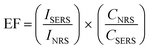Open Access Article
Open Access ArticleSuitability study of Ag nanosheet SERS substrates as a screening method for imidacloprid after QuEChERS extraction†
Felipe
Leyton-Soto
 a,
Zachary D.
Schultz
a,
Zachary D.
Schultz
 d,
Rodrigo
Ormazábal-Toledo
d,
Rodrigo
Ormazábal-Toledo
 e,
Domingo
Ruiz-León
f,
Ady
Giordano
*a and
Mauricio
Isaacs
e,
Domingo
Ruiz-León
f,
Ady
Giordano
*a and
Mauricio
Isaacs
 *abc
*abc
aDepartamento de Química Inorgánica, Facultad de Química y de Farmacia, Pontificia Universidad Católica de Chile, Av. Vicuña Mackenna 4860, Macul, Santiago, 7820436, Chile. E-mail: misaacs@uc.cl; agiordano@uc.cl
bCentro de Investigación en Nanotecnología y Materiales Avanzados (CIEN-UC), Pontificia Universidad Católica de Chile, Av. Vicuña Mackenna 4860, Macul, Santiago, 7820436, Chile
cMillennium Institute on Green Ammonia as Energy Vector, MIGA. Pontificia Universidad Católica de Chile. Avenida Vicuña Mackenna 4860, Macul, Santiago, 7820436, Chile
dDepartment of Chemistry and Biochemistry, The Ohio State University, 100 W 18th Ave., Columbus, OH 43210, USA
eCentro Integrativo de Biología y Química Aplicada (CIBQA), Universidad Bernardo O’Higgins, Santiago, 8370854, Chile
fDepartamento de Química de los Materiales, Facultad de Química y Biología, Universidad de Santiago de Chile, Av. Libertador Bernardo O’Higgins, 3363 Santiago, Chile
First published on 7th February 2024
Abstract
With the current food demand, pesticides have become some of the most important compounds to maintain food quality; however, this requires the development of methodologies that allow fast, sensitive, and low-cost screening for these contaminants. Surface-enhanced Raman spectroscopy is an emerging solution since the interaction between the analyte and metallic nanostructures increases the Raman signals, enabling trace detection and a chemical-specific measurement. In this way, the fast detection of contaminants is possible, and further advances may enable portable assays. In this work, SERS substrates with silver nanosheets (AgNSs) on a copper surface were synthesized, producing a strong SERS effect and a reproducible signal intensity from methylene blue probe molecules at an optimal reaction time of 1 min. A quantitative analysis of the pesticide imidacloprid was then performed by applying a PLSR chemometric model, revealing a high linear correlation between the reference values and the predicted values of the pesticide (Rcv2 = 0.9732 and RMSECV = 0.1239). AgNS substrates were used to determine the feasibility of using this methodology for screening imidacloprid in real bee honey samples obtained through QuEChERS extraction, and an average recovery of 75.5% ± 0.08 was obtained. In addition, density functional theory simulations were carried out to elucidate the possible molecular interaction with the SERS surface and to assign the observed vibrational modes of imidacloprid. SERS is thus demonstrated to be an alternative to conventional pesticide detection techniques.
1. Introduction
Pesticides are widely used to produce and maintain food quality, and it is estimated that their use will increase to 3.5 million tons per year.1,2 Since the 1990's, neonicotinoid pesticides, compounds derived from nicotine, have become some of the most widely used insecticides worldwide due to their systemic action, ease of use, and broad spectrum of action.3–5 Imidacloprid is the pesticide that has experienced the most significant growth, positioning itself among the most used pesticides in the world.6 These pesticides act as agonists of the nicotine acetylcholine receptors (nAChRs), located in the central nervous system of insects, preventing them from carrying out neuronal synapses.5,7One of the problems associated with these pesticides is the contamination of the environment, which has various repercussions for the ecosystem since non-target organisms are also affected.8 For example, due to their systemic action, these pesticides can translocate throughout plants, even reaching pollen and nectar, thus affecting pollinators such as bees. Several studies have associated these neurotoxic pesticides with colony collapse disorder (CCD) that has decimated bee populations globally, significantly affecting hives.9–14 The loss of bee colonies in the last twenty years has generated increased interest in monitoring pesticides within the research community.15
Currently, different analytical techniques are used for the detection of neonicotinoids, including electrochemical methods,16–18 immunoassays,19–21 fluorescence,22–24 HPLC, or GC coupled to mass detectors.25–30 These methods exhibit high selectivity and sensitivity for detecting these analytes. Still, they have a downside because most of the time, the treatment of the sample is very complex, the analysis times are long, and trained personnel are required to detect these analytes. Also, the associated high cost and low portability limit in situ analysis.
In the last 20 years, surface-enhanced Raman scattering (SERS) has emerged as a promising technique for detecting and identifying various trace contaminants through their unique spectroscopic fingerprints. In addition, it is a non-destructive technique with a high-speed analysis response and high sensitivity.31–33 SERS involves Raman signal amplification due to the analyte adsorption on a metallic nanostructure. This technique involves two enhancement mechanisms; the first corresponds to an electromagnetic mechanism (EM) produced by the excitation of localized surface plasmon resonance (LSPR) in the interstices of plasmonic materials. The second corresponds to the chemical mechanism (CM) produced by the interaction between the analyte and the metal substrate.32,34,35
The development of high-performance SERS substrates is crucial for detecting analytes. Over the years, several methodologies have been used, such as self-assembly,36 thermal evaporation,37 electrodeposition,38 lithography,39 and the use of semiconductors.40–42 Satisfactory SERS performances have been achieved from these substrates when used with probe molecules. However, these methodologies are quite time-consuming and have a high cost. The galvanic displacement reaction solves these problems since it is a simple, fast, low-cost technique.43 In this technique, Ag ions in a solution are reduced, forming isolated nanostructures, which act as seeds for continuous accumulation through deposition on the copper surface, which allows control of the composition and morphology of the nanoparticles, in addition to providing the possibility of generating a porous network to have a more significant number of hotspots.44 Due to the incredible versatility of this technique, it has been used in various studies to increase the sensitivity of SERS; Hu et al. synthesized silver nanoparticles on copper substrates with excellent sensitivity to detect rhodamine 6G in low concentrations.45 One of the difficulties associated with galvanic displacement is the low uniformity of the obtained substrates; to overcome this, capping agents have been used, given that these agents control the growth rate relationship between the (111) and (100) planes of silver, thus retarding the growth of the nanostructures;46 for example, Wu et al. used polyvinylpyrrolidone (PVP) as a capping agent to control the growth rate of the (100) facet of silver and thus, the uniformity of the obtained substrate was controlled. In this way, it was possible to obtain a substrate that improves SERS performance for detecting pesticides in water samples.47
In this work, easy-to-manufacture and low-cost silver nanosheets (AgNSs) were prepared through a galvanic displacement reaction, using sodium citrate as a capping agent to control the uniformity of the substrate. The SERS effect of this platform is tested using a probe molecule such as methylene blue. The SERS substrate was used to detect trace levels of the pesticide imidacloprid using the PLSR chemometric model and its feasibility was tested to evaluate imidacloprid at the screening level in honey samples. Additionally, density functional theory (DFT) calculations were performed to assign the characteristic vibrational modes of the molecule.
2. Materials and methods
2.1. Reagents and standard preparation
Aqueous solutions were prepared using ultrapure water (18.2 MΩ cm) purified using a Simplicity® system (Merck Millipore, Darmstadt, Germany). Imidacloprid (CHEM SERVICE, West Chester, PA, USA), copper foil of about 0.1 mm thickness, methylene blue, silver nitrate, sodium citrate tribasic dihydrate, ethanol, acetone, nitric acid, polyethylene glycol Mw 1000, sodium carbonate anhydrous, hydrogen peroxide, and methanol (Merck Millipore, Darmstadt, Germany) were purchased from the indicated vendor.A stock solution of 1000 μg mL−1 methylene blue in methanol was prepared. A stock solution of 1000 μg mL−1 in a mixture of methanol and ultrapure water at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) ratio was prepared for imidacloprid. From there, the working solutions were prepared for each analysis performed. Stock and working solutions were stored at 4 °C.
1 (v/v) ratio was prepared for imidacloprid. From there, the working solutions were prepared for each analysis performed. Stock and working solutions were stored at 4 °C.
2.2. Preparation of the SERS substrate
Silver nanosheets (AgNS) were synthesized by a process described by Li et al.48 using silver nitrate (AgNO3, 20 mM), sodium citrate (7 mM), and different reaction times (0.5, 1, 2, 5, and 10 min). The UV-vis absorption spectra were recorded using a spectrophotometer (UV-2450, Shimadzu), and the substrates were morphologically characterized by scanning electron microscopy (SEM, FEI Quanta 250), energy dispersive spectroscopy (EDS) and X-ray diffraction (XRD, Bruker D8 Advance X-ray diffractometer).2.3. DFT frequency computations
A conformational search on the structure of Imidacloprid was performed at the B3LYP/6-31G(d,p) level of theory to provide an appropriate description of dihedral angles between pyridinium and imidazolium rings. Next, a full geometry optimization in the gas phase was performed at the B3LYP/6-31 + G(d,p) level of theory. Anharmonic vibrational frequencies were obtained with the optimized geometry of imidacloprid at the same level of theory. All calculations were performed with Gaussian 16.492.4. Raman measurements
To achieve an efficient adsorption process for the analytes on the surface of the AgNS substrates, the platform is immersed in an analyte solution with different concentrations for 3 h, and then washed with water and air-dried before being measured by SERS.SERS spectra were obtained using an AFM-Raman confocal microscope (Witec Alpha 300 RA, Germany) equipped with a 532 nm laser. The microscope objective was 50× (0.8 NA). The spectra were obtained from the average of 3 measurements between 300 and 1800 cm−1, with an integration time of 10 s and 5 accumulations. Spectral data were analyzed using Witec Control FOUR software to facilitate analysis.
2.5. Statistical analysis
Partial least squares regression (PLSR) is used to quantitatively relate spectral data (variable X) to concentrations of imidacloprid in solution (variable Y). A complete cross-validation procedure determines the optimal number of factors to use within the PLSR regression. The model is evaluated using the coefficient of determination (R2) and the root mean square error (RMSE).2.6. QuEChERS extraction
5.0 g of honey was weighed into a 50 mL polypropylene tube. The extraction began by adding 7.5 mL of water and 10.0 mL of acetonitrile, shaking until complete homogenization. Then, 1.0 g of sodium chloride and 6.0 g of anhydrous magnesium sulfate were added and agitated vigorously. The tube was centrifuged for 5 minutes at 500 rpm, and 2.0 mL of supernatant was taken with a micropipette into a 15.0 mL polypropylene tube containing 50 mg of C18, 50 mg of PSA, and 150 mg of anhydrous magnesium sulfate, vortexed, and then centrifuged for 5 minutes at 3000 rpm. Finally, the supernatant was filtered through a 0.22 μm PVDF membrane.3. Results and discussion
3.1. SERS substrate characterization
The morphological characterization of the surfaces was carried out by scanning electron microscopy (SEM). Fig. 1 presents typical SEM images of the substrates prepared with different reaction times. Fig. 1(a) shows the morphology of the substrates with a reaction time of 0.5 minutes, indicating a rapid deposition rate; the formation of small isolated units corresponding to silver nanostructures was observed, which will serve as seeds during the process. | ||
| Fig. 1 SEM images of silver nanosheets (AgNSs) at different reaction times: (a) 0.5 min, (b) 1 min, (c) 2 min, (d) 5 min, and (e) 10 min and (f) elemental composition by EDS in SEM. | ||
Fig. 1(b) shows that when the reaction time increases to 1 min, it is observed that these nanostructures begin to accumulate on the copper surface, increasing the density of the nanosheets, which causes the separation in the AgNS to become smaller, thus generating a more significant number of hotspots. As the reaction time increases, it is observed that the deposition of these nanostructures becomes increasingly more prominent and much denser, as evident in the cases of the times of 2 and 5 minutes shown in Fig. 1(c) and (d). When the time reaches 10 minutes, much larger and thicker units are formed than the previous ones, as shown in Fig. 1(e).
Fig. 1(f) shows the atomic composition of the synthesized AgNS substrates at 0.5, 1, 2, 5, and 10 min through EDS analysis in SEM. Here, you can see that the ratio of Ag/Cu atoms on the surface of the platform changes with the course of the reaction since the amount of Cu decreases, and Ag increases as the reaction time increases, which suggests that the galvanic displacement reaction develops correctly.
To evaluate the optical properties of the substrates, they were analyzed by UV-visible spectroscopy (Fig. S1, ESI†), where it can be observed that on the AgNS platform, unlike the initial copper substrate, there is an absorption peak at 320 nm.
In relation to the composition of the substrates, XRD analysis was carried out to investigate the crystallinity of the surfaces. Fig. S2 (ESI†) shows the different patterns of each evaluated substrate, where the XRD spectrum of the copper foil (red) shows three diffraction peaks at 2θ of 43.6°, 50.7°, and 74.3°, corresponding to its network planes (111), (200) and (320), respectively. In the AgNS substrate (green), the three peaks associated with copper are observed and a discernible peak appears at a 2θ of 38.2°, which corresponds to the diffraction peak of the lattice plane (111) of silver, indicating that the preferential orientation of Ag was along the (111) plane and its low intensity is associated with the use of sodium citrate as a capping agent, controlling the rate of galvanic displacement, since it binds strongly to the (111) facets, making the growth much slower.
In SERS experiments, dye molecules play a crucial role because these compounds exhibit considerable Raman resonance effects. This is why a probe molecule, such as methylene blue dye, which shows a characteristic SERS signal, is used to study the performance of optimally synthesized substrates.50
Fig. 2(a) shows the SERS spectra of methylene blue (1 μg mL−1) on the AgNS platforms at 0.5, 1, 2, 5, and 10 minutes of reaction time. Herein, the characteristic signals of this molecule are observed, such as the C–N–C skeletal deformations (455 cm−1), the C–S–C skeletal deformations (615 cm−1), C–H in-plane bending (896 and 1033 cm−1), C–H in-plane ring deformation (1310 cm−1), the C–N symmetric stretching (1414 cm−1), and the C–C ring stretching (1610 cm−1).31,51 The behavior of the different SERS substrates used shows that as the reaction time increases, the intensity of the signal first increases and then decreases. During the initial times, there is little presence of the nanostructures on the surface, which reduces the presence of hotspots and the enhanced Raman signal; as the reaction time increases, an optimum structure is realized balancing the size and density of the nanostructures. Further reaction beyond this optimum point reduces the observed Raman enhancement. As a result, the optimum Raman signal enhancement is achieved with a reaction time of 1 min.
The reproducibility of the substrates plays a fundamental role in the SERS measurements, and this directly depends on the morphological and surface properties of the metallic nanostructures used. An area is delimited in the center of the AgNS platform at the optimal reaction time of 1 min to study reproducibility, and methylene blue spectra (1 μg mL−1) are collected at different points within this area. The SERS spectra of methylene blue in the region of the AgNS platform are shown in Fig. 2(b), where the characteristic signals of the molecule are observed. The intensity distribution histograms of the characteristic peaks of methylene blue at 1414 cm−1 and 1610 cm−1 are shown in Fig. 2(c) and (d). The relative standard deviation (RSD) is calculated for the spectra obtained at ten different points on the AgNS platform to quantify the point-to-point reproducibility. The resulting statistical evaluations showed an RSD value of 6.66% for the signal found at 1414 cm−1 and 3.58% for the sign at 1610 cm−1, which suggests that the platform has good performance and homogeneity given that the RSD value is below 15.00%, a value that is indicative of good reproducibility of the substrates as indicated by other studies,52–54 for example, Li determines the reproducibility of the substrate with %RSD values of 12.96 and 11.69%. The authors point out that the reproducibility is reasonable to guarantee the practical application of the substrate.52
A stability study of the AgNS platforms is carried out to observe if they oxidize over time. To achieve this, the SERS spectrum on the optimal AgNS substrate is determined in a certain area every 2 days without modifying the laser intensity. The results of the analysis are shown in Fig. S3 (ESI†), where when monitoring the characteristic signal of methylene blue at 1610 cm−1 in the AgNS substrate over time, it is observed that this signal gradually decreases during storage and no changes in observed sign are noted. Furthermore, there are no signals associated with oxides on the surface of the platforms, indicating good stability of the substrates.
The enhancement factor (EF) is a parameter that allows us to quantify the magnitude of the improvement that a SERS substrate presents; this is calculated from the following equation:47,48,55
3.2. Vibrational assignment of imidacloprid
Based on the results obtained for the AgNS platforms with the methylene blue probe molecule, we now seek to study the detection of the pesticide imidacloprid. To have a better understanding of the phenomena associated with the detection and monitoring of analytes at low concentrations, it is essential to combine SERS information with theoretical calculations such as DFT since this allows us to connect theoretical and experimental research, obtaining all the spectroscopic information relevant to the analytes. Initially, a DFT analysis was performed on the gas phase, and the optimized structure with a numbering scheme for imidacloprid is shown in Fig. 3(a). The molecule consists of a chloropyridine ring linked to an N-nitroimidazolidine ring. The experimental, theoretical unscaled, and SERS Raman spectrum of imidacloprid is shown in Fig. 3(b), where the characteristic peaks of the molecule can be seen. The assignment of these peaks and their corresponding vibrational modes are recorded in Table S1 (ESI†).3.3. Imidacloprid SERS detection
A quantitative analysis is performed by modifying the concentration of imidacloprid and applying the PLSR chemometric algorithm to see the relationship between the spectra and the concentration of the analyte. Fig. 4(a) shows the SERS spectra of the imidacloprid standard solution at different concentrations. The characteristic signals of the molecule are present and in accordance with the Raman spectrum. The intensity of the SERS spectrum decreases as the analyte concentration decreases, keeping the spectrum profile unchanged. Based on the characteristic bands of the SERS spectra, imidacloprid can be identified at levels as low as 0.02 μmol L−1. The SERS spectral information of imidacloprid was determined at different concentrations adsorbed on the platform surface (1.96–0.02 μmol L−1). For the construction of the model, an analysis was carried out in triplicate; the spectral data obtained were pretreated by applying the second derivative using the Savitzky–Golay algorithm with a second-order polynomial to reduce the deviations caused by dispersion effects, and cross-validation was employed to evaluate the quality of the model. Fig. 4b shows the concentration values predicted by the PLSR model as a function of the reference concentration of imidacloprid. The value of R2 for the calibration process corresponds to 0.9770, and for the cross-validation process, it is 0.9732. The RMSE is 0.1091, while the RMSECV corresponds to 0.1239. The results show high R2 values and low calibration errors; therefore, the model can perform a quantitative analysis for the pesticide imidacloprid.This methodology allows us to detect imidacloprid in concentrations as low as 0.02 μmol L−1, which is below the maximum residue limit (MRL) allowed by the Codex Alimentarius for the presence of pesticides in various foods (0.039–195.6 μmol L−1). The observed detection limit for imidacloprid with the AgNS platforms is compared with other studies in Table 1. The SERS methodology is an excellent alternative to classical analytical techniques with a lower associated cost and the potential for in situ detection.
Finally, the feasibility of using this methodology for screening analysis in a real sample will be studied. Honey will be used as a real sample since neonicotinoid pesticides have been detected to affect non-target organisms such as bees and have been considered one of the factors contributing to colony collapse disorder (CCD), leading to the disappearance of bees worldwide. Once the AgNS parameters in the solution have been optimized, a test is performed on extracts of honey samples intentionally enriched with imidacloprid at three concentration levels obtained by the QuEChERS method. Fig. 5 shows the comparison between the spectrum of imidacloprid in an aqueous solution and one of the honey extracts, where it is observed that the most important signal of imidacloprid presents a slight reduction compared to the aqueous solutions. This is mainly due to hotspot competition between the target molecule and honey components such as waxes, sugars, and pigments.73 The average recovery of the extracts is 75.5% ± 0.08, which is slightly lower than that obtained in previous tests using HPLC-MS./MS as a quantification method,74 demonstrating that AgNS substrates could detect contaminants in real samples at a screening level faster than chromatographic techniques.
 | ||
| Fig. 5 SERS performance of the AgNS platform for imidacloprid in aqueous solution and QuEChERS extract of honey. | ||
4. Conclusions
In the present work, easily fabricated and low-cost SERS AgNS platforms were synthesized. They exhibit good reproducibility and enhance Raman signals. These platforms enabled the spectroscopic characterization of the pesticide imidacloprid and the assignment of the experimental Raman and SERS bands to the corresponding vibrational modes through a DFT analysis. Finally, SERS successfully identified imidacloprid in solution at low concentrations with a LOD of 0.02 μmol L−1 and the applied chemometric model PLSR exhibited good linearity and sensitivity (Rcv2 = 0.9732 and RMSECV = 0.1239). Detecting this analyte as a screening method in complex matrices, such as honey after QuEChERS extraction, was proved to be feasible with an average percentage recovery of 75.5% ± 0.08. This methodology has been proven to be an alternative to conventional pesticide follow-up techniques.Author contributions
Felipe Leyton-Soto: investigation, methodology, formal analysis, and writing – original draft. Zachary D. Schultz: supervision and writing – review and editing. Rodrigo Ormazábal-Toledo: formal analysis and software. Domingo Ruiz-León: formal analysis and software. Ady Giordano: investigation, supervision, resources, and writing – review and editing. Mauricio Isaacs: conceptualization, supervision, writing – review and editing, and Funding acquisition.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
The authors acknowledge and appreciate the financial support from ANID Doctoral Fellowships 21181400, FONDEQUIP EQM 1500020, and FONDEQUIP EQM 150101. This work was funded by the Millenium Institute on Green Ammonia as Energy Vector MIGA, ANID/Millennium Science Initiative Program/ICN2021_023 and FONDECYT Regular #1221179. Powered@NLHPC: This research was partially supported by the supercomputing infrastructure of the NLHPC (ECM-02). ZDS acknowledges support from the National Science Foundation Award CHE-2107791.References
- A. Sharma, V. Kumar, B. Shahzad, M. Tanveer, G. P. S. Sidhu, N. Handa, S. K. Kohli, P. Yadav, A. S. Bali, R. D. Parihar, O. I. Dar, K. Singh, S. Jasrotia, P. Bakshi, M. Ramakrishnan, S. Kumar, R. Bhardwaj and A. K. Thukral, SN Appl. Sci., 2019, 1, 1446 CrossRef CAS.
- A. Sharma, A. Shukla, K. Attri, M. Kumar, P. Kumar, A. Suttee, G. Singh, R. P. Barnwal and N. Singla, Ecotoxicol. Environ. Saf., 2020, 201, 110812 CrossRef CAS PubMed.
- D. Browne, J. Levison, V. Limay-Rios, K. Novakowski and A. Schaafsma, Hydrogeol. J., 2021, 29, 651–666 CrossRef CAS.
- L. Bakker, W. van der Werf, P. Tittonell, K. A. G. Wyckhuys and F. J. J. A. Bianchi, Ecol. Soc., 2020, 25, 1–22 Search PubMed.
- B. Buszewski, M. Bukowska, M. Ligor and I. Staneczko-Baranowska, Environ. Sci. Pollut. Res., 2019, 26, 34723–34740 CrossRef CAS PubMed.
- C. M. Batikian, A. Lu, K. Watanabe, J. Pitt and R. M. Gersberg, Chemosphere, 2019, 223, 83–90 CrossRef CAS PubMed.
- A. J. Crossthwaite, A. Bigot, P. Camblin, J. Goodchild, R. J. Lind, R. Slater and P. Maienfisch, J. Pestic. Sci., 2017, 42, 67–83 CrossRef CAS PubMed.
- M. Pagano, A. Stara, V. Aliko and C. Faggio, J. Mar. Sci. Eng., 2020, 8, 1–14 CrossRef.
- M. S. Saleem, Z. Y. Huang and M. O. Milbrath, Front. Ecol. Evol., 2020, 8, 556856 CrossRef.
- D. A. Tavares, T. C. Roat, E. C. M. Silva-Zacarin, R. C. F. Nocelli and O. Malaspina, Ecotoxicol. Environ. Saf., 2019, 169, 523–528 CrossRef CAS PubMed.
- J. Wu-Smart and M. Spivak, Environ. Entomol., 2018, 47, 55–62 CrossRef CAS PubMed.
- M. Henry, N. Cerrutti, P. Aupinel, A. Decourtye, M. Gayrard, J.-F. Odoux, A. Pissard, C. Rüger and V. Bretagnolle, Proc. R. Soc. B, 2015, 282, 20152110 CrossRef PubMed.
- P. R. Whitehorn, S. O’Connor, F. L. Wackers and D. Goulson, Science, 1979, 336, 351–352 CrossRef PubMed.
- C. H. Lin, D. B. Sponsler, R. T. Richardson, H. D. Watters, D. A. Glinski, W. M. Henderson, J. M. Minucci, E. H. Lee, S. T. Purucker and R. M. Johnson, Environ. Toxicol. Chem., 2021, 40, 1212–1221 CrossRef CAS PubMed.
- K. M. Kasiotis, E. Zafeiraki, E. Kapaxidi, E. Manea-Karga, S. Antonatos, P. Anastasiadou, P. Milonas and K. Machera, Sci. Total Environ, 2021, 769, 145213 CrossRef CAS PubMed.
- H. Wang, L. Pan, Y. Liu, Y. Ye and S. Yao, J. Electroanal. Chem., 2020, 862, 113955 CrossRef CAS.
- N. Ajermoun, A. Farahi, S. Lahrich, M. Bakasse, S. Saqrane and M. A. el Mhammedi, J. Sci. Food Agric., 2019, 99, 4407–4413 CrossRef CAS PubMed.
- M. ben Brahim, M. F. Elahmadi, H. B. Ammar and Y. Samet, Global Nest J., 2018, 20, 628–636 Search PubMed.
- E. Watanabe, T. Yamasaki, Y. Hirakawa, A. Harada, S. Iwasa and S. Miyake, Anal. Methods, 2018, 10, 3162–3169 RSC.
- E. Watanabe and S. Miyake, J. Environ. Sci. Health, Part B, 2018, 53, 707–712 CrossRef CAS PubMed.
- E. Watanabe, Y. Hirakawa, T. Yamasaki, S. Iwasa and S. Miyake, Anal. Lett., 2020, 53, 174–187 CrossRef CAS.
- E. Fuentes, C. Cid and M. E. Báez, Talanta, 2015, 134, 8–15 CrossRef CAS PubMed.
- X. Yan, H. Li, Y. Li and X. Su, Anal. Chim. Acta, 2014, 852, 189–195 CrossRef CAS PubMed.
- J. Jiménez-López, P. Ortega-Barrales and A. Ruiz-Medina, J. Food Compos. Anal., 2016, 49, 49–56 CrossRef.
- D. Louca Christodoulou, O. Kourouzidou, M. Hadjigeorgiou, P. Hadjiloizou, M. Constantinou, P. Constantinou, K. Kika and M. Klavarioti, Accredit. Qual. Assur., 2018, 23, 145–175 CrossRef CAS.
- J. M. Montiel-león, S. Vo, G. Munoz, M. Verner, M. Y. Hendawi, H. Moya, M. Amyot and S. Sauvé, Food Control, 2019, 104, 74–82 CrossRef.
- C. I. Nannou, V. I. Boti and T. A. Albanis, Anal. Bioanal. Chem., 2018, 410, 1977–1989 CrossRef CAS PubMed.
- M. Gaweł, T. Kiljanek, A. Niewiadowska, S. Semeniuk, M. Goliszek, O. Burek and A. Posyniak, Food Chem., 2019, 282, 36–47 CrossRef PubMed.
- T. P. Ahammed Shabeer, R. Girame, S. Utture, D. Oulkar, K. Banerjee, D. Ajay, R. Arimboor and K. R. K. Menon, Chemosphere, 2018, 193, 447–453 CrossRef CAS PubMed.
- A. Lawal, R. C. S. Wong, G. H. Tan and L. B. Abdulra’uf, Anal. Lett., 2019, 52, 231–248 CrossRef CAS.
- T. Xu, X. Wang, Y. Huang, K. Lai and Y. Fan, Food Control, 2019, 106, 106720 CrossRef CAS.
- Z. Wang, Q. Zhu, Y. Wang, S. Dou, Q. Chen and N. Lu, Spectrochim. Acta, Part A, 2021, 251, 119447 CrossRef CAS PubMed.
- T. Janči, D. Valinger, J. Gajdoš Kljusurić, L. Mikac, S. Vidaček and M. Ivanda, Food Chem., 2017, 224, 48–54 CrossRef PubMed.
- Q. Tong, W. Wang, Y. Fan and L. Dong, TrAC, Trends Anal. Chem., 2018, 106, 246–258 CrossRef CAS.
- H. Zheng, D. Ni, Z. Yu and P. Liang, Food Chem., 2017, 217, 511–516 CrossRef CAS PubMed.
- G. Cabello, K. C. Nwoko, J. F. Marco, M. Sánchez-Arenillas, A. M. Méndez-Torres, J. Feldmann, C. Yáñez and T. A. D. Smith, J. Alloys Compd., 2019, 791, 184–192 CrossRef CAS.
- Z. Ye, G. Sun, C. Sui, B. Yan, F. Gao, P. Cai, B. Lv, Y. Li, N. Chen, F. Xu, K. Wang, G. Ye and S. Yang, Nanotechnology, 2018, 29, 375502 CrossRef PubMed.
- Y. Wang, Y. Yu, Y. Liu and S. Yang, ACS Sens., 2018, 3, 2343–2350 CrossRef CAS PubMed.
- K. Y. Hong, C. D. L. de Albuquerque, R. J. Poppi and A. G. Brolo, Anal. Chim. Acta, 2017, 982, 148–155 CrossRef CAS PubMed.
- K. Lai, T. Xu, Q. Ye, P. Xu, J. Xie, D. Yan, S. Zhu, T. Jiang, W. Xiong and C. Gu, Spectrochim. Acta, Part A, 2023, 300, 122971 CrossRef CAS PubMed.
- C. Zhou, L. Sun, F. Zhang, C. Gu, S. Zeng, T. Jiang, X. Shen, D. S. Ang and J. Zhou, ACS Appl. Mater. Interfaces, 2019, 11, 34091–34099 CrossRef CAS PubMed.
- S. Xie, K. Lai, C. Gu, T. Jiang, L. Zhou, X. Zheng, X. Shen, J. Han and J. Zhou, Mater. Today Nano, 2022, 18, 100179 CrossRef CAS.
- L. Hu, Y. J. Liu, Y. Han, P. Chen, C. Zhang, C. Li, Z. Lu, D. Luo and S. Jiang, J. Mater. Chem. C, 2017, 5, 3908–3915 RSC.
- R. Liu and A. Sen, Chem. Mater., 2012, 24, 48–54 CrossRef CAS.
- L. Hu, Y. J. Liu, S. Xu, Z. Li, J. Guo, S. Gao, Z. Lu, H. Si, S. Jiang and S. Wang, Chem. Phys. Lett., 2017, 667, 351–356 CrossRef CAS.
- J. Zeng, Y. Zheng, M. Rycenga, J. Tao, Z. Y. Li, Q. Zhang, Y. Zhu and Y. Xia, J. Am. Chem. Soc., 2010, 132, 8552–8553 CrossRef CAS PubMed.
- H. X. Wu, M. C. Rong, Y. Ma and S. D. Chen, Micro Nano Lett., 2020, 15, 590–594 CrossRef CAS.
- X. Li, X. Lin, B. Liu, X. Zhao, H. Zhao, L. Wang and S. Cong, Appl. Phys. A: Mater. Sci. Process., 2019, 125, 1–10 CrossRef.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery Jr., J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman and D. J. Fox, Gaussian 16, Rev. C. 01, 2016 Search PubMed.
- B. Sharma, R. R. Frontiera, A. I. Henry, E. Ringe and R. P. Van Duyne, Mater. Today, 2012, 15, 16–25 CrossRef CAS.
- C. Li, Y. Huang, K. Lai, B. A. Rasco and Y. Fan, Food Control, 2016, 65, 99–105 CrossRef CAS.
- L. Li and W. S. Chin, ACS Appl. Mater. Interfaces, 2020, 12, 37538–37548 CrossRef CAS PubMed.
- S. Kamal, A. Chowdhury and T. Chung-Kuang Yang, Spectrochim. Acta, Part A, 2022, 270, 120826 CrossRef CAS PubMed.
- J. Zhu, J. Liu, Y. Fan, M. Wu, C. Zhou, H. Fu and Y. She, Spectrochim. Acta, Part A, 2022, 282, 121646 CrossRef CAS PubMed.
- W. A. Tegegne, W. N. Su, M. C. Tsai, A. B. Beyene and B. J. Hwang, Appl. Mater. Today, 2020, 21, 100871 CrossRef.
- P. J. Larkin, Infrared and Raman Spectroscopy: Principles and Spectral Interpretation, 2017 Search PubMed.
- M. Kumar, M. Srivastava and R. A. Yadav, Spectrochim. Acta, Part A, 2013, 111, 242–251 CrossRef CAS PubMed.
- M. V. Cañamares and A. Feis, J. Raman Spectrosc., 2013, 44, 1126–1135 CrossRef.
- F. Zhang, Y. Zhang, H. Ni, K. Ma and R. Li, Spectrochim. Acta, Part A, 2014, 118, 162–171 CrossRef CAS PubMed.
- N. Creedon, P. Lovera, J. G. Moreno, M. Nolan and A. O’Riordan, J. Phys. Chem. A, 2020, 124, 7238–7247 CrossRef CAS PubMed.
- A. A. G. Moreira, P. de Lima-Neto, E. W. S. Caetano, I. L. Barroso-Neto and V. N. Freire, Spectrochim. Acta, Part A, 2017, 185, 245–255 CrossRef CAS PubMed.
- T. Polat and Ş. Yurdakul, Spectrochim. Acta, Part A, 2014, 133, 683–696 CrossRef CAS PubMed.
- M. Arivazhagan and S. Jeyavijayan, Spectrochim. Acta, Part A, 2011, 79, 161–168 CrossRef CAS PubMed.
- D. Singh, K. Vikram, D. K. Singh, W. Kiefer and R. K. Singh, J. Raman Spectrosc., 2008, 39, 1423–1432 CrossRef CAS.
- M. Stefanut, M. Dobrescu, A. Cata, F. Fitigau, G. Osser, I. Lile, L. Bondar and I. Ienascu, J. Sci. Arts, 2021, 1, 307–314 Search PubMed.
- D. Tomasini, M. Sampaio, S. Caldas, J. Buffon, F. Duarte and F. Duarte, Talanta, 2012, 99, 380–386 CrossRef CAS PubMed.
- L. Zhang, Z. Wang, Y. Wen, J. Shi and J. Wang, Anal. Methods, 2015, 5, 205–210 RSC.
- C. Chen, Z. Han, W. Lei, Y. Ding, J. Lv, M. Xia, F. Wang and Q. Hao, J. Electrochem. Sci. Eng., 2019, 9, 143–152 CrossRef CAS.
- X. Zhai, M. Zhang, P. Chen, T. Siriphithakyothin, J. Liu, H. Zhao, X. Yang, A. Abd El-Aty, D. Baranenko and A. Hacimüftüoğlu, J. Chin. Chem. Soc., 2020, 67, 1078–1088 CrossRef CAS.
- K. River-Guzman, L. Franco, O. García-Beltrán, J. Calderon and E. Nagles, Int. J. Electrochem. Sci., 2018, 13, 5775–5787 CrossRef CAS.
- U. Pham, Q. Phan, L. Nguyen, P. Luu, T. Doan, H. Trinh, C. Dinh, T. Van Nguyen, T. Tran, D. Le, T. Pham, T. Le and D. Nguyen, Processes, 2022, 10, 442 CrossRef CAS.
- R. Hou, S. Pang and L. He, Anal. Methods, 2015, 7, 6325–6330 RSC.
- D. Orso, M. L. Martins, F. F. Donato, T. M. Rizzetti, M. Kemmerich, M. B. Adaime and R. Zanella, J. Braz. Chem. Soc., 2014, 25, 1355–1364 CAS.
- R. Bridi, A. Larena, P. N. Pizarro, A. Giordano and G. Montenegro, Cienc. Agrotecnol., 2018, 42, 51–57 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nj05956a |
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2024 |