Open Access Article
Open Access ArticleGuanidine modification improves functions of natural RNA-targeting alkaloids†
Tamaki
Endoh
 *a,
Sagar
Satpathi
*a,
Sagar
Satpathi
 a,
Yutong
Chen
b,
Saki
Matsumoto
a,
Tatsuya
Ohyama
a,
Peter
Podbevšek
c,
Janez
Plavec
a,
Yutong
Chen
b,
Saki
Matsumoto
a,
Tatsuya
Ohyama
a,
Peter
Podbevšek
c,
Janez
Plavec
 cde,
Kazumitsu
Onizuka
cde,
Kazumitsu
Onizuka
 b,
Fumi
Nagatsugi
b,
Fumi
Nagatsugi
 b and
Naoki
Sugimoto
b and
Naoki
Sugimoto
 *af
*af
aFrontier Institute for Biomolecular Engineering Research (FIBER), Konan University, 7-1-20 Minatojima-minamimachi, Kobe, 650-0047, Japan. E-mail: t-endoh@konan-u.ac.jp; sugimoto@konan-u.ac.jp; Fax: +81-78-303-1495; Tel: +81-78-303-1416
bInstitute of Multidisciplinary Research for Advanced Materials, Tohoku University, 2-1-1 Katahira, Aoba-ku, Sendai, Miyagi 980-8577, Japan
cSlovenian NMR Centre, National Institute of Chemistry, Hajdrihova 19, Ljubljana, SI-1000, Slovenia
dEN → FIST Centre of Excellence, Trg OF 13, SI-1000 Ljubljana, Slovenia
eFaculty of Chemistry and Chemical Technology, University of Ljubljana, Večna pot 113, p. p. 537, SI-1000 Ljubljana, Slovenia
fGraduate School of Frontiers of Innovative Research in Science and Technology (FIRST), Konan University, 7-1-20 Minatojima-minamimachi, Kobe, 650-0047, Japan
First published on 8th April 2024
Abstract
One of the important issues for RNA-targeting drugs is to improve their binding properties. Herein, we modified berberine, which interacts with structured RNAs, with the guanidine moiety. Protein expression from target mRNA was efficiently reduced by the modification, suggesting a simple strategy to improve the therapeutic activity.
Drugs have been developed by mainly targeting proteins. However, a limited number of proteins are relevant to diseases and have structural properties to be recognized by small drugs.1 Since a variety of drugs have already been developed, the depletion of proteins as potential new drug targets has been a concern. RNA has been attracting attention as an alternative drug target.2,3 Although the building blocks for RNA are four ribonucleotides, which is much smaller than twenty amino acids composing proteins, the RNAs form complicated tertiary structures. The formation of tertiary structures in RNAs enables the modulation of gene expressions, some of which contribute to the development of diseases.4,5 Non-coding RNAs are also involved in diseases, whereas their detailed mechanisms are under investigation.6
Intramolecular base pairing, which forms secondary structure motifs such as hairpin, bulge, and internal loop, is the first step for the formation of complex tertiary structures in RNA. RNA structures are also known to be highly modular at the level of secondary structures including modules involved in simple tertiary organized units such as junction and turn.7 Based on the structure features of RNAs, the secondary structure is expected to be a potential target for RNA-targeting drugs.8 In addition, some biologically important RNAs such as microRNAs and disease-related repeat sequences are intrinsically forming secondary structures that can be targets of small chemicals.9
Natural chemical compounds, which show some biological activity, have been used as seeds for developing drug molecules. Natural antibiotics, aminoglycosides, and their derivatives have been demonstrated to target RNAs,10 suggesting the feasibility of drug discovery targeting disease-related RNAs.11 In the present field, the most critical challenge is identifying seed molecules capable of targeting RNA secondary structures and further developing strategies to modify these seed molecules to enhance interactions for improved RNA-binding properties.12
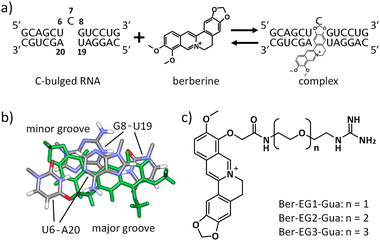
We have recently demonstrated that a natural alkaloid molecule, berberine, which has the potential to interact with several structured nucleic acids, such as triplexes,13 transfer RNA (tRNA),14 and G-quadruplexes,15 also interacts with the RNA secondary structure consisting of a single nucleotide bulge.16 The preferential binding was observed with the C-bulged RNA (Fig. 1a) consisting of 5′-UCG-3′ and 5′-UA-3′. The analysis of the tertiary structure of the complex between berberine and the C-bulged RNA elucidated the contribution of each chemical group of berberine to the interaction. The tertiary structure provides important information for further chemical modifications.16 In this study, we synthesized berberine derivatives modified with the guanidine moiety. Since the guanidine moiety provides positive charge and hydrogen bond-donor capabilities on berberine, the interaction with the C-bulged RNA would be enhanced by the electrostatic interactions and the formation of hydrogen bonds.17
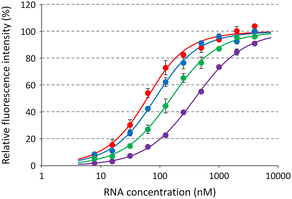
Based on the tertiary structure between berberine and the C-bulged RNA, one of the two methoxy groups of berberine is oriented toward the major groove side without direct interaction with RNA (Fig. 1b).16 We modified this methoxy group using the guanidine moiety with an ethylene glycol linker of different lengths (Fig. 1c). Three kinds of berberine derivatives, Ber-EG1-Gua, Ber-EG2-Gua, and Ber-EG3-Gua, were synthesized by coupling carboxy-modified berberine and guanidine having amino linkers with different lengths of ethylene glycol (Scheme S1 and Fig. S1–S8, ESI†). Interactions between the C-bulged RNA and synthesized berberine derivatives were evaluated based on their fluorescence changes after binding to the RNA (Fig. 2). Compared to the parental berberine, all derivatives showed higher association constants (KA) toward the C-bulged RNA (Table S1, ESI†). Ber-EG1-Gua, which has the shortest linker, showed a KA value that was more than 10-fold as compared to that of berberine. Electrostatic interactions have been suggested to be one of the important factors in enhancing the binding affinity because the decrease in binding affinity of PEG-EG1-Gua, depending on cation concentration, was more sensitive as compared to the parental berberine (Fig. S9 and Table S2, ESI†).
To evaluate the potential of the guanidine moiety to form hydrogen bonds with the C-bulged RNA, we have performed a molecular dynamics simulation based on the tertiary structure between RNA and parental berberine. The berberine complexed with the C-bulged RNA was replaced by the derivatives, and the dynamics of the complexes were simulated by GROMACS.18 From triplicated simulation at every 1 ps from 40 to 100 ns, the numbers of hydrogen bonds formed between the berberine derivatives and the RNA were analysed. Hydrogen bonds were formed between the guanidine donor and the phosphate acceptor (Fig. S10, ESI†). The number of hydrogen bonds was larger when the linker length was shorter (Table S3, ESI†), suggesting the contribution of the hydrogen bonds in enhancing the binding affinity. The nucleotide positions, where the hydrogen bonds were formed between the guanidine and the phosphate backbone, were more limited in the case of shorter linkers (Table S4, ESI†). The results suggest that the longer linker was too flexible and dynamic to enhance the binding affinity.
The berberine derivative, Ber-Gua (Fig. S11, ESI†), was also simulated as a model chemical, which has the guanidine moiety directly linked with berberine. The total number of hydrogen bonds formed between Ber-Gua and the C-bulged RNA was significantly less as compared to that formed between Ber-EG1-Gua and the C-bulged RNA (Fig. S11, ESI†) due to insufficient physical distance, suggesting that a too-short linker cannot contribute the additional interaction with the RNA. It would be important to estimate the appropriate linker length, e.g., by simulation, in order to efficiently enhance the binding affinity.
The structure of the complex between the C-bulged RNA and Ber-EG1-Gua was analysed by NMR. Similar to the parental berberine, the berberine unit of Ber-EG1-Gua was found to reside at the binding pocket formed by the cytosine bulge.16 Interaction with Ber-EG1-Gua induced the formation of the U–A and G–U base pairs adjacent to the cytosine bulge (Fig. S12, ESI†). The guanidine moiety of Ber-EG1-Gua was too dynamic and did not give rise to observable NOE contacts with the berberine unit or RNA. The result is in agreement with the molecular dynamics simulation, in which hydrogen bonds between various phosphate and guanidine moieties were observed.
The formation of the U–A and G–U base pairs caused by the interaction of Ber-EG1-Gua potentially stabilises the C-bulged RNA. The stabilities of the C-bulged RNA in the absence and presence of the berberine derivatives were analysed by UV melting profiles at 260 nm in a buffer containing magnesium and potassium, which corresponds to the buffer for the fluorescence titration study (Fig. 3a). The UV melting profile in the presence of parental berberine was also analysed. Thermodynamic parameters (ΔH°, ΔS°, and ΔG°25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (melting)) were calculated from the melting profiles (Table 1).19 The C-bulged RNA was stabilised in the presence of both berberine and its derivatives based on a favourable enthalpic contribution, which is likely provided by the induction of the U–A and G–U base pairs adjacent to the cytosine bulge. Berberine derivatives with shorter linkers caused greater stabilisation of the C-bulged RNA. Here, we calculated the interaction energy (ΔG°25
(melting)) were calculated from the melting profiles (Table 1).19 The C-bulged RNA was stabilised in the presence of both berberine and its derivatives based on a favourable enthalpic contribution, which is likely provided by the induction of the U–A and G–U base pairs adjacent to the cytosine bulge. Berberine derivatives with shorter linkers caused greater stabilisation of the C-bulged RNA. Here, we calculated the interaction energy (ΔG°25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (interaction)) between the C-bulged RNA and berberine and its derivatives from KA values obtained in the fluorescence titration (Table S1, ESI†) by using an equation ΔG°25
(interaction)) between the C-bulged RNA and berberine and its derivatives from KA values obtained in the fluorescence titration (Table S1, ESI†) by using an equation ΔG°25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (interaction) = −RTlnKA, where R represents the gas constant and T corresponds to the temperature. The ΔG°25
(interaction) = −RTlnKA, where R represents the gas constant and T corresponds to the temperature. The ΔG°25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (interaction) values were plotted against the differences in ΔG°25
(interaction) values were plotted against the differences in ΔG°25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (melting) (ΔΔG°25
(melting) (ΔΔG°25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (melting)) in the absence and the presence of the respective ligand. The plotted data showed a linear correlation (Fig. 3b), suggesting a similar mechanism of RNA stabilisation provided by the berberine derivatives depending on the binding affinity. The formation of U–A and G–U base pairs caused by the berberine unit and hydrogen bonding between the guanidine moiety and RNA were the main factors in stabilising the C-bulged RNA.
(melting)) in the absence and the presence of the respective ligand. The plotted data showed a linear correlation (Fig. 3b), suggesting a similar mechanism of RNA stabilisation provided by the berberine derivatives depending on the binding affinity. The formation of U–A and G–U base pairs caused by the berberine unit and hydrogen bonding between the guanidine moiety and RNA were the main factors in stabilising the C-bulged RNA.
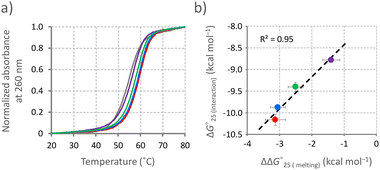 | ||
Fig. 3 Stabilisation of RNA by berberine derivatives. (a) UV melting profiles of the C-bulged RNA. Melting profiles of the C-bulged RNA (2 μM) at 260 nm were measured in the absence (gray) and presence of 10 μM berberine derivatives, berberine (purple), Ber-EG1-Gua (red), Ber-EG2-Gua (blue), or Ber-EG3-Gua (green), in a buffer containing 50 mM MES-LiOH (pH 7), 0.5 mM MgCl2, 100 mM KCl, and 0.1% DMSO. Melting profiles averaged from quadruplicated samples were normalized. (b) Correlation between the binding affinity and stabilisation effect provided by the berberine derivatives. ΔG°25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (interaction) values calculated from RNA titration in Fig. 2 were plotted against ΔΔG°25 (interaction) values calculated from RNA titration in Fig. 2 were plotted against ΔΔG°25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (melting) values calculated from the UV melting profiles. (melting) values calculated from the UV melting profiles. | ||
| ΔH° (kcal mol−1) | ΔS° (cal mol−1 K−1) | ΔG°25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (melting) (kcal mol−1) (melting) (kcal mol−1) |
|
|---|---|---|---|
| a Values are average ± S.D. calculated from quadruplicated samples. | |||
| Without chemical | −103 ± 2.7 | −288 ± 8.4 | −17.34 ± 0.23 |
| Berberine | −112 ± 1.5 | −314 ± 4.7 | −18.74 ± 0.13 |
| Ber-EG1-Gua | −120 ± 1.8 | −334 ± 5.3 | −20.47 ± 0.21 |
| Ber-EG2-Gua | −120 ± 1.0 | −335 ± 3.1 | −20.39 ± 0.04 |
| Ber-EG3-Gua | −118 ± 0.5 | −330 ± 1.7 | −19.84 ± 0.05 |
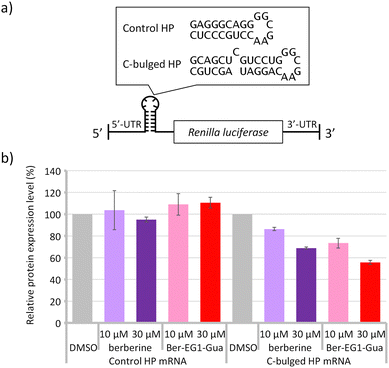
The structures and stabilities of messenger RNA (mRNA) are known to affect protein expression levels.5,20 When we consider RNA-targeting drug discovery, targeting the secondary structure region in the mRNA would be one of the strategies to control the expression level of proteins. Stabilisation of the secondary structure could potentially suppress the protein expression.21 Here, model mRNAs containing the canonical hairpin structure or secondary structure module of the C-bulged RNA were prepared (Fig. 4a).
In addition to the bulged secondary structure module, berberine is known to interact with the G-quadruplex formed by guanine-rich sequences and affect gene expression.22 As observed in the C-bulged RNA, the guanidine modification enhanced the interaction with the RNA G-quadruplex, although the affinity of Ber-EG1-Gua with the model RNA G-quadruplex (KA = 3.55 × 106 M−1) was relatively weaker than that with the C-bulged RNA (KA = 28.5 × 106 M−1) (Fig. S13, ESI†). Here, a reporter mRNA containing a module of the model RNA G-quadruplex (Fig. S14a, ESI†) was also prepared to evaluate the effect of the guanidine modification on the protein expression.
Protein expressions from the reporter mRNAs were evaluated in vitro in the absence and presence of berberine or Ber-EG1-Gua (Fig. 4b and Fig. S14b, ESI†). Protein expression from mRNA containing the usual hairpin structure module (control HP) was at similar levels irrespective of the additional berberine or Ber-EG1-Gua. In contrast, protein expression levels from mRNA containing the secondary structure module of the C-bulged RNA (C-bulged HP) were reduced by the addition of both berberine and Ber-EG1-Gua depending on their concentration. Ber-EG1-Gua showed more efficient suppression as compared to berberine, suggesting that the stabilisation of the secondary structure module resulted in reduced protein expression. In the case of reporter mRNA containing the G-quadruplex module, Ber-EG1-Gua also exhibited greater suppression of protein expression as compared to berberine at a concentration of 30 μM. However, the protein expression was less suppressed as compared to the mRNA containing the C-bulged HP module (Fig. S14b, ESI†). The relative protein expression levels, which correspond to suppression efficiency, depend on the binding affinity irrespective of the type of RNA module, C-bulged RNA or G-quadruplex (Fig. S15, ESI†), suggesting that the improvement of the affinity is important to construct a therapeutic material targeting RNAs.
Since the importance of RNAs in regulating biological processes has come to light, there has been a strong and active pursuit in developing molecules that specifically target these RNAs. Various strategies to obtain chemical compounds including natural chemicals that interact with structured regions of RNA have been proposed.23 The molecules obtained from these studies could serve as seed molecules for RNA-targeting drug discovery.3
In this study, we have modified berberine, which potentially targets structured RNAs, with a guanidine moiety to enhance its RNA binding affinity. The modification of guanidine with a linker of appropriate length has increased the binding affinity more than 10 times and stabilised the C-bulged RNA more than the parental berberine. The binding affinity with RNA G-quadruplex was also enhanced by the guanidine modification. The stabilisation effect of the C-bulged RNA, which resulted in the greater suppression of protein expression from reporter mRNA, was expected due to the formation of hydrogen bonds and electrostatic interactions between the guanidine moiety and RNA. Chemical functional groups such as carboxy and amine facilitate the modification of the guanidine moiety with different linker lengths. Therefore, as a strategy aimed at improving the RNA binding affinity of the seed chemicals, guanidine modification could be simple yet effective.
Conflicts of interest
There are no conflicts of interest.Acknowledgements
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and Japan Society for the Promotion of Science (JSPS) (KAKENHI Grant No. 18KK0164, 21H05108, and 23H02087), especially for Grant-in-Aid for Scientific Research (S) (22H04975), JSPS Core-to-Core Program (JPJSCCA20220005), The Hirao Taro Foundation of KONAN GAKUEN for Academic Research, the Chubei Itoh Foundation, Cooperative Research Program of “Network Joint Research Center for Materials and Devices”, and Slovenian Research and Innovation Agency (ARIS) Grant P1-0242. We thank Misa Kinoshita, Rika Manabe, Rie Sano, and Eri Miyaura, for their help with experiments. The authors acknowledge the CERIC-ERIC Consortium for access to experimental facilities.Notes and references
- K. D. Warner, C. E. Hajdin and K. M. Weeks, Nat. Rev. Drug Discov., 2018, 17, 547 CrossRef CAS PubMed; M. K. Sakharkar, K. R. Sakharkar and S. Pervaiz, Int. J. Biochem. Cell Biol., 2007, 39, 1156 CrossRef PubMed; M. K. Sakharkar and K. R. Sakharkar, Curr. Drug Discov. Technol., 2007, 4, 48 CrossRef PubMed.
- J. P. Falese, A. Donlic and A. E. Hargrove, Chem. Soc. Rev., 2021, 50, 2224 RSC; O. Khorkova, J. Stahl, A. Joji, C.-H. Volmar and C. Wahlestedt, Nat. Rev. Drug Discov., 2023, 22, 539 CrossRef CAS PubMed; C. M. Connelly, M. H. Moon and J. S. Schneekloth, Cell Chem. Biol., 2016, 23, 1077 CrossRef PubMed.
- A. L. Garner, ACS Med. Chem. Lett., 2023, 14, 251 CrossRef CAS PubMed.
- M. Zafferani and A. E. Hargrove, Cell Chem. Biol., 2021, 28, 594 CrossRef CAS PubMed.
- T. Endoh and N. Sugimoto, Chem. Rec., 2017, 17, 817 CrossRef CAS PubMed.
- M. Matsui and D. R. Corey, Nat. Rev. Drug Discovery, 2017, 16, 167 CrossRef CAS PubMed.
- M. A. Boerneke and K. M. Weeks, Biochemistry, 2018, 57, 6129 CrossRef CAS PubMed.
- A. J. Angelbello, J. L. Chen, J. L. Childs-Disney, P. Zhang, Z.-F. Wang and M. D. Disney, Chem. Rev., 2018, 118, 1599 CrossRef CAS PubMed; S. M. Meyer, C. C. Williams, Y. Akahori, T. Tanaka, H. Aikawa, Y. Tong, J. L. Childs-Disney and M. D. Disney, Chem. Soc. Rev., 2020, 49, 7167 RSC.
- R. Parkesh, J. L. Childs-Disney, M. Nakamori, A. Kumar, E. Wang, T. Wang, J. Hoskins, T. Tran, D. Housman, C. A. Thornton and M. D. Disney, J. Am. Chem. Soc., 2012, 134, 4731 CrossRef CAS PubMed.
- D. Moazed and H. F. Noller, Nature, 1987, 327, 389 CrossRef CAS PubMed.
- Q. Vicens and E. Westhof, Fac. Rev., 2022, 11, 39 CAS.
- M. D. Disney, J. Am. Chem. Soc., 2019, 141, 6776 CrossRef CAS PubMed; N. N. Patwardhan, L. R. Ganser, G. J. Kapral, C. S. Eubanks, J. Lee, B. Sathyamoorthy, H. M. Al-Hashimi and A. E. Hargrove, MedChemComm, 2017, 8, 1022 RSC.
- S. Das, G. S. Kumar, A. Ray and M. Maiti, J. Biomol. Struct. Dyn., 2003, 20, 703 CrossRef CAS PubMed.
- M. M. Islam, R. Sinha and G. S. Kumar, Biophys. Chem., 2007, 125, 508 CrossRef CAS PubMed.
- A. Arora, C. Balasubramanian, N. Kumar, S. Agrawal, R. P. Ojha and S. Maiti, FEBS J., 2008, 275, 3971 CrossRef CAS PubMed.
- S. Satpathi, T. Endoh, P. Podbevšek, J. Plavec and N. Sugimoto, Nucleic Acids Res., 2021, 49, 8449 CrossRef CAS PubMed.
- P. Gupta, O. Muse and E. Rozners, Biochemistry, 2012, 51, 63 CrossRef CAS PubMed; Y. Sato, T. Asami, Y. Toriyabe, T. Sato, N. Teramae and S. Nishizawa, Chem. Lett., 2016, 45, 982 CrossRef; L. Huang, J. Wang and D. M. J. Lilley, Cell Chem. Biol., 2017, 24, 695 CrossRef PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, SoftwareX, 2015, 1–2, 19 CrossRef.
- T. Endoh, H. Tateishi-Karimata and N. Sugimoto, in Handbook of Chemical Biology of Nucleic Acids, ed. N. Sugimoto, Springer Nature Singapore, Singapore, 2022 DOI:10.1007/978-981-16-1313-5_40-1.
- M. Kozak, Mol. Cell. Biol., 1989, 9, 5134; Y. Harigaya and R. Parker, Wiley Interdiscip. Rev. RNA, 2010, 1, 132 Search PubMed.
- T. Endoh, D. Hnedzko, E. Rozners and N. Sugimoto, Angew. Chem., Int. Ed., 2016, 55, 899 CrossRef CAS PubMed.
- T. Endoh and N. Sugimoto, Anal. Chem., 2013, 85, 11435 CrossRef CAS PubMed.
- W. J. Martin, P. Grandi and M. Marcia, Trends Pharmacol. Sci., 2021, 42, 758 CrossRef CAS PubMed; J. L. Childs-Disney, X. Yang, Q. M. R. Gibaut, Y. Tong, R. T. Batey and M. D. Disney, Nat. Rev. Drug Discov., 2022, 21, 736 CrossRef PubMed; A. Umuhire Juru and A. E. Hargrove, J. Biol. Chem., 2021, 296, 100191 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental methods, scheme to synthesize berberine derivatives (Scheme S1, and Fig. S1–S8), affinities between berberine derivatives and RNA (Table S1), potassium effects on RNA binding affinity (Fig. S9 and Table S2), analyses based on molecular dynamics simulation (Fig. S10 and Tables S3 and S4), berberine derivative for simulation (Fig. S11), structure analysis by NMR (Fig. S12), affinities between berberine derivatives and RNA G-quadruplex (Fig. S13), protein expression levels from mRNA containing G-quadruplex module (Fig. S14), and correlation between suppression of protein expression and interaction affinities (Fig. S15). See DOI: https://doi.org/10.1039/d3nj05833f |
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2024 |