in highly atom inefficient processes. The use of protective groups and frequent requirement for redox manipulations results in a chemical synthesis pathway that does not significantly approach the target over very many steps. Never-the-less the chemical synthesis represents a remarkable triumph of marshalling reactions, atoms, and selectivity towards a highly complex molecular goal.
9. Terpene: pleuromutilin
Pleuromutilin 123 is a tricyclic diterpene that is produced by a narrow family of basidiomycetes in the Clitopilus family.71 Bioactivity assays revealed the antimicrobial character of pleuromutilin. It inhibits protein synthesis by inhibiting the 50S subunit of the bacterial ribosome.72 Semi-synthetic derivatives find use as medicines for treatment of infections in poultry, swine and humans. These compounds include lefamulin,73 tiamulin,74 valnemulin and retapamulin75 which possess different 14-O-acyl groups compared with pleuromutilin 123. Activity vs. MRSA is particularly important as a response to the rapid growth of drug-resistant infections. Numerous independent chemical syntheses have been reported since Gibbons first 28-step racemic route reported in 1982,76 including Pronin's 16-step racemic route in 2022.77 Enantioselective total syntheses have also been reported by the Proctor,78 Herzon79 and Reisman groups,80 and the total biosynthesis has also been reported by Foster and co-workers.81The native host organisms produce 123 on solid substrates in low titres. Foster and co-workers, and the group of Oikawa,82 both delineated the biosynthetic pathway, and achieved total biosynthesis of pleuromutilin. Genome sequence of C. passeckerianus gave rapid access to the seven-gene pleuromutilin BGC and heterologous expression of the component genes in A. oryzae achieved the total biosynthesis. The BGC encodes a pathway-specific geranylgeranyl diphosphate synthase (Pl-GGS), a diterpene synthase (Pl-CYC), three P450 monooxygenases (Pl-P450-1, Pl-P450-2, and Pl-P450-3), a short-chain dehydrogenase/reductase (Pl-SDR) as well as an acetyl transferase (Pl-AT).81 Geranylgeranyl diphosphate 126 is not normally available as an intracellular component, so the pleuromutilin pathway starts with its synthesis from isopentenyl diphosphate 124 (IPP) and farnesyl diphosphate 125 (FPP). A terpene cyclase then creates the tricyclic alcohol 127 in a single step. Two cytochrome P450 monooxygenases can operate in either order, via diols 128 and 129 to form triol 130 (Scheme 8).
Oxidation to the ketone 131, 14-O-acetylation and final hydroxylation of the acetyl unit then provides pleuromutilin 123 after an overall 7-step process.83
A modular and enantioselective chemical synthesis of pleuromutilins was reported by Herzon and co-workers.84 The backbone was constructed by coupling of enimide 136 with neopentylic iodether 134. Compound 136 was achieved in eight steps from commercially available cyclohexanone 135. In parallel, fragment 134 was prepared via asymmetric alkylation of 133 with p-methoxybenzyl chloromethyl ether, and subsequent amide reduction as well as iodination.
Comins' reagent was used to convert methyl ketone 137 to acetylene 138. Deprotection of the PMB (para-methoxybenzyl) group followed by oxidation led to the formation of aldehyde 139. A nickel-based catalyst and triethylsilane was then used for the reductive cyclisation to give the eight-membered cyclic allylic alcohol 140. Oxidation with Dess–Martin periodinane and samarium diiodide reduction generated α-methyl-ketone 141 with the desired stereochemistry.
Birch reduction of 141 gave the corresponding alcohol, followed by ketal hydrolysis to provide epi-mutilin 142. Stepwise acylation with trifluoroacetylimidazole and O-trityl glycolic acid and routine methanolysis resulted in O-trityl-12-epi-pleuromutilin 143. When treated with Et2Zn, compound 143 was epimerized via a retroallylation–allylation process to afford (+)-pleuromutilin 123 in 33% yield, after in situ removal of the trityl group. Overall, the route requires 16 steps from cyclohexenone 135.
Analysis of the two routes in 3D chemical space highlights the similarities of the pathways. Both pathways approach the target monotonically over the first steps, in particular, reaching intermediates 142 and 131 effectively. Although the chemical synthesis appears to do this as effectively as the biosynthesis, this route is already 8-steps into the synthesis from cyclohexenone 135. As in previous comparisons the biosynthesis also continues to approach its target, the final two steps of acetylation and hydroxylation move the pathway only forwards because of the very high selectivity of the catalysts involved. The final two steps of the chemical synthesis are conceptually similar, involving O-acylation with the trityl-protected hydroxyacetate, then deprotection and adventitious 12-epimerization. The large step-lengths for the formation of 143 and 123 in the chemical synthesis reflect the addition and then removal of the large sp3-rich trityl protecting group, illustrating the low atom economy of this process (Fig. 8).
10. Conclusions
Nicolaou and Rigol defined total synthesis as “the art and science of making the molecules of living Nature in the laboratory, and by extension, their analogues”.85 This definition encapsulates both total chemical synthesis, total biosynthesis, and semisynthesis that lies somewhere between. In fact, there is a continuum of methods that nowadays integrate chemical and biological technologies. For example, the deployment of native or engineered enzymes from biosynthetic pathways into ‘traditional’ synthetic routes that often results in significant reduction in step-count of chemical pathways.86 In this review we have compared the extremes – total chemical synthesis vs. total biosynthesis. In each case the outcome is the same – production of SMs (and often their analogs) in the laboratory.Both total chemical synthesis and total biosynthesis can clearly be used for the production of SMs, but an important question is how much can be made? Direct comparison of absolute yields from synthesis and fermentation processes are not easy to make because they are reported in different units and depend on the scale of production. But reports in the academic literature generally stem from convenient laboratory scale processes (Table 1), yielding amounts between 1 mg and 100s of mg. Normally both processes are relatively easy to scale. For example, a lab-based chemical synthesis of sporothriolide yields more than 70 mg; a 10 L fermentation would also reach similar level. Likewise a 1 L fermentation to produce pleuromutilin would give 80 mg of the desired compound; total chemical synthesis would require a 40× scale-up to reach the same amount. For squalestatin S1, total biosynthesis only yields trace amount, but total chemical synthesis could obtain around 10 mg with an overall yield of 1.3%. In most cases, both sets of processes are within the achievable range.
| Compound | Chem. Synth. | Ref. | Biosynth. | Ref. |
|---|---|---|---|---|
| a * indicates unmentioned data. | ||||
| Sporothriolide | 73.5 mg (21%) | 23 | 8 mg L−1 | 22 |
| Strobilurin A | 202 mg (16.2%) | 26 and 27 | 2.6 mg L−1 | 28 |
| Citrinin | 229 mg (4.9%) | 33 | 19.1 mg L−1 | 31 |
| Squalestatin S1 | 9.9 mg (1.3%) | 44 and 45 | Trace | 40 and 41 |
| Bisorbicillinol | 8.4 mg (27.4%) | 51 | * | 47 |
| Tenellin | * (18.7%) | 61 | 20.4 mg L−1 | 57 |
| Communesin F | 8.6 mg (2.6%) | 70 | 8.3 mg L−1 | 68 and 69 |
| Pleuromutilin | 1.8 mg (0.4%) | 84 | 80 mg L−1 | 81 |
To compare the routes in more detail we adopted informatic methods pioneered by others15 that plot synthetic pathways as journeys through 3-dimensional chemical space. The selection of complexity (Cm), Mw (Da) and Fsp3 is convenient, if arbitrary, and different or better measures may be available. However, these three parameters have the benefit of being easily computed and understood, and their analysis does illuminate aspects of chemical- and bio-synthetic pathways that chemists have ‘felt’, if not quantified, for many years. Here, we have focussed on examples drawn from fungal SMs, as these compounds are often chemically complex and challenging to construct. However, the analysis could be easily extended to any other class or source of SM.
The analysis shows that in almost all cases the total biosynthetic processes are shorter in terms of step-count, and those steps progress the pathway more quickly towards to target. This is why the integration of enzymes from biosynthetic pathways find favour in chemical synthesis: they usually simultaneously reduce step-count, progress the pathway, and simplify routes. For example, the chemoenzymatic synthesis of bisorbicillinol 73 allows a dramatic simplification of the early synthetic steps (Scheme 5). In addition, the synthesis is more convergent and does not require protecting groups. In some cases, such as for squalestatin S1 50, communesin F 101 and pleuromutilin 123, the comparison is stark – the biosynthetic processes are dramatically shorter and more effective. In other cases, such as that of sporothriolide 1, the pathways are more comparable, although the biosynthesis is still shorter and more direct. The analysis in 3D space also allows identification of steps of low atom economy87 (even in the absence of analysis of the reagents and catalysts themselves). For example, steps from 64 to 67 during the synthesis of squalestatin S1 (already 27 steps ‘in’ from a complex starter, Scheme 4) involve addition of missing carbons and protecting group manipulations that move the pathway further away from the target. This is avoided in the biosynthetic routes as almost all carbons are present from the start, and protecting groups are not required.
Chemical synthesis is stepwise by its nature and design. It is clear that the step-count can be reduced, often dramatically so, by the introduction of biological catalysts. However, although the total biosynthetic pathways are represented on the page as consecutive synthetic steps, in reality the total biosynthesis is achieved in a single process. This is because all of the enzymes are expressed in a single host organism that produces both the required starting materials, cofactors and enzymes, as well as the mild biological conditions and structures required to support those catalysts: pleuromutilin 123 is produced in A. oryzae in a single process that requires a fermentation, an extraction and a purification.81–83 Such processes are already at the core of modern biotechnology. For example, filamentous fungal hosts produce penicillins at titres of 100 g L−1 in well-understood and controlled fermentations.8 In the case of pleuromutilin 123, the heterologous production also produces more than 20 times the material produced by the wild-type species because of the way in which the pathway expression is controlled.81 In the case of communesin F both lab-scale total chemical synthesis and lab scale total biosynthesis yield around 8 mg of the desired compound. However, the biosynthetic process requires a single fermentation, extraction and purification, while the total chemical synthesis requires 14 separate chemical processes. It is thus clear that total biosynthesis dramatically reduces step-counts compared to chemical synthesis, and it is also appropriate for scale-up because the technology for producing SMs by fermentation is already very mature.
Each synthetic step or process requires energy for heating or cooling, but also for the production of solvents, substrates, reagents, ligands and catalysts. Efficiency of synthesis will very likely become of the highest importance in the coming years as the imperative to reduce or remove carbon emissions becomes essential and as fossil-derived carbon becomes rarer and more expensive. Trost argued nearly 30 years ago that atom economy is of key importance,87 and since then focus on the development of new catalytic methodology has often improved the chemical economy of individual steps, and reduced the number of steps required. For example, Pronin's modern 16-step total chemical synthesis77 of pleuromutilin 123 is roughly half the length of Gibbons' 1982 route,76 representing a very significant improvement in efficiency. However, the chemical synthesis route is still 15-steps longer than the total biosynthesis. In a scenario where the requirement to decarbonise is urgent, total biosynthesis becomes a highly attractive way to produce the known SMs while minimising energy consumption and carbon emissions. A further advantage is that once a producing host has been constructed it need not be constructed again: it will continue to produce the desired SM continuously. Total chemical synthesis, on the other hand, has to be repeated to produce more material.
The second element of Nicolaou and Rigol's85 definition of total synthesis, however, is challenging: “and by extension their analogues” is something that total chemical synthesis can do easily. The measures and analysis of the synthetic pathways used here don't consider the flexibility of pathways. For example, Herzon's pleuromutilin 123 synthesis allows easy diversion to various structural analogs, many of which are biologically active.84 On the contrary, total biosynthesis produces pleuromutilin itself (and its precursors) very effectively, but while variation of the pathway to produce new derivatives is possible,88 it remains limited.
This requirement to produce new compounds is clearly a major challenge for the emerging practice of total biosynthesis. Total chemical synthesis can, in principle (and often in practice), produce any given target. It does this at the cost of energy and steps, but it is possible. At present, total biosynthesis is highly effective at making any given SM in a single process if the biosynthetic genes can be found, but making analogues will be the next major step. However, progress is being made in the area of fungal metabolites where the concept of mixing and matching genes from related gene clusters already creates libraries of related compounds,89,90 and where engineering of the core synth(et)ases to change their programmes is possible.62
For example, total biosynthesis has recently been used to produce focussed libraries of tropolone meroterpenoids that are currently difficult to obtain by total chemical synthesis.89 In the case of the parent compound xenovulene A,91 no total chemical synthesis has yet been reported, but combination of genes from the xenovulene, eupenifeldin and pycnidione BGCs allows rational total biosynthesis of new metabolites. Likewise, our group,90 and Oikawa, Minami and Liu and coworkers92 have recently reported the diversification of aristolochene-derived SMs using total biosynthesis as the key methodology. In these ways total biosynthesis starts to approach the flexibility of total chemical synthesis, while retaining the advantages of being single processes.
In the future, incorporation of engineered enzymes from directed evolution93 and the construction of entirely fabricated biosynthetic gene clusters should allow the construction of a much wider range of compounds by total biosynthesis. The goal will be for total biosynthesis to match the flexibility of total chemical synthesis while maintaining the advantages of efficiency.
11. Author contributions
The review was conceptualised by RJC and DST. DST made the calculations. DST and XZ prepared the initial draft and diagrams, and all authors contributed to the polishing of the MS.12. Conflicts of interest
There are no conflicts to declare.13. Acknowledgements
RJC thanks DFG (CO1328/10-1 & CO1328/4-2) and Leibniz Universität Hannover. XZ thanks the Scientific Research Starting Foundation of Southwest University (SWU020018), the National Natural Science Foundation of China (22101236), and the National Natural Science Foundation of China Excellent Young Scholars Fund (Overseas). DST thanks the Fundamental Research Funds for the Central Universities (SWU-KQ23009), and the Natural Science Foundation of Chongqing (CSTB2024NSCQ-MSX0656). Dr He Peng and Tim Alexander Mast (Medizinische Hochschule Hannover) are acknowledged for supports in calculations.14. Notes and references
- N. Fay, C. Kouklovsky and A. de la Torre, ACS Org. Inorg. Au, 2023, 3, 350–363 Search PubMed.
- R. J. Cox, Nat. Rev. Chem, 2024, 8, 61–78 CrossRef CAS.
- J. N. Denis, A. E. Greene, D. Guenard, F. Gueritte-Voegelein, L. Mangatal and P. Potier, J. Am. Chem. Soc., 1988, 110, 5917–5919 CrossRef CAS.
- J. Turconi, F. Griolet, R. Guevel, G. Oddon, R. Villa, A. Geatti, M. Hvala, K. Rossen, R. Göller and A. Burgard, Org. Process Res. Dev., 2014, 18, 417–422 CrossRef CAS.
- J. Kennedy, Nat. Prod. Rep., 2007, 25, 25–34 RSC.
- S. McAlister, Y. Ou, E. Neff, K. Hapgood, D. Story, P. Mealey and F. McGain, BMJ Open, 2016, 6, e013302 Search PubMed.
- V. Butsic, J. K. Carah, M. Baumann, C. Stephens and J. C. Brenner, Environ. Res. Lett., 2018, 13, 124017 CrossRef.
- C. Barreiro and C. García-Estrada, J. Proteomics, 2019, 198, 119–131 CrossRef CAS.
- P. Gabrielli, L. Rosa, M. Gazzani, R. Meys, A. Bardow, M. Mazzotti and G. Sansavini, One Earth, 2023, 6, 682–704 CrossRef.
- IPCC, 2023: Synthesis Report. Contribution of Working Groups I, II and III to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change, ed. H. Lee and J. Romero, IPCC, Geneva, Switzerland, pp. , pp. 35–115, DOI:10.59327/IPCC/AR6-9789291691647.
- F. Lovering, J. Bikker and C. Humblet, J. Med. Chem., 2009, 52, 6752–6756 CrossRef CAS PubMed.
- W. Wei, S. Cherukupalli, L. Jing, X. Liu and P. Zhan, Drug Discovery Today, 2020, 25, 1839–1845 Search PubMed.
- T. Böttcher, J. Chem. Inf. Model., 2016, 56, 462–470 CrossRef.
- T. Böttcher, J. Mol. Evol., 2018, 86, 1–10 CrossRef.
- E. M. Landwehr, M. A. Baker, T. Oguma, H. E. Burdge, T. Kawajiri and R. A. Shenvi, Science, 2022, 375, 1270–1274 CrossRef CAS PubMed.
- R. Schor and R. Cox, Nat. Prod. Rep., 2018, 35, 230–256 RSC.
- B. Reck and P. Spiteller, Synthesis, 2015, 47(19), 2885–2911 CrossRef CAS.
- Y. Matsuda and I. Abe, Nat. Prod. Rep., 2015, 33, 26–53 RSC.
- R. J. Cox, Nat. Rev. Chem, 2024, 1–18 Search PubMed.
- J. D. Hunter, Comput. Sci. Eng., 2007, 9(3), 90–95 Search PubMed.
- K. Krohn, K. Ludewig, H. J. Aust, S. Draeger and B. Schulz, J. Antibiot., 1994, 47, 113–118 CrossRef CAS PubMed.
- D.-S. Tian, E. Kuhnert, J. Ouazzani, D. Wibberg, J. Kalinowski and R. J. Cox, Chem. Sci., 2020, 11, 12477–12484 RSC.
- M. Kimura, T. Mohri, M. Enomoto, Y. Meguro, Y. Ogura and S. Kuwahara, J. Org. Chem., 2021, 86, 12475–12479 CrossRef CAS PubMed.
- H. Sauter, W. Steglich and T. Anke, Angew. Chem., Int. Ed., 1999, 38, 1328–1349 CrossRef.
- J. M. Clough, Nat. Prod. Rep., 1993, 10, 565–574 RSC.
- Ya. N. Grigorieva, V. Popovsky, A. Stepanov and E. Lubuzh, Russ. Chem. Bull., 2010, 59, 2086–2093 CrossRef.
- V. A. Popovsky, A. V. Stepanov and N. Grigorieva, Mendeleev Commun., 2013, 23, 190–192 CrossRef CAS.
- R. Nofiani, K. de Mattos-Shipley, K. E. Lebe, L.-C. Han, Z. Iqbal, A. M. Bailey, C. L. Willis, T. J. Simpson and R. J. Cox, Nat. Commun., 2018, 9, 3940 CrossRef PubMed.
- K. E. Lebe and R. J. Cox, RSC Adv., 2019, 9, 31527–31531 RSC.
- B. R. Clark, R. J. Capon, E. Lacey, S. Tennant and J. H. Gill, Org. Biomol. Chem., 2006, 4, 1520–1528 RSC.
- Y. He and R. J. Cox, Chem. Sci., 2015, 7, 2119–2127 RSC.
- Y. Xue, C. Kong, W. Shen, C. Bai, Y. Ren, X. Zhou, Y. Zhang and M. Cai, J. Biotechnol., 2017, 242, 64–72 CrossRef CAS PubMed.
- D. Bergmannniliuc, B. Cramer and H.-U. Humpf, Mycotoxin Res., 2018, 34, 141–150 CrossRef.
- P. Procopiou, E. Bailey, M. Bamford, A. Craven, B. Dymock, J. Houston, J. Hutson, B. Kirk, A. McCarthy and M. Sareen, J. Med. Chem., 1994, 37, 3274–3281 CrossRef CAS.
- J. D. Bergstrom, M. M. Kurtz, D. J. Rew, A. M. Amend, J. D. Karkas, R. G. Bostedor, V. S. Bansal, C. Dufresne, F. L. VanMiddlesworth and O. D. Hensens, Proc. Natl. Acad. Sci. U.S.A., 1993, 90, 80–84 CrossRef CAS PubMed.
- K. Hasumi, K. Tachikawa, K. Sakai, S. Murakawa, N. Yoshikawa, S. Kumazawa and A. Endo, J. Antibiot., 1993, 46, 689–691 CrossRef CAS PubMed.
- K. C. Nicolaou, A. Nadin, J. E. Leresche, E. W. Yue and S. L. Greca, Angew. Chem., Int. Ed., 1994, 33, 2190–2191 CrossRef.
- K. C. Nicolaou, E. W. Yue, S. la Greca, A. Nadin, Z. Yang, J. E. Leresche, T. Tsuri, Y. Naniwa and F. de Riccardis, Chem.–Eur. J., 1995, 1, 467–494 CrossRef CAS.
- R. J. Cox, F. Glod, D. Hurley, C. M. Lazarus, T. P. Nicholson, B. A. Rudd, T. J. Simpson, B. Wilkinson and Y. Zhang, Chem. Commun., 2004, 2260–2261 RSC.
- B. Bonsch, V. Belt, C. Bartel, N. Duensing, M. Koziol, C. Lazarus, A. Bailey, T. Simpson and R. Cox, Chem. Commun., 2016, 52, 6777–6780 RSC.
- K. E. Lebe and R. J. Cox, Chem. Sci., 2019, 10, 1227–1231 RSC.
- N. Liu, Y.-S. Hung, S.-S. Gao, L. Hang, Y. Zou, Y.-H. Chooi and Y. Tang, Org. Lett., 2017, 19, 3560–3563 CrossRef CAS.
- C. A. Jones, P. J. Sidebottom, R. J. P. Cannell, D. Noble and B. A. M. Rudd, J. Antibiot., 1992, 45, 1492–1498 CrossRef CAS PubMed.
- D. Stoermer, S. Caron and C. H. Heathcock, J. Org. Chem., 1996, 61, 9115–9125 CrossRef CAS.
- S. Caron, D. Stoermer, A. K. Mapp and C. H. Heathcock, J. Org. Chem., 1996, 61, 9126–9134 CrossRef CAS.
- A. al Fahad, A. Abood, K. M. Fisch, A. Osipow, J. Davison, M. Avramović, C. P. Butts, J. Piel, T. J. Simpson and R. J. Cox, Chem. Sci., 2013, 5, 523–527 RSC.
- L. Kahlert, E. F. Bassiony, R. J. Cox and E. J. Skellam, Angew. Chem., Int. Ed., 2020, 59, 5816–5822 CrossRef CAS.
- L. Kahlert, R. J. Cox and E. Skellam, Chem. Commun., 2020, 56, 10934–10937 RSC.
- K. C. Nicolaou, G. Vassilikogiannakis, K. B. Simonsen, P. S. Baran, Y.-L. Zhong, V. P. Vidali, E. N. Pitsinos and E. A. Couladouros, J. Am. Chem. Soc., 2000, 122, 3071–3079 CrossRef CAS.
- D. Barnes-Seeman and E. J. Corey, Org. Lett., 1999, 1, 1503–1504 CrossRef CAS.
- R. Hong, Y. Chen and L. Deng, Angew. Chem., Int. Ed., 2005, 44, 3478–3481 CrossRef CAS PubMed.
- A. Sib and T. A. M. Gulder, Angew. Chem., Int. Ed., 2017, 56, 12888–12891 CrossRef CAS PubMed.
- J. I. Müller and T. A. M. Gulder, Commun. Chem., 2024, 7, 39 CrossRef PubMed.
- R. Toshe, E. Charria-Girón, A. Khonsanit, J. J. Luangsa-ard, S. J. Khalid, H. Schrey, S. S. Ebada and M. Stadler, J. Fungi, 2024, 10, 69 CrossRef CAS.
- Z. Zhang, C. S. Jamieson, Y.-L. Zhao, D. Li, M. Ohashi, K. N. Houk and Y. Tang, J. Am. Chem. Soc., 2019, 141, 5659–5663 CAS.
- M. L. Shenouda, M. Ambilika and R. J. Cox, J. Fungi, 2021, 7, 1034 CAS.
- L. M. Halo, M. N. Heneghan, A. A. Yakasai, Z. Song, K. Williams, A. M. Bailey, R. J. Cox, C. M. Lazarus and T. J. Simpson, J. Am. Chem. Soc., 2008, 130, 17988–17996 CrossRef CAS.
- E. Skellam, Nat. Prod. Rep., 2017, 34, 1252–1263 RSC.
- Z. Song, R. J. Cox, C. M. Lazarus and T. J. Simpson, ChemBioChem, 2004, 5, 1196–1203 CrossRef CAS PubMed.
- K. Kleigrewe, F. Aydin, K. Hogrefe, P. Piecuch, K. Bergander, E.-U. Würthwein and H.-U. Humpf, J. Agric. Food Chem., 2012, 60, 5497–5505 CrossRef CAS PubMed.
- J. H. Rigby and M. Qabar, J. Org. Chem., 1989, 54, 5852–5853 CrossRef CAS.
- R. J. Cox, Nat. Prod. Rep., 2022, 40, 9–27 RSC.
- K. M. Fisch, W. Bakeer, A. A. Yakasai, Z. Song, J. Pedrick, Z. Wasil, A. M. Bailey, C. M. Lazarus, T. J. Simpson and R. J. Cox, J. Am. Chem. Soc., 2011, 133, 16635–16641 CrossRef CAS.
- X.-L. Yang, S. Friedrich, S. Yin, O. Piech, K. Williams, T. J. Simpson and R. J. Cox, Chem. Sci., 2019, 10, 8478–8489 RSC.
- K. Schmidt and R. J. Cox, RSC Adv., 2024, 14, 8963–8970 RSC.
- A. Numata, C. Takahashi, Y. Ito, T. Takada, K. Kawai, Y. Usami, E. Matsumura, M. Imachi, T. Ito and T. Hasegawa, Tetrahedron Lett., 1993, 34, 2355–2358 CrossRef CAS.
- P. Siengalewicz, T. Gaich and J. Mulzer, Angew. Chem., Int. Ed., 2008, 47, 8170–8176 CrossRef CAS PubMed.
- H. Lin, G. Chiou, Y. Chooi, T. C. McMahon, W. Xu, N. K. Garg and Y. Tang, Angew. Chem., Int. Ed., 2015, 54, 3004–3007 CrossRef CAS.
- H.-C. Lin, T. C. McMahon, A. Patel, M. Corsello, A. Simon, W. Xu, M. Zhao, K. Houk, N. K. Garg and Y. Tang, J. Am. Chem. Soc., 2016, 138, 4002–4005 CrossRef CAS PubMed.
- X. Liang, T.-Y. Zhang, X.-Y. Zeng, Y. Zheng, K. Wei and Y.-R. Yang, J. Am. Chem. Soc., 2017, 139, 3364–3367 CrossRef CAS PubMed.
- A. J. Hartley, K. D. Mattos-Shipley, C. M. Collins, S. Kilaru, G. D. Foster and A. M. Bailey, FEMS Microbiol. Lett., 2009, 297, 24–30 CrossRef CAS.
- F. Schlünzen, E. Pyetan, P. Fucini, A. Yonath and J. M. Harms, Mol. Microbiol., 2004, 54, 1287–1294 CrossRef PubMed.
- J. R. Covvey and A. J. Guarascio, J. Intern. Med., 2022, 291, 51–63 CrossRef CAS PubMed.
- G. Laber and E. Schütze, Antimicrob. Agents Chemother., 1975, 7, 517–521 CrossRef CAS PubMed.
- B. Boyd and J. Castaner, Drugs Future, 2006, 31(2), 0107 CrossRef CAS.
- E. G. Gibbons, J. Am. Chem. Soc., 1982, 104, 1767–1769 CrossRef CAS.
- N. J. Foy and S. V. Pronin, J. Am. Chem. Soc., 2022, 144(23), 10174–10179 CrossRef CAS.
- N. J. Fazakerley, M. D. Helm and D. J. Procter, Chem.–Eur. J., 2013, 19, 6718–6723 CrossRef CAS PubMed.
- O. Goethe, M. DiBello and S. B. Herzon, Nat. Chem., 2022, 14, 1270–1277 CrossRef CAS.
- E. P. Farney, S. S. Feng, F. Schäfers and S. E. Reisman, J. Am. Chem. Soc., 2018, 140, 1267–1270 CrossRef CAS PubMed.
- A. M. Bailey, F. Alberti, S. Kilaru, C. M. Collins, K. de Mattos-Shipley, A. J. Hartley, P. Hayes, A. Griffin, C. M. Lazarus, R. J. Cox, C. L. Willis, K. O'Dwyer, D. W. Spence and G. D. Foster, Sci. Rep., 2016, 6, 25202 CAS.
- M. Yamane, A. Minami, C. Liu, T. Ozaki, I. Takeuchi, T. Tsukagoshi, T. Tokiwano, K. Gomi and H. Oikawa, ChemBioChem, 2017, 18, 2317–2322 CrossRef CAS PubMed.
- T. Tsukagoshi, T. Tokiwano and H. Oikawa, Biosci., Biotechnol., Biochem., 2007, 71, 3116–3121 CAS.
- S. K. Murphy, M. Zeng and S. B. Herzon, Science, 2017, 356, 956–959 CAS.
- K. C. Nicolaou and S. Rigol, Nat. Prod. Rep., 2020, 37, 1404–1435 CAS.
- J. Li, F. Li, E. King-Smith and H. Renata, Nat. Chem., 2020, 12, 173–179 CAS.
- B. M. Trost, Angew Chem. Int. Ed. Engl., 1995, 34, 259–281 CrossRef CAS.
- F. Alberti, K. Khairudin, J. A. Davies, S. Sangmalee, C. L. Willis, G. D. Foster and A. M. Bailey, Chem. Sci., 2023, 14, 3826–3833 RSC.
- C. Schotte, L. Li, D. Wibberg, J. Kalinowski and R. J. Cox, Angew. Chem., Int. Ed., 2020, 59, 23870–23878 CrossRef CAS.
- Y. Sun, J. Gerke, K. Becker, E. Kuhnert, B. Verwaaijen, D. Wibberg, J. Kalinowski, M. Stadler and R. J. Cox, Chem. Sci., 2023, 14, 13463–13467 RSC.
- R. Schor, C. Schotte, D. Wibberg, J. Kalinowski and R. J. Cox, Nat. Commun., 2018, 9, 1963 CrossRef.
- Y. Sato, X. Shi, Y. Ye, S. Domon, J. Takino, T. Ozaki, C. Liu, H. Oikawa and A. Minami, ACS Chem. Biol., 2024, 19, 861–865 CrossRef CAS PubMed.
- H. Renata, J. Z. Wang and F. H. Arnold, Angew. Chem., Int. Ed., 2015, 54, 3351–3367 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |