Open Access Article
Open Access ArticleMarine natural products†
Anthony R.
Carroll
 *ab,
Brent R.
Copp
*ab,
Brent R.
Copp
 c,
Tanja
Grkovic
c,
Tanja
Grkovic
 d,
Robert A.
Keyzers
d,
Robert A.
Keyzers
 e and
Michèle R.
Prinsep
e and
Michèle R.
Prinsep
 f
f
aSchool of Environment and Science, Griffith University, Gold Coast, Australia. E-mail: A.Carroll@griffith.edu.au
bGriffith Institute for Drug Discovery, Griffith University, Brisbane, Australia
cSchool of Chemical Sciences, University of Auckland, Auckland, New Zealand
dNatural Products Branch, Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, and Molecular Targets Program, Center for Cancer Research, National Cancer Institute, Frederick, MD, USA
eCentre for Biodiscovery, and School of Chemical and Physical Sciences, Victoria University of Wellington, Wellington, New Zealand
fSchool of Science, University of Waikato, Hamilton, New Zealand
First published on 29th January 2024
Abstract
Covering: January to the end of December 2022
This review covers the literature published in 2022 for marine natural products (MNPs), with 645 citations (633 for the period January to December 2022) referring to compounds isolated from marine microorganisms and phytoplankton, green, brown and red algae, sponges, cnidarians, bryozoans, molluscs, tunicates, echinoderms, the submerged parts of mangroves and other intertidal plants. The emphasis is on new compounds (1417 in 384 papers for 2022), together with the relevant biological activities, source organisms and country of origin. Pertinent reviews, biosynthetic studies, first syntheses, and syntheses that led to the revision of structures or stereochemistries, have been included. An analysis of NP structure class diversity in relation to biota source and biome is discussed.
1 Introduction
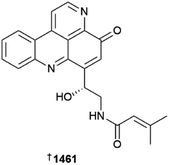
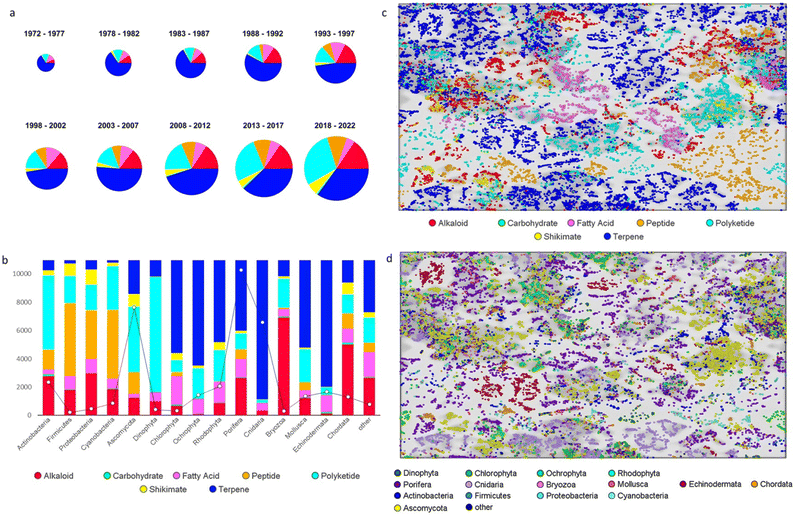
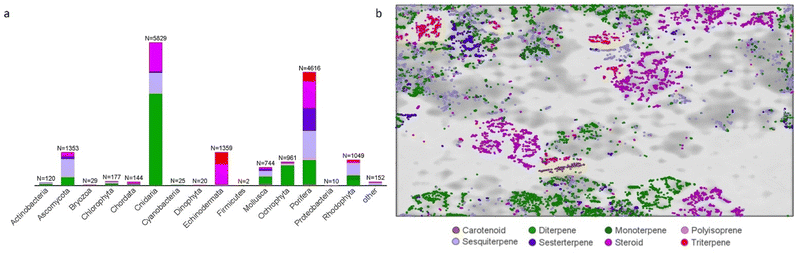
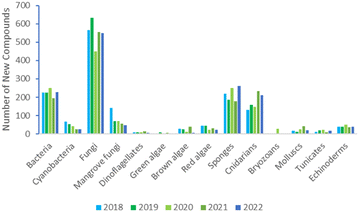
The annual review of marine natural products was first published in 1984 by John Falkner and the 2024 edition marks its 40th anniversary. This review is of the literature for 2022 and describes 1417 new compounds from 384 papers, compared to 1425 new compounds in 416 papers reported for 2021.1 In addition, 24 known NPs were reported from a marine source for the first time and 70 known MNPs had their structures revised. We have also introduced a new artefact category this year and this includes three compounds. Only new MNP structures or previously reported compounds where there has been a structural revision, or a newly established stereochemistry are shown in this review. The review also covers previously reported MNPs with significant new bioactivities or ones that have been synthesised for the first time, but their structures are generally not shown. A † symbol on the identifying diagram number is used to distinguish structures where the absolute configuration has been determined for all stereogenic centres, axes and/or planes in a compound. Reports of new MNPs that were identified based solely on a combination of gene cluster information, MS/MS data and/or Global Natural Products Social (GNPS)-based molecular networking, with compounds not isolated and no NMR data recorded, are excluded from the review. Only a selection of highlighted structures (89) is shown in the review. Compound numbers for structures not highlighted in the review are italicised, and all structures are available for viewing, along with their names, taxonomic origins, collection locations, and biological activities, in an associated ESI document.† Access to the curated MNP data held in the Marinlit database2 provides all the structural and literature data used to prepare this review. The 10 year anniversary of RSC running MarinLit is also being celebrated in 2024. This review welcomes Tanja Grkovic from the National Cancer Institute, USA as a new author into the team.Trends in the number of new MNPs reported annually over the semi-decade have returned to a pre-pandemic level. Decreasing trends in reporting of cyanobacterial, mangrove fungal and algal metabolites continues. However, this is compensated by increases in the reporting of sponge and cnidarian MNPs (Fig. 1).
 | ||
| Fig. 1 Trends in new MNPs. The bars represent the total number of new MNPs reported each year over the last five years. | ||
A paper describing the use of machine learning in combination with fast DU8+ hybrid density functional theory/parametric computations termed DU8ML provides an important additional tool for MNP chemists to accurately assign structures based on interpretation of NMR data. This new method has been applied to reassign the structures of previously published NPs, including a number from marine sources. We anticipate that implementing DU8ML routinely at the time of manuscript preparation and peer review should help to reduce the proliferation of incorrect structures appearing in the literature.3
2 Marine microorganisms and phytoplankton
2.1 Marine-sourced bacteria
Actinobacteria were the most common source of bacterial compounds with 182 new NPs reported in 2022. A new β-carboline, marinacarboline glucuronide 1 was isolated from a sponge-derived Actinoalloteichus cyanogriseus.4 The structure determination of the NP, including the absolute configuration, was aided by single-crystal X-ray diffraction (XRD) analysis. A deep-sea water-derived strain, Actinomadura sp. yielded seven new angucyclinone-class polyketides kumemicinones A–G 2–8 which included three novel structures, the 4-hydroxyspiro[4.5]deca-1,6-dione-containing 5 and two ether-bridged dimers 6 and 7.5 In a comprehensive study of the NP metabolome of a deep-sea sediment-derived Amycolatopsis sp., 22 compounds were reported, including four new structures, a linear peptide agrotetratide A 9, spermidine derivative 10, the furan 11 and agrocusin A 12.6 Six new chromones, amycolachromones A–F 13–18 were isolated from a deep-sea sediment-derived collection of Amycolatopsis sp.7 while a new 2,5-piperazinedione analogue, georgenione A 19 was reported from Georgenia sp.8 Continuing work on Gephyromycinifex aptenodytis derived from the gut microbiota of the Antarctic emperor penguin, Aptenodytes forsteri yielded an additional new angucyclinone analogue, 2-hydroxy-frigocyclinone 20.9 An aromatic glycoside 21 was identified from a sediment-derived collection of Nocardiopsis synnemataformans10 and four new fluvirucin-type macrolactams, fluvirucins B7–B10 22–25 were isolated from a sponge-derived rare actinomycete Nonomuraea sp.11Using metabolomic-guided microbial strain prioritisation and genome-mining strategies, a new 5-aminosalicylate containing siderophore, pseudonochelin 26 was isolated from a sponge-derived Pseudonocardia sp.12 Isotopic feeding studies with labelled p-aminobenzoate enabled annotation of the putative biosynthetic pathway and functional gene assignments for 26. Pseudonochelin showed moderate activity against methicillin-sensitive Staphylococcus aureus (MSSA) and methicillin-resistant S. aureus (MRSA), and a one log reduction of bacterial cell burden in MRSA-infected mice at an intraperitoneal dose of 160 mg kg−1. Four new macrolides, kongjuemycins A 27 B1 28, and B2–B3, 29, 30 were isolated from a coral-derived Pseudonocardia kongjuensis.13 While the structure of 27 has an unusual 2-hydroxy-2-methyl-4-methine-pyran-3,5-dione six membered O-heterocycle, in 28–30 it was substituted by a 2-hydroxy-2,5-dimethyl-pyrrol-3-one five membered N-heterocycle. Saccharomonospora sp. yielded a new indole dimer saccharobisindole 31, asterric acid analogue 32 and the quinolone 33.14
Seven new MNPs were isolated from the genus Salinispora in 2022. Salinispora arenicola yielded a new aminoquinone polyketide salinisporamine 34, and two lactones salinorcinol 35 and salinacetamide 36.15 The structure of proton-deficient 34 could not be assigned by NMR spectroscopy alone and elucidation was aided by XRD analysis. The structures of 35 and 36 have previously been reported from a mutated rifamycin producer A. mediterranei but never as NPs from a wild-type bacterial culture. S. pacifica yielded a new manumycin-type polyketide pacificamide 37.16 The candidate biosynthetic gene cluster (BGC) pac for 37 was identified and bioinformatic analysis showed that closely related BGC were present in other bacterial genera, opening further potential for the identification of this rare class of MNP. A mass spectrometry (MS)-guided screening method yielded three new phosphotriesters salinipostins L–N 38–40 from S. tropica with potent serine hydrolase inhibitory activity.17
As in previous years, the genus Streptomyces was the major source of new compounds from marine bacteria with 140 new MNPs reported in 2022. Enediyne-targeted genomic signature-based PCR screening of a bacterial DNA library of over 1000 strains resulted in the identification of a new tetracyclic NP, jejucarboside A 41 from a sediment-derived Streptomyces sp.18 The unusual structure of 41 contains a carbonate-bearing, chlorinated cyclopenta[a]indene skeleton and 3-methyl-4-methylamino-4,6-dideoxy-D-gulose. Upon comparison of the BGC of other nine-membered enediyne NPs, a putative biosynthetic pathway for jejucarboside was proposed. This involved cycloaromatisation of a nine-membered cyclic enediyne precursor. Notably, the initial yield of 41 in yeast malt extract liquid medium supplemented with sea salt was only 0.008 mg mL−1 and resulted in a total of 360 L of culture required to isolate 3 mg of the compound. To supply an adequate amount for biological testing, subcultures of the producing strain were screened for increased production of 41 and the titre was increased 150-fold to 1.25 mg mL−1. Jejucarboside A did not show any cytotoxic, antioxidant, or anti-inflammatory activity, demonstrating that the enediyne moiety is essential for the activity observed for this chemotype. Two chlorinated, pentacyclic polyketides, chlororesistoflavins A 42 and B 43, were reported from a marine sediment-derived Streptomyces sp., with 42 showing potent activity against MRSA (MIC 0.25 μg mL−1) which was eight-fold higher than that of the C-4 chloro isomer 43 (MIC 2.0 μg mL−1).19 The two compounds also had remarkably different ECD spectra, with 42 displaying a Cotton effect near the n–π* transition of the C-10 carbonyl group which was opposite to that observed in other resistoflavins. This difference was attributed to allylic-1,3 strain imposed by the chlorine substitution at the C-11 position of the cyclohexadiene ring in 42. Both new NPs were shown to transform into their respective C-10 phenolic resistomycin analogues upon prolonged exposure to UV light. A deep-sea hydrothermal vent-derived Streptomyces sp. yielded three new actinopyrone analogues, actinoketone 44 and actinopyrones E 45 and F 46, as well as three known MNPs, actinopyrone D 47, PM050463 48, and PM050511 49, the absolute configurations for which have been revised upon re-examination of the Mosher ester results and analysis of the BGC assembly data.20 The same strain also yielded polyethers, seco-salinomycins A–E 50–54, and minipyrone 55.21
A novel ring C-expanded angucyclinone oxemycin A 56 and seven new ring C-fragmented analogues 57–63 have been isolated from a sediment-derived Streptomyces sp.22 Other sediment-derived Streptomyces collections have yielded nine hexa-substituted benzothioate glycosides, suncheonosides E–M 64–72, and four other benzothioate MNPs 73–76,23 nine antifungal polyene macrolides filipins VI–XIV 77–85,24 and four polyketides with a unique 6/5/5 tricyclic ring system, streptoglycerides E–H 86–89.25 In addition, a xanthone sattahipmycin 90 was reported from a sediment-derived Streptomyces sp. and showed potent antibacterial activity, moderate anti-plasmodial activity, and weak antiproliferative activity against five human tumour cell lines (HTCLs).26 Butenolide 91 was reported as a NP for the first time from a sediment-derived collection of Streptomyces sp.27 and a deep-sea sediment-derived S. chumphonensis yielded six aromatic acids 92–97, and three oxime-containing leucine derivatives 98, 99, and 100. Compound 100 was reported as a MNP for the first time.28 Two dimeric benzoic polyene acids, youssoufenes A2 101 and A3 102 were reported from a dtlA activated mutant of a S. youssoufiensis strain.29 A sediment-derived Streptomyces strain yielded 11 napthalenic macrolides, hygrocins K–U 103–113 and one phenylpropanamide derivative, streptobenzenepropanamide A 114.30
Two peptides, polyoxyperuin A seco acid 115 and polyoxyperuin A 116 were reported from a marine sediment-derived Streptomyces sp.31 The putative BGC pop that is responsible for the production of the compounds, was identified, as well as the regulatory elements of the BGC. This enabled a strain that was engineered to overexpress the transcriptional activator to produce the compounds in significantly increased yield. The cyclic analogue 116 had potent antibacterial activity, while the ring opened 115 was inactive. Another sediment-derived Streptomyces sp. yielded chlorinated depsiheptapeptides, streptocinnamides A 117 and B 118.32 that each contain m,m-dichloro-p-hydroxy-cis-α-methoxycinnamic acid as well as two rare amino acids, 3-hydroxy-4-chlorovaline and 4-acetoxy-5-methyl-proline. Streptocinnamide A showed potent antibacterial activity against Micrococcus sp. with a MIC of 4 ng mL−1. Pyridapeptide A 119, a cyclohexapeptide containing the rare hexahydropyridazine-3-carboxylic acid and 5-hydroxytetrahydropyridazine-3-carboxylic acid residues, together with an additional four new glycosylated analogues pyridapeptides B–E 120–123, were reported from a sponge-derived Streptomyces sp.33 Bioinformatic analysis identified the putative BGC pdp to be responsible for the assembly of the compounds, including the production of the glycosidic bonds and the assembly of the sugar chains. Only 122 and 123 that contained a tetrasaccharide chain, showed weak to moderate antiproliferative activity against a panel of five HTCLs. Seven siderophore-related compounds 124–130 were isolated from a sponge-associated S. diastaticus.34 Other Streptomyces-sourced peptide NPs included the epoxy cinnamoyl-containing epoxinnamide 131,35 teanamides A 132 and B 133,36 xanthostatin B 134,37 cystargamides C 135 and D 136 (ref. 38) and levesquamide B 137,39 as well as four diketopiperazines 138–140 (ref. 40) and 141.41
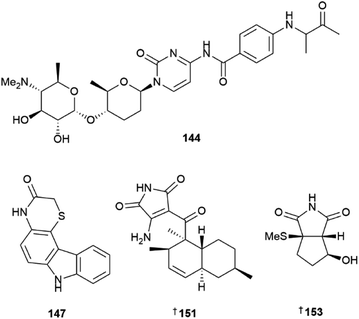
Three pyrimidine nucleosides streptcytosine P 142, and cytosaminomycins F 143, and G 144 were reported from a sediment-derived strain of Streptomyces sp.42 Cytosaminomycin G 144, with an amine instead of an amide bond between the p-aminobenzoic acid and the terminal side chain, represents the first such structural modification within the group. A sediment-derived Streptomyces sp. yielded two pyrazine alkaloids, actinopolymorphols E 145 and F 146 with 146 showing weak antibacterial activity against Kocuria rhizophilia.43 Four carbazoles, thiocarbazomycins A 147 and B 148, chlocarbazomycin E 149, and brocarbazomycin A 150 were reported from a coral-associated Streptomyces diacarni.44 Compounds 147 and 148 possess a rare thiomorpholinone group.
Microbial co-culture of two actinomycete strains, Streptomyces sp. and Achromobacter sp. derived from the gut microbiota of the isopod Ligia exotica, led to the identification of two 3-amino-1H-pyrrole-2,5-dione-containing alkaloids, ligiamycins A 151 and B 152.45 While the Streptomyces sp. strain was found to be the producer of the NPs, the yield of 151 was increased 24 times when co-cultured with Achromobacter sp., demonstrating the importance of using ecologically relevant microbial interactions when designing co-culture experiments. A methylsulfide substituted cyclopenta[c]pyrrole-1,3-dione bacillimide 153 and a pyrrole-carboxamide containing bacillapyrrole 154 were reported from a sediment-derived S. bacillaris.46 Other Streptomyces-sourced alkaloids included the strepotindoles A–D 155–158,47 streptocarbazoles F–H 159–161,48 penzonemycins A 162 and B 163 and demethylmycemycin A 164,49 as well as an indole, streptoprenylindole D 165, co-isolated with a diterpene, 15-hydroxycyclooctatin 166.50 Piercidins A5 167 and G1 168 were isolated from a sediment-derived Streptomyces sp.51 and two pyrrolosesquiterpenes, glaciapyrroles D 169 and E 170 were reported from a deep-sea sediment-derived Streptomyces sp.52 An ethyl acetate extract of the culture broth of two Streptomyces sp. strains yielded three flavonoid-like glycosides, actinoflavosides B–D 171–173.53 While their structures are similar to plant-sourced flavonoids, they are distinguished by additional alkylation at C-5 and the rare ristosamine amino sugar moiety. Two trehalose lipids, tsukalipids A 174 and B 175 were reported from a sediment-derived Tsukamurella pseudospumae.54
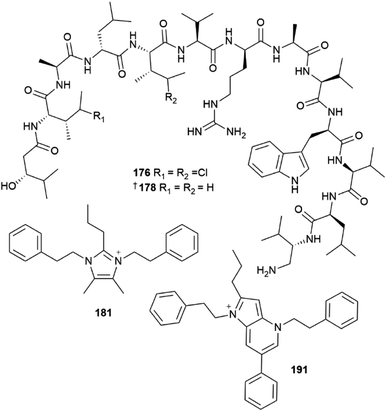
The phylum Bacteroidetes yielded 24 new MNPs in 2022. A sponge-derived Aquimarina sp. contained five antibacterial peptides, aquimarins A–E 176–180.55 These MNPs bear an unusual amino group at the C-terminus and isoleucine residues chlorinated at the γ-position for 176 and 177. Full structural assignment of 176–180, was carried out using conventional spectroscopic and spectrometric methods and the confirmation of the structures of 178 and 179 was complemented via total synthesis. The structures of three other, minor analogues, aquimarins F–H were only proposed based on the analysis of mass spectral fragmentation data and their structures are not included. Aquimarins A–D showed activity against a panel of Gram-positive bacteria. They most potently inhibited M. tuberculosis growth with IC50 values of 45 and 23 nM for 176 and 177 respectively. Fourteen bacteria-sourced, imidazolium-containing MNPs were reported in 2022. This is a remarkable statistic since prior to 2022, the positively charged 1,3-difunctionalised imidazolium structural motif had only been reported once in MNPs.2 A red alga-derived Tenacibaculum discolor yielded eight imidazolium alkaloids discolins A–H 181–188, together with two 1,2,3,5-alkylated pyridinium structures dispyridine A 189 and dispyridine 190, 1H-pyrrolo[3,2-b]pyridinium alkaloids dispyrrolopyridines A 191 and B 192, and the dispyrrole 193.56,57 Notably, dispyrrolopyridine A 191 showed potent antibacterial activity, moderate antifungal activity, and weak nematocidal activity. Hydroxamate-containing siderophores, tenacibactins K–M 194–196 were isolated from a coral-derived Tenacibaculum sp., with 196 showing moderate activity against rat embryonic fibroblasts and the P388 murine leukemia cell line.58
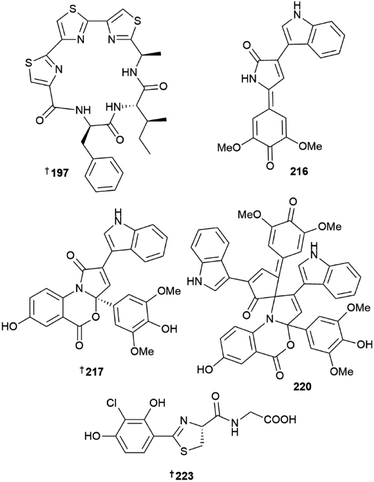
The phylum Fimicutes yielded 18 new MNPs in 2022. Eight tristhiazole-containing cyclic peptides, bathiapeptides A–G 197–204 were reported from a biofilm-derived Bacillus sp.59 A putative NRPS-encoding BGC bat was proposed to be responsible for the biosynthesis of the compounds. The cyclic hexapeptides 197–201 showed weak to moderate antiproliferative activity against four HTCLs, whereas the pentacyclic 202 and the two linear bathiapeptides 203 and 204 were inactive. Six additional imidazolium alkaloids bacillimidazole A–F 205–210 were reported from a sponge-associated Bacillus sp.,60 and a new glycosylated indole alkaloid pityriacitrin D 211 was isolated from B. siamensis.54 Other MNPs reported from the phylum Firmicutes included bacillamide F 212, a non-ribosomal peptide isolated from a sponge-derived B. atrophaeus,61 and (±)-bacillipyrrole A 213 and bacillipyrazine A 214 sourced from a Mariana Trench sediment-derived B. subtilis.62
The phylum Pseudomonadota yielded 22 new MNPs in 2022. Eight novel, 3-pyrrolin-2-one-containing alkaloids, sinacidins A 215, and B 216, sinartyptins A–C 217–219, and racemic mixtures of sinamicins A–C 220–222 were identified from the α-proteobacterium Phaeobacter inhibens.63 Structural assignment of 216, as well as 221 and 222 which were obtained in low yield and analysed as a mixture, was aided by microcrystal electron diffraction (microED). At the time, this was only the second report of this technique being used for the structural elucidation of a new NP. P. inhibens was grown in a sea-salt-containing medium enriched with tryptophan and sinapic acid, common microalgal metabolites present in the natural setting where these bacteria are found. The authors were able to demonstrate biosynthetic incorporation of isotopically-labelled tryptophan into the new bacterial NPs, and showed 215–217, and 220 to have weak algacidal activity against Emiliania huxleyi, suggesting they have a defensive role in the producing organism. Teredinibactin 223 and dechloroteredinibactin A 224 were isolated from a shipworm associated bacterium Teredinibacter turnerae.64 Dechloroteredinibactin A 223 represents the first instance of halogen incorporation in a phenolate-thiazoline siderophore and was shown to form complexes with copper, iron, and molybdenum in aqueous solution.
Other proteobacteria-sourced MNPs included eight new butanolides, deoxyenhygrolides C–J 225–232 isolated from the myxobacterium Plesiocystis pacifica,65 marinoquinolones A 233 and B 234 and marinobactoic acid 235 reported from a coral-derived Marinobacterium sp.,66 and a lipopolysaccharide 236 from a deep-sea collection of Idiomarina zobellii.67
As with previous years, a small number of MNPs published in the literature from marine bacteria did not have adequate spectrometric and spectroscopic data to support the proposed structures.68–71 Two structural revisions of bacterial MNPs were reported in 2022; the absolute configurations of the cyclic peptides ogipeptin A 237 and tumescenamide A 238 were corrected via total synthesis.72,73 Other total syntheses of bacterial NPs included cyanogramides A and C,74 (±)-nesteretal A,75 lucentamycin A together with barmumycin, oxotomaymycin and oxoprothracarcin,76 rakicidin F,77,78 anthracimycin and anthracimycin B,79 xiamycins C–F,80 seongsanamide E,81 salimabromide,82 (+)-nocardioazine B,83 the indolocarbazole alkaloid ZHD-0501,84 (±)-spiroindimicins A, D, G and H,85 bahamaolide A,86 aqabamycin G,87 and elmonin and pratenone A.88 Reviews focused on marine bacterial NPs published during 2022 included publications on the biosynthesis of microbial terpenoids,89 polyketides and non-ribosomal peptides,90 ribosomal peptides,91 and the anthracyclines.92 A comprehensive review of the biologically active NPs from the genus Micromonospora together with their mode of action, biosynthetic pathways and chemical syntheses was summarised,93 as was the potential of marine sponges as sources of diverse bacteria.94
Interestingly, most bacterial MNPs reported in 2022 were identified as a result of either genome- or metabolomics-guided studies of single bacterial strains. While some papers followed the bioassay-guided isolation paradigm, surprisingly few reported new NPs that were responsible for the observed extract bioactivity. Overall, sediment followed by marine invertebrate collections were the primary sources of new MNP producing bacterial strains. Unique ecological niches such as the deep sea,5,6,8,67 Mariana Trench,62 hydrothermal vents,20,21 and the gut microbiota of the Antarctic emperor penguin9 were also explored. In 2022, the biosynthetic potential of the open ocean microbiome was assessed by analysing more than 1000 seawater samples which resulted in the identification of over 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 putative BGCs, most of which were new,95 demonstrating there are many new niches and opportunities for new bacterial MNPs to be explored.
000 putative BGCs, most of which were new,95 demonstrating there are many new niches and opportunities for new bacterial MNPs to be explored.
2.2 Cyanobacteria
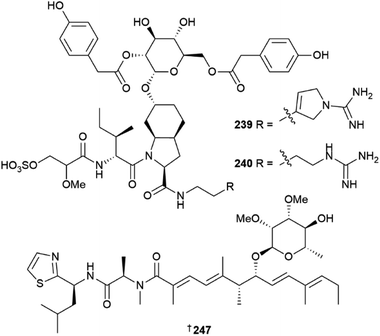
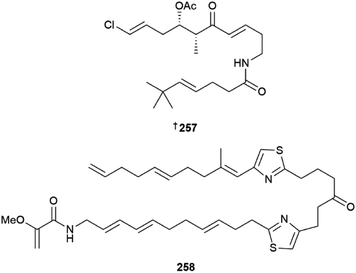
Thirty new cyanobacterial NP structures were reported in 2022, with the majority belonging to either the peptide or a mixed PKS/NRPS biosynthetic origin. Two aeruginosin-type linear peptides, varlaxins 1046A 239 and 1022A 240 were isolated from Nostoc sp.96 The compounds were shown to be potent inhibitors of porcine and human trypsins, with 239 one of the most potent cyanobacterial trypsin inhibitors found. Notably, the 1-amidino-3-(2-aminoethyl)-3-pyrroline amino acid moiety in 239 was shown to enhance activity and selectivity in three human trypsin isoforms over a hundred-fold compared to 240 that contained a 4-amidinobutylamide moiety. Other new cyanobacterial-sourced peptides included the linear peptides amantamide B 241 from Oscillatoria sp., identified via a MS/MS-based molecular networking approach,97 acetylene-containing odookeanynes A 242 and B 243 from Okeania sp.,98 as well as the cyclic depsipeptides triproamide 244 and pemukainalides A 245 and B 246 isolated from Symploca hydnoides.99A novel peptide-polyketide hybrid NP, iezoside 247, was reported from the marine benthic species Leptochromothrix valpauliae.100 Structurally novel features of 247 include an odd number of carbon atoms along the polyketide backbone, a β-branched methyl group at C-17, and a 2,3-O-dimethyl-α-L-rhamnose substitution positioned between an α,β,γ,δ-unsaturated amide and a conjugated diene group. Confirmation of the absolute configuration of 247 containing eight stereogenic centres, was achieved via total synthesis in over 16 steps and in a 4.4% overall yield. The compound showed potent antiproliferative activity against HeLa cells (IC50 of 6.8 nM) and caused spindle-type morphological changes and cell cycle delay, which led to it being identified as an inhibitor of the sarco/endoplasmic reticulum Ca2+-ATPase membrane protein. Luquilloamides A–G 248–254 of a mixed PKS/NRPS biosynthetic origin were reported from Oscillatoria sp.,101 while a novel strain of Hormoscilla sp. yielded anaenamides C 255 and D 256.102
The acyclic polyketide beru'amide 257 was isolated from Okeania sp.103 Configurational assignment was achieved via total synthesis, and the compound showed moderate activity against Trypanosoma brucei rhodesiense (IC50 1.2 μM) and weak activity against the HeLa cell line. Caldorazole 258, reported from Caldora sp., is a new polyketide with two thiazole rings and an O-methylenolpyruvamide end terminus. It showed potent antiproliferative activity towards three HTCLs and was found to inhibit mitochondrial electron transport.104 Heterologous expression of the columbamide BGC from Moorena bouilloni in Anabena yielded several known columbamide NPs as well as five chlorinated acyl amide analogues, including columbamide K 259.105 The gene cluster for this PKS/NRPS hybrid pathway is 28 kb in length and was not well tolerated in the host, limiting the isolated yields of the new NPs. Due to low mass of compounds available, complete NMR structural characterization was only achieved for columbamide K 259 while columbamides I, J, L and M were only characterised by high resolution MS, mass spectral fragmentation and 1H NMR data, so their structures are not included here. An azirine-containing NP, dysidazirine carboxylic acid 260 was isolated from a novel Caldora sp.106 Three eudesmane-type sesquiterpenes 261–263 were isolated from media extracts of Scytonema sp.107 While these sesquiterpene NPs are typically reported from fungi, no attempt was made to show the presence of the compounds in the cyanobacterial cell mass.
Reviews focused on marine cyanobacteria included a comprehensive review of the distribution, NP chemistry, and ecology of Moorena producens,108 and the anti-infective potential of the NPs reported from this genus.109 Cytotoxic, antiproliferative and antineoplastic activities110 and anti-inflammatory, antioxidant, antimicrobial, antiviral and anticancer activities of cyanobacterial NPs were also reviewed.111 In addition, the use of cyanobacteria and their toxins against fungal and oomycete phytopathogens was reviewed.112
Total syntheses of bastimolides A and B,113,114 and the 10-aza-9-oxakalkitoxin analogue of kalkitoxin115 were reported in 2022. Other notable work on cyanobacterial NPs included a report on the biosynthesis of the carbon skeleton of nocuolin,116 synthesis of coibamide A mimetics with improved cellular bioactivity,117 and an evaluation of antitrypanosomal activity of gallinamide and analogues.118
2.3 Marine-sourced fungi (excluding from mangroves)
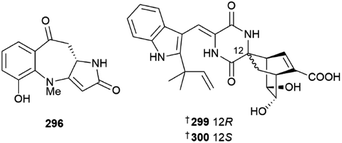
Cultures of Acremonium species led to isolation of sorbicillinoid derivatives 264–266,119 steroid 267,120 chlorinated orsellinic aldehyde derivatives 268–270 and brominated orsellinic acid 271, the last of which is a known synthetic compound but new NP.121Albifimbria verrucaria was the source of a modified γ-lactone 272 (ref. 122) and the genus Alternaria yielded a range of metabolites including the meroterpenoids tricycloalternarenes O–R, 273–276,123 dibenzo-α-pyrone derivatives 277–279,124 sulfated dibenzopyrones 280 and 281,125 phomalone derivatives 282–284 (ref. 126) and perylenequinone derivatives 285–287.127 Picoline-derived meroterpenoids, amphichoterpenoids D 288 and E 289 (C-10 epimers)128 and α-pyrones 290 and 291 (ref. 129) were obtained from Amphichorda felina, the last two via genome mining and heterologous expression and an Athrinium species yielded carboxamides 292 and 293 and polyketide 294.130As has been the case in previous years, the genus Aspergillus was overwhelmingly the most common source of fungal metabolites this year. A gorgonian-derived strain of Aspergillus candidus was the source of the pyrrolinone-fused benzoazepine alkaloids asperazepanones A 295 and B 296. Asperazepanone A 295 was first isolated as a racemate but further studies indicated that only (+)-295 was a NP. Total syntheses of both metabolites were achieved by employing an intramolecular Friedel–Crafts reaction and 296 exhibited potent LPS-induced expression of both TNF-α and IL-6.131 Co-culture of A. candidus and Beauveria felina yielded drimane sesquiterpenes 297 and 298 that were mixtures of stereoisomers.132 Chevalinulins A 299 and B 300, alkaloids with an unprecedented spiro[bicyclo[2.2.2]octane-diketopiperazine] skeleton, were isolated from A. chevalieri sourced from deep-sea cold-seep sediment,133 whilst another deep-sea-derived strain of A. chevalieri yielded indole diketopiperazine alkaloids 301 and 302.134 Other deep-sea-derived Aspergillus strains yielded indole alkaloids 303 and 304 (ref. 135) and polyketides 305,135306 and 307.136 Cytochalasins 308 and 309 were isolated from a culture of A. flavipes, although the possibility that they may be artefacts of isolation could not be excluded137 and A. flavus strains yielded triterpene 310 (ref. 138) and α-cyclopiazonic acid alkaloid 311.139
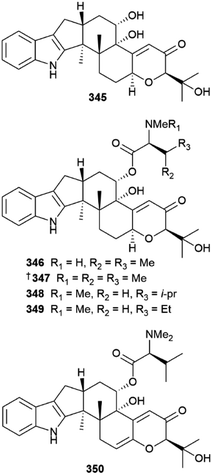
N-Methylated cyclic peptides, asperflomide 312 and asperflosamide 313 were isolated from A. flocculosus140 and A. fumigatus strains were the source of a steroid 314, a 2-oxofuranone derivative 315, indole alkaloids 316 and 317 (as an inseparable mixture), 318, pseurotin A derivative 319,141 alkaloids 320–324, and penibenzophenone E 325.142 Gorgonian-derived Aspergillus hiratsukae strains yielded α-pyrone meroterpenoids 326–331,143 cyclic peptide 332, ecdysteroid derivative 333 and sesquiterpene lactone 334,144 and nitrobenzoyl sesquiterpenoids 335–338,145 cyclohexadepsipeptides 339, 340 (ref. 146) and sesquiterpenoids 341, 342–344 (ref. 147) were obtained from sponge-derived Aspergillus strains. Isolation of indole diterpene amino acid conjugates 345–350 from A. noonimiae suggested that the corresponding BGC in the fungus contains an NRPS-like modifying enzyme able to incorporate multiple lipophilic amino acids.148
Culture of Aspergillus ochraceopetaliformis yielded polyketide 351 (ref. 149) and cyclic tripeptide 352,150 whilst co-cultivation of deep sea-derived A. ochraeus with a terrestrial soil-derived Penicillium yielded prenylated indole alkaloids 353 and 354.151Aspergillus strains derived from deep-sea sediment yielded a variety of metabolites including phenols 355, 356 (C-8 epimers), 357, 358,152 aromatic polyketides 359–361 and isoquinoline alkaloids 362, 363,153364–377,154 acremolin alkaloid 378,155 and cyclopropane acids 379–382.156 Sponge-derived strains were the source of 2,5-diketopiperazines 383–385,157 isocoumarin 386, propylpyridinium anthraquinone 387, resorcinol derivative 388,158 and dipyrroloquinones 389, 390,159 whilst notoamide alkaloids 391–396,160 prenylated notoamide-type alkaloids 397–406,161 butenolides 407–411 and p-hydroxybenzaldehyde derivative 412 (ref. 162) were obtained from gorgonian-derived strains. Pulvinone derivatives, aspulvinones S–V 413–416 were isolated from green alga-derived A. terreus,163 a coral-derived strain of A. unguis was the source of polyketides 417 and 418, (the latter a known synthetic compound and terrestrial metabolite but a new MNP).164 A shrimp-derived A. unguis strain yielded ergostane-type sterols 419–422,165 nitrogenous metabolites variotin B 423 and coniosulfide E 424,166 and phenolic polyketides 425, 426,167 whilst various seawater-derived Aspergillus strains yielded phenolic polyketide 427 (ref. 167) aromatic bisabolene sesquiterpenoids 428–431 and benzaldehyde derivative 432 (the last a known synthetic compound but new NP)168 and cyclohexapeptides 433–436.169 Other Aspergillus strains derived from sediment yielded nucleoside derivatives 437, 438,170 austocystin analogues 439, 440,171 diketopiperazines 441–444 (ref. 172) and indole diketopiperazine alkaloids 445–448,173 and gorgonian-derived strains were the source of indole diketopiperazine hybrids 449–452,174 benzodipyrans comprising eurotiumide G enantiomers 453, 454 (configurations revised to 1S,3R,4R and 1R,3S,4S respectively), 455–462 (all isolated as racemates but separated by chiral HPLC),175 asperbenzophenone A 463 and versicolamide C 464.176
Re-profiling of a fungal library derived from the gastrointestinal tract of commercially sourced mullet by various means including miniaturised 24-well plate cultivation (MATRIX), chemical profiling and precursor directed biosynthesis, identified an Aspergillus strain as a chrysosporazine producer and led to the isolation of chrysosporazines T, U, T1 and U1, 465–468.177 Other Aspergillus strains yielded phenolic glucosides 469, 470,178 dipeptides 471, 472,179 the unusual steroid-sorbicillinoid adduct 473,180 the unsaturated fatty acids, pantheric acids D–F 474–476,181p-terphenyl derivatives 477–481 (ref. 182) and depsidone 482.183
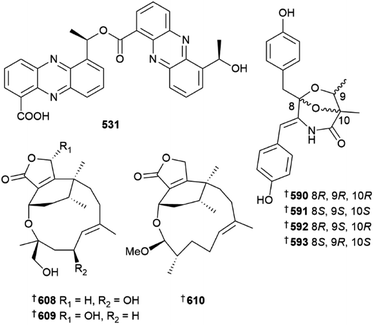
Destruxin hexadepsipeptides 483 and 484 were isolated from a culture of Beauveria felina184 and Byssochlamys spectabilis was the source of terpenoids 485–490 and polyketides 491–493, the last a known synthetic but new NP.185 A precursor-directed biosynthetic approach to culturing of a Chrysosporium species led to isolation of a range of metabolites. Supplementation with a range of precursors yielded neochrysosporazines A–L 494–505, chrysosporazines R 506 and S 507, neochrysosporazines M–Q 508–512 and the known terrestrial but new MNP, hancockiamide C 513. A SAR study of the new analogues indicated key structural requirements for reversal of P-glycoprotein efflux mediated doxorubicin resistance in a HTCL.186Cladosporium strains derived from deep-sea sediment were the sources of indole derivatives 514 and 515,187 sulfur and peroxy bridged macrolide 516 and iodinated dimeric naphtho-γ-pyrone 517 (ref. 188) and oxygenated fatty acids, seco-patulolides 518–520.189 Co-culture of a Cosmospora species with phytopathogenic fungus Magnaporthe oryzae yielded isochromanones 521 and 522, the latter as an inseparable mix of epimers190 and a culture of Curvularia verruculosa was the source of cytochalasin derivatives 523–525, the last a known synthetic compound but new NP.191Cystobasidium laryngis derived from deep-sea sediment yielded the diphenazines phenazostatins E–J 526–531, of which phenazostatin J 531 exhibited potent cytotoxicity to six HTCLs and potent antineuroinflammatory activity via inhibition of NO production whilst phenazostatins E–I were inactive in these assays.192 An ascidian-derived strain of Diaporthe was the source of a range of metabolites including xanthones 532–538,193 monoterpenes 539, 540, (C-3 epimers) 541 and α-pyrone 542.194
A large number of gabosine and/or chlorogentisyl alcohol metabolites 543–561 were obtained from an Epicoccum species,195 polyketides 562, 563, (C-14 epimers) and 564 were isolated from Eutypella scoparia196 and a tetrahydrocarbazol-1-one analogue, exophilone 565 was obtained from Exophiala oligosperma.197Fusarium strains yielded indole alkaloids 566–570,198 diketopiperazines 571, 572, polyketides 573–575 and isochromanone 576 (ref. 199) and prenylated glycine derivatives 577 and 578.200 The macrolides halosmysins B 579 and C 580 were isolated from a Halosphaeriaceae strain,201 cerebrosides 581 and 582 were obtained from Hortaea werneckii202 and Lecanicillium fusisporum was the source of 3-acyl tetramic acid derivatives 583–589.203 Culture of Leptosphaerulina chartarum derived from deep-sea sediment yielded the enantiomeric hydroxybenzyl dimers leptochartamides A 590/591 and B 592/593, which contain a dioxa-azabicyclo[3.2.1]octane core. Total synthesis of each enantiomeric pair was achieved in nine steps.204 Polyketides 594 and 595 and dendrodochol B derivatives 596 and 597 were isolated from a culture of Lopadostoma puzarii,205Metarhizium strains yielded a N-butenone spiroquinazoline alkaloid 598,206 macrolides 599, 600 (as an equilibrating mixture of C-2 epimers), 601 and aromatic glycosides 602 and 603,207 and supplementation of a Monascus albidus culture with amino acids led to isolation of γ-lactams; enantiomers 604/605 and C-3 epimers 606/607.208
Neocucurbins A–C, 608–610, phomactins featuring a polyoxygenated-hetero 5/6/12 or 5/6/13 fused tricyclic ring system were isolated from a strain of Neocucurbitaria unguis-hominis derived from deep-sea sediment, along with their open chain derivatives neocucurbins D–G 611–614 which contain a 5/6 fused bicyclic ring system.209 The same N. unguis-hominis strain also yielded neocucurbols A–H, 615–622, phomactin diterpene derivatives with either a 6/6/5/5/6 polycyclic ring system (615–618) or a 6/8/6 tricyclic ring system (619–622).210 Genome mining identified a BGC from Neosartorya pseudofischeri and determined that it encodes for the biosynthesis of pseudofisnin A 623, a 1-benzazepine-containing compound. The biosynthetic pathway was elucidated through in vivo and in vitro experiments.211 Culture of Ochroconis humicola resulted in isolation of N-(2-phenylacetyl)benzamide 624, a known synthetic compound but new NP212 and culture of various Paraconiothyrium strains yielded bergamotane sesquiterpenoid derivatives 625–629,213630–637 (ref. 214) and polyketide 638.215
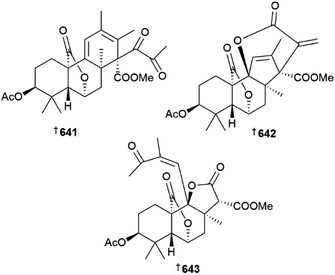
The Penicillium genus has again been extensively studied as a source of new metabolites. Culture of Penicillium aculeatum yielded sulfonyl metabolites 639 and 640 (ref. 216) and P. antarcticum was the source of meroantarctines A–C 641–643, meroterpenoids with unique 6/5/6/6, 6/5/6/5/6 and 6/5/6/5 skeletons respectively, all of which exhibited moderate inhibition of P-glycoprotein and resensitisation of resistant cancer cells to docetaxel.217
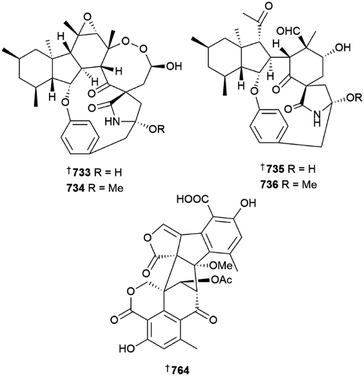
Further meroterpenoids were obtained from P. chermesinum (644–647)218 and P. chrysogenum (648–650),219 (the latter group via mutation with diethyl sulfate) and indole diterpenoids, paspalines C–D 651, 652 and paxillines B–D 653–655,220N-acetyl-D-glucosamine derivatives 656, 657,221 citreoviridins J–O 658–663,222 pentacyclic alkaloid 664,223 dipeptide 665 (ref. 224) and eremophilane-type sesquiterpenes 666–677 (ref. 225) were obtained from various other Penicillium strains. Heterologous expression of a nonreducing polyketide synthase (NR-PKS) gene from P. crustosum yielded 3-orsellinoxypropanoic acid 678 and indicated the role of the gene as an orsellinic acid (OA) synthase and a transferase,226 the 13-membered macrolides cyclopiumolides A 679 and B 680 were obtained from a P. cyclopium culture.227P. oxalicum cultures were the source of polyketide-amino acid hybrids 681 and 682 (ref. 228) and polyketides 683 and 684,229 while azaphilones 685 and 686 were isolated from a P. sclerotiorum strain.230 A deep-sea coral-derived strain of P. steckii yielded a number of tanzawaic acid derivatives 687–696 (ref. 231) and polyketides 697–704.232 A culture of P. sumatraense resulted in the isolation of 3-hydroxybutyric acid and glycolic acid derivatives 705–709, (706 a known synthetic but new NP)233 and cultures of various other Penicillium strains yielded diketopiperazine alkaloid 710 and polyketides 711 and 712,234 further tanzawaic acids 713–726 (719 and 720 known terrestrial fungal metabolites but new MNPs),235 citrinin dimers 727–730 (ref. 236) and meroterpenes 731, and 732.237 The decahydrofluorene alkaloids pyrrospirones K–Q 733–739 were isolated from a soft coral-derived Penicillium strain and of these, 733 and 734 possessed a 6/5/6/8/5/6/13 polycyclic skeleton whilst 735 and 736 possessed a 6/5/6/5/6/13 polycyclic skeleton.238 Co-culture of two deep-sea-derived Penicillium strains resulted in isolation of sesquiterpenes 740 and 741 from one strain and organic acid 742 from the other239 and other sediment-derived Penicillium strains were the source of meroterpenoid 743 (ref. 240) and alkaloids 744, 745,240746, 747.241
Polyketide derivatives 748–752, were isolated from a Pestalotiopsis strain242 and tyrosine derivative 753 and terezine derivatives 754 and 755 were obtained from Phoma herbarum.243Pseudopithomyces maydicus was the source of aromatic polyketides 756–760 (ref. 244) and a strain of Pyrenochaetopsis yielded pyrenosetin C (for which the configuration was revised to 761) and related compounds 762 and 763.245 Talaverrucin A 764, a heterodimeric oxaphenalenone with a unique 6/6/6/5/5/5/6 fused ring system, was isolated from an Antarctic sponge-derived strain of Talaromyces.246 Other Talaromyces strains yielded a diverse range of metabolites including thioester-containing benzoate derivatives 765–770,247 sulfur containing spiro alkaloid 771, unusual diacid 772 and alkaloids 773–777,248 aromatic polyketides 778–782,249 tripeptides 783 and 784, containing an N-trans-cinnamoyl group,250 chlorinated unsaturated alcohol 785 and coumarin derivative 786,251 decalin derivatives 787 and 788,252 meroterpenoid 789 (ref. 253) and oligophenalenone 790 and 791 and xanthoradone 792 dimers.254 Culture of a Trametes strain yielded spiromeroterpenoids 793 and 794 (ref. 255) and cultures of various Trichoderma strains were the source of trichothecanes 795–798,256 α-pyrone 799 and decalin 800,257 sesquiterpene glycoside 801 and sorbicillinoid glycosides 802 and 803,258 β-carboline alkaloids 804–807 (805 and 806, known synthetics but new NPs),259 and polyketides 808, 809,260810–814.261
A machine learning-augmented density functional theory method (DU8ML) was utilised to revise the stereochemistry of tersone E to 815, which is identical to the terrestrial fungal metabolite citridone A.3 Structural revision of penipacids A–E, originally isolated from Penicillium paneum, from amidines to hydrazones 816–820 through total synthesis also raised the possibility that they may be Schiff base adduct artefacts produced through conjugation with a putative NP precursor, the hydrazine N-aminoanthranilic acid with diacetone alcohol under extraction conditions.262 Total synthesis of trichomide D also resulted in revision of the stereochemistry to 821.263
A convergent synthetic strategy was utilised in the total synthesis of aspergillolide264 and a biomimetic route was employed in the synthesis of asperfloketal A which also provided evidence for the biosynthetic hypothesis proposed.265 Asymmetric total synthesis of asnovolins A and E utilised anionic fragment coupling to construct the sterically crowded skeleton266 and solid peptide synthesis was employed to assemble the cyclic pentapeptides, versicotides E and F.267 Indole diterpenoids (+)-shearinines G and D were prepared via a convergent route.268 The C-2 reverse prenyltransferase, NotF was characterised and it was then used in the first synthesis of eurotiumin A,269 synthesis of (−)-eurothiocin A was achieved in 14 linear steps from commercially available starting materials,270 the triazole penipanoid A was prepared from 4-methoxyphenyl acetic acid271 and total synthesis of the 13-membered macrolide (−)-melearoride A was achieved via a route including a Julia–Kocienski olefination.272 Synthesis of the polyketide penicyclone A was accomplished in ten steps using a double Grignard reaction,273 synthesis of raistrickindole A was achieved via two approaches to the diketopiperaine subunit,274 a divergent strategy was employed in the synthesis of polyketides heterocornol A and B275 and an Ir(III)-catalysed alkylation of acetophenone in aqueous medium was utilised in the total synthesis of cytosporones A and C.276
The cyclodepsipeptides, scopularides A and B displayed potent larvicidal activity against mosquito (Culex pipiens) larvae,277 alkaloids epi-aszonalenin A and aszonalenin were shown to inhibit angiogenesis by inhibition of inflammation and apoptosis278 and cyclodepsipeptide isaridin E was shown to downregulate the PI3K/Akt signalling pathway and thus possesses antiplatelet and antithrombotic effects.279 The alkaloid meleagrin had a protective effect in mice against pulmonary fibrosis induced by bleomycin280 and epidithiodiketopiperazine N-methylpretrichodermamide B displayed potent inhibition of P-glycoprotein and was able to resensitise drug resistant cells to docetaxel.281 Indolyl alkaloid oxaline and anthraquinone isorhodoptilometrin exerted weak anti-neuroinflammatory effects on murine cell lines282 and the polyketide citrinin exhibited weak anti-parasitic effects against Trichomonas vaginalis.283
Whole genome sequencing of a sponge-derived strain of Aspergillus niger indicated the presence of 69 BGCs for a wide array of secondary metabolites, highlighting the biosynthetic potential of the strain.284 Refactoring transcription factors in the biosynthetic pathway to the A. terreus metabolite terrein greatly enhanced production in mutant strains.285 Density Functional Theory (DFT) calculations were utilised in study of the asperterpenol/preasperterpenoid biosynthetic pathways in Aspergillus and led to proposal of a reaction cascade for construction of the molecular core which is consistent with experimental observations.286 The biosynthetic pathway to the homodimer phomoxanthone A from a Diaporthe fungus was elucidated and indicated that a cytochrome P450 enzyme was implicated in regioselective oxidative coupling to produce the dimer.287 Elucidation of the biosynthesis of the cyclase spiromaterpene A from a Spiromastix strain involved use of heterologous expression, chemical characterisation and incubation experiments.288
Ion mobility can be a useful tool to separate and distinguish coeluting molecules by mass spectrometry and was successfully employed in the case of the aphidicolane diterpenoids.289 There were many reviews on marine fungi and topics pertinent to them produced in 2022. A few of note include one on bioactive metabolites from extremophilic marine fungi290 and another on bioactive metabolites from deep-sea-derived strains,291 one on One Strain Many Compounds (OSMAC) and epigenetic approaches to cryptic metabolites from marine organisms which cited many fungal examples,292 a review on the use of fungal–fungal co-culture to generate chemical diversity293 and one covering halogenated metabolites from marine fungi with pharmacological activity.294
2.4 Fungi from mangroves
Fungi isolated from mangrove roots and their surrounding sediments continue to be a source of derivatives of commonly encountered NP structure classes associated with terrestrial fungi.295 None of the 47 new mangrove fungi-derived MNPs reported across 13 papers in 2022 contain novel ring systems and only three of the 45 compounds tested showed biological activity above the threshold criteria adopted in this review. Those MNPs that were isolated as single enantiomers (34) had their absolute configurations determined through DFT computational comparisons with experimentally determined ECD data 87% of the time.Drimane sesquiterpenes 822–825 were reported from a sediment-derived Aspergillus sp.296 and a butyrolactone 826 was isolated from Aspergillus terreus.297Daldinia eschscholtzii was the source of a simple macrocyclic ether, eschscholin B 827, benzoic acid derivatives dalditones A 828 and B 829, and naphthalene derivatives 830 and 831.298 Sorbicinol derivatives 832, 833, 834 and two dimeric sorbicinols 835 and 836 were isolated from sediment-derived Hypocrea jecorina.299 Sediment-derived Nigrospora camelliae-sinensis yielded two simple diketopiperazines nigrosporaamides A 837 and B 838.300 A thiodiketopiperazine, adametizine C 839 and alkane derivatives 840–844 were reported from sediment-derived Penicillium ludwigii. Adametizine C showed weak anti-inflammatory activity.301 A simple α-pyran 845 and tetrahydrofuran 846 were sourced from Penicillium polonicum,302 while sesquiterpenes containing either a benzofuran (citreobenzofurans D–F 847–849) or tetrahydronaphthalenone moiety (phomenones A 850 and B 851), were found in a sediment-derived Penicllium sp. collected in Wenchang, China.303 Phomenone B was the only compound possessing an epoxide and it showed moderate antibacterial activity towards Bacillus subtilis but was inactive towards four other bacterial strains. None of the related compounds showed any antibacterial activity. The roots of the Chinese mangrove Xylocarpus granatum yielded a Penicillium sp. endophyte that contained two isocoumarins penicimarins L 852 and M 853 but neither compound showed antioxidant activity.304 Two prenylated indole diketopiperazines, penicilamides A 854 and B 855, and three simple polyketides penicinones A–C 856–858, were reported from a sediment-derived Penicillium sp. The NPs were screened for cytotoxic activity and only penicinone A had weak activity towards one HTCL.305 Two cyclodecadepsipeptides, phaeosphamides A 859 and B 860 from Phaeosphaeriopsis sp. growing in the rhizosphere of the Chinese mangrove Bruguiera gymnorrhiza were tested for cytotoxicity against five HTCLs but only 859 weakly arrested AGS cells in the G2 phase through induction of apoptosis.306Phomopsis asparagi obtained from the roots of the red mangrove, Rhizophora mangle contained cytochalasin derivatives, phomoparagins A–C 861–863,307 while Talaromyces sp. contained talarobenzofurans A–C 864, 865, 866 that are either thioester or carboxylic acid derivatives of a dihydrobenofuran, and two α-pyrones, talaropyrones A 867, and B 868. None of these MNPs possessed antibacterial or α-glucosidase activity.308
2.5 Dinoflagellates
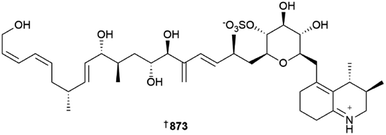
The number of new compounds reported from dinoflagellates and other microalgae plummeted in 2022; the number of newly reported MNPs (only six) was approximately half that of the average for the preceding nine years.1 As is the norm for dinoflagellates, polyketide-derived compounds remain the main biosynthetic class of metabolites reported. Voratins A–C 869–871 are spiro-cyclic compounds from Effrenium voratum, a symbiotic dinoflagellate associated with the coral Alveopora japonica.309 while an anti-fungal amphidinol congener 872 was reported from an Irish isolate of Amphidinium carteri.310Ovataline 873 is a linear carbohydrate-bearing polyketide isolated from cultures of a Korean strain of Ostreopsis ovata, the absolute stereo-structure of which was assigned using standard methodologies. Encouragingly, 873 is non-toxic against brine shrimp and human RWPE-1 epithelial prostate cells, but it moderately suppresses RWPE-1 type II 5α-reductase involved in benign prostatic hyperplasia, making it a useful lead for prostate cancer therapies. Ovataline is also the first non-toxic compound reported from strains of O. ovata that also produce the potent toxins palytoxin-like ovatoxins and ostreols.311
The final new compound reported from dinoflagellates was a saxitoxin congener 874 from Alexandrium pacifichem;312 the structure of a maitotoxin congener was only partially characterised and proposed using MS and NMR data and consequently is not shown here.313
The synthesis of the structures proposed for iriomoteolide-1a and 1b, and three possible diastereomers, has been achieved yet none of the spectroscopic data for the synthetic materials matches that reported for the natural isolates, hence the structures of these MNPs require revision.314 Amphidinolide C2 has been synthesised for the first time.315
Notable dinoflagellate-related reviews published in 2022 include a summary of the taxonomy, distribution, genetics and toxin content of Gambierdiscus spp.,316 a compilation of microalgal compounds that influence β-amyloid and τ-protein formation and hence may have application in Alzheimer's disease treatments,317 and an assessment of the methodology used to establish toxicity and regulatory limits of algal biotoxins, along with the implications of climate change on production levels of these important environmental metabolites.318
A study has shown not all brevetoxin metabolites (BTXs) bind to voltage-gated sodium channels, the commonly accepted mechanism of action for these biotoxins. Rather, aldehyde-containing BTXs are more immunotoxic, with the conclusion from the study being that different BTX pharmacophores can elicit different toxicity profiles.319 Another study has evaluated the levels of paralytic shellfish toxins (PSTs; in saxitoxin equivalents) in Arctic food webs during the anomalously warm 2019 period. Notably, high levels of PSTs in benthic clams and the Pacific walruses who predate upon them were detected, at levels high enough to cause observable physiological effects if consumed by humans in comparable amounts.320 GNPS has been used to profile the chemical diversity of MNPs present in Gambierdiscus spp. from the Philippines, indicating a high level of biosynthetic potential in the strains studied,321 while a new LCMS-based method has been developed to detect PSTs, including C-11 hydroxyl (“M”-class) and benzoate (“GC”-class) saxitoxin analogues that current methods do not pick up.322
A stable isotope labelling study using 13CO2 has indicated that a polyketide pathway is involved in polyunsaturated fatty acid biosynthesis in Tisochrysis lutea, with remarkably fast production of the fatty acids meaning 13C incorporation into early components of the pathway is undetectable.323 The anaerobic transformation of domoic acid (Pseudonitzschia spp.) by a consortium of benthic microbes (Dalian, China) was profiled, indicating the role other microorganisms could play in the fate and effects of this important biotoxin in marine environments.324 In a separate study, the biotic and abiotic factors that control toxin production in Ostreopsis spp. were examined. Both the ovatoxins and the liguriatoxins were rapidly degraded following exposure to sunlight or other bacteria, which calls into question their effective role in presentation of dermatitis and toxic inhalation following blooms of Ostreopsis.325
Found in marine sediments, 4-methyl steranes are “molecular fossils” that have been used to unravel eukaryotic evolutionary history. An accepted paradigm in steroidal biosynthesis is that 4β-methyl steranes are formed exclusively from isomerism of their 4α-stereoisomers within sedimentary deposits. On the other hand, 4α-methyl steranes are believed to be formed from oxidative demethylation of a triterpenoid precursor. A study of methyl sterane biosynthesis in the coral endosymbiont Breviolum minutum has uncovered a methyltransferase that can produce both 4α- and 4β-methyl steranes, as opposed to the sterol methyl oxidases that were assumed to produce the previously known demethylated compounds. This mechanism of formation of 4-methyl steranes is present not only in dinoflagellates but also in higher eukaryotes of various phyla, identifying a previously unknown source of these biomarkers and could provide further insight into our understanding of the evolution of eukaryotic life on earth.326
3 Green algae
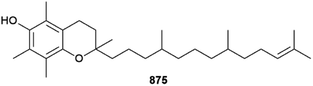
Although the Chlorophyta (green algae) are always a minor contributor to the number of new MNPs each year,1 2022 was exceptional in that only one new compound was reported. 11′α-Tocomonoenol 875 is a common terrestrial plant metabolite, but its presence in green alga Tetraselmis sp. means it has been detected from the marine environment for the first time.327Other structures were claimed but they are inconsistent with the spectroscopic data provided and are not shown here.328,329
A review of the biotechnological application of edible green alga Ulva lactuca has been published.330 A combined MS-based metabolomics and bioactivity profiling approach using supervised discriminatory statistical methods has been used to highlight various fatty acids, terpenoids and phenolics as the main bioactive components within Ulva fasciata and Spirulina platensis from the Mediterranean Sea.331
The lipidome of a plant is an important element in the organism's resilience to environmental stresses. Betaine lipid diacyglycerl-N,N,N-trimethylhomoserine (DGTS) is exclusively found in green algae and non-flowering plants. A study of the effects of phosphorus deficiency upon the green alga Chlorella kessleri showed that various zwitterionic phospholipids were degraded to release cellular phosphorus, while DGTS synthesis was promoted at the same time. This remodelling of the lipidome under these conditions to replace phosphorus-containing lipids with an equivalent non-phosphorus containing zwitterionic metabolite can help explain how lower plants respond to conditions of phosphorus deficiency as an acclimation strategy.332
4 Brown algae
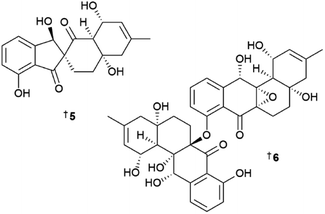
The number of new compounds reported from the Ochrophyta (brown algae) in 2022 was the fewest by a significant margin, being roughly one third of the rolling average over a ten-year period.1 Somewhat surprisingly, all the newly reported compounds were terpenoid in origin. One meroterpenoid 876 was reported from a Japanese Sargassum.333 A confusing situation has arisen relating to naming of several nor-diterpenoid compounds. A butenolide 877, named sargassumin A, was reported from a South Korean (Jeju Island) Sargassum micracanthum; this manuscript was submitted in mid-2021 and accepted for publication in December of that year.334 The name sargassumin A was then used for a different lactone 878 along with sargassumin B 879, from a Jeju Island Sargassum macrocarpum, published by a different group with their manuscript submitted and accepted well after the former paper was published.335 Butenolide sargassumin C 880 was subsequently reported from S. micracanthum and is related to 877,336 although this publication shows the authors were obviously aware of the others publication and therefore they did not use the name sargassumin B. Given the timing precedent, the S. macrocarpum metabolites should be given a different trivial name, although this may create more confusion in the field than it solves. Two sterols 881 and 882 were also reported from an Australian (Tasmania) Cystophora xiphocarpa.337 Several other structures were claimed but are inconsistent with the spectroscopic data provided and their structures are not shown here.338,339Only one first total synthesis of a brown algal MNP was reported in 2022, that of carotenoid loroxanthin.340 Reviews focusing on brown algae and their metabolites published in 2022 include summaries of the anticancer activity of various eckols and fucoxanthin, respectively,341,342 phlorotannins as sedative aids for promoting sleep,343 and catalogues of the compounds found from species Ecklonia stolonifera,344 and the genera Dictyota and Dictyopteris, respectively.345,346
An in silico study of various brown algal phlorotannins as SARS-COV-2 protease inhibitors identified several lead compounds, which were then validated as weak to moderate inhibitors using in vitro bioassays.347 One study has examined the effects of temperature and light on lipid production in two different Streblonema algae, with the concentrations of various polar glycerol-derived metabolites varying significantly following exposure to low or high light intensities.348 The structure of a tetraprenylated chromane has been revised to 883 following in-depth J-based configurational analysis coupled with comparison of experimental and calculated chiro-optical properties.349 A seasonal study of the volatilome of both fresh and dried Croatian Dictyota dichotomahas indicated that the main volatile components are short chain lipids or are diterpenoid in origin, with distinct variation throughout the months of the study facilitating statistical differentiation of the sample treatments.350
5 Red algae
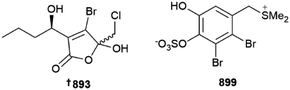
Acetogenins are common metabolites of red algae, with nine C15 variants being reported from investigations of various Laurencia species from Japan 884 (Katsura)351 and Greece 885–892, (Tinos Island),352 respectively. The tongalides 893–898 are halogenated butenolides isolated from an Antarctic (Bonaparte Point) Delisea sp. Although all six isolates were inactive against the ESKAPE panel of clinically relevant microbial pathogens, they are related to the co-isolated known compound Z-acetoxyfimbrolide A, which was responsible for the antibacterial activity of the algal extract, showing the importance of the exocyclic alkene functionality for bioactivity.353 An Irish Vertebrata lanosa was the source of six brominated amino acid derivatives 899–904 along with a glycolipid 905; 899 is the first algal compound to incorporate a dimethylsulfonium moiety on a phenolic ring.354 Other structures were put forward although these were inconsistent with the spectroscopic data provided and are not shown here.355Several first total syntheses of red algal metabolites were published in 2022, including those of C15 acetogenins dehydroitomanallene B,356Z- 906 and E-ocellenyne 907, which also established their absolute configurations,357 sesquiterpenoid laurencenone C358 and thysiferol-type triterpenoid saiyacenol A.359
A review of the metabolites from the possibly invasive genus Acanthophora and of Asparagopsis, respectively, have been published,360,361 as has a summary of red algal compounds with skin whitening cosmeceutical potential.362
The role of carotenoids in the adaptation of the intertidal red alga Neopyropia yezoensis towards daily desiccation has been investigated. Upregulation of carotenoid biosynthesis was observed once the thalli of the alga reached the critical 60% dehydration point, presumably to act as antioxidants against damage caused in the air.363
Domoic acid is an important diatom-derived neurotoxin, although it was originally discovered from the red alga Chondria armata. Analysis of the C. armata BGC responsible for domoic acid production showed that the rad genes responsible for its production are organised in a similar fashion to those found in the diatom Pseudonitzschia multiseries, including the presence of a key cytochrome P450 that is absent in other red algal BGCs that encode for the related, but chemically simpler, neurotoxin kainic acid. Detailed investigation of the domoic acid biosynthesis in C. armata suggests a slightly different route to its formation when compared to that found in the diatom. Overall, this study has shown that domoic acid production in C. armata incorporates aspects of algal and microbial biosynthesis and hints at a complex evolutionary history.364 A timely review of the biosynthesis of domoic and kainic acids have also been published.365
6 Sponges
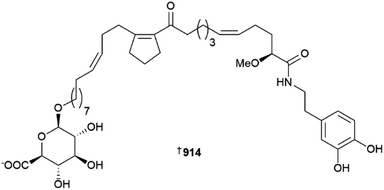
Various glycerolipids and butenolides have been isolated from Pericharax heteroraphis 908, 909 and Scopalina hapalia910–913, respectively; the synthesis of the Pericharax metabolites was also achieved.366,367 The toporosides A–D 914–917 are unusual cyclopentene-containing fatty acid amides from a deep-sea (dredged at 476–519 m) Stelodoryx toporoki. Three of the isolates prevent TNF-α-induced damage to rat cardiac cells at non-toxic concentrations; based upon SAR analysis, the C-11 carbonyl is important for cardio–protective activity.368 A series of alkynynols 918–934 and polyacetylenes 935–937 have been reported from Cribrochalina and Petrosia sponges, respectively,369,370 while plakilactone J 938 is an ethyl-branched polyketide γ-lactone and its absolute configuration was determined using VCD data.371As always, many peptidic MNPs were reported from sponges in 2022. Such isolates include subarmigeride A 939 from an Indonesian Suberea,372 various friomaramides 940, 941 and shagamides 942–947 from an Antarctic Inflatella,373 ectyoplasin 948 from a Mexican Ectyoplasia,374 cyclopsammocinamide A 949 and B 950 which are of the opposite enantiomeric series to known cyclocinamide A,375 and a new aciculitin 951 from a Poecillastra dredged from a Japanese sea-mount.376 Two separate samples of the prolific genus Theonella yielded three theonellapeptolides 952–954 and theonellamide/theopalauamides 955–957, respectively.377,378
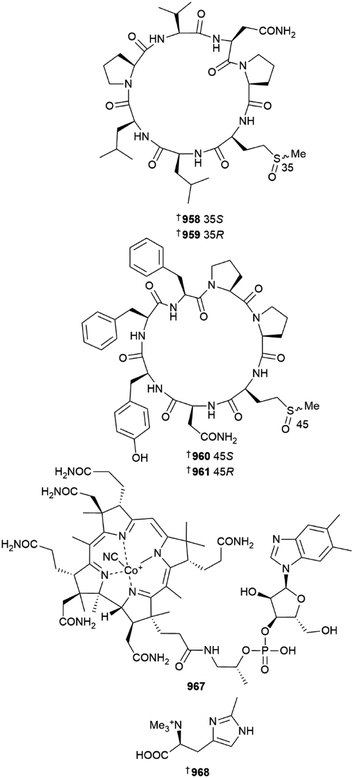
A Chinese (Yongxing Island) Axinella was the source of four peptides named axinellasin A–D 958–961, found as two pairs of diastereomeric methionine oxides. The isolation of the peptides was facilitated by use of precursor ion-directed supercritical fluid chromatography targeting sulfoxide-containing metabolites. All four compounds were inactive against five HTCLs but were active as immunosuppressive agents at 10 μM via inhibition of B and T cell proliferation. The total synthesis of axinellasin D was also achieved.379
Three enigmazole congeners 962–964 were reported from a Homophymia sponge, one of which (enigmazole E) is an artefact of isolation; this was also the only report of a new sponge-derived macrolide in 2022.380 Two diketopiperazines 965 and 966 were reported from a Haliclona baeri specimen.381
A known terrestrial vitamin B12 derivative, cyanocobalamine 967, was isolated from a deep-sea (809 m) Characella pachastrelloides dredged from an Irish submarine canyon. This species has previously been the source of unique anti-inflammatory glycolipopeptides. Trace amounts of B12 were detected by LCMS in extracts of the sponge. In addition, an unusual and weakly cytotoxic amino acid, 6-methylhercynine 968 was isolated during the same study.382
Sponge-derived alkaloids include haliclorensin D 969 and two haliclonacyclamines 970 and 971 from a Neopetrosia and a Callyspongia, respectively,383,384 while related zamamiphidin alkaloids 972 and 973 were isolated from a Japanese Amphimedon sponge.385 An N-oxide congener of manzamine A 974 has been isolated from Neopetrosia proxima,386 while N. chaliniformis yielded four indole-C-mannopyranosides, neopetrosins A–D 975–978, most of which have weak hepatoprotective activity.383
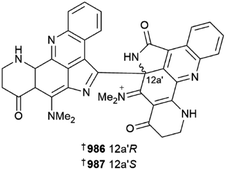
Various aaptamines 979–985 have been reported from two separate collections of Aaptos aaptos.387,388 A racemic mixture of an aromatic alkaloid, plakoramine A 986/987 was isolated from a Plakortis sponge (Kingdom of Tonga). Chiral separation of the enantiomers provided both metabolites, which are products of photochemical dimerisation of co-isolated and commonly found metabolite plakinidine B. Both enantiomers are weak inhibitors of Casitas B-lineage lymphoma proto-oncogene-b (CLB-B), but are inactive in the NCI 60-cell line panel, while surprisingly monomer plakinidine B is potently cytotoxic vs. the NCI panel but inactive against CLB-B. The ubiquitin ligase activity of CLB-B requires its dimerisation, hence it would be interesting to investigate the differential SAR of plakoramine and plakinidine B in this context.389
Pyrrolo-amide and -lactam alkaloids are commonly encountered in sponge extracts. Reports of sponge alkaloids of the class include a structural revision of mukanadin C 988 (Agelas nakamurai),390 alongside cyclic 989–1010 and linear monomers and dimers 1011–1020 reported from Axinella, Stylissa and Agelas sponges.391–393 Guanine-containing metabolites included 1021–1023, and additionally 1024–1031 that are the first metabolites reported from genus Ernsta.394–396 The sponge genera Suberea and Pseudoceratina yielded eleven bromotyrosines 1032–1042 between them.397,398
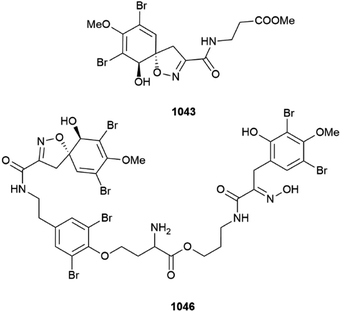
A Western Australian (Ningaloo Reef) specimen of Pseudoceratina verrucosa was the source of several bromotyrosines, methyl purpurocerate A 1043 and B 1044, possible artefacts of methanolic isolation from known purpuroceratic acids, along with purpuroceratic acid C 1045 and bromotyrosine trimer nialamide A 1046 and dimer B 1047. A new method for determining the enantiomeric purity (% ee) of chiral bromotyrosines using a combination of chiral chromatography with ECD detection was established. The % ee of all the new isolates varied from almost full racemic mixtures (∼5%ee) to almost enantiomeric purity (∼84% ee).399
Unsurprisingly, most sponge-derived metabolites reported in 2022 were of terpenoid origin. Merosesquiterpenoids were isolated from sponges of genus Dactylospongia1048–1051, Dysidea1052–1055, and Spongia1056–1058, respectively.400,401 Note 1057, named pelorol A, is a methylated congener of known pelorol, and so logically should be termed pelorol B.402 Meroditerpenoid alkaloids 1059–1062 were discovered from a Mexican Agelas citrina sponge, with some showing antimicrobial properties.403
Various sesquiterpenoid structures were disclosed in 2022. The use of computational-aided structure elucidation facilitated the assignment of agelasidine G–I 1063–1065 from a specimen of Agelas nakamurai,390 which was also the source of related sulfone cyclohexagelasidine A 1066 reported in a separate study.404 A series of monomeric and dimeric bisabolene-class sesquiterpenoids, bubaridin A–F 1067–1073 and several theonellin congeners 1074–1076, were obtained from a Futuna Island sponge of order Bubarida.405 Other sesquiterpenoids were reported from Acanthella (1077–1081) and Myrmekioderma (1082–1084) sponges, respectively.406,407 Isolation of a number of nitrogenous sesquiterpenoids 1085–1104 from a Chinese Acanthella cavernosa also required revision of two pairs of known compounds as 1105–1108, respectively.408
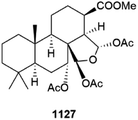
Diterpenoid metabolites have been reported from specimens of dictyoceratid Spongia from China 1109–1121 and from Saudi Arabia 1122, respectively.409–411 A Japanese Halichondria yielded aromatic peroxide amitorin 1123 that exhibits weak cytotoxicity,412 while a different Saudi specimen of Spongia was the source of three spongian diterpenoids 1124–1126 that were inactive in various assays.413 Spongionellol A 1127 is a further spongian diterpenoid from a dredged (Gulf of Sakhalin) Spongionella which exhibits weak to moderate cytotoxicity vs. seven HTCLs, in particular vs. prostate cancer cells, but was less active against non-cancerous lines with a mean selectivity index of 5.2. The mechanism of action of spongionellol A involves caspase-dependent apoptosis. In addition, 1127 inhibits drug efflux by P-glycoprotein multi-drug resistance protein-1 (MDR-1), a main detoxification effector in cancer cells that helps them avoid the effects of standard chemotherapeutics, making spongionellol an exciting anti-cancer lead compound.414
Marine sponges remain one of the main reservoirs of new sesterterpenoids reported annually. A Hippospongia fistulosa specimen was the source of four linear furospinulosin-type sesterterpenoids 1128–1131, while two unstable ircinianin butenolides 1132 and 1133 were obtained from an Australian (Great Barrier reef) Ircinia.415,416Phorbas sponges from Howe Sound, British Columbia, continue to yield ansellone-class metabolites, including 1134–1138, which have activity as HIV-1 latency reversal agents.417 Isomalabaricane sesterterpenoids were reported from Rhabdastrella globostellata from China 1139–1147 and Vietnam 1148–1150, respectively,418,419 while scalaranes were found from specimens of Phyllospongia1151–1154, Hyrtios1155–1162 and Spongia1163–1174; the Phyllospongia metabolites were already known but their structures revised in the current study.420–422 New sterols were reported from Halichondria 1175, Neopetrosia 1176, 1177, and Spongia 1178, 1179; the latter study also reported a fatty acid 1180.411,423,424 Other manuscripts reported a number of new metabolites that were either not isolated,425 or the structures presented are inconsistent with the spectroscopic data provided and hence are dubious in nature and not shown as part of this review.426–430
Numerous first total syntheses of sponge metabolites were published in 2022. These include the syntheses of ethyl branched polyketides plakortone Q and plakdiepoxide,431 and peptides stylissatin B,432 cyclotheonellazole A,433 and stylopeptide II, the last of which was prepared synthetically on the gram scale.434 Hemicalide is an extremely potent (sub nM IC50) cytotoxin, although only the planar structure was originally published within a patent application. It contains 21 stereocentres with over two million possible stereoisomers. The potent bioactivity and structural complexity of hemicalide has prompted many syntheses of parts of the compound to try and establish the stereochemistry of the molecule, the latest of which has refined the structure as 1181, leaving only eight possible stereoisomers for the compound.435 Two reviews of the struggle to identify this compound were also published in 2022.436,437
Five bromophenol NPs were synthesised for the first time,438 as was pyridinium alkaloid epi-tetrahydrohalicyclamine B.439 The structure of echinosulfonic acid D has been synthesised, one year after it was revised independently by three groups as reported in last year's review.1,440 Aaptamine-type alkaloids suberitines A–D were synthesised for the first time,441 as were pyrrole-containing metabolites denigrin D and E.442 Somewhat surprising given its age, the first (racemic) synthesis of archetypical bromotyrosine compound psammaplysin A, which was first reported in 1982, was accomplished in 2022,443 as was the synthesis of related spiroisoxazolines clavatadine D and E.444 The first total syntheses of merosesquiterpenoid dysiherbol C and D,445 dactyloquinone A,446 and amino-sesquiterpenoids boneratamide A–C,447 and two halichonic acids were reported.448 Both the relative and absolute configurations of lamellodysidine A were confirmed following its synthesis.449 Use of computer-assisted retro-synthetic analysis of the pupukeanane class of sesquiterpenoid MNPs identified several routes with unusual disconnections beyond those already precedented for manufacture of the compounds.450 Diterpenoids ansellone G and hamigeran F,451,452 eight phorbaketal congeners,453 aromatic sterol myrmenaphthol A, and seco-sterols glaciasterol B and 6-ketoaplidiasterol B have been synthesised for the first time.454,455 Complex tropolone-containing triterpenoid gukulenin B has also been prepared.456
Reviews of MNPs from class Demospongiae,457 the genera Agelas,458Oceanapia,459Smenospongia,460 and the species Dactylospongia elegans have been published,461 as have summaries of the development of the bengamides as antibiotics,462 the quintessential alkaloid aaptamine,463 and the scalaranes, a common class of sponge sesterterpenoid.464
High-throughput screening (HTS) of 3764 marine-derived fractions has identified macrolide leiodolide A as a potent anti-Cryptosporidium agent with high selectivity for the parasite over mammalian cell lines.465 The cytotoxic mode of action of polyketide psymberin (a.k.a. irciniastatin A) was determined to be via inhibition of protein translation leading to upregulation of the p38 cellular stress response.466 Manzamine A inhibits Sine oculus homeobox homolog 1 (SIX1) gene expression, which is highly expressed in osteoblasts and is involved in craniofacial development; this famous alkaloid may therefore have importance for development of new promoters of bone formation.467 The marine alkaloid thiaplakortone B was found to suppress RANKL-induced osteoclast formation, and therefore may have value in reducing bone-reabsorption and hence be a lead for treatment of osteoporosis.468 Promiscuous alkaloid aaptamine is a dual inhibitor of both acetyl- and butyryl-cholinesterase and therefore has potential as an Alzheimer's disease therapeutic lead.469 HTS of 300 marine invertebrate extracts from Australia has revealed that the known guanidine-based, plant growth promoter asterubine binds to α-synuclein, therefore making it a lead for Parkinson's disease; asterubine structurally resembles the β-amyloid aggregation inhibitors drugs tramiprosate and ALZ-801.470 The same HTS campaign also identified (+)-aerothionin-1 and -2 as α-synuclein inhibitors; interestingly both inhibit protein aggregation but while (+)-aerothionin-1 is toxic to dopaminergic neuronal cells, −2 is not.471 Bromotyrosine aerophobin-1 shows low toxicity in a zebrafish model of development but promoted osteogenesis at 100 nM so may have application in promoting bone regeneration.472
A report has described a virtual structure-based approach to highlight likely bioactive bromotyrosine metabolites. Following the in silico portion of the study, 12 alkaloids were then isolated using a molecular networking-guided approach from a Pseudoceratina durissima sponge with one (aeroplysinin-1) showing antibiotic activity, five showing toxicity to zebrafish and potentially having effects on the central nervous system, and four inhibiting the parasitic nematode Haemonchus contortus. This study highlights the value of using in silico predictive models with modern chemical screening methodologies to identify new lead compounds.473 In other reports, the mode of anti-colorectal cancer action of merosesquiterpenoid ilimaquinone is via mitochondrial-mediated apoptosis, caused by activation of caspase-9/-3 and the subsequent DNA damage caused,474 while spongian diterpenoid membranolide G irreversibly inhibits relaxation of supercoiled DNA via inhibition of topoisomerase 1B and it may therefore be a new anti-cancer lead compound.475 The biosynthesis of macrolide lasonolide A (Forcepia sp.) has been accomplished using a metagenomic approach, suggesting the potent (nM) cytotoxin is made by the first member of new bacterial genus Candidatus Thermopylae lasonolidus, whose nearest taxonomic neighbour is Pedosphaera parvula.476
Use of the DU8ML approach has resulted in the reassignment of two compounds from the Red Sea sponge Negombata magnifica as the common and known metabolites epi-loliolide and loliolide; the structures of the “new” compounds magnificines A and B were not shown in our previous review as they were not consistent with the spectroscopic data provided, and the revision shows that the structures were misassigned due to incorrect interpretation of MS data leading to erroneous molecular formulae being proposed.1 In addition, the structure of cinerol A was also revised to 1182 as part of the same study; the amidine unit in the polycyclic aromatic system is “flipped” in the revision.3
A chemical ecological study of the Caribbean sponge Agelas tubulata has shown that specimens across multiple sites and depths (down to 61 m) in both Belize and the Grand Cayman Islands, including reciprocal transplanted samples, possessed different secondary metabolic profiles driven mainly by nine compounds, and using regression analysis that various metabolites showed targeted antibacterial activity against different pathogenic strains. The results of this study examine the disease resistance and resilience of sponges within tropical reef systems.477
Sponges are notorious for being difficult to taxonomically classify. A study to investigate isomalabaricane triterpenoids from the sponge genera Jaspis, Geodia, Stelletta and Rhabdastrella provided an opportunity to re-examine voucher specimens of the sponge sources of the compounds. Of the 21 vouchers re-examined, which came from 27 unique sponge sources described in 44 articles, over 50% (11) were mis-identified; all of the sponges described as members of the Jaspis and Stelletta genera were in fact Rhabdastrella globostellata specimens. The authors of the study recommend stricter guidelines for identification of sponges and also for the storage of voucher specimens.478
7 Cnidarians
Two landmark publications have described for the first time the presence of BGCs for terpene biosynthesis in soft corals. In one of the studies,479 sequencing of the metatranscriptome of Erythropodium caribaeorum, a source of eunicellane and briarane diterpenes, led to the identification of eight putative terpene synthase (TPS) genes. The majority of these genes were considered of coral origin, with the sequences found in all published octocoral genomes. The eleutherobin genes were clustered with the genes of tailoring enzymes including cytochrome P450 and acyltransferases. Expression in E. coli afforded soluble protein, that when incubated with geranylgeranyl pyrophosphate afforded klysimplexin R. Similar methodology identified TPS associated with the biosynthesis of cembrene C, the prototypical precursor of cembranoid MNPs. The second study480 utilised an untargeted genome mining approach, querying publicly available genomes, to identify a set of 14 genes that harbored motifs typical of plant and microbial terpene cyclases. Expression in E. coli followed by precursor incubation studies identified enzymes producing cembrene A, cembrene C, elisabethatriene, a previously unreported xeniaphyllene scaffold and klysimplexin R, with four producing mixtures of sesquiterpenes. The cembrene A producing enzyme ErTC-2 from Eleutherobia rubra, was successfully crystallised. In a further demonstration of the utility of this newfound knowledge of soft coral terpene cyclase genes, the genome of Capnella imbricata was sequenced and queried, identifying a terpene cyclase homologue that was expressed and functionally validated as being a capnellane terpene cyclase. Both groups speculated on the timing of the origin of terpenes in soft corals, noting that they appear to coincide with the evolutionary divergence of hard (stony) corals and soft corals, supporting the premise that these MNPs represent a means of chemical defence.To explore the pharmaceutical potential of a set of 5600 cnidarian NPs, a systematic approach was used to calculate and compare descriptors of chemical space, drug-likeness and predicted toxicity and to virtually screen the compound set against a panel of cancer-relevant protein targets.481 The most favourable metabolites spanned 210–265 Da, were often sesquiterpenes and possessed moderate complexity. The structure of butanolide 1183 from Cladiella conifera has been previously reported as a synthetic compound.482 It exhibited weak ability to inhibit the release of COX-2 from stimulated macrophage cells. A deep-sea collection of the soft coral Duva florida afforded the meroterpene tuaimenal A 1184.483In silico docking experiments suggested it could bind to SARS-CoV-2 3CLpro, with a biochemical assay identifying it as a very weak inhibitor of the enzyme, with an IC50 of 21 μM. A set of eight diacylated zoanthoxanthin derivatives, dentithecamides A–H 1185–1192, were isolated from the hydroid Dentitheca habereri.484 The biosynthetic origin of the alkaloids could be Parazoanthus sp. zoanthids that the hydroid associates with. While not all compounds were tested, dentithecamide A was the most active (IC50 12 μM) at suppressing PAX3-FOXO1-driven transcription, a driver of soft tissue rhabdomyosarcoma in young people. While this level of activity is considered inactive by the criteria set for this review, the value is presented here to indicate that dentithecamide A is in fact an inhibitor, albeit weakly, of a transcription factor fusion which some view as undruggable. The zoanthamine alkaloid norzoanthamine demonstrates anti-osteoporotic activity in an in vivo model.485
Three norsesquiterpenes pathyspirolactone A 1193 and B 1194 and napalilactone 1195 were isolated from cultured specimens of Paralemnalia thyrsoides.486 The structure and relative configuration proposed for pathyspirolactone A is identical to that reported for a synthetic compound epi-pathylactone A. Notable differences in chemical shift and coupling constants for the H-1 methine resonance between the two compounds were attributed to the unlikely scenario that the NP was a new conformational isomer that did not interconvert when heated. Absolute configuration was assigned to known co-metabolite napalilactone using XRD. Sesquiterpene alkaloid echinoflorine 1196 and lactones echinofloranolide A–C 1197–1199 were isolated from a South China Sea collection of Echinogorgia flora487 and their absolute configurations were assigned by standard methods. Four acetylated sesquiterpene lipids, crannenol A–D 1200–1203 were reported from an Irish deep-sea collection of Acanella arbuscula.488 In addition to the related acetal and methylacetal analogues, parathyrsoidin K 1204 was isolated from a Malaysian collection of a soft coral from the genus Lemnelia.489 Three rearranged sesquiterpenes lemnalemnane A–C 1205–1207, were isolated from Paralemnalia thyrsoides and Lemnalia sp.490 In a bioassay using transgenic zebrafish, lemnalemnanes A and B reduced the number of macrophages surrounding the neuromast, while lemnalemnane C exhibited strong angiogenesis effects. In addition to a highly oxidised sterol (alcyosterone discussed later), three illudane sesquiterpenes, alcyopterosins T–V 1208–1210 were purified from extracts of an Antarctic collection of Alcyonium sp.491 Although inactive against a panel of ESKAPE microorganisms, alcyopterosin V was weakly active against Clostridium difficile (MIC 8.1 μM) and Leishmania donovani (IC50 7.0 μM) with low toxicity towards HEK293T and HepG2 cells and J774.A1 macrophages. Two studies have reported new examples of nardosinane-type sesquiterpenoids in addition to unsymmetrical bis-sesquiterpenoids. In the first study, investigation of the constituents of Xisha Is. collections of Lemnalia sp. afforded parathyrsoidins H–J 1211–1213, linardosinenes D 1214 and E 1215 and nardosinoids A 1216 and B 1217.492 Bioactivity of the compounds ranged from weak to inactive against a variety of viruses, bacteria, and tumour cell lines. In the second study, specimens of Litophyton nigrum, also from the Xisha Is. region of the South China Sea yielded, in addition to several known MNPs, bis-sesquiterpenoid linardosinene H 1218 and the nardosinane sesquiterpene linardosinene I 1219.493 Structure confirmation and assignment of absolute configuration of the previously reported sesquiterpene 1220 was achieved by XRD analysis. Of the three compounds, only linardosinene I exhibited biological activity, being a very weak inhibitor of PTP1B, while all compounds were inactive towards a panel of four HTCLs. Subergorgines A–E 1221–1225 are C-6/N-linked suberosanone-purine hybrids reported from extracts of Subergorgia suberosa.494 Defining the head-to-head and head-to-tail orientations, respectively, of the indanone-dimers weizhouochrones A 1226 and B 1227, isolated from Anthogorgia ochracea, required analysis of an extensive set of experimental data including NMR residual dipolar couplings, residual chemical shift anisotropy, computational chemistry including DP4+ probability analysis and computer-assisted 3D structure elucidation.495 In the case of DP4+ analysis, the study provided more evidence that successful assignment of relative configuration using this workflow requires the use of both 1H and 13C NMR chemical shift data. Weizhouochrone A was racemic, while the optical purity status of the co-metabolite could not be determined due to its instability. In addition to seven xenicane-type diterpenoids described later, an extract of Sinularia hirta afforded norcaryophyllenes sinuhirtins F 1228 and G 1229.496 The structure of oxyfungiformin 1230, originally isolated from Capnella fungiformis, was confirmed by total synthesis, with the study also establishing its absolute configuration.497 A biomimetic synthetic approach was used to define the structure and absolute configuration of the disesquiterpene rumphellolide J 1231.498 The target was prepared by ester formation between two building blocks of (+)-rumphellaoic acid A, starting from caryophyllene oxide, and 4β,8β-epoxycaryophyllan-5-ol, derived from (−)-caryophyllene.
A new example of a prenylelemane-type diterpene, lobatate 1232, was isolated from the soft coral Sinularia nanolobata collected from Ximao Is., South China Sea.499 A total of nine diterpenes were purified from extracts of Clavularia inflata comprised of five dolabellanes clavularinlides A–E 1233–1237 and four racemic elemane alkaloids, clavulacylides A–D each of which were separated by chiral HPLC prior to characterisation.500 The structures of clavulacylides A and C were assigned as being 1238–1241, bearing a trans C-1 Me/C-6 H relationship. The congeners clavulacylides B and D were originally proposed to bear a cis Me/H relationship, based upon the claimed observation of NOESY correlations. A subsequent study however, by Novitskiy and Kutateladze3 that made use of machine learning DU8+ hybrid density functional theory calculations suggested that the structures were more likely to be the trans substituted stereoisomers 1242–1245. Closer examination of the ESI of the original isolation report failed to indicate the presence of the claimed NOESY correlations, leading to the conclusion that the structures of clavulacylides B and D should indeed be revised to those shown. A Xisha Is. collection of Clavularia viridis was the source of 12 examples of dolabellane diterpenoids, including clavurols A 1246 and B 1247, peroxide-containing clavuperoxylides A 1248 and B 1249, clavurols C–E 1250–1253, furan-ring containing clavufuranolides A–C 1254–1256 and epoxide clavirolide I 1257.501 In addition to confirming the structure and assigning the absolute configuration of clavurol E and clavirolide I by the use of XRD, absolute configuration was also established for the known co-metabolite clavudiol A 1258.
New examples of capnosane diterpenes, sarboettgerins A–E 1259–1263 were isolated from Weizhou Is., South China Sea collections of Sarcophyton boettgeri.502 XRD was used to confirm the structure and assign absolute configuration to sarboettgerin A. The structurally-related sinuhumilol A 1264, a non-cytotoxic capnosane, was purified from Sinularia humilis collected at Xiamo Is., also in the South China Sea.503 In a relatively rare example, diterpenes belonging to two different bicyclic skeletons, mililatensol A 1265 (sarsolenane) and mililatensols B 1266 and C 1267 (capnosanes), were reported from a collection of Sarcophyton mililatensis.504
Unusual tricyclic diterpenoids sinuaustones A 1268 and B 1269 and a spartane-type diterpenoid isolobophytumin E 1270 were isolated from a Hainan collection of Sinularia australiensis.505 The structures and absolute configurations of both sinuaustones were secured by XRD. Mercury lamp irradiation of a methanolic solution of co-metabolite lobophytumin A afforded a mixture containing the three new NPs, suggesting they either derive from photochemical reactions in nature or could be considered artefacts of isolation. Biselisabethoxanes A 1271 and B 1272 are unusual dimeric diterpenoids, with the former representing amphilectane and elisapterane diterpenes linked by an ether, while the latter appears to be derived from hetero-Diels–Alder cycloaddition between two amphilectane-type diterpenes.506 The structure and absolute configuration of biselisabethoxane A was secured by XRD. Despite being given every opportunity to exhibit biological activity against a diverse panel of targets, including Mycobacterium tuberculosis and Plasmodium falciparum, cytotoxicity, neurodegenerative and neuroinflammation, cyclin-dependent kinases and antiviral assays, no activity was observed for either MNP.
A comprehensive review of the structures, naming, and NMR assignments of pseudopterosin and seco-pseudopterosin has been published, highlighting inconsistencies in the primary literature.507 The introduction of primary amino groups onto Gram-positive only antibiotics of the pseudopterosin aglycone family has been demonstrated to convert the compounds into broad spectrum antibiotics with activity towards Gram-negative bacteria.508 The structure of the intriguing Pseudopterogorgia elisabethae metabolite (+)-aberrarone 1273 has been confirmed by synthesis, with the core 5-5-5 fused ring system constructed using a Au-catalysed cascade.509 The structure of the final NP was further confirmed by XRD. The synthetic material exhibited a relatively strong specific rotation ([α]D +85.0 (c = 0.4, CHCl3)) compared to the isolated NP ([α]D +2.2 (c = 1.4, CHCl3)), suggesting the NP was isolated as either the racemate or a nearly equal scalemic mixture.
Many cembranoid-related diterpenes were reported from cnidarians in 2022. Xishaglaucumins A–J 1274–1283 were isolated from a Xisha Is. South China Sea collection of Sarcophyton glaucum.510 XRD was used to define the structure and absolute configuration of the known co-metabolites sarcophytol Q 1284, iso-sarcophytol Q 1285, and 13-acetoxy-7,8-epoxycembra-1(15),3,11-trien-2,16-olide 1286. Amongst a set of five cembranoids, sarcomililatols C–G 1287–1291, purified from Sarcophyton mililatensis, sarcomililatol D was the only compound to inhibit (weakly) TNF-α.511 XRD analysis of a sixth cembranoid 1292 from the coral led to its structure revision. There are only a few reports of investigation of the NP chemistry of Sarcophyton boettgeri; a new study has led to the characterisation of sarcoboettgerols A–C 1293–1295 and 12-epi-humilisin D 1296 from a Ximao Is. collection.512 Eight cembranoids, sarcophytembranoids A–H 1297–1304, were isolated from a Ximao Island collection of the soft coral Sarcophyton trocheliophorum; the study also reported three terpenoid compounds 1305–1307 as MNPs for the first time.513
Biosynthetically, the metabolites ximaonanolobatin A–I 1308–1316, isolated from Sinularia nanolobata, likely all derive from the common precursor cembrene A.514 The structure of ximaonanolobatin A was secured by XRD. Three terpenoids, comprised of two cembranoids 1317 and 1318 and casbane-type diterpene 1319 were isolated from a Ximao Is. collection of Sinularia pedunculata; all were inactive in a TNF-α release assay and did not inhibit the enzyme PTP1B.515 An unusual 13,15-ether bridged cembranoid, odosinularol 1320, was reported from extractions of a collection of Sinularia sp. collected from the Odo Coast, Okinawa.516 In addition to the venerable furanocembranoid pukalide, a new congener, 11-hydroxy-Δ12(13)-pukalide 1321, also isolated from an Okinawan collection of Sinularia sp.517 No activity was observed against a bacterial phytopathogen and the MNP was inactive towards brine shrimp. A bisepoxy derivative of cembrene A 1322, and two chlorine-containing cembranoids 1323 and 1324 were purified from extracts of Sinularia sp. collected at Turtle Is., Taiwan.518,519 The structures and absolute configurations of the former two compounds were secured by XRD. Furanone-containing cembranoids sarcoconvolutums F 1325 and G 1326, isolated from Sarcophyton convolutum, were proposed to be biosynthetically-derived from sarcophine by a series of steps including oxidation and dehydration.520 Additional examples of furanone cembranoids were also reported, including ehrenbergols F 1327 and G 1328, isolated from a Vietnamese collection Sarcophyton ehrenbergi,521 sarcoeleganolides C–G 1329–1333 from South China Sea samples of S. elegans,522 and sarcoroseolides A–D 1334–1337 from a Red Sea collection of S. roseum.523 Cembranoid pyran 1338 from a Red Sea collection of Sarcophyton trocheliophorum was inactive when tested against a panel of HTCLs and Leishmania major.524 Two studies reported new examples of 8,20-lactonised cembranoids, including cinerenolides A–C 1339–1341 from Sarcophyton cinereum525 and tortuolides A 1342 and B 1343 isolated from S. tortuosum.526 The latter study also described a diterpene epi-sarcophytonolide Q 1344. Nine C19-norcembranoids comprised of sinudenoids A–E 1345–1349, isolated from Sinularia densa,527 and sinuscalides A–D 1350–1353, from S. scabra,528 were isolated from the respective soft corals collected from Xisha Island, South China Sea. The latter study also led to the determination of the structure of the diterpenoid sinuscatone A 1354. Sinuscalide A exhibited weak antiviral activity towards enterovirus 71 and all the new MNPs from S. scabra inhibited RANKL-induced osteoclastogenesis at a single dose of 10 μM.
An unusual tricyclic cembranoid cinerelolide 1355 was reported from a Taiwanese collection of Sarcophyton cinereum.529 Its absolute configuration was assigned by comparison of experimental and calculated specific rotation values. In addition, a weakly cytotoxic sarsolenone analogue 1356 was characterised along with known congeners sarsolenone 1357 and 7-deacetylsarsolenone 1358. Structure elucidation of 1356 prompted re-examination of the latter two MNPs, leading to revision of their relative configurations and assignment of absolute configurations. Seven biscembranoids were reported from either open water collections of Sarcophyton sereni from Xisha Island, or from S. trocheliophorum that was originally collected from the coast of Pingtung, Taiwan but had been subjected to aquaculture. Bistrochelides H–L 1359–1363 were isolated from S. sereni specimens with the structure and absolute configuration of bistrochelide H being secured by XRD.530 Bistrochelides H and J were moderate inhibitors of RANKL-induced osteoclastogenesis.
The remaining two biscembranoids, sarcotrochelides A 1364 and B 1365 were identified using a molecular networking-directed study of extracts of S. trocheliophorum.531 Both biscembranoids were inactive in testing for anti-inflammatory responses related to superoxide anion or elastase release from induced human neutrophils. First syntheses of cembranoid MNPs molestin E,532 (+)-ineleganolide and (−)-sinulochmodin C,533 and scabrolide F 1366 have been reported, with the latter study establishing the absolute configuration of the molecule.534 The structure assigned to scabrolide B has been called into question as the result of a total synthesis of the purported structure.535 An asymmetric total synthesis of havellockate afforded a product that exhibited opposite sign and different magnitude of specific rotation to those reported for the isolated NP, leaving the authors unsure if there was an error in the original measurement or whether they had synthesised the enantiomer of the NP.536 A racemic synthesis of the cembranoid rameswaralide, originally isolated from Sinularia dissecta, has been reported.537 Further investigation of the biological properties of cembranoid MNPs has identified that crassolide induces cell death in human non-small-cell lung cancer cells by reactive oxygen species accumulation leading to cell cycle arrest at G2/M,538 and sinularin is also cytotoxic towards gliobastoma multiforme via a mechanism involving ROS generation leading to mitochondria-mediated apoptosis.539
Details regarding the isolation of new briarane diterpenes from Briareum stechei have been reported over three publications. Specimens grown in aquaculture were the source of congeners briastecholides D–F 1367–1369,540 a Taiwanese collection was the source of briastecholides K 1370 and L 1371 (ref. 541) while Okinawan specimens afforded briastecholides G–J 1372–1375.542 Between them, these studies also led to the confirmation of structure and assignment of absolute configuration by XRD analysis of brianodin A 1376, and excavatolides E 1377, and M 1378. Additional examples of briarane diterpenoids, briavioids A–C 1379–1381 were reported from cultured specimens of Briareum violaceum.543 The structure and relative configuration of the former metabolite was secured by XRD analysis. As noted earlier, for some cembranoid MNPs, briarane excavatolide C also exhibits cytotoxicity, this time towards bladder cancer cell lines, via a mechanism involving modulation of oxidative stress.544 Two publications from the same research group described new xenicane diterpenes, coniferains A–D 1382–1385, isolated from a Taiwanese collection of Cladiella conifera.545,546 The structure of coniferain A matched that reported in 2010 for australin F, prompting reexamination of NMR data leading in turn to reassignment of the structure of the latter to 1386. Litophynols C 1387 and D 1388 were isolated from a Ximao Island collection of Cladiella krempfi.547 Their structures and absolute configurations were established by a combination of single crystal XRD analysis and chemical conversion. Clakrefielin A 1389, also isolated from a Ximao Island collection of Cladiella krempfi, represents a des-butyrate ester variant of australin E.548 Reaction of clakrefielin A with butyrylchloride afforded a product identical to australin E 1390, single crystal XRD analysis of which further confirmed the structure and established the absolute configuration of both NPs. An extract of Sinularia ornata was the source of ximaoornatins A–C 1391–1393.549 The structures likely derive from oxidation and ring-closures of a eunicellin diterpene precursor. Also purified from the extract was litophynin K 1394.
In addition to two norcaryophyllenes noted earlier, Sinularia hirta was also the source of eight xenicane-related diterpenes sinuhirfuranones A–C 1395–1397, sinuhirtins C–E 1398–1400 and sinuhirtones A 1401 and B 1402.496 Absolute configuration was assigned to the known xenicane 13-epi-9-deacetylxenicin 1403 by a combination of XRD analysis and modified Mosher's methodology, and a number of acylated derivatives were prepared semi-synthetically, identifying many derivatives with weak to moderate cytotoxicity towards a panel of four HTCLs.550 All of the examples of a library of lipophilic semisynthetic analogues of the xenicane plumisclerin A were effectively inactive as cytotoxins.551 The synthesis of racemic alcyonolide has been reported using an inverse electron demand hetero-Diels–Alder reaction to construct the core of the NP.552
A series of 12 sterols were reported from cnidarians, including seco-sterol cladiellasterol A 1404 from Cladiella krempfi,548 epoxides 1405 (ref. 553) and 1406 (ref. 554) from Sinularia variabilis and Isis hippuris, respectively, ketones and enones 1407–1410 from Lobophytum pauciflorum,555 alcyosterone 1411 from Alcyonium sp.491 and dienones 1412–1415 from Dendronephthya sp.556 In addition to these new NPs, samples of previously reported MNPs hippuristerones A 1416 and I 1417 were isolated from the I. hippuris specimens as well as nanjiol A 1418 from the Dendronephthya sp. sample and their respective absolute configurations assigned by single crystal XRD analysis. A gram-scale total synthesis of pinnisterol E 1419 from ergosterol has been reported.455
A Kunitz peptide purified from the venom of the sea anemone Anemonia sulcata activates G protein-coupled inward-rectifier potassium channels with only minor effect on voltage-gated Kv1.6 channels.557 Hcr 1b-2, a peptide from Heteractis crispa, exhibits interactions with a diverse set of voltage-gated ion channels including those for potassium, sodium and calcium, making it of interest as a tool to pharmacological studies.558 Analysis of the transcriptomes from six species of clownfish hosting anemones, including Cryptodendrum adhaesivum, Stichodactyla haddoni, Heterodactyla hemprichii, Heteractis crispa, Macrodactyla doreensis and Entacmaea quadricolor identified a large number of toxins with reported hemolytic and hemorrhagic properties.559
8 Bryozoans
Once again in 2022, there were very few papers published on metabolites from bryozoans. An undescribed bryozoan of the genus Calyptotheca was the source of the steroid sulfates 1420 and 1421 (ref. 560) and the β-carboline alkaloids, orthoscuticellines A–E were synthesised via a straightforward route from tryptamine via pavettine (1-vinyl-β-carboline) which co-occurs in the producing organism (Orthoscuticella ventricosa) so is likely also the biosynthetic precursor.5619 Molluscs
A weakly cytotoxic, 20-residue lysine-rich peptide was isolated from the hemolymph of the bivalve mollusc Arca inflata and demonstrated the ability to induce apoptosis in colorectal carcinoma cells by a mechanism involving activation of the p38-MAPK pathway.562In vivo activity towards CRC HT-29 cells in a xenograft model was observed.In a study designed to evaluate the antifeedant/deterrent properties of nudibranchs, extracts of different body parts of 30 species collected in Australian waters were evaluated for antifeedant activities against three generalist fish predators and toxicity towards brine shrimp.563 No clear relationship was observed between palatability and toxicity.
Asymmetric total synthesis has established that the sea hare metabolite aplysiol B 1422 and the red algal metabolite laurenmariannol are in fact identical, with the structure shown.359 Two brominated sesquiterpenes dactylomelanins A 1423 and B 1424 were isolated from a Vietnamese collection of the seahare Aplysia dactylomela.564 The absolute configurations of both molecules were assigned using DP4+ and calculated ECD analysis. Two new halogenated acetogenins 1425 and 1426 were reported from Okinawan collections of Aplysia dactylomela.565 The metabolites contain a terminal enyne-moiety, more commonly contained in MNPs reported from Laurencia species of red algae.
The structures and absolute configurations of azuriaplysins A 1427 and B 1428, isolated from the sea hare Aplysia kurodai, were confirmed and established via total synthesis starting from farnesol.566 Azuriaplysin B and its enantiomer exhibited weak cytotoxicity. Endo-peroxide containing γ-pyrones ocellatuperoxides A–F 1429–1440, were isolated as their respective racemates.567 Absolute configurations were ascribed to the structures following separation using chiral HPLC and configurations assigned using standard methods. Racemate 1433/1434 exhibited weak cytotoxicity towards the A549 cell line, which upon further examination determined the activity was due solely to the (+)-enantiomer. Gene expression studies suggested a mechanism of action related to regulation of cell proliferation and migration. Proline betaine 1441 was reported from a marine organism, the seahare Elysia crispata, for the first time.568 Two sterol disulfates, 1442 and 1443, were isolated from a Vietnamese collection of the marine spider conch Lambis lambis.569 A new bioinspired route for the synthesis of aplaminal has been reported, enabling the discovery of a new phenolic analogue with increased cytotoxic potency.570 A late-stage diversification methodology has been used to synthesise, and confirm the structure of ocellatusone C, recently reported from Placobranchus ocellatus.571 Exploration of the antifouling activity of the cyclic depsipeptide dolastatin 16 has identified smaller fragments with activity in a barnacle cyprid settlement assay, but the level of potency is significantly reduced compared to the NP.572 RNA-Seq and shotgun proteomic studies of venomous gastropods of the genus Vexillum has identified them as a source of conotoxin-like, short chain, cysteine containing cyclic peptides.573
The pharmacological potential, and possible future trends in engineering methods to improve the properties, of conotoxins isolated from Conus regius have been reviewed.574 Transcriptomic sequencing of Conus quercinus led to the identification of α-CTx QuIA, a new conotoxin which demonstrates preferential antagonism of neuronal rα3β2 nAChR and rα6/α3β4 nAChR's making it of some interest.575 Genomic analysis of the fish hunting cone snail Conus striatus identified a novel αM-superfamily sequence SIIID which was prepared by solid-phase synthesis.576 Disulfide bond formation gave three isomers with one being more active in patch-clamp studies and demonstrating selective inhibition of α7 nAChR's. Exploration of the structure–activity relationship of the α-conotoxin AuIB, originally isolated from Conus aulicus, has identified a variant with the Pro7 residue changed to arginine and an alternative disulfide connectivity that is a potent inhibitor of GABABR-coupled CaV 2.2 channels and exhibits in vivo analgesic activity.577 Comparative evaluation of μ-conotoxins SxIIIC, SmIIIA, and KIIIA in patch-clamp and receptor-based studies has identified that the number of residues in loop 3 can influence the peptides ability to inhibit human voltage-gated sodium hNaV 1.7 channels.578 Five peptides were purified from the venom of Conus marmoreus – notably, none of the peptides contained disulfide bonds, and they were inactive towards a panel of rat nAChR subtypes.579
Five additional publications claim to report new mollusc-derived MNPs, however the structures are inconsistent with spectroscopic evidence and are not included in this review.580–584
10 Tunicates (ascidians)
A new congener of the azadecalin family of alkaloids, lepadin L 1444 was isolated from a Mediterranean Sea collection of the ascidian Clavelina lepadiformis.585 In comparative testing, the related analogue lepadin A was found to exhibit weak levels of cytotoxicity towards a panel of HTCLs, while lepadin F was inactive. Using 1H NMR spectroscopy and a zebrafish embryo development assay for guidance, investigation of an Antarctic collection of Synoicum sp. afforded australindolones A–D (1445–1448), and in addition to the known meridianins A–G, a new example, meridianin H 1449.586 Australindolones A–D represent C-2/C-3 oxidation products of meridianins G, C, D and F respectively. With the observation of small magnitude laevorotatory specific rotations ([α]D −7 to −12), and the finding that australindolone B crystallised as a racemate, the authors were unable to determine whether the NPs were in fact racemic or scalemic mixtures. Meridianins A–G were the more active NPs in the zebrafish embryo assay, causing a range of developmental defects, while the australindolones were considerably less active.New examples of bis-spiroketal-containing butenolides, prunolides D–I 1450–1455, and cis-prunolide C 1456 and a β-carboline sulfamate, pityriacitrin C 1457 were isolated from an Australian collection of Synoicum prunum.587 The structure of cis-prunolide C is notable as being the first congener, in what is now an expanding set of prunolides, to feature a cis-configuration bis-spiroketal at its core. Also notable is that the structure of pityriacitrin C is closely related to pityricitrins A and B, isolated from the pathogenic yeast Malassezia furfur. In an extensive set of biological evaluations related to the discovery of novel methods to limit the progression of Parkinson's disease, the prunolides formed α-synuclein protein–ligand complexes as judged by mass spectrometry and inhibited α-synuclein aggregation in an in vitro assay, with prunolide B also found to reduce the number of tyrosine hydroxylase positive dopamine neurons that contained phosphorylated α-synuclein aggregates in an embryonic mouse midbrain model.
In a search for new antimycobacterial compounds, lissoclinotoxin F sulfoxide 1458 was isolated from an Indonesian collection of Lissoclinum sp.384 While it and known co-metabolite lissoclibadin 2 were barely active (MIC 10 μM) and with poor to low selectivity vs. HEK293 cells, a third metabolite from the extract, lissoclinotoxin F, was weakly active against M. tuberculosis with moderate selectivity (SI = 19) and did inhibit bacterial growth in infected macrophages. The compound exhibited some degree of selectivity towards mycobacteria vs. other Gram-positive and Gram-negative bacteria. The unusual cyclopentanone end group of the carotenoid roretziaxanthin 1459 was proposed to be derived by the action of bacterial symbionts in Halocynthia roretzi on the Flavobacterium-sourced carotenoid rubixanthin.588 The absolute configuration of (+)-(R)-tiruchanduramine 1460 has been determined by enantioselective total synthesis.589 The synthetic material exhibited only mM α-glucosidase inhibiting properties, with one synthetic model compound being more active (IC50 7.5 μM). Bioinspired syntheses of cystodytins A–K have been reported, leading to assignment of 12R absolute configuration for cystodytins D–I 1461–1466 and K 1467, and clarifying that the fatty acid present as an ester at C-12 of cystodytins H and I is oleic acid.590
The first total syntheses of (−)-cylindricine H 1468 (ref. 591) and iheyamine B 1469 (ref. 592) have been reported, leading to assignment of their respective absolute configurations.
Further investigation of ascidian-derived NPs continues to reveal new and intriguing biological activities. The previously noted abilities of the meridianins to inhibit glycogen synthase kinase 3β (GSK3β) were the basis for evaluation in a mouse model of depression caused by chronic stress where a mixture of the alkaloids showed some beneficial alteration in behaviour.593 GSK3β inhibitors have the potential to act as treatments for diabetes – optimisation of meridianin C led to the identification of new sulfonamide analogues that modelling suggested had retained kinase binding, that exhibited increased glucose uptake in L6 cells and had significantly improved in vivo bioavailability.594
In the search for new anticancer therapy adjuvants and immune-based anticancer candidates from Mediterranean marine organisms, lepadin H was found to be an activator of innate immune cells.595 Testing of a library of cadiolide analogues, including alkyne-containing synthetic intermediates, identified a number of compounds with the ability to inhibit quorum sensing in Vibrio harveyi, and that could inhibit biofilm formation by Escherichia coli RP347 at concentrations that had no effect on planktonic growth of the microorganism.596 Conversely, some of the compounds were found to inhibit the growth of planktonic Pseudomonas aeruginosa PAO1 while also acting to promote biofilm formation by the microorganism. The mechanism by which didemnin B inhibits translation has been studied using cryoelectron microscopy and single molecule fluorescence, identifying that the cyclic depsipeptide is able to trap GTPase elongation factor-1 alpha (eEF1A) in an intermediate state during amino acid-tRNA selection, ultimately preventing tRNA accommodation in the ribosome binding site and eEF1A release.597
The mandelalides are cytotoxic macrolactones known to inhibit mitochondrial ATP synthase, leading to cellular depletion of ATP. This mechanism of action have been studied in more detail, identifying a consequential activation of AMP-activated protein kinase (AMPK), which normally acts to maintain cellular ATP balance in response to energy stress.598 When evaluated for cytotoxicity towards a panel of glioblastoma cell lines, mandelalide A was potently (nM) active towards some cell lines and essentially inactive towards others, presumably due to their degree of glycolytic metabolic phenotype, when run in a standard three day (72 h) end-point assay format. When the assays were performed using an extended six day run time, the survival pathways were overwhelmed, and cytotoxicity was restored to all cell lines.
11 Echinoderms
A Red Sea, Egypt collection of the brittle star Ophiocoma dentata afforded two sesquiterpene ethers 1470 and 1471.599 The metabolites isolated from starfish are usually either ceramides/cerebrosides or steroid derivatives. A total of six new lipids, ceramides 1472–1474 and cerebrosides 1475–1477, were isolated from extracts of a Far Eastern collection of the orange cookie starfish Ceramaster patagonicus.600 Although several of the MNPs were evaluated against a panel of four HTCLs and for the ability to inhibit colony formation by MDA-MB-231 human breast cancer cells, the compounds were either inactive or failed to reach a level of activity that the authors of this review consider active.Two simple sterol derivatives, 1478 and 1479, were isolated from Red Sea collections of the starfish Acanthaster planci,601 with four disulfated steroids 1480–1483 being reported from a Far Eastern deep sea trawling collection of Pteraster marsippus.602 Steroids 1480 and 1481 were characterised as a mixture. Two polyhydroxylated sterols, including the glycoside regulusoside D 1484 and cholestane 1485 isolated from a Vietnamese collection of Pentaceraster regulus exhibited weak inhibition of LPS-induced NO production in RAW264.7 cells.603
The C-3/C-26 diglycosides 1486 and 1487, purified from Xisha Is. specimens of Culcita novaeguineae, were found to be inactive when tested against three HTCLs in vitro.604 In addition to eight triterpene glycosides, pacificusosides D–K 1488–1495, extracts of the starfish Solaster pacificus also afforded the known related glycoside cucumarioside D, previously reported from the sea cucumber Eupentacta fraudatrix.605 Variable levels of cytotoxicity towards a normal human cell line and three HTCLs were observed, with all compounds also exhibiting haemolytic properties. Pacificusoside A and cucumarioside D were the most active of the purified compounds at inhibiting the neoplastic transformation of non-cancerous mouse epidermal cells when exposed to chemical (EGF, TPA) or ionising radiation (X-ray or UVB) carcinogens. A Vietnamese collection of the sea cucumber Stichopus herrmanni was the source of sulfated hydrocarbons herrmananes A 1496 and B 1497, both of which were inactive towards a panel of five HTCLs.606 A Troitsa Bay, Japan collection of Paracaudina chilensis afforded the triterpene pentaosides chilensosides A, A1, B, C and D 1498–1502, all of which, except for the last, exhibited haemolytic properties towards erythrocytes with weak to no activity towards a panel of five HTCLs.607 Additional examples of triterpene glycosides, chitonoidosides I, J, K, K1 and L 1503–1507 are hexaosides with all but chitonoidoside K1 exhibiting haemolytic properties and variable levels of cytotoxicity towards three HTCLs.608
A chemoenzymatic synthesis of LLG-5, a neuritogenic ganglioside first isolated from the starfish Linckia laevigata has been reported, achieved in 10 steps with an overall yield of about 12%.609 The methodology is flexible enough to allow preparation of a variety of analogues. The structures of the complex steroidal C-3/C-6 trisaccharide linked starfish metabolites luzonicosides A 1508 and D 1509 and sepositoside A 1510 have been confirmed by total synthesis, with absolute configuration assigned.610
A plausible biosynthetic precursor to the echinochrome family of naphthoquinone sea urchin pigments is 2-acetyl-1,3,6,8-tetrahydroxynaphthalene, synthesis of which has now been directly linked by genetic and biochemical studies to a unique class of nonreducing aromatic polyketide synthases.611 Phylogenetic analysis identified the presence of the gene SpPks1 and its homologues are widespread amongst echinoderms and close relatives the acorn worms, providing further support for the importance of this structural class to the evolutionary fitness of these organisms. The anti-inflammatory activities of echinoderm metabolites in general have been reviewed.612 The biological activities of specifically quinonoid sea urchin pigments, including echinochrome A, have also been reviewed,613 with a number of new studies reporting a variety of additional biological activities for echinochrome A including regulation of inflammation related to myocardial dysfunction,614 reduced blood pressure in a rat model of preeclampsia,615 as a treatment for menopausal dry mouth,616 suppression of melanin synthesis,617 and maintenance and regeneration of intestinal epithelium cells.618 The ecological roles played by anthraquinone pigments have been widened by the finding that host selection between crinoids and the symbiotic snapping shrimp Synalpheus stimpsonii is mediated by the pigments.619 In addition to antioxidant properties, ovothiol appears to play a central role in the control of cell proliferation in the early stages of embryo development in the sea urchin Paracentrotus lividus.620
The multi-step biosynthesis of sterols in animals involves the enzyme lanosterol synthase to achieve cyclisation of a squalene epoxide to form lanosterol. Sea cucumbers lack this enzyme, relying instead upon two oxidosqualene cyclase enzymes that appear to have evolved from an ancestral lanosterol synthase.621 With the common occurrence of steroidal glycosides in sea cucumbers, and the knowledge that saponins act as membrane disruptors, the authors noted the ability of the marine organisms to produce larger quantities of lathosterol and related sterols that appear to confer protection from saponin self-poisoning. Further biological evaluation of sea cucumber metabolites has identified the simple fatty acid decanoic acid as being active in a number of Parkinson's disease related models and assays,622 desulfated echinoside A has much reduced haemolytic activities and can still act to lower lipid levels in vitro and in vivo,623 and the saponin stichloroside C2 is cytotoxic towards triple-negative breast cancer cells via mechanisms that include inhibition of epithelial–mesenchymal transition and the induction of apoptosis via the MAPK signalling pathway.624
12 Miscellaneous
A new member of the zosterabisphenone family of diarylheptanoid dimers, zosterabisphenone C 1511 was reported from Northern Germany collections of the seagrass Zostera marina.625 The structure contains an unusual spiro-fused γ-lactone moiety, proposed to be biosynthetically derived by rearrangement of co-metabolite zosterabisphenone A. Weak cytotoxicity towards the HCT-116 cell line was observed.Two new tetrodotoxin-related NPs have been characterised, with 9-epi-tetrodotoxin isolated from Takifugu flavipterus as a mixture of hemilactal 1512 and 10,8-lactone 1513 forms, and spiro-cyclic guanidine Tb-242B 1514 isolated from Dichotomyctere ocellatus.626 Tb-242B was found to convert in neutral buffer to the previously reported 9-epimer NP Tb-242A, whereas the latter did not convert to the former under the same conditions. 9-epi-Tetrodotoxin did not exhibit any toxic effects in mice, identifying the stereochemistry at C-9 as being important for the biological activity of tetrodotoxin. The first asymmetric syntheses of four spiro-cyclic guanidines related to tetrodotoxin, Tb-210B, Tb-226, Tb-242C, and Tb-258, have been reported, with the methodology utilising a common intermediate.627 LC-MS analysis of flatworms collected on the New South Wales, Australia coastline has identified most to contain tetrodotoxin analogues, with only Stylochus cf. mcgrathi containing tetrodotoxin itself.628 Imaging MS was attempted – using a fixing/dehydration method revealed no tetrodotoxin-like signals, while freeze-dried specimens of Stylochus sp. only revealed the most dominant metabolite, 11-deoxy-TTX, which was mainly located in the rostral intestinal region.
The mechanism of antibacterial and antibiofilm activity of the 20-residue peptide fragment capitellacin, derived from whole genome sequencing and gene expression studies of the polychaete worm Capitella teleta, is attributed to detergent-like membrane destabilisation.629 A mixture of eight amino alcohol-containing betaines were isolated from the stinging fireworm Hermodice carunculata.630 Although structures of the NPs were presented in the publication, the reliance placed upon LC-MS/MS fragment analysis and NMR spectroscopy of the mixture, means the decision has been made to not show the structures in this review.
Nereistoxin, 4-dimethylamino-1,2-dithiolane, is a structurally simple insecticidal nAChR antagonist originally isolated from the annelid worm Lumbriconereis heteropoda. A set of analogues, comprised of quaternised N-methyl-nereistoxin, 5-dimethylamino-1,3-dithiane and its quaternised N-methyl derivative, were prepared and evaluated against three mammalian nAChR's.631 The quaternised analogues, while displaying lower affinity for α4β2 nAChRs, bound with higher affinities than their secondary amine variants to Torpedo muscle nAChR and rat α7 brain receptors.
Genome mining of the nematode Anisakis sp. has led to the identification of eight new antimicrobial peptides, the anisaxins, which exhibit preferential activity towards Gram-negative bacteria, with a membranolytic mechanism that leads to membrane bulging and lipid extraction.632 Only mild cytotoxicity to a mammalian cell line was observed but the peptides were unstable in serum.
Methoxy-bromodiphenyl ethers (MeO-BDEs) are susceptible to CYP-450 oxidative demethylation by hepatic microsomes of the red snapper Lutjanus campechanus, with the phenolic products being similar to those formed by oxidation of BDE flame retardants.633 The phenols are in turn susceptible to glucuronidation and sulfation excretion mechanisms by the same fish species.
13 Conclusion
The analysis of NP chemical diversity through cheminformatics techniques has benefited from the availability of extensive databases housing a wide range of NP structures and their meta data. We previously used principal component analysis of physicochemical properties to analyse MNPs and drugs and this showed that MNPs from most marine phyla are physicochemically different from each other.634 An alternative method to assess NP diversity uses Tanimoto indices derived from pre-defined substructure fragments.635 A 2017 study using a Tanimoto index cut-off of 0.4 between pairs of compounds highlighted that the discovery of novel NPs as a proportion of total NPs isolated annually is becoming less frequent.636 However, a paired Tanimoto index can be a blunt instrument if one is trying to establish if compounds have similar or different molecular frameworks since pairs of NPs with Tanimoto indices >0.7 can often belong to different structure classes, while compounds sharing the same biosynthetic origin can have indices <0.4.Rather than simply relying on the paired Tanimoto similarities, the matrix of paired similarities of compounds in large data sets can be mapped using self-organising maps (SOM) that use artificial neural networks to group objects with similar attributes based on features such as the Tanimoto index. For chemical diversity analysis, this more powerfully discriminates molecules that have similar Tanimoto indices into clusters of compounds that are more likely to share common structural frameworks (including both rings and sidechains). MNP structure class diversity vs. phyla were last reported in the 2004 edition of this review.637 Another study in 2020 looked at structure class diversity across all taxa but was limited in the scope of its analysis.638 We have now used SOMs derived from analysis of extended connectivity-based fingerprints to map MNP structure class diversity. Firstly, most MNPs (n = 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 357) found in Marinlit up to the end of 2022 were categorised using NP classifier, a deep neural network-based structural classification tool that is available as part of the GNPS toolbox hosted at the Scripps Institute for Oceanography in the USA.639 NP classifier categorises NPs at three levels: pathway, superclass and class.
357) found in Marinlit up to the end of 2022 were categorised using NP classifier, a deep neural network-based structural classification tool that is available as part of the GNPS toolbox hosted at the Scripps Institute for Oceanography in the USA.639 NP classifier categorises NPs at three levels: pathway, superclass and class.
A time series (Fig. 2a) shows the change in the proportion of structure classes (pathway classification: alkaloid, carbohydrate, fatty acid, peptide, polyketide, shikimate, and terpene) reported each semi-decade since the early 1970s. Over the last 10 years, a shift to more polyketide derived MNPs compared to terpenoid MNPs has occurred. This shift is largely because marine fungi (and to a lesser extent Actinobacteria) have recently become prolific sources of new MNPs. The proportion of different structure classes within individual phyla supports this trend (Fig. 2b).
Applying a SOM (200 × 200 neurons using the skelspheres descriptor in Osiris DataWarrior640 (DW)) and colour coding the map according to structure classes shows how effective it is at grouping structurally related molecules (Fig. 2c). By comparison, colour coding the map according to phylogeny shows that specific groups of compounds within structure classes are phylogenetically separated (Fig. 2c) (the grey scale background on the SOM shows areas of higher similarity are darker). Structure subclass groupings are further illustrated when the terpenoid pathway (representing ∼41% of all MNPs) is colour coded by terpene super-classes (monoterpene, sesquiterpene, diterpene, sesterterpene, triterpene, steroid and polyisoprene) (Fig. 3). Interestingly, there are discrete differences in the occurrence of terpenes within different phyla (Fig. 3a) and limited overlap of MNPs within discrete super-classes vs. phyla (Fig. 2d and 3b). Cnidarians (producers of the most terpene NPs) mainly yield diterpenes, echinoderms contain triterpene and steroid NPs and sesterterpenes are almost exclusively produced by sponges.
Marine fungi are the main source of peptides and have yielded 78% of all diketopiperazine MNPs, while linear and cyclic oligopeptides and depsipeptides have mostly been isolated from sponges (510), cyanobacteria (354), fungi (278), actinobacteria (208) and tunicates (123). Polyketides represent 23% of MNPs and marine fungi contribute 44% of this total. More than half of these fungal MNPs are simple aromatic phenols, xanthones, naphthoquinones, anthraquinones, benzopyrones or benzofurans. Polyketide macrolides are mostly reported from actinobacteria (385), sponges (140), fungi (196) cyanobacteria (81) and dinoflagellates (53). Polyether molecules are produced by dinoflagellates (99), red algae (70), sponges (31) and sequestered by molluscs (26). Diphenyl ethers (DPE) are found in brown algae (157), fungi (134) and sponges (52). While the fungi-derived diphenyl ethers are mainly acyl/alkyl resorcinol derivatives, the sponge DPEs are polybrominated phenols and the brown algae DPEs are phlorotannins. Endoperoxides are almost exclusively found in sponges (see the ESI† for more figures showing the MNP SOM filtered by super-class and/or class categories).
Additional structure class diversity comparisons were undertaken amongst a wider range of NPs including terrestrial microbial NPs from NPAtlas641 (n = 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 706) and plant NPs from the Universal NP Database (UNPD)642 (n = 114
706) and plant NPs from the Universal NP Database (UNPD)642 (n = 114![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 257) compared to the MNPs (n = 38
257) compared to the MNPs (n = 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 357). All 172
357). All 172![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 320 NPs were assigned to structure classes using NP classifier and the dataset was analysed to generate a SOM (200 × 200 neuron) using the skelspheres descriptor in DW. Colour coding by structure class again highlighted discrete areas of pathway specific diversity (Fig. 4a). Colour coding the NPs by biome (Fig. 4b) also showed segregation of many NPs as specifically terrestrial or marine in origin. A 2022 study showed that NPs reported from marine and terrestrial microorganisms are structurally closely related and the SOM mirrored this result (see Fig S4 and S5 in the ESI†).295
320 NPs were assigned to structure classes using NP classifier and the dataset was analysed to generate a SOM (200 × 200 neuron) using the skelspheres descriptor in DW. Colour coding by structure class again highlighted discrete areas of pathway specific diversity (Fig. 4a). Colour coding the NPs by biome (Fig. 4b) also showed segregation of many NPs as specifically terrestrial or marine in origin. A 2022 study showed that NPs reported from marine and terrestrial microorganisms are structurally closely related and the SOM mirrored this result (see Fig S4 and S5 in the ESI†).295
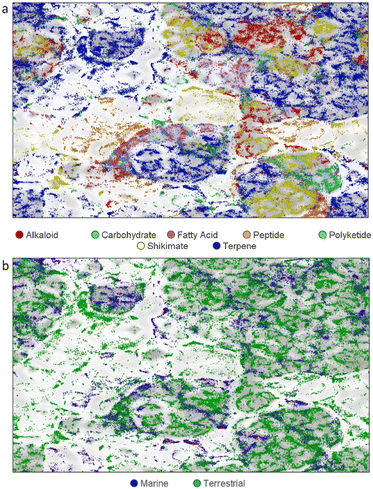 | ||
Fig. 4 SOM (200 × 200 neuron) of 172![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 320 NPs (114 320 NPs (114![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 257 plant NPs, 19 257 plant NPs, 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 706 terrestrial microbe NPs, 38 706 terrestrial microbe NPs, 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 357 MNPs) (a) colour coded by structure class, (b) colour coded by biome. 357 MNPs) (a) colour coded by structure class, (b) colour coded by biome. | ||
Cyanobacteria are the exception with greater differences in their structural diversity between freshwater and marine species (see Fig S6 ESI†). Sponges are regularly reported to contain NPs derived from microorganisms and several studies that have investigated the origins of some NPs in sponge tissues have shown that they are produced by symbionts.643–645 These investigations use genome mining techniques to support the microbial origins of some sponge MNPs. However little overlap exists between newly published sponge and marine microorganism chemical diversity, particularly for alkaloid, terpene, and polyketide NPs. Peptide NPs reported from sponges, bacteria and fungi are more closely related. A caveat to using NP databases is that they often only include the first reported source of NPs and so there may be more examples of MNPs first reported from marine macroorganisms being later found in marine microorganisms. One might expect new MNPs isolated from cultured marine microorganisms to overlap with known macroorganism MNPs if the symbiotic microbiomes of host organisms are being efficiently accessed but the lack of overlap suggests otherwise.
Marine macroorganism and plant-derived NPs are structurally different. Bromoindole, bromotyrosine, guanidine, imidazole, purine, pyrazine, pyridine, pyridoacridine, and pyrrole alkaloids are mostly produced by marine macroorganisms (85%) and are structurally different from plant alkaloids in the categories (Fig. 5a). Likewise, acetogenins, 4-pyrones, DPEs, endoperoxides, isonitriles, isothiocyanates, oximes, linear polyunsaturated compounds and polyethers are also predominately reported from marine species (Fig. 5a). Plant tryptophan-derived alkaloids are mainly monoterpene indole alkaloids (75%), but in marine macroorganisms they are more structurally diverse and occupy different areas of chemical space (Fig. 5b).
The terpene super-classes: diterpene, sesterterpene, triterpene and steroid all possess compounds that are separated into either plant or marine macroorganism-derived groups (Fig. 6). Marine specific classes occupying unique region of the SOM are briarane, cembrane, eunicellin, spongiane and xenicane diterpenes, cholestane steroids, sesterterpenes, many meroterpenes and lanostane triterpenes. These groups of terpenes represent ∼30% of all MNPs (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 474) and the combined total of uniquely marine structures represents more than 50% of MNPs isolated from macroorganisms.
474) and the combined total of uniquely marine structures represents more than 50% of MNPs isolated from macroorganisms.
This structure class analysis further highlights the unique chemical diversity of MNPs. Even within well studied and categorised groups of NP structure classes, there is a significant difference in the chemical diversity reported from marine macroorganisms compared to plants, while marine and terrestrial microorganism NPs (except cyanobacteria) share much in common with each other.
14 Conflicts of interest
There are no conflicts to declare.15 Acknowledgements
We thank Adrian Robinson and Sarah Rogers (Royal Society of Chemistry) for the provision of data used in this review, adapted from the MarinLit database with permission from the Royal Society of Chemistry.216 References
- A. R. Carroll, B. R. Copp, R. A. Davis, R. A. Keyzers and M. R. Prinsep, Nat. Prod. Rep., 2023, 40, 275–325, 10.1039/d2np00083k.
- http://pubs.rsc.org/marinlit, accessed July 2023.
- I. M. Novitskiy and A. G. Kutateladze, J. Org. Chem., 2022, 87, 4818–4828, DOI:10.1021/acs.joc.2c00169.
- D. Zhang, J. Xu, Q. Qi, F. An, S. Wang, L. Li and H. Lin, J. Antibiot., 2022, 75, 523–525, DOI:10.1038/s41429-022-00552-4.
- Z. Zhang, Y. In, K. Fukaya, T. Yang, E. Harunari, D. Urabe, C. Imada, N. Oku and Y. Igarashi, J. Nat. Prod., 2022, 85, 1098–1108, DOI:10.1021/acs.jnatprod.1c01205.
- W. Ding, Y. Li, X. Tian, M. Chen, Z. Xiao, R. Chen, H. Yin and S. Zhang, Mar. Drugs, 2022, 20, 431, DOI:10.3390/md20070431.
- J. Chen, J. Chen, S. Wang, X. Bao, S. Li, B. Wei, H. Zhang and H. Wang, Mar. Drugs, 2022, 20, 162, DOI:10.3390/md20030162.
- J. Chen, S. Wang, Y. Xu, H. Zhang and H. Wang, Chem. Biodiversity, 2022, 19, e202200037, DOI:10.1002/cbdv.202200037.
- W.-Z. Zhu, S.-H. Wang, H.-M. Gao, Y.-M. Ge, J. Dai, X.-L. Zhang and Q. Yang, Mar. Drugs, 2022, 20, 34, DOI:10.3390/md20010034.
- V. T. Quyen, L. T. H. Minh, V. T. T. Huyen, N. M. Anh, N. T. Hue, P. T. Dao, N. T. Linh, P. V. Cuong and D. T. M. Huong, Chem. Nat. Compd., 2022, 58, 704–707, DOI:10.1007/s10600-022-03772-0.
- H. Yu, S. Chen, H. Li, R. Wang, Y. Jiang, Y. Lan and P. Sun, RSC Adv., 2022, 12, 15479–15485, 10.1039/D2RA01701F.
- F. Zhang, R. F. R. Alvarenga, K. Throckmorton, S. Chanana, D. R. Braun, J. Fossen, M. Zhao, S. McCrone, M. K. Harper, S. R. Rajski, W. E. Rose, D. R. Andes, M. G. Thomas and T. S. Bugni, Org. Lett., 2022, 24, 3998–4002, DOI:10.1021/acs.orglett.2c01408.
- Z. Fang, Q. Zhang, L. Zhang, J. She, J. Li, W. Zhang, H. Zhang, Y. Zhu and C. Zhang, Org. Lett., 2022, 24, 3482–3487, DOI:10.1021/acs.orglett.2c01089.
- S. Kim, T. C. Le, S.-A. Han, P. F. Hillman, A. Hong, S. Hwang, Y. E. Du, H. Kim, D.-C. Oh, S.-S. Cha, J. Lee, S.-J. Nam and W. Fenical, Mar. Drugs, 2022, 20, 35, DOI:10.3390/md20010035.
- D. E. Williams, K. D. Morgan, D. S. Dalisay, T. Matainaho, E. Perrachon, N. Viller, M. Delcroix, J. Gauchot, H. Niikura, B. O. Patrick, K. S. Ryan and R. J. Andersen, Molecules, 2022, 27, 3569, DOI:10.3390/molecules27113569.
- G. Castro-Falcón, K. E. Creamer, A. B. Chase, M. C. Kim, S. Douglas, E. Glukhov, W. Fenical and P. R. Jensen, J. Nat. Prod., 2022, 85, 980–986, DOI:10.1021/acs.jnatprod.1c01117.
- Y. Kudo, K. Konoki and M. Yotsu-Yamashita, Biosci., Biotechnol., Biochem., 2022, 86, 1333–1342, DOI:10.1093/bbb/zbac131.
- J. H. Im, D. Shin, Y. H. Ban, W. S. Byun, E. S. Bae, D. Lee, Y. E. Du, J. Cui, Y. Kwon, S.-J. Nam, S. Cha, S. K. Lee, Y. J. Yoon and D.-C. Oh, Org. Lett., 2022, 24, 7188–7193, DOI:10.1021/acs.orglett.2c02934.
- M. C. Kim, Z. Li, R. Cullum, T. F. Molinski, M. A. G. Eid, A. M. S. Hebishy, A. H. I. Faraag, A. E. A. Moneim, M. S. Abdelfattah and W. Fenical, J. Nat. Prod., 2022, 85, 270–275, DOI:10.1021/acs.jnatprod.1c01084.
- H. Zhang, X. Zhang, Y. Huang, J. Yuan, X. Wei and J. Ju, J. Nat. Prod., 2022, 85, 625–633, DOI:10.1021/acs.jnatprod.1c00901.
- H. Zhang, Y. Chen, Y. Li, Y. Song, J. Ma and J. Ju, Mar. Drugs, 2022, 20, 393, DOI:10.3390/md20060393.
- X.-Z. Fu, S.-M. Zhang, G.-F. Wang, Q.-L. Yang, L. Guo, G. Pescitelli and Z.-P. Xie, J. Org. Chem., 2022, 87, 15998–16010, DOI:10.1021/acs.joc.2c02134.
- M. Jiang, Y. Zhang, Y. Zhang, Z. Ma and J. Wang, J. Nat. Prod., 2022, 85, 1771–1778, DOI:10.1021/acs.jnatprod.2c00320.
- Y. Bao, H. Li, Y. Dong, D. He, H. Li and W. Li, J. Nat. Prod., 2022, 85, 365–374, DOI:10.1021/acs.jnatprod.1c00952.
- H. J. Shin, C.-S. Heo, C. V. Anh, Y. D. Yoon and J. S. Kang, Mar. Drugs, 2022, 20, 44, DOI:10.3390/md20010044.
- K. Leetanasaksakul, W. Koomsiri, T. Suga, H. Matsuo, R. Hokari, P. Wattana-Amorn, Y. Takahashi, K. Shiomi, T. Nakashima, Y. Inahashi and A. Thamchaipenet, J. Nat. Prod., 2022, 85, 1211–1217, DOI:10.1021/acs.jnatprod.1c00870.
- M. Gao, S. B. Lee, J.-E. Lee, G. J. Kim, J. Moon, J.-W. Nam, J.-S. Bae, J. Chin, Y. H. Jeon and H. Choi, Appl. Sci., 2022, 12, 4510, DOI:10.3390/app12094510.
- Z. Su, K. Li, X. Luo, Y. Zhu, S.-Y. Mai, Q. Zhu, B. Yang, X. Zhou and H. Tao, Mar. Drugs, 2022, 20, 259, DOI:10.3390/md20040259.
- J. Liu, H. Li, Z. Liu, L. Tong, F. Xiao and W. Li, Mar. Drugs, 2022, 20, 394, DOI:10.3390/md20060394.
- W. Yi, A. W. Newaz, Y. Kuo, M. Ma, X.-Y. Lian and Z. Zhang, Antibiotics, 2022, 11, 1455, DOI:10.3390/antibiotics11111455.
- I. D. M. Kresna, Z. G. Wuisan, J.-M. Pohl, U. Mettal, V. L. Otoya, M. Gand, M. Marner, L. L. Otoya, N. Böhringer, A. Vilcinskas and T. F. Schäberle, J. Nat. Prod., 2022, 85, 888–898, DOI:10.1021/acs.jnatprod.1c01018.
- T. N. Makarieva, L. A. Romanenko, K. S. Mineev, L. K. Shubina, E. B. Guglya, N. I. Kalinovskaya, N. V. Ivanchina, A. G. Guzii, O. A. Belozerova, S. I. Kovalchuk, R. S. Popov, V. A. Denisenko, V. V. Mikhailov, V. V. Babenko, E. N. Ilina, M. V. Malakhova, S. S. Terekhov, A. M. Kudzhaev, P. S. Dmitrenok, I. V. Yampolsky and V. A. Stonik, Org. Lett., 2022, 24, 4892–4895, DOI:10.1021/acs.orglett.2c01714.
- S. Zhao, Y. Xia, H. Liu, T. Cui, P. Fu and W. Zhu, Org. Lett., 2022, 24, 6750–6754, DOI:10.1021/acs.orglett.2c02520.
- Y. Liu, L. Ding, Y. Deng, X. Wang, W. Cui and H. Shan, Phytochemistry, 2022, 196, 113078, DOI:10.1016/j.phytochem.2021.113078.
- S. Kang, J. Han, S. C. Jang, J. S. An, I. Kang, Y. Kwon, S.-J. Nam, S. H. Shim, J.-C. Cho, S. K. Lee and D.-C. Oh, Mar. Drugs, 2022, 20, 455, DOI:10.3390/md20070455.
- J. Cui, E. Kim, D. H. Moon, T. H. Kim, I. Kang, Y. Lim, D. Shin, S. Hwang, Y. E. Du, M. C. Song, M. Bae, J.-C. Cho, J. Jang, S. K. Lee, Y. J. Yoon and D.-C. Oh, Mar. Drugs, 2022, 20, 400, DOI:10.3390/md20060400.
- S. Chen, Q. Zhang, X. Zhang, X. Jiang, H. Zhang, Y. Zhu, C. Zhang and L. Zhang, Nat. Prod. Res., 2022, 36, 3529–3537, DOI:10.1080/14786419.2020.1867131.
- J. Seo, Y.-H. Shin, S. J. Jo, Y. E. Du, S. Um, Y. R. Kim and K. Moon, Front. Microbiol., 2022, 13, 904954, DOI:10.3389/fmicb.2022.904954.
- M. M. LeClair, Z. A. Maw, A. L. Grunwald, J. R. Kelly, B. A. Haltli, R. G. Kerr and C. Cartmell, Molecules, 2022, 27, 7794, DOI:10.3390/molecules27227794.
- Y.-Y. Chen, L.-Y. Chen, P.-J. Chen, M. El-Shazly, B.-R. Peng, Y.-C. Chen, C.-H. Su, J.-H. Su, P.-J. Sung, P.-T. Yen, L.-S. Wang and K.-H. Lai, Metabolites, 2022, 12, 320, DOI:10.3390/metabo12040320.
- M.-M. Song, Y.-H. Xie, W.-H. Chen, Y.-W. Hu, K. Zhao, Y.-H. Liu, X.-L. Huang, Q.-C. Liu and J.-F. Wang, Nat. Prod. Res., 2022, 36, 1197–1204, DOI:10.1080/14786419.2020.1864632.
- N. Aryal, J. Chen, K. Bhattarai, H. Oliver, I. Handayani, M. Kramer, S. Jan, T. Wommer, A. Berscheid, S. Peter, N. Reiling, H. Brötz-Oesterhelt, C. Geibel, M. Lämmerhofer, Y. Mast and H. Gross, J. Nat. Prod., 2022, 85, 530–539, DOI:10.1021/acs.jnatprod.1c01051.
- S. Kim, P. F. Hillman, Ji Y. Lee, J. Lee, J. Lee, S.-S. Cha, D.-C. Oh, S.-J. Nam and W. Fenical, J. Antibiot., 2022, 75, 619–625, DOI:10.1038/s41429-022-00562-2.
- Q. Wu, H. Zhu, C. Sun, L. Zhou, H. Wang, S. Shi, X. Tian and J. Ju, Mar. Drugs, 2022, 20, 537, DOI:10.3390/md20080537.
- H.-J. Lim, J. S. An, E. Seo Bae, E. Cho, S. Hwang, S.-J. Nam, K.-B. Oh, S. K. Lee and D.-C. Oh, Mar. Drugs, 2022, 20, 83, DOI:10.3390/md20020083.
- B. Chung, J.-Y. Hwang, S. C. Park, O.-S. Kwon, E. Cho, J. Lee, H.-S. Lee, D.-C. Oh, J. Shin and K.-B. Oh, Mar. Drugs, 2022, 20, 138, DOI:10.3390/md20020138.
- A. W. Newaz, Y. Kuo, X.-Y. Lian and Z. Zhang, Tetrahedron, 2022, 104, 132598, DOI:10.1016/j.tet.2021.132598.
- T. Cui, S. Lin, Z. Wang, P. Fu, C. Wang and W. Zhu, Front. Microbiol., 2022, 13, 957473, DOI:10.3389/fmicb.2022.957473.
- W. Liu, M. Liang, L. Zhang, Y. Chen, Q. Zhang, H. Zhang, W. Zhang, C. Zhang and W. Zhang, Mar. Drugs, 2022, 20, 449, DOI:10.3390/md20070449.
- Y. Xie, Y. Song, Z. Cong, K. Zhao, X. Pang, Y. Liu, X. Huang and J. Wang, Chem. Biodiversity, 2022, 19, e202200731, DOI:10.1002/cbdv.202200731.
- J. Peng, Q. Zhang, X. Jiang, M. Liang, T. Long, Z. Cheng, C. Zhang and Y. Zhu, Nat. Prod. Res., 2022, 36, 2458–2464, DOI:10.1080/14786419.2021.1901699.
- K. Ko, S.-H. Kim, S. Park, H. S. Han, J. K. Lee, J. W. Cha, S. Hwang, Ki Y. Choi, Y.-J. Song, S.-J. Nam, J. Shin, S.-I. Nam, H. C. Kwon, J.-S. Park and D.-C. Oh, Mar. Drugs, 2022, 20, 281, DOI:10.3390/md20050281.
- H. Jeong, S. J. Jo, M. Bae, Y. R. Kim and K. Moon, Mar. Drugs, 2022, 20, 565, DOI:10.3390/md20090565.
- C. V. Anh, J. S. Kang, H.-S. Lee, P. T. H. Trinh, C.-S. Heo and H. J. Shin, Mar. Drugs, 2022, 20, 464, DOI:10.3390/md20070464.
- C. L. Dieterich, S. I. Probst, R. Ueoka, I. Sandu, D. Schäfle, M. D. Molin, H. A. Minas, R. Costa, A. Oxenius, P. Sander and J. Piel, Angew. Chem., Int. Ed., 2022, 61, e202115802, DOI:10.1002/anie.202115802.
- L. Wang, V. Linares-Otoya, Y. Liu, U. Mettal, M. Marner, L. Armas-Mantilla, S. Willbold, T. Kurtán, L. Linares-Otoya and T. F. Schäberle, J. Nat. Prod., 2022, 85, 1039–1051, DOI:10.1021/acs.jnatprod.1c01173.
- L. Wang, M. Marner, U. Mettal, Y. Liu and T. F. Schäberle, Mar. Drugs, 2022, 20, 620, DOI:10.3390/md20100620.
- Y. Igarashi, Y. Ge, T. Zhou, A. R. Sharma, E. Harunari, N. Oku and A. Trianto, Beilstein J. Org. Chem., 2022, 18, 110–119, DOI:10.3762/bjoc.18.12.
- C. Wu, J. Tang, J. J. L. Malit, R. Wang, H. H.-Y. Sung, I. D. Williams and P.-Y. Qian, J. Nat. Prod., 2022, 85, 1751–1762, DOI:10.1021/acs.jnatprod.2c00290.
- J.-X. Yan, Q. Wu, J. Eric, N. Helfrich, M. G. Chevrette, D. R. Braun, H. Heyman, G. E. Ananiev, S. R. Rajski, C. R. Currie, J. Clardy and T. S. Bugni, Mar. Drugs, 2022, 20, 43, DOI:10.3390/md20010043.
- S. Zhang, G. Croppi, H. Hu, Y. Li, C. Zhu, F. Wu, F. Zhang and Z. Li, Biology, 2022, 11, 1712, DOI:10.3390/biology11121712.
- L. Qin, Y. Kuo, X.-Y. Lian and Z. Zhang, Phytochem. Lett., 2022, 49, 79–82, DOI:10.1016/j.phytol.2022.03.010.
- J.-D. Park, Y. Li, K. Moon, E. J. Han, S. R. Lee and M. R. Seyedsayamdost, Angew. Chem., Int. Ed., 2022, 61, e202114022, DOI:10.1002/anie.202114022.
- B. W. Miller, E. W. Schmidt, G. P. Concepcion and M. G. Haygood, J. Nat. Prod., 2022, 85, 479–484, DOI:10.1021/acs.jnatprod.1c01049.
- J. J. Hug, L. Kjaerulff, R. Garcia and R. Müller, Mar. Drugs, 2022, 20, 72, DOI:10.3390/md20010072.
- M. R. U. Karim, K. Fukaya, Y. In, A. R. Sharma, E. Harunari, N. Oku, D. Urabe, A. Trianto and Y. Igarashi, J. Nat. Prod., 2022, 85, 1763–1770, DOI:10.1021/acs.jnatprod.2c00281.
- M. S. Kokoulin, P. S. Dmitrenok and L. A. Romanenko, Mar. Drugs, 2022, 20, 700, DOI:10.3390/md20110700.
- K. Chakraborty, V. K. Kizhakkekalam, M. Joy and R. D. Chakraborty, Appl. Microbiol. Biotechnol., 2022, 106, 329–340, DOI:10.1007/s00253-021-11632-0.
- C. Katif, T. Chilczuk, B. Sabour, Z. Belattmania, A. Hilmi, T. H. J. Niedermeyer and M. Barakate, Biointerface Res. Appl. Chem., 2022, 12, 5647–5662, DOI:10.33263/BRIAC124.56475662.
- F. Song, J. Hu, X. Zhang, W. Xu, J. Yang, S. Li and X. Xu, Mar. Drugs, 2022, 20, 214, DOI:10.3390/md20030214.
- S.-R. Ko, Y. Jeong, S.-H. Cho, E. Lee, B.-S. Jeong, S. H. Baek, B.-H. Oh, C.-Y. Ahn, H.-M. Oh, B.-K. Cho and S. Cho, Chemosphere, 2022, 300, 134535, DOI:10.1016/j.chemosphere.2022.134535.
- S. Takiguchi, Y. Hirota-Takahata and T. Nishi, Org. Lett., 2022, 24, 4935–4938, DOI:10.1021/acs.orglett.2c01863.
- H. Xue, S. Fan, J. Xu and S. Liu, Org. Chem. Front., 2022, 9, 1675–1679, 10.1039/D1QO01700D.
- D. M. Sarnes, P. G. Jones and T. Lindel, Org. Lett., 2022, 24, 2479–2482, DOI:10.1021/acs.orglett.2c00510.
- T. Dentani, A. Kawachi, M. Kato, T. Yoshimura and J. Matsuo, Org. Chem. Front., 2022, 9, 3786–3793, 10.1039/D2QO00740A.
- Q.-Z. Li, S.-H. Hou, J.-C. Kang, P.-F. Lian, H. Yu, C. Chen, Z. Jia, T.-M. Ding and S.-Y. Zhang, Angew. Chem., Int. Ed., 2022, 61, e202207088, DOI:10.1002/anie.202207088.
- F. Han, G. Liu, X. Zhao, S. Du, Y. Ding, Q. Zhang, H. Deng, L. Wang and Y. Chen, Org. Biomol. Chem., 2022, 20, 4135–4140, 10.1039/D2OB00692H.
- C. P. Bold, K. Yeung, F. Pape, D. Kaiser and V. K. Aggarwal, Org. Lett., 2022, 24, 9398–9402, DOI:10.1021/acs.orglett.2c03716.
- P. Tian, W. Ye, X. Zhang, Y. Tong, P.-Y. Qian and R. Tong, Chem. Sci., 2022, 13, 12776–12781, 10.1039/D2SC05049H.
- M. Munda, R. Nandi, V. R. Gavit, S. Kundu, S. Niyogi and A. Bisai, Chem. Sci., 2022, 13, 11666–11671, 10.1039/D2SC03479D.
- J. T. Brazeau-Henrie, A. R. Paquette, A. Q. O'Rourke, M. G. Darnowski and C. N. Boddy, Org. Lett., 2022, 24, 6369–6373, DOI:10.1021/acs.orglett.2c02271.
- H.-H. Lu, K.-J. Gan, F.-Q. Ni, Z. Zhang and Y. Zhu, J. Am. Chem. Soc., 2022, 144, 18778–18783, DOI:10.1021/jacs.2c08337.
- T. M. Khopade, K. Ajayan, D. M. Vincent, A. L. Lane and R. Viswanathan, J. Org. Chem., 2022, 87, 11519–11533, DOI:10.1021/acs.joc.2c01120.
- L. Wang, Y. Zhang, Z. Xu, L. Jing and W. Zhu, Org. Chem. Front., 2022, 9, 4110–4117, 10.1039/D2QO00844K.
- X. Zheng, Y. Li, M. Guan, L. Wang, S. Wei, Y.-C. Li, C.-Y. Chang and Z. Xu, Angew. Chem., Int. Ed., 2022, 61, e202208802, DOI:10.1002/anie.202208802.
- S. G. Aiken, J. M. Bateman, H.-H. Liao, A. Fawcett, T. Bootwicha, P. Vincetti, E. L. Myers, N. Adam and V. K. Aggarwal, Nat. Chem., 2022, 15, 248–256, DOI:10.1038/s41557-022-01087-9.
- D. F. Vargas, T. S. Kaufman and E. L. Larghi, J. Org. Chem., 2022, 87, 13494–13500, DOI:10.1021/acs.joc.2c00063.
- M. T. Uiterweerd and A. J. Minnaard, Org. Lett., 2022, 24, 9361–9365, DOI:10.1021/acs.orglett.2c03449.
- M. Avalos, P. Garbeva, L. Vader, G. P. van Wezel, J. S. Dickschat and D. Ulanova, Nat. Prod. Rep., 2022, 39, 249–272, 10.1039/D1NP00047K.
- R. F. Little and C. Hertweck, Nat. Prod. Rep., 2022, 39, 163–205, 10.1039/D1NP00035G.
- L. Sukmarini, Mar. Drugs, 2022, 20, 544, DOI:10.3390/md20090544.
- M. B. Hulst, T. Grocholski, J. J. C. Neefjes, G. P. van Wezel and M. Metsä-Ketelä, Nat. Prod. Rep., 2022, 39, 814–841, 10.1039/D1NP00059D.
- S. Yan, M. Zeng, H. Wang and H. Zhang, J. Med. Chem., 2022, 65, 8735–8771, DOI:10.1021/acs.jmedchem.2c00626.
- Y. S. Anteneh, Q. Yang, M. H. Brown and C. M. M. Franco, Appl. Microbiol. Biotechnol., 2022, 106, 1729–1744, DOI:10.1007/s00253-022-11791-8.
- P. Lucas, H.-J. Ruscheweyh, C. C. Forneris, F. Hubrich, S. Kautsar, A. Bhushan, A. Lotti, Q. Clayssen, G. Salazar, A. Milanese, C. I. Carlström, C. Papadopoulou, D. Gehrig, M. Karasikov, H. Mustafa, M. Larralde, L. M. Carroll, P. Sánchez, A. A. Zayed, D. R. Cronin, S. G. Acinas, B. Peer, C. Bowler, T. O. Delmont, J. M. Gasol, A. D. Gossert, A. Kahles, M. B. Sullivan, P. Wincker, G. Zeller, S. L. Robinson, J. Piel and S. Sunagawa, Nature, 2022, 607, 111–118, DOI:10.1038/s41586-022-04862-3.
- L. M. P. Heinilä, J. Jokela, M. N. Ahmed, M. Wahlsten, S. Kumar, P. Hrouzek, P. Permi, H. Koistinen, D. P. Fewer and K. Sivonen, Org. Biomol. Chem., 2022, 20, 2681–2692, 10.1039/D1OB02454J.
- T. Li, C. Xi, Y. Yu, N. Wang, X. Wang, A. Iwasaki, F. Fang, L. Ding, S. Li, W. Zhang, Y. Yuan, T. Wang, X. Yan, S. He, Z. Cao and C. B. Naman, J. Nat. Prod., 2022, 85, 493–500, DOI:10.1021/acs.jnatprod.1c00983.
- A. Yamano, Y. Asato, N. Natsume, A. Iwasaki, K. Suenaga and T. Teruya, J. Nat. Prod., 2022, 85, 169–175, DOI:10.1021/acs.jnatprod.1c00915.
- M. Y. Phyo, J. X. Goh and L. T. Tan, J. Nat. Prod., 2022, 85, 485–492, DOI:10.1021/acs.jnatprod.1c00996.
- N. Kurisawa, A. Iwasaki, K. Teranuma, S. Dan, C. Toyoshima, M. Hashimoto and K. Suenaga, J. Am. Chem. Soc., 2022, 144, 11019–11032, DOI:10.1021/jacs.2c04459.
- G. J. Kim, S. J. Mascuch, E. Mevers, P. D. Boudreau, W. H. Gerwick and H. Choi, J. Org. Chem., 2022, 87, 1043–1055, DOI:10.1021/acs.joc.1c02340.
- D. A. Brumley, S. P. Gunasekera, T. Sauvage, A. Larissa, H. dos Santos, Q.-Y. Chen, V. J. Paul and H. Luesch, J. Nat. Prod., 2022, 85, 581–589, DOI:10.1021/acs.jnatprod.1c01073.
- R. Taguchi, A. Iwasaki, A. Ebihara, G. Jeelani, T. Nozaki and K. Suenaga, Org. Lett., 2022, 24, 4710–4714, DOI:10.1021/acs.orglett.2c02013.
- O. Ohno, A. Iwasaki, K. Same, C. Kudo, E. Aida, K. Sugiura, S. Sumimoto, T. Teruya, E. Tashiro, S. Simizu, K. Matsuno, M. Imoto and K. Suenaga, Org. Lett., 2022, 24, 4547–4551, DOI:10.1021/acs.orglett.2c01566.
- T. Arnaud, S. Rohrer, B. Diaz, R. Reher, A. M. C. Rodriguez, M. L. Pierce, P. C. Dorrestein, L. Gerwick, W. H. Gerwick and J. W. Golden, ACS Chem. Biol., 2022, 17, 1910–1923, DOI:10.1021/acschembio.2c00347.
- S. P. Gunasekera, S. Kokkaliari, R. Ratnayake, T. Sauvage, A. Larissa, H. dos Santos, H. Luesch and V. J. Paul, Molecules, 2022, 27, 1717, DOI:10.3390/molecules27051717.
- T. J. O'Donnell, Y. Luo, W. Y. Yoshida, S. Suzuki, R. Sun and P. G. Williams, J. Nat. Prod., 2022, 85, 415–425, DOI:10.1021/acs.jnatprod.1c01014.
- E. Curren, C. P. Leaw, P. T. Lim and S. C. Y. Leong, Environ. Sci. Pollut. Res., 2022, 29, 78178–78206, DOI:10.1007/s11356-022-23096-4.
- D. T. A. Youssef, S. J. Mufti, A. A. Badiab and L. A. Shaala, Mar. Drugs, 2022, 20, 768, DOI:10.3390/md20120768.
- B. Robles-Bañuelos, L. M. Durán-Riveroll, E. Rangel-López, H. I. Pérez-López and L. González-Maya, Molecules, 2022, 27, 4814, DOI:10.3390/molecules27154814.
- M. H. Saad, E. M. El-Fakharany, M. S. Salem and N. M. Sidkey, J. Biomol. Struct. Dyn., 2022, 40, 2828–2850, DOI:10.1080/07391102.2020.1838948.
- H. Righini, O. Francioso, A. Martel Quintana and R. Roberti, Horticulturae, 2022, 8, 58, DOI:10.3390/horticulturae8010058.
- J. B. Cox, A. A. Kellum, Y. Zhang, B. Li and A. B. Smith, Angew. Chem., Int. Ed., 2022, 61, e202204884, DOI:10.1002/anie.202204884.
- D. Fiorito, S. Keskin, J. M. Bateman, M. George, N. Adam and V. K. Aggarwal, J. Am. Chem. Soc., 2022, 144, 7995–8001, DOI:10.1021/jacs.2c03192.
- K. Upadhyaya and D. Crich, Org. Lett., 2022, 24, 1833–1836, DOI:10.1021/acs.orglett.2c00349.
- T. P. Martins, N. R. Glasser, D. J. Kountz, P. Oliveira, E. P. Balskus and P. N. Leão, ACS Chem. Biol., 2022, 17, 2528–2537, DOI:10.1021/acschembio.2c00464.
- T. Kitamura, R. Suzuki, S. Inuki, H. Ohno, K. L. McPhail and S. Oishi, ACS Med. Chem. Lett., 2022, 13, 105–110, DOI:10.1021/acsmedchemlett.1c00591.
- E. B. D. Silva, V. Sharma, L. Hernandez-Alvarez, A. H. Tang, A. Stoye, A. J. O'Donoghue, W. H. Gerwick, R. J. Payne, J. H. McKerrow and L. M. Podust, J. Med. Chem., 2022, 65, 4255–4269, DOI:10.1021/acs.jmedchem.1c02063.
- C. Duan, S. Wang, R. Huo, E. Li, M. Wang, J. Ren, Y. Pan, L. Liu and G. Liu, J. Fungi, 2022, 8, 530, DOI:10.3390/jof8050530.
- Y.-P. Feng, H.-K. Wang, J.-L. Wu, S. Peng, W.-L. Zhou, Q.-L. Lai, H.-W. Lin, C. B. Naman, T.-T. Wang and H. Shan, Chem. Biodiversity, 2022, 19, e202200028, DOI:10.1002/cbdv.202200028.
- H. Lu, Y. Tan, Y. Zhang, Z. Li, J. Chen, C. Gao, Y. Liu and X. Luo, Fitoterapia, 2022, 159, 105201, DOI:10.1016/j.fitote.2022.105201.
- D.-L. Zhao, H.-S. Wang, L.-W. Gao and P. Zhang, Front. Mar. Sci., 2022, 9, 923128, DOI:10.3389/fmars.2022.923128.
- G. Liu, D. Liu, Z. Li, J. Jiao, X. Hou, X. Zhang, Q. Che, T. Zhu, D. Li and G. Zhang, Front. Mar. Sci., 2022, 9, 1015684, DOI:10.3389/fmars.2022.1015684.
- J. Zhang, B. Zhang, L. Cai and L. Liu, Mar. Drugs, 2022, 20, 778, DOI:10.3390/md20120778.
- Y. Chen, C. Liu, K. Kumaravel, L. Nan and Y. Tian, Front. Microbiol., 2022, 13, 879674, DOI:10.3389/fmicb.2022.879674.
- T.-H. Zhong, X.-M. Zeng, S.-B. Feng, H.-T. Zhang, Y.-H. Zhang, Z.-H. Luo, W. Xu and X.-H. Ma, Nat. Prod. Res., 2022, 36, 414–418, DOI:10.1080/14786419.2020.1771706.
- Z. Hou, C. Sun, X. Chen, G. Zhang, Q. Che, D. Li and T. Zhu, Tetrahedron Lett., 2022, 96, 153778, DOI:10.1016/j.tetlet.2022.153778.
- M. Jiang, H. Guo, Q. Wu, S. Yuan and L. Liu, Molecules, 2022, 27, 5076, DOI:10.3390/molecules27165076.
- S. Yuan, L. Chen, Q. Wu, M. Jiang, H. Guo, Z. Hu, S. Chen, L. Liu and Z. Gao, Mar. Drugs, 2022, 20, 294, DOI:10.3390/md20050294.
- J. She, Y. Chen, Y. Ye, X. Lin, B. Yang, J. Xiao, Y. Liu and X. Zhou, Mar. Drugs, 2022, 20, 475, DOI:10.3390/md20080475.
- L. Xu, F.-W. Guo, X.-Q. Zhang, T.-Y. Zhou, C.-J. Wang, M.-Y. Wei, Y.-C. Gu, C.-Y. Wang and C.-L. Shao, Commun. Chem., 2022, 5, 80, DOI:10.1038/s42004-022-00696-2.
- O. I. Zhuravleva, E. B. Belousova, G. K. Oleinikova, A. S. Antonov, Y. V. Khudyakova, A. B. Rasin, R. S. Popov, E. S. Menchinskaya, P. T. H. Trinh, A. N. Yurchenko and E. A. Yurchenko, Mar. Drugs, 2022, 20, 584, DOI:10.3390/md20090584.
- L.-H. Yan, P.-H. Li, X.-M. Li, S.-Q. Yang, K.-C. Liu, B.-G. Wang and X. Li, Org. Lett., 2022, 24, 2684–2688, DOI:10.1021/acs.orglett.2c00781.
- D. Lv, J. Xia, X. Guan, Q. Lai, B. Zhang, J. Lin, Z. Shao, S. Luo, D. Zhangsun, J.-J. Qin and W. Wang, Front. Microbiol., 2022, 13, 950857, DOI:10.3389/fmicb.2022.950857.
- A. Xu, X.-N. Xu, M. Zhang, C.-L. Li, L. Liu and D.-Y. Fu, Front. Microbiol., 2022, 13, 959754, DOI:10.3389/fmicb.2022.959754.
- Z. Liu, S. Li, Y. Chen, M. Li, H. Liu and W. Zhang, Nat. Prod. Res., 2022, 36, 5701–5707, DOI:10.1080/14786419.2021.2015595.
- S. Miao, M. Liu, S. Qi, Y. Wu, K. Sun, Z. Zhang, K. Zhu, G. Cai and K. Gong, J. Antibiot., 2022, 75, 410–414, DOI:10.1038/s41429-022-00527-5.
- D.-d. Li, Y. Wang, E. L. Kim, J. Hong and J. H. Jung, Mar. Drugs, 2022, 20, 203, DOI:10.3390/md20030203.
- J. Wang, Z. Li, Y. Zhang, C. Chen, W. Chen, C. Gao, Y. Liu, Y. Tan and X. Luo, J. Ocean Univ. China, 2022, 21, 1307–1312, DOI:10.1007/s11802-022-4959-5.
- F.-R. Jiao, H.-R. Zhu, B.-B. Gu, Y. Wu, F. Sun, S.-P. Wang, A. Zhang, W.-H. Jiao, S.-H. Xu and H.-W. Lin, Tetrahedron, 2022, 109, 132579, DOI:10.1016/j.tet.2021.132579.
- L.-H. Yan, X.-M. Li, L.-P. Chi, X. Li and B.-G. Wang, Mar. Drugs, 2022, 20, 4, DOI:10.3390/md20010004.
- R. Zhang, H. Wang, B. Chen, H. Dai, J. Sun, J. Han and H. Liu, Mar. Drugs, 2022, 20, 302, DOI:10.3390/md20050302.
- X.-Y. Chen, Z. Qi, Y.-C. Chen, W.-M. Zhong, X. Yao, J.-F. Wang, X.-F. Shi, W.-M. Zhang, S. Zhang and F.-Z. Wang, Mar. Drugs, 2022, 20, 71, DOI:10.3390/md20010071.
- Z. Qi, Y. Chen, J. Wang, X. Shi, Y. Che, X. Chen, W. Zhong, W. Zhang, X. Wei, F. Wang and S. Zhang, Mar. Drugs, 2022, 20, 150, DOI:10.3390/md20020150.
- C. Sun, X. Liu, N. Sun, X. Zhang, M. Shah, G. Zhang, Q. Che, T. Zhu, L. Jing and D. Li, J. Nat. Prod., 2022, 85, 987–996, DOI:10.1021/acs.jnatprod.1c01118.
- H. Wang, R. Zhang, B. Ma, W. Wang, Y. Chong, J. Han, L. Zhu, X. Zhang, H. Dai, H. Liu and B. Chen, J. Fungi, 2022, 8, 1058, DOI:10.3390/jof8101058.
- R.-Y. Shang, J. Cui, J.-X. Li, X.-X. Miao, L. Zhang, D.-D. Xie, L. Zhang, H.-W. Lin and W.-H. Jiao, Tetrahedron, 2022, 104, 132599, DOI:10.1016/j.tet.2021.132599.
- S. Kankanamge, Z. G. Khalil, P. V. Bernhardt and R. J. Capon, Mar. Drugs, 2022, 20, 698, DOI:10.3390/md20110698.
- C. Chen, X. Ren, H. Tao, W. Cai, Y. Chen, X. Luo, P. Guo and Y. Liu, Mar. Drugs, 2022, 20, 295, DOI:10.3390/md20050295.
- Y. Liao, X. Hong, J. Yang, J.-J. Qin, B. Zhang, J. Lin, Z. Shao and W. Wang, Nat. Prod. Res., 2022, 36, 3572–3578, DOI:10.1080/14786419.2020.1869969.
- X. Xiao, T. Zhou, Y. Zhang, H. Zhou, M. Luo, T. Hu, P. Hu, L. Kong, Z. Liu, Y. Chan, Z. Huang and L. Hu, Mar. Drugs, 2022, 20, 191, DOI:10.3390/md20030191.
- J. He, X. Wu, S. Huang, J. Wang, S. Niu, M. Chen, G. Zhang, S. Cai, J. Wu and B. Hong, Mar. Drugs, 2022, 20, 575, DOI:10.3390/md20090575.
- C.-M. Liu, X. Liang, F.-H. Yao and S.-H. Qi, Tetrahedron, 2022, 126, 133067, DOI:10.1016/j.tet.2022.133067.
- C.-M. Liu, F.-H. Yao, X.-H. Lu, X.-X. Zhang, L.-X. Luo, X. Liang and S.-H. Qi, Mar. Drugs, 2022, 20, 78, DOI:10.3390/md20010078.
- S. Niu, Z. Chen, S. Pei, Z. Shao, G. Zhang and B. Hong, Nat. Prod. Res., 2022, 36, 4936–4942, DOI:10.1080/14786419.2021.1913587.
- S. Niu, S. Huang, B. Hong, Q. Huang, X. Liu, Z. Shao and G. Zhang, Mar. Drugs, 2022, 20, 410, DOI:10.3390/md20070410.
- C.-Y. Wang, X.-H. Liu, Y.-Y. Zheng, X.-Y. Ning, Y.-H. Zhang, X.-M. Fu, X. Li, C.-L. Shao and C.-Y. Wang, Front. Microbiol., 2022, 13, 808532, DOI:10.3389/fmicb.2022.808532.
- F. P. Machado, I. C. Rodrigues, L. Gales, J. A. Pereira, P. M. Costa, T. Dethoup, S. Mistry, M. Artur, S. Silva, V. Vasconcelos and A. Kijjoa, Mar. Drugs, 2022, 20, 672, DOI:10.3390/md20110672.
- L. Niveditha, P. Fu, T. F. Leao, T. Li, T. Wang, R. X. Poulin, L. R. Gaspar, C. B. Naman and S. T. Puthiyedathu, Planta Med., 2022, 88, 774–782, DOI:10.1055/a-1769-8480.
- X. Guo, Q. Meng, J. Liu, J. Wu, H. Jia, L. Dong, Y. Gu, J. Liu, J. Huang, A. Fan and W. Lin, J. Nat. Prod., 2022, 85, 1067–1078, DOI:10.1021/acs.jnatprod.1c01194.
- Q. Meng, X. Guo, J. Wu, L. Dong, Y. Gu, J. Huang, A. Fan and W. Lin, Phytochemistry, 2022, 203, 113424, DOI:10.1016/j.phytochem.2022.113424.
- Q. Peng, W. Chen, X. Lin, J. Xiao, Y. Liu and X. Zhou, Mar. Drugs, 2022, 20, 212, DOI:10.3390/md20030212.
- Y.-J. F. Chiang and T.-H. Lee, Phytochemistry, 2022, 200, 113229, DOI:10.1016/j.phytochem.2022.113229.
- Y. Zhang, Z. Li, B. Huang, K. Liu, S. Peng, X. Liu, C. Gao, Y. Liu, Y. Tan and X. Luo, Mar. Drugs, 2022, 20, 178, DOI:10.3390/md20030178.
- V. A. Cao, J.-H. Kwon, J. S. Kang, H.-S. Lee, C.-S. Heo and H. J. Shin, J. Nat. Prod., 2022, 85, 2177–2183, DOI:10.1021/acs.jnatprod.2c00398.
- C. V. Anh, Y. D. Yoon, J. S. Kang, H.-S. Lee, C.-S. Heo and H. J. Shin, Mar. Drugs, 2022, 20, 217, DOI:10.3390/md20030217.
- C. V. Anh, J.-H. Kwon, J. S. Kang, H.-S. Lee, C.-S. Heo and H. J. Shin, Pharmaceuticals, 2022, 15, 74, DOI:10.3390/ph15010074.
- H.-Z. Weng, J.-Y. Zhu, F.-Y. Yuan, Z.-Y. Tang, X.-Q. Tian, Y. Chen, C.-Q. Fan, G.-H. Tang and S. Yin, Mar. Drugs, 2022, 20, 322, DOI:10.3390/md20050322.
- H. Zhang, Y. Kang, X. Qi, J. Wu, L. Dong, A. Fan, J. Huang and W. Lin, Bioorg. Chem., 2022, 129, 106114, DOI:10.1016/j.bioorg.2022.106114.
- H. Li, J. Guo, R. Zhang, J. Wang, Z. Hu and Y. Zhang, Nat. Prod. Res., 2022, 36, 3346–3352, DOI:10.1080/14786419.2020.1858409.
- H. Lv, J. Zhang, Y. Xue, S. Li, X. Sun, J. Jia, H. Bi, S. Wang, H. Su, M. Zhu, H. Wang, K. Hong and X. Li, Chem. Biodiversity, 2022, 19, e202200207, DOI:10.1002/cbdv.202200207.
- L. Li, Q.-H. Chang, S.-S. Zhang, K. Yang, F.-L. Chen, H.-J. Zhu, F. Cao and Y.-F. Liu, J. Mol. Struct., 2022, 1261, 132904, DOI:10.1016/j.molstruc.2022.132904.
- Z. Liu, Y. Chen, S. Li, C. Hu, H. Liu and W. Zhang, Nat. Prod. Res., 2022, 36, 5213–5221, DOI:10.1080/14786419.2021.1925271.
- W. Xia, J.-C. Su, J.-S. Hu, X.-X. He, S.-J. Lin, D.-M. Zhang, W.-C. Ye, M.-F. Chen, H.-W. Lin and C.-X. Zhang, Org. Lett., 2022, 24, 158–163, DOI:10.1021/acs.orglett.1c03795.
- W. Xia, F.-T. Wang, M.-X. Si-Tu, H. Fan, J.-S. Hu, H. Yang, S.-Y. Guan, A. Lin-Kun and C.-X. Zhang, Mar. Drugs, 2022, 20, 211, DOI:10.3390/md20030211.
- J. Long, X. Pang, X. Lin, S. Liao, X. Zhou, J. Wang, B. Yang and Y. Liu, Chem. Biodiversity, 2022, 19, e202100925, DOI:10.1002/cbdv.202100925.
- A. A. Dewa, Z. G. Khalil, A. H. Elbanna and R. J. Capon, Molecules, 2022, 27, 3172, DOI:10.3390/molecules27103172.
- H. Wen, Y. Zang, Q. Zhu, S. Ouyang, J. Luo, N. Luo, H. Zhu and Y. Zhang, Nat. Prod. Res., 2022, 36, 3255–3261, DOI:10.1080/14786419.2020.1851226.
- D. T. A. Youssef, L. A. Shaala and G. Genta-Jouve, Mar. Drugs, 2022, 20, 451, DOI:10.3390/md20070451.
- D.-L. Zhao, H.-S. Wang, L.-W. Gao and P. Zhang, Front. Mar. Sci., 2022, 9, 923128, DOI:10.3389/fmars.2022.923128.
- T. Zhao, X.-Y. Zhang, R.-S. Deng, Z. Tan, G.-Y. Chen and X.-H. Nong, Nat. Prod. Res., 2022, 36, 3965–3971, DOI:10.1080/14786419.2021.1903002.
- Z. Guo, A. Abulaizi, L. Huang, Z. Xiong, S. Zhang, T. Liu and R. Wang, Mar. Drugs, 2022, 20, 679, DOI:10.3390/md20110679.
- J. Yang, L. Zhou, Z. Zhou, Y. Song and J. Ju, Biochem. Syst. Ecol., 2022, 102, 104415, DOI:10.1016/j.bse.2022.104415.
- F.-Y. Du, A. Mándi, X.-M. Li, L.-H. Meng, T. Kurtán and B.-G. Wang, Chin. J. Chem., 2022, 40, 378–384, DOI:10.1002/cjoc.202100586.
- X.-H. Liu, Y.-P. Song, X.-L. Yin and N.-Y. Ji, J. Agric. Food Chem., 2022, 70, 4658–4666, DOI:10.1021/acs.jafc.2c00259.
- A. A. Dewa, A. H. Elbanna, Z. G. Khalil and R. J. Capon, J. Med. Chem., 2022, 65, 2610–2622, DOI:10.1021/acs.jmedchem.1c01989.
- Z.-B. Zou, L.-H. Chen, M.-Y. Hu, L. Xu, Y.-J. Hao, Q.-X. Yan, C.-F. Wang, C.-L. Xie and X.-W. Yang, Chem. Biodiversity, 2022, 19, e202200538, DOI:10.1002/cbdv.202200538.
- C.-P. Li, Y.-P. Song, B.-G. Wang and N.-Y. Ji, Tetrahedron Lett., 2022, 93, 153689, DOI:10.1016/j.tetlet.2022.153689.
- M.-M. Xie, J.-Y. Jiang, Z.-B. Zou, L. Xu, Y. Zhang, C.-F. Wang, C.-B. Liu, Q.-X. Yan, L. Zhu and X.-W. Yang, Chem. Biodiversity, 2022, 19, e202200963, DOI:10.1002/cbdv.202200963.
- E. Oppong-Danquah, M. Blümel, S. Scarpato, A. Mangoni and D. Tasdemir, Int. J. Mol. Sci., 2022, 23, 782, DOI:10.3390/ijms23020782.
- X. -Y. Hu, X. -M. Li, S. -Q. Yang, B. -G. Wang and L. -H. Meng, Chem. Biodiversity, 2022, 19, e202200550, DOI:10.1002/cbdv.202200550.
- H.-S. Lee, J. S. Kang, D.-Y. Cho, D.-K. Choi and H. J. Shin, J. Nat. Prod., 2022, 85, 857–865, DOI:10.1021/acs.jnatprod.1c00985.
- S. Chen, H. Guo, M. Jiang, Q. Wu, L. Jing, H. Shen and L. Liu, Mar. Drugs, 2022, 20, 51, DOI:10.3390/md20010051.
- G. Zhai, S. Chen, H. Shen, H. Guo, M. Jiang and L. Liu, Mar. Drugs, 2022, 20, 553, DOI:10.3390/md20090553.
- Y. Ge, Y. Ma, M. Zhao, J. Wei, X. Wu, Z. Zhang, Y. Han, H. Lei and B. Wu, Eur. J. Med. Chem., 2022, 242, 114699, DOI:10.1016/j.ejmech.2022.114699.
- Y.-H. Zhang, H.-F. Du, W.-B. Gao, L. Wan, F. Cao and C.-Y. Wang, Mar. Drugs, 2022, 20, 486, DOI:10.3390/md20080486.
- M.-J. Hong, M.-J. Hao, G.-Y. Zhang, H.-J. Li, Z.-Z. Shao, X.-P. Liu, W.-Z. Ma, J. Xu, T. Mahmud and W.-J. Lan, Mar. Drugs, 2022, 20, 448, DOI:10.3390/md20070448.
- X.-M. Dai, H.-L. Pan, W.-J. Lan, L.-P. Chen, G.-K. Feng, R. Deng, X.-F. Zhu and H.-J. Li, Phytochemistry, 2022, 204, 113456, DOI:10.1016/j.phytochem.2022.113456.
- K. H. A. U. Zaman, A. M. Sarotti, X. Wu, L. DeVine and S. Cao, Phytochemistry, 2022, 198, 113138, DOI:10.1016/j.phytochem.2022.113138.
- X. Chen, J. Wei, J. Tang and B. Wu, Chem. Biodiversity, 2022, 19, e202100899, DOI:10.1002/cbdv.202100899.
- T. Yamada, K. Yoshida, T. Kikuchi and T. Hirano, Mar. Drugs, 2022, 20, 226, DOI:10.3390/md20040226.
- Y.-H. Chen, X. Wu, L. Xu, M. El-Shazly, C. Ma, S. Yuan, P. Wang and L. Luo, Chem. Biodiversity, 2022, 19, e202200008, DOI:10.1002/cbdv.202200008.
- X. Xu, Y. Tan, C. Gao, K. Liu, Z. Tang, C. Lu, H. Li, X. Zhang and Y. Liu, Mar. Drugs, 2022, 20, 255, DOI:10.3390/md20040255.
- M. Cheng, X. Cui, Y. Tang, Z. Shao, X. Liu, J. Su, J. Zhang, Q. Wang and G. Li, J. Org. Chem., 2022, 87, 11090–11096, DOI:10.1021/acs.joc.2c01347.
- P. T. H. Trinh, A. N. Yurchenko, O. O. Khmel, T. V. T. Dieu, N. T. D. Ngoc, E. V. Girich, A. S. Menshov, N. Y. Kim, E. A. Chingizova, T. T. T. Van, J. S. Lee, H.-S. Lee and E. A. Yurchenko, Molecules, 2022, 27, 7650, DOI:10.3390/molecules27217650.
- G.-S. Yao, Z.-L. Ma, Y.-Y. Zheng, L. Lv, J.-Q. Mao and C.-Y. Wang, J. Fungi, 2022, 8, 1218, DOI:10.3390/jof8111218.
- O. G. Mohamed, Z. G. Khalil, V. Santiago and R. J. Capon, Tetrahedron, 2022, 113, 132759, DOI:10.1016/j.tet.2022.132759.
- Q. Guo, X.-M. Dai, W.-J. Lan, L.-P. Chen, C.-K. Lam, G.-K. Feng, R. Deng, X.-F. Zhu and H.-J. Li, Nat. Prod. Res., 2022, 36, 2534–2541, DOI:10.1080/14786419.2021.1915308.
- J. Hu, W. Zhang, H. Tan, S. Li, X. Gao, Z. Liu, Y. Wang, H. Liu and W. Zhang, Org. Biomol. Chem., 2022, 20, 4376–4384, 10.1039/D2OB00585A.
- J. Hu, Z. Zou, Y. Chen, S. Li, X. Gao, Z. Liu, Y. Wang, H. Liu and W. Zhang, J. Nat. Prod., 2022, 85, 1967–1975, DOI:10.1021/acs.jnatprod.2c00249.
- X.-X. Xue, L. Chen and M.-C. Tang, Antibiotics, 2022, 11, 1444, DOI:10.3390/antibiotics11101444.
- H. Wu, X. You, J. Chen, K. Zhang and X. Li, Chem. Nat. Compd., 2022, 58, 780–781, DOI:10.1007/s10600-022-03794-8.
- W. Wang, Y. Shi, Y. Liu, Y. Zhang, J. Wu, G. Zhang, Q. Che, T. Zhu, M. Li and D. Li, Mar. Drugs, 2022, 20, 338, DOI:10.3390/md20050338.
- Y.-H. Pei, Li-H. Zhang, X.-L. Wu, H.-H. Wu, H.-F. Wang, Y.-N. Wang and G. Chen, Phytochemistry, 2022, 194, 113000, DOI:10.1016/j.phytochem.2021.113000.
- W. Xing, Y. Ding and F. An, Nat. Prod. Commun., 2022, 17 DOI:10.1177/1934578X221075986.
- U. W. Hawas, L. T. A. El-Kassem, E. F. Ahmed and R. A. Alghamdi, Nat. Prod. Res., 2022, 36, 2713–2721, DOI:10.1080/14786419.2021.1917571.
- E. V. Leshchenko, A. S. Antonov, S. A. Dyshlovoy, D. V. Berdyshev, J. Hauschild, O. I. Zhuravleva, G. V. Borkunov, A. S. Menshov, N. N. Kirichuk, R. S. Popov, A. V. Gerasimenko, A. A. Udovenko, M. Graefen, C. Bokemeyer, G. v. Amsberg and A. N. Yurchenko, J. Nat. Prod., 2022, 85, 2746–2752, DOI:10.1021/acs.jnatprod.2c00677.
- X.-Y. Hu, X.-M. Li, H. Liu, B.-G. Wang and L.-H. Meng, Bioorg. Chem., 2022, 128, 106021, DOI:10.1016/j.bioorg.2022.106021.
- H. Qiao, S.-H. Zhang, D. Yuan, Y. Yang, R. Xu, B. Chen, Y. Wang, T.-J. Zhu, C.-B. Cui, G.-G. Zhang and C.-W. Li, Nat. Prod. Res., 2022, 36, 1834–1841, DOI:10.1080/14786419.2020.1819271.
- W. Lin, H. Li, Z. Wu, J. Su, Z. Zhang, L. Yang, X. Deng and Q. Xu, Mar. Drugs, 2022, 20, 684, DOI:10.3390/md20110684.
- J. Chen, Li-N. Huo, Y. Gao, Y.-L. Zhang and Y. Chen, Nat. Prod. Res., 2022, 36, 3988–3991, DOI:10.1080/14786419.2021.1889543.
- Z.-B. Zou, G. Zhang, Y.-Q. Zhou, C.-L. Xie, M.-M. Xie, L. Xu, H. You-Jia, L.-Z. Luo, X.-K. Zhang, X.-W. Yang and J.-S. Wang, Mar. Drugs, 2022, 20, 736, DOI:10.3390/md20120736.
- J. Jiang, H. Jiang, D. Shen, Y. Chen, H. Shi and F. He, J. Antibiot., 2022, 75, 301–303, DOI:10.1038/s41429-022-00516-8.
- D. T. A. Youssef, L. A. Shaala, A. Almohammadi, S. S. Elhady, T. A. Alzughaibi and K. Z. Alshali, Lett. Org. Chem., 2022, 19, 144–149, DOI:10.2174/1570178618666210617112441.
- J. Zhang, L. Dong, A. Fan, J. Huang and W. Lin, Mar. Drugs, 2022, 20, 712, DOI:10.3390/md20110712.
- X. Pan and S.-M. Li, Org. Lett., 2022, 24, 462–466, DOI:10.1021/acs.orglett.1c04189.
- Y.-H. Li, S.-Q. Yang, X.-M. Li, X. Li, B.-G. Wang and H.-L. Li, Bioorg. Chem., 2022, 128, 106104, DOI:10.1016/j.bioorg.2022.106104.
- H. Li, W. Zhang, X. Zhang, S. Tang, P. Men, M. Xiong, Z. Li, Y. Zhang, X. Huang and X. Lu, Mar. Drugs, 2022, 20, 523, DOI:10.3390/md20080523.
- W. Weng, R. Li, Y. Zhang, X. Pan, S. Jiang, C. Sun, C. Zhang and X. Lu, Front. Microbiol., 2022, 13, 1033823, DOI:10.3389/fmicb.2022.1033823.
- S. Wang, Y. Zeng, J. Yin, W. Chang, X. Zhao and Y. Mao, Phytochem. Lett., 2022, 47, 76–80, DOI:10.1016/j.phytol.2021.11.006.
- X.-Y. Hu, X.-M. Li, B.-G. Wang and L.-H. Meng, J. Nat. Prod., 2022, 85, 1398–1406, DOI:10.1021/acs.jnatprod.2c00211.
- X.-Y. Hu, X.-M. Li, B.-G. Wang and L.-H. Meng, Int. J. Mol. Sci., 2022, 23, 6332, DOI:10.3390/ijms23116332.
- H.-Y. Hsi, S.-W. Wang, C.-H. Cheng, K.-L. Pang, J.-Y. Leu, S.-H. Chang, Y.-T. Lee, Y.-H. Kuo, C.-Y. Huang and T.-H. Lee, Molecules, 2022, 27, 8940, DOI:10.3390/molecules27248940.
- D. Tian, X. Gou, J. Jia, J. Wei, M. Zheng, W. Ding, H. Bi, B. Wu and J. Tang, Fitoterapia, 2022, 156, 105095, DOI:10.1016/j.fitote.2021.105095.
- J. Wang, T. Li, P. Wang, W. Ding and J. Xu, J. Nat. Prod., 2022, 85, 1218–1228, DOI:10.1021/acs.jnatprod.1c01020.
- H. Fan, Z.-M. Shi, Y.-H. Lei, M.-X. Si-Tu, F.-G. Zhou, F. Chan, W. Xia, X.-H. Shao, Y. Chen and C.-X. Zhang, Mar. Drugs, 2022, 20, 443, DOI:10.3390/md20070443.
- J. Zou, J. Wu, L. Ding, W. Wang, Y. Liu, Y. Feng, Q. Lai, W. Lin, T. Wang and H. Shan, Chem. Biodiversity, 2022, 19, e202200724, DOI:10.1002/cbdv.202200475.
- F.-H. Yao, X. Liang, X.-H. Lu, X. Cheng, L.-X. Luo and S.-H. Qi, J. Nat. Prod., 2022, 85, 2071–2081, DOI:10.1021/acs.jnatprod.2c00473.
- X. Li, Y. Ge, Y. Ma, S. Wang, S. Li, Q. Yin, X. Liu, J. Wie, X. Wu and B. Wu, Chem. Biodiversity, 2022, 19, e202200055, DOI:10.1002/cbdv.202200055.
- Y. Kuo, S. Kaleem, M. Ma, X. Lian and Z. Zhang, Molecules, 2022, 27, 7099, DOI:10.3390/molecules27207099.
- S. Li, Y. Ma, L. Wang, D. Lan, L. Fu and B. Wu, Chem. Biodiversity, 2022, 19, e202200310, DOI:10.1002/cbdv.202200310.
- P. Jiang, J. Luo, Y. Jiang, L. Zhang, L. Jiang, B. Teng, H. Niu, D. Zhang and H. Lei, Mar. Drugs, 2022, 20, 711, DOI:10.3390/md20110711.
- Z. Hu, J. V. Gopal, L. Liu and Z. Gao, Nat. Prod. Res., 2022, 36, 4003–4008, DOI:10.1080/14786419.2021.1892671.
- B. N. S. Ningsih, V. Rukachaisirikul, S. Pansrinun, S. Phongpaichit, S. Preedanon and J. Sakayaroj, Nat. Prod. Res., 2022, 36, 4982–4989, DOI:10.1080/14786419.2021.1915309.
- B. Fan, L. Grauso, F. Li, S. Scarpato, A. Mangoni and D. Tasdemir, Mar. Drugs, 2022, 20, 210, DOI:10.3390/md20030210.
- C. Sun, Q. Liu, M. Shah, Q. Che, G. Zhang, T. Zhu, J. Zhou, X. Rong and D. Li, Org. Lett., 2022, 24, 3993–3997, DOI:10.1021/acs.orglett.2c01394.
- M. Li, S. Li, J. Hu, X. Gao, Y. Wang, Z. Liu and W. Zhang, Mar. Drugs, 2022, 20, 33, DOI:10.3390/md20010033.
- K. Zhang, X. Zhang, R. Lin, H. Yang, F. Song, X. Xu and L. Wang, Mar. Drugs, 2022, 20, 79, DOI:10.3390/md20020079.
- S. Qi, S.-Q. Yang, X.-M. Li, X.-Y. Hu, X. Li and B.-G. Wang, Mar. Drugs, 2022, 20, 529, DOI:10.3390/md20080529.
- C. Zhou, X. Cao, Y. Ge, X. Wu, Z. Zhang, Y. Ma, J. S. Dickschat and B. Wu, J. Nat. Prod., 2022, 85, 2620–2625, DOI:10.1021/acs.jnatprod.2c00638.
- M. Ma, W. Yi, L. Qin, X.-Y. Lian and Z. Zhang, Nat. Prod. Res., 2022, 36, 460–465, DOI:10.1080/14786419.2020.1779265.
- G. Liu, R. Huo, S. Niu, F. Song and L. Liu, Chem. Biodiversity, 2022, 19, e202100990, DOI:10.1002/cbdv.202100990.
- X. Hong, X. Guan, Q. Lai, D. Yu, Z. Chen, X. Fu, B. Zhang, C. Chen, Z. Shao, J. Xia, J.-J. Qin and W. Wang, Appl. Microbiol. Biotechnol., 2022, 106, 2927–2935, DOI:10.1007/s00253-022-11914-1.
- F. Song, Y. Dong, S. Wei, X. Zhang, K. Zhang and X. Xu, Antibiotics, 2022, 11, 222, DOI:10.3390/antibiotics11020222.
- Z. Ren, L. Yang, Q. Ma, Q. Xie, H. Dai, K. Sun and Y. Zhao, Molecules, 2022, 27, 8782, DOI:10.3390/molecules27248782.
- S. Safwan, S.-W. Wang, G. Hsiao, S.-W. Hsiao, Su-J. Hsu, T.-H. Lee and C.-K. Lee, Mar. Drugs, 2022, 20, 80, DOI:10.3390/md20020080.
- S. Nuansri, V. Rukachaisirikul, N. Rungwirain, S. Kaewin, C. Yimnual, S. Phongpaichit, S. Preedanon, J. Sakayaroj and C. Muanprasat, Nat. Prod. Res., 2022, 36, 5462–5469, DOI:10.1080/14786419.2021.2015593.
- Y. Wang, X.-M. Li, S.-Q. Yang, F.-Z. Zhang, B.-G. Wang, H.-L. Li and L.-H. Meng, Mar. Drugs, 2022, 20, 177, DOI:10.3390/md20030177.
- H. Meng-Jiao, P.-N. Chen, H.-J. Li, F. Wu, G.-Y. Zhang, Z.-Z. Shao, X.-P. Liu, W.-Z. Ma, J. Xu, T. Mahmud and W.-J. Lan, Front. Microbiol., 2022, 13, 947226, DOI:10.3389/fmicb.2022.947226.
- K. H. A. U. Zaman, X. Wu, A. M. Sarotti and S. Cao, Nat. Prod. Res., 2022, 36, 5984–5990, DOI:10.1080/14786419.2022.2056890.
- C. Lai, J. Chen, J. Liu, D. Tian, D. Lan, T. Liu, B. Wu, H. Bi and J. Tang, Mar. Drugs, 2022, 20, 720, DOI:10.3390/md20110720.
- Z. G. Khalil, S. Kankanamge and R. J. Capon, Mar. Drugs, 2022, 20, 339, DOI:10.3390/md20060339.
- M. Yoshida, T. Matsushita, S. Kondo, H. Isoda and H. Kigoshi, J. Nat. Prod., 2022, 85, 1850–1860, DOI:10.1021/acs.jnatprod.2c00440.
- S. Subba, S. Saha, S. Mandal, A. J. Ghosh and T. Saha, Tetrahedron Lett., 2022, 106, 154081, DOI:10.1016/j.tetlet.2022.154081.
- M. Alekseychuk and P. Heretsch, J. Am. Chem. Soc., 2022, 144, 21867–21871, DOI:10.1021/jacs.2c10735.
- F. Yang and J. A. Porco Jr, J. Am. Chem. Soc., 2022, 144, 12970–12978, DOI:10.1021/jacs.2c05366.
- L. Posada, L. Rey, J. Villalba, S. Colombo, L. Aubriot, N. Badagian, B. Brena and G. Serra, ChemistrySelect, 2022, 7, e202201956, DOI:10.1002/slct.202201956.
- N. Hauser, M. A. Imhof, S. S. Eichenberger, K. Tomas and E. M. Carreira, Angew. Chem., Int. Ed., 2022, 61, e202112838, DOI:10.1002/anie.202112838.
- S. P. Kelly, V. V. Shende, A. R. Flynn, Q. Dan, Y. Ye, J. L. Smith, S. Tsukamoto, M. S. Sigman and D. H. Sherman, J. Am. Chem. Soc., 2022, 144, 19326–19336, DOI:10.1021/jacs.2c06631.
- G. Zhang, X. Pan, B. Yang, L. Li and Z. Liu, J. Nat. Prod., 2022, 85, 997–1005, DOI:10.1021/acs.jnatprod.1c01151.
- A. S. Reddy, V. K. Reddy, G. N. Rao and K. Basavaiah, Tetrahedron Lett., 2022, 90, 153612, DOI:10.1016/j.tetlet.2021.153612.
- B. K. Yasam and S. Pabbaraja, SynOpen, 2022, 06, 227–237, DOI:10.1055/a-1942-6969.
- T. Gregor, E. Topić, J. Meštrović and N. Cindro, J. Org. Chem., 2022, 87, 16054–16062, DOI:10.1021/acs.joc.2c02200.
- T. L. Pham, P. Sae-Lao, H. H. M. Toh, D. Csókás and R. W. Bates, J. Org. Chem., 2022, 87, 16111–16114, DOI:10.1021/acs.joc.2c02033.
- M. Markovič, K. Peter, S. Sokoliová, N. Boháčiková, T. Vyskočil, J. Moncoľ and T. Gracza, J. Org. Chem., 2022, 87, 15947–15962, DOI:10.1021/acs.joc.2c02092.
- P. Patel, J. Org. Chem., 2022, 87, 4852–4862, DOI:10.1021/acs.joc.2c00197.
- A. N. B. Singab, N. M. Mostafa, Y. A. Elkhawas, E. Al-Sayed, M. M. Bishr, A. M. Elissawy, M. S. Elnaggar, I. M. Fawzy, O. M. Salama, Yi-H. Tsai and F.-R. Chang, Mar. Drugs, 2022, 20, 331, DOI:10.3390/md20050331.
- Y. Liu, Y. Li, M. Chen, Y. Liu, J. Liang, Y. Zhang and Z.-J. Qian, Int. Immunopharmacol., 2022, 109, 108931, DOI:10.1016/j.intimp.2022.108931.
- N. Pan, Z.-C. Li, Z.-H. Li, S.-H. Chen, M.-H. Jiang, H.-Y. Yang, Y.-S. Liu, R. Hu, Y.-W. Zeng, L.-H. Dai, L. Liu and G.-L. Wang, Mar. Drugs, 2022, 20, 23, DOI:10.3390/md20010023.
- S. S. Elhady, M. S. Goda, E. T. Mehanna, M. A. Elfaky, A. E. Koshak, A. O. Noor, H. A. Bogari, R. T. Malatani, R. F. A. Abdelhameed and A. S. Wahba, Biomedicines, 2022, 10, 1164, DOI:10.3390/biomedicines10051164.
- S. A. Dyshlovoy, B. Tobias, J. Hauschild, E. V. Girich, M. Kriegs, K. Hoffer, M. Graefen, A. N. Yurchenko, C. Bokemeyer and G. v. Amsberg, Mar. Drugs, 2022, 20, 597, DOI:10.3390/md20100597.
- D.-C. Kim, T. H. Quang, N. T. Tien, K.-W. Kim, Y.-C. Kim, N. T. T. Ngan, N. X. Cuong, N. H. Nam and H. Oh, Arch. Pharmacal Res., 2022, 45, 90–104, DOI:10.1007/s12272-022-01370-w.
- C. T. Endres, G. V. Rigo, L. A. Loges, M. F. Landell, D. B. Silva, A. J. Macedo and T. Tasca, Mar. Biotechnol., 2022, 24, 1014–1022, DOI:10.1007/s10126-022-10164-6.
- P. Wang, S. Xu, Y. Tang, H. Wang, X. Bai and J. Huawei Zhang, Fungi, 2022, 8, 591, DOI:10.3390/jof8060591.
- G. Yao, X. Bai, B. Zhang, L. Wang, S. Chen and Z. Wang, Microb. Cell Fact., 2022, 21, 136, DOI:10.1186/s12934-022-01859-5.
- K. Sakamoto, H. Sato and M. Uchiyama, J. Org. Chem., 2022, 87, 6432–6437, DOI:10.1021/acs.joc.2c00291.
- S.-W. Yuan, S.-H. Chen, H. Guo, L.-T. Chen, H.-J. Shen, L. Liu and Z.-Z. Gao, Org. Lett., 2022, 24, 3069–3074, DOI:10.1021/acs.orglett.2c01050.
- J. Liu, X. Guo, X. Guo, B. Zhong, T. Wang, L. Dong, H. Jin, J. Ren, Z. Liu, J. Gao, S.-M. Li, A. Fan and W. Lin, J. Nat. Prod., 2022, 85, 2723–2730, DOI:10.1021/acs.jnatprod.2c00614.
- J. Xia, W. Xiao, X. Lin, Y. Zhou, P. Qiu, H. Si, X. Wu, S. Niu, Z. Luo and X. Yang, Mar. Drugs, 2022, 20, 541, DOI:10.3390/md20090541.
- L.-A. Giddings and D. J. Newman, Mar. Drugs, 2022, 20, 62, DOI:10.3390/md20010062.
- G. Zhao, W. Tang, J. Zhang, P. Shi, Y. Li, J. Wang, Q. Shen, H. Si, L. Jiang, X. Yu, H. Zhu, G. Chen, X. Zhang and H. Jia, Front. Mar. Sci., 2022, 9, 929561, DOI:10.3389/fmars.2022.929561.
- C. Pinedo-Rivilla, J. Aleu and R. Durán-Patrón, Mar. Drugs, 2022, 20, 84, DOI:10.3390/md20020084.
- S. L. Knowles, H. A. Raja, C. D. Roberts and N. H. Oberlies, Nat. Prod. Rep., 2022, 39, 1557–1573, 10.1039/D1NP00070E.
- Y. Chen, L.-C. Xu, S. Liu, Z.-X. Zhang and G.-Y. Cao, Front. Microbiol., 2022, 13, 1038487, DOI:10.3389/fmicb.2022.1038487.
- T. M. Voser, M. D. Campbell and A. R. Carroll, Nat. Prod. Rep., 2022, 39, 7–19, 10.1039/D1NP00051A.
- P. Gui, J. Fan, T. Zhu, P. Fu, K. Hong and W. Zhu, Mar. Drugs, 2022, 20, 408, DOI:10.3390/md20070408.
- S. Tilvi, R. Parvatkar, A. Awashank and S. Khan, ChemistrySelect, 2022, 7, e202203742, DOI:10.1002/slct.202203742.
- G. Wang, Z. Yin, S. Wang, Y. Yuan, Y. Chen and W. Kang, Front. Microbiol., 2022, 13, 900227, DOI:10.3389/fmicb.2022.900227.
- S.-Z. Liu, G.-X. Xu, F.-M. He, W.-B. Zhang, Z. Wu, M.-Y. Li, X.-X. Tang and Y.-K. Qiu, Mar. Drugs, 2022, 20, 213, DOI:10.3390/md20030213.
- D.-Y. Huang, X.-H. Nong, Y.-Q. Zhang, W. Xu, L.-Y. Sun, T. Zhang, G.-Y. Chen and C.-R. Han, Nat. Prod. Res., 2022, 36, 3651–3656, DOI:10.1080/14786419.2021.1878168.
- J. Cai, X. Wang, Z. Yang, Y. Tan, B. Peng, Y. Liu and X. Zhou, Front. Microbiol., 2022, 13, 857041, DOI:10.3389/fmicb.2022.857041.
- S.-Z. Liu, X.-X. Tang, F.-M. He, J.-X. Jia, H. Hu, B.-Y. Xie, M.-Y. Li and Y.-K. Qiu, Nat. Prod. Res., 2022, 36, 2370–2378, DOI:10.1080/14786419.2020.1837811.
- Q. Wu, Y. Chang, Q. Che, D. Li, G. Zhang and T. Zhu, Mar. Drugs, 2022, 20, 137, DOI:10.3390/md20020137.
- R.-Q. Mei, B. Wang, W.-N. Zeng, G.-L. Huang, G.-Y. Chen and C.-J. Zheng, Nat. Prod. Res., 2022, 36, 1260–1265, DOI:10.1080/14786419.2021.1873981.
- R. Huo, J. Zhang, S. Niu and L. Liu, Front. Mar. Sci., 2022, 9, 1097594, DOI:10.3389/fmars.2022.1097594.
- S. Niu, J. He, S. Huang, S. Wu, L. Zeng, J. Wang, B. Hong and Z. Chen, Mar. Drugs, 2022, 20, 591, DOI:10.3390/md20100591.
- F. Zhao, X. Zhang, J. Wu, C. Wei, F. Ting, D. Zhou, Z. Wen and J. Xu, Mar. Drugs, 2022, 20, 526, DOI:10.3390/md20080526.
- R. Zhang, J. Zhang, R. Huo, Y. Xue, K. Hong and L. Liu, Front. Mar. Sci., 2022, 9, 1034945, DOI:10.3389/fmars.2022.1034945.
- H. Lee, S. J. Moon, Y. D. Yoo, E. J. Jeong and J.-R. Rho, J. Nat. Prod., 2022, 85, 1495–1502, DOI:10.1021/acs.jnatprod.1c01190.
- E. Murphy, M. Elena Barone, F. Campanile, N. Touzet and O. P. Thomas, Phytochem. Lett., 2022, 51, 104–108, DOI:10.1016/j.phytol.2022.08.001.
- S. Lee, S. J. Moon, Y. D. Yoo, B. S. Hwang, E. J. Jeong and J.-R. Rho, Org. Lett., 2022, 24, 4182–4186, DOI:10.1021/acs.orglett.2c01443.
- M. Akamatsu, R. Hirozumi, Y. Cho, Y. Kudo, K. Konoki, Y. Oshima and M. Yotsu-Yamashita, Mar. Drugs, 2022, 20, 166, DOI:10.3390/md20030166.
- J. S. Murray, S. C. Finch, E. M. Mudge, A. L. Wilkins, J. Puddick, D. T. Harwood, L. L. Rhodes, R. van Ginkel, F. Rise and M. R. Prinsep, Mar. Drugs, 2022, 20, 453, DOI:10.3390/md20070453.
- A. K. Ghosh and H. Yuan, Mar. Drugs, 2022, 20, 587, DOI:10.3390/md20100587.
- L. Ferrié, I. Ciss, J. Fenneteau, S. Vallerotto, M. Seck and F. Bruno, J. Org. Chem., 2022, 87, 1110–1123, DOI:10.1021/acs.joc.1c02458.
- D.-Z. Wang, Y.-H. Xin and M.-H. Wang, Toxins, 2022, 14, 485, DOI:10.3390/toxins14070485.
- M. J. Botelho, J. Milinovic, N. M. Bandarra and C. Vale, Mar. Drugs, 2022, 20, 253, DOI:10.3390/md20040253.
- M. C. Louzao, N. Vilariño, C. Vale, C. Costas, A. Cao, S. Raposo-Garcia, M. R. Vieytes and L. M. Botana, Mar. Drugs, 2022, 20, 198, DOI:10.3390/md20030198.
- J. R. McCall, K. T. Sausman, D. M. Keeler, A. P. Brown and S. L. Turrise, Mar. Drugs, 2022, 20, 233, DOI:10.3390/md20040233.
- K. A. Lefebvre, E. Fachon, E. K. Bowers, D. G. Kimmel, J. A. Snyder, R. Stimmelmayr, J. M. Grebmeier, S. Kibler, D. R. Hardison, D. M. Anderson, D. Kulis, J. Murphy, J. C. Gann, D. Cooper, L. B. Eisner, J. T. Duffy-Anderson, G. Sheffield, R. S. Pickart, A. Mounsey, M. L. Willis, P. Stabeno and E. Siddon, Harmful Algae, 2022, 114, 102205, DOI:10.1016/j.hal.2022.102205.
- Z. B. L. Malto, G. A. Benico, J. D. Batucan, J. D. Cruz, M. L. J. Romero, R. V. Azanza and L. A. Salvador-Reyes, Front. Mar. Sci., 2022, 8, 767024, DOI:10.3389/fmars.2021.767024.
- S. Lage, P. R. Costa, A. V. M. Canário and J. P. D. Silva, Mar. Drugs, 2022, 20, 680, DOI:10.3390/md20110680.
- M. Remize, F. Planchon, M. Garnier, A. N. Loh, F. Le Grand, B. Antoine, C. Lambert, R. Corvaisier, A. Volety and P. Soudant, Mar. Drugs, 2022, 20, 22, DOI:10.3390/md20010022.
- M. Du, Z. Li, J. Wang, F. Wang, S. Zan and C. Gu, J. Hazard. Mater., 2022, 421, 126798, DOI:10.1016/j.jhazmat.2021.126798.
- E. Ternon, O. P. Thomas, R. Lemée and W. H. Gerwick, Mar. Drugs, 2022, 20, 748, DOI:10.3390/md20120748.
- W. Zhou, X. Zhang, A. Wang, L. Yang, Q. Gan, Y. Liang, R. E. Summons, J. K. Volkman and Y. Lu, J. Am. Chem. Soc., 2022, 144, 9023–9032, DOI:10.1021/jacs.2c01401.
- A. Montoya-Arroyo, K. Lehnert, P. E. Lux, V. M. Jiménez, A. Esquivel, A. M Silva-Benavides, J. Vetter and J. Frank, J. Food Compos. Anal., 2022, 107, 104325, DOI:10.1016/j.jfca.2021.104325.
- K. Chakraborty, S. Dhara and A. E. Mani, Nat. Prod. Res., 2022, 36, 4114–4124, DOI:10.1080/14786419.2021.1976173.
- A. H. Elmaidomy, E. M. Zahran, R. Soltane, A. Alasiri, H. Saber, C. J. Ngwa, G. Pradel, F. Alsenani, A. M. Sayed and U. R. Abdelmohsen, Molecules, 2022, 27, 5617, DOI:10.3390/molecules27175617.
- A. H. Shobier and E. S. H. El Ashry, Russ. J. Mar. Biol., 2022, 47, 425–439, DOI:10.1134/S1063074021060122.
- D. S. Ghallab, E. Shawky, R. S. Ibrahim and M. M. Mohyeldin, Sci. Rep., 2022, 12, 8094, DOI:10.1038/s41598-022-12265-7.
- Y. Oishi, R. Otaki, Y. Iijima, E. Kumagai, M. Aoki, M. Tsuzuki, S. Fujiwara and N. Sato, Commun. Biol., 2022, 5, 19, DOI:10.1038/s42003-021-02927-z.
- M. Sekiguchi, S. Shinoda, S. Kurimoto and T. Kubota, Heterocycles, 2022, 104, 1141–1147, DOI:10.3987/COM-22-14651.
- J.-Y. Kim, J. M. Lee, H.-S. Kim, D.-W. Ki, M.-J. Yim, S.-C. Ko, J. M. Shin, M. S. Lee, Y. G. Park and D.-S. Lee, Nat. Prod. Commun., 2022, 17 DOI:10.1177/1934578X211068606.
- J. Kwon, K. Lee, H. Hwang, S.-H. Kim, S. E. Park, P. Durai, K. Park, H.-S. Kim, D. S. Jang, J. S. Choi and H. C. Kwon, Plants, 2022, 11, 1998, DOI:10.3390/plants11151998.
- Ji-Y. Kim, G.-W. Oh, J. M. Lee, H.-S. Kim, D.-W. Ki, S.-C. Ko, Mi-J. Yim, K. W. Kim, D.-S. Lee and K. Baek, Nat. Prod. Commun., 2022, 17 DOI:10.1177/1934578X221137411.
- Y. M. Bakri, I. P. Holland, C. A. Motti and I. A. van Altena, Biochem. Syst. Ecol., 2022, 101, 104392, DOI:10.1016/j.bse.2022.104392.
- S. Dhara and K. Chakraborty, Chem. Biodiversity, 2022, 19, e202100723, DOI:10.1002/cbdv.202100723.
- K. Chakraborty and S. Dhara, Phytochemistry, 2022, 195, 113024, DOI:10.1016/j.phytochem.2021.113024.
- Y. Yamano, M. Tanabe, A. Shimada and A. Wada, Mar. Drugs, 2022, 20, 658, DOI:10.3390/md20110658.
- P. Monteiro, S. Lomartire, J. Cotas, J. C. Marques, L. Pereira and A. M. M. Gonçalves, Mar. Drugs, 2022, 20, 387, DOI:10.3390/md20060387.
- T.-Y. Lau and H.-Y. Kwan, Mar. Drugs, 2022, 20, 370, DOI:10.3390/md20060370.
- S. Kim, D. Kim, M. Y. Um, M. Yoon, J.-S. Choi, Y. H. Choi and S. Cho, Mar. Drugs, 2022, 20, 774, DOI:10.3390/md20120774.
- M. Xiao, X. Han, S.-J. Lee, G. Oh, H. Jin, H.-J. Oh, E. Kim, J. Kim, B.-Y. Lee, S.-I. Choi and O.-H. Lee, Mar. Drugs, 2022, 20, 607, DOI:10.3390/md20100607.
- M. I. Rushdi, I. A. M. Abdel-Rahman, E. Z. Attia, H. Saber, A. A. Saber, G. Bringmann and U. R. Abdelmohsen, Molecules, 2022, 27, 672, DOI:10.3390/molecules27030672.
- M. I. Rushdi, I. A. M. Abdel-Rahman, H. Saber, E. Z. Attia and U. R. Abdelmohsen, J. Mex. Chem. Soc., 2022, 66, 1639, DOI:10.29356/jmcs.v66i1.1639.
- D. P. Nagahawatta, N. M. Liyanage, J.-G. Je, H. H. A. C. K. Jayawardhana, T. U. Jayawardena, S.-H. Jeong, H.-J. Kwon, C. S. Choi and Y.-J. Jeon, Mar. Drugs, 2022, 20, 786, DOI:10.3390/md20120786.
- O. Chadova, A. Skriptsova and P. Velansky, Mar. Drugs, 2022, 20, 428, DOI:10.3390/md20070428.
- J. C. C. Fuentes-Monteverde, N. Nath, A. M. Forero, E. M. Balboa, A. Navarro-Vázquez, C. Griesinger, C. Jiménez and J. Rodríguez, Mar. Drugs, 2022, 20, 462, DOI:10.3390/md20070462.
- S. Radman, M. Čagalj, V. Šimat and I. Jerković, Molecules, 2022, 27, 3012, DOI:10.3390/molecules27093012.
- M. Yu, H. Matsuura, T. Ishii, M. Miyagi, Y. Shinjo, K. Sato, T. Kamada, Y. Mihara, I. Togashi, K. Sugimoto, T. Abe, N. Kikuchi and M. Suzuki, Nat. Prod. Bioprospect., 2022, 12, 10, DOI:10.1007/s13659-022-00328-1.
- M. Harizani, D.-I. Diakaki, S. Perdikaris, V. Roussis and E. Ioannou, Molecules, 2022, 27, 1866, DOI:10.3390/molecules27061866.
- J. Bracegirdle, S. J. Kennedy, C. Shan, L. Wojtas, L. N. Shaw, C. D. Amsler, J. B. McClintock and B. J. Baker, J. Nat. Prod., 2022, 85, 1886–1891, DOI:10.1021/acs.jnatprod.2c00344.
- J. Jacobtorweihen, M. Schmitt and V. Spiegler, Mar. Drugs, 2022, 20, 420, DOI:10.3390/md20070420.
- S. S. Elhady, E. S. Habib, R. F. A. Abdelhameed, M. S. Goda, R. M. Hazem, E. T. Mehanna, M. A. Helal, K. M. Hosny, R. M. Diri, H. A. Hassanean, A. K. Ibrahim, E. E. Eltamany, U. R. Abdelmohsen and S. A. Ahmed, Mar. Drugs, 2022, 20, 63, DOI:10.3390/md20010063.
- R. A. Fernandes, A. Kumar and R. S. Pathare, Chem. Commun., 2022, 58, 11921–11924, 10.1039/D2CC04574E.
- H. B. Hicks, D. S. Brown, H. S. S. Chan, B. A. Sousa, K. E. Christensen and J. W. Burton, Org. Lett., 2022, 24, 9174–9178, DOI:10.1021/acs.orglett.2c03524.
- S. Ghosh, J. E. Erchinger, R. Maji and B. List, J. Am. Chem. Soc., 2022, 144, 6703–6708, DOI:10.1021/jacs.2c01971.
- K. Nishikibe, K. Nishikawa, M. Kumagai, M. Doe and Y. Morimoto, Chem.–Asian J., 2022, 17, e202101137, DOI:10.1002/asia.202101137.
- F. Budiyanto, M. A. Ghandourah, N. O. Bawakid, H. S. Alorfi, A. Abdel-Lateff and W. M. Alarif, Algal Res., 2022, 65, 102751, DOI:10.1016/j.algal.2022.102751.
- J. M. S. Ponte, A. M. L. Seca and M. C. Barreto, Molecules, 2022, 27, 1787, DOI:10.3390/molecules27061787.
- S. R. David, N. B. Baharulnizam and R. Rajabalaya, J. Herb. Med., 2022, 35, 100585, DOI:10.1016/j.hermed.2022.100585.
- W. Zhao, C.-M. Hu, W. Zhou, Y.-Y. Deng, G.-P. Xu, C.-C. Tian, Q.-Q. Lu, S. Lu, M.-R. Zhang and L.-E. Yang, Algal Res., 2022, 61, 102606, DOI:10.1016/j.algal.2021.102606.
- T. S. Steele, J. K. Brunson, Y. Maeno, R. Terada, A. E. Allen, M. Yotsu-Yamashita, J. R. Chekan and B. S. Moore, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2117407119, DOI:10.1073/pnas.2117407119.
- P. -L. Brassart, O. P. Thomas, C. Vincent and N. Papon, ChemBioChem, 2022, 23, e202200223, DOI:10.1002/cbic.202200223.
- C. J. de Muizon, C. Moriou, S. Petek, M. Ekins, M. Rousseau and A. A. Mourabit, Mar. Drugs, 2022, 20, 635, DOI:10.3390/md20100635.
- C. S. Hassane, G. Herbette, E. Garayev, F. Mabrouki, P. Clerc, N. J. de Voogd, S. Greff, I. P. Trougakos, J. Ouazzani, M. Fouillaud, L. Dufossé, B. Baghdikian, E. Ollivier and A. Gauvin-Bialecki, Mar. Drugs, 2022, 20, 186, DOI:10.3390/md20030186.
- A. G. Guzii, T. N. Makarieva, S. N. Fedorov, A. S. Menshov, V. A. Denisenko, R. S. Popov, E. A. Yurchenko, E. S. Menchinskaya, B. B. Grebnev, V. V. Iarotsckaia, N. Y. Kim and V. A. Stonik, J. Nat. Prod., 2022, 85, 1186–1191, DOI:10.1021/acs.jnatprod.2c00130.
- D. Kovalerchik, A. Zovko, P. Hååg, A. Sierakowiak, K. Viktorsson, R. Lewensohn, M. Ilan and S. Carmeli, Mar. Drugs, 2022, 20, 265, DOI:10.3390/md20040265.
- X. -L. Liu, Y. -F. Ding, S. -P. Wang, L. Liu, J. Wang and Y. Fan, Chem. Biodiversity, 2022, 19, e202200159, DOI:10.1002/cbdv.202200159.
- A. N. L. Batista, M. F. Brito, J. M. Batista Jr and A. L. Valverde, Tetrahedron Lett., 2022, 112, 154241, DOI:10.1016/j.tetlet.2022.154241.
- A. Castaldi, R. Teta, G. Esposito, M. A. Beniddir, N. J. de Voogd, S. Duperron, V. Costantino and M.-L. Bourguet-Kondracki, Mar. Drugs, 2022, 20, 673, DOI:10.3390/md20110673.
- J. Bracegirdle, D. Casandra, J. R. Rocca, J. H. Adams and B. J. Baker, J. Nat. Prod., 2022, 85, 2454–2460, DOI:10.1021/acs.jnatprod.2c00684.
- A. Ortiz-Celiseo, G. Valerio-Alfaro, J. Sosa-Rueda, F. C. López-Fentanes, V. Domínguez-Melendez and F. Cen-Pacheco, Nat. Prod. Res., 2022, 36, 3957–3964, DOI:10.1080/14786419.2021.1902326.
- A. H. H. El-Desoky, Y. Hitora, K. Onodera, Y. Ise, F. Losung, E. Remy, P. Mangindaan and S. Tsukamoto, Chem. Pharm. Bull., 2022, 70, 818–822, DOI:10.1248/cpb.c22-00523.
- K. Sugawara, D. Kanki, R. Watanabe, R. Matsushima, Y. Ise, H. Yokose, Y. Morii, N. Yamawaki, A. Ninomiya, S. Okada and S. Matsunaga, Tetrahedron, 2022, 119, 132859, DOI:10.1016/j.tet.2022.132859.
- J. R. Haedar, A. R. Uria, S. Lallo, D. F. Dibwe and T. Wakimoto, Mar. Drugs, 2022, 20, 661, DOI:10.3390/md20110661.
- O. Hasin, S. Shoham, Y. Kashman, M. Ilan and S. Carmeli, Mar. Drugs, 2022, 20, 31, DOI:10.3390/md20010031.
- Y. Wu, Z.-M. Wu, S.-S. Zhang, Li-Y. Liu, F. Sun, W.-H. Jiao, S.-P. Wang and H.-W. Lin, Org. Lett., 2022, 24, 934–938, DOI:10.1021/acs.orglett.1c04309.
- G. Tarazona, R. Fernández, M. Pérez, R. E. Millán, C. Jiménez, J. Rodríguez and C. Cuevas, J. Nat. Prod., 2022, 85, 1059–1066, DOI:10.1021/acs.jnatprod.1c01179.
- C. Wang, Y. Lei, X. Liao, S. Xu and B. Zhao, Chin. J. Org. Chem., 2022, 42, 1839–1842, DOI:10.6023/cjoc202201024.
- S. Afoullouss, A. R. Sanchez, L. K. Jennings, Y. Kee, A. L. Allcock and O. P. Thomas, Mar. Drugs, 2022, 20, 52, DOI:10.3390/md20010052.
- D. Zhang, Y. Li, X. Li, X. Han, Z. Wang, W. Zhang, B. Dou, Z. Lu, P. Li and G. Li, J. Nat. Prod., 2022, 85, 1626–1633, DOI:10.1021/acs.jnatprod.2c00292.
- H. A. Sahile, D. E. Williams, N. J. de Voogd, M. Ko, R. J. Andersen and Y. Av-Gay, J. Antibiot., 2022, 75, 213–225, DOI:10.1038/s41429-022-00507-9.
- S. Kurimoto, S. Suzuki, M. Ueno, J. Fromont, J. Kobayashi and T. Kubota, J. Nat. Prod., 2022, 85, 2226–2231, DOI:10.1021/acs.jnatprod.2c00395.
- T.-H. Huynh, N.-F. Chen, J.-R. Weng, S.-Y. Chien, Y.-H. Liu, Y.-C. Tsai and P.-J. Sung, Tetrahedron Lett., 2022, 95, 153748, DOI:10.1016/j.tetlet.2022.153748.
- Q.-Q. He, Y.-Q. Man, K.-L. Sun, L.-J. Yang, Y. Wu, J. Du, W.-W. Chen, J.-J. Dai, N. Ni, S. Miao and K.-K. Gong, Nat. Prod. Res., 2022, 36, 6215–6223, DOI:10.1080/14786419.2021.2024533.
- D. T. Trang, B. H. Tai, D. T. T. Hang, P. H. Yen, N. X. Nhiem and P. V. Kiem, Nat. Prod. Res., 2022, 36, 5022–5031, DOI:10.1080/14786419.2021.1917572.
- Q. T. Khong, D. Li, B. A. P. Wilson, K. Ranguelova, M. Dalilian, E. A. Smith, W. Antony, E. I. Goncharova, T. Grkovic, D. Voeller, S. Lipkowitz, M. J. Schnermann, B. R. O'Keefe and L. Du, Org. Lett., 2022, 24, 9468–9472, DOI:10.1021/acs.orglett.2c03922.
- Y.-C. Lin, C.-H. Chao, C.-W. Fu, S.-F. Chiou, T.-Y. Huang, Y.-J. Yang, S.-H. Wu, S.-L. Chen, H.-C. Wang, M.-C. Yu, H.-C. Huang and J.-H. Sheu, Tetrahedron, 2022, 126, 133077, DOI:10.1016/j.tet.2022.133077.
- L. Xu, P. Wang, S. Yuan, L. Yu, J. Zhao, G. Li, G. Zhang and L. Luo, Chem. Biodiversity, 2022, 19, e202200311, DOI:10.1002/cbdv.202200311.
- Q. Wang, C. Gao, Z. Wei, X. Tang, L. Ji, X. Luo, X. Peng, G. Li and H. Lou, Mar. Drugs, 2022, 20, 454, DOI:10.3390/md20070454.
- V. F. Freire, J. R. Gubiani, T. M. Spencer, E. Hajdu, A. G. Ferreira, D. A. S. Ferreira, E. V. de Castro Levatti, J. E. Burdette, C. H. Camargo, A. G. Tempone and R. G. S. Berlinck, J. Nat. Prod., 2022, 85, 1340–1350, DOI:10.1021/acs.jnatprod.2c00094.
- S. A. Dyshlovoy, L. K. Shubina, T. N. Makarieva, A. G. Guzii, J. Hauschild, N. Strewinsky, D. V. Berdyshev, E. K. Kudryashova, A. S. Menshov, R. S. Popov, P. S. Dmitrenok, M. Graefen, C. Bokemeyer and G. v. Amsberg, Mar. Drugs, 2022, 20, 738, DOI:10.3390/md20120738.
- D. W. Prebble, T. M. Voser, S. Er, I. Hlushchuk, A. Domanskyi, M. Airavaara, M. G. Ekins, G. D. Mellick and A. R. Carroll, Results Chem., 2022, 4, 100302, DOI:10.1016/j.rechem.2022.100302.
- P.-E. Campos, G. Herbette, L. Fougère, P. Clerc, F. Tintillier, N. J. de Voogd, G. Le Goff, O. Jamal and A. Gauvin-Bialecki, Mar. Drugs, 2022, 20, 637, DOI:10.3390/md20100637.
- S. Kurimoto, A. Okamoto, S. Seino, J. Fromont, J. Kobayashi and T. Kubota, Tetrahedron Lett., 2022, 103, 153985, DOI:10.1016/j.tetlet.2022.153985.
- M. N. Salib, R. Hendra and T. F. Molinski, J. Org. Chem., 2022, 87, 12831–12843, DOI:10.1021/acs.joc.2c01415.
- R. Hendra, M. N. Salib and T. F. Molinski, J. Nat. Prod., 2022, 85, 2207–2216, DOI:10.1021/acs.jnatprod.2c00595.
- B. Chen, Q. Zhao, Y.-C. Gu, L. Lan, C.-Y. Wang and Y.-W. Guo, Mar. Drugs, 2022, 20, 118, DOI:10.3390/md20020118.
- H.-Y. Liu, M. Zhou, R.-Y. Shang, L.-L. Hong, G.-H. Wang, W.-J. Tian, W.-H. Jiao, H.-F. Chen and H.-W. Lin, Chin. J. Nat. Med., 2022, 20, 148–154, DOI:10.1016/S1875-5364(22)60161-4.
- W.-Z. Tang, H.-M. Zhao, T. Yuan, S.-W. Dai, A. Zhang, H.-W. Lin, C.-X. Zhang and Y. Fan, Tetrahedron Lett., 2022, 93, 153690, DOI:10.1016/j.tetlet.2022.153690.
- D. Pech-Puch, A. M. Forero, J. C. Fuentes-Monteverde, C. Lasarte-Monterrubio, M. Martinez-Guitian, C. González-Salas, S. Guillén-Hernández, H. Villegas-Hernández, A. Beceiro, C. Griesinger, J. Rodríguez and C. Jiménez, Mar. Drugs, 2022, 20, 298, DOI:10.3390/md20050298.
- C.-W. Fu, Y.-C. Lin, S.-F. Chiou, T.-Y. Huang, Y.-J. Yang, S.-H. Wu, S.-L. Chen, C.-C. Lin, H.-C. Wang, M.-C. Yu and J.-H. Sheu, Tetrahedron Lett., 2022, 103, 153964, DOI:10.1016/j.tetlet.2022.153964.
- M. Miguel-Gordo, M. M. Reddy, P. Sánchez, J. J. Buckley, T. A. Mackenzie, L. K. Jennings, F. Reyes, K. Calabro and O. P. Thomas, Org. Biomol. Chem., 2022, 20, 1031–1040, 10.1039/D1OB02297K.
- S.-M. Shen, Q. Yang, Y. Zang, L. Jia, X. Liu and Y.-W. Guo, Beilstein J. Org. Chem., 2022, 18, 916–925, DOI:10.3762/bjoc.18.91.
- M.-H. Zhou, X.-C. Luo, H.-M. Zhao, J.-R. Lu, Y. Dai, Y. Yu, L. Zhang, H.-W. Lin and Y. Fan, Chem. Biodiversity, 2022, 19, e202200455, DOI:10.1002/cbdv.202200455.
- S.-M. Shen, Z.-Y. Zhang, L.-G. Yao, J.-R. Wang, Y.-W. Guo and X.-W. Li, Chin. J. Chem., 2022, 40, 235–246, DOI:10.1002/cjoc.202100676.
- Y. Liang, X. Liao, L. Ling, Y. Yang, B. Zhao and S. Xu, Chin. J. Org. Chem., 2022, 42, 901–904, DOI:10.6023/cjoc202109024.
- T. Jin, P. Li, C. Wang, X. Tang, X. Yu, F. Sun, L. Luo, H. Ou and G. Li, Phytochemistry, 2022, 194, 113006, DOI:10.1016/j.phytochem.2021.113006.
- C.-J. Tai, A. F. Ahmed, C.-H. Chao, C.-H. Yen, T.-L. Hwang, F.-R. Chang, Y. M. Huang and J.-H. Sheu, Mar. Drugs, 2022, 20, 241, DOI:10.3390/md20040241.
- T. Wauke, N. Hanif, S. Ohlinger, N. J. de Voogd, V. Kurnianda, T. Jomori and J. Tanaka, Chem. Lett., 2022, 51, 1080–1082, DOI:10.1246/cl.220371.
- C.-J. Tai, A. F. Ahmed, C.-H. Chao, C.-H. Yen, T.-L. Hwang, F.-R. Chang, Y. M. Huang and J.-H. Sheu, Mar. Drugs, 2022, 20, 498, DOI:10.3390/md20080498.
- S. A. Dyshlovoy, L. K. Shubina, T. N. Makarieva, J. Hauschild, N. Strewinsky, A. G. Guzii, A. S. Menshov, R. S. Popov, B. B. Grebnev, B. Tobias, Su J. Oh-Hohenhorst, T. Maurer, D. Tilki, M. Graefen, C. Bokemeyer, V. A. Stonik and G. v. Amsberg, Sci. Rep., 2022, 12, 13570, DOI:10.1038/s41598-022-17447-x.
- D. T. Hang, D. T. Trang, B. H. Tai, P. H. Yen, V. K. Thu, N. X. Nhiem and P. Van Kiem, Nat. Prod. Res., 2022, 36, 5247–5254, DOI:10.1080/14786419.2021.1929973.
- T. Majer, K. Bhattarai, S. Jan, J. Pohlmann, P. Cahill, M. O. Zimmermann, M. P. Hübner, M. Kaiser, J. Svenson, M. Schindler, H. Brötz-Oesterhelt, F. M. Boeckler and H. Gross, Mar. Drugs, 2022, 20, 532, DOI:10.3390/md20080532.
- M. Wang, A. Sciorillo, S. Read, D. N. Divsalar, K. Gyampoh, G. Zu, Z. Yuan, K. Mounzer, D. E. Williams, L. J. Montaner, N. de Voogd, I. Tietjen and R. J. Andersen, J. Nat. Prod., 2022, 85, 1274–1281, DOI:10.1021/acs.jnatprod.1c01225.
- B. Chen, P. Qiu, B. Xu, Q. Zhao, Y.-C. Gu, L. Fu, S. Bi, L. Lan, C.-Y. Wang and Y.-W. Guo, J. Nat. Prod., 2022, 85, 1799–1807, DOI:10.1021/acs.jnatprod.2c00348.
- D. T. Trang, D. T. Dung, N. X. Nhiem, N. T. Cuc, P. H. Yen, D. T. T. Hang, T. M. Linh, N. C. Mai, P. T. T. Huong, B. H. Tai and P. V. Kiem, Tetrahedron Lett., 2022, 89, 153607, DOI:10.1016/j.tetlet.2021.153607.
- S. Hayes, A. C. Taki, K. Y. Lum, J. J. Byrne, J. M. White, M. G. Ekins, R. B. Gasser and R. A. Davis, J. Nat. Prod., 2022, 85, 1723–1729, DOI:10.1021/acs.jnatprod.2c00229.
- H. N. K. Tran, M. J. Kim and Y.-J. Lee, Mar. Drugs, 2022, 20, 604, DOI:10.3390/md20100604.
- A.-Y. Shin, H.-S. Lee and J. Lee, Mar. Drugs, 2022, 20, 726, DOI:10.3390/md20110726.
- P. V. Kiem, D. T. Trang, B. H. Tai, N. X. Nhiem, D. T. T. Hang, D. C. Thung, D. T. Thao, P. H. Yen and P. T. T. Huong, Nat. Prod. Commun., 2022, 17 DOI:10.1177/1934578X221088061.
- B. Chen, Y.-C. Gu, N. J. de Voogd, C.-Y. Wang and Y.-W. Guo, Nat. Prod. Res., 2022, 36, 1941–1947, DOI:10.1080/14786419.2020.1837820.
- S. A. Dyshlovoy, S. N. Fedorov, V. I. Svetashev, T. N. Makarieva, A. I. Kalinovsky, O. P. Moiseenko, V. B. Krasokhin, L. K. Shubina, A. G. Guzii, G. von Amsberg and V. A. Stonik, Mar. Drugs, 2022, 20, 409, DOI:10.3390/md20070409.
- P. Francis and K. Chakraborty, Chem. Biodiversity, 2022, 19, e202100838, DOI:10.1002/cbdv.202100838.
- R. F. Angawi, M. M. Alamri, S.-E. N. Ayyad, A. A. Alotaibi, M. M. Aly and W. M. Alarif, Chem. Nat. Compd., 2022, 58, 301–306, DOI:10.1007/s10600-022-03664-3.
- P. Francis and K. Chakraborty, Nat. Prod. Res., 2022, 36, 3069–3077, DOI:10.1080/14786419.2021.1956491.
- K. Chakraborty and P. Francis, Nat. Prod. Res., 2022, 36, 3786–3795, DOI:10.1080/14786419.2021.1887177.
- K. Chakraborty and P. Francis, Nat. Prod. Res., 2022, 36, 5676–5687, DOI:10.1080/14786419.2021.2013216.
- S. Okazaki, K. Senda, A. Tokuta, M. Inagaki, K. Kamaike, K. Ota and H. Miyaoka, Org. Biomol. Chem., 2022, 20, 6771–6775, 10.1039/D2OB01032A.
- H. Konno, A. Tan, K. Takamatsu, F. Xu and S. Osanai, Heterocycles, 2022, 105, 397–405, DOI:10.3987/COM-21-S(R)6.
- Y. Cui, M. Zhang, H. Xu, T. Zhang, S. Zhang, X. Zhao, P. Jiang, L. Jing, B. Ye, Y. Sun, M. Wang, Y. Deng, Q. Meng, Y. Liu, Q. Fu, J. Lin, L. Wang and Y. Chen, J. Med. Chem., 2022, 65, 2971–2987, DOI:10.1021/acs.jmedchem.1c01583.
- C. Bérubé, A. Borgia and N. Voyer, Tetrahedron Lett., 2022, 92, 153677, DOI:10.1016/j.tetlet.2022.153677.
- T. P. Stockdale, N. Y. S. Lam and I. Paterson, Chem. Commun., 2022, 58, 5729–5732, 10.1039/D2CC01802K.
- N. Y. S. Lam and I. Paterson, Eur. J. Org Chem., 2022, 2022, e202200467, DOI:10.1002/ejoc.202200467.
- L. Marcourt and G. Massiot, Eur. J. Org Chem., 2022, 2022, e202200208, DOI:10.1002/ejoc.202200208.
- L.-X. Liu, R.-R. Gu, Y. Jin, X.-Q. Chen, X.-W. Li, Y.-M. Zheng, Z.-B. Gao and Y.-W. Guo, Bioorg. Chem., 2022, 126, 105909, DOI:10.1016/j.bioorg.2022.105909.
- A. G. Dalling, G. Späth and A. Fürstner, Angew. Chem., Int. Ed., 2022, 61, e202209651, DOI:10.1002/anie.202209651.
- T. Abe, N. Ren, T. Yamashiro and D. Sawada, J. Nat. Prod., 2022, 85, 2122–2125, DOI:10.1021/acs.jnatprod.2c00559.
- S. Tang, Z. Wu, M. Gao, G. Li and Z. -J. Yao, Chem. - Eur. J., 2022, 28, e202200644, DOI:10.1002/chem.202200644.
- Y. Chen, P. Lan and M. G. Banwell, Org. Lett., 2022, 24, 2931–2934, DOI:10.1021/acs.orglett.2c00955.
- J. Paciorek, D. Höfler, K. R. Sokol, K. Wurst and T. Magauer, J. Am. Chem. Soc., 2022, 144, 19704–19708, DOI:10.1021/jacs.2c10010.
- K. Maxfield, M. Payne and S. Chamberland, ACS Omega, 2022, 7, 22915–22929, DOI:10.1021/acsomega.2c02913.
- S. Hu and Y. Tang, J. Am. Chem. Soc., 2022, 144, 19521–19531, DOI:10.1021/jacs.2c08435.
- Y. Wu, X. Du, X. Wang, H. Liu, L. Zhou, Y. Tang and D. Li, Org. Chem. Front., 2022, 9, 4705–4711, 10.1039/D2QO00792D.
- K. Ooka, K. Nakanishi, Y. Udagawa, Y. Ichikawa and S. Hosokawa, Org. Biomol. Chem., 2022, 20, 8236–8242, 10.1039/D2OB00486K.
- K. P. Reber and E. L. Niner, Beilstein J. Org. Chem., 2022, 18, 1629–1635, DOI:10.3762/bjoc.18.174.
- S. Kamo, H. Kurosawa, A. Matsuzawa and K. Sugita, Org. Lett., 2022, 24, 921–923, DOI:10.1021/acs.orglett.1c04289.
- M. A. Hardy, B. Nan, O. Wiest and S. Richmond, Tetrahedron, 2022, 104, 132584, DOI:10.1016/j.tet.2021.132584.
- M. Yanagihara, K. Nakahara, N. Kishimoto, T. Abe, S. Miura, S. Misumi, M. Sako, M. Arisawa and K. Murai, J. Org. Chem., 2022, 87, 16913–16917, DOI:10.1021/acs.joc.2c02278.
- Y. Xiong, Y.-H. Chen, T. Li, J.-H. Xie and Qi-L. Zhou, Org. Lett., 2022, 24, 5161–5165, DOI:10.1021/acs.orglett.2c01997.
- J. Lee, S. Joung and H. -Y. Lee, Asian J. Org. Chem., 2022, 11, e202200533, DOI:10.1002/ajoc.202200533.
- M. Zhang, H. Fan, Y. Wang and J. Gui, Org. Lett., 2022, 24, 7383–7387, DOI:10.1021/acs.orglett.2c02910.
- Y. Miao, X. Li, M. Zhang, H. Fan and J. Gui, Org. Lett., 2022, 24, 1684–1688, DOI:10.1021/acs.orglett.2c00281.
- K. C. Nicolaou, R. Yu, Z. Lu and F. G. Alvarez, J. Am. Chem. Soc., 2022, 144, 5190–5196, DOI:10.1021/jacs.2c01305.
- R. Esposito, S. Federico, M. Bertolino, V. Zupo and M. Costantini, Mar. Drugs, 2022, 20, 244, DOI:10.3390/md20040244.
- M.-J. Chu, L. Meng, M. He, P.-L. Li and G.-Q. Li, RSC Adv., 2022, 12, 7789–7820, 10.1039/D1RA08765G.
- M.-J. Xu, L.-J. Zhong, J.-K. Chen, Q. Bu and L.-F. Liang, Mar. Drugs, 2022, 20, 144, DOI:10.3390/md20020144.
- S. R. M. Ibrahim, S. A. Fadil, H. A. Fadil, R. H. Hareeri, H. M. Abdallah and G. A. Mohamed, Molecules, 2022, 27, 5969, DOI:10.3390/molecules27185969.
- S. R. M. Ibrahim, S. A. Fadil, H. A. Fadil, R. H. Hareeri, S. O. Alolayan, H. M. Abdallah and G. A. Mohamed, Mar. Drugs, 2022, 20, 221, DOI:10.3390/md20040221.
- C. Porras-Alcalá, F. Moya-Utrera, M. García-Castro, A. Sánchez-Ruiz, J. M. López-Romero, M. S. Pino-González, A. Díaz-Morilla, S. Kitamura, D. W. Wolan, J. Prados, C. Melguizo, C.-S. Iván and F. Sarabia, Mar. Drugs, 2022, 20, 373, DOI:10.3390/md20060373.
- V. M. Nadar, S. Manivannan, R. Chinnaiyan, M. Govarthanan and P. Kumar, Chem. Biol. Drug Des., 2022, 99, 103–110, DOI:10.1111/cbdd.13932.
- H.-B. Yu, H.-Y. Chen, S. Duan, Y.-P. Zhu, B. Hu, Y. He, S. T. Cheng, B.-H. Jiao and X.-Y. Liu, Chem. Biodiversity, 2022, 19, e202200049, DOI:10.1002/cbdv.202200049.
- R. M. B. Relat, P. L. Winder, G. D. Bowden, E. A. Guzmán, T. A. Peterson, S. A. Pomponi, J. C. Roberts, A. E. Wright and R. M. O’Connor, Mar. Drugs, 2022, 20, 240, DOI:10.3390/md20040240.
- D. L. Dayanidhi, J. A. Somarelli, J. B. Mantyh, G. Rupprecht, R. S. Roghani, S. Vincoff, I. Shin, Y. Zhao, S. Y. Kim, S. McCall, J. Hong and D. S. Hsu, Front. Med., 2022, 9, 999004, DOI:10.3389/fmed.2022.999004.
- S. Hardy, Y.-M. Choo, M. Hamann and C. James, Mar. Drugs, 2022, 20, 647, DOI:10.3390/md20100647.
- Q. Wang, D. Chen, Y. Wang, C. Dong, J. Liu, K. Chen, F. Song, C. Wang, J. Yuan, R. A. Davis, V. Kuek, H. Jin and J. Xu, Biomed. Pharmacother., 2022, 154, 113622, DOI:10.1016/j.biopha.2022.113622.
- S. Miao, Q. He, L. Chen, Y. Wu, M. Liu, Y. Chen, S. Qi and K. Gong, Pharm. Biol., 2022, 60, 1502–1510, DOI:10.1080/13880209.2022.2102657.
- D. W. Prebble, S. Er, I. Hlushchuk, A. Domanskyi, M. Airavaara, M. G. Ekins, G. D. Mellick and A. R. Carroll, Bioorg. Med. Chem. Lett., 2022, 64, 128677, DOI:10.1016/j.bmcl.2022.128677.
- D. W. Prebble, S. Er, M. Xu, I. Hlushchuk, A. Domanskyi, M. Airavaara, M. G. Ekins, G. D. Mellick and A. R. Carroll, Results Chem., 2022, 4, 100472, DOI:10.1016/j.rechem.2022.100472.
- M. Carnovali, M. L. Ciavatta, E. Mollo, V. Roussis, G. Banfi, M. Carbone and M. Mariotti, Mar. Drugs, 2022, 20, 135, DOI:10.3390/md20020135.
- J. Lever, F. Kreuder, J. Henry, A. Hung, P.-M. Allard, R. Brkljača, C. Rix, A. Taki, R. Gasser, K. Jan, D. Wlodkowic, J.-L. Wolfender and S. Urban, Mar. Drugs, 2022, 20, 554, DOI:10.3390/md20090554.
- M. Surti, M. Patel, A. Redhwan, L. A. Al-Keridis, A. Mohd, N. Alshammari and M. N. Reddy, Mar. Drugs, 2022, 20, 582, DOI:10.3390/md20090582.
- A. Ottaviani, J. Welsch, K. Agama, Y. Pommier, A. Desideri, B. J. Baker and J. Paola Fiorani, J. Enzyme Inhib. Med. Chem., 2022, 37, 1404–1410, DOI:10.1080/14756366.2022.2078320.
- S. Uppal, J. L. Metz, R. K. M. Xavier, K. K. Nepal, D. Xu, G. Wang, J. C. Kwan and J. Ravel, mBio, 2022, 13, e01524, DOI:10.1128/mbio.01524-22.
- A. C. Abraham, D. J. Gochfeld, B. Avula, K. J. Macartney, M. P. Lesser and M. Slattery, Mar. Ecol.: Prog. Ser., 2022, 690, 51–64, DOI:10.3354/meps14042.
- P. Cárdenas, J. Gamage, C. M. Hettiarachchi and S. Gunasekera, Mar. Drugs, 2022, 20, 190, DOI:10.3390/md20030190.
- P. D. Scesa, Z. Lin and E. W. Schmidt, Nat. Chem. Biol., 2022, 18, 659–663, DOI:10.1038/s41589-022-01027-1.
- I. Burkhardt, T. de Rond, P. Y.-T. Chen and B. S. Moore, Nat. Chem. Biol., 2022, 18, 664–669, DOI:10.1038/s41589-022-01026-2.
- C. Laguionie-Marchais, A. L. Allcock, B. J. Baker, E.-A. Conneely, S. G. Dietrick, F. Kearns, K. McKeever, R. M. Young, C. A. Sierra, S. Soldatou, H. L. Woodcock and M. P. Johnson, Mar. Drugs, 2022, 20, 42, DOI:10.3390/md20010042.
- P.-J. Sung, C.-C. Liaw, J.-S. Zeng, S.-H. Chen, Y.-H. Lo, N.-F. Chen, Z.-H. Wen and J.-J. Chen, Heterocycles, 2022, 104, 178–184, DOI:10.3987/COM-21-14565.
- N. E. Avalon, N. Jordan, C. de Marco Verissimo, L. C. Warrensford, S. G. Dietrick, A. R. Pittman, R. M. Young, F. L. Kearns, T. Smalley, J. M. Binning, J. P. Dalton, M. P. Johnson, H. L. Woodcock, A. Louise Allcock and B. J. Baker, J. Nat. Prod., 2022, 85, 1315–1323, DOI:10.1021/acs.jnatprod.2c00054.
- W. Jiang, X. Tian, D. Wang, H. R. Bokesch, C. L. Thomas, G. M. Woldemichael, B. E. Gryder, J. S. Wei, Y. K. Song, H.-C. Chou, J. Khan, B. R. O'Keefe and K. R. Gustafson, J. Nat. Prod., 2022, 85, 1419–1427, DOI:10.1021/acs.jnatprod.2c00246.
- P. García-García, R. Reyes, C. Évora, A. Delgado, J. J. Fernández and A. H. Daranas, Biomed. Pharmacother., 2022, 147, 112631, DOI:10.1016/j.biopha.2022.112631.
- G. H. Phan, H.-C. Hu, F.-R. Chang, Z.-H. Wen, J.-J. Chen, H.-M. Chung, Yu-C. Tsai and P.-J. Sung, RSC Adv., 2022, 12, 27970–27976, 10.1039/D2RA05015C.
- X. Luo, R. Wu, X. Han, X. Tang, Q. Wang, P. Li and G. Li, RSC Adv., 2022, 12, 2662–2667, 10.1039/D1RA08631F.
- J. T. Welsch, R. M. Young, A. Louise Allcock, M. P. Johnson and B. J. Baker, J. Nat. Prod., 2022, 85, 2395–2398, DOI:10.1021/acs.jnatprod.2c00602.
- C. S. Vairappan, K. Tani, T. Kamada and C.-S. Phan, Heterocycles, 2022, 104, 1469–1476, DOI:10.3987/COM-22-14667.
- X. Han, Q. Wang, X. Luo, X. Tang, Z. Wang, D. Zhang, S. Cao, P. Li and G. Li, Org. Lett., 2022, 24, 11–15, DOI:10.1021/acs.orglett.1c03386.
- A.-C. D. Limon, H. M. L. W. Patabendige, A. Ala, X. Sun, D. E. Kyle, N. G. Wilson and B. J. Baker, Mar. Drugs, 2022, 20, 576, DOI:10.3390/md20090576.
- J. Liu, F. Xia, O. Han, W. Wang, T. Li, Y. Shi, X. Yan, X. Yan and H. Shan, Phytochemistry, 2022, 196, 113088, DOI:10.1016/j.phytochem.2022.113088.
- Y. Fan, H. Qiang, L.-G. Yao, L.-F. Liang, Y.-X. Lou, Y.-H. Lu, F.-L. An and Y.-W. Guo, Tetrahedron Lett., 2022, 88, 153571, DOI:10.1016/j.tetlet.2021.153571.
- G. Zhang, X. Tang, L. Luo, X. Zhang, P. Li and G. Li, Bioorg. Chem., 2022, 128, 106040, DOI:10.1016/j.bioorg.2022.106040.
- X.-L. Li, R. Tong, A. Navarro-Vázquez, P. Lindemann, M. Nazaré, X.-W. Li, Y.-W. Guo and S. Han, J. Nat. Prod., 2022, 85, 1730–1737, DOI:10.1021/acs.jnatprod.2c00257.
- Z.-H. Chen, S.-Q. Lu, G.-Y. Han, X.-W. Li and Y.-W. Guo, Mar. Drugs, 2022, 20, 272, DOI:10.3390/md20040272.
- M. P. Rahelivao, I. Bauer, T. Lübken, O. Kataeva, A. Vehlow, N. Cordes and H. -J. Knölker, Eur. J. Org Chem., 2022, 2022, e202200809, DOI:10.1002/ejoc.202200809.
- G. Stakanovs, S. Belyakov, A. Jirgensons and D. Rasina, Org. Biomol. Chem., 2022, 20, 2455–2461, 10.1039/d2ob00238h.
- Z. Zeng, M. Zhang, H. Wang, L. Jia, Y. Guo and M. Su, Chin. J. Org. Chem., 2022, 42, 891–895, DOI:10.6023/cjoc202109004.
- X. Han, X. Luo, L. Xue, L. van Ofwegen, W. Zhang, K. Liu, Y. Zhang, X. Tang, P. Li and G. Li, J. Nat. Prod., 2022, 85, 276–283, DOI:10.1021/acs.jnatprod.1c01103.
- Y. Gao, Y.-Q. Du, Y. Zang, H.-C. Liu, H.-Y. Wan, L. Jia, X.-W. Li and Y.-W. Guo, ACS Omega, 2022, 7, 3052–3059, DOI:10.1021/acsomega.1c06156.
- Y.-Q. Du, J. Chen, M.-J. Wu, H.-Y. Zhang, L.-F. Liang and Y.-W. Guo, Mar. Drugs, 2022, 20, 602, DOI:10.3390/md20100602.
- J. Li, X.-J. Huan, M.-J. Wu, Z.-H. Chen, B. Chen, Z.-H. Miao, Y.-W. Guo and X.-W. Li, Nat. Prod. Res., 2022, 36, 3324–3330, DOI:10.1080/14786419.2020.1855645.
- Q. Bu, M. Yang, X.-Y. Yan, S.-W. Li, Z.-Y. Ge, L. Zhang, L.-G. Yao, Y.-W. Guo and L.-F. Liang, Mar. Drugs, 2022, 20, 566, DOI:10.3390/md20090566.
- M.-J. Wu, D.-D. Yu, M.-Z. Su, J.-R. Wang, L. Gong, Z.-Y. Zhang, H. Wang and Y.-W. Guo, Org. Chem. Front., 2022, 9, 5921–5928, 10.1039/D2QO01265K.
- I. I. Rodríguez, A. D. Rodríguez and C. L. Barnes, Molecules, 2022, 27, 7879, DOI:10.3390/molecules27227879.
- J. C. Toledano, Nat. Prod. Commun., 2022, 17 DOI:10.1177/1934578X221079415.
- C. E. Schumacher, M. Rausch, T. Greven, J. -M. Neudörfl, T. Schneider and H. -G. Schmalz, Eur. J. Org Chem., 2022, 2022, e202200058, DOI:10.1002/ejoc.202200058.
- W. M. Amberg and E. M. Carreira, J. Am. Chem. Soc., 2022, 144, 15475–15479, DOI:10.1021/jacs.2c07150.
- J.-F. Li, Y.-B. Zeng, W.-S. Li, H. Luo, H.-Y. Zhang and Y.-W. Guo, Chin. J. Chem., 2022, 40, 79–90, DOI:10.1002/cjoc.202100564.
- Q. Bu, M. Yang, X.-Y. Yan, L.-G. Yao, Y.-W. Guo and L.-F. Liang, Int. J. Biol. Macromol., 2022, 222, 880–886, DOI:10.1016/j.ijbiomac.2022.09.210.
- J. Liu, T. Xu, L.-G. Yao, S.-W. Li and Y.-W. Guo, Fitoterapia, 2022, 162, 105299, DOI:10.1016/j.fitote.2022.105299.
- Z.-R. Zeng, J. Chen, H. Wang, H.-Y. Zhang, L. Jia, B. Xu and Y.-W. Guo, ACS Omega, 2022, 7, 41678–41686, DOI:10.1021/acsomega.2c05687.
- N. Lin, H. Li, J.-R. Wang, W. Tang, M.-Y. Zheng, H. Wang, C.-S. Jiang and Y.-W. Guo, Chin. J. Chem., 2022, 40, 28–38, DOI:10.1002/cjoc.202100597.
- P.-P. Fu, Y. Jin, L.-G. Yao, Y.-W. Guo and X.-W. Li, Tetrahedron Lett., 2022, 97, 153792, DOI:10.1016/j.tetlet.2022.153792.
- T. Kamada, T. Ishii, K. Sato, G. Ito, K. Jin, W. Takabe, C.-S. Phan and S. Ishigami, Heterocycles, 2022, 104, 797–803, DOI:10.3987/COM-21-14613.
- M. Nagasaka, K. Tani, K. Nishikawa, R. Kinjo and T. Ishii, Nat. Prod. Bioprospect., 2022, 12, 7, DOI:10.1007/s13659-022-00330-7.
- H.-J. Tseng, S.-Y. Chien, Z.-H. Wen, J. Kuo, L.-S. Fang, Y.-C. Wu, T.-Y. Wu and P.-J. Sung, Tetrahedron Lett., 2022, 90, 153628, DOI:10.1016/j.tetlet.2021.153628.
- H.-J. Tseng, L.-M. Kuo, P.-J. Chen, S.-H. Chen, C.-J. Liu, S.-Y. Chien, Y.-C. Tsai, Y.-J. Wu, T.-R. Su and P.-J. Sung, Tetrahedron, 2022, 119, 132851, DOI:10.1016/j.tet.2022.132851.
- T. A. Mohamed, A. I. Elshamy, M. H. A. El-Razek, A. M. Abdel-Tawab, S. K. Ali, A. Mohamed, M. Suenaga, P. W. Pare, A. Umeyama and M.-E. F. Hegazy, Molecules, 2022, 27, 5835, DOI:10.3390/molecules27185835.
- N. N. Thi, H. T. T. Hong, Q. T. Hong, C. N. Xuan, N. N. Hoai, D. T. Thao, P. V. Cuong, T. D. Cong, K. P. Van and C. V. Minh, Nat. Prod. Res., 2022, 36, 5517–5523, DOI:10.1080/14786419.2021.2018587.
- C. Wang, J. Zhang, X. Shi, K. Li, F. Li, X. Tang, G. Li and P. Li, Mar. Drugs, 2022, 20, 574, DOI:10.3390/md20090574.
- M. M. A. Ahmed, M. A. Albadry, E. A. Ragab, E. M. El-Ghaly, S. K. Ismail, Z. Ali, S. I. Khan, A. G. Chittiboyina and I. A. Khan, Nat. Prod. Res., 2022, 36, 1842–1850, DOI:10.1080/14786419.2020.1821015.
- S. A. H. Zidan, R. A. Abdelhamid, A. Alian, M. A. Fouad, K. Matsunami and M. A. A. Orabi, J. Asian Nat. Prod. Res., 2022, 24, 794–802, DOI:10.1080/10286020.2021.1979522.
- C.-H. Chao, Y.-J. Chen, C.-Y. Huang, F.-R. Chang, C.-F. Dai and J.-H. Sheu, Molecules, 2022, 27, 1760, DOI:10.3390/molecules27061760.
- C.-H. Chao, K.-H. Lin, C.-Y. Huang, T.-L. Hwang, C.-F. Dai, H.-C. Huang and J.-H. Sheu, Mar. Drugs, 2022, 20, 297, DOI:10.3390/md20050297.
- C.-L. Wang, T.-Y. Jin, X.-H. Liu, J.-R. Zhang, X. Shi, M.-F. Wang, R. F. Huang, Y. Zhang, K.-C. Liu and G.-Q. Li, Org. Lett., 2022, 24, 9007–9011, DOI:10.1021/acs.orglett.2c03631.
- J. Liu, T. Qi, J. Huang, T. Li, H. Ouyang, W. Lin, X. Yan, X. Yan and S. He, J. Org. Chem., 2022, 87, 9806–9814, DOI:10.1021/acs.joc.2c00784.
- Y.-J. Chen, C.-H. Chao, C.-Y. Huang, T.-L. Hwang, F.-R. Chang, C.-F. Dai and J.-H. Sheu, Bull. Chem. Soc. Jpn., 2022, 95, 374–379, DOI:10.1246/bcsj.20210393.
- X. Yan, J. Liu, J. Huang, Y. Wang, L. Xue, T. Li, O. Han, X. Yan and H. Shan, Phytochemistry, 2022, 204, 113438, DOI:10.1016/j.phytochem.2022.113438.
- N. B. A. Nguyen, L.-Y. Chen, P.-J. Chen, M. El-Shazly, T.-L. Hwang, J.-H. Su, C.-H. Su, P.-T. Yen, B.-R. Peng and K.-H. Lai, Int. J. Mol. Sci., 2022, 23, 15464, DOI:10.3390/ijms232415464.
- O. Hoff, N. Kratena, D. Aynetdinova, K. E. Christensen and T. J. Donohoe, Chem. - Eur. J., 2022, 28, e202202464, DOI:10.1002/chem.202202464.
- J. P. Tuccinardi and J. L. Wood, J. Am. Chem. Soc., 2022, 144, 20539–20547, DOI:10.1021/jacs.2c09826.
- H. Takamura, Y. Sugitani, R. Morishita and I. Kadota, Org. Lett., 2022, 24, 7845–7849, DOI:10.1021/acs.orglett.2c03263.
- Z. Meng and A. Fürstner, J. Am. Chem. Soc., 2022, 144, 1528–1533, DOI:10.1021/jacs.1c12401.
- N. J. Hafeman, M. Chan, T. J. Fulton, E. J. Alexy, S. A. Loskot, S. C. Virgil and B. M. Stoltz, J. Am. Chem. Soc., 2022, 144, 20232–20236, DOI:10.1021/jacs.2c09583.
- N. J. Truax, S. Ayinde, J. O. Liu and D. Romo, J. Am. Chem. Soc., 2022, 144, 18575–18585, DOI:10.1021/jacs.2c08245.
- K.-M. Lai, J.-H. Wang, S.-C. Lin, Y. Wen, C.-L. Wu, J.-H. Su, C.-C. Chen and C.-C. Lin, Int. J. Mol. Sci., 2022, 23, 5624, DOI:10.3390/ijms23105624.
- S.-Y. Hsu, Z.-H. Wen, P.-C. Shih, H.-M. Kuo, S.-C. Lin, H.-T. Liu, Y.-H. Lee, Y.-J. Wang, W.-F. Chen and N.-F. Chen, Antioxidants, 2022, 11, 1433, DOI:10.3390/antiox11081433.
- P.-J. Sung, Y.-C. Tsai, Y.-Y. Chen, S.-N. Yang, Z.-H. Wen, S.-Y. Chien, J.-J. Chen, J.-H. Su and C.-C. Liaw, Heterocycles, 2022, 104, 1423–1434, DOI:10.3987/COM-22-14680.
- Y.-T. Yeh, C.-Y. Chen, P.-J. Chen, Y.-H. Hsieh, S.-Y. Chien, C.-J. Liu, Z.-H. Wen, T.-L. Hwang, N.-F. Chen and P.-J. Sung, Tetrahedron Lett., 2022, 103, 153997, DOI:10.1016/j.tetlet.2022.153997.
- T. H. Huynh, S.-Y. Chien, Z.-H. Wen, C.-C. Liaw, Y.-C. Tsai and P.-J. Sung, Tetrahedron, 2022, 120, 132909, DOI:10.1016/j.tet.2022.132909.
- T. H. Huynh, Z.-H. Wen, S.-Y. Chien, H.-M. Chung, J.-H. Su, L.-S. Fang, Y.-J. Wu, S.-H. Lin and P.-J. Sung, Tetrahedron, 2022, 125, 133037, DOI:10.1016/j.tet.2022.133037.
- C.-W. Yang, T.-M. Chien, C.-H. Yen, W.-J. Wu, J.-H. Sheu and H.-W. Chang, Pharmaceuticals, 2022, 15, 917, DOI:10.3390/ph15080917.
- J.-S. Zeng, C.-C. Tseng, Y.-C. Tsai, S.-H. Chen, P.-J. Chen, N.-F. Chen, Z.-H. Wen, T.-L. Hwang and P.-J. Sung, Phytochem. Lett., 2022, 49, 88–92, DOI:10.1016/j.phytol.2022.03.008.
- P.-J. Sung, S.-H. Chen, L.-M. Kuo, J.-S. Zeng, T.-Y. Wu, P.-J. Chen, W.-C. Chi, N.-F. Chen, Y.-C. Wu and L.-S. Fang, Heterocycles, 2022, 104, 1461–1468, DOI:10.3987/COM-22-14692.
- Y. Jin, L.-G. Yao, Y.-W. Guo and X.-W. Li, Mar. Drugs, 2022, 20, 381, DOI:10.3390/md20060381.
- N. Lin, M.-M. Zhang, C.-S. Jiang, L. Jia, H. Wang, Y.-R. Shen and Y.-W. Guo, Tetrahedron Lett., 2022, 95, 153719, DOI:10.1016/j.tetlet.2022.153719.
- L.-L. Sun, X.-W. Li and Y.-W. Guo, Mar. Drugs, 2022, 20, 218, DOI:10.3390/md20030218.
- Z.-Y. Lin, K.-C. Cheng, H.-J. Su, C.-I. Chang, J.-L. Hsu, M.-f. Hsu, M.-C. Lu and Y.-S. Lin, Chem. Nat. Compd., 2022, 58, 659–668, DOI:10.1007/s10600-022-03765-z.
- M. Gao, Y. Bao-Bao, C. Jia and Z.-J. Yao, Org. Biomol. Chem., 2022, 20, 4553–4558, 10.1039/D2OB00539E.
- H. Fumiyama, A. Takahashi, Y. Suzuki, N. Fujioka, H. Matsumoto and S. Hosokawa, J. Org. Chem., 2022, 87, 15492–15498, DOI:10.1021/acs.joc.2c02031.
- P. M. Pour, A. Yegdaneh, M. Aghaei, Z. Ali, I. A. Khan and M. Ghanadian, Nat. Prod. Res., 2022, 36, 3796–3805, DOI:10.1080/14786419.2021.1887178.
- Y.-Q. Huang, Po-J. Chen, S.-N. Yang, S.-Y. Chien, L.-G. Zheng, Z.-H. Wen, L.-M. Kuo and P.-J. Sung, Tetrahedron Lett., 2022, 108, 154142, DOI:10.1016/j.tetlet.2022.154142.
- D. Zhang, Z. Wang, X. Han, X.-L. Li, Z.-Y. Lu, B.-B. Dou, W.-Z. Zhang, X.-L. Tang, P.-L. Li and G.-Q. Li, Beilstein J. Org. Chem., 2022, 18, 374–380, DOI:10.3762/bjoc.18.42.
- T. Wang, J. Zou, T. Li, S. Peng, W. Zhou, Q. Lai, Y. Feng, C. B. Naman, X. Yan and S. He, Aquaculture, 2022, 549, 737727, DOI:10.1016/j.aquaculture.2021.737727.
- D. An, E. L. Pinheiro-Junior, L. Béress, I. Gladkikh, E. Leychenko, E. A. B. Undheim, S. Peigneur and J. Tytgat, Mar. Drugs, 2022, 20, 140, DOI:10.3390/md20020140.
- E. L. Pinheiro-Junior, R. Kalina, I. Gladkikh, E. Leychenko, T. Jan and S. Peigneur, Mar. Drugs, 2022, 20, 147, DOI:10.3390/md20020147.
- A. Delgado, C. Benedict, J. Macrander and M. Daly, Mar. Drugs, 2022, 20, 730, DOI:10.3390/md20120730.
- S. R. Roy, A. Minei, P. Ahmadi, I. Hermawan, V. Kurnianda, M. H. Dick and J. Tanaka, Nat. Prod. Res., 2022, 36, 742–747, DOI:10.1080/14786419.2020.1800695.
- L. Yi, Y.-T. He, S. Tan, L. V. White, P. Lan, M. G. Gardiner, Z. Pei, M. L. Coote and M. G. Banwell, J. Org. Chem., 2022, 87, 12287–12296, DOI:10.1021/acs.joc.2c01477.
- C. Li, S. Zhang, J. Zhu, W. Huang, Y. Luo, H. Shi, D. Yu, L. Chen, L. Song and R. Yu, Mar. Drugs, 2022, 20, 110, DOI:10.3390/md20020110.
- A. E. Winters, W. Chan, A. M. White, C. P. Berg, M. J. Garson and K. L. Cheney, J. Anim. Ecol., 2022, 91, 831–844, DOI:10.1111/1365-2656.13643.
- P. T. Binh, D. T. Trang, N. P. Thao, N. C. Mai, N. X. Cuong, N. H. Nam and N. Van Thanh, J. Mol. Struct., 2022, 1259, 132744, DOI:10.1016/j.molstruc.2022.132744.
- K. Matsuyama, T. Inoue, T. Muroga, N. Arima, M. Doe, F. Tani, Y. Ookawa, Y. Okamoto, S. Onitsuka, H. Okamura, T. Iwagawa and T. Hamada, Tetrahedron, 2022, 120, 132889, DOI:10.1016/j.tet.2022.132889.
- T. Ohyoshi, Y. Zhao and H. Kigoshi, J. Nat. Prod., 2022, 85, 2082–2089, DOI:10.1021/acs.jnatprod.2c00476.
- S.-W. Li, Q. Wu, H. Xu, L. G. Yao, C. Luo, H. Wang, H. Zhang, X.-W. Li and Y.-W. Guo, Mar. Drugs, 2022, 20, 590, DOI:10.3390/md20100590.
- D. Arrieche, A. Ugarte, F. Salazar, J. E. Villamizar, N. Rivero, M. Caballer, L. Llovera, J. Montañez, L. Taborga and Q. Alberto, Nat. Prod. Res., 2022, 36, 4013–4016, DOI:10.1080/14786419.2021.1895147.
- N. T. Dan, Le T. Giang, C. N. Dinh, T. B. Hai, N. D. Hoi, V. T. Loan, D. T. T. Hang, N. X. Nhiem, B. H. Tai and P. V. Kiem, Nat. Prod. Commun., 2022, 17 DOI:10.1177/1934578X221113184.
- T. Ohyoshi, Y. Zhao, K. Akemoto, T. Ishihara, A. Taniguchi, M. Zhang and H. Kigoshi, Bull. Chem. Soc. Jpn., 2022, 95, 1242–1249, DOI:10.1246/bcsj.20220149.
- A. Sanchez and T. J. Maimone, J. Am. Chem. Soc., 2022, 144, 7594–7599, DOI:10.1021/jacs.2c02627.
- L. O. Casalme, K. Katayama, Y. Hayakawa, K. Nakamura, A. Yamauchi, Y. Nogata, E. Yoshimura, F. Matsuda and T. Umezawa, Mar. Drugs, 2022, 20, 124, DOI:10.3390/md20020124.
- K. G. Kuznetsova, S. S. Zvonareva, R. Ziganshin, E. S. Mekhova, P. Dgebuadze, T. Dinh, H. Yen, H. Thanh, T. Nguyen, S. A. Moshkovskii and A. E. Fedosov, Proc. Royal Soc. B, 2022, 289, 20221152, DOI:10.1098/rspb.2022.1152.
- F. Margiotta, L. Micheli, C. Ciampi, C. Ghelardini, J. M. McIntosh and L. D. C. Mannelli, Mar. Drugs, 2022, 20, 773, DOI:10.3390/md20120773.
- L. Wang, X. Wu, X. Zhu, D. Zhangsun, Y. Wu and S. Luo, Mar. Drugs, 2022, 20, 146, DOI:10.3390/md20020146.
- H. Wang, Y. Li, M. Yang and M. Zhou, Toxicon, 2022, 210, 141–147, DOI:10.1016/j.toxicon.2022.03.002.
- Y. Wei, M. Zhang, S. Yu, Q. Huang, R. Chen, S. Xu, Y. Huang, Y. Yu, M. Liao and Q. Dai, Mar. Drugs, 2022, 20, 750, DOI:10.3390/md20120750.
- K. L. McMahon, H. N. T. Tran, J. R. Deuis, D. J. Craik, I. Vetter and C. I. Schroeder, Toxins, 2022, 14, 600, DOI:10.3390/toxins14090600.
- Y. Fu, Y. Zhang, S. Ju, B. Ma, W. Huang and S. Luo, J. Venomous Anim. Toxins Incl. Trop. Dis., 2022, 28, e20210116, DOI:10.1590/1678-9199-jvatitd-2021-0116.
- S. K. Paulose and K. Chakraborty, Med. Chem. Res., 2022, 31, 462–473, DOI:10.1007/s00044-022-02846-6.
- S. K. Paulose and K. Chakraborty, Steroids, 2022, 182, 108995, DOI:10.1016/j.steroids.2022.108995.
- K. Chakraborty and S. K. Paulose, Nat. Prod. Res., 2022, 36, 5688–5700, DOI:10.1080/14786419.2021.2013841.
- K. P. Silpa and K. Chakraborty, Nat. Prod. Res., 2022, 36, 3002–3012, DOI:10.1080/14786419.2021.1938041.
- S. K. Paulose and K. Chakraborty, Chem. Biodiversity, 2022, 19, e202200277, DOI:10.1002/cbdv.202200277.
- M. Casertano, M. Genovese, P. Paoli, A. Santi, A. Aiello, M. Menna and C. Imperatore, Mar. Drugs, 2022, 20, 65, DOI:10.3390/md20010065.
- S. Kokkaliari, K. Pham, N. Shahbazi, L. Calcul, L. Wojtas, N. G. Wilson, A. D. Crawford and B. J. Baker, Mar. Drugs, 2022, 20, 196, DOI:10.3390/md20030196.
- D. C. Holland, D. W. Prebble, S. Er, J. B. Hayton, L. P. Robertson, V. M. Avery, A. Domanskyi, M. J. Kiefel, J. N. A. Hooper and A. R. Carroll, J. Nat. Prod., 2022, 85, 441–452, DOI:10.1021/acs.jnatprod.1c01172.
- T. Maoka and C. Tode, Mar. Drugs, 2022, 20, 732, DOI:10.3390/md20120732.
- Z. S. Al-Taie, B. Bartholomew, C. Cartmell, R. T. Froom, R. G. Kerr, R. Kraehenbuehl, P. J. Murphy, R. J. Nash, Y. B. Penkova and A. van Teijlingen, Molecules, 2022, 27, 1338, DOI:10.3390/molecules27041338.
- D. Jiang, Y. Chen and S. Wang, J. Org. Chem., 2022, 87, 11063–11072, DOI:10.1021/acs.joc.2c01317.
- M. Piccichè, A. Pinto, R. Griera, J. Bosch and M. Amat, Org. Lett., 2022, 24, 5356–5360, DOI:10.1021/acs.orglett.2c02004.
- J. Jeon and C.-H. Cheon, Org. Lett., 2022, 24, 7128–7133, DOI:10.1021/acs.orglett.2c02788.
- A. Sancho-Balsells, E. García-García, F. Flotta, W. Chen, J. Alberch, M. J. Rodríguez, C. Avila and A. Giralt, Mar. Drugs, 2022, 20, 648, DOI:10.3390/md20100648.
- S. Han, W. Zhou, C. Zhuang and F. Chen, Bioorg. Chem., 2022, 119, 105537, DOI:10.1016/j.bioorg.2021.105537.
- G. Nuzzo, C. Gallo, F. Crocetta, L. Romano, G. Barra, G. Senese, M. dell'Isola, D. Carbone, V. Tanduo, F. Albiani, V. Guido, G. d'Ippolito, E. Manzo and A. Fontana, Biomolecules, 2022, 12, 246, DOI:10.3390/biom12020246.
- T. A. Moreira, I. V. Antolínez, W. O. Valença, S. Roy, I. Ramirez, L. C. A. Barbosa and D. Ren, Bioorg. Med. Chem. Lett., 2022, 57, 128498, DOI:10.1016/j.bmcl.2021.128498.
- M. F. Juette, J. D. Carelli, E. J. Rundlet, A. Brown, S. Shao, A. Ferguson, M. R. Wasserman, M. Holm, T. Jack and S. C. Blanchard, eLife, 2022, 11, e81608, DOI:10.7554/eLife.81608.
- D. R. Mattos, X. Wan, J. D. Serrill, M. H. Nguyen, I. R. Humphreys, B. Viollet, A. B. Smith, K. L. McPhail and J. E. Ishmael, Mar. Drugs, 2022, 20, 418, DOI:10.3390/md20070418.
- S. E. El Feky, M. S. M. A. El Hafez, N. A. A. El Moneim, H. A. H. Ibrahim, M. A. Okbah, A. Ata, A. S. El Sedfy and A. Hussein, Sci. Rep., 2022, 12, 8209, DOI:10.1038/s41598-022-12192-7.
- T. V. Malyarenko, V. M. Zakharenko, A. A. Kicha, A. S. Kuzmich, O. S. Malyarenko, A. I. Kalinovsky, R. S. Popov, V. I. Svetashev and N. V. Ivanchina, Mar. Drugs, 2022, 20, 641, DOI:10.3390/md20100641.
- M. S. M. A. El Hafez, M. A. El Aziz Okbah, H. A. H. Ibrahim, A. A. E. R. Hussein, N. A. A. El Moneim and A. Ata, Nat. Prod. Res., 2022, 36, 5545–5552, DOI:10.1080/14786419.2021.2021200.
- A. A. Kicha, A. I. Kalinovsky, T. V. Malyarenko, O. S. Malyarenko, S. P. Ermakova, R. S. Popov, V. A. Stonik and N. V. Ivanchina, Mar. Drugs, 2022, 20, 164, DOI:10.3390/md20030164.
- L. T. Vien, T. T. H. Hanh, T. H. Quang, N. X. Cuong, Do T. Thao, N. H. Nam, D. C. Thung, P. V. Kiem and C. V. Minh, Nat. Prod. Res., 2022, 36, 2223–2229, DOI:10.1080/14786419.2020.1826478.
- Y.-Y. Lu, M.-C. Wang, K. Liu, W. Zhang, Y. Liu and H.-F. Tang, Nat. Prod. Res., 2022, 36, 2118–2124, DOI:10.1080/14786419.2020.1845676.
- T. V. Malyarenko, O. S. Malyarenko, A. A. Kicha, A. I. Kalinovsky, P. S. Dmitrenok and N. V. Ivanchina, Mar. Drugs, 2022, 20, 216, DOI:10.3390/md20030216.
- L. T. Vien, T. T. H. Hanh, T. H. Quang, N. V. Thanh, N. X. Cuong and N. H. Nam, Vietnam J. Chem., 2022, 60, 685, DOI:10.1002/vjch.202200095.
- A. S. Silchenko, S. A. Avilov, P. V. Andrijaschenko, R. S. Popov, E. A. Chingizova, B. B. Grebnev, A. B. Rasin and V. I. Kalinin, Molecules, 2022, 27, 7655, DOI:10.3390/molecules27217655.
- A. S. Silchenko, S. A. Avilov, P. V. Andrijaschenko, R. S. Popov, E. A. Chingizova, P. S. Dmitrenok, A. I. Kalinovsky, A. B. Rasin and V. I. Kalinin, Mar. Drugs, 2022, 20, 369, DOI:10.3390/md20060369.
- Y.-T. Cho, A. K. Adak, Y.-Y. Su, T.-W. Chang and C.-C. Lin, Adv. Synth. Catal., 2022, 364, 3573–3588, DOI:10.1002/adsc.202200724.
- D. Zhu, M. Geng and B. Yu, Angew. Chem., Int. Ed., 2022, 61, e202203239, DOI:10.1002/anie.202203239.
- F. Li, Z. Lin, J. P. Torres, E. A. Hill, D. Li, C. A. Townsend and E. W. Schmidt, J. Am. Chem. Soc., 2022, 144, 9363–9371, DOI:10.1021/jacs.2c01416.
- H. Ghelani, M. Khursheed, T. E. Adrian and R. K. Jan, Mar. Drugs, 2022, 20, 693, DOI:10.3390/md20110693.
- N. V. Ageenko, K. V. Kiselev and N. A. Odintsova, Mar. Drugs, 2022, 20, 611, DOI:10.3390/md20100611.
- B.-W. Song, S. Kim, R. Kim, S. Jeong, H. Moon, H. Kim, E. A. Vasileva, N. P. Mishchenko, S. A. Fedoreyev, V. A. Stonik, M. Y. Lee, J. Kim, H. K. Kim, J. Han and W. Chang, Mar. Drugs, 2022, 20, 756, DOI:10.3390/md20120756.
- H. Cui, J. Liu, E. A. Vasileva, N. P. Mishchenko, S. A. Fedoreyev, V. A. Stonik and Y. Zhang, Mar. Drugs, 2022, 20, 722, DOI:10.3390/md20110722.
- J.-M. Kim, S.-C. Shin, Y.-I. Cheon, H.-S. Kim, G.-C. Park, H.-K. Kim, J. Han, J.-E. Seol, E. A. Vasileva, N. P. Mishchenko, S. A. Fedoreyev, V. A. Stonik and B.-J. Lee, Mar. Drugs, 2022, 20, 729, DOI:10.3390/md20120729.
- M. R. Choi, H. Lee, H. K. Kim, J. Han, J. E. Seol, E. A. Vasileva, N. P. Mishchenko, S. A. Fedoreyev, V. A. Stonik, W. S. Ju, D.-J. Kim and S.-R. Lee, Mar. Drugs, 2022, 20, 555, DOI:10.3390/md20090555.
- J.-S. Ahn, Y. Y. Shin, S.-J. Oh, M.-H. Song, M.-J. Kang, S. Y. Park, P. T. Nguyen, D. K. Nguyen, H. K. Kim, J. Han, E. A. Vasileva, N. P. Mishchenko, S. A. Fedoreyev, V. A. Stonik, Y. Seo, B.-C. Lee and H.-S. Kim, Mar. Drugs, 2022, 20, 715, DOI:10.3390/md20110715.
- G. Caulier, A. Lourtie, L. Brasseur, J. Mallefet, G. Pascal, P. Flammang and I. Eeckhaut, Chemoecology, 2022, 32, 95–104, DOI:10.1007/s00049-022-00368-6.
- A. Milito, M. Cocurullo, A. Columbro, S. Nonnis, G. Tedeschi, I. Castellano, M. I. Arnone and A. Palumbo, Open Biol., 2022, 12, 210262, DOI:10.1098/rsob.210262.
- R. Thimmappa, S. Wang, M. Zheng, R. C. Misra, A. C. Huang, G. Saalbach, Y. Chang, Z. Zhou, V. Hinman, Z. Bao and A. Osbourn, Nat. Chem. Biol., 2022, 18, 774–781, DOI:10.1038/s41589-022-01054-y.
- T. Sanguanphun, N. Sornkaew, N. Malaiwong, P. Chalorak, P. Jattujan, N. Niamnont, P. Sobhon and K. Meemon, Front. Pharmacol, 2022, 13, 1004568, DOI:10.3389/fphar.2022.1004568.
- X. Li, B. Zeng, W. Lu, Y. Zhao, Z. Li, C. Xue, T. Zhang and Y. Wang, Mar. Drugs, 2022, 20, 703, DOI:10.3390/md20110703.
- C. Cui, C.-H. Ding, F.-F. Liu, J.-R. Lu, S.-Y. Zheng, H.-W. Lin, W.-K. Zhu, Y. Fan, L. He and Y. Qin, J. Oncol., 2022, 2022, 6449984, DOI:10.1155/2022/6449984.
- L. Grauso, Y. Li, S. Scarpato, N. A. Cacciola, P. de Cicco, C. Zidorn and A. Mangoni, J. Nat. Prod., 2022, 85, 2468–2473, DOI:10.1021/acs.jnatprod.2c00796.
- Y. Yaegashi, Y. Kudo, N. Ueyama, K. Onodera, Y. Cho, K. Konoki and M. Yotsu-Yamashita, J. Nat. Prod., 2022, 85, 2199–2206, DOI:10.1021/acs.jnatprod.2c00588.
- M. Nishiumi, T. Miyasaka, M. Adachi and T. Nishikawa, J. Org. Chem., 2022, 87, 9023–9033, DOI:10.1021/acs.joc.2c00704.
- J. M. McNab, M. T. Briggs, J. E. Williamson, P. Hoffmann, J. Rodriguez and P. Karuso, Mar. Drugs, 2022, 20, 788, DOI:10.3390/md20120788.
- V. N. Safronova, P. V. Panteleev, S. V. Sukhanov, I. Y. Toropygin, I. A. Bolosov and T. V. Ovchinnikova, Mar. Drugs, 2022, 20, 167, DOI:10.3390/md20030167.
- S. Righi, L. Forti, R. Simonini, V. Ferrari, D. Prevedelli and A. Mucci, Mar. Drugs, 2022, 20, 585, DOI:10.3390/md20090585.
- W. R. Kem, K. Andrud, G. Bruno, H. Xing, F. Soti, T. T. Talley and P. Taylor, Mar. Drugs, 2022, 20, 49, DOI:10.3390/md20010049.
- T. Rončević, M. Gerdol, M. Mardirossian, M. Maleš, S. Cvjetan, M. Benincasa, A. Maravić, G. Gajski, L. Krce, I. Aviani, J. Hrabar, Ž. Trumbić, M. Derks, P. Alberto, M. Weingarth, L. Zoranić, A. Tossi and I. Mladineo, Acta Biomater., 2022, 146, 131–144, DOI:10.1016/j.actbio.2022.04.025.
- A. Sultan, C. Hindrichs, K. V. Cisneros, C. J. Weaver, L. R. Faux, V. Agarwal and M. O. James, Chemosphere, 2022, 286, 131620, DOI:10.1016/j.chemosphere.2021.131620.
- J. W. Blunt, A. R. Carroll, B. R. Copp, R. A. Davis, R. A. Keyzers and M. R. Prinsep, Nat. Prod. Rep., 2018, 35, 8–53, 10.1039/C7NP00052A.
- D. Rogers and J. Mathew Hahn, J. Chem. Inf. Model., 2010, 50, 742–754, DOI:10.1021/ci100050t.
- C. R. Pye, M. J. Bertin, R. S. Lokey, W. H. Gerwick and R. G. Linington, Proc. Natl. Acad. Sci. U.S.A., 2017, 114, 5601–5606, DOI:10.1073/pnas.1614680114.
- J. W. Blunt, B. R. Copp, M. H. G. Munro, P. T. Northcote and M. R. Prinsep, Nat. Prod. Rep., 2004, 21, 1–49, 10.1039/B305250H.
- F. Ntie-Kang and D. Svozil, Phys. Sci. Rev., 2020, 5, 20180121, DOI:10.1515/psr-2018-0121.
- H. W. Kim, M. Wang, C. A. Leber, L.-F. Nothias, R. Reher, J. J. Kyo Bin Kang, J. van der Hooft, P. C. Dorrestein, W. H. Gerwick and G. W. Cottrell, J. Nat. Prod., 2021, 84, 2795–2807, DOI:10.1021/acs.jnatprod.1c00399.
- T. Sander, J. Freyss, M. von Korff and C. Rufener, J. Chem. Inf. Model., 2015, 55, 460–473, DOI:10.1012/ci500588j.
- J. A. van Santen, G. Jacob, A. L. Singh, V. Aniebok, M. J. Balunas, D. Bunsko, F. C. Neto, L. Castaño-Espriu, C. Chang, T. N. Clark, J. L. Cleary Little, D. A. Delgadillo, P. C. Dorrestein, K. R. Duncan, J. M. Egan, M. M. Galey, F. P. J. Haeckl, A. Hua, A. H. Hughes, D. Iskakova, A. Khadilkar, J.-H. Lee, S. Lee, N. LeGrow, D. Y. Liu, J. M. Macho, C. S. McCaughey, M. H. Medema, R. P. Neupane, T. J. O’Donnell, J. S. Paula, L. M. Sanchez, A. F. Shaikh, S. Soldatou, B. R. Terlouw, T. A. Tran, M. Valentine, J. J. J. van der Hooft, D. A. Vo, M. Wang, D. Wilson, K. E. Zink and R. G. Linington, ACS Cent. Sci., 2019, 5, 1824–1833, DOI:10.1021/acscentsci.9b00806.
- J. Gu, Y. Gui, L. Chen, Y. Gu, H.-Z. Lu and X. Xu, PLoS One, 2013, 8, 62839, DOI:10.1371/journal.pone.0062839.
- A. Bhushan, E. E. Peters and J. Piel, Entotheonella Bacteria as Source of Sponge-Derived Natural Products: Opportunities for Biotechnological Production, in Blue Biotechnology-Progress in Molecular and Subcellular Biology, ed. W. Müller, H. Schröder and X. Wang, Springer, 2017, vol. 55, DOI:10.1007/978-3-319-51284-6_9.
- E. E. Peters, J. K. B. Cahn, A. Lotti, A. Gavriilidou, U. A. E. Steffens, C. Loureiro, M. A. Schorn, P. Cárdenas, N. Vickneswaran, P. Crews, D. Sipkema and J. Piel, ISME Commun., 2023, 3, 50, DOI:10.1038/s43705-023-00259-z.
- M. Kogawa, R. Miyaoka, F. Hemmerling, M. Ando, K. Yura, K. Ide, Y. Nishikawa, M. Hosokawa, Y. Ise, J. K. B. Cahn, K. Takada, S. Matsunaga, T. Mori, J. Piel and H. Takeyama, PNAS Nexus, 2022, 1, 1–13, DOI:10.1093/pnasnexus/pgab007.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3np00061c |
| This journal is © The Royal Society of Chemistry 2024 |