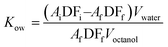Open Access Article
Open Access ArticleInvestigation of the inherent characteristics of copper(II) Schiff base complexes as antimicrobial agents†
Thasnim P
Mohammed
a,
Abinaya Sushana
Thennarasu
a,
Ravi
Jothi
b,
Shanmugaraj
Gowrishankar
b,
Marappan
Velusamy
c,
Suman
Patra
d and
Muniyandi
Sankaralingam
 *a
*a
aBioinspired & Biomimetic Inorganic Chemistry Laboratory, Department of Chemistry, National Institute of Technology Calicut, Kozhikode, Kerala 673601, India. E-mail: msankaralingam@nitc.ac.in; sankarjan06@gmail.com
bDepartment of Biotechnology, Science Campus, Alagappa University, Karaikudi–630 003, Tamil Nadu, India
cDepartment of Chemistry, North-Eastern Hill University, Shillong-793022, India
dSchool of Chemical Sciences, Indian Association for the Cultivation of Science, Kolkata 700032, India
First published on 29th May 2024
Abstract
We synthesised a sequence of structurally related copper(II) complexes of Schiff base ligands, 2-[(2-benzylamino-ethylimino)-methyl]-phenol [L1(H)], 2-[(2-benzylamino-ethylimino)-methyl]-4-methyl-phenol [L2(H)], and 2-[(2-benzylamino-ethylimino)-methyl]-4-bromo-phenol [L3(H)]. The complexes were characterized by ATR, UV-vis, EPR, ESI-MS, single crystal XRD, and elemental analyses. Based on the crystal structure, the ligand was coordinated to the copper(II) centre in a tridentate mode through phenolate(oxygen), imine nitrogen and amine nitrogen in an NNO fashion and the azide moiety served as an auxiliary ligand. Furthermore, we have investigated the antimicrobial activities of copper(II) complexes against clinically relevant fungal strains, including Candida albicans, Candida glabrata, and Candida tropicalis, as well as bacteria such as Pseudomonas aeruginosa and methicillin-resistant Staphylococcus aureus (MRSA), representing Gram-negative and Gram-positive pathogenic strains. Comparing the activity of complex 2 (128–512 μg mL−1) to complex 3 (64–128 μg mL−1), the latter demonstrated more potent effectiveness against all of the tested fungal strains. The activity of the complexes against MRSA reveals the inhibitory concentrations of 1024 μg mL−1 for 1, 128 μg mL−1 for 2, and 256 μg mL−1 for 3. It is worth mentioning that, at their minimum inhibitory concentrations (MICs), the complexes are not toxic to Galleria mellonella. Furthermore, the spot and broth microdilution assay results were in good agreement with the actual metabolic viability of bacterial and fungal strains, in which the complexes were active while having no effect on the metabolite resazurin. Additionally, in silico studies have been employed to elucidate both non-bonding interactions and the mode of action across protein sequences of experimentally studied microbes. The performance of complexes closely mirrored the reactivity order determined in in vitro studies. The impeding properties of complexes can be due to the dysfunctioning of virulent genes and signalling proteins and also affect the activities of transferase and transcriptase proteins. Overall, the complexes are promising candidates for antimicrobial drug development. We anticipate utilising these molecules for clinical purposes.
1. Introduction
During World War II, several metal-based compounds with biological applications were discovered and gained wide acceptance. However, the excessive use and misuse of antimicrobial drugs lead to resistance to pathogenic diseases. Considering this, the World Health Organisation (WHO) announced that antimicrobial resistance (AMR) is one of the biggest menace to the entire global community. Therefore, to combat microbial illnesses, chemists and biologists came together to develop medicines. In line with this, the main emphasis of modern bioinorganic chemistry research is on developing coordination compounds with exceptional functionalities for biological applications.1–3 Substantially, the applications of transition metal complexes have been extended to a wide range, owing to the ease of formation of these complexes with high stability and potentiality in different oxidation states.4 Concomitantly, the complexes of Schiff base ligands are of great scientific interest as they play a crucial role in medicinal and pharmaceutical fields.5–7For instance, there are reports on Schiff base complexes with varied denticity comprising N and O donor sites exhibiting excellent properties that include the ability to adjust auxiliary ligands, tunable steric and electronic environments on the metal core, etc.8,9 Furthermore, a variety of Schiff base complexes are employed with different transition metals such as Cu, Co, Fe, Ni, Mn, Pd, Au, Pt, V, and Zn to enhance their exceptional antimicrobial activity.9 The basic potentiality of the Schiff base copper(II) complexes in biological applications lies in their ability to bind and cleave the DNA under physiological conditions, which makes them potent agents for the treatment of many diseases.10,11 Among them, copper complexes have consistently been studied and proven as effective antimicrobial agents.12–18 A recent work reported a cellulose-based Schiff base copper(II) complex, which exhibited enhanced activity against S. aureus, which was 823% higher than the respective Schiff base ligand.15 Another work reported the use of quinoline based Schiff base copper(II) complexes, which exhibited good activity against bacteria and fungi (ZOI 31 mm mg−1 for S. aureus and 3 mm mg−1 for C. albicans).16 Following this, the anti-aminopyrine linked Schiff base copper(II) complex exhibited prominent antibacterial activity [minimum inhibitory concentration (MIC): 32 μg mL−1 for S. aureus].17 Lately, a copper(II) complex formed from a Schiff base prepared by dehydrative condensation of 3-acetyl-6-methyl-2H-pyran-2,4(3H)-dione and benzohydrazide showed different activities under various solvent conditions. Among them, greater activity against bacteria was found when aniline derivatives were used as co-solvents [MIC: S. aureus, 11.89 μM]. Additionally, an improved activity was observed when the same complex in benzhydrylamine was used with 70% ethanol with a >6-log reduction in microbial recovery.18
Based on previous research, we can infer that the selection of a ligand system is crucial for designing the metallodrugs. In this context, the metal complexes made up of Schiff bases derived from the reaction of salicylaldehyde and its derivatives with different diamines such as ethylenediamine, o-phenylenediamines, etc. were found to have prominent antibacterial and antifungal properties.19 The imine nitrogen itself can function as a Lewis base or an electron donor towards metal ions. Stable coordination is often achieved when metal ions are coordinated with other electron-donating groups of the molecule in such a way as to form stable five- or six-membered chelate rings. This requirement is often accomplished when groups like phenolate are present in the vicinity of other electron-donating groups. On top of that, phenolic compounds are known to exhibit good antibacterial properties and are universal inhibitors of free radical reactions. When coordinated with the metal ions, these compounds are believed to cause damage to the cytoplasmic membrane, contributing to their microbicidal properties.20 These findings offer a solid foundation for attempts to synthesise novel bioactive metal complexes using basic phenolic ligands.21 Despite these extensive studies on antimicrobial properties, seeking out new complexes to find more effective agents with a better understanding of the mechanism is still essential.
In pursuit of profound knowledge regarding the biological activities of Schiff base copper(II) complexes, we have moved forward to tune the electronic factors of the ligand by introducing various substituents. Therefore, we synthesized and characterized three structurally related Schiff base copper(II) complexes. All three complexes were characterized using modern spectroscopic techniques and tested against clinically relevant fungal strains Candida albicans (C. albicans), Candida glabrata (C. glabrata) and Candida tropicalis (C. tropicalis) as well as bacterial strains such as Pseudomonas aeruginosa (P. aeruginosa) and methicillin-resistant Staphylococcus aureus (MRSA). These microbes are chosen as they cause a majority of bloodstream infections that end up causing morbidity, particularly in immunocompromised patients. Interestingly, complexes 2 and 3 exhibited low MIC values against the tested bacterial and fungal strains, respectively. Furthermore, the in silico studies of microbial strains prove that the complexes are efficient in inhibiting the growth of microorganisms, by affecting cell stability activity, and lipophilicity, and causing thymidine-less cell death, as well as by inhibiting other biological functions.
2. Experimental section
2.1. Materials
All reagents and chemicals were of analytical grade (AR), and they were used without further purification. 5-Bromosalicylaldehyde and N-benzylethylenediamine were purchased from Sigma Aldrich. 5-Methylsalicylaldehyde was purchased from TCI Chemicals. Reagent grade copper(II) perchlorate hexahydrate was purchased from Alfa Aesar. AR grade sodium azide was purchased from Spectrochem. Salicylaldehyde and diethyl ether (laboratory grade) were purchased from Fisher Scientific. Dimethylformamide (ACS grade) was purchased from Qualigens. Dimethyl sulfoxide (HPLC grade, 99.8%) was purchased from Sisco Research Laboratories. Commercial grade methanol purchased from Fisher Scientific was distilled over magnesium turnings and iodine and was stored in an air-tight bottle until further use.Caution! It is known that metallic perchlorates and azide compounds are potential explosives. Only small quantities of these compounds should be prepared and suitable precautions should be taken when they are handled.
2.2. Physical measurements
An Agilent 8454 diode array spectrophotometer was used to collect the UV-visible spectra of complexes at room temperature. The CHN analysis was performed using a PerkinElmer 2400 Series II CHN analyser. Infrared spectra of the samples as KBr pellets were recorded at room temperature in a Jasco 4700 ATR-FTIR spectrometer. Mass data of the samples were recorded on a LC-MS-MS, XEVO-TQD 2000 (Waters) spectrometer. X-band EPR measurements were performed at 77 K on a JEOL X-band spectrometer (JESFA100). The g value was calibrated using the manganese marker. Magnetic measurements viz. zero-field cooling (ZFC) and field cooling (FC) were performed on a Quantum Design Versa Lab Vibrating Sample Magnetometer (VSM) in the range of 50–300 K. EPR data of the complexes 1–3 were recorded in MeOH:DMF (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) at 70 K.
1) at 70 K.
2.3. Synthesis of ligands
The Schiff base ligands were prepared by following the published procedure developed by us.22 For example, the Schiff-base ligand L1(H) was prepared by refluxing N-benzylethylenediamine (4 mmol) with salicylaldehyde (4 mmol) in hot methanol (10 mL) for 1 h, and a very good yield of yellow oily ligands was obtained after evaporating the solvent at low pressure.Ligand L1(H) (yield: 0.93 g, 90(2) %), IR data (ATR, cm−1) (Fig. S1(L1) in ESI†): 3302 ν(N–H), 1635 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). Ligand L2(H) (yield: 0.97 g, 91(1) %), IR data (ATR, cm−1) (Fig. S2(L2) in ESI†): 3322 ν(N–H), 1631 ν(C
N). Ligand L2(H) (yield: 0.97 g, 91(1) %), IR data (ATR, cm−1) (Fig. S2(L2) in ESI†): 3322 ν(N–H), 1631 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). Ligand L3(H) (yield: 1.2 g, 90(1), IR data (ATR, cm−1) (Fig. S3(L3) in ESI†): 3316 ν(N–H), 1633 ν(C
N). Ligand L3(H) (yield: 1.2 g, 90(1), IR data (ATR, cm−1) (Fig. S3(L3) in ESI†): 3316 ν(N–H), 1633 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N).
N).
2.4. Synthesis of copper complexes
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. The crystal data of 1 and 2 are provided in the subsequent section.
1 ratio. The crystal data of 1 and 2 are provided in the subsequent section.
Complex 1: (yield: 0.25 g, 70(2)%). ATR-IR (cm−1) (Fig. S1(1) in ESI†): 1635 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 2047 ν(N3); 3211 ν(N–H); UV/Vis (Fig. 1) (MeOH): λmax (nm) (ε, M−1 cm−1) 360 (5283), 617 (240); ESI-MS (Fig. S4 in ESI†) (+ve mode): m/z = 316.38 [Cu(L1)]+ (calc. 316.06); elemental analysis calcd for (C16H17CuN5O): C 53.55, H 4.77, N 19.51; found: C 53.56, H 4.75, N 19.46.
N); 2047 ν(N3); 3211 ν(N–H); UV/Vis (Fig. 1) (MeOH): λmax (nm) (ε, M−1 cm−1) 360 (5283), 617 (240); ESI-MS (Fig. S4 in ESI†) (+ve mode): m/z = 316.38 [Cu(L1)]+ (calc. 316.06); elemental analysis calcd for (C16H17CuN5O): C 53.55, H 4.77, N 19.51; found: C 53.56, H 4.75, N 19.46.
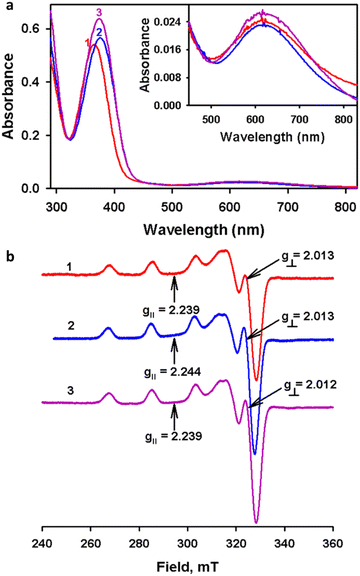 | ||
Fig. 1 (a) UV-vis spectra of complexes 1–3 (0.1 mM) recorded in methanol. (b) EPR spectra of 1–3 recorded at 77 K in methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMF (4 DMF (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solution. 1) solution. | ||
Complex 2: (yield: 0.25 g, 68(3)%). ATR-IR (cm−1) (Fig. S2(2) in ESI†): 1640 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 2028 ν(N3); 3244 ν(N–H); UV/Vis (Fig. 1) (MeOH): λmax (nm) (ε, M−1 cm−1) 376 (5649), 610 (230); ESI-MS (Fig. S5 in ESI†) (+ve mode): m/z = 330.42 [Cu(L2)]+ (calc. 330.08); elemental analysis calcd for (C17H19CuN5O): C 54.75, H 5.14, N 18.78; found: C 54.68, H 5.12, N 18.73.
N); 2028 ν(N3); 3244 ν(N–H); UV/Vis (Fig. 1) (MeOH): λmax (nm) (ε, M−1 cm−1) 376 (5649), 610 (230); ESI-MS (Fig. S5 in ESI†) (+ve mode): m/z = 330.42 [Cu(L2)]+ (calc. 330.08); elemental analysis calcd for (C17H19CuN5O): C 54.75, H 5.14, N 18.78; found: C 54.68, H 5.12, N 18.73.
Complex 3: (yield: 0.35 g, 81(2)%). ATR-IR (cm−1) (Fig. S3(3) in ESI†): 1636 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 2025 ν(N3); 3248 ν(N–H); UV/Vis (Fig. 1) (MeOH): λmax (nm) (ε, M−1 cm−1) 374 (6371), 620 (260); ESI-MS (Fig. S6 in ESI†) (+ve mode): m/z = 394.29 [Cu(L3)]+ (calc. 394.29); elemental analysis calcd for (C16H16BrCuN4O): C 43.90, H 3.68, N 16.00; found: C 43.78, H 3.67, N 15.94.
N); 2025 ν(N3); 3248 ν(N–H); UV/Vis (Fig. 1) (MeOH): λmax (nm) (ε, M−1 cm−1) 374 (6371), 620 (260); ESI-MS (Fig. S6 in ESI†) (+ve mode): m/z = 394.29 [Cu(L3)]+ (calc. 394.29); elemental analysis calcd for (C16H16BrCuN4O): C 43.90, H 3.68, N 16.00; found: C 43.78, H 3.67, N 15.94.
2.5. Single-crystal X-ray diffraction studies
An appropriate single crystal of complexes 1 and 2 was chosen from the mother liquor and bathed in paraffin oil before being placed on the top of a glass fiber that was further sealed with epoxy resin. The intensity data for the crystals are gathered using MoKα (λ = 0.71073 Å) radiation on a Bruker SMART APEX II diffractometer connected to a CCD area detector at 296 K. The data collection, reflection indexing, and lattice parameter determination were done using the SMART23 program. Integration of the intensity of reflections and scaling was done by utilizing the SAINT23 program. The absorption correction was done using the SADABS24 program. The structure determination, space group resolution, and least square refinement on F2 were performed using the SHELXTL25 program. The positions of all the atoms were obtained by direct methods. The metal atom in the complex was located from the E-map, and non-hydrogen atoms were refined anisotropically. The hydrogen atoms bound to the carbon atoms were placed in geometrically constrained positions and refined with isotropic temperature factors, generally 1.2 Ueq of their parent atoms. The crystallographic details of data collection for 1 and 2 are given in Table 1. The CCDC numbers 2263468 and 2263469† contain the supplementary crystallographic data for 1 and 2.| Details | 1 | 2 |
|---|---|---|
a
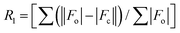 .
b .
b
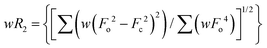 . .
|
||
| Empirical formula | C32H34Cu2N10O2 | C17H19CuN5O |
| Molecular weight (g mol−1) | 717.77 | 372.91 |
| Crystal habit and color | Needle and blue | Needle and blue |
| Crystal system | Triclinic | Orthorhombic |
| Space group |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
Pccn |
| a (Å) | 9.489(2) | 13.7472(19) |
| b (Å) | 9.759(3) | 28.684(4) |
| c (Å) | 10.043(3) | 8.5539(13) |
| α (°) | 63.432(9) | 90 |
| β (°) | 89.480(9) | 90 |
| γ (°) | 71.598(9) | 90 |
| V (Å3) | 779.6(4) | 3373.1(8) |
| ρ calc (g cm−3) | 1.529 | 1.469 |
| F(000) | 370 | 1544 |
| Temperature (K) | 296 | 296 |
| No. of reflections collected | 3924 | 4156 |
| No. of unique reflections | 3097 | 2226 |
| Radiation [MoKα] (Å) | 0.71073 | 0.71073 |
| Residuals [I > 2σ(I)] | ||
| R 1 | 0.0378 | 0.068 |
| wR 2 | 0.0937 | 0.1638 |
2.6. Lipophilicity
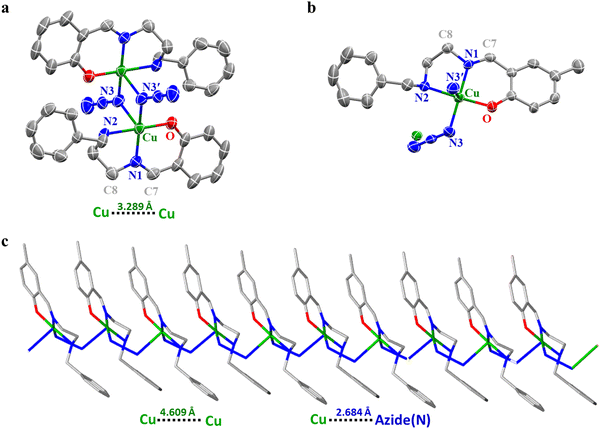
The lipophilicity of complexes 1–3 was determined by calculating the partition coefficient, P (or log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kow). It is determined by the shake-flask method, which gives an idea of the relative distribution of the complexes between 1-octanol and water.26 Herein, we have used 1-octanol as a mimic of the hydrophobic bilayer. Briefly, 1 mg of the complexes was mixed with water (5 mL) presaturated with 1-octanol. The UV-vis absorption spectra of the solution were recorded to obtain Ai. Then the mixture was treated with 1-octanol presaturated with water and the layers were separated. The spectrum of the aqueous layer was recorded to get Af. The equilibrium constant, Kow, was obtained using the equation:
Kow). It is determined by the shake-flask method, which gives an idea of the relative distribution of the complexes between 1-octanol and water.26 Herein, we have used 1-octanol as a mimic of the hydrophobic bilayer. Briefly, 1 mg of the complexes was mixed with water (5 mL) presaturated with 1-octanol. The UV-vis absorption spectra of the solution were recorded to obtain Ai. Then the mixture was treated with 1-octanol presaturated with water and the layers were separated. The spectrum of the aqueous layer was recorded to get Af. The equilibrium constant, Kow, was obtained using the equation:DFi and DFf are the dilution factors used to obtain Ai and Af respectively. Vwater and Voctanol are the volumes of the water and octanol phases.
2.7. Toxicity analysis
Toxicity analysis of complexes used in this study was tested using the invertebrate animal model Galleria mellonella. Larvae weighing between 0.2 and 0.4 g were used in the studies. Experiments were divided into five groups, with 10 larvae selected from each group. The test substances were injected into the final left proleg using a U-100 syringe. Group I acted as the positive control and was injected with methanol. Group II acted as the vehicle control and was injected with DMSO. Group III was injected with 1024 μg mg−1 complex 1. Group IV was injected with 512 μg mg−1 complex 2. Group V was injected with 256 μg mg−1 complex 3. Larva groups were incubated at 37 °C for 5 days. Every 24 hours, survival was measured.272.8. Antimicrobial activity of complexes 1–3
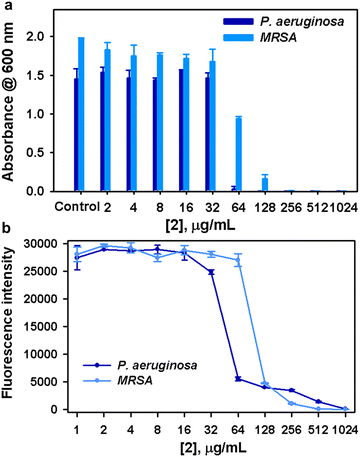
(A) Disk diffusion assay. In order to screen the antimicrobial potency of 1–3, the agar well diffusion technique was performed using the aforementioned test microorganisms by following the protocol previously described in the literature with minor modifications.28 Initially, the standard cell suspensions (3 h culture) of fungal and bacterial strains containing 1 × 106 CFU mL−1 cells were prepared. Then, 100 μL of 3 h culture corresponding to fungal and bacterial strains were swabbed accordingly on YEPD and MHI agar plates using a sterile cotton swab and allowed to dry for 5 min. Next, three representative wells were made using a well puncher on agar plates. The wells were then loaded with a maximum concentration of complexes 1–3i.e., 500 μg mL−1 (10 μL) and the plates were incubated at 37 °C for 24 h. After incubation, the inhibitory propensity of complexes on the growth of fungal and bacterial species was ascertained through the formation of a zone of inhibition around the wells.
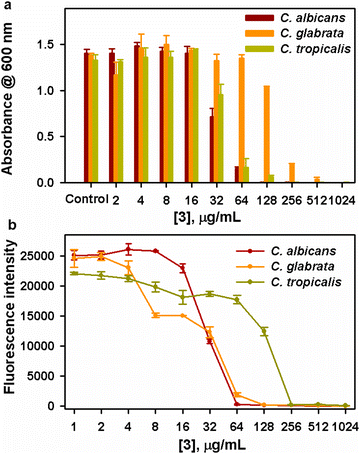
(B) Determination of minimum inhibitory concentration (MIC). The MIC of complexes against the test organisms was determined by broth microdilution assay following the CLSI guidelines described in the literature with required modifications.29 In brief, 1% standard culture suspensions of test organisms were inoculated in 96 well microtiter plates (MTPs) containing 200 μL of suitable broth (either YEPD or BHI) with varying concentrations of active complexes from 0 to 1024 μg mL−1. The plates were then incubated at 37 °C for 24 h. After incubation, the MIC of complexes was determined by measuring the optical density at 600 nm. MIC is defined as minimal concentrations of drugs that inhibit visible growth of microorganisms in the assay medium.
(C) Determination of minimum fungicidal/bactericidal concentration (MFC/MBC). To determine the effective concentrations of complexes that completely block the growth of test organisms, a spot assay was performed. Two microlitres of the aliquots from each well in broth microdilution assay were spotted on the YEPD/BHI agar plates and incubated at 37 °C for 48 h. As a result, MFC/MBC was defined as the lowest concentration of drug that showed no viable growth on YEPD/BHI agar plates to obtain approximately 99–99.5% killing activity.
(D) Metabolic viability assay. Additionally, the Alamar blue assay was employed to determine the actual metabolic state of test microorganisms during manifestation with the active complexes as previously described in the literature with slight modifications.30 Briefly, all the test pathogens were exposed to the active complexes for 24 h at 37 °C. The wells added with test organisms without any compound exposure served as a control. After incubation, the cell pellets were obtained through centrifugation of culture suspension at 8000 rpm for 10 min and resuspended with 200 μL of sterile phosphate-buffered saline (PBS). Alamar blue at 20 μg mL−1 concentration was added to each cell pellet suspended in PBS and incubated in the dark for 4–8 h. Subsequently, the fluorescence intensity was taken at the excitation and emission wavelengths of 560 nm and 590 nm, respectively.
2.9. In silico studies
Molecular docking analysis was performed to computationally identify the predominant binding modes of compounds with the proteins of known three-dimensional structures. Exploring the optimal binding modes of compounds to the target of interest is a crucial step in the process. The analysis was carried out using the molecular docking program MGL tools 1.5.7 and AutoDock Vina.31 The protein sequences were downloaded from the protein data bank (PDB; https://www.rcsb.org./pdb). Docking studies were performed by considering the protonation states, removing interfering water molecules, setting appropriate atom and bond types, and accounting for the flexibility of amino acid side chains. The geometry optimization of the complexes was carried out using the Gaussian 09 software package at the B3LYP level of theory with the basis sets 6-31G for C, H, O, and N and LANL2DZ for the metal center.32 The grid box dimensions (35 Å × 35 Å × 35 Å) were developed in a way that encompasses the active site. The output from AutoDock Vina was rendered with Autodock tools 1.5.7 and Discovery studio.333. Results and discussion
3.1. Synthesis of Schiff base ligands and copper(II) complexes
Schiff base ligands L1(H)–L3(H) were synthesized by the condensation of N-benzylethylenediamine with salicylaldehyde derivatives as per the literature methods.22 Complexes 1–3 were prepared by treating the ligands with copper(II) perchlorate hexahydrate followed by the addition of sodium azide in methanol. The dark green complexes were filtered and washed with cold methanol and diethyl ether. The complexes were characterized by ESI-MS, and the molecular ion peaks of complexes 1–3, [(L1)Cu]+, [(L2)Cu]+ and [(L3)Cu]+, were observed at m/z 316.38, 330.42 and 394.29, respectively. The d–d bands observed in the UV-vis spectrum in the range of 610–620 nm and the axial symmetry EPR spectra confirm the square-based geometry of complexes 1–3 in solution. Moreover, the molecular structures of complexes 1 and 2 were characterized by a single crystal X-ray diffraction study, and both 1 and 2 were found to possess a distorted square pyramidal geometry upon considering a mono copper centre. 1 is a dimeric complex with azide bridges and formulated as [Cu(L1)(N3)]2 and complex 2 is a 1D polymeric form with azide ion coordinates in a 1,3-end-to-end mode as represented in [Cu(L2)N3]n.3.2. Vibrational and electronic properties of complexes
The IR spectra of the ligands and complexes were recorded in ATR and FT-IR modes, respectively. The comparison between the IR spectra of ligands and complexes is presented in Fig. S1–S3 in the ESI.† In the IR spectra of L1(H)–L3(H), the bands located at 1631–1635 cm−1 indicate the imine formation in the ligands. Upon comparing the IR spectra of the ligands and complexes, it was observed that the N–H stretching frequencies of the complexes were shifted to a lower frequency than the corresponding ligands due to the weakening of the N–H bond after nitrogen donates its lone pair to the copper(II) centre for forming a complex. The bands observed between 2025 and 2047 cm−1 in the complexes confirm the coordination of azide to the copper(II) center.22,34 Particularly, the absorption band at 2028 cm−1 and 2025 cm−1 for 2 and 3 strongly indicates the occurrence of azide as a terminal ion. In contrast, an intense absorption band for complex 1 at 2047 cm−1 suggests the possibility of an azide bridge in the structure that was later confirmed using single crystal XRD. Nevertheless, the respective peak was present at a relatively lower frequency than the μ2-1,1-azide bridge, which can be attributed to the asymmetric nature of the azide bridges in complex 1.34The electronic spectra of complexes 1–3 were recorded at 0.1 mM in methanol at room temperature (Fig. 1). The UV-vis spectrum of the complexes showed two absorptions corresponding to ligand to metal charge transfer (LMCT) and d–d transitions. The strong peak around 360–376 nm was due to the LMCT transition from phenolate oxygen to copper(II). The d–d transitions were observed around 610–620 nm; the observations are consistent with the square-based geometry of copper(II) complexes.22,35,36 Moreover, all the complexes exhibit similar UV-vis spectral patterns in solution, which suggests similar coordination around the copper centre in solution.
Furthermore, complexes 1–3 were characterized by EPR spectroscopy in methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMF (4
DMF (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) at 77 K. A well-defined hyperfine splitting in the g‖ area (Fig. 1(b) and Table 2) was observed in all cases.37 The axial EPR signals are consistent with the copper(II) complexes in a ground state of dx2−y2 as revealed by g‖ > g⊥ > 2.011 (g‖ = 2.239–2.244 and g⊥ = 2.012–2.013).38,39 The observed g-values are within the range of g‖ > 2.1 > g⊥ > 2.0 revealing the square planar coordination geometry around the copper(II) centre.39 The coupling constant (A‖) values of the unpaired electron with the copper(II) centre depends on the Fermi contact, spin and orbital dipolar terms.40 Moreover, the g-tensors and the hyperfine coupling constant values do not show much deviation, which clearly suggests that the present complex geometries were not altered much in solution.
1) at 77 K. A well-defined hyperfine splitting in the g‖ area (Fig. 1(b) and Table 2) was observed in all cases.37 The axial EPR signals are consistent with the copper(II) complexes in a ground state of dx2−y2 as revealed by g‖ > g⊥ > 2.011 (g‖ = 2.239–2.244 and g⊥ = 2.012–2.013).38,39 The observed g-values are within the range of g‖ > 2.1 > g⊥ > 2.0 revealing the square planar coordination geometry around the copper(II) centre.39 The coupling constant (A‖) values of the unpaired electron with the copper(II) centre depends on the Fermi contact, spin and orbital dipolar terms.40 Moreover, the g-tensors and the hyperfine coupling constant values do not show much deviation, which clearly suggests that the present complex geometries were not altered much in solution.
3.3. Molecular structure description of copper(II) complexes 1 and 2
The crystal structures of complexes [Cu(L1)(N3)]2, 1, and [Cu(L2)N3]n, 2 are shown in Fig. 2, together with the atom numbering scheme. Complex 1 features an azido-bridged copper(II) dimer complex, whereas complex 2 features a 1D coordination polymer where the azide ion bridges between two copper(II) centers and coordinates in a 1,3-end-to-end mode (Fig. 2c). Furthermore, complex 1 crystallizes in the space group P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) in a triclinic crystal system, whereas complex 2 crystallizes in the space group Pccn in an orthorhombic crystal system as shown in Table 1. The significant bond lengths and bond angles are provided in Table 3.
in a triclinic crystal system, whereas complex 2 crystallizes in the space group Pccn in an orthorhombic crystal system as shown in Table 1. The significant bond lengths and bond angles are provided in Table 3.
| Bond description | 1 | 2 |
|---|---|---|
| Cu(1)–O(1) | 1.9020(17) | 1.910(4) |
| Cu(1)–N(1) | 1.9281(19) | 1.946(4) |
| Cu(1)–N(3) | 1.987(2), 2.534(38) | 1.978(5) |
| Cu(1)–N(2) | 2.047(2) | 2.049(4) |
| N(1)–C(7) | 1.284(3) | 1.282(6) |
| N(1)–C(8) | 1.462(3) | 1.458(6) |
| O(1)–Cu(1)–N(1) | 94.54(8) | 92.19(16) |
| O(1)–Cu(1)–N(3) | 92.44(8), 92.64(38) | 89.17(18) |
| N(1)–Cu(1)–N(3) | 165.79(8), 104.58(77) | 174.28(19) |
| O(1)–Cu(1)–N(2) | 176.55(7) | 176.04(15) |
| N(1)–Cu(1)–N(2) | 84.16(9) | 84.52(17) |
| N(3)–Cu(1)–N(2) | 89.53(9), 84.61(4) | 93.90(18) |
Complex 1: the structural characterization clarifies that complex 1 formed a centrosymmetric dimer with five-coordinate bonds on each copper(II) center where the three bonds are from the Schiff base ligand and two from bridging azide anions as shown in Fig. 2a. The structure reveals that the basal plane contains three nitrogen atoms and one oxygen atom. The remaining fifth coordination site was occupied by the nitrogen atom from the other azide group. Correspondingly, the symmetry-related azide group on the equatorial side, along with the azide from the basal plane, forms a Cu–Cu bridge (3.289 Å). The smaller internuclear distance between the copper atoms (3.289 Å) may favor the end-on fashion of the azide bridge.35
The coordination environment around complex 1 possesses a distorted square pyramidal geometry. The distortion in the coordination geometry from square pyramidal to trigonal bipyramidal was evaluated using Addison's trigonality index (τ5) parameter41 value of 0.179, [τ = (β − α/60), where β = O(1) − Cu(1) − N(2) = 176.55° and α = N(1) − Cu(1) − N(3) = 165.79°]. The τ value is zero for a perfect square pyramidal geometry, while it becomes unity for a perfect trigonal bipyramidal geometry. The Cu–Nazide–Cu angle was similar to those of the reported end-on double azido bridged binuclear complexes. Moreover, the difference in the bond lengths of Cu–Nazide (2.534 Å) and Cu–Nazide (1.987 Å) was also observed in the previously reported complexes having the Cu–Nazide bridge.34,42
Complex 2: each unit of the 1D polymer of complex 2 surrounding copper(II) possesses a distorted square pyramidal geometry with the trigonality index (τ5)41 value of 0.029, [τ = (β − α/60), where β = O(1) − Cu(1) − N(2) = 176.04° and α = N(1) − Cu(1) − N(3) = 174.28°]. The ascertained bond angles are in the ranges of 84.52–93.90° and 174.28–176.04°, which indicate the deviation from the ideal square pyramidal geometry where the angles are 90° and 180°. Preferably, the Cu–N2amine (2.049(4) Å) bond is slightly longer than the Cu–N1imine (1.946(4) Å) and Cu–N1azide bonds (1.978(5) Å), owing to the sp3 and sp2 hybridization, respectively. Moreover, the bond distance between two adjacent copper(II) centres of the polymeric form is observed as 4.609 Å. Furthermore, the Cu–O bond (1.910(4) Å) is shorter than the Cu–N bond (2.049–1.946 Å), indicating a stronger coordination of phenolate oxygen than the nitrogen atom. Furthermore, the Cu–N and Cu–O bond lengths are in the ranges observed for copper(II) complexes reported previously.22,34,42
3.4. Lipophilicity
The relative lipophilicity of the complex is one of the significant physicochemical properties that is highly required for the complexes to enter into the bloodstream. An ideal drug/compound should pass through the hydrophobic lipid bilayer present in the cell membrane in one part of the cell and the hydrophilic aqueous cytosol layer in the other part. The lipophilicity of complexes 1–3 was determined by the shake-flask method. The obtained partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kow) reveals that complexes 2 (0.7896) and 3 (0.6028) are more lipophilic than complex 1 (−0.0882) (Fig. S7 in ESI†).43
Kow) reveals that complexes 2 (0.7896) and 3 (0.6028) are more lipophilic than complex 1 (−0.0882) (Fig. S7 in ESI†).43
3.5. Antimicrobial activity
| Test strains | Zone of inhibition (mm) | MIC (μg mL−1) | MBC/MFC (μg mL−1) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | |
| C. albicans | 3 | 12 | 18 | 512 | 128 | 64 | 1024 | 256 | 128 |
| C. glabrata | NA | 12 | 15 | NA | 512 | 128 | NA | 512 | 256 |
| C. tropicalis | NA | 10 | 15 | NA | 512 | 64 | NA | 512 | 128 |
| P. aeruginosa | 10 | 15 | 18 | 512 | 64 | 128 | 512 | 64 | 256 |
| MRSA | 4 | 13 | 23 | 1024 | 128 | 256 | NA | 256 | 256 |
Based on the study, we can assert that the activity of complexes varied with the substituents on the donor moiety. Notably, the previous studies showed that the systems with substitutions like Br, Cl, OMe, Me, etc are better antimicrobial agents than the respective parent (H) systems.45 Furthermore, the reports on quinazoline, morpholine, and BODIPY have proved that the systems with brominated derivatives exhibited greater activity against distinct fungi owing to their electron-withdrawing nature and heavy atom effect.46 It is worth mentioning that we also observed a similar trend in the activity and this may be explained using Overtone's concept.47 This concept shows the importance of lipophilicity in controlling antimicrobial activity and asserts that the lipid membrane that surrounds the cell favors the passage of only the lipid-soluble materials. When it comes to our studies, complexes 2 and 3 are more lipophilic than parent complex 1, which is reflected in their activity. To evaluate further, the analysis based on the mode of interactions by each moiety with the different protein strains was carried out by the in silico method.
3.6. In silico mechanistic investigation
The in silico studies were performed to simulate the interaction and affinity of the newly studied complexes 1–3 toward different bacterial and fungal strains. To validate the mode of action and predict the binding abilities of complexes, different drug target proteins were selected for the studied strains. In particular, N-myristoyl transferase (PDB ID 1iyl), dihydrofolate reductase (DHFR) (PDB ID 4hof), secreted aspartic protease (PDB ID 3q70) from C. albicans, oxidoreductase (PDB ID 5jlc) from C. glabrata, and telomerase reverse transcriptase (PDB ID 6zd2) from C. tropicalis were selected as antifungal targets, and transcriptional activator protein (PDB ID 2uv0), aminopeptidase P (PDB ID 5wze), nucleotidyl transferase (PDB ID 6p8j), protein binding sites (PDB IDs 6p8o and 6p8s), nucleotidyltransferase (PDB ID 6p8u) from P. aeruginosa and transferase (PDB IDs 5tz8, 5tzi, and 5tze) from MRSA were selected as the antibacterial targets. The potencies of the complexes were analyzed computationally by evaluating their docking score. Interestingly, all these proteins showed good interactions that agree with the experimental results. The docking scores obtained against tested active sites are presented in Table 5. Molecular docking results suggest that these complexes inhibited the target proteins effectively, with the appearance of a substantial number of hydrogen bonds and electrostatic interactions.| Sl no | Protein strains | 1 | 2 | 3 |
|---|---|---|---|---|
| Candida albicans | ||||
| 1 | 1iyl | −8.6 | −17 | −9.2 |
| 2 | 4hof | −5 | −5.2 | −7.3 |
| 3 | 3q70 | −6.1 | −6.4 | −6.6 |
| Candida glabrata | ||||
| 4 | 5jlc | −5.4 | −6 | −7.5 |
| Candida tropicalis | ||||
| 5 | 6zd2 | −4.7 | −5.1 | −8.8 |
| Pseudomonas aeruginosa | ||||
| 6 | 2uv0 | −4.4 | −6.4 | −5 |
| 7 | 5wze | −6.7 | −7.8 | −6.8 |
| 8 | 6p8j | −5.5 | −6.2 | −6 |
| 9 | 6p8o | −5.7 | −5.8 | −5.6 |
| 10 | 6p8s | −5.4 | −6.7 | −6.5 |
| 11 | 6p8u | −6.2 | −6.4 | −6.3 |
| MRSA | ||||
| 12 | 5tz8 | −7.7 | −7.9 | −7.7 |
| 13 | 5tzi | −7.5 | −7.8 | −7.7 |
| 14 | 5tze | −6.2 | −7.1 | −6.4 |
| 15 | 5tzj | −6.1 | 0 | 0 |
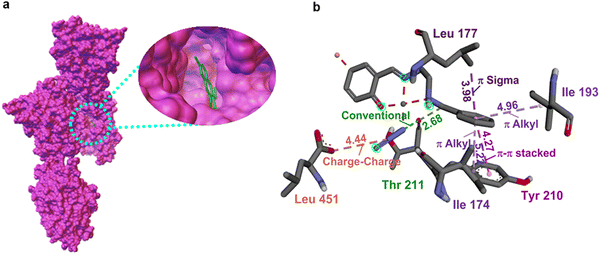
Inhibiting the activity of the dihydrofolate reductase (DHFR) enzyme was demonstrated as an effective approach for both unicellular and multicellular organisms. The inhibition causes a blockade in thymidine synthesis leading to cell death.51 While considering the interactions of the complexes with the dihydrofolate reductase protein (4hof), complex 3 exhibits two hydrogen bonds of N1–Asn(123) and phenolate(O)–Asn(123), whereas no such bond was observed in the case of complexes 1 and 2. Instead, a conventional bond observed within the complex, which occurred between azide(N) and phenolate(O) with the bond distances of 2.84 Å for 1 and 2.92 Å for 2, might have resulted in the reduction of their affinity towards the protein strains. Additionally, π–alkyl interactions have also been observed between benzyl groups with Lys(150) and Pro(152) in complex 3 and a single benzyl–Pro(46) and benzyl–Lys(158) interaction in complexes 1 and 2. Upon close analysis of the interactions of complexes with the aspartic protease (3q70) unit, we can see that the system exhibits various forms of hydrogen bonding interactions. Complex 3 (−6.6 kcal mol−1) alone can exhibit such interactions, which include N1–Asn(160), O–Asn(160), N1–Asn(160), N1–Thr(160), N2–Thr(160) and N1–phenolate(O).
In the case of 2, two types of hydrogen bonding interactions were observed; one was found between N1 and Gln(295) and the other one was found within the complex itself, which was between phenolate(O) and azide(N). A similar hydrogen bonding interaction was observed for complex 1 as well, where it was directed towards azide(N) from N2–H. Moreover, π–anionic interactions are observed between benzyl carbon and Asp(245) and Asp(299) in complex 2 and also a charge–charge correlation between azide(N)–Asp(299) and Asp(58) was observed for 2 and 1. As shown in Table 5, we can find that complex 3 shows high binding affinity towards the significant C. albicans strains (PDB IDs 1iyl, 4hof, and 3q70) via forming effective interaction with the negative energy scores of −9.2, −7.3, and −6.6 kcal mol−1. Altogether, the complexes exhibit the trend in the affinity values as 3 > 2 > 1, and the order of molecular interactions is very much in line with the experimental observations.
Analogous to the previous study, complex 3 displayed more negative docking scores of −7.5 kcal mol−1 against C. glabrata and −8.8 kcal mol−1 against C. tropicalis as the domains used are (oxidoreductase and telomerase reverse transcriptase) prominent for cell stability and activity.52 When the protein oxidoreductase (5jlc) was used, complex 3 displayed a hydrogen bonding interaction between N1 and Pro(202) along with a charge–charge interaction between azide(N) and Asp(234). A π–σ interaction exists in the complex between phenyl carbon and Ile(206). Complex 2 forms hydrogen bonding with N2–H–Asp(234). Moreover, we have not detected any effective conventional form of interactions in complex 1 towards the protein strains. All the complexes showed different π–alkyl correlations between the phenyl groups of the complexes and amino acids like Ala(227), Leu(222), Pro(202), Ala(321), and Pro(365). Analogously, the treatment with telomerase reverse transcriptase (6zd2) results in the formation of different hydrogen bonds like phenolate(O)–Arg(181)/Tyr(177)/azide(N) using 1 and 2 and Arg(181), N2–H–Val(397) using 3 along with the π–alkyl correlations. The affinity data and the interactions of 2 and 3 substantiate that the in silico studies are very much in correlation with the experimental results. Although we have obtained moderate binding energy values for complex 1, the in vitro studies prove that the complex has a less inhibiting nature towards fungi, specifically towards C. glabrata and C. tropicalis. In a way, this emphasizes the significance of various substituents on the ligand moiety in tuning the inhibitory properties.48,49 In fact, there always exist speculations regarding the relationship between activity and the inhibitor size or lipophilicity, variations in transport, or the method followed that might be crucial for antifungal efficacy.53 A German company has developed a class of powerful benzyl(oxy)pyrimidine-based inhibitors against C. albicans. We have also verified that the complexes are not vulnerable to efflux; instead, we speculated that the complexes are not able to enter C. albicans.54
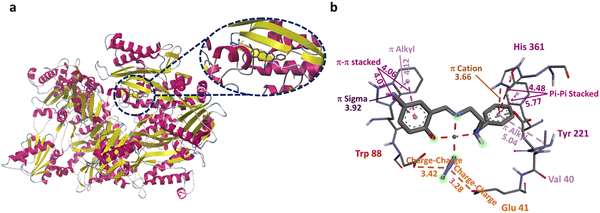 | ||
| Fig. 6 (a) The two-dimensional depiction of molecular docking of 2 in the active site of Pseudomonas aeruginosa aminopeptidase (5wze). (b) The interaction distance is given in Å. | ||
Another significant inhibition strategy was blocking the correlation between proteins and carbohydrates. It was substantiated that when complexes are tried against the strand 2uvo, carbohydrate binding protein,56 complex 2 showed good interaction, in line with the experimental data. No effective hydrogen bonding interactions are found in complex 1 except an internal one like N2(H)–azide(N), whereas complex 2 showed phenolate(O)–Tyr(157) interaction and complex 3 exhibited N2(H)–Phen(167)/Glu(168), and Azide(N)–Phen(167) interactions. Apart from these interactions, π–alkyl interactions were observed in 1 and 3, whereas complex 2 was involved in π–cation and π–donor interactions within the complex, that is, between phenyl and azide(N). Considering the interaction of complexes against 6p8j, a signalling protein,57 complex 2 displayed strong interaction by forming hydrogen bonds with phenolate(O)–Tyr(124), azide(N)–Tyr(124), phenolate(O)–azide(N), and azide(N)–OH2, whereas complex 3 formed azide(N)–Tyr(124) and phenolate(O)–azide(N) interactions and less interactive complex 1 formed phenolate(O)–azide(N) and N2(H)–azide(N) correlations. There are also different forms of π–alkyl interactions of phenyl groups with Arg(20), Arg(14) (2 and 3) and Ala(1) (1). In the case of 6p8o,57 conventional binding was observed between phenolate(O) and azide(N) for all complexes and additionally N2(H)–azide(N) for complex 1. Apart from these, π–alkyl and π–cationic connections were also observed between the phenyl group and Lys(104)/Lys(100). The complexes show similar activity against 6p8s as well.58 Interestingly, complex 2 exhibits relatively higher interrelations using the methyl moiety with amino acids like Leu(30), Lys(104), and Ileu(103). Other than these interactions, many predominant interactions are detected using 2 like phenolate(O)–Arg(102) and N2(H)–Asp(192) along with phenyl group–H2O (π-donor) connections. Complexes 1 and 3 showed N2(H)–azide(N) and phenolate(O)–azide(N) interactions apart from the phenolate(O)–Arg(102) interaction for 1 and phenolate(O)–Lys(296) interaction for 3. Analogously, the complexes showed similar interactions with MRSA as well.58 It is noteworthy to mention that complex 2 showed better activity against all the bacterial strains (vide supra), which is in line with the experimental results. Intrinsically, the methyl group in 2 is also involved in additional alkyl interactions (Fig. S21 in ESI†) compared to complexes 1 and 3. The observed interaction of the methyl moiety in 2 with distinct amino acid groups might be the reason for their higher affinity value in bacterial strains. These findings reveal the importance of substitution at the salicylaldehyde scaffold. Additionally, the experimental data of complex 1 show no activity against MRSA, but the docking results show a favorable affinity towards the transferase strands. Upon careful analysis, we can see that complex 1 only participated in weak hydrophobic interactions. The negative impact of antibacterial activities of 1 may be either due to the inability of this complex to penetrate through the bacterial cell wall, thereby preventing the interactions in biological processes, or due to dissemination and inactivation by some unknown cellular mechanisms involving bacterial enzymes.59 To confirm whether complex 1 is capable of interacting with any membrane-bound proteins, we used a membrane-bound transglycolase strain (PDB ID 3vmt).55 As expected, no interactions were observed with the complex.60,61
Overall, in silico studies with different types of protein domains reveal that complexes 1–3 are capable of stimulating inhibition to virulent microbes. More precisely, considering the entire studies performed against fungal strains, we can collectively deduce that the complexes especially 3 can either affect the interactions of the hydrophobic part of proteins owing to their increased lipophilicity or else reduce the stability as well as the proper functioning of the cell by inhibiting various processes. When it comes to the activity of complexes against bacteria, the complexes can interact well with virulent genes, signalling proteins, and carbohydrate binding proteins and also affect the activities of transferase proteins. Altogether, the complexes can impede the functioning of unwanted microbial strains even though their mode of action is different at each section of the cell. Since the complexes operate differently in each domain, a careful analysis of their activity is needed for the selective functioning of the complexes.
4. Conclusion
In the present study, a series of Schiff-base copper(II) complexes were synthesised and characterized using modern techniques. In addition, three different bioassays were adapted for evaluating the antimicrobial efficacy of complexes. The MIC, MBC and MFC values determined for 1, 2 and 3 are comparable with those values reported in the literature (250 to 500 μg mL−1). Alamar blue assay confirmed the inability of 1 to lyse the cells and showed the significance of substitutions at the 2nd and 5th positions in the aryl moiety. In support of these experimental results, the in silico studies were performed and verified with distinct protein microbial strains. The interactions and reactivity patterns are as expected and consistent with the in vitro studies. As alluded to above, the complexes show specific inhibitory functions towards each strain of microbes. Such mechanistic details are profoundly studied further using active virulent microbes with great attention.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
M. S. sincerely thanks the Department of Science and Technology for the DST-Inspire Faculty Award (IFA17-CH286) and the Science and Engineering Research Board (SERB) for the CRG grant (CRG/2023/002850). T. M. sincerely acknowledges the Ministry of Education and National Institute of Technology (NIT) Calicut for her PhD (GATE) fellowship. We would like to sincerely thank Prof. Abhishek Dey, Indian Association for the Cultivation of Science, Kolkata for helping us to perform mass spectrometry and CHN analysis. We would like to thank the Instrumentation facility, Central Leather Research Institute for helping us to record mass and EPR spectra. Furthermore, we sincerely acknowledge the NIT Calicut for providing other research facilities.References
- (a) I. Yousuf, M. Bashir, F. Arjmand and S. Tabassum, Advancement of Metal Compounds as Therapeutic and Diagnostic Metallodrugs: Current Frontiers and Future Perspectives, Coord. Chem. Rev., 2021, 445, 214104 CrossRef CAS; (b) E. J. Anthony, E. M. Bolitho, H. E. Bridgewater, O. W. L. Carter, J. M. Donnelly, C. Imberti, E. C. Lant, F. Lermyte, R. J. Needham, M. Palau, P. J. Sadler, H. Shi, F.-X. Wang, W.-Y. Zhang and Z. Zhang, Metallodrugs are Unique: Opportunities and Challenges of Discovery and Development, Chem. Sci., 2020, 11, 12888–12917 RSC; (c) A. Frei, Metal Complexes, an Untapped Source of Antibiotic Potential?, Antiobiotics, 2020, 9, 90 CrossRef CAS; (d) I. Salah, I. P. Parkin and E. Allan, Copper as an Antimicrobial Agent: Recent Advances, RSC Adv., 2021, 11, 18179–18186 RSC; (e) G. Gasser, I. Ott and N. Metzler-Nolte, Organometallic Anticancer Compounds, J. Med. Chem., 2011, 54, 3–25 CrossRef CAS; (f) S. D. Roughley and A. M. Jordan, The Medicinal Chemist's Toolbox: An Analysis of Reactions used in the Pursuit of Drug Candidates, J. Med. Chem., 2011, 54, 3451–3479 CrossRef CAS.
- (a) S. S. Harmalkar, R. J. Butcher, V. V. Gobre, S. K. Gaonkar, L. R. D'Souza, M. Sankaralingam, I. Furtado and S. N. Dhuri, Synthesis, Characterization and Antimicrobial Properties of Mononuclear Copper(II) Compounds of N,N′-di(quinolin-8-yl)cyclohexane-1,2-diamine, Inorg. Chim. Acta, 2019, 496, 119020 CrossRef; (b) A. Das, A. Rajeev, S. Bhunia, M. Arunkumar, N. Chari and M. Sankaralingam, Synthesis, Characterization and Antimicrobial Activity of Nickel(II) Complexes of Tridentate N3 Ligands, Inorg. Chim. Acta, 2021, 526, 120515 CrossRef CAS; (c) A. Das, T. P. Mohammed, R. Kumar, S. Bhunia and M. Sankaralingam, Carbazole Appended Trans-Dicationic Pyridinium Porphyrin finds Supremacy in DNA Binding/Photocleavage over a Non-carbazolyl Analogue, Dalton Trans., 2022, 51, 12453–12466 RSC; (d) A. Das and M. Sankaralingam, Are Zn(II) pincer complexes efficient apoptosis inducers? a deep insight into their activity against A549 lung cancer cells, Dalton Trans., 2023, 52, 14465–14476 RSC; (e) A. Das, T. P. Mohammed and M. Sankaralingam, Biological Activity of Copper Porphyrins, Coord. Chem. Rev., 2024, 506, 215661 CrossRef CAS.
- (a) Y. Jia and J. Li, Molecular Assembly of Schiff Base Interactions: Construction and Application, Chem. Rev., 2015, 115, 1597–1621 CrossRef CAS; (b) M. Pervaiz, S. Sadiq, A. Sadiq, U. Younas, A. Ashraf, Z. Saeed, M. Zuber and A. Adnan, Azo-Schiff Base Derivatives of Transition Metal Complexes as Antimicrobial Agents, Coord. Chem. Rev., 2021, 447, 214128 CrossRef CAS; (c) L. H. Abdel-Rahman, A. M. Abu-Dief, M. Basha and A. A. H. Abdel-Mawgoud, Three novel Ni(II), VO(II) and Cr(III) Mononuclear Complexes Encompassing Potentially Tridentate Imine Ligand: Synthesis, Structural Characterization, DNA Interaction, Antimicrobial Evaluation and Anticancer Activity, Appl. Organomet. Chem., 2017, e3750 CrossRef; (d) S. Adsule, V. Barve, D. Chen, F. Ahmed, Q. P. Dou, S. Padhye and F. H. Sarkar, Novel Schiff Base Copper Complexes of Quinoline-2 Carboxaldehyde as Proteasome Inhibitors in Human Prostate Cancer Cells, J. Med. Chem., 2006, 49, 7242–7246 CrossRef CAS.
- (a) W. Kaim and B. Schwedersk. Bioinorganic Chemistry: Inorganic Elements in the Chemistry of Life, Wiley, 1st edn, 1994 Search PubMed; (b) A. Rajeev, M. Balamurugan and M. Sankaralingam, Rational Design of First-Row Transition Metal Complexes as the Catalysts for Oxidation of Arenes: A Homogeneous Approach, ACS Catal., 2022, 12, 9953–9982 CrossRef CAS; (c) T. P. Mohammed and M. Sankaralingam, Reactivities of High Valent Manganese-Oxo Porphyrins in Aqueous Medium, Tetrahedron, 2022, 103, 132483 CrossRef CAS; (d) M. Sankaralingam, M. Balamurugan and M. Palaniandavar, Alkane and Alkene Oxidation Reactions Catalyzed by Nickel(II) Complexes: Effect of Ligand Factors, Coord. Chem. Rev., 2020, 403, 213085 CrossRef; (e) M. Sankaralingam, Y.-M. Lee, W. Nam and S. Fukuzumi, Amphoteric Reactivity of Metal–Oxygen Complexes in Oxidation Reactions, Coord. Chem. Rev., 2018, 365, 41–59 CrossRef CAS; (f) A. Rajeev and M. Sankaralingam, Highlights of Oxygen Atom Transfer Reactions Catalysed by Nickel Complexes, Oxygen Atom Transfer Reactions, 2023, 1, 62–90 Search PubMed.
- (a) L. H. Abdel-Rahman, A. M. Abu-Dief, M. S. S. Adam and S. K. Hamdan, Some New Nano-Sized Mononuclear Cu(II) Schiff Base Complexes: Design, Characterization, Molecular Modeling and Catalytic Potentials in Benzyl Alcohol Oxidation, Catal. Lett., 2016, 146, 1373–1396 CrossRef CAS; (b) M. Montazerozohori, S. Farrokhiyani, A. Masoudiasl and J. M. White, Crystal Structures, Hirshfeld Surface Analyses and Thermal Behavior of Two New Rare Tetrahedral Terminal Zinc(II) Azide and Thiocyanate Schiff Base Complexes, RSC Adv., 2016, 6, 23866–23878 RSC.
- (a) J. Valentová, S. Varényi, P. Herich, P. Baran, A. Bilková, J. Kozíšek and L. Habala, Synthesis, Structures and Biological Activity of Copper(II) and Zinc(II) Schiff Base Complexes Derived from Aminocyclohexane-1-Carboxylic Acid. New Type of Geometrical Isomerism in Polynuclear Complexes, Inorg. Chim. Acta, 2018, 480, 16–26 CrossRef; (b) E. L. de Araújo, H. F. G. Barbosa, E. R. Dockal and É. T. G. Cavalheiro, Synthesis, Characterization and Biological Activity of Cu(II), Ni(II) and Zn(II) Complexes of Biopolymeric Schiff Bases of Salicylaldehydes and Chitosan, Int. J. Biol. Macromol., 2017, 95, 168–178 CrossRef; (c) T.-J. Khoo, M. K. Break, K. A. Crouse, M. I. M. Tahir, A. M. Ali, A. R. Cowley, D. J. Watkin and M. T. H. Tarafder, Synthesis, Characterization and Biological Activity of Two Schiff Base Ligands and their Nickel(II), Copper(II), Zinc(II) and Cadmium(II) Complexes Derived from S-4-Picolyldithiocarbazate and X-Ray Crystal Structure of Cadmium(II) Complex Derived From Pyridine-2-Carboxaldehyde, Inorg. Chim. Acta, 2014, 413, 68–76 CrossRef CAS; (d) W.-J. Lian, X.-T. Wang, C.-Z. Xie, H. Tian, X.-Q. Song, H.-T. Pan, X. Qiao and J.-Y. Xu, Mixed-ligand Copper(II) Schiff Base Complexes: The Role of the Co-Ligand in DNA Binding, DNA Cleavage, Protein Binding and Cytotoxicity, Dalton Trans., 2016, 45, 9073–9087 RSC.
- (a) T. Ueno, M. Ohashi, M. Kono, K. Kondo, A. Suzuki, T. Yamane and Y. Watanabe, Crystal Structures of Artificial Metalloproteins: Tight Binding of FeIII (Schiff-Base) by Mutation of Ala71 to Gly in Apo-Myoglobin, Inorg. Chem., 2004, 43, 2852–2858 CrossRef CAS; (b) S. Pal, A. K. Barik, S. Gupta, A. Hazra, S. K. Kar, S.-M. Peng, G.-H. Lee, R. J. Butcher, M. S. E. Fallah and J. Ribas, Copper(II) Mediated Anion Dependent Formation of Schiff Base Complexes, Inorg. Chem., 2005, 44, 3880–3889 CrossRef CAS; (c) K. A. Maher, and S. R. Mohammed, Metal Complexes of Schiff Base Derived from Salicylaldehyde - A Review, Int. J. Cur. Res. Rev., 2015, 7, 6–16 CAS.
- (a) K. C. Gupta and A. K. Sutar, Catalytic Activities of Schiff Base Transition Metal Complexes, Coord. Chem. Rev., 2008, 252, 1420–1450 CrossRef CAS; (b) S. Thakurta, J. Chakraborty, G. Rosair, R. J. Butcher and S. Mitra, The Interplay of O–H⋯O Hydrogen Bonding in the Generation of Three New Supramolecular Complexes of CuII, NiII and CoIII: Syntheses, Characterization and Structural Aspects, Inorg. Chim. Acta, 2009, 362, 2828–2836 CrossRef CAS; (c) B. Weber, Spin Crossover Complexes with N4O2 Coordination Sphere – The Influence of Covalent Linkers on Cooperative Interactions, Coord. Chem. Rev., 2009, 253, 2432–2449 CrossRef CAS; (d) I. P. Ejidike and P. A. Ajibade, Transition Metal Complexes of Symmetrical and Asymmetrical Schiff Bases as Antibacterial, Antifungal, Antioxidant, and Anticancer Agents: Progress and Prospects, Rev. Inorg. Chem., 2015, 35, 191–224 CAS; (e) M. S. More, P. G. Joshi, Y. K. Mishra and P. K. Khanna, Metal complexes driven from Schiff bases and semicarbazones for biomedical and allied applications: a review, Mater. Today Chem., 2019, 14, 100195 CrossRef CAS.
- (a) M. Pervaiz, S. Sadiq, A. Sadiq, U. Younas, A. Ashraf, Z. Saeed, M. Zuber and A. Adnan, Azo-Schiff base derivatives of transition metal complexes as antimicrobial agents, Coord. Chem. Rev., 2021, 447, 214128 CrossRef CAS; (b) C. Mohan, V. Kumar, N. Kumari, S. Kumari, J. Yadav, T. Gandass and S. Yadav, Synthesis, Characterization and Antibacterial Activity of Semicarbazide based Schiff Bases and their Pb(II), Zr(IV) and U(VI) Complexes, Adv. J. Chem. Sec. B, 2020, 2, 187–196 Search PubMed; (c) A. Hameed, M. Al-Rashida, M. Uroos, S. Abid Ali and K. M. Khan, Schiff Bases in Medicinal Chemistry: A Patent Review (2010-2015), Exp. Opin. Ther. Pat., 2017, 27, 63–79 CrossRef CAS; (d) H. B. Howsaui, A. S. Basaleh, M. H. Abdellattif, W. M. Hassana and M. A. Hussien, Synthesis, Structural Investigations, Molecular Docking, and Anticancer Activity of Some Novel Schiff Bases and their Uranyl Complexes, Biomolecules, 2021, 11, 1138 CrossRef CAS; (e) M. T. Kaczmarek, M. Zabiszak, M. Nowak and R. Jastrzab, Lanthanides: Schiff Base Complexes, Applications in Cancer Diagnosis, Therapy, and Antibacterial Activity, Coord. Chem. Rev., 2018, 370, 42–54 CrossRef CAS.
- A. Sengul, B. Coban, P. Türkoğlu and P. Tutkun, DNA Binding Studies of Platinum(II) and Palladium(II) Complexes with Planar Tridentate 2,6-Bis(N-R-benzimidazol-2-yl)pyridine Ligands, Pharm. Chem. J., 2017, 4, 15–27 CAS.
- (a) M. Krátký, M. Dzurková, J. Janoušek, K. Konečná, F. Trejtnar, J. Stolaříková and J. Vinšová, Sulfadiazine Salicylaldehyde-Based Schiff Bases: Synthesis, Antimicrobial Activity and Cytotoxicity, Molecules, 2017, 22, 1573 CrossRef; (b) A. B. Begum, N. D. Rekha, B. C. V. Kumar, V. L. Ranganatha and S. A. Khanum, Synthesis, Characterization, Biological and Catalytic Applications of Transition Metal Complexes Derived from Schiff Base, Bioorg. Med. Chem. Lett., 2014, 24, 3559–3564 CrossRef; (c) B. K. Singh, H. K. Rajour and A. Prakash, Synthesis, Characterization and Biological Activity of Transition Metal Complexes with Schiff Bases Derived from 2-Nitrobenzaldehyde with Glycine and Methionine, Spectrochim. Acta, Part A, 2012, 94, 143–151 CrossRef CAS; (d) B. S. Creaven, E. Czeglédi, M. Devereux, É. A. Enyedy, A. F.-A. Kia, D. Karcz, A. Kellett, S. McClean, N. V. Nagy, A. Noble, A. Rockenbauer, T. Szabó-Plánka and M. Walsh, Biological Activity and Coordination Modes of Copper(II) Complexes of Schiff Base-Derived Coumarin Ligands, Dalton Trans., 2010, 39, 10854–10865 RSC.
- (a) J. Ceramella, D. Iacopetta, A. Catalano, F. Cirillo, R. Lappano and M. S. Sinicropi, A Review on the Antimicrobial Activity of Schiff Bases: Data Collection and Recent Studies, Antibiotics, 2022, 11, 191 CrossRef CAS; (b) Q. U. A. Sandhu, M. Pervaiz, A. Majid, U. Younas, Z. Saeed, A. Ashraf, R. R. M. Khan, S. Ullah, F. Ali and S. Jelani, Schiff Base Metal Complexes as Anti-Inflammatory Agents, J. Coord. Chem., 2023, 76, 1094–1118 CrossRef CAS.
- A. Das, R. Sangavi, S. Gowrishankar, R. Kumar and M. Sankaralingam, Deciphering the Mechanism of MRSA Targeting Copper(II) Complexes of NN2 Pincer-Type Ligands, Inorg. Chem., 2023, 62, 18926–18939 CrossRef CAS.
- (a) X. Tai, X. Yin, Q. Chen and M. Tan, Synthesis of Some Transition Metal Complexes of a Novel Schiff Base Ligand Derived from 2,2'-bis(p-methoxyphenylamine) and Salicylicaldehyde, Molecules, 2003, 8, 439–443 CrossRef CAS; (b) S. Slassi, A. El-Ghayoury, M. Aarjane, K. Yamni and A. Amine, New Copper(II) and Zinc(II) Complexes Based on Azo Schiff Base Ligand: Synthesis, Crystal Structure, Photoisomerization Study and Antibacterial Activity, Appl Organometal. Chem., 2020, e5503 CrossRef CAS; (c) S. Mahato, N. Meheta, M. Kotakonda, M. Joshi, P. Ghosh, M. Shit, A. R. Choudhury and B. Biswas, Ligand Directed Synthesis of a Unprecedented Tetragonalbipyramidal Copper(II) Complex and its Antibacterial Activity and Catalytic Role in Oxidative Dimerisation of 2-Aminophenol, Appl. Organomet. Chem., 2020, e5935 CrossRef CAS.
- Y. Xua, Y. Shia, F. Leia and L. Dai, A Novel and Green Cellulose-based Schiff Base-Cu(II) Complex and its Excellent Antibacterial Activity, Carbohydr. Polym., 2020, 230, 115671 CrossRef.
- (a) H. A. Althobiti and S. A. Zabin, New Schiff Bases of 2-(quinolin-8-yloxy) acetohydrazide and their Cu(II), and Zn(II) Metal Complexes: their in vitro antimicrobial potentials and in silico physicochemical and pharmacokinetics properties, Open Chem. J., 2020, 18, 591–607 CAS; (b) P. Kavitha, M. Saritha and K. L. Reddy, Synthesis, Structural Characterization, Fluorescence, Antimicrobial, Antioxidant and DNA Cleavage Studies of Cu(II) Complexes of Formyl Chromone Schiff bases, Spectrochim. Acta, Part A, 2013, 102, 159–168 CrossRef CAS.
- (a) H. Kargar, F. A. Meybodi, R. B. Ardakani, M. R. Elahifard, V. Torabi, M. F. Mehrjardi, M. N. Tahir, M. Ashfa and K. S. Munawar, Synthesis, Crystal Structure, Theoretical Calculation, Spectroscopic and Antibacterial Activity Studies of Cu(II) Complexes Bearing Bidentate Schiff Base Ligands Derived from 4-aminoantipyrine: Influence of Substitutions on Antibacterial Activity, J. Mol. Struct., 2021, 1230, 129908 CrossRef CAS; (b) H. Kargar, F. A. Meybodi, M. R. Elahifard, M. N. Tahir, M. Ashfaq and K. S. Munawar, Some New Cu(II) Complexes Containing O, N-donor Schiff Base Ligands Derived from 4-aminoantipyrine: Synthesis, Characterization, Crystal Structure and Substitution Effect on Antimicrobial Activity, J. Coord. Chem., 2021, 74, 1534–1549 CrossRef CAS.
- N. Richa, S. Kushwaha, A. Negi, E. Kumar, R. Zangrando, V. Kataria and Saini, Synthesis, Characterization and Utility of a Series of Novel Copper(II) Complexes as an Excellent Surface Disinfectant against Nosocomial Infections, Dalton Trans., 2021, 50, 13699–13711 RSC.
- (a) S. M. Mandal, R. O. Dias and O. L. Franco, Phenolic Compounds in Antimicrobial Therapy, J. Med. Food, 2017, 20, 1031–1038 CrossRef CAS; (b) K. Ecevit, A. A. Barros, J. M. Silva and R. L. Reis, Preventing Microbial Infections with Natural Phenolic Compounds, Future Pharmacol., 2022, 2, 460–498 CrossRef.
- (a) H. Kargar, M. Ashfaq, M. F. Mehrjardi, R. B. Ardakani, K. S. Munawar and M. N. Tahir, Synthesis, Crystal Structure, Spectral Characterization, Theoretical and Computational Studies of Ni(II), Cu(II) and Zn(II) Complexes Incorporating Schiff Base Ligand Derived from 4-(diethylamino)salicylaldehyde, Inorg. Chim. Acta, 2022, 536, 120878 CrossRef CAS; (b) H. Kargar, A. A. Ardakani, M. N. Tahir, M. Ashfaq and K. S. Munawar, Synthesis, Spectral Characterization, Crystal Structure And Antibacterial Activity Of Nickel(II), Copper(II) and Zinc(II) Complexes Containing ONNO Donor Schiff Base Ligands, J. Mol. Struct., 2021, 1233, 130112 CrossRef CAS; (c) B. Dutta, S. Jana, R. Bera, P. K. Saha and S. Koner, Immobilization of Copper Schiff Base Complexes in Zeolite Matrix: Preparation, Characterization and Catalytic Study, Appl. Catal., A, 2007, 318, 89–94 CrossRef CAS.
- (a) M. Rajasekar, S. Sreedaran, R. Prabhu, V. Narayanan, R. Jegadeesh, N. Raaman and A. K. Rahiman, Synthesis, Characterization and Antimicrobial Activities of Nickel(II) and Copper(II) Schiff Base Complexes, J. Coord. Chem., 2010, 63, 136–146 CrossRef CAS; (b) N. V. Loginova, T. Y. V. Koval’chuk, R. A. Zheldakova, A. A. Chernyavskaya, N. P. Osipovich, G. K. Glushonok, H. I. Polozov, V. L. Sorokin and O. I. Shadyro, Copper(II) Complexes of Sterically Hindered o-diphenol Derivatives: Synthesis, Characterization and Microbiological Studies, Cent. Eur. J. Chem., 2006, 4, 440–457 CAS; (c) N. Yamashita, H. Tanemura and S. Kawanishi, Mechanism of Oxidative DNA Damage Induced by Quercetin in the Presence of Cu(II), Mutat. Res. -Fundam. Mol., 1999, 425, 107–115 CrossRef CAS.
- (a) A. S. Thennarasu, T. P. Mohammed and M. Sankaralingam, Mononuclear Copper(II) Schiff Base Complexes as Effective Models for Phenoxazinone Synthase, New J. Chem., 2022, 46, 21684–21694 RSC; (b) T. P. Mohammed, A. George, M. P. Sivaramakrishnan, P. Vadivelu, S. Balasubramanian and M. Sankaralingam, J. Inorg. Biochem., 2023, 247, 112309 CrossRef CAS.
- SMART & SAINT Software Reference manuals, version 5.0, Bruker AXS Inc., Madison, WI, 1998.
- Sheldrick, G. M. SADABS software for empirical absorption correction, University of Göttingen, Germany, 2000.
- SHELXTL Reference Manual, version 5.1, Bruker AXS Inc., Madison, WI, 1998.
- M. F. Harris and J. L. Logan, Determination of log Kow Values for Four Drugs, J. Chem. Educ., 2014, 91, 915–918 CrossRef CAS.
- A. Priya, A. Selvaraj, D. Divya, K. R. Raja and S. K. Pandian, In Vitro and In Vivo Anti-Infective Potential of Thymol Against Early Childhood Caries Causing Dual Species Candida Albicans and Streptococcus Mutans, Front. Microbiol., 2021, 12, 760768 CAS.
- Y. Y. Loo, Y. Rukayadi, M. A. R. Nor-Khaizura, C. H. Kuan, B. W. Chieng, M. Nishibuchi and S. Radu, In Vitro Antimicrobial Activity of Green Synthesized Silver Nanoparticles against Selected Gram-negative Foodborne Pathogens, Front. Microbiol., 2018, 9, 1555 CrossRef.
- R. Jothi, R. Sangavi, P. Kumar, S. K. Pandian and S. Gowrishankar, Catechol Thwarts Virulent Dimorphism in Candida Albicans and Potentiates the Antifungal Efficacy of Azoles and Polyenes, Sci. Rep., 2021, 11, 21049 CrossRef CAS.
- A. Priya and S. K. Pandian, Piperine Impedes Biofilm Formation and Hyphal Morphogenesis of Candida Albicans, Front. Microbiol., 2020, 11, 756 CrossRef.
- O. Trott and A. J. Olson, AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading, J. Comput. Chem., 2010, 31, 455–461 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg Williams; F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell Jr., J. A. Montgomery, J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman and D. J. Fox, Gaussian 16; Rev. C.01, Gaussian Inc., Wallingford, CT, 2016 Search PubMed.
- Accelrys Software Inc, Discovery Studio Modeling Environment, Release 4.0, Accelrys Software Inc., San Diego, 2013.
- (a) S. Koner, S. Saha, T. Mallah and K.-I. Okamoto, Unprecedented Low Cu−N (azide)−Cu Angles in End-On Double Azido Bridged Copper(II) Complex, Inorg. Chem., 2004, 43, 840–842 CrossRef CAS; (b) P. P. Chakrabarty, S. Giri, D. Schollmeyer, H. Sakiyama, M. Mikuriya, A. Sarkar and S. Saha, Double Azido-Bridged and Mixed-Bridged Binuclear Copper(II) and Nickel(II) Compounds with N,N,O-Donor Schiff Bases: Synthesis, Structure, Magnetic and DFT Study, Polyhedron, 2015, 89, 49–54 CrossRef; (c) M. S. Ray, A. Ghosh, R. Bhattacharya, G. Mukhopadhyay, M. G. B. Drew and J. Ribas, Different Supramolecular Hydrogen Bond Structures and Significant Changes in Magnetic Properties in Dinuclear μ2-1,1-N3 Copper(II) Complexes with Very Similar Tridentate Schiff Base Blocking Ligands, Dalton Trans., 2004, 252–259 RSC.
- A. Mandal, A. Sarkar, A. Adhikary, D. Samanta and D. Das, Structure and Synthesis of Copper-Based Schiff Base and Reduced Schiff Base Complexes: A Combined Experimental and Theoretical Investigation of Biomimetic Catalytic Activity, Dalton Trans., 2020, 49, 15461–15472 RSC.
- W. P. Sohtun, S. Muthuramalingam, M. Sankaralingam, M. Velusamy and R. J. Mayilmurugan, Copper(II) Complexes of Tripodal Ligand Scaffold (N3O) as Functional Models for Phenoxazinone Synthase, J. Inorg. Biochem., 2021, 216, 11313 CrossRef.
- (a) E. I. Solomon, R. K. Szilagyi, S. D. George and L. Basumallick, Electronic Structures of Metal Sites in Proteins and Models: Contributions to Function in Blue Copper Proteins, Chem. Rev., 2004, 104, 419–458 CrossRef CAS; (b) E. I. Solomon, Dioxygen Binding, Activation, and Reduction to H2O by Cu Enzymes, Inorg. Chem., 2016, 55, 6364–6375 CrossRef CAS; (c) C. H. Kjaergaard, S. M. Jones, S. Gounel, N. Mano and E. I. Solomon, Two-Electron Reduction versus One-Electron Oxidation of the Type 3 Pair in the Multicopper Oxidases, J. Am. Chem. Soc., 2015, 137, 8783–8794 CrossRef CAS.
- E. Garribba and G. Micera, The Determination of the Geometry of Cu(II) Complexes: An EPR Spectroscopy Experiment, J. Chem. Educ., 2006, 83, 1229–1232 CrossRef CAS.
- B. J. Hathaway and D. E. Billing, The Electronic Properties and Stereochemistry of Mono-Nuclear Complexes of The Copper(II) Ion, Coord. Chem. Rev., 1970, 5, 143–207 CrossRef CAS.
- (a) E. I. Solomon, Spectroscopic Methods in Bioinorganic Chemistry: Blue to Green to Red Copper Sites, Inorg. Chem., 2006, 45, 8012–8025 CrossRef CAS; (b) B. J. Hathaway, G. Wilkinsons, R. G. Gillard and J. A. McCleverty, Comprehensive Coordination Chemistry, Pergamon, Oxford, 1987, 5, p. 861 Search PubMed.
- A. W. Addison, T. N. Rao, J. Reedijk, J. Rijn and G. C. Verschoor, Synthesis, Structure, and Spectroscopic Properties of Copper(II) Compounds Containing Nitrogen–Sulphur Donor Ligands; The Crystal and Molecular Structure of Aqua[1,7-bis(N-methylbenzimidazol-2′-yl)-2,6-dithiaheptane]copper(II) Perchlorate, J. Chem. Soc., Dalton Trans., 1984, 7, 1349–1356 RSC.
- (a) M. Zbiri, S. Saha, C. Adhikary, S. Chaudhuri, C. Daul and S. Koner, Asymmetric μ2-1,1-Azido Bridged Copper(II) Complex: Synthesis, X-Ray Structure, Magnetic Study and DFT Calculations, Inorg. Chim. Acta, 2006, 359, 1193–1199 CrossRef CAS; (b) S. Koner, S. Saha, T. Mallah and K.-I. Okamoto, End-on Double Azido Bridged Copper(II) Complex with (N, N, O) Schiff Base: Synthesis, Structure and Magnetic Study, J. Phys. Chem. Solids, 2004, 65, 697–700 CrossRef CAS; (c) J. P. Costes, F. Dahan, J. Ruiz and J. P. Laurent, New Examples of Dinuclear Copper Complexes with Ferromagnetic Interactions Mediated by μ-1,1-bridging azido and nitrito groups: Structures and Magnetic Properties of [L2(N3)2Cu2] and [L2(NO2)2Cu2]· H2O (L= 7-amino-4-methyl-5-aza-3-hepten-2-onato (1−)), Inorg. Chim. Acta, 1995, 239, 53–59 CrossRef CAS; (d) T. C. W. Mak and M. A. S. Goher, Synthesis and Structural Determination of Di-μ(1,1)-Azido-bis[azido(2-benzoyl-pyridine)] dicopper(II) and Catena-di-μ(1,3)-azido[di-μ(1,1)-azido-bis(ethyl nicotinate) dicopper(II)], Inorg. Chim. Acta, 1986, 115, 17–23 CrossRef CAS.
- C. A. Lipinski, F. Lombardo, B. W. Dominy and P. J. Feeney, Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings, Adv. Drug Delivery Rev., 2012, 64, 4–17 CrossRef.
- D. K. Singh, R. Tóth and A. Gácser, Mechanisms of Pathogenic Candida Species to Evade the Host Complement Attack, Front. Cell. Infect. Microbiol., 2020, 10, 94 CrossRef CAS.
- (a) P. Przybylski, A. Huczynski, K. Pyta, B. Brzezinski and F. Bartl, Biological Properties of Schiff Bases and Azo Derivatives of Phenols, Curr. Org. Chem., 2009, 13, 124–148 CrossRef CAS; (b) M. Claudel, J. V. Schwarte and K. M. Fromm, New Antimicrobial Strategies based on Metal Complexes, Chemistry, 2020, 2, 849–899 CrossRef CAS.
- (a) Y. Murti, A. K. Singh and D. Pathak, New 6-Bromo-2-methyl-3-(substituted phenyl)-(3H)-quinazolin-4-ones with Antimicrobial and Antiinflammatory Activities, Indian J. Pharm. Sci., 2011, 73, 333–337 CAS; (b) D. Prasannan, D. Raghav, S. Sujatha, H. H. Kumar, K. Rathinasamy and C. Arunkumar, Synthesis, Structure, Photophysical, Electrochemical Properties and Antibacterial Activity of Brominated BODIPYs, RSC Adv., 2016, 6, 80808–80824 RSC; (c) S. Balaji, M. Rajarajan, R. Vijayakumar, V. Manikandan, R. Senbagam, G. Vanangamudi and G. Thirunarayanan, Synthesis, Evaluation of Substituent Effect and Antimicrobial Activities of Substituted (E)-1-(3-bromo-4-morpholinophenyl)-3-phenylprop-2-en-1-one Compounds, Orbital: The Electro. J. Chem., 2017, 9, 1–17 CAS.
- (a) S. Belaid, A. Landreau, S. Djebbar, O. Benali-Baitich, G. Bouet and J.-P. Bouchara, Synthesis, Characterization and Antifungal Activity of a Series of Manganese(II) and Copper(II) Complexes with Ligands Derived from Reduced N,N′-O-Phenylenebis(Salicylideneimine), J. Inorg. Biochem., 2008, 102, 63–69 CrossRef CAS; (b) J. Joseph, K. Nagashri and G. A. B. Rani, Synthesis, Characterization and Antimicrobial Activities of Copper Complexes Derived from 4-Aminoantipyrine Derivatives, J. Saudi Chem. Soc., 2013, 17, 285–294 CrossRef CAS.
- (a) S. Sogabe, M. Masubuchi, K. Sakata, T. A. Fukami, K. Morikami, Y. Shiratori, H. Ebiike, K. Kawasaki, Y. Aoki, N. Shimma, A. D'Arcy, F. K. Winkler, D. W. Banner and T. Ohtsuka, Crystal Structures of Candida Albicans N-Myristoyltransferase with Two Distinct Inhibitors, Chem. Biol., 2002, 9, 1119–1128 CrossRef CAS; (b) D. A. Towler, J. I. Gordon, S. P. Adams and L. Glaser, The Biology and Enzymology of Eukaryotic Protein Acylation, Annu. Rev. Biochem., 1988, 57, 69–99 CrossRef CAS; (c) D. R. Johnson, R. S. Bhatnagar, L. J. Knoll and J. I. Gordon, Genetic and Biochemical Studies of Protein N-Myristoylation, Annu. Rev. Biochem., 1994, 63, 869–914 CrossRef CAS.
- (a) A. Frei, A. G. Elliott, A. Kan, H. Dinh, S. Bräse, A. E. Bruce, M. R. Bruce, F. Chen, D. Humaidy, N. Jung, A. P. King, P. G. Lye, H. K. Maliszewska, A. M. Mansour, D. Matiadis, M. P. Muñoz, T.-Y. Pai, S. Pokhrel, P. J. Sadler, M. Sagnou, M. Taylor, J. J. Wilson, D. Woods, J. Zuegg, W. Meyer, A. K. Cain, M. A. Cooper and M. A. T. Blaskovich, Metal Complexes as Antifungals? From a Crowd-Sourced Compound Library to the First In Vivo Experiments, JACS Au., 2022, 2, 2277–2294 CrossRef CAS; (b) M. A. Ghannoum and L. B. Rice, Antifungal Agents: Mode of Action, Mechanisms of Resistance, and Correlation of these Mechanisms with Bacterial Resistance, Clin. Microbiol. Rev., 1999, 12, 501–517 CrossRef CAS.
- (a) A. Sethiya, D. Joshi, A. Manhas, N. Sahiba, D. K. Agarwal, P. C. Jha and S. Agarwal, Glycerol Based Carbon Sulfonic Acid Catalyzed Synthesis, In Silico Studies and In Vitro Biological Evaluation of Isonicotinohydrazide Derivatives as Potent Antimicrobial and Anti-Tubercular Agents, Heliyon, 2023, 9, e13226 CrossRef CAS; (b) B. O. Aljohny, A. Rauf, Y. Anwar, S. Naz and A. Wadood, Antibacterial, Antifungal, Antioxidant, and Docking Studies of Potential Dinaphthodiospyrols from Diospyros lotus Linn Roots, ACS Omega, 2021, 6, 5878–5885 CrossRef CAS.
- N. G-Dayanandan, J. L. Paulsen, K. Viswanathan, S. Keshipeddy, M. N. Lombardo, W. Zhou, K. M. Lamb, A. E. Sochia, J. B. Alverson, N. D. Priestley, D. L. Wright and A. C. Anderson, Propargyl-Linked Antifolates are Dual Inhibitors of Candida Albicans and Candida Glabrata, J. Med. Chem., 2014, 57, 2643–2656 CrossRef CAS.
- (a) M. V. Keniya, M. Sabherwal, R. K. Wilson, M. A. Woods, A. A. Sagatova, J. D. A. Tyndall and B. C. Monk, Crystal Structures of Full-Length Lanosterol 14α-Demethylases of Prominent Fungal Pathogens Candida Albicans and Candida Glabrata Provide Tools for Antifungal Discovery, Antimicrob. Agents Chemother., 2018, 62, e01134–18 Search PubMed; (b) L.-T. Zhai, S. Rety, W.-F. Chen, Z.-Y. Song, D. Auguin, B. Sun, S.-X. Dou and X.-G. Xi, Crystal Structures of N-Terminally Truncated Telomerase Reverse Transcriptase from Fungi, Nucleic Acids Res. Spec. Publ, 2021, 49, 4768–4781 CrossRef CAS.
- L. F. Kuyper, D. P. Baccanari, M. L. Jones, R. N. Hunter, R. L. Tansik, S. S. Joyner, C. M. Boytos, S. K. Rudolph, V. Knick, H. R. Wilson, J. M. Caddell, H. S. Friedman, J. C. W. Comley and J. N. Stables, High-Affinity Inhibitors of Dihydrofolate Reductase: Antimicrobial and Anticancer Activities of 7,8-Dialkyl-1, 3-diaminopyrrolo[3,2-f] Quinazolines with Small Molecular Size, J. Med. Chem., 1996, 39, 892–903 CrossRef CAS.
- T. Otzen, E. G. Wempe, B. Kunz, R. Bartels, G. Lehwark-Yvetot, W. Hänsel, K.-J. Schaper and J. K. Seydel, Folate-Synthesizing Enzyme System as Target for Development of Inhibitors and Inhibitor Combinations against Candida albicans- Synthesis and Biological Activity of New 2,4-Diaminopyrimidines and 4‘Substituted 4-Aminodiphenyl Sulfones, J. Med. Chem., 2004, 47, 240–253 CrossRef CAS.
- C.-T. Peng, L. Liu, C.-C. Li, L.-H. He, T. Li, Y.-L. Shen, C. Gao, N.-Y. Wang, Y. Xia, Y.-B. Zhu, Y.-J. Song, Q. Lei, L.-T. Yu and R. Bao, Structure–Function Relationship of Aminopeptidase P from Pseudomonas Aeruginosa, Front. Microbiol., 2017, 8, 2385 CrossRef.
- D. Schwefel, C. Maierhofer, J. G. Beck, S. Seeberger, K. Diederichs, H. M. Möller, W. Welte and V. Wittmann, Structural Basis of Multivalent Binding to Wheat Germ Agglutinin, J. Am. Chem. Soc., 2010, 132, 8704–8719 CrossRef CAS.
- Q. Ye, R. K. Lau, I. T. Mathews, E. A. Birkholz, J. D. Watrous, C. S. Azimi, J. Pogliano, M. Jain and K. D. Corbett, HORMA Domain Proteins and a Trip13-Like ATPase Regulate Bacterial cGAS-Like Enzymes to Mediate Bacteriophage Immunity, Mol. Cell, 2020, 77, 709–722 CrossRef CAS.
- S. Sobhanifar, L. J. Worrall, D. T. King, G. A. Wasney, L. Baumann, R. T. Gale, M. Nosella, E. D. Brown, S. G. Withers and N. C. J. Strynadka, Structure and Mechanism of Staphylococcus Aureus TarS, The Wall Teichoic Acid β-Glycosyltransferase Involved in Methicillin Resistance, PLoS Pathog., 2016, 12, 1006067 CrossRef.
- C.-Y. Huang, H.-W. Shih, L.-Y. Lin, Y.-W. Tien, T.-J. R. Cheng, W.-C. Cheng, C.-H. Wong and C. Ma, Crystal Structure of Staphylococcus Aureus Transglycosylase in Complex with a Lipid II Analog and Elucidation of Peptidoglycan Synthesis Mechanism, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 6496–6501 CrossRef CAS.
- G. M. de Tejada, S. Sánchez-Gómez, I. Kowalski, Y. Kaconis, J. Andrä, T. Schürholz, M. Hornef, A. Dupont, P. Garidel, T. Gutsmann, S. A. David and K. Brandenburg, Bacterial Cell Wall Compounds as Promising Targets of Antimicrobial Agents I. Antimicrobial Peptides and Lipopolyamines, Curr. Drug Targets, 2012, 13, 1121–1130 CrossRef.
- T. A. Fayed, M. Gaber, G. M. A. El-Reash and M. M. El-Gamil, Structural, DFT/B3LYP and Molecular Docking Studies of Binuclear Thiosemicarbazide Copper(II) Complexes and their Biological Investigations, Appl. Organomet. Chem., 2020, 34, e5800 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1–S21, which include IR spectra of L1(H)–L3(H) and 1–3, ESI mass-spectra of 1–3, UV-vis spectral profile of the lipophilicity study, images and plots of toxicity results and antimicrobial results, and images showing the interactions of complexes 1–3 obtained from the in silico study. CCDC 2263468 and 2263469 contain the supplementary crystallographic data for 1 and 2. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d4nj01271b |
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2024 |