Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Identification of key active residues and solution conditions that affect peptide-catalyzed ester hydrolysis†
Kyle B.
Meerbott
a and
Marc R.
Knecht
 *ab
*ab
aDepartment of Chemistry, University of Miami, Coral Gables, Florida 33146, USA
bDr. J. T. Macdonald Foundation Biomedical Nanotechnology Institute, University of Miami, Miami, Florida 33136, USA. E-mail: knecht@miami.edu
First published on 10th April 2024
Abstract
Peptides respresent intriguing materials to achieve sustainable catalytic reactivity that mimic the natural functions of enzymes, but without the limitations of temperature/solvent sensitivity. They could also be applicable to a wide variety of substrates, thus expanding their potential use at different reaction levels ranging from the benchtop to industrial. Unfortunately, significant use of catalytic peptides remains limited due to the general lack of understanding of the fundamental basis of their inherent reactivity. In this contribution, we examine the reactivity of a peptide (termed CPN3) previously isolated with ester hydrolysis reactivity. It is demonstrated that the system is most reactive under slightly basic conditions. While the system is slower than comparable enzymes, it demonstrates significant reactivity across multiple substrates and different reaction conditions that could likely lead to enzymatic denaturation. In addition, key active site residues were identified to begin to elucidate the fundamental basis of the reactivity. Such results could be used to design new sequences with enhanced reactivity under sustainable conditions.
Introduction
Enzymes provide incredible opportunities for highly precise catalytic reactivity.1–10 They drive a vast swath of different catalytic reactions, ranging from the synthesis of chiral alcohols to interfacial hydrolysis of ester bonds.11,12 In general, the specificity of the reaction is precisely tuned to substrate structure, which is critically important for biological processes. While this specificity is required for biological systems, the broad application of enzymes for industrially important reactions remains limited for a variety of reasons. First: they are highly susceptible to changes in reaction conditions, which leads to denaturation and loss of reactivity. For instance, just slight changes away from physiological temperature and/or pH can result in complete loss of enzymatic reactivity. Second: the specificity required by biology can greatly limit commercial application of the catalytic enzymatic process. To this end, broad application of the enzyme for catalytic conversion over a great number of substrates is desired, but could be prohibited. Taken together, these two aspects have limited the application of enzymes in commercially viable catalytic processes.As an alternative to protein-based enzymes, catalytic peptides provide exciting new opportunities to employ sustainable biological conditions for technologically important reactions.13–17 Peptides are intriguing as they are derived from the twenty canonical amino acids and can be designed to incorporate enzyme active sites to drive reactions. From their small sequence, secondary structure requirements are less stringent to achieve their catalytic reactivity, thus potentially allowing for a greater breadth of reaction conditions for catalysis than what can be achieved using enzymes. In addition, this lack of significant secondary structure requirements potentially also facilitates reactivity across more substrates, expanding the scope of the reaction, which is highly desirable.
One specific catalytic peptide is the CPN3 sequence (DLRSCTACAVNA), previously reported by Matsui, Ulijn, and colleagues.18 This sequence was isolated from a unique phage display selection process, where it was found with the ability to catalyze amide condensation reactions. To this end, the peptides drove the reaction, from which the product was able to aggregate in solution. This aggregation process led to the catalytic phage being encapsulated and removed from the mixture, allowing for identification of the peptide sequences that drove the reaction. Beyond amide condensation, the CPN3 peptide has also demonstrated the ability to drive ester hydrolysis,18 showing its ability to catalyze complementary reactions, both of which could be important for different catalytic systems. While the peptide has demonstrated significant reactivity, it remains poorly understood the basis of the reactivity, nor how the reaction conditions affect the catalytic process. Such information could prove to be invaluable, confirming the breadth of scope of the catalytic reactivity, as well as identifying structural features that comprise the active site of the system.
In this contribution, the basis for CPN3-driven ester hydrolysis is examined, identifying key reaction criteria and residues necessary for the catalytic process. To monitor the reactivity of the system, the hydrolysis of 4-nitrophenylacetate (4-NPA) to 4-nitrophenol (4-NP) driven via the peptide was monitored (Scheme 1). The results indicate that the reactivity is sensitive to the solution pH, solvent composition, and substrate concentration. In addition, experimental analysis suggests that the serine and cysteines of the biomolecule are key residues for the catalytic functionality, potentially working in concert to drive the reaction. Taken together, this study provides key results in understanding the basis of catalytic peptide functionality, which could be used to design new sequences with either enhanced reactivity or new catalytic functionality.
Experimental
Materials
KCl, NaCl, HCl, and tris(hydroxymethyl)amino methane (TRIS) buffer were purchased from VWR. KH2PO4 was acquired from Mallinckrodt Chemicals. Na2HPO4 was sourced from Acros. NaOH pellets were purchased from Macron Fine Chemicals. 4-NPA, 4-nitrophenyl palmitate, 3-indoxyl acetate, RbCl, and LiCl were purchased from Thermo Fisher Scientific. 4-Nitrophenyl butyrate was acquired from Cayman Chemical, while 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) buffer was purchased from Alfa Aesar. 4-Morpholinepropanesulfonic acid (MOPS) sodium salt was purchased from Amresco and MgCl2·6H2O was sourced from Calbiochem. All peptides were commercially sourced from either Genscript (CPN3, CPN3-A4, and AuBP1) or Biomatik (CPN3-A6 and CPN3-A4,A6). Methanol was purchased from EMD Millipore. Finally, water was acquired from a Milli-Q water system by Millipore at 18 mΩ cm. All reagents were used as received without additional processing.Buffer preparation
Stock phosphate buffer and saline solutions (containing NaCl and KCl) were prepared. For the buffer, 1.42 g of Na2HPO4 and 0.24 g of KH2PO4 were co-dissolved into 100 mL of water, resulting in a total phosphate concentration of 0.12 M. In a separate solution, 8 g of NaCl and 0.2 g of KCl were co-dissolved in 100 mL of water to generate the stock saline solution. To prepare the standard reaction buffer, 5 mL of both the phosphate buffer stock and saline stock were commixed and subsequently diluted with 40 mL of water. The new buffer solution was then adjusted to the appropriate pH using concentrated NaOH or HCl, as needed, to reach the intended reaction pH.Ester hydrolysis reaction
For the reaction pH analysis, 1 mg of CPN3 peptide was dissolved in 14.715 mL of freshly prepared buffer solution to reach a concentration of 55.6 μM at the intended reaction pH. In a separate vial, 3 mL of an 8 mM 4-NPA solution was freshly prepared in MeOH. Once the two components were prepared, 1.8 mL of the peptide solution was added to a quartz cuvette. Next, 80 μL of MeOH was added and the system was allowed to reach a reaction temperature of 25 °C. Once at temperature, 120 μL of the 4-NPA solution was added and the reaction was monitored via UV-vis spectroscopy where the absorbance of the sample was measured for 10 min at 405 nm. Under these conditions, a peptide concentration of 50 μM, a substrate concentration of 480 μM, and a saline concentration of 123.2 mM NaCl and 2.4 mM KCl was achieved. Identical conditions were employed for the additional studies; however, for catalytic analysis of various substrates, the saline concentration was 100 mM NaCl, while a solution of 98 mM NaCl and 2 mM KCl was employed for the mutation studies.Results and discussion
While the CPN3 has been identified with intriguing catalytic capabilities, the basis for this reactivity remains unclear. Furthermore, the effects of reaction solution conditions also are generally unknown. To explore these effects, CPN3-driven ester hydrolysis was initially examined as a function of pH ranging from 6.5–8.0, as shown in Fig. 1. For the reaction process, the hydrolysis of 4-NPA to 4-NP was examined using UV-vis spectroscopy. To this end, the 4-NP product generates an absorbance at 405 nm, which can be monitored as a function of time to determine the first order reaction rate constant (k). Two controls were also studied: a peptide free system and a system with a non-catalytic peptide included. These two systems were important to confirm the reactivity associated with the CPN3. For the non-catalytic peptide, the AuBP1 sequence (WAGAKRLVLRRE) was employed, which was previously identified with affinity to bind Au surfaces.19,20 For these initial reactions, a phosphate buffer (10.7 mM) was used at the indicated pH value. The buffer contained a saline component (123 mM NaCl, 3 mM KCl) as well.Fig. 1a presents the absorbance at 405 nm as a function of time for the reaction driven using the CPN3 peptide at pH 8 (red plot). As is evident, two reaction domains were present: an initial rapid burst in 4-NP product formation for the first ∼10 s of the reaction followed by slow product release over time for the rest of the catalytic process. This effect is quite similar to the reactivity previously observed for ester hydrolysis catalyzed by chymotrypsin, a serine-based protease.21 For this enzyme, rapid product formation is observed, due to nucleophilic attack by the active serine residue. However, this first step, stabilized by additional residues at the active site, acylates the active site serine residue. Subsequent serine hydrolysis, aided again by additional residues at the active site, regenerates the catalyst for further catalytic turnover. This second step of the process is notably slower than the first, resulting in slow product release after the initial burst, similar to what was observed using the CPN3-catalyzed reaction. To compare the inherent reactivity of the peptide-driven process, k values associated with the second step of the reaction after the initial burst of product were used, consistent with prior results.18 As such, fitting of the data for the kinetic analysis to determine k values began after completion of the burst, thus not including any of the initial first step.
Determination of the actual k values for the selected reaction (with or without peptide) was processed using the graph of Fig. 1b over the linear regime of the plot (first ∼200 s of the reaction after the initial burst). From the reaction analysis, at pH 6.5, a k value of (2.2 ± 2.0) × 10−4 s−1 was determined for the CPN3-catalyzed reaction that was generally similar to both the peptide free ((0.4 ± 1.2) × 10−4 s−1) and AuBP1-catalyzed ((2.6 ± 2.4) × 10−4 s−1) controls. This suggests that at this low pH condition, the CPN3 peptide is not catalyzing the reaction above the background associated with the buffer. When the pH was raised to 7.0, CPN3-catalyzed reactivity was noted with a rate constant of (6.8 ± 1.4) × 10−4 s−1. This value was higher than for the two controls: (2.6 ± 0.9) × 10−4 and (2.2 ± 0.9) × 10−4 s−1 for the peptide free and AuBP1 systems, respectively. As the pH of the system increased, enhanced reactivity was noted, giving the highest rate constant for the CPN3-catalyzed reactions at pH 8.0 ((3.1 ± 0.2) × 10−3 s−1). Such values were notably higher than the two controls, thus pH 8.0 was used for all subsequent reactions as the optimized pH value.
While phosphate buffer is commonly used in this pH range, other buffers are available. Since the buffer can drive the hydrolysis reaction, changes to this composition could be used to abate background reactivity. To explore such effects, the buffer composition was varied across three different species beyond phosphate: Tris, HEPES, and MOPs. All of the reactions were processed at the optimal pH value of 8.0. Fig. 2a presents the absorbance intensity over time for the four buffered systems where similar degrees of reactivity were noted for the Tris-, HEPES-, and MOPS-based systems; for these reactions, they were slightly lower in absorbance at longer time points compared to the phosphate-buffered system. While this may suggest greater reactivity for the reactions processed in phosphate buffer, it could also indicate enhanced buffer-driven hydrolysis as well.
Fig. 2b compares the calculated k values for all four buffered systems and their controls. For each control, the reaction was processed in the indicated buffer in the absence of any peptide. To compare the different systems, subtraction of the buffer control k values from the CPN3-driven rate constants in the selected medium was employed. This value represents the actual catalytic effect of the peptide in the selected buffered system. In this regard, for the phosphate-buffered system, a k value of (3.1 ± 0.2) × 10−3 s−1 was noted when CPN3 was present in the reaction, but a value of (2.0 ± 0.3) × 10−3 s−1 was observed for the peptide-free control. Subtraction of these two values results in a k of (1.1 ± 0.4) × 10−3 s−1, which arises from the peptide-driven reaction in the absence of background reactivity. Using this approach for the Tris-, HEPES-, and MOPS-based reactions, k values of (1.1 ± 0.3) × 10−3, (1.0 ± 0.1) × 10−3, and (1.0 ± 0.3) × 10−3 s−1 were noted, respectively, which were equivalent to the phosphate-driven system.
From the buffer analysis, it was evident that nearly identical degrees of CPN3 catalytic reactivity were noted over the selected buffer systems. This suggests that the composition of the buffer has a negligible role in controlling the peptide-derived reactivity. It is interesting to note, however, that increased background reactivity was observed for the phosphate buffer. This is likely why higher degrees of 4-NP absorbance were observed in this system (Fig. 2a) as compared to the other buffers. Since the buffer does not affect the peptide catalytic capabilities, continued use of the phosphate buffered system at pH 8.0 was employed.
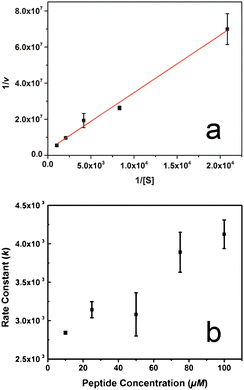
To explore the effect of substrate concentration on the reactivity of the CPN3-peptide, standard enzyme kinetics analyses were employed. Fig. 3a presents the Lineweaver–Burk plot of CPN3 catalysed ester hydrolysis at selected concentrations of 4-NPA at pH 8. From this analysis, a Km value of (1.2 ± 0.4) × 10−3 M was found, along with a kcat of (7.2 ± 2.1) × 10−3 s−1. From these two values, an enzyme efficiency of 5.3 ± 1.4 M−1 s−1 could be determined. The catalytic character of the peptide can be put into perspective by comparing it to a well-studied serine protease such as chymotrypsin, with Km and kcat values of 1.35 × 10−4 M and 2.54 × 10−2 s−1, respectively.21 Since Km is inversely proportional to the affinity of the enzyme to the substrate, it is evident that the CPN3 peptide has significantly lower affinity for the substrate than the enzyme. From the comparison of kcat values, it is demonstrated that the peptide converts the substrate at a slower rate than an enzyme of similar mechanistic pathways. These differences indicate diminished catalytic efficiency of the peptide as compared to the enzyme. While diminished efficiency is observed for CPN3, it may be possible that enhanced breadth of reactivity for more substrates could be achieved.
Additional analysis of the reaction was processed to determine the effect of peptide concentration on the reaction (Fig. 3b). For this reaction, the system was prepared under the model conditions; however, the concentration of the CPN3 peptide in solution varied from 10–100 μM. As anticipated, the k values increased as the amount of the peptide in the reaction mixture also increased. For instance, at a CPN3 concentration of 10 μM, a rate constant of (2.8 ± 0.02) × 10−3 s−1 was observed that increased to (4.1 ± 0.2) × 10−3 s−1 at a peptide concentration of 100 μM. This dependency of the rate constant on the peptide concentration proved to be relatively moderate, increasing by 1.5 fold at an increased peptide concentration of 10 fold. Such effects may be due to the fact that the initial substrate concentration (480 μM) was notably lower than the calculated Km value (1.2 mM).
With identification of notable reaction solution condition effects on CPN3-catalyzed ester hydrolysis, the effect of substrate structure was also explored. Such effects are critical as many enzymes are highly specific to the substrate. In this case, four different ester-containing substrates were explored (Fig. 4a): 4-NPA, 4-nitrophenylbutyrate (4-NPB), 4-nitrophenyl palmitate (4-NPP), and 3-indoxyl acetate. It is important to note that two different cosolvents (MeOH or DMF) had to be used due to different substrate solubilities. While 4-NPA, 4-NPB, and indoxyl acetate were soluble in MeOH, 4-NPP was not, thus DMF was used for the 4-NPP substrate.
Fig. 4b compares the rate constants observed for the different systems. When using MeOH as the co-solvent, hydrolysis of the 3-NPB was observed catalyzed by the CPN3 peptide; however, significant buffer-driven hydrolysis was also indicated. Subtraction of the buffer-driven k value from the peptide catalyzed value gave a normalized rate constant of (1.8 ± 0.6) × 10−3 s−1, which was similar to the value noted for 4-NPA. Interestingly, when the 3-indoxyl acetate system was employed as the substrate using the MeOH co-solvent, the reactivity of the peptide free control and the CPN3-based system were essentially the same. This suggests that hydrolysis of this substrate was not possible using the CPN3 peptide.
Since 4-NPP was not soluble in MeOH, DMF was used for this specific substrate. To confirm and compare the reactivity effects, ester hydrolysis of the primary 4-NPA substrate using DMF as the co-solvent was also explored. In this system, CPN3-catalyzed 4-NPA hydrolysis was observed with a k value of (1.5 ± 0.04) × 10−3 s−1. For the peptide free control in DMF, a rate constant of (8.9 ± 0.7) × 10−4 s−1 was noted, thus giving rise to a net k value of (5.7 ± 0.8) × 10−4 s−1. Such values are substantially lower than those observed for the MeOH system, indicating that the co-solvent plays a significant role in controlling the observed catalytic activity, including the background peptide-free reactivity. Using this system with 4-NPP as the substrate, a net k value of (4.0 ± 3.0) × 10−4 s−1 was observed after subtraction of the peptide free control rate constant from the CPN3-catalyzed k value. Such a value is generally consistent with the rate constant observed for 4-NPA hydrolysis with DMF as the co-solvent.
From the substrate analysis, it is clear that expanded reactivity across different substrates is possible; however, the substrate structure does play a role in controlling/achieving reactivity. In addition, the effect of co-solvent on the reactivity is substantial. Changing from MeOH to DMF resulted in notably diminished catalytic functionality, but also the background reactivity of the buffer system was also decreased. This is an important factor when designing reaction systems, which may play a role in controlling each step of the reaction.
While the different reaction conditions are highly important to the CPN3-based reactivity, identification of the key active residues in the sequence is critical. From these residues, understanding of the fundamental basis for the reactivity can be achieved. To address this question, two modifications to the peptide were employed focusing on the serine, threonine, and cysteine residues (Fig. 5a). These three residues were specifically chosen as they play significant effects in protease-driven reactions.22 First, to probe the effects of the serine and threonine, mutation of the residues to an alanine was achieved, generating the CPN3-S4A and CPN3-T6A peptides that mutated the serine and threonine, respectively. In addition, a third peptide, CPN3-S4A,T6A was also prepared that mutated both the serine and threonine residues to alanines. Second, derivatization of the cysteine free thiol residues via the Ellman's reagent was employed,23 generating the CPN3-DTNB peptide.
Fig. 5b presents the correlation of the ln(A/A0) as a function of time for the reactions catalyzed with the indicated biomolecules. Note that all reactions were processed in the same phosphate buffer conditions at pH 8.0, which was identified to display maximal catalytic activity. As is evident, greater reactivity was observed from the parent CPN3 system as compared to the modified biomolecules. For this system, two controls were again presented, including the peptide free control in buffer. The second control was the same as the peptide free system; however, extraneous, unreacted Ellman's reagent was present in the mix (termed DTNB). In this case, similar reactivity as compared to the peptide-free control was noted, suggesting that the derivatization reagent does not affect the reactivity.
Fig. 5c compares the k values achieved from the five different biomolecules and the two controls. Consistent with the results above, the parent CPN3 peptide presents a rate constant of (3.7 ± 0.1) × 10−3 s−1, while the two controls displayed rate constants of (2.2 ± 0.3) × 10−3 s−1 for the peptide free system and (1.8 ± 0.1) × 10−3 s−1 for the system with free Ellman's reagent and no peptide. This provides a baseline for comparing maximal (CPN3-driven reaction) and negligible (controls) reactivity. For the CPN3-S4A mutant-catalyzed reaction, a rate constant of (2.5 ± 0.1) × 10−3 s−1 was determined. This value was notably lower than the parent peptide, and nearly equivalent to the peptide-free control. This indicates that the serine is highly important to the reaction to facilitate the catalytic process. When the threonine mutated peptide was employed (CPN3-T6A) to drive the reaction, a k value of (3.5 ± 0.1) × 10−3 s−1 was observed. This value is essentially equivalent to the reactivity observed for the parent CPN3 peptide. This suggests that the threonine residue is not overly important in the ester hydrolysis reactivity. Unexpectedly, upon mutation of both the serine and threonine residues (CPN3-S4A,T6A), using this peptide in the reaction gave rise to a k value of (3.1 ± 0.1) × 10−3 s−1. Such a value is diminished compared to the parent CPN3 sequence; however, it does demonstrate reactivity above the controls. This suggests that other residues beyond the hydroxylated species are now available to catalyze the reaction. In this situation, the thiol-bearing cysteine residues are likely the basis of the reactivity.
To further probe the roles of the different residues, modification of both cysteines was processed to explore the effects of the free sulfhydryl groups (CPN3-DTNB). Using this biomolecule in the reaction, a k value of (1.7 ± 0.2) × 10−3 s−1 was observed, which is nearly identical to the two control values. This suggests that the cysteine residues of the peptide are highly important to the observed reactivity of the CPN3 peptide for ester hydrolysis.
From the mutation analysis, it is evident that the serine and cysteine residues are key active site species for driving the reaction. In many enzymatic-driven processes,22,24,25 secondary amino acids in the active site are required to interact with the active site species, thus making it more nucleophilic to attack the substrate. Similar effects may be happening here between the hydroxylated and sulfhydryl containing residues. In this regard, assuming that the serine is the actual active site, one of the cysteine residues could be interacting with the serine hydroxyl group to make it more reactive to drive 4-NPA hydrolysis. This is supported by the two reaction systems that mutated the serine residue (CPN3-S4A) or derivatized the cysteines (CPN3-DTNB), both of which completely lost reactivity. The lack of diminished reactivity for the threonine mutated peptide (CPN3-T6A) indicates that this residue has only a minimal effect on the reaction.
The more interesting observation is that reactivity was generally observed when both the serine and threonine residues were removed (CPN3-S4A, T6A). In this situation, it is possible that the two cysteine residues in the peptide could form a new catalytic active site to facilitate ester hydrolysis. In this case, diminished reactivity was noted, compared to the CPN3 parent, demonstrating a difference based upon the changes in the active site species.
Conclusions
In conclusion, key new insights concerning the catalytic reactivity of peptides were observed, specifically for ester hydrolysis driven via the CPN3 peptide. These results demonstrated specific reaction conditions that altered the reactivity and can be used to tune the overall catalytic capability of the sequence. In addition, specific residues of the biomolecule were identified as likely active sites for the reaction, suggesting that both the serine and cysteines of the peptide were critical for the reactivity. These different effects work synergistically to achieve the catalytic capabilities of the biomolecule. While it is true that the peptide's reactivity is modestly greater than the background reactivity of the solvent, these results present important criteria for the design of new catalytic peptides, based upon active site residues, which could be translated for either enhanced reactivity or reactivity for different reactions based upon the specific system. Future studies will focus on these effects and how the overall reaction conditions can be exploited to control the desired catalytic process under sustainable conditions.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This material is based upon work supported by the National Science Foundation under grant no. 2203862 (MRK).Notes and references
- J. M. Woodley, Curr. Opin. Green Sustainable Chem., 2020, 21, 22–26 CrossRef CAS.
- M. Bilal and H. M. N. Iqbal, Crit. Rev. Food Sci. Nutr., 2020, 60, 2052–2066 CrossRef PubMed.
- M. Mathesh, J. Sun and D. A. Wilson, J. Mater. Chem. B, 2020, 8, 7319–7334 RSC.
- J.-Y. Park and K.-M. Park, J. Chem., 2022, 2022, 7609019 Search PubMed.
- V. Tournier, S. Duquesne, F. Guillamot, H. Cramail, D. Taton, A. Marty and I. André, Chem. Rev., 2023, 123, 5612–5701 CrossRef CAS PubMed.
- M. C. R. Franssen, P. Steunenberg, E. L. Scott, H. Zuilhof and J. P. M. Sanders, Chem. Soc. Rev., 2013, 42, 6491–6533 RSC.
- E. A. Osifalujo, B. N. Rutkowski, L. R. Satterwhite, P. C. Betts, A. K. Nkosi and J. T. Froese, Catal. Sci. Technol., 2023, 13, 3784–3790 RSC.
- A. Colacicco, G. Catinella, C. Pinna, A. Pellis, S. Farris, L. Tamborini, S. Dallavalle, F. Molinari, M. L. Contente and A. Pinto, Catal. Sci. Technol., 2023, 13, 4348–4352 RSC.
- C. Gastaldi, V. Hélaine, M. Joly, A. Gautier, C. Forano and C. Guérard-Hélaine, Catal. Sci. Technol., 2023, 13, 1623–1627 RSC.
- J. Gao, Z. Wang, R. Guo, Y. Hu, X. Dong, Q. Shi and Y. Sun, Catal. Sci. Technol., 2023, 13, 991–999 RSC.
- L. Sarda and P. Desnuelle, Biochim. Biophys. Acta, 1958, 30, 513–521 CrossRef CAS PubMed.
- D. Zhu, Y. Yang and L. Hua, J. Org. Chem., 2006, 71, 4202–4205 CrossRef CAS PubMed.
- I. W. Hamley, Biomacromolecules, 2021, 22, 1835–1855 CrossRef CAS PubMed.
- J. Han, H. Gong, X. Ren and X. Yan, Nano Today, 2021, 41, 101295 CrossRef CAS.
- D. J. Mikolajczak, A. A. Berger and B. Koksch, Angew. Chem., Int. Ed., 2020, 59, 8776–8785 CrossRef CAS PubMed.
- S. Carvalho, D. Q. Peralta Reis, S. V. Pereira, D. Kalafatovic and A. S. Pina, Isr. J. Chem., 2022, 62, e202200029 CrossRef CAS.
- A. J. Metrano, A. J. Chinn, C. R. Shugrue, E. A. Stone, B. Kim and S. J. Miller, Chem. Rev., 2020, 120, 11479–11615 CrossRef CAS PubMed.
- Y. Maeda, N. Javid, K. Duncan, L. Birchall, K. F. Gibson, D. Cannon, Y. Kanetsuki, C. Knapp, T. Tuttle, R. V. Ulijn and H. Matsui, J. Am. Chem. Soc., 2014, 136, 15893–15896 CrossRef CAS PubMed.
- M. Hnilova, E. E. Oren, U. O. S. Seker, B. R. Wilson, S. Collino, J. S. Evans, C. Tamerler and M. Sarikaya, Langmuir, 2008, 24, 12440–12445 CrossRef CAS PubMed.
- Z. Tang, J. P. Palafox-Hernandez, W.-C. Law, Z. E. Hughes, M. T. Swihart, P. N. Prasad, M. R. Knecht and T. R. Walsh, ACS Nano, 2013, 7, 9632–9646 CrossRef CAS PubMed.
- H. Gutfreund and J. M. Sturtevant, Biochem. J., 1956, 63, 656–661 CrossRef CAS PubMed.
- F. J. Gisdon, E. Bombarda and G. M. Ullmann, J. Phys. Chem. B, 2022, 126, 4035–4048 CrossRef CAS PubMed.
- C. K. Riener, G. Kada and H. J. Gruber, Anal. Bioanal. Chem., 2002, 373, 266–276 CrossRef CAS PubMed.
- R. D. Kidd, P. Sears, D.-H. Huang, K. Witte, C.-H. Wong and G. K. Farber, Protein Sci., 1999, 8, 410–417 CrossRef CAS PubMed.
- C. N. Fuhrmann, M. D. Daugherty and D. A. Agard, J. Am. Chem. Soc., 2006, 128, 9086–9102 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nj00977k |
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2024 |