Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synthesis of sulfated lactosyl glycosides for evaluation in vaccine adjuvant formulations†
Tarasha
Sharma
a,
Sophie
Régnier
b,
Lise
Deschatelets
c,
Felicity C.
Stark
c,
Vinicio
Vasquez
b,
Camilo F.
Martinez-Farina
d,
Renu
Dudani
c,
Blair A.
Harrison
 c,
Bassel
Akache
c,
Yimei
Jia
c,
Michael J.
McCluskie
*c and
Usha D.
Hemraz
c,
Bassel
Akache
c,
Yimei
Jia
c,
Michael J.
McCluskie
*c and
Usha D.
Hemraz
 *a
*a
aHuman Health Therapeutics, National Research Council of Canada, 6100 Royalmount Avenue, Montreal, Quebec, H4P 2R2, Canada. E-mail: Usha.Hemraz@nrc-cnrc.gc.ca
bAquatic and Crop Resource Development, National Research Council of Canada, 6100 Royalmount Avenue, Montreal, Quebec, H4P 2R2, Canada
cHuman Health Therapeutics, National Research Council of Canada, 1200 Montreal Road, Ontario, Ottawa K1A 0R6, Canada. E-mail: Michael.McCluskie@nrc-cnrc.gc.ca
dAquatic and Crop Resource Development, National Research Council of Canada, 1411 Oxford Street, Halifax, Nova Scotia B3H 3Z1, Canada
First published on 25th September 2024
Abstract
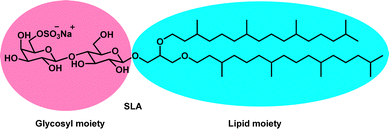
Adjuvants are essential components of vaccines as they enable protection against multiple pathogens by enhancing the duration, magnitude and or quality of immune responses. SLA Archaeosomes, a type of liposome composed of sulfated lactosyl archaeol (SLA) glycolipids, are highly stable vaccine adjuvants that have been shown to induce strong immune responses in preclinical models of infectious disease and cancer. To better understand the mechanism of activity behind SLA archaeosomes strong immunogenic properties, we studied the effect of structural change on vaccine adjuvanticity. Herein, we report the synthesis of three new sulfated lactosyl glycosides (SLGs) by replacing the archaeol moiety with various side-chains. These derivatives were characterized using nuclear magnetic resonance, mass spectrometry, and thin layer chromatography for identity and purity assessment. The SLGs were co-assembled in the presence of DMPC and cholesterol to produce lipid vesicles. The abilities of the SLG-based liposomes to act as vaccine adjuvants were compared to SLA archaeosomes in an in vivo murine vaccine model.
Introduction
Adjuvants are crucial components of vaccines that are combined with antigens to enhance the magnitude, quality and/or duration of immune responses. Many modern-day vaccines, such as those containing recombinant or synthetic antigens are safer but less immunogenic than whole killed or attenuated vaccines. Therefore, they require adjuvants to stimulate protective and long-lasting immunity against multiple pathogens.1 Adjuvants can allow comparable immunity while significantly reducing vaccine dosages and boosters – an important consideration during pandemics and in areas where vaccine production facilities are limited.2Despite their importance, only a few adjuvants and delivery systems are licensed for human use despite two centuries of vaccine use.1 Aluminum salts (alum) are the most common, however, they are insufficient at inducing protection against more challenging pathogens and in certain subpopulations such as diabetics and the elderly, as shown in the case with the hepatitis B surface antigen (HBsAg) vaccine EngerixB.3 Alum was also shown to be a weak inducer of cellular immune responses that are necessary in fighting against intracellular pathogens such as viruses, bacteria, and cancer-based diseases.4 To address this, commercial adjuvants such as squalene-based oil-in-water emulsions (e.g. MF59) and toll-like receptor (TLR) agonists (e.g. monophosphoryl lipid A (MPL) and CpG) have been developed that are often used in vaccine formulations today.5 However, there remains a need for additional adjuvants that are both safe and effective to address unmet medical needs and to improve our understanding of their mechanisms of action.
Archaeosomes are a type of liposome derived from archaeal lipids that have been shown to induce strong cellular and humoral immune responses to antigens. These liposomes contain characteristic ether linkages between the fully saturated isoprenoid carbon chains and the sn-2, 3 carbons of glycerol backbone. These structural features are associated with their higher stabilities in extreme pH, bile salts, high temperatures and resistance against lipase hydrolysis.6 The ability of archaea membrane lipids to adapt to harsh environments has made archaeal lipids an interesting target for developing nano-drug delivery systems capable of overcoming biological, biophysical, and biomedical barriers of the body that conventional ester-linked liposomes are prone to. Along with their stability, archaeosomes can recruit immune cells and activate robust antigen-specific responses, making them an attractive vaccine delivery vehicle.7 Archaeosomes can induce strong humoral and cell-mediated immune responses to entrapped antigens to protect immunized mice from several evasive pathogens such as Listeria monocytogenes, and Mycobacterium tuberculosis.8,9 They also provide protection against solid and metastatic tumors in murine models.10 However, total polar lipid archaeosome formulations can be relatively complex since they are comprised of multiple lipid species which can pose a challenge in quality control during scale-up.11 To simplify the formulation, archaeosomes based on the single sulfated lactosyl archaeol (SLA) glycolipid have been developed which can be simply admixed with antigen in comparison to traditional methods requiring antigen entrapment. This greatly reduces antigen loss and entrapment variability during formulation. The SLA glycolipid consists of a sulfated saccharide group covalently linked to the backbone of an archaeal core lipid. The SLA archaeosomes are highly stable vaccine adjuvants that safely induce strong humoral and cellular immune responses to multiple antigens in preclinical studies. For example, they showed superior adjuvanticity and strong antigen-specific immune responses compared to other commercially available immunological adjuvants such as TLR3/4/9 agonists, oil-in-water and water-in-oil emulsions and aluminum hydroxide when used in vaccine formulations with ovalbumin (Ova) and hepatitis B surface antigen (HBsAg).12 SLA archaeosomes were also shown to induce strong and specific immunity against H1N1 Influenza A virus in young, aged, and pregnant mice providing them and the pups with strong protection.13 More recently in response to the COVID-19 pandemic, it was shown that vaccine formulations containing a resistin-trimerized spike antigen and SLA archaeosomes adjuvant provided robust antigen-specific immune responses and that the resulting immunogenicity was sufficient to mediate cross-neutralization activity against the spike proteins of multiple variants for SARS-CoV-2.14
Ideally, a vaccine adjuvant should be able to produce strong and long-lasting immune responses rapidly. Its production should be economically appealing, scalable and meet quality control assessments. SLA lipids can be produced semi-synthetically where the glycosyl moiety, synthesized from a chemically modified lactose is coupled to the archaeol core lipid derived from the total polar lipid extract of the archaebacteria Halobacterium salinarum (Fig. 1).15 Although this method provides high yields and the pure stereoisomer of archaeol, the dependence on microbial growth and subsequent purification steps are time consuming.16 Recently, our group has reported a fully synthetic method for producing SLA using commercially available reagents, an economically more viable process that yields a mixture of stereoisomers of the archaeol core lipid.17 In spite of substantial difference in the chiral purities of archaeol, no significant differences were observed on the adjuvanticity of SLA archaeosomes and all SLA archaeosomes of different chiral purities induced comparable humoral and cell-mediated antigen-specific responses.17 Considering these results, we became interested in exploring the effect that replacing the archaeol moiety with other lipids had on vaccine adjuvanticity, which could help us better understand the mechanism of activity behind SLA's strong immunogenic properties. It could also assist in finding alternate vaccine adjuvant candidates, which would potentially lower the cost of SLA adjuvant systems and enable large-scale production if SLGs demonstrate similar responses in terms of safety and efficacy.
Herein, we synthesized three new sulfated lactosyl glycosides (SLGs) by replacing the archaeol moiety with various sidechains while keeping the glycosyl moiety intact. Previous investigations have demonstrated that neutrally charged archaeal glycolipids including non-sulfated lactosyl archaeal glycolipids were unable to form liposomes and were ineffective as adjuvants.11 In this structure activity relationship (SAR) study, we probed whether the performance of SLA vaccine adjuvant depended on the structure of lipid portion of SLA, archaeol, and whether a simpler moiety could produce a comparable effect. Three commercially available fatty alcohols with varying chain length and functional groups were coupled to the glycosyl moiety to create a small library of SLGs. The resulting derivatives were characterized by nuclear magnetic resonance, mass spectrometry and thin layer chromatography for identity and purity assessment. They were subsequently co-assembled into liposomes and their abilities to act as vaccine adjuvants were studied in mice models and compared to SLA archaeosomes.
Experimental
General methods
Unless stated otherwise, all reactions were performed under an inert atmosphere using argon. All commercially available solvents and reagents were purchased from Sigma Aldrich, unless stated otherwise and used without further purification. Mass spectrometry, NMR, and TLC were used to confirm the identity and purity of synthesized compounds. Reactions were monitored through TLC analysis using UV254 pre-coated plates and visualized under UV light or by staining with sulfuric acid (H2SO4) in methanol or potassium permanganate (KMnO4). 1H and 13C NMR spectra were obtained in the specified deuterated solvents using a Bruker AVANCE III 700 MHz spectrometer equipped with a 5 mm TCI cryoprobe, as indicated. The NMR data are presented in the following order: chemical shift δ (ppm), multiplicity, integration, and coupling constant. The following abbreviations were used to explain the multiplicities: s = singlet, d = doublet, t = triplet, m = multiplet, br s = broad singlet. Residual resonance from the deuterated solvent was used to reference the 1H NMR and 13C NMR spectra.Synthetic procedures
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 hexanes/ethyl acetate/dichloromethane. The reaction mixture was diluted with dichloromethane, filtered by vacuum filtration, and rinsed with additional dichloromethane. The resulting filtrate was washed with 10% sodium thiosulfate (50 mL), a saturated sodium bicarbonate solution (50 mL) and brine (50 mL). The organic layer was dried over MgSO4, filtered by vacuum filtration, and evaporated to dryness. The residue was purified by flash chromatography by eluting with 8
1 hexanes/ethyl acetate/dichloromethane. The reaction mixture was diluted with dichloromethane, filtered by vacuum filtration, and rinsed with additional dichloromethane. The resulting filtrate was washed with 10% sodium thiosulfate (50 mL), a saturated sodium bicarbonate solution (50 mL) and brine (50 mL). The organic layer was dried over MgSO4, filtered by vacuum filtration, and evaporated to dryness. The residue was purified by flash chromatography by eluting with 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 7
1, 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 7
1 and 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 hexanes/ethyl acetate/dichloromethane to yield compounds 7A and 7B, and DCM to 5% MeOH to yield 7C.
1 hexanes/ethyl acetate/dichloromethane to yield compounds 7A and 7B, and DCM to 5% MeOH to yield 7C.
Compound 7A. Following GP1, from 500 mg (0.5 mmol) of donor 6, 330 mg of 7A was collected as a white waxy solid (68% yield). 1H NMR (700 MHz, CDCl3) δ 8.00 (d, 2H, J = 7.8 Hz), 7.91–7.88 (m, 8H), 7.56 (t, 1H, J = 7.4 Hz), 7.46 (m, 5H), 7.38–7.27 (m, 12H), 7.17 (t, 2H, J = 7.6 Hz), 5.84 (t, 1H, J = 9.1 Hz), 5.79 (dd, 1H, J1 = 10.4 Hz, J2 = 7.9 Hz), 5.32 (dd, 1H, J1 = 9.5 Hz, J2 = 7.8 Hz), 5.29 (s, 1H), 5.15 (dd, 1H, J1 = 10.4 Hz, J2 = 3.5 Hz), 4.84 (d, 1H, J = 7.9 Hz), 4.61 (dd, 1H, J1 = 11.7, J2 = 1.9 Hz), 4.36 (dd, 1H, J1 = 11.7, J2 = 4.2 Hz), 4.30 (d, 1H, J = 3.3 Hz), 4.21 (t, 1H J = 9.2 Hz), 3.84 (d, 1H J = 9.4 Hz), 3.77 (d, 1H, J = 12.0 Hz), 3.70 (dd, 1H, J1 = 9.6 Hz, J2 = 5.2 Hz), 3.57 (d, 1H, J = 11.8 Hz), 3.21 (dd, 1H, J1 = 9.6 Hz, J2 = 6.5 Hz), 2.96 (s, 1H), 1.24 (m, 2H), 1.13 (m, 8H), 0.99 (m, 16H), 0.85 (t, 3H, J = 7.3 Hz), 0.79 (t, 3H, J = 7.2 Hz); 13C NMR (176 MHz, CDCI3) δ 166.3, 165.8, 165.4, 165.2, 165.0, 137.6, 133.5, 133.3, 133.1, 133.1, 130.0, 129.9, 129.8, 129.8, 129.8, 129.7, 129.0, 128.9, 128.9, 128.6, 128.5, 128.4, 128.4, 128.1, 126.5, 101.6, 101.2, 100.7, 74.2, 73.2, 72.8,73.0, 72.8,72.7, 69.5, 68.1, 66.5, 62.5, 38.0, 32.0, 31.9, 31.1, 30.9, 30.0, 29.7, 29.4, 26.9, 26.8, 26.6, 26.5, 22.8, 22.8, 14.3; HRMS (ESI+) calcd for C70H78O16 [M + NH4]+: 1192.5634 found: 1192.5583.
Compound 7B. Following GP1, from 1000 mg (1 mmol) of donor 6, 580 mg of 7B was collected as a white waxy solid (56% yield). 1H NMR (700 MHz, CDCl3) δ 8.00 (ddd, 2H, J1 = 8.3 Hz, J2 = 3.5 Hz, J3 = 1.4 Hz), 7.91–7.88 (m, 8H), 7.56 (td, 1H, J1 = 7.9 Hz, J2 = 1.3 Hz), 7.52–7.40 (m, 5H), 7.38–7.27 (m, 12H), 7.17 (t, 2H, J = 7.7 Hz), 5.83 (t, 1H, J = 9.1 Hz), 5.78 (dd, 1H, J = 10.4 Hz), 5.31 (ddd, 1H, J1 = 9.7 Hz, J2 = 7.7 Hz, J3 = 2.4 Hz), 5.29 (s, 1H), 5.15 (ddd, 1H, J1 = 10.5 Hz, J2 = 3.6 Hz, J3 = 1.2 Hz), 4.84 (d, 1H, J = 7.9 Hz), 4.66 (t, 1H, J = 7.6 Hz), 4.61 (d, 1H, J = 11.3 Hz), 4.37 (dd, 1H, J1 = 11.7 Hz, J2 = 4.3 Hz), 4.30 (d, 1H, J = 3.4 Hz), 4.21 (t, 1H J = 9.2 Hz), 3.83 (m, 2H), 3.77 (t, 1H, J = 6.0 Hz), 3.58 (d, 1H, J = 11.5 Hz), 3.44 (m, 1H), 2.97 (s, 1H), 1.16 (m, 26H), 0.85 (d, 6H, J = 6.5 Hz), 0.82 (d, 3H, J = 6.5 Hz), 0.75 (d, 3H, J = 6.5 Hz), 0.63 (d, 3H, 6.6 Hz); 13C NMR (176 MHz, CDCI3) δ 166.3, 165.8, 165.5, 165.2, 165.0, 143.4,137.6, 133.5, 133.3, 133.2, 133.1, 130.0, 129.9, 129.8, 129.8, 129.7, 129.6, 129.0, 128.9, 128.5, 128.5, 128.4, 128.4, 128.4, 128.1, 126.5, 101.6, 100.7, 74.2, 73.2, 72.9,72.7, 72.5, 70.3, 69.5, 68.1, 66.5, 39.5, 37.6, 37.6, 32.9, 29.4, 28.1,25.0, 24.6, 22.9, 22.8, 19.9, 19.8; HRMS (ESI+) calcd for C74H86O16 [M + NH4]+: 1248.6316, found: 1248.6254.
Compound 7C. Following GP1, from 1000 mg (1 mmol) of donor 6, 856 mg of 7C was collected as a white waxy solid (67% yield). 1H NMR (700 MHz, CDCl3) δ 8.00 (d, 2H, J = 7.8 Hz), 7.91–7.86 (m, 8H), 7.56 (t, 1H, J = 7.4 Hz), 7.46 (m, 5H), 7.38–7.27 (m, 12H), 7.16 (t, 2H, J = 7.6 Hz), 5.84 (t, 1H, J = 9.1 Hz), 5.78 (dd, 1H, J1 = 2.0 Hz, J2 = 18.3 Hz), 5.32 (t, 1H, J = 17.3 Hz), 5.29 (s, 1H), 5.16 (d, 1H, J = 3.4 Hz), 5.14 (d, 1H, J = 3.4 Hz), 4.84 (d, 1H, J = 7.9 Hz), 4.79 (d, 1H, J = 7.8 Hz), 4.62 (d, 1H, J = 11.3 Hz,), 4.36 (dd, 1H, J1 = 11.8 Hz J2 = 4.0 Hz), 4.30 (d, 1H, J = 3.3 Hz), 4.21 (t, 1H J = 9.2 Hz), 3.84 (d, 1H J = 10.9 Hz), 3.84 (d, 2H, J = 3.5 Hz), 3.77 (d, 2H, J = 12.6 Hz), 3.64 (m, 32H), 3.57 (d, 1H, J = 11.8 Hz), 3.44 (t, 2H, J = 3.4 Hz), 3.21 (d, 1H, J = 16.0 Hz), 2.96 (s, 1H), 1.25 (s, 19H), 0.87 (t, 3H, J = 7.0 Hz); 13C NMR (176 MHz, CDCI3) δ 166.2, 165.8, 165.5, 165.2, 165.0, 137.6, 133.3, 130.0, 130.0, 129.8, 129.8, 129.7, 129.7, 129.7, 129.6, 129.0, 128.9, 128.9, 128.6, 128.5, 128.5, 128.4, 128.1, 126.5, 125.0, 101.6, 101.0, 100.7, 74.2, 73.2, 72.9,72.7, 72.4, 71.7, 70.7, 70.4, 70.2, 69.5, 68.1, 66.5, 62.5, 38.0, 32.1, 29.8, 29.8, 29.8, 29.7, 29.5, 26.2, 22.8, 14.331.9, 31.1, 30.9, 30.0, 29.7, 29.4, 26.9, 26.8, 26.6, 26.5, 22.8, 22.8, 14.3; HRMS (ESI+) calcd for C84H106O15 [M + H]+: 1515.7023, found: 1515.7034.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 6
1, 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 5
1, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.5
3.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5 and 4
1.5 and 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) to yield waxy solids 8A and 8B. Eluting with DCM to 5% MeOH yielded compound 8C.
2) to yield waxy solids 8A and 8B. Eluting with DCM to 5% MeOH yielded compound 8C.
Compound 8A. Following GP2, from 280 mg (0.24 mmol) of 7A, 260 mg of 8A was collected as a white waxy solid (>95% yield). 1H NMR (700 MHz, CDCl3) δ 8.03 (d, 2H, J = 7.4 Hz), 7.98–7.89 (m,8H), 7.59 (t, 1H, J = 7.4 Hz), 7.56 (t, 1H, J = 7.4 Hz), 7.46 (m, 5H), 7.34 (m, 5H), 7.24 (t, 2H, J = 7.8 Hz), 5.74 (t, 1H, J = 9.1 Hz), 5.41 (t, 1H, J = 5.8 Hz), 5.07 (dd, 1H, J1 = 13.4 Hz, J2 = 7.2 Hz), 4.78 (d, 1H, J = 7.9 Hz), 4.62 (d, 1H, J = 7.8 Hz), 4.59 (d, 1H, J = 1.6 Hz), 4.42 (dd, 1H, J1 = 11.9, J2 = 4.8 Hz), 4.18 (m, 2H), 3.83 (dd, 1H, J1 = 11.5 Hz, J2 = 4.7 Hz), 3.72 (dd, 1H, J1 = 9.5 Hz, J2 = 5.2 Hz), 3.35 (m, 2H), 3.25 (m, 2H), 1.24 (m, 2H), 1.13 (m, 8H), 0.99 (m, 16H), 0.85 (t, 3H, J = 7.3 Hz), 0.79 (t, 3H, J = 7.3 Hz); 13C NMR (176 MHz, CDCI3) δ 166.1, 165.9, 165.6, 165.3, 165.2, 133.6, 133.5, 133.4, 133.3, 130.0, 129.9, 129.89, 129.85, 129.7, 129.5, 129.1, 129.0, 128.8, 128.6, 128.5, 128.45, 101.4, 101.3, 74.4, 74.2, 73.7, 73.1, 73.0, 72.0, 69.8, 68.5, 62.8, 62.75, 38.0, 32.0, 31.9, 31.1, 30.9, 30.0, 29.7, 29.4, 26.9, 26.8, 26.6, 26.5, 22.8, 22.75, 14.3, 29.6, 29.6, 29.5, 29.4, 29.4, 25.6, 22.8, 14.3, 14.2; HRMS (ESI+) calcd for C63H74O16 [M + NH4]+: 1104.5377, found: 1104.5315.
Compound 8B. Following GP2, from 560 mg (0.24 mmol) of 7B, 378 mg of 8B was collected as a white waxy solid (73% yield). 1H NMR (700 MHz, CDCl3) δ 8.03 (dd, 2H, J1 = 7.3 Hz, J2 = 3.5 Hz), 7.99–7.88 (m, 8H), 7.59 (t, 1H, J = 7.4 Hz),), 7.56 (t, 1H, J = 7.4 Hz), 7.53–7.29 (m, 11H), 7.23 (t, 2H, J = 7.7 Hz), 5.73 (dd, 2H, J1 = 10.1 Hz, J2 = 8.1 Hz), 5.40 (ddd, 1H, J1 = 9.8 Hz, J2 = 7.7 Hz, J3 = 2.4 Hz), 5.07 (dd, 1H, J = 10.3 Hz, J = 2.5 Hz), 4.78 (dd, 1H, J1 = 7.9 Hz, J2 = 1.2 Hz), 4.66 (t, 1H, J = 7.7 Hz), 4.60 (d, 1H, J = 10.7 Hz), 4.42 (dd, 1H, J1 = 11.9 Hz, J2 = 4.8 Hz), 4.18 (m, 2H), 3.82 (m, 2H), 3.45 (m, 1H), 3.36 (m, 2H), 3.28 (dd, 1H, J1 = 11.8 Hz, J2 = 4.8 Hz), 1.52–0.88 (m, 26H), 0.86 (d, 6H, J = 6.5 Hz), 0.83 (d, 3H, J = 6.6 Hz), 0.75 (d, 3H, J = 6.5 Hz), 0.68 (d, 2H, J = 6.6 Hz), 0.61 (d, 2H, J = 6.5 Hz); 13C NMR (176 MHz, CDCI3) δ 166.1, 165.9, 165.6, 165.4, 165.2, 133.6, 133.5, 133.4, 133.3, 133.3, 129.98, 128.5, 101.4, 101.2, 100.8, 74.4, 74.2, 73.7, 73.0, 72.0, 69.8, 68.7, 68.5, 38.5, 62.9, 62.7, 39.5, 37.6, 37.61, 37.55, 37.4, 37.3, 36.5, 32.9, 29.7, 29.4, 28.1, 24.96, 24.94, 24.6, 24.4, 22.9, 22.8, 19.9, 19.5, 19.1; HRMS (ESI−) calcd for C67H82O16 [M − H]+: 1141.5603, found: 1141.5530.
Compound 8C. Following GP2, from 1100 mg (0.73 mmol) of 7C, 750 mg of 8C was collected as a white waxy solid (72% yield). 1H NMR (700 MHz, CDCl3) δ 8.03 (d, 2H, J = 7.4 Hz), 7.95–7.89 (m,8H), 7.59 (t, 1H, J = 7.4 Hz), 7.56 (t, 1H, J = 7.4 Hz), 7.51 (t, 1H, J = 7.4 Hz), 7.45 (m, 5H), 7.38 (t, 2H, J = 7.8 Hz), 7.31 (m, 3H), 7.23 (t, 2H, J = 7.7 Hz), 5.74 (m, 2H), 5.39 (t, 1H, J = 8.6 Hz), 5.06 (dd, 1H, J1 = 3.1 Hz, J2 = 10.4 Hz), 4.81 (d, 1H, J = 7.8 Hz), 4.78 (d, 1H, J = 7.9 Hz), 4.60 (d, 1H, J = 10.4 Hz,), 4.40 (dd, 1H, J1 = 4.7 Hz, J2 = 11.9 Hz), 4.18 (m, 2H), 3.85 (m, 2H), 3.63 (m, 35H), 3.49 (m, 3H), 3.26 (m, 1H), 1.56 (m, 2H), 1.25 (s, 18H), 0.87 (t, 3H, J = 7.1 Hz); 13C NMR (176 MHz, CDCI3) δ 166.0, 165.9, 165.5, 165.4, 165.2, 133.7, 133.5, 133.4, 130.0, 129.9, 129.9, 129.8,129.7, 129.7, 129.4, 129.1, 128.9, 128.9, 128.8, 128.6, 128.5, 125.0, 101.4, 101.0, 100.7, 74.3, 74.2, 73.7, 72.9,72.0, 70.7, 70.7, 70.6, 70.5, 70.2, 69.8, 69.3, 68.3, 62.7, 32.1, 29.8, 29.8, 29.7, 29.7, 29.6, 29.5, 26.2, 22.8, 14.3; HRMS (ESI+) calcd for C77H102O25Na [M + Na]+: 1449.6602, found: 1449.6634.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90
90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.2 methanol/dichloromethane/pyridine to yield waxy solids 9A–9C.
0.2 methanol/dichloromethane/pyridine to yield waxy solids 9A–9C.
Compound 9A. Following GP3, from 300 mg (0.28 mmol) of 8A, 310 mg of 9A was collected as a pale white waxy solid (96% yield). 1H NMR (700 MHz, CDCl3) δ 8.00–7.90 (m, 8H), 7.86 (d, 2H, J = 7.6 Hz), 7.59 (t, 1H, J = 7.4 Hz), 7.60–7.27 (m, 15H), 7.24 (t, 2H, J = 7.7 Hz), 5.71 (t, 1H, J = 9.3 Hz), 5.66 (m, 1H), 5.29 (t, 1H, J = 5.8 Hz), 5.07 (dd, 1H, J1 = 13.7 Hz, J2 = 7.2 Hz), 4.78 (d, 1H, J = 7.9 Hz), 4.62 (m, 2H), 4.38 (dd, 1H, J1 = 4.6, J2 = 11.8 Hz), 4.27 (d, 1H, J = 3.2 Hz), 4.18 (t, 2H, J = 9.4 Hz), 3.78 (m, 4H), 3.72 (dd, 1H, J1 = 14.7 Hz, J2 = 4.4 Hz), 3.57 (t, 1H, J = 6.6 Hz), 3.21 (m, 1H), 3.16 (m, 2H), 1.26–0.90 (m, 26H), 0.96 (m, 8H), 0.99 (m, 16H), 0.85 (t, 3H, J = 7.3 Hz), 0.79 (t, 3H, J = 7.3 Hz); 13C NMR (176 MHz, CDCI3) δ 166.1, 165.9, 165.6, 165.3, 165.2, 133.6, 133.5, 133.4, 133.3, 130.0, 129.9, 129.89, 129.85, 129.7, 129.5, 129.1, 129.0, 128.8, 128.6, 128.5, 128.45, 101.4, 101.3, 74.4, 74.2, 73.7, 73.1, 73.0, 72.0, 69.8, 68.5, 62.8, 62.75, 38.0, 32.0, 31.9, 31.1, 30.9, 30.0, 29.7, 29.4, 26.9, 26.8, 26.6, 26.5, 22.8, 22.75, 14.3, 29.6, 29.6, 29.5, 29.4, 29.4, 25.6, 22.8, 14.3, 14.2; HRMS (ESI−) calcd for C63H73O19S− [M–Na + H]−: 1165.4472, found: 1165.4466.
Compound 9B. Following GP3, from 280 mg (0.25 mmol) of 8B, 280 mg of 9B was collected as a pale white waxy solid (94% yield). 1H NMR (700 MHz, CDCl3) δ 7.98 (dd, 2H, J = 7.3 Hz, J = 3.3 Hz), 7.94–7.89 (m, 6H), 7.86 (d, 2H, J = 7.5 Hz), 7.59 (t, 2H, J = 7.4 Hz) 7.53–7.28 (m, 11H), 7.23 (t, 2H, J = 7.5 Hz), 5.71 (td, 1H, J = 9.3, J = 2.7 Hz), 5.64 (dd, 1H, J1 = 10.4 Hz, J2 = 7.9 Hz), 5.28 (t, 1H, J = 10.0 Hz), 5.07 (dd, 1H, J1 = 10.4 Hz, J2 = 3.2 Hz), 4.78 (d, 1H, J = 7.9 Hz), 4.66 (t, 1H, J = 7.1 Hz), 4.62 (d, 1H, J = 11.7 Hz), 4.42 (dd, 1H, J1 = 11.8 Hz, J2 = 4.6 Hz), 4.26 (d, 1H, J = 3.1 Hz), 4.20 (t, 1H, J = 9.4 Hz), 3.82 (m, 4H), 3.60 (t, 1H, J = 6.5 Hz), 3.36 (m, 2H), 1.55–0.95 (m, 26H), 0.86 (d, 6H, J = 6.5 Hz), 0.83 (d, 3H, J = 6.6 Hz), 0.75 (d, 3H, J = 6.5 Hz), 0.68 (d, 2H, J = 6.6 Hz), 0.61 (d, 2H, J = 6.6 Hz); 13C NMR (176 MHz, CDCI3) δ 166.1, 165.9, 165.8, 165.4, 165.2, 133.8, 133.5, 133.43, 133.40, 133.35, 129.93, 129.91, 129.89, 129.87, 129.83, 129.76, 129.6, 129.5, 129.3, 129.0, 128.8, 128.7, 128.6, 128.55, 128.50, 128.46, 128.44, 127.3, 125.0, 101.3, 101.2, 100.8, 76.3, 73.9, 73.3, 73.12, 73.10, 72.8, 72.4, 69.9, 68.7, 68.5, 66.4, 64.7, 62.7, 45.7, 39.5, 37.6, 37.53, 37.50, 37.49, 37.3, 36.5, 32.9, 29.7, 29.4, 28.1, 24.96, 24.94, 24.6, 24.3, 22.9, 22.8, 19.9, 19.5, 19.1; HRMS (ESI−) calcd for C67H81O19S− [M–Na + H]−: 1221.5098, found: 1221.5098.
Compound 9C. Following GP3, from 750 mg (0.53 mmol) of 8C, 487 mg of 9C was collected as a pale white waxy solid (62% yield). 1H NMR (700 MHz, CDCl3) δ 7.98 (m, 6H), 7.87 (d, 2H, J = 6.9 Hz), 7.77 (d, 2H, J = 7.7 Hz), 7.60–7.26 (m, 15H), 7. 09 (t, 2H, J = 7.8 Hz), 5. 83 (m, 1H), 5.63 (m, 1H), 5.33 (m, 1H), 5.14 (d, 1H, J = 10.0 Hz), 4.99–4.77 (m, 3H), 4.38 (m, 2H), 4.24 (s, 1H), 4.09 (m, 2H), 3.63 (m, 27H), 3.56 (m, 6H), 3.43 (m, 5H), 1.56 (m, 2H), 1.25 (s, 18H), 0.87 (t, 3H, J = 7.1 Hz); 13C NMR (176 MHz, CDCI3) δ 165.8, 165.6, 165.3, 133.3, 133.1, 130.1, 130.0, 129.9, 129.7, 129.2, 129.1, 128.8, 128.7, 128.5, 128.4, 128.3, 124.9, 100.9, 72.9, 71.7, 70.5, 70.3, 70.2, 70.0, 32.0, 29.8, 29.8, 29.7, 29.6, 29.5, 26.2, 22.8, 14.3; HRMS (ESI−) calcd for C77H101O28S− [M–Na + H]−: 1505.6206, found: 1505.6198.
Compound 10A. Following GP4, from 247 mg (0.22 mmol) of 9A, 50 mg of 10A was collected as a white waxy solid (37% yield). 1H NMR (700 MHz, 1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CD3OD/CD2Cl2) δ 4.32 (d, 1H, J = 7.7 Hz), 4.26 (d, 1H, J = 7.8 Hz), 4.25 (dd, 1H, J1 = 10.7 Hz, J2 = 8.8 Hz), 4.11 (dd, 1H, J1 = 10.7 Hz, J2 = 3.8 Hz), 3.84 (m, 4H), 3.76 (dd, 1H, J1 = 8.9 Hz, J2 = 6.7 Hz), 3.53 (m, 4H), 3.40 (m, 2H), 3.27 (t, 1H, J1 = 8.5 Hz) 1.60 (m, 1H), 1.26 (m, 24H), 0.87 (t, 6H, J = 7.0 Hz); 13C NMR (176 MHz, 1
1 CD3OD/CD2Cl2) δ 4.32 (d, 1H, J = 7.7 Hz), 4.26 (d, 1H, J = 7.8 Hz), 4.25 (dd, 1H, J1 = 10.7 Hz, J2 = 8.8 Hz), 4.11 (dd, 1H, J1 = 10.7 Hz, J2 = 3.8 Hz), 3.84 (m, 4H), 3.76 (dd, 1H, J1 = 8.9 Hz, J2 = 6.7 Hz), 3.53 (m, 4H), 3.40 (m, 2H), 3.27 (t, 1H, J1 = 8.5 Hz) 1.60 (m, 1H), 1.26 (m, 24H), 0.87 (t, 6H, J = 7.0 Hz); 13C NMR (176 MHz, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CD3OD/CD2Cl2) 105.0, 103.6, 82.9, 75.4, 74.3, 74.1, 73.9, 71.8, 69.2, 67.4, 62.1, 39.0, 32.59, 32.58, 31.7, 31.6, 30.7, 30.4, 30.3, 30.0, 23.3, 14.3; HRMS (ESI−) calcd for C28H53O14S− [M–Na + H]−: 645.3162, found: 645.3163.
1 CD3OD/CD2Cl2) 105.0, 103.6, 82.9, 75.4, 74.3, 74.1, 73.9, 71.8, 69.2, 67.4, 62.1, 39.0, 32.59, 32.58, 31.7, 31.6, 30.7, 30.4, 30.3, 30.0, 23.3, 14.3; HRMS (ESI−) calcd for C28H53O14S− [M–Na + H]−: 645.3162, found: 645.3163.
Compound 10B. Following GP4, from 408 mg (0.33 mmol) of 9B, collected 72 mg of 10B as a white waxy solid (31% yield). 1H NMR (700 MHz, 1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CD3OD/CD2Cl2) δ 4.32 (d, 1H, J = 7.7 Hz), 4.29 (d, 1H, J = 7.9 Hz), 4.25 (dd, 1H, J1 = 8.8 Hz, J2 = 10.7 Hz), 4.16 (dd, 1H, J1 = 10.7 Hz, J2 = 3.7 Hz), 3.91 (m, 1H), 3.84 (m, 4H), 3.53 (m, 4H), 3.41 (m, 1H), 3.26 (t, 1H, J = 8.4 Hz), 1.70–1.02 (m, 26H), 0.89 (d, 3H, J = 6.6 Hz), 0.86 (d, 6H, J = 6.6 Hz), 0.84 (d, 6H, J = 6.6 Hz); 13C NMR (176 MHz, 1
1 CD3OD/CD2Cl2) δ 4.32 (d, 1H, J = 7.7 Hz), 4.29 (d, 1H, J = 7.9 Hz), 4.25 (dd, 1H, J1 = 8.8 Hz, J2 = 10.7 Hz), 4.16 (dd, 1H, J1 = 10.7 Hz, J2 = 3.7 Hz), 3.91 (m, 1H), 3.84 (m, 4H), 3.53 (m, 4H), 3.41 (m, 1H), 3.26 (t, 1H, J = 8.4 Hz), 1.70–1.02 (m, 26H), 0.89 (d, 3H, J = 6.6 Hz), 0.86 (d, 6H, J = 6.6 Hz), 0.84 (d, 6H, J = 6.6 Hz); 13C NMR (176 MHz, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CD3OD/CD2Cl2) δ 105.0, 103.2, 82.8, 75.5, 75.4, 73.5, 74.3, 74.1, 71.8, 69.2, 67.5, 62.1, 40.0, 38.09, 38.6, 38.0, 37.9, 33.5, 31.4, 30.6, 28.7, 25.4, 25.1, 25.0, 22.9, 22.8, 22.7, 20.0, 19.9; HRMS (ESI−) calcd for C32H61O14S− [M–Na + H]−: 701.3788, found: 701.3777.
1 CD3OD/CD2Cl2) δ 105.0, 103.2, 82.8, 75.5, 75.4, 73.5, 74.3, 74.1, 71.8, 69.2, 67.5, 62.1, 40.0, 38.09, 38.6, 38.0, 37.9, 33.5, 31.4, 30.6, 28.7, 25.4, 25.1, 25.0, 22.9, 22.8, 22.7, 20.0, 19.9; HRMS (ESI−) calcd for C32H61O14S− [M–Na + H]−: 701.3788, found: 701.3777.
Compound 10C. Following GP4, from 485 mg (0.32 mmol) of 9C, collected 253 mg of 10C as a white waxy solid (80% yield). 1H NMR (700 MHz, 1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CD3OD/CD2Cl2) δ 4.36 (d, 1H, J = 7.9 Hz), 4.32 (d, 1H, J = 7.7 Hz), 4.22 (t, 1H, J = 9.6 Hz), 4.12 (d, 1H, J = 4.9 Hz), 4.01 (m, 1H), 3.84 (m, 4H), 3.75 (m, 1H), 3.63 (m, 33H), 3.56 (m, 3H), 3.49 (m, 6H), 1.56 (m, 2H), 1.26 (m, 20H), 0.87 (t, 3H, J = 7.0 Hz); 13C NMR (176 MHz, 1
1 CD3OD/CD2Cl2) δ 4.36 (d, 1H, J = 7.9 Hz), 4.32 (d, 1H, J = 7.7 Hz), 4.22 (t, 1H, J = 9.6 Hz), 4.12 (d, 1H, J = 4.9 Hz), 4.01 (m, 1H), 3.84 (m, 4H), 3.75 (m, 1H), 3.63 (m, 33H), 3.56 (m, 3H), 3.49 (m, 6H), 1.56 (m, 2H), 1.26 (m, 20H), 0.87 (t, 3H, J = 7.0 Hz); 13C NMR (176 MHz, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CD3OD/CD2Cl2) δ 104.8, 103.4, 82.3, 75.7, 75.5, 74.4, 74.1, 72.1, 71.8, 70.9, 70.84, 70.77, 70.6, 62.0, 32.6, 30.29, 30.28, 30.2, 30.1, 30.0, 26.7, 23.3, 14.3; HRMS (ESI−) calcd for C42H81O23S− [M–Na + H]−: 985.4895, found: 985.4889.
1 CD3OD/CD2Cl2) δ 104.8, 103.4, 82.3, 75.7, 75.5, 74.4, 74.1, 72.1, 71.8, 70.9, 70.84, 70.77, 70.6, 62.0, 32.6, 30.29, 30.28, 30.2, 30.1, 30.0, 26.7, 23.3, 14.3; HRMS (ESI−) calcd for C42H81O23S− [M–Na + H]−: 985.4895, found: 985.4889.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol 3
Chol 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, DMPC
1, DMPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol: 10A (or 10B or 10C) in a 3
Chol: 10A (or 10B or 10C) in a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio, and DMPC
2 ratio, and DMPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol
Chol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SLA in a 3
SLA in a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio were dried under nitrogen and lyophilized overnight under vacuum. The hydration of the dried lipid film was done in Milli-Q water at 40 °C under stirring conditions followed by vesicle size reduction to about 50–150 nm diameter using sonication and high-pressure extrusion. The vesicles were annealed by incubation at 4 °C for 18 h and then sterilized (0.22 μm Millex-HV, non-pyrogenic, low protein binding filter, Millipore). The resulting liposomes were characterized for size (Z-average diameter), polydispersity index (PDI) and zeta potentials using Zetasizer NanoZS (Malvern Instruments, Malvern, UK) and the dry weight was measured based on an aliquot.
2 ratio were dried under nitrogen and lyophilized overnight under vacuum. The hydration of the dried lipid film was done in Milli-Q water at 40 °C under stirring conditions followed by vesicle size reduction to about 50–150 nm diameter using sonication and high-pressure extrusion. The vesicles were annealed by incubation at 4 °C for 18 h and then sterilized (0.22 μm Millex-HV, non-pyrogenic, low protein binding filter, Millipore). The resulting liposomes were characterized for size (Z-average diameter), polydispersity index (PDI) and zeta potentials using Zetasizer NanoZS (Malvern Instruments, Malvern, UK) and the dry weight was measured based on an aliquot.
Immunization and mice
Female C57BL6NCrl mice (6–8 weeks old) were obtained from Charles River Laboratories (Saint-Constant, Canada). All mice were maintained at the small animal facility of the National Research Council Canada (NRC) in accordance with the guidelines of the Canadian Council on Animal Care. All procedures performed on animals in this study were in accordance with regulations and guidelines reviewed and approved in animal use protocol 2020.10 by the NRC Human Health Therapeutics Animal Care Committee. Ovalbumin protein, Ova, (Sigma-Aldrich) alone or in combination with liposomes as described in Table 1 were diluted in Dulbecco's phosphate buffered saline (DPBS; Cytiva, Marlborough, Massachusetts, USA) prior to administration. Mice were first anesthetized with isofluorane then immunized by intramuscular injection (50 μL into the left tibialis anterior muscle) on days 0, 21. On day 20, 150 μL of blood was collected and assessed to quantify antibody titers (see below). On day 27, caryboxyfluorescein succinimidyl ester (CFSE) stained target and non-target cells (as described below) diluted in Hank's balanced salt solution (HBSS; GE Life Sciences) to a final volume of 200 μL were injected intravenously via the tail vein. On day 28, all mice were euthanized by cervical dislocation under isoflurane anaesthesia, blood was collected to quantify antibody titers by ELISA and spleens were collected to quantify T cell cytotoxicity by in vivo cytotoxicity assay.| Sample | Size (nm) | Polydispersity index | Zeta potential (mV) |
|---|---|---|---|
| DMPC–Chol | 149 ± 1 | 0.28 ± 0.01 | −35 ± 1 |
| 10A (DMPC–Chol) | 80 ± 1 | 0.31 ± 0.04 | −82 ± 2 |
| 10B (DMPC–Chol) | 94 ± 1 | 0.43 ± 0.01 | −68 ± 1 |
| 10C in DMPC–Chol | 93 ± 1 | 0.28 ± 0.01 | −57 ± 1 |
| SLA (water) | 92 ± 1 | 0.26 ± 0.01 | −69 ± 1 |
| SLA (DMPC–Chol) | 71 ± 1 | 0.24 ± 0.01 | −87 ± 6 |
Antibody titer quantification
Anti-Ova total IgG antibody titers in serum were quantified by enzyme-linked immunosorbent assay (ELISA), as described previously.21 Briefly, 96-well high-binding ELISA plates (Thermo Fisher Scientific) were coated overnight at room temperature (RT) with 100 μL of 10 μg mL−1 Ova protein diluted in DPBS (Sigma-Aldrich, St. Louis, MO, USA). Plates were washed five times with DPBS/0.05% Tween20 (DPBS-T; Sigma-Aldrich, St. Louis, MO, USA). To prevent non-specific binding, plates were blocked for 1 hour at 37 °C with 200 μL 10% fetal bovine serum (FBS; Thermo Fisher Scientific) in DPBS. After the plates were washed five times with DPBS-T, 3.162-fold serially diluted samples in DPBS-T with 10% FBS were added in 100 μL volumes and incubated for 1 hour at 37 °C. After five washes with DPBS-T (Sigma-Aldrich), 100 μL of goat anti-mouse IgG-horse radish peroxidase (HRP) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4000, Southern Biotech, Birmingham, AL, USA) was added for 1 hour at 37 °C. After five washes with DPBS-T, 100 μL per well of the substrate o-phenylenediamine dihydrochloride (OPD, Sigma-Aldrich) diluted in 0.05 M citrate buffer (pH 5.0) was added. Plates were developed for 30 minutes at RT in the dark. The reaction was stopped with 50 μL per well of 4N H2SO4. Ova-specific IgG antibody, that bound to the plate, was detected spectrophotometrically at 450 nm. Antibody titers for IgG in serum were defined as the dilution that resulted in an absorbance value (OD 450) of 0.2 and were calculated using XLfit software (ID Business Solutions, Guildford, UK). Samples that do not reach the target OD were assigned the value of the lowest tested dilution (i.e. 10) for analysis purposes.
4000, Southern Biotech, Birmingham, AL, USA) was added for 1 hour at 37 °C. After five washes with DPBS-T, 100 μL per well of the substrate o-phenylenediamine dihydrochloride (OPD, Sigma-Aldrich) diluted in 0.05 M citrate buffer (pH 5.0) was added. Plates were developed for 30 minutes at RT in the dark. The reaction was stopped with 50 μL per well of 4N H2SO4. Ova-specific IgG antibody, that bound to the plate, was detected spectrophotometrically at 450 nm. Antibody titers for IgG in serum were defined as the dilution that resulted in an absorbance value (OD 450) of 0.2 and were calculated using XLfit software (ID Business Solutions, Guildford, UK). Samples that do not reach the target OD were assigned the value of the lowest tested dilution (i.e. 10) for analysis purposes.
In vivo cytotoxicity assay
In vivo cytolytic activity in immunized mice was enumerated as described previously.22,23 Donor spleen-cell suspensions from syngeneic mice were prepared. Cells were split into two aliquots. One aliquot was designated ‘non-targets’ and left as is and the other aliquot was designated ‘targets’ and was incubated with the Ova CD8+ T cell epitope peptide SIINFEKL (JPT Peptide Technologies GmbH) in R10 media. After 30 min of incubation, the ‘non-target’ aliquot was stained with a low concentration of CFSE (0.25 μM; Thermo Fisher Scientific, Waltham, MA, USA), and the ‘target’ aliquot was stained with a 10-fold higher concentration of CFSE (2.5 μM). The two cell aliquots were mixed 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and injected (total of 20 × 106 cells per mouse) into all study mice. At ∼20 to 22 h after the donor cell transfer, spleens were removed from recipients, single cell suspensions were prepared and cells were analyzed by flow cytometry on a BD Fortessa flow cytometer (Becton Dickenson, Franklin Lakes, NJ, US). The fraction of ‘target’ to ‘non-target’ cells were measured using the equation below to determine the in vivo cytotoxicity and is a measure of CD8 T cell activity.
1 and injected (total of 20 × 106 cells per mouse) into all study mice. At ∼20 to 22 h after the donor cell transfer, spleens were removed from recipients, single cell suspensions were prepared and cells were analyzed by flow cytometry on a BD Fortessa flow cytometer (Becton Dickenson, Franklin Lakes, NJ, US). The fraction of ‘target’ to ‘non-target’ cells were measured using the equation below to determine the in vivo cytotoxicity and is a measure of CD8 T cell activity.To determine the % of target cells killed in an individual mouse as compared to the average of a group of naïve mice the above equation was used. Target/non-targetexp: percent “target” cells divided by percent “non-target” cells in an experimental mouse. ![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif) (Target/non-target)naive: the average (target/non-target) of all naive mice used in the experiment.
(Target/non-target)naive: the average (target/non-target) of all naive mice used in the experiment.
Statistical analysis
Data were analysed using GraphPad Prism (GraphPad Software, San Diego, CA). Statistical significance of the differences between groups were calculated by one-way ANOVA followed by post hoc analysis using Tukey's multiple comparison tests. Antibody titres were log-transformed prior to statistical analysis.Results and discussion
Three alcohols (A: 2-hexyl-1-decanol; B: dihydrophytol; C: nonaethylene glycol monododecyl ether) were selected to create a small library of SLGs that could be obtained at a low-cost to better understand the need for specific moieties on bioactivity (Fig. 2). The SLGs were designed to keep the highly stable properties of archaeal lipids such as pH resistance, thermal stability, and resistance against oxidative degradation intact. A defining characteristic of archaeal lipids is the ether linkage between the phytanyl chains and the glycerol backbone, which ensures stability over a broad range of pH.7 It was important to test how the fatty alcohols A and B which lacked that feature would fare. On the other hand, the alcohol C had abundant ether linkages. The incorporation of fully saturated branched carbon chains in the SLGs mirrors the approach used to enhance stability by preventing oxidative degradation in archaeal lipids. The branched structure is also attributed to membrane fluidity due to steric hindrance.24 These properties guided our design of cost-effective and stable library of SLG compounds. The alcohol A (2-hexyl-1-decanol), which is branched but has a shorter carbon chain than archaeol and lacks 2 ether linkages, was coupled to the glycosyl moiety to help us understand the influence of carbon chain length on vaccine adjuvanticity. Compared to archaeol, the dihydrophytol B has only 1 isoprenoid (phytanyl chain) and lacks the ether linkage between the glycerol and phytanyl chain. This compound was used to generate SLG 10B to see whether the number of isoprenoid units or lack of the glycerol backbone played a role in vaccine adjuvanticity. PEGylated liposomes are known for offering more stability and drug-like properties such as water solubility and long blood circulation time.25 Barbeau and coworkers have prepared PEGylated archaeosomes and demonstrated these stealth archaeosomes display significantly higher stability than PEGylated conventional liposomes.26 Considering this, we were interested in seeing how a PEG chain linked to the sulfated glycosyl moiety would impact vaccine adjuvanticity while offering the high stability associated with PEGylated lipids. Nonaethylene glycol monododecyl ether C was therefore used to generate SLG 10C. In all three cases, unlike SLA, the resulting SLGs were devoid of the glycerol backbone connecting the lipid to the sugar moiety. Overall, this study aimed to shed light on how specific structural features of the archaeol lipid could contribute to SLA's robust immunogenic properties, all the while introducing potential cost-effective novel adjuvants.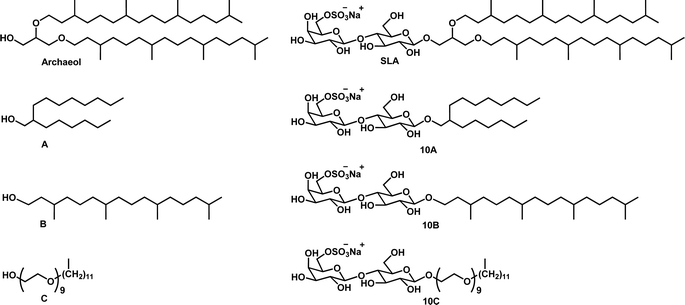 | ||
| Fig. 2 Structure of archaeol, SLA, and fatty alcohols A: 2-hexyl-1-decanol, B: dihydrophytol and C: nonaethylene glycol monododecyl ether, and their corresponding SLGs 10A, 10B and 10C, respectively. | ||
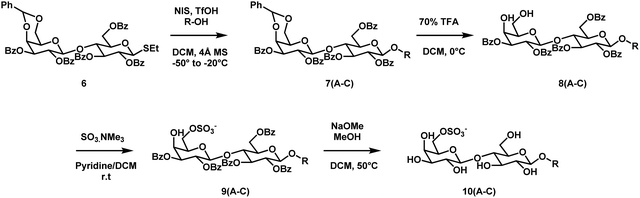
The SLGs 10A–C were prepared according to the synthetic route adapted from Régnier et al. (Scheme 1).17 The common starting material required for the 3 SLGs is the thioglycoside 6. Its synthesis has been previously reported and involves subjecting lactose 1 to a series of reactions involving protection, nucleophilic substitution and deprotection through intermediates 2–5 (Scheme S1, ESI†).17 Using the procedures described by Fraser-Reid and co-workers, the glycosylation step between the thioglycoside 6 and the three commercially available fatty alcohols (2-hexyl-1-decanol, dihydrophytol and nonaethylene glycol monododecyl ether) was performed using N-iodosuccinimide and triflic acid to afford compounds 7A–C.27 Following the glycosylation step, the benzylidene protective group was selectively removed under acidic conditions to yield compounds 8A–C. The latter was subjected to sulfation to produce the derivatives 9A–C. Deprotection of the benzoyl groups in dilute basic conditions yielded the final glycosides 10A, 10B, and 10C. The final step's yields were relatively lower for compounds 10A and 10B in comparison to 10C. This could be attributed to the shorter hydrophobic chain lengths in 10A and 10B, making the glycolipids more polar and susceptible to remaining in the aqueous phase during work-up.
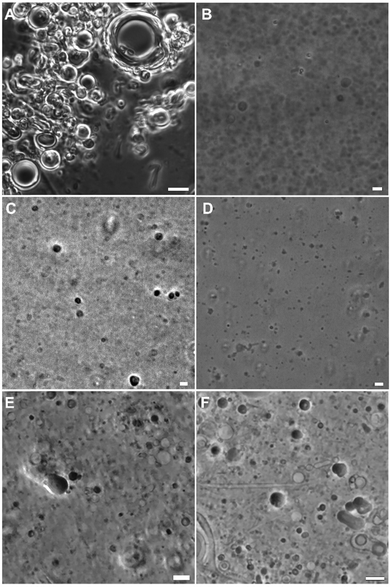
SLA self-assembles readily in water to form a multilamellar liposomal suspension which upon sonication undergoes a size reduction to produce unilamellar vesicles.20 The SLGs 10A–C did not self-assemble in water readily, possibly stemming from the lower stability due to the shorter lipid chain in 10A, single phytanyl chain for 10B and the increased hydrophilicity of the single chain nonaethylene glycol monododecyl ether in 10C. As such, they were each co-assembled in presence of a cholesterol (Chol) and 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) mixture (Table S1, ESI†). DMPC is a synthetic zwitterionic phospholipid that is used to prepare liposomes through bilayer formation and has been used to deliver biomacromolecules. On the other hand, cholesterol is used in several clinically approved liposomal nanomedicines due to its ability to form ordered bilayers with saturated phospholipids like DMPC.28 Once the SLGs 10A–C were co-assembled in DMPC:Chol and subjected to size reduction, the hydrodynamic radii of the vesicles were measured using dynamic light scattering (DLS). In general, they were around 100 nm, close to the dimensions of SLA self-assembled in water (Table 1). Polydispersity is a measure of size heterogeneity of a suspension. A polydispersity index (PI) of 0.3 and under is usually desirable and indicate monodispersed particles. In this case, all samples showed a PI of a 0.2–0.4, implying that most of them were fairly monodispersed, with some degree of aggregation as the PI increased. Optical microscopy was used to visualize the morphology of the samples and when co-assembled with DMPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol, they all produced vesicles (Fig. 3).
Chol, they all produced vesicles (Fig. 3).
Assessment of SLGs’ adjuvanticity
The ability of SLGs to act as vaccine adjuvants, when co-assembled with DMPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
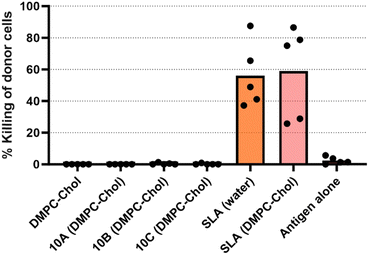
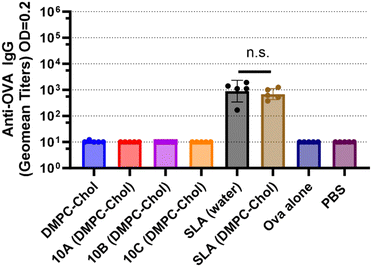
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol, was evaluated in a mouse model and compared to SLA. The effectiveness of an SLG-based vaccine to induce antigen-specific CD8+ T cell responses was assessed using an in vivo cytotoxicity assay. SLA archaeosomes, when combined with full-length protein Ova antigen, has previously been demonstrated to induce robust anti-Ova cytotoxic T cell responses, as evidenced by high cytotoxic responses in mice.22,29 In this study, we explored the ability of SLGs and SLA in DMPC:Chol archaeosomes to induce antigen-specific immune responses in C57BL/6NCrl mice, assessing the responses 3 weeks after a single immunization (Fig. 4) and 1 week following the second immunization (Fig. 5).
Chol, was evaluated in a mouse model and compared to SLA. The effectiveness of an SLG-based vaccine to induce antigen-specific CD8+ T cell responses was assessed using an in vivo cytotoxicity assay. SLA archaeosomes, when combined with full-length protein Ova antigen, has previously been demonstrated to induce robust anti-Ova cytotoxic T cell responses, as evidenced by high cytotoxic responses in mice.22,29 In this study, we explored the ability of SLGs and SLA in DMPC:Chol archaeosomes to induce antigen-specific immune responses in C57BL/6NCrl mice, assessing the responses 3 weeks after a single immunization (Fig. 4) and 1 week following the second immunization (Fig. 5).
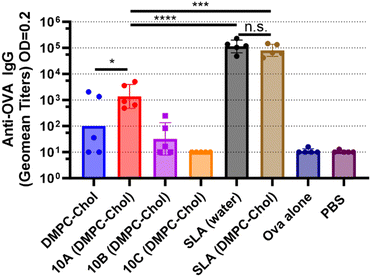
The addition of DMPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol to SLA did not alter the cytotoxic T lymphocyte (CTL) response (Fig. 4) or the antibody response (Fig. 5 and 6) observed with SLA alone. After a single immunization, no anti-Ova IgG antibodies were detected in the mice immunized with any of the SLGs’ formulations (Fig. 5), suggesting that replacing the archaeal lipid moiety of SLA with alcohols A–C compromises the adjuvanticity. However, on day 28, which is 7 days after a booster immunization, SLG 10A, but not SLG 10B or 10C, was able to generate an anti-Ova IgG antibody response, although this response was lower than that induced by SLA (Fig. 6). This data suggests that while SLA remains a robust adjuvant even when formulated with DMPC:Chol, the SLGs, particularly 10A, show some limited potential, but their overall efficacy in inducing antibody responses is significantly lower compared to SLA. This highlights the critical role of the lipid composition in the adjuvanticity of SLA-based formulations and underscores the need for further optimization of SLG structures to enhance their immunogenic potential.
Chol to SLA did not alter the cytotoxic T lymphocyte (CTL) response (Fig. 4) or the antibody response (Fig. 5 and 6) observed with SLA alone. After a single immunization, no anti-Ova IgG antibodies were detected in the mice immunized with any of the SLGs’ formulations (Fig. 5), suggesting that replacing the archaeal lipid moiety of SLA with alcohols A–C compromises the adjuvanticity. However, on day 28, which is 7 days after a booster immunization, SLG 10A, but not SLG 10B or 10C, was able to generate an anti-Ova IgG antibody response, although this response was lower than that induced by SLA (Fig. 6). This data suggests that while SLA remains a robust adjuvant even when formulated with DMPC:Chol, the SLGs, particularly 10A, show some limited potential, but their overall efficacy in inducing antibody responses is significantly lower compared to SLA. This highlights the critical role of the lipid composition in the adjuvanticity of SLA-based formulations and underscores the need for further optimization of SLG structures to enhance their immunogenic potential.
Conclusions
SLA archaeosomes has been demonstrated to be an effective vaccine adjuvant, eliciting both antibody and cellular immune responses in numerous disease models. To better understand the mechanism of action (MOA) underlying SLA's adjuvanticity, we engineered SLG variants that preserved SLA's sulfated headgroup while incorporating different lipid moieties. Among these, SLG 10A was able to initiate an immune response, as evidenced by the presence of antibodies in the serum of immunized mice by day 28. However, the antibody response was significantly weaker compared to that induced by SLA, suggesting that removing the glycerol backbone or shortening the lipid moiety adversely affects the adjuvant's efficacy. For variants 10B and 10C, there was a complete absence of adjuvant effect indicating that they substantially compromised the adjuvant's effectiveness. Moving forward, we plan to generate additional SLG variants to further investigate the mechanism of action behind SLA's adjuvanticity. This will help us understand the critical structural components of SLA and guide the design of more effective adjuvant formulations.Author contributions
The manuscript was written through contributions from all authors. Tarasha Sharma: conceptualization, methodology, investigation, visualization, writing – original draft. Sophie Régnier: methodology, investigation, visualization. Lise Deschatelets: conceptualization, methodology, investigation, visualization, writing – original draft. Felicity C. Stark: conceptualization, methodology, formal analysis, supervision, investigation, visualization, writing – original draft. Vinicio Vasquez: methodology, investigation, formal analysis, visualization. Camilo F. Martinez-Farina: methodology, validation, investigation. Renu Dudani: methodology, investigation. Blair A. Harrison: methodology, investigation. Bassel Akache: conceptualization, methodology, validation, supervision. Yimei Jia: conceptualization, methodology. Michael J. McCluskie: conceptualization, writing – review & editing, supervision, project administration, funding acquisition. Usha D. Hemraz: conceptualization, methodology, validation, writing – original draft, writing – review & editing, supervision, project administration. All authors have reviewed and given approval to the final version of the manuscript.Data availability
The data supporting this article have been included in the Experimental section of the manuscript and in the ESI.†Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was funded by the National Research Council of Canada, a Federal Research and Development Organization of the Government of Canada.References
- A. Pashine, N. M. Valiante and J. B. Ulmer, Nat. Med., 2005, 11, S63–68 CrossRef PubMed
.
- A. Banzhoff, R. Gasparini, F. Laghi-Pasini, T. Staniscia, P. Durando, E. Montomoli, P. L. Capecchi, P. di Giovanni, L. Sticchi, C. Gentile, A. Hilbert, V. Brauer, S. Tilman and A. Podda, PLoS One, 2009, 4, e4384 CrossRef PubMed
.
- P. J. Wismans, J. van Hattum, G. C. de Gast, K. P. Bouter, R. J. Diepersloot, T. Maikoe and G. C. Mudde, J. Med. Virol., 1991, 35, 216–222 CrossRef PubMed
.
- E. Oleszycka and E. C. Lavelle, Curr. Opin. Immunol., 2014, 28, 1–5 CrossRef PubMed
.
- N. Garcon and A. Di Pasquale, Hum. Vaccines Immunother., 2017, 13, 19–33 CrossRef PubMed
.
- G. B. Patel and G. D. Sprott, Crit. Rev. Biotechnol., 1999, 19, 317–357 CrossRef PubMed
.
- G. Kaur, T. Garg, G. Rath and A. K. Goyal, Drug Delivery, 2016, 23, 2497–2512 CrossRef PubMed
.
- M. A. Ansari, S. Zubair, A. Mahmood, P. Gupta, A. A. Khan, U. D. Gupta, A. Arora and M. Owais, PLoS One, 2011, 6, e22889 CrossRef CAS PubMed
.
- M. A. Ansari, S. Zubair, S. Tufail, E. Ahmad, M. R. Khan, Z. Quadri and M. Owais, Int. J. Nanomed., 2012, 7, 2433–2447 CrossRef CAS PubMed
.
- L. Krishnan, L. Deschatelets, F. C. Stark, K. Gurnani and G. D. Sprott, Clin. Dev. Immunol., 2010, 2010, 578432 Search PubMed
.
- M. J. McCluskie, L. Deschatelets and L. Krishnan, Hum. Vaccines Immunother., 2017, 13, 2772–2779 CrossRef PubMed
.
- B. Akache, F. C. Stark, Y. Jia, L. Deschatelets, R. Dudani, B. A. Harrison, G. Agbayani, D. Williams, M. P. Jamshidi, L. Krishnan and M. J. McCluskie, PLoS One, 2018, 13, e0208067 CrossRef PubMed
.
- F. C. Stark, B. Akache, A. Ponce, R. Dudani, L. Deschatelets, Y. Jia, J. Sauvageau, D. Williams, M. P. Jamshidi, G. Agbayani, K. Wachholz, B. A. Harrison, X. Li, L. Krishnan, W. Chen and M. J. McCluskie, Vaccine, 2019, 37, 7108–7116 Search PubMed
.
- B. Akache, T. M. Renner, A. Tran, L. Deschatelets, R. Dudani, B. A. Harrison, D. Duque, J. Haukenfrers, M. A. Rossotti, F. Gaudreault, U. D. Hemraz, E. Lam, S. Regnier, W. Chen, C. Gervais, M. Stuible, L. Krishnan, Y. Durocher and M. J. McCluskie, Sci. Rep., 2021, 11, 21849 CrossRef CAS
.
- G. D. Sprott, A. Yeung, C. J. Dicaire, S. H. Yu and D. M. Whitfield, Archaea, 2012, 2012, 513231 Search PubMed
.
- D. M. Whitfield, S. H. Yu, C. J. Dicaire and G. D. Sprott, Carbohydr. Res., 2010, 345, 214–229 Search PubMed
.
- S. Regnier, E. Lam, V. Vasquez, C. F. Martinez-Farina, F. C. Stark, G. Agbayani, L. Deschatelets, R. Dudani, B. A. Harrison, B. Akache, M. J. McCluskie and U. D. Hemraz, J. Med. Chem., 2022, 65, 8332–8344 CrossRef CAS
.
- D. M. Whitfield, S. H. Yu, C. J. Dicaire and G. D. Sprott, Carbohydr. Res., 2010, 345, 214–229 CrossRef CAS PubMed
.
-
D. M. Whitfield, G. Dennis Sprott and L. Krishnan, Sulfated-Glycolipids as Adjuvants for Vaccines, WO2016004512A1, 2016
.
- Y. Jia, B. Akache, L. Deschatelets, H. Qian, R. Dudani, B. A. Harrison, F. C. Stark, V. Chandan, M. P. Jamshidi, L. Krishnan and M. J. McCluskie, Int. J. Pharm., 2019, 561, 187–196 CrossRef CAS PubMed
.
- B. Akache, F. C. Stark and M. J. McCluskie, Methods Mol. Biol., 2021, 2183, 537–547 CrossRef CAS PubMed
.
- F. C. Stark, R. Dudani, G. Agbayani and M. J. McCluskie, Methods Mol. Biol., 2021, 2183, 549–558 CrossRef CAS PubMed
.
- D. L. Barber, E. J. Wherry and R. Ahmed, J. Immunol., 2003, 171, 27–31 CrossRef CAS PubMed
.
- P. B. Santhosh and J. Genova, ACS Omega, 2023, 8, 1–9 CrossRef CAS PubMed
.
- S. Adepu and S. Ramakrishna, Molecules, 2021, 26, 5905 CrossRef CAS PubMed
.
- J. Barbeau, S. Cammas-Marion, P. Auvray and T. Benvegnu, J. Drug Delivery, 2011, 2011, 396068 Search PubMed
.
- P. Konradsson, D. R. Mootoo, R. E. McDevitt and B. Fraser-Reid, J. Chem. Soc., Chem. Commun., 1990, 270–272 RSC
.
- K. Juszkiewicz, A. F. Sikorski and A. Czogalla, Int. J. Mol. Sci., 2020, 21, 9559 CrossRef CAS PubMed
.
- Y. Jia, B. Akache, L. Deschatelets, H. Qian, R. Dudani, B. A. Harrison, F. C. Stark, V. Chandan, M. P. Jamshidi, L. Krishnan and M. J. McCluskie, Int. J. Pharm., 2019, 561, 187–196 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nj03019b |
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2024 |