Open Access Article
Open Access ArticleHarnessing the potential: advances in cyanobacterial natural product research and biotechnology†
Martin
Baunach
 ab,
Arthur
Guljamow
ab,
Arthur
Guljamow
 a,
María
Miguel-Gordo
a,
María
Miguel-Gordo
 a and
Elke
Dittmann
a and
Elke
Dittmann
 *a
*a
aUniversity of Potsdam, Institute of Biochemistry and Biology, Karl-Liebknecht-Str. 24/25, 14476 Potsdam, Germany. E-mail: editt@uni-potsdam.de
bUniversity of Bonn, Institute of Pharmaceutical Biology, Nußallee 6, 53115 Bonn, Germany
First published on 13th December 2023
Abstract
Covering: 2000 to 2023
Cyanobacteria produce a variety of bioactive natural products that can pose a threat to humans and animals as environmental toxins, but also have potential for or inspire pharmaceutical use. As oxygenic phototrophs, cyanobacteria furthermore hold great promise for sustainable biotechnology. Yet, the necessary tools for exploiting their biotechnological potential have so far been established only for a few model strains of cyanobacteria, while large untapped biosynthetic resources are hidden in slow-growing cyanobacterial genera that are difficult to access by genetic techniques. In recent years, several approaches have been developed to circumvent the bottlenecks in cyanobacterial natural product research. Here, we summarize current progress that has been made in unlocking or characterizing cryptic metabolic pathways using integrated omics techniques, orphan gene cluster activation, use of genetic approaches in original producers, heterologous expression and chemo-enzymatic techniques. We are mainly highlighting genomic mining concepts and strategies towards high-titer production of cyanobacterial natural products from the last 10 years and discuss the need for further research developments in this field.
1. Introduction
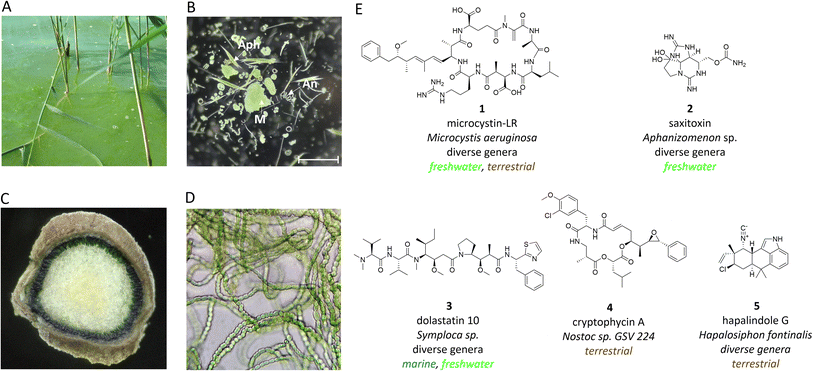
Cyanobacteria are a manifold group of oxygenic phototrophic prokaryotes that are thought to be responsible for transforming the Earth's atmosphere from anoxic to oxic conditions.1 However, cyanobacteria are also notorious for producing environmental toxins and are repeatedly associated with poisoning in humans and animals.2 It was primarily the toxins that attracted the interest of natural product researchers to cyanobacteria early on. Attention was initially focused on freshwater genera such as Microcystis, which form macroscopically visible colonies and can form dense surface blooms in eutrophic lakes (Fig. 1A and B).3Microcystis is now recognized as one of the most widespread producers of the potent hepatotoxin microcystin (1).4 Another threat in freshwater lakes comes from the production of neurotoxins such as saxitoxin (2) and anatoxin by different species of filamentous cyanobacteria (Fig. 1E).2 Cyanobacteria were also associated with severe skin irritations after swimming in marine environments. Such symptoms can be caused by mat-forming cyanobacteria such as Moorena in coastal habitats and were connected to the production of the dermatotoxins lyngbyatoxin and aplysiatoxin.5 The adverse effects on humans and animals have led to extensive bioactivity-guided screening programs of cyanobacteria that also included cyanobacteria of terrestrial origin in addition to freshwater and marine strains.6 Of particular importance in terrestrial habitats is the filamentous genus Nostoc, which can fix nitrogen and live both freely in the soil and in association with diverse plant hosts, such as cycads, where the cyanobacteria reside in coralloid roots (Fig. 1C and D).7,8Widespread interest in cyanobacteria has increasingly revealed the structural uniqueness of cyanobacterial natural products and their versatile bioactive potential.9–11 Notably, cyanobacteria produce a number of highly active cytotoxic compounds that have attracted pharmaceutical interest such as dolastatins (3) and cryptophycins (4).12,13 There are also cyanobacterial natural products with antibiotic properties, such as the hapalindole family of compounds (5).14 The significant bioactive potential of cyanobacteria has been confirmed by genome sequencing programs over the past two decades, and furthermore, a large untapped biosynthetic potential has been revealed.15,16 In particular, cyanobacterial genomes frequently contain biosynthetic gene clusters (BGCs) encoding nonribosomal peptide synthetases (NRPS), polyketide synthases (PKS) or hybrids thereof, as well as ribosomally produced and posttranslationally modified peptide (RiPPs) and terpene biosynthetic pathways. Despite all progress made in assigning of BGCs to known families of cyanobacterial natural products (detailed in previous reviews9–11,17), to date 80% or more of BGCs are still orphan.
Besides being a rich source of unique natural products; cyanobacteria also hold great promise for sustainable production of chemical compounds.18 Cyanobacterial biotechnology has long outgrown its infancy. An increasing number of companies are cultivating cyanobacteria at large scale as food supplements, cosmetics or fertilizers or develop them as chassis for the production of biopolymers, biofuels and pigments.19 The autotrophic metabolism of cyanobacteria enables the production of organic chemicals using only sunlight, CO2 and minerals. Production through green biotechnology is CO2 neutral or even CO2 negative and meets the aim of establishing a climate-neutral circular bioeconomy.19 Accordingly, there are extensive efforts to further develop methods of synthetic biology for cyanobacteria and to expand the product range. Currently, synthetic biology and biotechnology of cyanobacteria is focused on unicellular model strains of the genera Synechococcus and Synechocystis and on the edible genus Spirulina.20 Considering both the richness in untapped BGCs with great pharmaceutical and biosynthetic value and the possibility for exploitation by sustainable biotechnology, cyanobacteria could become a mainstay of microbial natural product research.
Yet, both the assignment of unknown BGCs to unknown metabolites and the high-titer production of complex specialized metabolites of cyanobacterial origin in cyanobacterial hosts are not well advanced. From our microbiological perspective, similar obstacles are currently limiting both the genomic mining of new compound families in cyanobacteria and their high-titer production in cyanobacteria. Understanding the bottlenecks that limit growth of cyanobacteria, production of specialized metabolites as well as amenability to genetic manipulation can potentially advance both the field of natural product discovery as well as provide avenues for the production of specialized compounds or new-to-nature compounds in cyanobacteria. In the present review, we have therefore set ourselves the goal not only to compile the current state of knowledge on method development of natural product research in cyanobacteria, but also to identify gaps that should be addressed for efficient natural product exploitation.
2. Estimating the hidden potential of cyanobacteria to produce natural products
As in many bacterial groups, cyanobacteria include highly adapted specialists that thrive primarily in habitats with stable conditions as well as versatile adapted generalists that can exhibit a highly variable lifestyle.21 The range extends from unicellular minimalists to multicellular organisms with pronounced cellular differentiation and genome sizes between 1 and 12 Mbp.21 Genome-streamlined specialists are especially abundant in marine habitats, but versatile multicellular genera thrive in different niches of terrestrial, freshwater as well as marine environments. A comparative analysis has recently shown that gene family expansion is a common strategy in cyanobacteria and is particularly observed in terrestrial cyanobacteria which are exposed to a fluctuating environment and also reach the largest genome sizes. Hundreds of cyanobacterial genes were found to be highly habitat specific.21 Unsurprisingly, this fact is also reflected in the potential for natural product biosynthesis.Following the CyanoGEBA initiative that aimed to improve the sequence coverage by sequencing diverse axenic strains of the well curated Pasteur Culture Collection of Cyanobacteria (PCC),15 Gugger and coworkers carried out a phylum-wide analysis of the presence of NRPS and PKS gene clusters. This analysis revealed a burst of these genes in late branching lineages of cyanobacteria which are dominated by multicellular genera of marine, freshwater and terrestrial origin. Moreover, the study indicated that 80% or more of the NRPS and PKS BGCs were still not assigned to their products.16 A comprehensive analysis covering all types of BGCs identified by AntiSMASH (in particular NRPS, PKS, RiPPs and terpenes)22 was recently presented by Ziemert, Medema and coworkers, who created a compendium for the potential to produce specialized metabolites for bacterial genomes and metagenomes.23 This involved adapting the BiG-SLiCE software tool,24 which subdivides unique BGCs into distinguishable gene cluster families (GCFs) that were adapted to the hierarchical clustering of compounds in the NPAtlas database.25 The study revealed a significant dissimilarity of GCFs at the genus level rather than species level as well a correlation with the habitats.23 To reliably rank the biosynthetic diversity among phyla, the study applied a common phylogenetic metric, relative evolutionary divergence (RED) groups, which were roughly consistent with the genus range across phyla.26 For cyanobacteria, 536 genomes were included in the comparative part of the study and classified into 142 RED groups. For these genomes, 1.867 actual unique GCFs were predicted ranking cyanobacteria fifth behind actinobacteria, proteobacteria, firmicutes and bacteriodota.23 Since the necessary sequence depth to cover all cyanobacterial GCFs is far from being reached, the authors also estimated the diversity potential of GCFs that could be reached if the sequencing was saturated. Thereby, a diversity of approximately 5.000 unique GCFs was indicated for cyanobacteria in total. Both the actual and the estimated GCF diversity are distributed very differently among phyla. Fig. 2 zooms into the 30 richest RED groups of cyanobacteria, which are responsible for 87% of the cyanobacterial actual GCF diversity. Seventeen of the groups are assigned to the order Nostocales comprising a diversity of different genera including amongst others Nostoc, Fischerella, Calothrix, Scytonema, Nodularia and Cylindrospermopsis while in the orders Chroococcales, Oscillatoriales and Synechococcales rather single genera such as Microcystis, Moorena, Planktothrix and Leptolyngbya, stand out, respectively (Fig. 2). However, there is a bias with regard to the number of genomes representing different RED groups. Thus, at the present time, one can read a trend rather than make a ranking among phyla. Nevertheless, it is possible to draw valuable conclusions from the analysis. (1) The known prolific natural product producing genera such as Nostoc, Microcystis and Moorena are indeed among the bacterial groups with the greatest actual and estimated genomic potential. (2) The current 144 entries of characterized BGCs of cyanobacteria included in the MIBiG database (ref. 27, as of August 2023) occupy only a small fraction of the actual 1.867 GCFs identified for cyanobacteria. Although it must be assumed that the MIBiG database does not cover all characterized BGCs, there is a large untapped potential for cyanobacteria to produce natural products which can be estimated at about 90%. The order Nostocales shows by far the largest hidden potential to produce natural products. (3) For some genera or RED groups, a significant number of GCFs is matched by only a few entries in the NPAtlas database.25 This is particularly true for the genus Pleurocapsa solely representing the poorly investigated order Pleurocapsales for which there is only one entry in NPAtlas. The analysis by Ziemert, Medema and coworkers also revealed a low sequence coverage for this genus.23 There are other rich genera for which there are few entries in the NPAtlas database (<15).25 These include, for example, the genus Calothrix. This is particularly interesting because three of the five largest cyanobacterial genomes sequenced to date were assigned to the genus Calothrix.21 Further notable genera include Cylindrospermopsis and Crocosphera which are not intensively investigated yet. (4) Members of the orders Synechococcales and Spirulinales have only a small number of predicted GCFs. Although six Synechococcales RED groups made it to the list of most productive groups, they represent only a small subgroup of a much larger phylum; the majority of all Prochlorococcus and Synechococcus strains barely have predictable GCFs.23 Also, for the genus Synechocystis, to which the model strain Synechocystis sp. PCC 6803 belongs, a total of only 8 GCFs are predicted. From this analysis, one can conclude that unicellular cyanobacteria rarely have a great potential for natural product biosynthesis (except colony formers such as Microcystis or Crocosphera). The situation is similar for the filamentous order Spirulinales, for which there is also only one entry in NPAtlas.25 One can therefore also state that the strains currently used for cyanobacterial synthetic biology and biotechnology generally have a very low natural product biosynthetic potential. We will discuss this fact again later when we talk about available tools and growth of cyanobacteria.
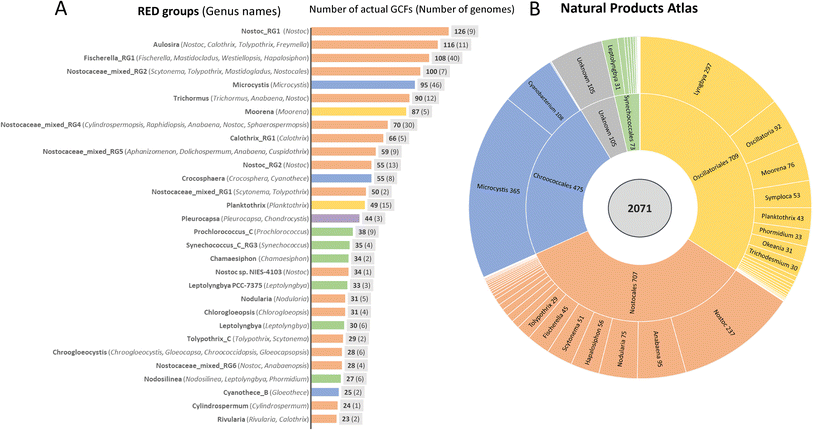 | ||
| Fig. 2 Number of distinct biosynthetic gene cluster families (GCFs) in different phylogenetic groups of cyanobacteria and number of cyanobacterial entries in the Natural Product Atlas. (A) Number of GCFs in standardized relative evolutionary divergence (RED) groups as defined in Gavriilidou et al.23 RED groups correspond to single genera of cyanobacteria or may include strains of multiple genera of cyanobacteria (as indicated in parentheses). The number of genomes used for the analysis in individual RED groups is given in parentheses next to the GCF number. Data were adapted from Gavrilidou et al.23 (B) Number of cyanobacterial chemical entities in different orders and genera of cyanobacteria according to Natural Product Atlas (ref. 25 as of August 2023). The color coding corresponds to the assignment of the genera or RED groups to the orders Nostocales (orange), Chroococcales (blue) Oscillatoriales (yellow), Synechococcales (green) and Pleurocapsales (purple). | ||
While the integration of different databases such as the MiBIG database and NPAtlas is very useful,25,27 some of the conclusions that can be drawn when comparing the GCF analysis and the NPAtlas database must be viewed with caution. Many of the entries in NPAtlas go back to older studies. Cyanobacterial taxonomy has undergone many changes since then, and many genus names were created only in the course of more detailed 16S rRNA analysis and taxon refinement.28 It is therefore unsurprising that certain genus names with high GCF potential rarely appear in NPAtlas. These include, for example, the genera Aulosira, Trichormus or Dolichospermum, which were more commonly used recently and previously classified under Anabaena or Nostoc.28–30 Well-known examples of name changes in the field of natural products also include the change of the species name Lyngbya majuscula, first in Moorea producens and later to Moorena producens31,32 and Oscillatoria agardhii to Planktothrix agardhii.33 While taxon refinement is a general trend in modern microbiology, the situation is even more complicated in cyanobacteria. Longer than for other bacterial phyla, cyanobacterial taxonomy was largely based on morphology and reference strains were described according to the rules of the International Code of Botanical Nomenclature (ICBN) rather than the International Code of Nomenclature of Prokaryotes (ICNP).34 The problem of the morphology-based phylogeny is at least two-fold. Not all morphological features used for classification correspond to the molecular phylogenetic classifications and the morphological classifications require a specific expertise which is not always available. Therefore, one must also assume false annotations and genus names may occur several times in different polyphyletic groups.28 Even if the genus and species name chaos is gradually being resolved by careful polyphasic analyses that consider both morphology and molecular phylogeny and increasing sequencing depth, the problem is still reflected in the databases. The genus name is therefore often not a useful identifier in the cyanobacterial phylum.
3. Challenges and limitations of cyanobacterial natural products research
There are currently three main reasons that complicate natural product research in cyanobacteria. The first of these is the comparably poor growth of the autotrophic bacteria, which slows down all approaches in both basic research and applied biotechnology.35 Another obstacle is the fact that heterotrophic bacteria associate themselves very stably to the carbon- and often also nitrogen-fixing cyanobacteria.36–38 Even though diverse protocols for cyanobacterial axenization exist, cyanobacteria are frequently kept as xenic isolates even in strain collections.37,38 This makes isolation of the bacteria very laborious and exposes research work and biotechnology to a high risk of contamination. Last but not least, an efficient exploitation of cyanobacterial natural products and their biosyntheses is hindered by the frequent lack of accessibility for genetic manipulations.39 This limits the characterization of biosynthetic pathways in the cyanobacteria themselves and the development of production strains. Some of the challenges are general to work with cyanobacteria, and many are particularly applicable to work with prolific natural product producers. We highlighted in the previous section that these are primarily multicellular strains, such as those in the genera Nostoc, Moorena, Fischerella, or Microcystis. These genera are characterized by a massive mucus layer and form a particularly close association with heterotrophic bacteria. The pronounced sheath makes not only axenic isolation but also genetic manipulation a particular challenge.The growth rates of cyanobacteria differ considerably. While some model strains achieve doubling times between 2–6 hours under ideal conditions, many cyanobacteria divide only about once a day even under optimal conditions.35 The first group mainly includes model strains of the genera Synechococcus and Synechocystis, with the strain Synechococcus elongatus UTEX 2973 being the frontrunner with a doubling time of 2 h under autotrophic conditions.40 The latter group, on the other hand, includes genera with high natural product potential such as Nostoc or Microcystis.41 Thus, all work with these non-model genera is automatically associated with a high time expenditure. The growth of cyanobacteria depends mainly on photosynthetic rates and CO2 availability.35 Photosynthetic rates, in turn, depend on light intensities, whereby cyanobacteria stand in their own way because they attenuate light through shading at higher cell densities. Higher light intensities again trigger photoinhibition, so there is limited scope for cyanobacteria to grow.35 CO2 concentrations cannot be increased arbitrarily either, because this decreases the pH in the medium. A simple way to increase the biomass production of cyanobacteria, which has proven particularly useful for natural product producers, is the so-called high-density cultivation (HDC). The technology was developed by the CellDEG company in Berlin and enables bubble-free gas exchange and turbulent mixing on a vibrating membrane. HDC minimizes photobleaching and photoinhibition leading to a high quantum yield even at high light intensities.42 For the model strain Nostoc punctiforme PCC 73102, not only much higher cell densities and biomass accumulation is achieved by HDC, but growth rates are also increased by a factor of 2–3.43 Some of the natural product-rich genera can accumulate biomass very efficiently in nature. This is especially true for bloom-forming genera such as Microcystis, Planktothrix, Nodularia or Dolichospermum,4 but also for mat-formers such as Moorena and Symploca.31,44 A considerable number of cyanobacterial natural products have been isolated directly from field material, such as from Microcystis45 or Symploca,46 which are relatively easy to harvest. However, many downstream applications and a detailed BGC characterization are more difficult without cultivation.
Looking at the success of cyanobacteria in nature, there is obviously quite some potential for optimizing growth rates even for genera like Microcystis or Nostoc. However, this also requires an understanding of the bottlenecks that currently impede rapid growth in the laboratory. The heterotrophic microbiome of bacteria seems to play a key role in this context. For Microcystis in particular, there are studies showing that the relationship with heterotrophic bacteria is predominantly growth-promoting for the cyanobacteria, although the reasons are not well understood.47 As mentioned above, heterotrophic bacteria are an obstacle to reproducible research and biotechnology. Yet, one could certainly harness the contribution of heterotrophic bacteria to cyanobacterial growth by supplementing axenic bacterial isolates with selected heterotrophic partners in synthetic communities. This is an emerging trend in the synthetic biology of cyanobacteria,48 which could also have a lasting impact on the development of model production strains in natural product research.
Not all questions to be addressed in cyanobacterial natural product research necessarily require axenic isolates. The assignment of BGCs to natural products or vice versa does not necessarily have to be carried out with pure strains. The analysis of metagenomes has now progressed to the point where assignment to cyanobacterial genomes is reliably possible.37 The assignment of biosynthetic pathways is in any case usually supported by further analyses, such as feeding studies or in vitro characterizations of enzymes. Despite the progress in purifying strains some genera of cyanobacteria are only accessible as xenic isolates. However, this does not diminish the value of axenic cultures, especially for non-model genera such as Microcystis or Nostoc. Physiological and molecular biological experiments are dependent on axenic strains. Genetic manipulations are currently limited to a few strains. This problem has changed little in recent years. However, there has been a breakthrough in the biotechnologically important genus Spirulina, which is being cultivated at large scale but was not amenable to genetic manipulation. Recently, Spirulina has been efficiently established as a chassis for high-titer expression of therapeutic proteins. It was shown that Spirulina could only be manipulated in coculture with selected heterotrophic bacteria.49 These findings may point the way forward for cyanobacterial natural product producers. Again, synthetic consortia could provide a solution. Doudna and colleagues have recently also demonstrated the possibility of species- and site-specific genome editing in complex bacterial communities via an RNA-guided CRISPR-Cas transposase (DART) system.50 Although this technique has not been applied to cyanobacteria yet, it opens up new possibilities for the analysis of cyanobacterial consortia. We will revisit the issue of genetic manipulation later when we discuss in detail the use of genetic methods in the natural products field.
4. Designing a strategy for the genome-based discovery of novel cyanobacterial natural products
Many of the well-studied cyanobacterial species produce a limited number of specialized metabolites largely constitutively. Individual strains of the genus Microcystis, for example, commonly produce 2–5 classes of peptides, such as microcystins, cyanopeptolins, aeruginosins, anabaenopeptins, microginins, aeruginoguanidins, cyanobactins, and microviridins, in different combinations.51 Terrestrial symbiotic bacteria of the genus Nostoc also produce various families of peptides, including microcystins, aeruginosins, anabaenopeptins, and also nostopeptolides and nostocyclopeptides.8 These constitutively produced peptides are relatively easy to detect and are recurrently discovered. Numerous analogues of many of these compound families have been described and their initial description was usually by bioactivity-based purification (Fig. S1†).25 In contrast, genome-based discovery of entirely new classes of cyanobacterial natural products requires the development of rational strategies for assigning metabolites to their BGCs, characterizing BGCs, and optimizing production titers.In addition to an appropriate dereplication strategy for the known natural product classes, it is central to this effort to understand the reasons for the lack of assignment of compounds to orphan BGCs. In the search for causes, the continuously improved bioinformatic analysis of BGCs, especially by the AntiSMASH platform,22 is a very good basis, not only because BGCs are reliably predicted, but also because structural prediction is often possible through increasing insights into the role and specificity of enzymes.
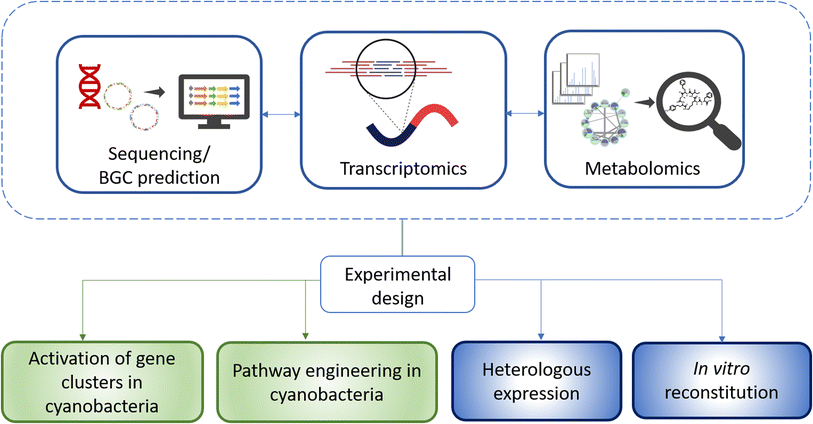
A first step into developing a strategy for the discovery of new natural products is often transcriptional analysis.52 This analysis can reveal whether cyanobacterial BGCs are silent and whether production of the corresponding metabolites can be expected under the given cultivation conditions. These findings can be paired with metabolomic studies. For the model strain N. punctiforme PCC 73102, it was shown that the majority of cryptic BGCs are actively transcribed, but only at very low levels.53 Moreover, a transcriptional reporter analysis revealed that expression is often restricted to a few cells within the multicellular consortium.53 Low levels of transcription in turn can be assumed to lead to low metabolite production levels. Based on these findings, one can consider different methodological options and develop workflows depending on the purpose of the analysis and properties of the investigated strains (Fig. 3). (1) Can transcription of BGCs and metabolite production be stimulated in the cyanobacterial strains? In principle, different abiotic and biotic conditions can be tested randomly. This approach is commonly designated as OSMAC approach (one strain – many compounds).54 However, a more rational approach can also be taken, for example if there are regulatory factors or transport proteins that have already been characterized in other bacteria in the vicinity of BGCs and when a functional hypothesis can be deduced. (2) Is the strain amenable to genetic manipulation? Accessibility to genetic engineering offers many possibilities such as overexpression of regulatory components and characterization of biosynthetic intermediates and tailoring enzymes. (3) Is the purpose of the analysis primarily to characterize a BGC and develop a production strategy? Then, heterologous expression is an increasingly successful choice for cyanobacterial BGCs.55 It depends on the ultimate goal which production hosts are suitable, with the focus being especially on E. coli and well manipulable model strains of cyanobacteria. (4) Are biochemical reactions and precursors for the assembly of compounds well predictable? Then in vitro reconstitution offers an elegant way to characterize BGCs or even to produce bioactive compounds. All methodological approaches are associated with individual challenges. Very often, different methodological options are being combined. Combining alternative technologies is particularly important because of the named bottlenecks in cyanobacterial natural product research, their slow growth, frequent contamination with heterotrophic bacteria, and poor accessibility for genetic manipulation. Hereafter, we will dissect the progress in the different methodological fields.
5. Use of multi-omics technologies for mining of cyanobacterial natural products
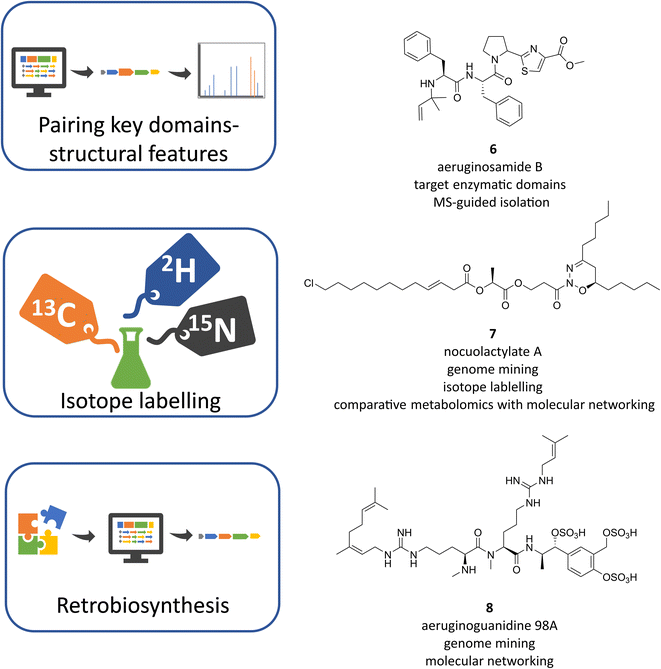
The -omics revolution in the last decades has significantly changed the cyanobacterial natural products research. Similar to other microbial phyla, natural product discovery is gradually shifting from the traditional bioactivity-guided screening strategies to genome mining and metabolomic approaches, or a combination of both for better dereplication and prioritisation. Additionally, the advances in transcriptomics and proteomics are key to facilitating the link between the BGCs and the specialized metabolites.56,57 Workflows on genomic-metabolomics have been reviewed recently, all having their challenges and limitations, but also numerous benefits, leaving the decision of the strategy of choice to the researcher.58–60 So far, genome mining and untargeted metabolomics, the most common metabolomics in secondary metabolites discovery, rely on the comparison of experimental data with databases to identify known/unknown BGCs or molecules, respectively. Therefore, the creation of publicly available databases of genomic data, chemical structures and their properties, spectral libraries, and metabolomics is essential for the progress in dereplication analysis, but also for biological and taxonomic studies. Databases for microbial research have been reviewed until 2020 (ref. 61) and are continuously expanding as exemplified by the construction of specific repositories like CyanoMetDB,62 dedicated to cyanobacterial toxins and secondary metabolites and its incorporation into the NPAtlas database25 and the recently created LOTUS collaborative database of natural products.63 In addition, new initiatives are arising under the necessity to link the emergent multi-omics data, like the Paired Omics Data Platform (PoDP) whose aim is linking the genomic and metabolomic data deposited in public repositories.64In this context, a number of studies on cyanobacteria have already implemented advanced genomics-metabolomics strategies to dig into the genomes, prioritise and connect the biosynthetic machineries with their products.11,17 Genome-guided methodologies can expand chemical families by interrogating the genome in different strains and searching for variants of known BGCs or enzymes, which coupled with mass spectrometry (MS)-based strategies, allows the assignment of BGCs to products. For example, new cyanobactin linear peptides were found after genome mining of 126 cyanobacterial strains. Using BLAST searches for the signature proteases PatA and PatG, 31 putative cyanobactin BGCs were identified. A closer inspection of these sequences predicted the presence of cyanobactins containing not only the characteristic thiazoles but also methylation(s) and prenylation(s) as further posttranslational modifications in Microcystis aeruginosa PCC 9432 and Oscillatoria nigro-viridis PCC7112. The liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis identified peptides with these key features and guided the discovery of aeruginosamide B (Fig. 4, 6) and C and viridisamide A.65 The pairing between particular enzymatic domains within BGCs and MS or NMR signatures is an effective tool that can work in both ways, finding the characteristic metabolites using BGC prediction in complex extracts or deorphanizing BGCs with known distinctive compounds. For instance, halogenases can be linked with the MS isotopic patterns of halogens as exemplified in the discovery of columbamides A, B and C in Moorena bouillonii PNG. A paired genomic and metabolomic comparison of three Moorena strains correlated a novel BGC in M. bouillonii that encoded cryptic halogenase domains with two clusters of halogenated compounds found exclusively in M. bouillonii using Global Natural Products Social Molecular Networking (GNPS) analysis.66,67 Compounds of these clusters were selected for isolation leading to the structure elucidation of the columbamides and allowing assignment of the previously unknown col BGC.68
The capacity to identify the isotopic patterns can be further exploited by stable isotope-labelled supplementation experiments based on the BGC features. Applying a lipid-version of the genomisotopic approach, new fatty acids (FA)-derived compounds were discovered using supplementation experiments with deuterated FA and comparative metabolomics. After proving the absence of a beta-oxidation pathway in cyanobacteria, which supports the prevalence of FA-derived compounds, supplementation experiments were designed to detect labelled metabolites in different cyanobacterial strains containing FA-incorporating enzymes. Comparative metabolomics between supplemented and non-supplemented cultures was performed by LC-HRESIMS and combined with data processing in MZmine,69 whose modules allow filtering the peak list, for example by adducts. Feature-based molecular networking using GNPS and MS-guided isolation focusing on unknown features led to the identification of new hapalosin analogues in Fischerella sp. and the nocuolactylates A (Fig. 4, 7) and B in Nodularia sp.70 In another experiment, labelling with stable 15N was performed in Nostoc sp. UIC 10630 to correlate BGCs with their metabolites by pairing the prediction of nitrogen atoms according to the amino acid specificity of adenylation domains with the number of nitrogen atoms identified by comparative metabolomics between labelled and unlabelled experiments. This approach was able to deorphanize three of the six BGCs predicted to contain nitrogen in their products, a new anabaenopeptin analogue, a new compound named nostopyrrolidonamide and also the known aeruginosin 865 whose cluster was formerly unidentified.71
As can be seen, retro-biosynthetic strategies can be useful to connect known compounds with their BGCs provided that the order of BGC modules shows collinearity with the order of building blocks in the compounds, as is common in type I PKS, NRPS and hybrid PKS–NRPS pathways. This tactic was applied to assign the biosynthetic pathway of aeruginoguanidines (AGDs), and then the microguanidines (MGDs) that were co-assigned to the same BGC in Microcystis strains.51 According to the structural features of AGD the predicted pathway was expected to involve an NRPS with specificity for L-arginine and tailoring enzymes such as a prenyltransferase and a sulfatase/sulfotransferase. A candidate BGC fulfilling the expected features was found in the AGD-producing strain M. aeruginosa NIES-98 and screened in other public Microcystis genomes. Subsequent analysis of metabolomes using LC-MS/MS revealed the presence of AGD in eleven strains of Microcystis. Molecular networking analysis expanded the AGD compound family with new variants AGD-98A (Fig. 4, 8), AGD-98B and AGD-98D, but unexpectedly also revealed the presence of another cluster in the same strains, comprising shorter MGD variants. Isolation and structure elucidation of the MGD metabolites, revealed new variants and new intermediates, the microguanidines amides MGA-771, and MGA-787. Analysis of the presence/absence of the compounds and genes in the different strains by a phylogenetic approach uncovered the unprecedented biosynthetic versatility of the AGD/MGD/MGA pathway and showed that the production was mutually exclusive with microcystin.51
(Meta)-genomics has boosted the exploration of cyanobionts with several successful examples linking biosynthesis and chemistry, as it can be seen in the recent review of D'Agostino.11 The development of these technologies also facilitates the study of collections of microorganisms, rather than single organisms, and computational networking approaches enable the visualization and data analysis of large datasets. Following a phylogenetics-guided multi-omics study, the genome and metabolome of 24 tropical filamentous cyanobacteria were compared to evaluate the natural product potential and to prioritise strains.72 The sequence similarity networking of BGCs was created from the meta-genomic assemblies of the environmental cyanobacteria using antiSMASH,22 BIG-SCAPE73 and the MIBIG27 database to assign the GCFs. The BGC similarity network was integrated with a classical GNPS molecular networking generated using LC-MS/MS and in silico annotation tools such as Network Annotation Propagation,74 DEREPLICATOR+,75 and MolNetEnhancer76 for the chemical classification. Interestingly, the information of both networks highlighted, aside from the metabolic potential of the strains, some of the challenges in the assignment of cyanobacterial BGCs, as despite the fact that the majority of the molecular families had a spectral match, many BGCs were still orphans. Specifically, a variety of peptides were predicted in the BGC analysis, while in the metabolomics analysis, they were underrepresented compared to lipidic molecules. This pointed out the need for advanced genomic engineering approaches to enhance the production of the cryptic peptides or to develop strategies to facilitate their detection.
The power of genome and metabolome mining strategies can be complemented with further omics technologies. Peptidogenomics and de novo peptide sequencing are emerging as new approaches linking peptides with MS data through automated processes in tools like NRPquest77 for NRPS or MetaMiner78 and DeepRipp79 for RiPPs. Although proteomics has been less integrated within the workflows, it can be useful to leverage the information of the enzymes involved in the biosynthesis of the secondary metabolites. Available metadata about the samples can inspire experiments and guide the prioritisation. This can include taxonomy and phylogenetics, as we have seen, but also bioactivity, growth conditions, and phenotypes of mutant strains. As shown, the majority of studies on cyanobacteria follow the pattern-based approach, focusing on the presence/absence of metabolites and BGCs across strains. However, other methods can further explore cyanobacteria, such as the correlation-based approach, which adds metrics to score a given BGC-metabolite connection (metabologenomics),80 and the feature-based approach, supported by in silico tools and metabolite prediction, as the multi-omics study previously described.72 The above mentioned peptidogenomics approach also falls into this category. Another emerging method is the combination of feature and correlation-based approaches like applied by the NPLinker software framework.81 Through many advances in omics technologies, bioinformatics and machine learning, the opportunities for mining the untapped biosynthetic potential of cyanobacteria natural products are steadily increasing.
6. Endogenous activation of cryptic biosynthetic gene clusters in native cyanobacterial hosts
As emphasized above, studies on the model strain N. punctiforme PCC 73102 have shown that a major reason for the lack of assignment of the cryptic specialized metabolites is the low expression level of the majority of BGCs under standard growth conditions.43,53 It is likely that this is also true for many other talented cyanobacterial strains. A simple way for genome-based discovery for previously cryptic metabolites is to compare different cultivation conditions (OSMAC – one strain-many compounds).82 This can involve, for example, changing the composition of macro-, micro- or trace elements in the medium, altering the physical parameters, or testing different chemical elicitors and environmental cues.82 In addition, there are numerous examples of stimulation of BGCs by biotic interactions.82 In this context, interactions with other bacteria as well as interactions with eukaryotic organisms can induce or alter the expression of specialized metabolites. In the field of cyanobacterial research, there are few systematic or comprehensive studies on the influence of growth conditions; rather, it is individual studies that show the potential of this methodological approach.A group of metabolites in cyanobacteria that can be induced by changing the macro elemental composition in the growth medium are siderophores that are stimulated by iron deprivation. Until recently, very few cyanobacterial siderophores were known such as the hydroxamate siderophores schizokinen and synechobactin and the catecholate siderophore anachelin.83 A recent study that has used a genome-based approach led to the discovery of cyanochelins, a family of NRPS-derived siderophores containing β-hydroxyaspartate moieties (Fig. 5, 9).84 Further, changing the trace elemental composition can elicit production of specialized metabolites. An impressive example was recently shown for the eagle-killing toxin aetokthonotoxin (AETX).85 While the toxin was detected in environmental samples growing on the invasive water plant Hydrilla verticillata, it could not be identified in isolated laboratory strains. The addition of potassium bromide to the medium resulted in stimulation of AETX production.85 However, the reasons for the stimulating influence are different for the siderophore example and the AETX example. While the former are transcriptionally stimulated by iron deficiency, since the corresponding BGCs are typically controlled by specific transcription factors of the Fur family,83 bromide is a direct precursor of AETX, without which production of the toxin is not possible.85
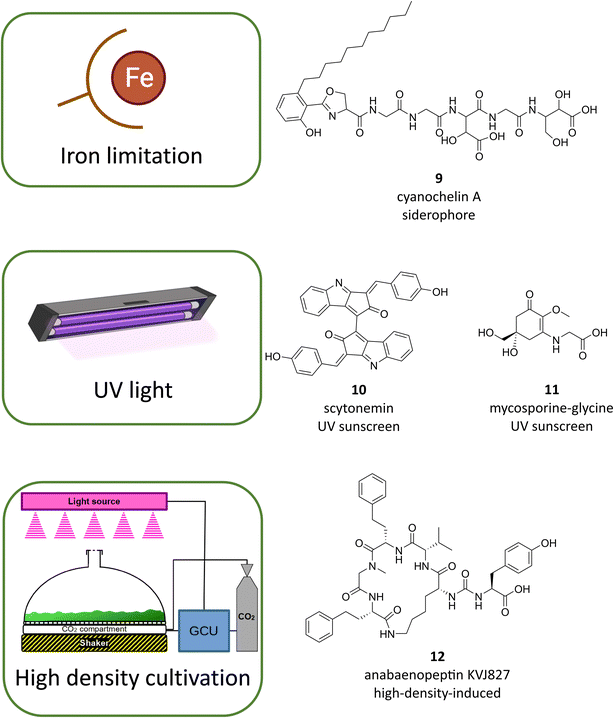 | ||
| Fig. 5 Examples of cultivation-based discovery of cyanobacterial metabolites following BGC activation by iron limiting cultivation conditions, UV induction or high-density cultivation. | ||
Changing physical parameters during growth can also induce cyanobacterial specialized metabolites. Well-known examples of such inducible metabolites are the sunscreen compounds scytonemin (Fig. 5, 10) and mycosporic acids (MAAs, Fig. 5, 11).86 While MAAs are also known from other phyla such as fungi, microalgae, macroalgae and heterotrophic bacteria, scytonemin is exclusively synthesized by cyanobacteria.86 Scytonemin responds rather specifically to induction by UV-A and B,87 while MAAs are induced by various stress conditions including UV stress, salt stress and desiccation,88 MAAs are widespread among cyanobacteria and have been described in numerous variants. The inducibility of sunscreen compounds has been investigated both using laboratory strains and in field studies and is generally linked to the induction of transcription.89,90 Production of scytonemin, in particular, seems to be highly relevant for the adaptation to the extreme light conditions in deserts and is able to filter out damaging UV light portions. Scytonemin production was also linked to localized warming in soil crust communities.90 In addition to their ecological significance, scytonemin and MAAs are also among the cyanobacterial metabolites with potential for application in cosmetics and medicine.86
A combination of chemical and physical parameters again is changed by the above-mentioned HD cultivation. In this cultivation apparatus, both high light irradiation and higher amounts of the carbon source CO2 are being used. In addition, cyanobacteria grow as a dense biofilm. The use of this cultivation technology in N. punctiforme PCC 73102 and other Nostoc strains led not only to efficient growth but also reprogramming of specialized metabolism, both qualitatively and quantitatively.43,53 Transcriptional analysis of BGCs revealed that at least 50% of BGCs were strongly stimulated under these conditions and that a combination of high light conditions, high CO2 availability and medium factors are triggering both transcription and production of compounds.53 As proof of principle, HD cultivation of the strain Nostoc sp. KVJ2 led to the discovery and structural elucidation of three new anabaenopeptin variants, KVJ827 (Fig. 5, 12), KVJ841 and KVJ811 which showed allelopathic activity against other Nostoc strains from the same habitat.43
The influence of biotic interactions on the production of specialized compounds in cyanobacteria is generally poorly studied. The fact that many talented producers still have accompanying heterotrophic bacteria and that the effects of specific interactions are thus difficult to dissect certainly plays a role here. A comparison of axenic toxic and nontoxic Microcystis strains in mono- and coculture revealed an intraspecific quantitative variation of major peptide classes and led to the discovery of new cyanopeptolin analogues.91 Co-cultivation of Microcystis with a heterotrophic bacterial community again resulted in negligible effects on the intracellular specialized metabolome. Remarkably, no extracellular levels of the known peptide classes in Microcystis were detected when the cyanobacterium was accompanied by a heterotrophic consortium suggesting an ability of the community to degrade structurally different compounds or a physiological suppression of their secretion.92 Hence, while intra- and interspecific biotic interactions certainly have an impact on the intra- and extracellular metabolome of Microcystis, they did not lead to the genome-based discovery of entirely new classes of compounds. A comparison of the metabolome of the symbiotic cyanobacterium N. punctiforme in the free-living state and in physical association with the plant host Gunnera manicata using MALDI-imaging showed major differences in the metabolite profiles with nostopeptolides being downregulated and a number of novel metabolites being upregulated in planta.93 Since only limited amounts of symbiotic tissue were available the study did not lead to the description of a new metabolite family either. Yet, it gives hints that biotic interactions with eukaryotic partners may lead to a more pronounced impact on the specialized metabolome than interaction with other prokaryotes.
In summary, cultivation-induced biodiscovery is a promising approach for cyanobacteria. However, there are only incidental studies with a focus on single factors. To generalize conclusions regarding the impact of abiotic and biotic factors on the specialized metabolome high throughput approaches are needed using a highly parallelized approach. Development of such approaches requires minimization of culture volumes and ideally includes an automated control of cultivation conditions.82 One methodology that is now commonly used for other microorganisms is the microfluidics technology.82 Although this technology requires optimization for filamentous cyanobacteria, it could provide a means for semi-automated analysis of cyanobacteria in a miniaturized format. Thereby, both the impact of abiotic factors as well as biotic factors could be tested in a larger format. There are certainly additional ways to rationalize metabolite induction. The AntiSMASH platform already gives some hints on the possible specificity of transcription factors and their regulation.22 There is also much evidence that environmental cues commonly present in the habitat of the given organisms may play a particular role in the induction of specialized metabolites. Ultimately, it depends on the range of biological and metabolic properties of individual strains, which conditions could lead to an altered special metabolite profile. Since variations in cultivation conditions are easily feasible for cyanobacteria of all genera and habitats, the general potential of cultivation-based approaches is far from exhausted.
7. Use of genetic approaches for the discovery and manipulation of cyanobacterial natural products
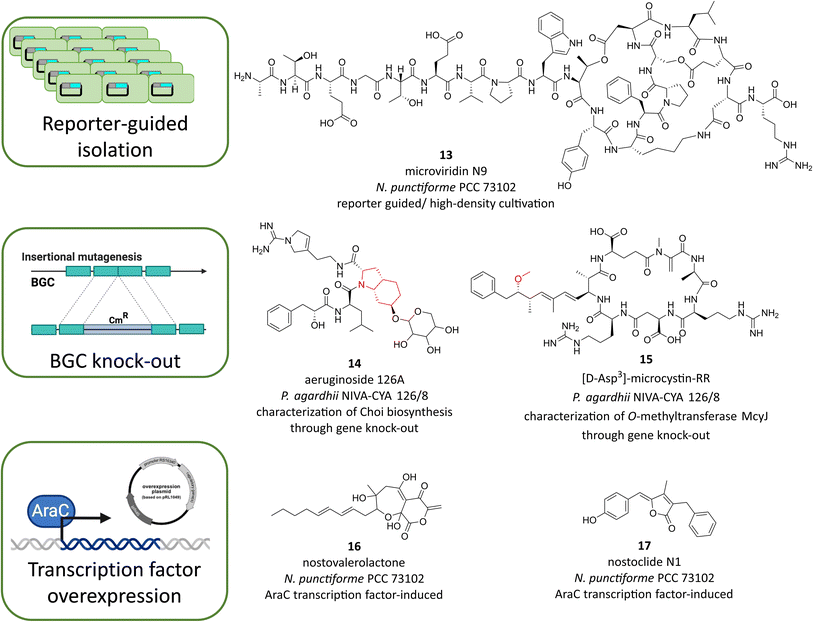
Genetic techniques have contributed to significant progress in microbial natural product research, ultimately advancing natural product discovery and enabling the development of production strains, e.g. for Streptomycetes and filamentous fungi.90,94,95 In principle, BGCs and their products can be assigned by forward genetic approaches with untargeted random mutagenesis or by reverse genetic approaches with targeted gene manipulation. Genetic techniques can also enable the activation of silent BGCs or be used to generate transcriptional reporters.96 Since only a few cyanobacterial strains have been amenable to genetic manipulation to date, the scope of related studies in cyanobacteria is comparatively limited. However, endogenous activation of BGCs offers several advantages over heterologous expression. When BGCs are being activated in the native host, it can be assumed that precursors should be available even for biosynthetic pathways that are difficult to predict or that require enzymes encoded in trans of the BGC. Activation or inactivation of BGCs in the native cyanobacteria also allows the study of the biological function of their products. In addition, studies on the regulation of BGCs may also inspire future development of cyanobacterial production strains.One of the few strains with great potential for natural products production that is accessible to genetic manipulation is the symbiotic strain N. punctiforme PCC 73102. For this strain, a library of transcriptional reporter mutants could be generated, in each case fusing the 5′UTR region containing the putative promoter region of BGCs of the NRPS, PKS, or RiPP types with the CFP reporting gene.53 As mentioned above, this reporter library was used to test the transcriptional response to HD cultivation in high throughput format. Because several RiPP reporting strains showed a pronounced upregulation after HD cultivation, the study was combined with a targeted metabolomic search for the hypothetical RiPP products. Thereby, the new microviridin N1–N9 variants were discovered (Fig. 6, 13).53 Microviridins had previously been described primarily in Microcystis and Planktothrix, but microviridins of N. punctiforme are distinguished by a variable chain length at the N-terminus and were overlooked prior to the reporter-guided study. While this is an example how transcriptional reporters can facilitate genomic mining of novel compounds there are certainly further ways to use the library in the future, such as BGC elicitor screening or screening of biotic interactions with either prokaryotic or eukaryotic organisms.
Strain N. punctiforme PCC 73102 (alternatively designated as N. punctiforme ATCC 29133) was also the only strain in which a cyanobacterial natural product BGC could be assigned using a forward genetic approach. The biosynthetic pathway for the sunscreen compound scytonemin was originally discovered using random transposon mutagenesis.87 Besides, several BGCs could be inactivated in non-model cyanobacteria using a directed reverse genetic approach. First, the microcystin biosynthetic pathway was knocked-out in the genus Microcystis.97 In the same genus, the BGCs for aeruginosin,98 micropeptin,99 and MAA88 were also successfully manipulated. Notably, mutagenesis has only been successful in two Microcystis strains: M. aeruginosa PCC 7806 and M. viridis S-70.99 Other cyanobacterial BGCs that could be inactivated include the anabaenopeptilide BGC in Anabaena sp.91,100 the microcystin and aeruginoside BGCs in Planktothrix NIVA-Cya126 (ref. 101 and 102) and the pks2 BGC in N. punctiforme PCC 73102.103 In all cases, mutagenesis contributed to the assignment of natural products and their BGCs. Only in the case of the cryptic pks2 BGC of N. punctiforme PCC 73102, no product could be assigned so far, probably due to the low expression level of the BGC.53 In some cases, mutagenesis has contributed essential insights into the biosynthetic mechanism. For example, mutagenesis of the aerD, E, and F genes and subsequent feeding experiments demonstrated their involvement in the biosynthesis of the characteristic Choi moiety of aeruginosins in P. agardhii NIVA-Cya 126 (Fig. 6, 14).102 Mutagenesis of the tailoring enzyme McyJ in P. agardhii NIVA-Cya126 allowed isolation of microcystins lacking the O-methylation at the ADDA moiety (Fig. 6, 15), thereby identifying McyJ as responsible O-methyl transferase.101
As mentioned earlier, a positive side aspect of biosynthetic mutants is their potential for studies of biological function. In particular, the microcystin-free ΔmcyB mutant has been intensively used for years for research on microcystin function. In this context, a close connection of microcystin with primary carbon exchange could be demonstrated. In cyanobacteria, the toxin binds to key proteins of the Calvin–Benson–Bassham cycle including the CO2-fixing enzyme RubisCO.104 MAA mutants were also used for functional analyses. It was shown that the loss of MAA did not affect growth under UV stress, but did affect the structure of the EPS layer of strain M. aeruginosa PCC 7806. This observation strengthens the hypothesis that MAA is not produced primarily for UV protection and that the sunscreen effect may be a useful side effect of the compound family.88 The pks2-mutant was used as an indirect tool to explore the role of nostopeptolide in N. punctiforme PCC 73102. Since nostopeptolide was greatly reduced in the mutant, it was shown that nostopeptolide represses or stimulates the development of motile hormogonia filaments in a concentration-dependent manner, thereby having a significant impact on the symbiotic interaction of N. punctiforme with the host Gunnera manicata.93 Loss of constitutively produced cyanopeptides often leads to increased production of other cyanopeptides, apparently because unused resources become available. For example, loss of microcystin in M. aeruginosa PCC 7806 leads to increased production of cyanopeptolin and microcyclamides,91 and loss of anabaenopeptilide in strain Anabaena 90 leads to increased production of anabaenopeptin.105 These observations may also contribute to the optimization of production strains in the future.
The strain N. punctiforme PCC 73102 is also suitable for overexpression of transcriptional regulators. There are shuttle plasmids that replicate in both E. coli and N. punctiforme. Applying a shuttle plasmid technology, it has recently been possible to overexpress an AraC type positive regulator for the previously cryptic pks1 pathway.106 Upregulation of the pks1 pathway, in turn, led to the global stimulation of the specialized metabolism in N. punctiforme, in particular under high density cultivation conditions. To identify the responsible signals, a reporter mutant was used to evaluate the response of the pks1-dependent cryptic lanthipeptide pathway ripp4. The bioactivity-targeted strategy led to the discovery and high titer production of two signal molecules: nostovalerolactone (Fig. 6, 16), representing a new family of compounds derived from a tetronate-like pathway in N. punctiforme, and nostoclide(s) N1 and N2, representing new members of the cyanobacterin family (Fig. 6, 17).106
Although only a few strains are amenable to genetic manipulation, insights gained through genetic analyses can be very helpful in making screening of cyanobacterial extracts more efficient and in developing individual strains for synthetic biology applications. However, further research and development is needed to catch up to known microbial model systems such as in Streptomyces or Aspergillus strains.94,95 For the further development of individual strains as production strains, knowledge on the regulation of secondary metabolism must be further deepened. The power of engineering strategies that exploit underlying regulatory mechanisms for the targeted overproduction of individual substances has been demonstrated, for example, for proteobacteria of the genera Xenorhabdus and Pseudomonas. Once it was known that BGC expression was primarily controlled post-transcriptionally, knocking out the RNA chaperone Hfq was able to abolish global production of specialized compounds. Against this background, individual BGCs can be specifically activated and optimized.107 Similar strategies are also conceivable for individual cyanobacterial strains in the future and can greatly advance the biotechnological potential of cyanobacteria for the production of bioactive natural products.
8. Heterologous expression: E. coli vs. cyanobacterial hosts
Despite advances in the development of genetic tools for cyanobacteria, their application is limited to very few strains. Furthermore, most of the bioactive natural products can only be isolated at low yields thus hampering economical large-scale production and development of routines that ultimately could lead to commercial applications. The aforementioned problem of the enormous untapped potential hidden away in “orphan” or “silent” BGCs further exacerbates the need for alternative methods to make cyanobacteria live up to their expectations. In this regard, heterologous expression of cyanobacterial BGCs is an increasingly successful avenue (most recently reviewed in ref. 55), albeit the number of relevant studies is small and lags significantly behind the in silico identification of novel BGCs. In particular, the list of heterologous host organisms is short and can roughly be divided in cyanobacterial and non-cyanobacterial hosts. While the latter group is dominated by the biotechnological mainstay E. coli, the former includes the cyanobacterial “models” Synechocystis sp. PCC 6803, Synechococcus elongatus PCC 7942 and Anabaena PCC 7120. Organisms from other clades have also been used, most notably yeast108 and Streptomyces venezuelae DHS 2001,109 but these are clearly the exception. Choosing the host best suitable for the expression of a given BGC is critical, however, owing to the complex requirements of individual biosynthetic pathways, there is currently no reliable method to predict the outcome before experimental validation. Therefore, it is important to gather all available information about the target BGC and its product range to infer specific requirements regarding, among others, substrate availability, medium composition or genetic compatibility.Unsurprisingly, early attempts at heterologous expression of cyanobacterial natural products have focused on RiPPs as their BGCs are comparably small and as such easily handled by established molecular biological techniques. Moreover, RiPPs solely rely on the presence of proteinogenic amino acids and the ribosomal translational machinery,110 their BGCs normally encode all enzymes necessary for post-translational product modification and they do not require activation by off-site factors. Consequently, RiPPs make up a large proportion of cyanobacterial natural products that were reported to be produced heterologously.111–118 In these pioneering works, E. coli was the obvious choice of expression host as the BGCs were introduced on established vectors with little or no modifications necessary to the genetic sequences or the host metabolism. This situation is drastically different when BGCs of the NRPS and/or PKS type are to be analyzed. Not only do these gene clusters tend to be much larger than those of RiPPs, which makes cloning more difficult,110 but there is also a certain degree of incompatibility between elements of transcriptional regulation, most prominently promoters, from cyanobacteria and E. coli.110,119–121 The most important impediment, however, is that the metabolism of E. coli needs to be primed for the expression of NRPS/PKS type of BGCs, primarily because they require the activity of a suitable phosphopantetheinyl transferase (PPTase) to activate their cognate carrier proteins.122,123 In connection to that, the introduction of additional factors is often needed to meet the substrate and co-factor requirements of each individual BGC as they tend to rely on the availability of rare non-proteinogenic amino acids or their precursors. In a case-dependent manner, these problems can be solved either by supplying the necessary factors with the growth medium or by genetically engineering the host metabolism to provide off-site auxiliary enzymes. Despite this, E. coli has undeniable advantages that still uphold its attractiveness as a host for BGC heterologous expression. The most convincing arguments for E. coli are the plethora of well-established genetic tools, the straightforward cultivation and the short doubling times. Accordingly, many attempts have been made to adapt E. coli to the expression of large BGCs and to rationalize workflows. Recent advances in DNA assembly, for instance, have facilitated the handling of large genetic constructs. A method based on Gibson assembly termed “direct pathway cloning” (DiPaC,124–126 and the transformation-associated recombination (TAR) approach)127,128 have successfully been employed to capture large BGCs. The necessity to provide a constitutive PPTase has also been addressed by the generation of two widely used E. coli strains, BAP1 (ref. 129) and GB05-MtaA,120 which express promiscuous Sfp-type PPTases from Bacillus subtilis and the myxobacterium Stigmatella aurantiaca, respectively. With these tools in hand, a number of NRPS/PKS derived cyanobacterial compounds have been successfully produced in E. coli. Among those are the dermatotoxin lyngbyatoxin (Fig. 7, 18 (ref. 130)), the infamous hepatotoxin microcystin-LR (1),120,131 the depsipeptide hapalosin (Fig. 7, 19 (ref. 132)) and, most recently, the natural herbicide cyanobacterin124 and the protease inhibitor microginin.125 Frequently, the products detected in E. coli did not fully reflect the compound spectrum found in the natural producers, however, these expression systems still can provide very valuable mechanistic insight (e.g. with cyanobacterin124) or can serve in establishing an accessible platform for biocombinatorial production of novel congeners (e.g. with microcystin-LR131). Therefore, E. coli will doubtlessly continue to maintain its status of a major heterologous expression system, as evidenced by the high number of current research efforts in that field.
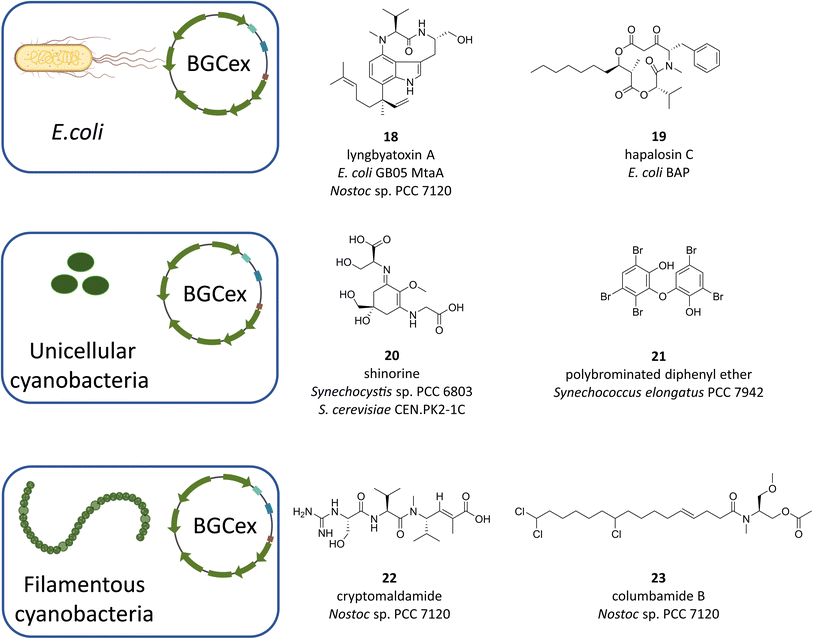 | ||
| Fig. 7 Examples of cyanobacterial metabolites heterologously produced in either E. coli strains or cyanobacterial model strains. | ||
Despite the accessibility and adaptability of the E. coli system, there have been many examples where the production of cyanobacterial natural product proved to be very troublesome or downright impossible. The resulting demand of alternative expression systems is best illustrated with the case of lyngbyatoxin A, an NRPS-type compound and promising drug lead.133 While its heterologous production was altogether unsuccessful in Streptomyces coelicolor134 and required the exchange of the cyanobacterial native promoters in E. coli,130 it was easily achieved in the filamentous cyanobacterium Anabaena sp. strain PCC7120 (Anabaena 7120,128). This follows the widely accepted notion that heterologous expression of complex metabolites is best carried out in hosts more closely related to the original producer. As a number of cyanobacterial model strains with fairly advanced genetic tools have been available for more than two decades, scientists have started to investigate these strains for their suitability as expression hosts for natural product BGCs. The three most widely used strains of cyanobacteria are Synechocystis sp. PCC 6803 (Synechocystis), Synechococcus elongatus PCC 7942 (S. elongatus) and Anabaena 7120 and indeed, heterologous expression of cyanobacterial BGCs has been attempted in all of them (Fig. 7). Gauging the success of these approaches, however, Anabaena 7120 has emerged as the clear favorite. While there are two reported cases where Synechocystis and S. elongatus were employed for the successful production of small or partial BGCs such as the MAA shinorine (Fig. 7, 20 (ref. 135)) and a sponge-derived polybrominated diphenyl ether (Fig. 7, 21 (ref. 136)), in recent years the products of complex, full-size NRPS/PKS-type BGCs were isolated with comparably high yields from Anabaena 7120. In retrospect, this does not seem very surprising, because the genus Anabaena displays a number of traits that appear to be beneficial for the production of NRPS/PKS type compounds and that are absent from the two other strains. First and foremost, while Anabaena encodes several NRPS/PKS BGCs15 together with an active, very promiscuous Sfp-type PPTase,123 it does not seem to produce any bioactive compounds that may potentially disturb expression of foreign BGCs.128 Additionally, many cyanobacterial promoters seem to work well in Anabaena 7120 and its genetic toolbox is constantly being expanded.128,137–140 With their groundbreaking work, two groups in particular have paved the way in establishing Anabaena 7120 as a promising host for the heterologous expression of cyanobacterial natural products. Implementing their lyngbyatoxin expression platform, Philmus and coworkers were not only the first to report the successful heterologous production of a complex NRPS-type compound in Anabaena 7120, they were also able to take their system a step further, employing biocombinatorics to produce additional indolactam-V derivatives by swapping in tailoring enzymes from different genera.127,128 Most recently, the group reported the successful production of the meroterpenoid tolypodiol and some of its analogs from a 21 kb BGC.141 While in these cases the BGCs were expressed from replicative plasmids, Golden and colleagues have successfully produced cyanobacterial compounds from BGCs that were integrated into a neutral site of the chromosome of Anabaena 7120. In their first reported case, they initially attempted to have S. elongatus produce the putative anti-infective compound cryptomaldamide (Fig. 7, 22) from a 28.7 kb NRPS/PKS megasynthase BGC.142 Despite many efforts to induce biosynthesis, they were only successful once they changed the heterologous host to Anabaena 7120. More recently, the group utilized their expression system for the production of columbamides (Fig. 7, 23), a family of chlorinated acyl amides, from a 28.5 kb NRPS/PKS hybrid BGC.143 What makes this study stand out is the fact that apart from the discovery of new high potential compound analogs, the BGC in question was merely proposed, at the time, to code for the columbamide pathway and was thus experimentally confirmed by heterologous expression.
Evidently, substantial progress has been made in recent years, with both the E. coli and the cyanobacterial system. Both offer certain advantages over the other, with E. coli being better established and more accessible and cyanobacteria holding more promise with regard to enzyme fidelity and product biochemistry. Both systems suffer from a lack of predictability, as there are still no clear indicators available from which to infer host compatibility or to estimate production levels. Indeed, so far neither system has proven to be advantageous in this respect, as examples for very high and very low yields have been reported for both.55 The most convincing argument in favor of cyanobacteria as heterologous host, not only for natural product research, is the great potential for sustainable, green biotechnology. From the progress that has been made in recent years and with cyanobacterial expression systems being now in the focus of many efforts to optimize their handling and performance, the development of potentially carbon-negative production strains that can offset the advantages that E. coli still holds, seems to be only a matter of time. This could also go a long way towards fully unlocking the cyanobacterial natural product potential by finally enabling the systematic introduction of orphan BGCs into heterologous hosts and the identification of their cognate natural products.
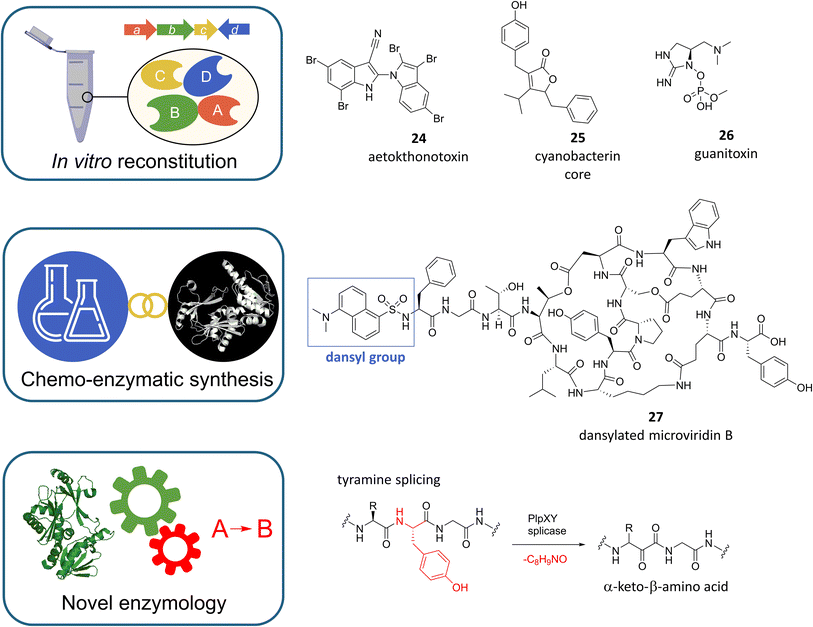
9. From in vitro reconstitution of cyanobacterial pathways to chemo-enzymatic exploitation
In view of the challenges discussed above for the mining and study of cyanobacterial natural products, the in vitro reconstitution of key biosynthetic steps or even entire biosynthetic pathways, sometimes referred to as “total biosynthesis”,144 offers an elegant solution for the characterization of BGCs or the production of bioactive compounds when biochemical reactions and precursors for the assembly of compounds are known or well predictable. Similar to the approaches discussed earlier, this emerging field is benefiting greatly from continued advances in biotechnology, computation, and analytics. In addition, innovation is fueled by the chemical and pharmaceutical industries' growing appetite for biocatalysts as drivers of green and sustainable chemistry.145Recent elegant examples of in vitro reconstitution of cyanobacterial pathways are the aforementioned eagle-killing toxin aetokthonotoxin (Fig. 8, 24),146 the core of the bioherbicide cyanobacterin (Fig. 8, 25),124 or the potent anticholinesterase neurotoxin guanitoxin (Fig. 8, 26).147 For the biosynthesis of aetokthonotoxin (AETX) Adak and Lukowski et al. showed that the freshwater cyanobacterium Aetokthonos hydrillicola uses an efficient, five-enzyme biosynthetic pathway to convert two molecules of tryptophan to yield this structurally unique pentabrominated biindole nitrile by assembling two functionalized indole monomers that are then linked by biaryl coupling.146 Of the five biosynthetic enzymes, two stand out for their potential as biocatalysts, namely the unique single-component, flavin-dependent tryptophan halogenase AetF, which does not require an accompanying reductase partner, and the iron-dependent nitrile synthase AetD, which builds a nitrile functional group via unprecedented rearrangement chemistry. In addition to revealing novel chemistry, the identification of the AETX BGC may help to identify related BGCs and thus help to discover new environmental toxins.
When the potent photosynthesis inhibitor cyanobacterin was isolated from Scytonema hofmanni in 1982 it was the first chlorinated natural product identified from freshwater cyanobacteria.148 Four decades later D'Agostino et al. took advantage of this property and identified the cyanobacterin (cyb) biosynthetic gene cluster by targeted bioinformatic screening for halogenase-encoding genes.124 After validation of the BGC by heterologous expression in E. coli, the authors were able to break down the biosynthesis of the furanolide core structure to four enzymes by gene knockouts and the development of a one-pot biocatalytic in vitro synthesis. At the heart of the reaction sequence the furanolide synthase CybF fuses two molecular building blocks, which are provided by CybB, CybC and CybE to form the furanolide core. Since members of the furanolide natural product family such as nostoclides106,149 or maculalactones150 are more widespread in cyanobacteria, the elucidation of furanolide biosynthesis paves the way for their targeted discovery, biosynthetic engineering and enzymatic synthesis.
Guanitoxin and cyanobacterin have in common, that their BGCs have remained elusive for decades. To identify candidate guanitoxin BGCs in Sphaerospermopsis torquesreginae ITEP-024, Lima et al. focused on genes associated with the known guanitoxin precursors (S)-4-hydroxy-L-arginine and L-enduracididine.147 This led to the identification of a candidate gnt BGC whose involvement in guanitoxin biosynthesis was confirmed by the impressive in vitro reconstitution of a nine-step metabolic pathway starting from L-arginine. The elucidation of the genetic basis of guanitoxin biosynthesis now allows environmental biosynthetic gene monitoring of this lethal neurotoxin, which shares an identical mechanism of action with the synthetic chemical warfare agent sarin and is one of the most potent cyanotoxins known today.
These three studies are prime examples of how in vitro reconstitution can help to elucidate biosynthetic pathways. However, the in vitro reconstitution of individual enzymatic steps or biosynthetic sequences also offers the possibility of exploiting these pathways chemo-enzymatically. Among the diverse compound classes found in cyanobacteria, RiPPs are particularly well suited for this approach due to the synthetic accessibility of precursor and core peptides by solid-phase peptide synthesis (SPPS), the hypervariability of the peptides, and the relaxed substrate specificity and modularity reported for many RiPP-modifying enzymes.
A recent study perfectly exemplifies the great potential of combining hypervariable precursor peptides with highly promiscuous RiPP-modifying enzymes in vitro. By incubating 120 peptide substrates from a synthetic positional scanning library with the macrocyclase PagG and the prenyltransferase PagF from the cyanobactin prenylagaramide biosynthetic pathway Sarkar et al. generated a library of more than 100 prenylagaramide-like cyclic peptides and their prenylated derivatives.151 Interestingly, the promiscuity of cyanobactin prenyltransferases towards the prenyl donor can also be exploited chemo-enzymatically. For this, a synthetic alkyl pyrophosphate analogue was used to introduce a reactive moiety into a tryptophan-containing cyclic peptide. This reaction was catalysed by the N1-tryptophan prenyltransferase AcyF from the anacyclamide A8P pathway. After the conversion, click chemistry was used to fluorescently label the enzymatically modified peptide.152
Another cyanobacterial RiPP family that has been the subject of multiple chemo-enzymatic studies are the graspetides. These compounds, which include microviridins, are defined by the presence of ester or amide side-chain linkages that are installed by ATP-grasp ligases, resulting in peptide macrocycles.153 While the ATP-grasp ligases in microviridin biosynthesis catalyse cyclisation reactions in strict order and with stringent ring size requirements, they tolerate variations in the amino acid composition of the core peptide.154 This property was exploited by Reyna-González et al. in the synthesis of a microviridin-based library of protease inhibitors.155 In one-pot chemo-enzymatic reactions synthetic microviridin K and microviridin B core peptides, varied by all 20 proteinogenic amino acids at the position responsible for protease specificity, were transformed by two ATP-grasp ligases that were constitutively activated by covalently attached leader peptides, and a GNAT-type N-acetyltransferase. This leader peptide-free approach, which was inspired by a study from lantibiotic biosynthesis154 is highly efficient, since the relatively short core peptides can be synthesized much easier and at reduced costs compared to full-length RiPP precursor peptides. Later, the chemo-enzymatic synthesis platform was used to synthesize four microviridins encoded by the cyanobacterium Cyanothece sp. PCC 7822, providing a powerful example of culture-independent genome mining.156 In addition, Scholz et al. used the platform to introduce functional tags into different microviridin variants with the help of modified core peptides, yielding biotinylated, dansylated or propargylated congeners as tool compounds to further investigate the biology of microviridins.157 As a proof of concept, dansylated microviridin B (Fig. 8, 27) was used as a diagnostic tool to selectively label elastase in protease mixtures.
The examples given in this section illustrate how in vitro enzyme reconstitution facilitates the study and exploitation of cyanobacterial metabolic pathways. However, cyanobacteria also exhibit amazing enzymology that could be exploited beyond single pathways. A remarkable example is the discovery of non-canonical protein splicing by the radical S-adenosylmethionine (rSAM) enzyme PlpX from Pleurocapsa sp. PCC 7319. In a recent study Morinaka et al. showed that PlpX, together with the helper protein PlpY, catalyzes backbone carbon–carbon bond cleavage and the net excision of tyramine, resulting in the formation of α-keto-β-amino acids (Fig. 8).158 The reaction has been used to incorporate diverse and multiple β-amino acids into various RiPP precursors and recombinant proteins in E. coli.158,159 In addition to expanding the set of basic amino acid building blocks in peptidic natural products and proteins, tyramine splicing yields keto functions that can be readily exploited as orthogonal reaction sites for chemical diversification.158,160
Another impressive example of unprecedented enzyme catalysis in the biosynthesis of secondary metabolites comes from Cylindrospermum licheniforme ATCC 29412. Nakamura et al. were able to show that in cylindrocyclophane biosynthesis, chlorination of an unactivated carbon center by the halogenase CylC sets the stage for an enzymatic dimerization reaction catalyzed by CylK.161 This reaction features sequential, stereospecific alkylations of resorcinol aromatic rings by the alkyl halide electrophiles.161,162 Notably, alkylation of aromatic rings with alkyl halides is an important transformation in organic synthesis. However, an enzymatic equivalent was unknown. Interestingly, homologs of CylC and CylK cluster together with a number of additional enzymes in other unrelated cyanobacterial gene clusters, suggesting that related C–C bond formation strategies involving cryptic chlorination may be more widespread in the biosynthesis of cyanobacterial secondary metabolites.161 This, together with the prominent role of aryl–alkyl bonds in pharmaceuticals and industrial chemicals, makes CylC and CylK promising targets for future efforts in biocatalysis and metabolic engineering.
The selected enzymatic studies underline the extraordinary biosynthetic potential of cyanobacteria. As emphasized already, in vitro reconstitution is independent of cultivation and therefore applicable to the broad mass of cyanobacterial natural product producers. If the transformation of substrates succeeds in vitro, a wide range of possibilities for diversification and functionalization of metabolites arise. However, the amounts of substrates and their conversion cannot be arbitrarily upscaled. Individual chemical reactions, for example in the chemical synthesis of peptide precursors by solid-phase peptide synthesis, are also not very environmentally friendly. Therefore, in vitro (bio)synthesis is not always the appropriate method of choice. However, its importance for the study of biosynthetic mechanisms is central. In particular, linkage with structural biology analyses of enzymes can rationalize the production of new-to-nature libraries of metabolites. Enzymes are at the core of cyanobacterial natural product research and provide a hub for further exploitation.
10. Conclusions
In the last two decades, cyanobacterial natural products research has evolved on many levels. The majority of known natural product families could now be assigned to their dedicated GCFs. Public access to this information has enabled expansion and refinement of methods for dereplication of cyanobacterial datasets and provides avenues for prioritization of metabolite families. At the same time, technologies for cyanobacterial cultivation and genetic manipulation have advanced. Above all, heterologous expression of BGCs has made much progress in recent years. However, there are major areas of cyanobacterial natural products research that lag significantly behind comparable research in other microbial phyla. In particular, few entirely new classes of natural products have been discovered and described by genome-based approaches. Also, the development of production strains for natural products from cyanobacteria is still in its infancy. This is particularly striking because synthetic biology and biotechnology of cyanobacteria is a successful research and business field, and cyanobacteria are expected to contribute significantly to the development of a circular bioeconomy.This discrepancy is due to the strong focus in engineering tool development on a few unicellular cyanobacterial model strains. Future cyanobacterial natural products research needs to drive method development for appropriate filamentous strains themselves. This path is already being followed for the strain Nostoc sp. PCC 7120 and N. punctiforme PCC 73102. So far, these attempts are hardly on par with heterologous expression of cyanobacterial BGCs in E. coli. However, studies on production strains in e.g. Actinobacteria and Proteobacteria have shown that there is still much room for improvement in the optimization of production titers in original producers. This effort is definitely worthwhile because it can give new impulses to the whole cyanobacterial natural product research and biotechnology. To this end, the spectrum of suitable strains should be expanded and insights into the regulation of BGCs should be further gained and explored. These efforts also depend on understanding microbial physiology and biotic interactions among strains. In this way, microbiology potentially plays a key role in future cyanobacterial natural products research.
11. Author contributions
E. D. conceived the review, E. D., M. B., A. G. and M. M. G. wrote the review and contributed illustrations.12. Conflicts of interest
There are no conflicts to declare.13. Acknowledgements
This study was supported by Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – Project-ID 239748522 – SFB 1127 to E. D. and by the DFG-funded graduate school RTG 2473 (Bioactive Peptides) to E. D. M. B. is supported by Germany's joint federal and state program supporting early-career researchers (WISNA).14. References
- P. Sanchez-Baracaldo, G. Bianchini, J. D. Wilson and A. H. Knoll, Trends Microbiol., 2022, 30, 143–157 CrossRef CAS PubMed.
- E. Dittmann, D. P. Fewer and B. A. Neilan, FEMS Microbiol. Rev., 2013, 37, 23–43 CrossRef CAS PubMed.
- K. Harada, M. Suzuki, A. M. Dahlem, V. R. Beasley, W. W. Carmichael and K. L. Rinehart Jr, Toxicon, 1988, 26, 433–439 CrossRef CAS PubMed.
- J. Huisman, G. A. Codd, H. W. Paerl, B. W. Ibelings, J. M. H. Verspagen and P. M. Visser, Nat. Rev. Microbiol., 2018, 16, 471–483 CrossRef CAS PubMed.
- P. Rzymski and B. Poniedzialek, Postepy Dermatol. Alergol., 2012, 29, 47–50 Search PubMed.
- R. E. Moore, J. Ind. Microbiol., 1996, 16, 134–143 CrossRef CAS PubMed.
- P. Hrouzek, P. Tomek, A. Lukesova, J. Urban, L. Voloshko, B. Pushparaj, S. Ventura, J. Lukavsky, D. Stys and J. Kopecky, Environ. Toxicol., 2011, 26, 345–358 CrossRef CAS PubMed.
- A. Liaimer, J. B. Jensen and E. Dittmann, Front. Microbiol., 2016, 7, 1693 Search PubMed.
- J. C. Kehr, D. Gatte Picchi and E. Dittmann, Beilstein J. Org. Chem., 2011, 7, 1622–1635 CrossRef CAS PubMed.
- K. Kleigrewe, L. Gerwick, D. H. Sherman and W. H. Gerwick, Nat. Prod. Rep., 2016, 33, 348–364 RSC.
- P. M. D’Agostino, Nat. Prod. Rep., 2023, 40, 1701–1717 RSC.
- J. Demay, C. Bernard, A. Reinhardt and B. Marie, Mar. Drugs, 2019, 17, 320 CrossRef CAS PubMed.
- M. Schwark, J. A. Martinez Yerena, K. Rohrborn, P. Hrouzek, P. Divoka, L. Stenclova, K. Delawska, H. Enke, C. Vorreiter, F. Wiley, W. Sippl, R. Sobotka, S. Saha, S. B. Wilde, J. Mares and T. H. J. Niedermeyer, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2219230120 CrossRef CAS PubMed.
- R. M. Hohlman and D. H. Sherman, Nat. Prod. Rep., 2021, 38, 1567–1588 RSC.
- P. M. Shih, D. Wu, A. Latifi, S. D. Axen, D. P. Fewer, E. Talla, A. Calteau, F. Cai, N. Tandeau de Marsac, R. Rippka, M. Herdman, K. Sivonen, T. Coursin, T. Laurent, L. Goodwin, M. Nolan, K. W. Davenport, C. S. Han, E. M. Rubin, J. A. Eisen, T. Woyke, M. Gugger and C. A. Kerfeld, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 1053–1058 CrossRef CAS PubMed.
- A. Calteau, D. P. Fewer, A. Latifi, T. Coursin, T. Laurent, J. Jokela, C. A. Kerfeld, K. Sivonen, J. Piel and M. Gugger, BMC Genomics, 2014, 15, 977 CrossRef PubMed.
- E. Dittmann, M. Gugger, K. Sivonen and D. P. Fewer, Trends Microbiol., 2015, 23, 642–652 CrossRef CAS PubMed.
- A. Hitchcock, C. N. Hunter and D. P. Canniffe, Microb. Biotechnol., 2020, 13, 363–367 CrossRef PubMed.
- C. J. Knoot, J. Ungerer, P. P. Wangikar and H. B. Pakrasi, J. Biol. Chem., 2018, 293, 5044–5052 CrossRef CAS PubMed.
- T. T. Selao, J. Exp. Bot., 2022, 73, 3057–3071 CrossRef CAS PubMed.
- M. Y. Chen, W. K. Teng, L. Zhao, C. X. Hu, Y. K. Zhou, B. P. Han, L. R. Song and W. S. Shu, ISME J., 2021, 15, 211–227 CrossRef PubMed.
- K. Blin, S. Shaw, H. E. Augustijn, Z. L. Reitz, F. Biermann, M. Alanjary, A. Fetter, B. R. Terlouw, W. W. Metcalf, E. J. N. Helfrich, G. P. van Wezel, M. H. Medema and T. Weber, Nucleic Acids Res., 2023, 51, W46–W50 CrossRef PubMed.
- A. Gavriilidou, S. A. Kautsar, N. Zaburannyi, D. Krug, R. Muller, M. H. Medema and N. Ziemert, Nat. Microbiol., 2022, 7, 726–735 CrossRef CAS PubMed.
- S. A. Kautsar, J. J. J. van der Hooft, D. de Ridder and M. H. Medema, Gigascience, 2021, 10, giaa154 CrossRef PubMed.
- J. A. van Santen, E. F. Poynton, D. Iskakova, E. McMann, T. A. Alsup, T. N. Clark, C. H. Fergusson, D. P. Fewer, A. H. Hughes, C. A. McCadden, J. Parra, S. Soldatou, J. D. Rudolf, E. M. Janssen, K. R. Duncan and R. G. Linington, Nucleic Acids Res., 2022, 50, D1317–D1323 CrossRef CAS PubMed.
- D. H. Parks, M. Chuvochina, D. W. Waite, C. Rinke, A. Skarshewski, P. A. Chaumeil and P. Hugenholtz, Nat. Biotechnol., 2018, 36, 996–1004 CrossRef CAS PubMed.
- B. R. Terlouw, K. Blin, J. C. Navarro-Munoz, N. E. Avalon, M. G. Chevrette, S. Egbert, S. Lee, D. Meijer, M. J. J. Recchia, Z. L. Reitz, J. A. van Santen, N. Selem-Mojica, T. Torring, L. Zaroubi, M. Alanjary, G. Aleti, C. Aguilar, S. A. A. Al-Salihi, H. E. Augustijn, J. A. Avelar-Rivas, L. A. Avitia-Dominguez, F. Barona-Gomez, J. Bernaldo-Aguero, V. A. Bielinski, F. Biermann, T. J. Booth, V. J. Carrion Bravo, R. Castelo-Branco, F. O. Chagas, P. Cruz-Morales, C. Du, K. R. Duncan, A. Gavriilidou, D. Gayrard, K. Gutierrez-Garcia, K. Haslinger, E. J. N. Helfrich, J. J. J. van der Hooft, A. P. Jati, E. Kalkreuter, N. Kalyvas, K. B. Kang, S. Kautsar, W. Kim, A. M. Kunjapur, Y. X. Li, G. M. Lin, C. Loureiro, J. J. R. Louwen, N. L. L. Louwen, G. Lund, J. Parra, B. Philmus, B. Pourmohsenin, L. J. U. Pronk, A. Rego, D. A. B. Rex, S. Robinson, L. R. Rosas-Becerra, E. T. Roxborough, M. A. Schorn, D. J. Scobie, K. S. Singh, N. Sokolova, X. Tang, D. Udwary, A. Vigneshwari, K. Vind, S. Vromans, V. Waschulin, S. E. Williams, J. M. Winter, T. E. Witte, H. Xie, D. Yang, J. Yu, M. Zdouc, Z. Zhong, J. Collemare, R. G. Linington, T. Weber and M. H. Medema, Nucleic Acids Res., 2023, 51, D603–D610 CrossRef CAS PubMed.
- O. Strunecky, A. P. Ivanova and J. Mares, J. Phycol., 2023, 59, 12–51 CrossRef CAS PubMed.
- P. Rajaniemi, P. Hrouzek, K. Kastovska, R. Willame, A. Rantala, L. Hoffmann, J. Komarek and K. Sivonen, Int. J. Syst. Evol. Microbiol., 2005, 55, 11–26 CrossRef CAS PubMed.
- J. Osterholm, R. V. Popin, D. P. Fewer and K. Sivonen, Toxins, 2020, 12, 248 CrossRef PubMed.
- N. Engene, E. C. Rottacker, J. Kastovsky, T. Byrum, H. Choi, M. H. Ellisman, J. Komarek and W. H. Gerwick, Int. J. Syst. Evol. Microbiol., 2012, 62, 1171–1178 CrossRef PubMed.
- A. E. Tronholm and N. Engene, Not. algarum, 2019, 122, 1–2 Search PubMed.
- S. Suda, M. M. Watanabe, S. Otsuka, A. Mahakahant, W. Yongmanitchai, N. Nopartnaraporn, Y. Liu and J. G. Day, Int. J. Syst. Evol. Microbiol., 2002, 52, 1577–1595 CAS.
- A. Oren, Int. J. Syst. Evol. Microbiol., 2011, 61, 10–15 CrossRef PubMed.
- R. L. Burnap, Front. Bioeng. Biotechnol., 2015, 3, 1 CrossRef PubMed.
- R. Rippka, Methods Enzymol., 1988, 167, 3–27 CAS.
- D. O. Alvarenga, M. F. Fiore and A. M. Varani, Front. Microbiol., 2017, 8, 809 CrossRef PubMed.
- K. Heck, G. S. Machineski, D. O. Alvarenga, M. Vaz, A. M. Varani and M. F. Fiore, J. Microbiol. Methods, 2016, 129, 55–60 CrossRef CAS PubMed.
- B. M. Berla, R. Saha, C. M. Immethun, C. D. Maranas, T. S. Moon and H. B. Pakrasi, Front. Microbiol., 2013, 4, 246 Search PubMed.
- J. Yu, M. Liberton, P. F. Cliften, R. D. Head, J. M. Jacobs, R. D. Smith, D. W. Koppenaal, J. J. Brand and H. B. Pakrasi, Sci. Rep., 2015, 5, 8132 CrossRef CAS PubMed.
- D. H. Baracho and A. T. Lombardi, Microb. Cell Fact., 2023, 22, 36 CrossRef CAS PubMed.
- L. Bahr, A. Wustenberg and R. Ehwald, J. Appl. Phycol., 2016, 28, 783–793 CrossRef.
- A. Guljamow, M. Kreische, K. Ishida, A. Liaimer, B. Altermark, L. Bahr, C. Hertweck, R. Ehwald and E. Dittmann, Appl. Environ. Microbiol., 2017, 83, e01510–e01517 CrossRef PubMed.
- I. Echenique-Subiabre, A. Villeneuve, S. Golubic, J. Turquet, J. F. Humbert and M. Gugger, Microb. Ecol., 2015, 69, 234–244 CrossRef PubMed.
- S. Elkobi-Peer and S. Carmeli, Mar. Drugs, 2015, 13, 2347–2375 CrossRef CAS PubMed.
- L. A. Salvador, J. S. Biggs, V. J. Paul and H. Luesch, J. Nat. Prod., 2011, 74, 917–927 CrossRef CAS PubMed.
- K. A. Berg, C. Lyra, K. Sivonen, L. Paulin, S. Suomalainen, P. Tuomi and J. Rapala, ISME J., 2009, 3, 314–325 CrossRef CAS PubMed.
- C. Zuniga, T. Li, M. T. Guarnieri, J. P. Jenkins, C. T. Li, K. Bingol, Y. M. Kim, M. J. Betenbaugh and K. Zengler, Nat. Commun., 2020, 11, 3803 CrossRef CAS PubMed.
- B. W. Jester, H. Zhao, M. Gewe, T. Adame, L. Perruzza, D. T. Bolick, J. Agosti, N. Khuong, R. Kuestner, C. Gamble, K. Cruickshank, J. Ferrara, R. Lim, T. Paddock, C. Brady, S. Ertel, M. Zhang, A. Pollock, J. Lee, J. Xiong, M. Tasch, T. Saveria, D. Doughty, J. Marshall, D. Carrieri, L. Goetsch, J. Dang, N. Sanjaya, D. Fletcher, A. Martinez, B. Kadis, K. Sigmar, E. Afreen, T. Nguyen, A. Randolph, A. Taber, A. Krzeszowski, B. Robinett, D. B. Volkin, F. Grassi, R. Guerrant, R. Takeuchi, B. Finrow, C. Behnke and J. Roberts, Nat. Biotechnol., 2022, 40, 956–964 CrossRef CAS PubMed.
- B. E. Rubin, S. Diamond, B. F. Cress, A. Crits-Christoph, Y. C. Lou, A. L. Borges, H. Shivram, C. He, M. Xu, Z. Zhou, S. J. Smith, R. Rovinsky, D. C. J. Smock, K. Tang, T. K. Owens, N. Krishnappa, R. Sachdeva, R. Barrangou, A. M. Deutschbauer, J. F. Banfield and J. A. Doudna, Nat. Microbiol., 2022, 7, 34–47 CrossRef CAS PubMed.
- C. Pancrace, K. Ishida, E. Briand, D. G. Pichi, A. R. Weiz, A. Guljamow, T. Scalvenzi, N. Sassoon, C. Hertweck, E. Dittmann and M. Gugger, ACS Chem. Biol., 2019, 14, 67–75 CrossRef CAS PubMed.
- G. C. A. Amos, T. Awakawa, R. N. Tuttle, A. C. Letzel, M. C. Kim, Y. Kudo, W. Fenical, B. S. Moore and P. R. Jensen, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E11121–E11130 CrossRef CAS PubMed.
- D. Dehm, J. Krumbholz, M. Baunach, V. Wiebach, K. Hinrichs, A. Guljamow, T. Tabuchi, H. Jenke-Kodama, R. D. Sussmuth and E. Dittmann, ACS Chem. Biol., 2019, 14, 1271–1279 CrossRef CAS PubMed.
- H. Wei, Z. Lin, D. Li, Q. Gu and T. Zhu, Weishengwu Xuebao, 2010, 50, 701–709 CAS.
- D. Dhakal, M. Chen, H. Luesch and Y. Ding, J. Ind. Microbiol. Biotechnol., 2021, 48, kuab003 CrossRef CAS PubMed.
- J. J. R. Louwen and J. J. J. van der Hooft, mSystems, 2021, 6, e0072621 CrossRef PubMed.
- N. Ziemert, M. Alanjary and T. Weber, Nat. Prod. Rep., 2016, 33, 988–1005 RSC.
- L. K. Caesar, R. Montaser, N. P. Keller and N. L. Kelleher, Nat. Prod. Rep., 2021, 38, 2041–2065 RSC.
- T. Hautbergue, E. L. Jamin, L. Debrauwer, O. Puel and I. P. Oswald, Nat. Prod. Rep., 2018, 35, 147–173 RSC.
- J. J. J. van der Hooft, H. Mohimani, A. Bauermeister, P. C. Dorrestein, K. R. Duncan and M. H. Medema, Chem. Soc. Rev., 2020, 49, 3297–3314 RSC.
- J. A. van Santen, S. A. Kautsar, M. H. Medema and R. G. Linington, Nat. Prod. Rep., 2021, 38, 264–278 RSC.
- M. R. Jones, E. Pinto, M. A. Torres, F. Dorr, H. Mazur-Marzec, K. Szubert, L. Tartaglione, C. Dell'Aversano, C. O. Miles, D. G. Beach, P. McCarron, K. Sivonen, D. P. Fewer, J. Jokela and E. M. Janssen, Water Res., 2021, 196, 117017 CrossRef CAS PubMed.
- A. Rutz, M. Sorokina, J. Galgonek, D. Mietchen, E. Willighagen, A. Gaudry, J. G. Graham, R. Stephan, R. Page, J. Vondrasek, C. Steinbeck, G. F. Pauli, J. L. Wolfender, J. Bisson and P. M. Allard, Elife, 2022, 11, e70780 CrossRef CAS PubMed.
- M. A. Schorn, S. Verhoeven, L. Ridder, F. Huber, D. D. Acharya, A. A. Aksenov, G. Aleti, J. A. Moghaddam, A. T. Aron, S. Aziz, A. Bauermeister, K. D. Bauman, M. Baunach, C. Beemelmanns, J. M. Beman, M. V. Berlanga-Clavero, A. A. Blacutt, H. B. Bode, A. Boullie, A. Brejnrod, T. S. Bugni, A. Calteau, L. Cao, V. J. Carrion, R. Castelo-Branco, S. Chanana, A. B. Chase, M. G. Chevrette, L. V. Costa-Lotufo, J. M. Crawford, C. R. Currie, B. Cuypers, T. Dang, T. de Rond, A. M. Demko, E. Dittmann, C. Du, C. Drozd, J. C. Dujardin, R. J. Dutton, A. Edlund, D. P. Fewer, N. Garg, J. M. Gauglitz, E. C. Gentry, L. Gerwick, E. Glukhov, H. Gross, M. Gugger, D. G. Guillen Matus, E. J. N. Helfrich, B. F. Hempel, J. S. Hur, M. Iorio, P. R. Jensen, K. B. Kang, L. Kaysser, N. L. Kelleher, C. S. Kim, K. H. Kim, I. Koester, G. M. Konig, T. Leao, S. R. Lee, Y. Y. Lee, X. Li, J. C. Little, K. N. Maloney, D. Mannle, H. C. Martin, A. C. McAvoy, W. W. Metcalf, H. Mohimani, C. Molina-Santiago, B. S. Moore, M. W. Mullowney, M. Muskat, L. F. Nothias, E. C. O'Neill, E. I. Parkinson, D. Petras, J. Piel, E. C. Pierce, K. Pires, R. Reher, D. Romero, M. C. Roper, M. Rust, H. Saad, C. Saenz, L. M. Sanchez, S. J. Sorensen, M. Sosio, R. D. Sussmuth, D. Sweeney, K. Tahlan, R. J. Thomson, N. J. Tobias, A. E. Trindade-Silva, G. P. van Wezel, M. Wang, K. C. Weldon, F. Zhang, N. Ziemert, K. R. Duncan, M. Crusemann, S. Rogers, P. C. Dorrestein, M. H. Medema and J. J. J. van der Hooft, Nat. Chem. Biol., 2021, 17, 363–368 CrossRef CAS PubMed.
- N. Leikoski, L. Liu, J. Jokela, M. Wahlsten, M. Gugger, A. Calteau, P. Permi, C. A. Kerfeld, K. Sivonen and D. P. Fewer, Chem. Biol., 2013, 20, 1033–1043 CrossRef CAS PubMed.
- A. T. Aron, E. C. Gentry, K. L. McPhail, L. F. Nothias, M. Nothias-Esposito, A. Bouslimani, D. Petras, J. M. Gauglitz, N. Sikora, F. Vargas, J. J. J. van der Hooft, M. Ernst, K. B. Kang, C. M. Aceves, A. M. Caraballo-Rodriguez, I. Koester, K. C. Weldon, S. Bertrand, C. Roullier, K. Sun, R. M. Tehan, P. C. Boya, M. H. Christian, M. Gutierrez, A. M. Ulloa, J. A. Tejeda Mora, R. Mojica-Flores, J. Lakey-Beitia, V. Vasquez-Chaves, Y. Zhang, A. I. Calderon, N. Tayler, R. A. Keyzers, F. Tugizimana, N. Ndlovu, A. A. Aksenov, A. K. Jarmusch, R. Schmid, A. W. Truman, N. Bandeira, M. Wang and P. C. Dorrestein, Nat. Protoc., 2020, 15, 1954–1991 CrossRef CAS PubMed.
- R. Schmid, D. Petras, L. F. Nothias, M. Wang, A. T. Aron, A. Jagels, H. Tsugawa, J. Rainer, M. Garcia-Aloy, K. Duhrkop, A. Korf, T. Pluskal, Z. Kamenik, A. K. Jarmusch, A. M. Caraballo-Rodriguez, K. C. Weldon, M. Nothias-Esposito, A. A. Aksenov, A. Bauermeister, A. Albarracin Orio, C. O. Grundmann, F. Vargas, I. Koester, J. M. Gauglitz, E. C. Gentry, Y. Hovelmann, S. A. Kalinina, M. A. Pendergraft, M. Panitchpakdi, R. Tehan, A. Le Gouellec, G. Aleti, H. Mannochio Russo, B. Arndt, F. Hubner, H. Hayen, H. Zhi, M. Raffatellu, K. A. Prather, L. I. Aluwihare, S. Bocker, K. L. McPhail, H. U. Humpf, U. Karst and P. C. Dorrestein, Nat. Commun., 2021, 12, 3832 CrossRef CAS PubMed.
- K. Kleigrewe, J. Almaliti, I. Y. Tian, R. B. Kinnel, A. Korobeynikov, E. A. Monroe, B. M. Duggan, V. Di Marzo, D. H. Sherman, P. C. Dorrestein, L. Gerwick and W. H. Gerwick, J. Nat. Prod., 2015, 78, 1671–1682 CrossRef CAS PubMed.
- T. Pluskal, S. Castillo, A. Villar-Briones and M. Orešič, MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data, BMC Bioinformatics, 2010, 11, 395 CrossRef PubMed.
- S. A. C. Figueiredo, M. Preto, G. Moreira, T. P. Martins, K. Abt, A. Melo, V. M. Vasconcelos and P. N. Leao, Angew Chem. Int. Ed. Engl., 2021, 60, 10064–10072 CrossRef CAS PubMed.
- D. S. May, C. M. Crnkovic, A. Krunic, T. A. Wilson, J. R. Fuchs and J. E. Orjala, ACS Chem. Biol., 2020, 15, 758–765 CrossRef CAS PubMed.
- T. Leao, M. Wang, N. Moss, R. da Silva, J. Sanders, S. Nurk, A. Gurevich, G. Humphrey, R. Reher, Q. Zhu, P. Belda-Ferre, E. Glukhov, S. Whitner, K. L. Alexander, R. Rex, P. Pevzner, P. C. Dorrestein, R. Knight, N. Bandeira, W. H. Gerwick and L. Gerwick, Mar. Drugs, 2021, 19, 20 CrossRef CAS PubMed.
- J. C. Navarro-Munoz, N. Selem-Mojica, M. W. Mullowney, S. A. Kautsar, J. H. Tryon, E. I. Parkinson, E. L. C. De Los Santos, M. Yeong, P. Cruz-Morales, S. Abubucker, A. Roeters, W. Lokhorst, A. Fernandez-Guerra, L. T. D. Cappelini, A. W. Goering, R. J. Thomson, W. W. Metcalf, N. L. Kelleher, F. Barona-Gomez and M. H. Medema, Nat. Chem. Biol., 2020, 16, 60–68 CrossRef CAS PubMed.
- R. R. da Silva, M. Wang, L. F. Nothias, J. J. J. van der Hooft, A. M. Caraballo-Rodriguez, E. Fox, M. J. Balunas, J. L. Klassen, N. P. Lopes and P. C. Dorrestein, PLoS Comput. Biol., 2018, 14, e1006089 CrossRef PubMed.
- H. Mohimani, A. Gurevich, A. Shlemov, A. Mikheenko, A. Korobeynikov, L. Cao, E. Shcherbin, L. F. Nothias, P. C. Dorrestein and P. A. Pevzner, Nat. Commun., 2018, 9, 4035 CrossRef PubMed.
- M. Ernst, K. B. Kang, A. M. Caraballo-Rodriguez, L. F. Nothias, J. Wandy, C. Chen, M. Wang, S. Rogers, M. H. Medema, P. C. Dorrestein and J. J. J. van der Hooft, Metabolites, 2019, 9, 144 CrossRef CAS PubMed.
- H. Mohimani, W. T. Liu, R. D. Kersten, B. S. Moore, P. C. Dorrestein and P. A. Pevzner, J. Nat. Prod., 2014, 77, 1902–1909 CrossRef CAS.
- L. Cao, A. Gurevich, K. L. Alexander, C. B. Naman, T. Leao, E. Glukhov, T. Luzzatto-Knaan, F. Vargas, R. Quinn, A. Bouslimani, L. F. Nothias, N. K. Singh, J. G. Sanders, R. A. S. Benitez, L. R. Thompson, M. N. Hamid, J. T. Morton, A. Mikheenko, A. Shlemov, A. Korobeynikov, I. Friedberg, R. Knight, K. Venkateswaran, W. H. Gerwick, L. Gerwick, P. C. Dorrestein, P. A. Pevzner and H. Mohimani, Cell Syst., 2019, 9, 600–608 CrossRef CAS PubMed.
- N. J. Merwin, W. K. Mousa, C. A. Dejong, M. A. Skinnider, M. J. Cannon, H. Li, K. Dial, M. Gunabalasingam, C. Johnston and N. A. Magarvey, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 371–380 CrossRef CAS PubMed.
- L. K. Caesar, F. A. Butun, M. T. Robey, N. J. Ayon, R. Gupta, D. Dainko, J. W. Bok, G. Nickles, R. J. Stankey, D. Johnson, D. Mead, K. B. Cank, C. E. Earp, H. A. Raja, N. H. Oberlies, N. P. Keller and N. L. Kelleher, Nat. Chem. Biol., 2023, 19, 846–854 CrossRef CAS PubMed.
- G. Hjorleifsson Eldjarn, A. Ramsay, J. J. J. van der Hooft, K. R. Duncan, S. Soldatou, J. Rousu, R. Daly, J. Wandy and S. Rogers, PLoS Comput. Biol., 2021, 17, e1008920 CrossRef CAS PubMed.
- S. Romano, S. A. Jackson, S. Patry and A. D. W. Dobson, Mar. Drugs, 2018, 16, 244 CrossRef PubMed.
- E. Arstol and M. F. Hohmann-Marriott, Mar. Drugs, 2019, 17, 281 CrossRef PubMed.
- T. Galica, N. Borbone, J. Mares, A. Kust, A. Caso, G. Esposito, K. Saurav, J. Hajek, K. Rehakova, P. Urajova, V. Costantino and P. Hrouzek, Appl. Environ. Microbiol., 2021, 87, e0312820 CrossRef PubMed.
- S. Breinlinger, T. J. Phillips, B. N. Haram, J. Mares, J. A. Martinez Yerena, P. Hrouzek, R. Sobotka, W. M. Henderson, P. Schmieder, S. M. Williams, J. D. Lauderdale, H. D. Wilde, W. Gerrin, A. Kust, J. W. Washington, C. Wagner, B. Geier, M. Liebeke, H. Enke, T. H. J. Niedermeyer and S. B. Wilde, Science, 2021, 371, eaax9050 CrossRef CAS PubMed.
- Q. Gao and F. Garcia-Pichel, Nat. Rev. Microbiol., 2011, 9, 791–802 CrossRef CAS PubMed.
- T. Soule, V. Stout, W. D. Swingley, J. C. Meeks and F. Garcia-Pichel, J. Bacteriol., 2007, 189, 4465–4472 CrossRef CAS PubMed.
- C. Hu, G. Voller, R. Sussmuth, E. Dittmann and J. C. Kehr, Environ. Microbiol., 2015, 17, 1548–1559 CrossRef CAS PubMed.
- C. Hu, S. A. Ludsin, J. F. Martin, E. Dittmann and J. Lee, Harmful Algae, 2018, 77, 1–10 CrossRef CAS PubMed.
- E. Couradeau, U. Karaoz, H. C. Lim, U. Nunes da Rocha, T. Northen, E. Brodie and F. Garcia-Pichel, Nat. Commun., 2016, 7, 10373 CrossRef CAS PubMed.
- E. Briand, M. Bormans, M. Gugger, P. C. Dorrestein and W. H. Gerwick, Environ. Microbiol., 2016, 18, 384–400 CrossRef CAS PubMed.
- E. Briand, J. F. Humbert, K. Tambosco, M. Bormans and W. H. Gerwick, Microbiologyopen, 2016, 5, 469–478 CrossRef CAS PubMed.
- A. Liaimer, E. J. Helfrich, K. Hinrichs, A. Guljamow, K. Ishida, C. Hertweck and E. Dittmann, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 1862–1867 CrossRef CAS PubMed.
- C. Y. Chiang, M. Ohashi and Y. Tang, Nat. Prod. Rep., 2023, 40, 89–127 RSC.
- M. Myronovskyi and A. Luzhetskyy, Nat. Prod. Rep., 2019, 36, 1281–1294 RSC.
- B. C. Covington, F. Xu and M. R. Seyedsayamdost, Annu. Rev. Biochem., 2021, 90, 763–788 CrossRef CAS PubMed.
- E. Dittmann, B. A. Neilan, M. Erhard, H. von Dohren and T. Borner, Mol. Microbiol., 1997, 26, 779–787 CrossRef CAS PubMed.
- K. Ishida, M. Welker, G. Christiansen, S. Cadel-Six, C. Bouchier, E. Dittmann, C. Hertweck and N. Tandeau de Marsac, Appl. Environ. Microbiol., 2009, 75, 2017–2026 CrossRef CAS PubMed.
- T. Nishizawa, A. Ueda, T. Nakano, A. Nishizawa, T. Miura, M. Asayama, K. Fujii, K. Harada and M. Shirai, J. Biochem., 2011, 149, 475–485 CrossRef CAS PubMed.
- L. Rouhiainen, L. Paulin, S. Suomalainen, H. Hyytiainen, W. Buikema, R. Haselkorn and K. Sivonen, Mol. Microbiol., 2000, 37, 156–167 CrossRef CAS PubMed.
- G. Christiansen, J. Fastner, M. Erhard, T. Borner and E. Dittmann, J. Bacteriol., 2003, 185, 564–572 CrossRef CAS PubMed.
- K. Ishida, G. Christiansen, W. Y. Yoshida, R. Kurmayer, M. Welker, N. Valls, J. Bonjoch, C. Hertweck, T. Borner, T. Hemscheidt and E. Dittmann, Chem. Biol., 2007, 14, 565–576 CrossRef CAS PubMed.
- A. Liaimer, H. Jenke-Kodama, K. Ishida, K. Hinrichs, J. Stangeland, C. Hertweck and E. Dittmann, Environ. Microbiol. Rep., 2011, 3, 550–558 CrossRef CAS PubMed.
- T. Barchewitz, A. Guljamow, S. Meissner, S. Timm, M. Henneberg, O. Baumann, M. Hagemann and E. Dittmann, Environ. Microbiol., 2019, 21, 4836–4851 CrossRef CAS PubMed.
- S. Repka, M. Koivula, V. Harjunpa, L. Rouhiainen and K. Sivonen, Appl. Environ. Microbiol., 2004, 70, 4551–4560 CrossRef CAS PubMed.
- J. Krumbholz, K. Ishida, M. Baunach, J. E. Teikari, M. M. Rose, S. Sasso, C. Hertweck and E. Dittmann, Angew Chem. Int. Ed. Engl., 2022, 61, e202204545 CrossRef CAS PubMed.
- E. Bode, A. K. Heinrich, M. Hirschmann, D. Abebew, Y. N. Shi, T. D. Vo, F. Wesche, Y. M. Shi, P. Grun, S. Simonyi, N. Keller, Y. Engel, S. Wenski, R. Bennet, S. Beyer, I. Bischoff, A. Buaya, S. Brandt, I. Cakmak, H. Cimen, S. Eckstein, D. Frank, R. Furst, M. Gand, G. Geisslinger, S. Hazir, M. Henke, R. Heermann, V. Lecaudey, W. Schafer, S. Schiffmann, A. Schuffler, R. Schwenk, M. Skaljac, E. Thines, M. Thines, T. Ulshofer, A. Vilcinskas, T. A. Wichelhaus and H. B. Bode, Angew Chem. Int. Ed. Engl., 2019, 58, 18957–18963 CrossRef CAS PubMed.
- S. H. Park, K. Lee, J. W. Jang and J. S. Hahn, ACS Synth. Biol., 2019, 8, 346–357 CrossRef CAS PubMed.
- E. J. Kim, J. H. Lee, H. Choi, A. R. Pereira, Y. H. Ban, Y. J. Yoo, E. Kim, J. W. Park, D. H. Sherman, W. H. Gerwick and Y. J. Yoon, Org. Lett., 2012, 14, 5824–5827 CrossRef CAS PubMed.
- J. J. Zhang, X. Tang and B. S. Moore, Nat. Prod. Rep., 2019, 36, 1313–1332 RSC.
- M. S. Donia, B. J. Hathaway, S. Sudek, M. G. Haygood, M. J. Rosovitz, J. Ravel and E. W. Schmidt, Nat. Chem. Biol., 2006, 2, 729–735 CrossRef CAS PubMed.
- M. S. Donia, J. Ravel and E. W. Schmidt, Nat. Chem. Biol., 2008, 4, 341–343 CrossRef CAS PubMed.
- N. Leikoski, D. P. Fewer, J. Jokela, M. Wahlsten, L. Rouhiainen and K. Sivonen, Appl. Environ. Microbiol., 2010, 76, 701–709 CrossRef CAS PubMed.
- P. F. Long, W. C. Dunlap, C. N. Battershill and M. Jaspars, ChemBioChem, 2005, 6, 1760–1765 CrossRef CAS PubMed.
- E. W. Schmidt, J. T. Nelson, D. A. Rasko, S. Sudek, J. A. Eisen, M. G. Haygood and J. Ravel, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 7315–7320 CrossRef CAS PubMed.
- W. Tang and W. A. van der Donk, Biochemistry, 2012, 51, 4271–4279 CrossRef CAS PubMed.
- A. R. Weiz, K. Ishida, K. Makower, N. Ziemert, C. Hertweck and E. Dittmann, Chem. Biol., 2011, 18, 1413–1421 CrossRef CAS PubMed.
- N. Ziemert, K. Ishida, A. Liaimer, C. Hertweck and E. Dittmann, Angew Chem. Int. Ed. Engl., 2008, 47, 7756–7759 CrossRef CAS PubMed.
- D. Liu and H. B. Pakrasi, Microb. Cell Fact., 2018, 17, 48 CrossRef PubMed.
- T. Liu, R. Mazmouz, S. E. Ongley, R. Chau, R. Pickford, J. N. Woodhouse and B. A. Neilan, ACS Chem. Biol., 2017, 12, 2021–2029 CrossRef CAS PubMed.
- K. N. Wells, P. Videau, D. Nelson, J. E. Eiting and B. Philmus, FEMS Microbiol. Lett., 2018, 365, fny164 CrossRef CAS PubMed.
- R. H. Lambalot, A. M. Gehring, R. S. Flugel, P. Zuber, M. LaCelle, M. A. Marahiel, R. Reid, C. Khosla and C. T. Walsh, Chem. Biol., 1996, 3, 923–936 CrossRef CAS PubMed.
- G. Yang, Y. Zhang, N. K. Lee, M. A. Cozad, S. E. Kearney, H. Luesch and Y. Ding, Sci. Rep., 2017, 7, 11888 CrossRef PubMed.
- P. M. D'Agostino, C. J. Seel, X. Ji, T. Gulder and T. A. M. Gulder, Nat. Chem. Biol., 2022, 18, 652–658 CrossRef PubMed.
- N. Eusebio, R. Castelo-Branco, D. Sousa, M. Preto, P. D'Agostino, T. A. M. Gulder and P. N. Leao, ACS Synth. Biol., 2022, 11, 3493–3503 CrossRef CAS PubMed.
- C. Greunke, E. R. Duell, P. M. D'Agostino, A. Glockle, K. Lamm and T. A. M. Gulder, Metab. Eng., 2018, 47, 334–345 CrossRef CAS PubMed.
- P. Videau, K. N. Wells, A. J. Singh, J. Eiting, P. J. Proteau and B. Philmus, ACS Synth. Biol., 2020, 9, 63–75 CrossRef CAS PubMed.
- P. Videau, K. N. Wells, A. J. Singh, W. H. Gerwick and B. Philmus, ACS Synth. Biol., 2016, 5, 978–988 CrossRef CAS PubMed.
- H. Zhang, L. Fang, M. S. Osburne and B. A. Pfeifer, Methods Mol. Biol., 2016, 1401, 121–134 CrossRef CAS PubMed.
- S. E. Ongley, X. Bian, Y. Zhang, R. Chau, W. H. Gerwick, R. Muller and B. A. Neilan, ACS Chem. Biol., 2013, 8, 1888–1893 CrossRef CAS PubMed.
- T. Liu, R. Mazmouz, L. A. Pearson and B. A. Neilan, ACS Synth. Biol., 2019, 8, 1187–1194 CrossRef CAS PubMed.
- P. M. D'Agostino and T. A. M. Gulder, ACS Synth. Biol., 2018, 7, 1702–1708 CrossRef PubMed.
- J. H. Cardellina 2nd, F. J. Marner and R. E. Moore, Science, 1979, 204, 193–195 CrossRef CAS PubMed.
- A. C. Jones, S. Ottilie, A. S. Eustaquio, D. J. Edwards, L. Gerwick, B. S. Moore and W. H. Gerwick, FEBS J., 2012, 279, 1243–1251 CrossRef CAS PubMed.
- G. Yang, M. A. Cozad, D. A. Holland, Y. Zhang, H. Luesch and Y. Ding, ACS Synth. Biol., 2018, 7, 664–671 CrossRef CAS PubMed.
- V. Agarwal, J. M. Blanton, S. Podell, A. Taton, M. A. Schorn, J. Busch, Z. Lin, E. W. Schmidt, P. R. Jensen, V. J. Paul, J. S. Biggs, J. W. Golden, E. E. Allen and B. S. Moore, Nat. Chem. Biol., 2017, 13, 537–543 CrossRef CAS PubMed.
- S. Baldanta, G. Guevara and J. M. Navarro-Llorens, Microb. Cell Fact., 2022, 21, 103 CrossRef CAS PubMed.
- B. Bishe, A. Taton and J. W. Golden, iScience, 2019, 20, 216–228 CrossRef CAS PubMed.
- T. C. Niu, G. M. Lin, L. R. Xie, Z. Q. Wang, W. Y. Xing, J. Y. Zhang and C. C. Zhang, ACS Synth. Biol., 2019, 8, 170–180 CrossRef CAS PubMed.
- J. Svoboda, B. Cisneros and B. Philmus, Synth. Biol., 2021, 6, ysab019 CrossRef PubMed.
- D. Back, T. J. O'Donnell, K. K. Axt, J. R. Gurr, J. M. Vanegas, P. G. Williams and B. Philmus, ACS Chem. Biol., 2023, 18, 1797–1807 CrossRef CAS PubMed.
- A. Taton, A. Ecker, B. Diaz, N. A. Moss, B. Anderson, R. Reher, T. F. Leao, R. Simkovsky, P. C. Dorrestein, L. Gerwick, W. H. Gerwick and J. W. Golden, ACS Synth. Biol., 2020, 9, 3364–3376 CrossRef CAS PubMed.
- A. Taton, S. Rohrer, B. Diaz, R. Reher, A. M. Caraballo Rodriguez, M. L. Pierce, P. C. Dorrestein, L. Gerwick, W. H. Gerwick and J. W. Golden, ACS Chem. Biol., 2022, 17, 1910–1923 CrossRef CAS PubMed.
- E. S. Sattely, M. A. Fischbach and C. T. Walsh, Nat. Prod. Rep., 2008, 25, 757–793 RSC.
- R. A. Sheldon and J. M. Woodley, Chem. Rev., 2018, 118, 801–838 CrossRef CAS PubMed.
- S. Adak, A. L. Lukowski, R. J. B. Schäfer and B. S. Moore, J. Am. Chem. Soc., 2022, 144, 2861–2866 CrossRef CAS PubMed.
- S. T. Lima, T. R. Fallon, J. L. Cordoza, J. R. Chekan, E. Delbaje, A. R. Hopiavuori, D. O. Alvarenga, S. M. Wood, H. Luhavaya, J. T. Baumgartner, F. A. Dörr, A. Etchegaray, E. Pinto, S. M. K. McKinnie, M. F. Fiore and B. S. Moore, J. Am. Chem. Soc., 2022, 144, 9372–9379 CrossRef CAS PubMed.
- C. P. Mason, K. R. Edwards, R. E. Carlson, J. Pignatello, F. K. Gleason and J. M. Wood, Science, 1982, 215, 400–402 CrossRef CAS PubMed.
- X. Yang, Y. Shimizu, J. R. Steiner and J. Clardy, Tetrahedron Lett., 1993, 34, 761–764 CrossRef CAS.
- S. C. Lee and G. D. Brown, J. Nat. Prod., 1998, 61, 29–33 CrossRef CAS PubMed.
- S. Sarkar, W. Gu and E. W. Schmidt, ACS Catal., 2020, 10, 7146–7153 CrossRef CAS PubMed.
- A. Colombano, L. Dalponte, S. Dall'Angelo, C. Clemente, M. Idress, A. Ghazal and W. E. Houssen, Angew Chem. Int. Ed. Engl., 2023, 62, e202215979 CrossRef CAS PubMed.
- B. Choi and A. J. Link, Trends Chem., 2023, 5, 620–633 CrossRef CAS PubMed.
- T. J. Oman, P. J. Knerr, N. A. Bindman, J. E. Velásquez and W. A. van der Donk, J. Am. Chem. Soc., 2012, 134, 6952–6955 CrossRef CAS PubMed.
- E. Reyna-González, B. Schmid, D. Petras, R. D. Süssmuth and E. Dittmann, Angew Chem. Int. Ed. Engl., 2016, 55, 9398–9401 CrossRef PubMed.
- M. N. Ahmed, E. Reyna-González, B. Schmid, V. Wiebach, R. D. Süssmuth, E. Dittmann and D. P. Fewer, ACS Chem. Biol., 2017, 12, 1538–1546 CrossRef CAS PubMed.
- S. Scholz, S. Kerestetzopoulou, V. Wiebach, R. Schnegotzki, B. Schmid, E. Reyna-González, L. Ding, R. D. Süssmuth, E. Dittmann and M. Baunach, ChemBioChem, 2022, 23, e202200345 CrossRef CAS PubMed.
- B. I. Morinaka, E. Lakis, M. Verest, M. J. Helf, T. Scalvenzi, A. L. Vagstad, J. Sims, S. Sunagawa, M. Gugger and J. Piel, Science, 2018, 359, 779–782 CrossRef CAS PubMed.
- E. Lakis, S. Magyari and J. Piel, Angew Chem. Int. Ed. Engl., 2022, 61, e202202695 CrossRef CAS PubMed.
- D. Richter, E. Lakis and J. Piel, Nat. Chem., 2023, 1422–1430 CrossRef CAS PubMed.
- H. Nakamura, E. E. Schultz and E. P. Balskus, Nat. Chem. Biol., 2017, 13, 916–921 CrossRef CAS PubMed.
- N. R. Braffman, T. B. Ruskoski, K. M. Davis, N. R. Glasser, C. Johnson, C. D. Okafor, A. K. Boal and E. P. Balskus, eLife, 2022, 11, e75761 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3np00045a |
| This journal is © The Royal Society of Chemistry 2024 |