Open Access Article
Open Access ArticleDoxorubicin loaded thermostable nanoarchaeosomes: a next-generation drug carrier for breast cancer therapeutics†
Kaviya Vijayalakshmi
Babunagappan
a,
Abirami
Seetharaman
a,
Subastri
Ariraman
a,
Poornima Budime
Santhosh
b,
Julia
Genova
b,
Natasa Poklar
Ulrih
 c and
Swathi
Sudhakar
*a
c and
Swathi
Sudhakar
*a
aDepartment of Applied Mechanics and Biomedical Engineering, Indian Institute of Technology Madras, Chennai, India. E-mail: swathi.s@iitm.ac.in
bInstitute of Solid State Physics, Bulgarian Academy of Sciences, Tzarigradsko Chausee, Sofia, Bulgaria
cDepartment of Food Science and Technology, Biotechnical Faculty, University of Ljubljana, Ljubljana, Slovenia
First published on 2nd January 2024
Abstract
Breast cancer has a poor prognosis due to the toxic side effects associated with high doses of chemotherapy. Liposomal drug encapsulation has resulted in clinical success in enhancing chemotherapy tolerability. However, the formulation faces severe limitations with a lack of colloidal stability, reduced drug efficiency, and difficulties in storage conditions. Nanoarchaeosomes (NA) are a new generation of highly stable nanovesicles composed of the natural ether lipids extracted from archaea. In our study, we synthesized and characterized the NA, evaluated their colloidal stability, drug release potential, and anticancer efficacy. Transmission electron microscopy images have shown that the NA prepared from the hyperthermophilic archaeon Aeropyrum pernix K1 was in the size range of 61 ± 3 nm. The dynamic light scattering result has confirmed that the NA were stable at acidic pH (pH 4) and high temperature (70 °C). The NA exhibited excellent colloidal stability for 50 days with storage conditions at room temperature. The cell viability results have shown that the pure NA did not induce cytotoxicity in NIH 3T3 fibroblast cells and are biocompatible. Then NA were loaded with doxorubicin (NAD), and FTIR and UV-vis spectroscopy results have confirmed high drug loading efficiency of 97 ± 1% with sustained drug release for 48 h. The in vitro cytotoxicity studies in MCF-7 breast cancer cell lines showed that NAD induced cytotoxicity at less than 10 nM concentration. Fluorescence-activated cell sorting (FACS) results confirmed that NAD induced late apoptosis in nearly 92% of MCF-7 cells and necrosis in the remaining cells with cell cycle arrest at the G0/G1 phase. Our results confirmed that the NA could be a potential next-generation carrier with excellent stability, high drug loading efficiency, sustained drug release ability, and increased therapeutic efficacy, thus reducing the side effects of conventional drugs.
1. Introduction
Over the past few decades, different varieties of drug carriers have been developed to improve the therapeutic index of various drugs. Self-assembled lipid bilayer structures called liposomes are among the most explored drug delivery vehicles.1 However, conventional or first-generation liposomes prepared from synthetic or natural phospholipids suffer from severe drawbacks such as physical and chemical instability in the biological environment. The colloidal instability resulted in uncontrolled or rapid drug release at the non-targeted region, associated side effects, poor drug availability, and low therapeutic index.2,3 Several methods were implemented to increase the liposome's colloidal stability by including cholesterol, charged components (anionic/cationic lipids), and high molecular weight polymers like polyethylene glycol. However, those studies resulted in limited success.4,5The thermostable lipids extracted from the domain archaea consist of single-celled microorganisms that are evolutionarily distinct from the other two domains, namely bacteria and Eukarya. Archaeal membrane lipids contain highly branched isoprenoid side chains, that are ether-linked to glycerol-1-phosphate backbone.6–8 These ether linkages are more stable than the ester linkages found in bacteria and Eukarya. They are resistant to oxidation, chemical hydrolysis, activity of enzymes like lipases, acidic, alkaline pH, and high temperatures.9 This extreme stability of the archaeal lipids renders them suitable to deliver drugs safely, thereby increasing the bioavailability, biodistribution, controlled drug release at the target region, and reduced side effects.10 Hence, there is a significant interest to use archaeal lipids in developing stable drug delivery systems in cancer theranostics.
Due to their amphipathic nature, the archaeal lipids form closed bilayer membrane nanovesicles called “nanoarchaeosomes (NA)” in polar media, similar to liposomes.11 However, NA differs from the conventional liposomes in various aspects such as high rigidity, less permeability to ions and other molecules, reactivity to different physical and chemical agents, lipid stereoisomerism, and heterogeneous lipid composition.12 Few studies reported that NA are biocompatible for various theragnostic applications, including imaging, drug delivery, and therapy.13,14 Additionally, they are cost-effective and simple to prepare in lab conditions, and also at an industrial scale. Reports have shown that NA exhibits superior properties in delivering anticancer peptides and drugs.15–17 However, the mechanism by which the drug-loaded archaeosomes induce cell death in cancer cells remains unclear. It is essential to properly understand NA-mediated cytotoxicity before their in vivo administration and applications. Moreover, only a handful of studies have been performed to elucidate the role of NA as a stable drug carrier for cancer therapeutics.15–17
In this aspect, in our study, we have synthesized and characterized the NA as a thermostable drug carrier system for cancer therapeutics, investigated their sustained drug release potential, and unravelled the cytotoxic mechanism in MCF-7 breast cancer cells. The NA was prepared by using archaeal lipid derived from the hyperthermophilic archaeon Aeropyrum pernix K1. The NA exhibited excellent thermal and pH stability and were stable at room temperature for 50 days with a highly biocompatible nature. The NA loaded with the anticancer drug doxorubicin (NAD) exhibited high drug entrapment efficiency and sustained drug release over 48 h. The NAD showed in vitro cytotoxicity in MCF-7 breast cancer cell lines with IC50 value less than 10 nM. Fluorescence-activated cell sorting (FACS) results confirmed that NAD induced late apoptosis in nearly 92% of MCF-7 cells with cell cycle arrest at the G0/G1 phase. Our studies conclude that NA could be a potential novel biomaterial-based nanotherapeutic tool with high colloidal stability and excellent therapeutic efficacy.
2. Experimental section
2.1 Growth of Aeropyrum pernix K1
A. pernix K1 was purchased from the Japan Collection of Microorganisms (number 9820; Wako-shi, Japan). The culture medium comprised (per litre): 34.0 g marine broth 2216 (Difco Becton, Dickinson & Co., Franklin Lakes, NJ, USA), 5.0 g trypticase peptone (Becton, Dickinson and Company, Sparks, USA), 1.0 g yeast extract (Becton, Dickinson and Company, Sparks, USA) and 1.0 g Na2S2O3·5H2O (Sigma-Aldrich, St. Louis, USA). The buffer system used was 20 mM HEPES [4-(2-hydroxyethyl)-1-piperazine ethane sulfonic acid] (Sigma-Aldrich Chemie GmbH, Steinheim, Germany) for archaeal growth at pH 7.0. The A. pernix cells were grown in 800 mL growth medium in 1000 mL heavy-walled flasks, with a hot plate magnetic stirrer and forced aeration (0.5 L min−1) at 92 °C, as described previously.182.2 Isolation and purification of archaeal lipids
The polar lipids extracted from the lyophilized hyperthermophilic archaeon A. pernix K1 consist of C25,25-archaeol (2,3-di-sesterpanyl-sn-glycerol), with C25,25-archetidyl(glucosyl)inositol (AGI) accounting for 91 mol%, and C25,25-archetidylinositol (AI) accounting for 9 mol%. These lipids were extracted from archaea using the protocol described by Gmajner et al.19 The lipids were fractionated using adsorption chromatography and analyzed by thin layer chromatography (TLC) with the chloroform/methanol/acetic acid/water (85/30/15/5) solvent. The TLC plate was developed and sprayed with 20% H2SO4. Lipid spots were visualized by heating at 180 °C for 20 min. TLC plates were analyzed using Just TLC software (version 3.5.3. https://www.sweday.com/).2.3 Preparation and characterization of NA
NA were prepared by the thin film method, as reported previously.20 The 1-stearoyl-2-oleoyl-sn-glycero-3-phosphocholine (SOPC) lipid and archaeal lipid in 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 (% w/v) ratio was dissolved in chloroform to achieve a final concentration of 1 mg mL−1 and vacuum-dried. The dry lipid film was hydrated with Milli-Q water and vortexed for 3 min to facilitate the formation of multilamellar vesicles (MLVs). The MLVs were further transformed into small unilamellar vesicles (SUVs) by sonication at 45 °C for 30 min, using an ultrasonic bath sonicator (BR Biochem Life Sciences). The sample was centrifuged at 13
20 (% w/v) ratio was dissolved in chloroform to achieve a final concentration of 1 mg mL−1 and vacuum-dried. The dry lipid film was hydrated with Milli-Q water and vortexed for 3 min to facilitate the formation of multilamellar vesicles (MLVs). The MLVs were further transformed into small unilamellar vesicles (SUVs) by sonication at 45 °C for 30 min, using an ultrasonic bath sonicator (BR Biochem Life Sciences). The sample was centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min (Eppendorf Centrifuge 5415C) to separate the SUVs from debris. The size and surface charge of the synthesized NA were analyzed using a DLS and a zeta analyzer (Horiba Scientific). About 5 μL of NA was spotted on the TEM copper grids, dried at room temperature, and imaged using a high-resolution transmission electron microscopy (JEOL JEM 3010, filament-LaB6).
000 rpm for 30 min (Eppendorf Centrifuge 5415C) to separate the SUVs from debris. The size and surface charge of the synthesized NA were analyzed using a DLS and a zeta analyzer (Horiba Scientific). About 5 μL of NA was spotted on the TEM copper grids, dried at room temperature, and imaged using a high-resolution transmission electron microscopy (JEOL JEM 3010, filament-LaB6).
2.4 Drug loading efficiency
Doxorubicin loaded NA (NAD) were prepared by adding 500 μL of NA (1 mg mL−1) with 500 μL of 100 μM doxorubicin and mixed (Eppendorf Thermo-Mixture C) at 300 rpm for 12 h at room temperature as previously described.21 Then the co-incubated nanoarchaeosomes and the doxorubicin molecules were subjected to a substantial freeze–thaw method. The freeze–thaw cycle was carried out five times via freezing the sample mixture under −80 °C and subsequent heating in a water bath at 65 °C. Due to this repeated process, the interlayer distance within the liposomes increases. The ice crystals formed during this process, induce temporary holes and pores within the liposomal structure, facilitating the passive entrapment of doxorubicin molecules inside the nanoarchaeosomes. Then the drug loaded NA were subjected to ultrafiltration using Amicon Ultra centrifugal filters, MWCO 30 kDa for 30 min at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm as previously described.22 Then the separated free drug in the filtrate was quantified by calculating the absorption at 478 nm using a UV-vis spectrophotometer (Nanodrop One, Thermo Scientific). With the intervention of a standard calibration curve, the amount of drug loaded, (i.e.) the drug loading efficiency (DLE) in NA was calculated using the below formula,23
000 rpm as previously described.22 Then the separated free drug in the filtrate was quantified by calculating the absorption at 478 nm using a UV-vis spectrophotometer (Nanodrop One, Thermo Scientific). With the intervention of a standard calibration curve, the amount of drug loaded, (i.e.) the drug loading efficiency (DLE) in NA was calculated using the below formula,232.5 In vitro drug release
The drug release kinetics of NAD was experimentally investigated using the dialysis membrane method. The NAD-filled dialysis membrane (12–14 kDa, HiMedia), was immersed in a beaker with phosphate buffer (pH 7.4 and 4.5), and stirred at 150 rpm for 48 h continuously. The sample was withdrawn and analyzed using a UV-vis spectrophotometer at 478 nm in predetermined intervals with dilution factor of 0.025.2.6 Fourier transform infrared spectroscopy
The interaction between the doxorubicin and NA was characterized using FTIR spectroscopy (Nicolet iS5 FT-IR). The individual samples (SOPC, archaeal lipid, NA, doxorubicin, and NAD) were mixed with KBr in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (% w/w), using a pellet-making mould administered under the pressure range 0.5–1 Pa of a hydraulic press. The pellet was mounted on a FTIR sample mount, and the transmittance spectrum was recorded (500–4000 cm−1) for all the samples.
100 (% w/w), using a pellet-making mould administered under the pressure range 0.5–1 Pa of a hydraulic press. The pellet was mounted on a FTIR sample mount, and the transmittance spectrum was recorded (500–4000 cm−1) for all the samples.
2.7 Cell line and cell culture
The human breast cancer cell line MCF-7 and the embryonic mouse fibroblast cell line NIH 3T3 were obtained from NCCS (National Centre for Cell Science, Pune). The cells were free of mycoplasma as evidenced by a negative result using agar culture method. The cells were cultured in 5% CO2 at 37 °C with the supplementation of DMEM (Dulbecco's modified Eagle's medium) along with 10% FBS (Fetal Bovine Serum) and 1% penicillin–streptomycin solution (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 units of penicillin and 10 mg of streptomycin in 0.9% NaCl) solution for routine sub-culturing and all further experiments.
000 units of penicillin and 10 mg of streptomycin in 0.9% NaCl) solution for routine sub-culturing and all further experiments.
2.8 Cell viability and MTT assay
The biocompatibility of NA and NAD was evaluated in NIH 3T3 cells using di-methyl thiazolyl diphenyl tetrazolium (MTT) assay based on the detection of mitochondrial dehydrogenase activity. NIH 3T3 cells were seeded and treated with NA (0.02 mg mL−1), and NAD (100 nM) once it reached 80% of confluency. The cytotoxic effect of NAD on MCF-7 cells was also assessed by the MTT assay. Briefly, cells (MCF-7 and NIH 3T3) were plated in 96 well tissue culture plates at a density of 5000 cells per mL and allowed to attach overnight. Following this, the cells were treated with different concentrations of NA alone, doxorubicin alone, and NAD alone (10 nM, 50 nM, 100 nM, 500 nM, and 1000 nM) for 24 h. Then the media was removed, and cells were washed with 1× PBS. After washing, MTT (5 mg mL−1) was added to the plate and incubated for 4 h in the dark. After incubation, the MTT was removed, and 100 μL DMSO (Dimethyl Sulfoxide) was added to dissolve the formazan crystals. Then the optical density was recorded using a microplate reader (Biotek H1m synergy) at 570 nm. Results were analyzed with the Origin Pro 8.5 software. The percentage of cell viability was determined using the following formula.23The control (MCF-7 and NIH 3T3), NA, doxorubicin, and NAD-treated cells were imaged using an inverted phase contrast microscope (Magnus Invi) after 24 h to assess the morphological changes.
2.9 Live/dead cell assay
The apoptotic effect of doxorubicin and NAD on breast cancer cells was analyzed using a fluorescent microscope (Nikon Inverted Ti U Eclipse). The acridine orange/ethidium bromide (AO/EB) staining can differentiate between live and dead cells. AO diffuses into cells binds with DNA and emits green fluorescence whereas EB doesn't cross intact cell membrane and it can diffuse across compromised cell membrane of dead cells indicated by red or orange cells. In brief, cells were seeded at a density of 1 × 105 cells per mL in 6 well cell culture plates for 24 h and treated with doxorubicin alone (500 nM) and NAD (100 nM), then incubated for 24 h. After incubation, the medium was removed, and the cells were washed with 1× PBS, pH 7.4. Then, equal volumes (100 μg mL−1) of AO/EB fluorescent dyes were added to the cells and incubated for 30 min. Then, the cells were washed with 1× PBS.2.10 Apoptosis and necrosis assay
To elucidate the mechanism of action of NAD on cell cytotoxicity, apoptosis, and necrosis assay were carried out using a flow cell cytometry. MCF-7 cells were seeded in a 6-well plate at the density of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well and waited till 80% confluency was reached. Then, the cells were treated with NAD (100 nM) and incubated for 24 h, and the supernatant from each well was transferred to collect the non-adherent cells. Then, the remaining adherent cells were trypsinized and collected. Next, treated and untreated (negative control) cells were harvested by centrifuging the sample at 1000 rpm for 5 min, and then they were washed twice using cold 1× PBS. The pellet was resuspended in a 1× binding buffer containing HEPES (10 mM), sodium chloride (140 mM), and calcium chloride (2.5 mM) to favour the efficient binding of Annexin V. Then, 2 μL of Annexin V-APC (Allophycocyanin: 4 μg mL−1) and 2 μL of propidium iodide (PI) (1 mg mL−1) were added to each suspension to stain the cells. Suspensions were then gently vortexed and incubated in the dark at 4 °C for 15 min. To eliminate doxorubicin interferences and fluorochrome spillover, single controls for Annexin V, PI and doxorubicin were employed along with the unstained control. The cells suspended in the 1× binding buffer were analyzed using flow cell cytometry (Beckman Coulter). Unstained negative control sample was acquired first to consider autofluorescence followed by single control samples then proper gating was done ensuring fluorochrome compensation and results were analyzed in CytExpert software version 2.4.
000 cells per well and waited till 80% confluency was reached. Then, the cells were treated with NAD (100 nM) and incubated for 24 h, and the supernatant from each well was transferred to collect the non-adherent cells. Then, the remaining adherent cells were trypsinized and collected. Next, treated and untreated (negative control) cells were harvested by centrifuging the sample at 1000 rpm for 5 min, and then they were washed twice using cold 1× PBS. The pellet was resuspended in a 1× binding buffer containing HEPES (10 mM), sodium chloride (140 mM), and calcium chloride (2.5 mM) to favour the efficient binding of Annexin V. Then, 2 μL of Annexin V-APC (Allophycocyanin: 4 μg mL−1) and 2 μL of propidium iodide (PI) (1 mg mL−1) were added to each suspension to stain the cells. Suspensions were then gently vortexed and incubated in the dark at 4 °C for 15 min. To eliminate doxorubicin interferences and fluorochrome spillover, single controls for Annexin V, PI and doxorubicin were employed along with the unstained control. The cells suspended in the 1× binding buffer were analyzed using flow cell cytometry (Beckman Coulter). Unstained negative control sample was acquired first to consider autofluorescence followed by single control samples then proper gating was done ensuring fluorochrome compensation and results were analyzed in CytExpert software version 2.4.
2.11 Cell cycle analysis
For the cell cycle analysis, the treated and untreated (negative control) cells were collected and centrifuged at 1000 rpm for 10 min. The cell pellet was washed with ice-cold 1× PBS and resuspended in 500 μL PBS. 1 mL of 70% cold ethanol was added drop by drop and fixed for at least 2 h at −20 °C. Then, the cells were washed with cold 1× PBS by centrifuging at 1000 rpm for 5 min. Then the cells were incubated with 10 μL ribonuclease A (10 μg mL−1) in 100 μL of 0.1% Triton X buffer and incubated for 15 min at 4 °C. Before being analyzed, the cells were stained with a mixture containing 200 μL of propidium iodide (12 μg mL−1) in 0.1% Triton X buffer. The cell cycle profile (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events per sample) was determined using flow cell cytometry (Beckman Coulter). Results were analyzed in CytExpert software version 2.4. Student's t-test was performed to identify the statistical significance between control and NAD using SPSS software version 27.
000 events per sample) was determined using flow cell cytometry (Beckman Coulter). Results were analyzed in CytExpert software version 2.4. Student's t-test was performed to identify the statistical significance between control and NAD using SPSS software version 27.
3. Results and discussion
3.1 Development and characterization of thermostable NA
The NA were prepared by mixing SOPC and archaeal lipids in 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 (% w/v) ratio followed by vortexing, sonicating, and centrifuging the sample, as mentioned in the ESI methods section.† Since the archaeal lipids have extreme stability and less permeability, drug release from the liposomes synthesized entirely using pure archaeal lipids will be difficult. Hence, the archaeosomes intended for drug delivery are generally prepared by mixing a small proportion of archaeal lipids with different types of phospholipids.24 The NA size, shape, and surface charge were characterized using TEM, DLS, and Zeta analyzer. The TEM images of NA were shown in Fig. 1A. The TEM images confirm the spherical NA with a size range of 64 ± 9.8 nm (mean ± SD). We also observed NA with a size range of 61 ± 3 nm (mean ± SEM from Gauss fit) (Fig. 1B(i)) from DLS analysis, which coincides with the TEM results. Then in order to characterize their charge densities, we have measured the zeta potential. The zeta potential of SOPC, archaeal lipids, and nanoarchaeosomes (NA) was found to be −18 ± 3, −72 ± 6, and −55 ± 3 mV, which is in accordance with previous reports.25 The negative charge in NA can contribute to the electrostatic and steric stabilization, resulting in excellent stability of the NA. Next, we checked the stability of NA at 70 °C and acidic pH 4 by measuring the size of the NA. The size remained around 64 ± 3 nm and 60 ± 3 nm (mean ± SEM from Gauss fit) (Fig. 1B(ii) and (iii)). This confirms that the NA exhibits colloidal stability in acidic and high-temperature environments. Next, we studied the stability of NA at room temperature for 50 days. The DLS results have shown that NA displays excellent stability at room temperature with a size range of 60 ± 3 nm, even at day 50 (Fig. 1B(iv)).
20 (% w/v) ratio followed by vortexing, sonicating, and centrifuging the sample, as mentioned in the ESI methods section.† Since the archaeal lipids have extreme stability and less permeability, drug release from the liposomes synthesized entirely using pure archaeal lipids will be difficult. Hence, the archaeosomes intended for drug delivery are generally prepared by mixing a small proportion of archaeal lipids with different types of phospholipids.24 The NA size, shape, and surface charge were characterized using TEM, DLS, and Zeta analyzer. The TEM images of NA were shown in Fig. 1A. The TEM images confirm the spherical NA with a size range of 64 ± 9.8 nm (mean ± SD). We also observed NA with a size range of 61 ± 3 nm (mean ± SEM from Gauss fit) (Fig. 1B(i)) from DLS analysis, which coincides with the TEM results. Then in order to characterize their charge densities, we have measured the zeta potential. The zeta potential of SOPC, archaeal lipids, and nanoarchaeosomes (NA) was found to be −18 ± 3, −72 ± 6, and −55 ± 3 mV, which is in accordance with previous reports.25 The negative charge in NA can contribute to the electrostatic and steric stabilization, resulting in excellent stability of the NA. Next, we checked the stability of NA at 70 °C and acidic pH 4 by measuring the size of the NA. The size remained around 64 ± 3 nm and 60 ± 3 nm (mean ± SEM from Gauss fit) (Fig. 1B(ii) and (iii)). This confirms that the NA exhibits colloidal stability in acidic and high-temperature environments. Next, we studied the stability of NA at room temperature for 50 days. The DLS results have shown that NA displays excellent stability at room temperature with a size range of 60 ± 3 nm, even at day 50 (Fig. 1B(iv)).
 | ||
| Fig. 1 (A) TEM images of as-synthesized nanoarchaeosomes (NA) (B) DLS measurement of the NA dispersion at (i) room temperature (pH 7), (ii) 70 °C, (iii) pH 4, (iv) day 50 at room temperature. | ||
The therapeutic efficacy of the drug mainly depends on the stability of the carrier molecule. There are several studies that exist with liposomes as drug delivery agents.26,27 However, conventional liposomes lack colloidal stability, a major problem associated with their application at the commercial level. Most liposomal formulations are generally stable only for a few days when stored in a refrigerated condition. When exposed to in vivo environments, due to varied pH, enzymatic activity, and acidic environment, they lose their stability and release the drug at non-targeted regions, which causes deleterious effects.28 In our study, the next-generation NA displays excellent stability at high temperatures, acidic pH, and was stable at room temperature for more than 50 days. This property makes NA an excellent candidate for drug delivery applications by overcoming the limitations faced by the existing liposomal systems. Studies reported that nanomaterials less than 100 nm interact readily with cell surfaces and are taken up by the cells more efficiently through various cellular uptake mechanisms, including endocytosis, receptor-mediated endocytosis, and passive diffusion through the cell membrane.29,30 Reports also have shown that smaller nanoparticles can extravasate more easily through leaky tumor vasculature and penetrate deeper into tissues, increasing their chances of encountering cancer cells.31,32 We speculate that the novel NA with a size range of less than 100 nm will have a higher cellular uptake rate, enhanced permeability, and retention effect as previously reported by various nanosystems. Since NA are stable at acidic pH, they can resist the acidic environment of endosomes preventing premature drug release and ensure that the cargo reaches the cytoplasm where the drug can exert its therapeutic effect with less concentration, thus reducing drastic side effects.
3.2 Drug entrapment efficiency and release rate of NA loaded with doxorubicin (NAD)
Next, we tested the NA drug loading and release efficiency by loading doxorubicin to NA. The NA (1 mg mL−1) and doxorubicin (100 μM) were mixed for 12 h, and drug loading efficiency was measured using a UV-vis spectrometer. The drug loading efficiency of NA was found to be 97 ± 1%. Encapsulating doxorubicin in the NA is crucial to achieve a better therapeutic efficacy. Yoncheva et al., reported that encapsulation of doxorubicin results in chemical stability of doxorubicin, reducing side effects by decreasing undesired toxicity and by overcoming cellular resistance.33 Bandak et al., found that doxorubicin loaded liposomes significantly decreased the photodegradation.34 O'Brien et al., reported that liposomal doxorubicin exhibited lower cardiac toxicity compared to the free doxorubicin.35 Onafuye et al., reported that doxorubicin loaded albumin nanomaterials overcome an ABC transporter mediated drug resistance in neuroblastoma cells.36 Moreover, the nanomaterials loaded with doxorubicin will have high accumulation and drug delivery into a leaky tumor vasculature preserving the normal cells from cytotoxicity.33 The encapsulation of doxorubicin in NA helps to maintain the stability of doxorubicin molecules, protects it from degradation even at room and higher temperature, and thereby provides superior therapeutic effects, than free doxorubicin alone. The NAD can prevent rapid clearance, increases drug bioavailability, and helps to achieve sustained drug release thus reducing toxicity towards normal cells.Next, the drug release rate was evaluated using a standard assay at physiological (pH 7.4) and acidic pH (pH 4.5). From Fig. 2, we observed the sustained drug release at both pH. However, faster drug release was observed at an acidic pH than the physiological pH. At 12 h, around 63 ± 3% of drug release occurred at pH 4.5, whereas only 39 ± 3% of the drug was released at pH 7.4. At the end of 50 h, almost 100 ± 3% of drugs have been released at pH 4.5, while only 63 ± 2% drug release was observed at 50 h at pH 7.4.
 | ||
| Fig. 2 Cumulative drug release profile of doxorubicin released from NA in phosphate buffer at pH 7.4 (red) and 4.5 (green) (N = 3, mean ± SD). | ||
In general, at acidic pH (5.5–6.5) the liposomes undergo a phase transition from a lamellar phase to an inverted hexagonal phase, leading to slight variations in their spherical structure and membrane permeability, leading to the faster release of encapsulated molecules. Rehman et al., reported that the common feature observed in most of the liposomes, which tend to be stable at the physiological pH, but disrupt in slightly acidic media such as the tumor microenvironment to release the loaded drug.37 It is also worth to note that, doxorubicin is a weakly basic drug (pKa = 9.9) in both protonated and unprotonated forms.38 We hypothesize that the drug becomes predominantly protonated in an acidic environment. This change in the drug's ionization state can affect its interactions with the NA and the surrounding environment, facilitating its faster release from the NA at an acidic pH than the physiological pH. The tumors create an acidic microenvironment due to increased glucose metabolism, known as the “Warburg effect”.39 Thus the unique properties of NA are certainly advantageous for doxorubicin release because these vesicles have the ability to release the drug in a slow and controlled fashion due to their thermostable and rigid nature. On the other hand, if the drug carriers are not stable, continuous drug leakage or sudden drug release at the non-targeted region may occur due to the instability of normal lipid-based vesicles, which in turn may drastically affect the rate of drug release, leading to undesirable health impacts. Moreover, NAD has specific faster drug release properties at acidic pH, through which we can achieve targeted drug delivery specifically at the tumor site, minimizing the exposure of healthy tissues to the drug, reducing potential side effects, and enhancing the therapeutic index.
3.3 FTIR analysis of NAD
Next, we investigated the interaction of encapsulated doxorubicin molecules with archaeal lipids in NAD using the FTIR method. Fig. 3 shows the FTIR spectra of NAD, doxorubicin, pure SOPC, NA, and pure archaeal lipid. The FTIR spectra of pure archaeal lipid have shown characteristic peaks for the presence of amide group N–H stretching (3418 cm−1), CH2 asymmetric stretching of methylene group in fatty acid (2919 cm−1), symmetric CO2 stretching (1467 cm−1), aromatic ether band (1375 cm−1) and C–O ether linkage (1034 cm−1). The spectra corresponding to pure SOPC lipids have shown peaks related to the C–H stretching (3438 cm−1), methylene C–H stretching (2854 cm−1), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching/ester bonds (1733 cm−1), PO2 anti-symmetric stretching (1232 cm−1) and choline C–N peak height position (971 cm−1). Similar values were obtained in our previous work for pure SOPC lipids.40 The NA prepared by mixing the archaeal and SOPC lipids, has shown intermittent peak values for both lipids.
O stretching/ester bonds (1733 cm−1), PO2 anti-symmetric stretching (1232 cm−1) and choline C–N peak height position (971 cm−1). Similar values were obtained in our previous work for pure SOPC lipids.40 The NA prepared by mixing the archaeal and SOPC lipids, has shown intermittent peak values for both lipids.
To confirm the loading of doxorubicin to NA, the comparison of FTIR peaks of doxorubicin and NAD were examined. FTIR spectrum of doxorubicin was taken as a control to analyze NAD. FTIR spectrum of NAD has shown peaks corresponding to N–H stretch (3440 cm−1), C–H stretch (2925 cm−1), ester bond linkage (1741 cm−1), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C ring stretch/symmetric CO2 stretching (1465 cm−1), PO2 anti-symmetric stretching (1239 cm−1). Interestingly, the FTIR spectrum of NAD has clearly shown characteristic peaks of both doxorubicin and NA and the bonds corresponding to doxorubicin remain intact in the NA signals. These results indicate the successful encapsulation of doxorubicin molecules in NA.
C ring stretch/symmetric CO2 stretching (1465 cm−1), PO2 anti-symmetric stretching (1239 cm−1). Interestingly, the FTIR spectrum of NAD has clearly shown characteristic peaks of both doxorubicin and NA and the bonds corresponding to doxorubicin remain intact in the NA signals. These results indicate the successful encapsulation of doxorubicin molecules in NA.
3.4 Biocompatibility of NA
The use of NA and NAD for anticancer therapy mandates an improved understanding of its safety and biological effects. So, we checked the biocompatibility of NA and NAD in NIH 3T3 fibroblast cell lines using the MTT assay. Fig. 4A shows that NA and NAD do not exhibit cellular cytotoxicity in NIH 3T3 cell lines. The T-test with p > 0.05 showed no significant difference between the control (100% viability), NA and NAD treated cells. Fig. 4B(i)–(iii) correspond to the optical microscopic images of control, NA and NAD-treated cells. The results showed a healthy cell with an intact cell membrane and cytoplasm with prominent nucleoli for control, NA and NAD-treated samples. Next, live/dead assay was performed using AO/EB stain. Fig. 4B(iv)–(vi) show the fluorescence microscopic images of control, NA and NAD-treated cells using AO/EB staining. The fluorescence microscopic imaging with green fluorescence for control, NA and NAD treated cells confirmed the live nucleated cells with integrated membranes. We observed that AO/EB results were consistent with the MTT assays confirming the biocompatible nature of both the NA and NAD.3.5 In vitro cytotoxic analysis of NAD on MCF-7 breast cancer cells
Next, we assessed and compared the in vitro cytotoxic properties of NA alone, doxorubicin alone and NAD of various concentrations (10, 100, 500, 1000 nM and 2000 nM) on MCF-7 breast cancer cells after 24 h incubation by the MTT assay. From Fig. 5A, we observed that NA alone didn't exhibit any cytotoxic effect on MCF-7 cancer cell lines. We found that NAD exhibited the dose dependent cytotoxic effect. NAD reduced the cell viability by less than 50%, even at 10 nM concentration. The cell viability decreased from 31 ± 3% to 10 ± 3% with an increase in NAD concentration from 10–1000 nM. Then in order to calculate the half maximal (50%) inhibitor concentration (IC50) value, we performed MTT assays and tested the effect of NAD on MCF-7 at various lower concentration ranging from 0.01 to 10 nM. The IC50 value for NAD was found to be 6 ± 2 nM (Fig. 5B). By comparing the NAD cytotoxicity of doxorubicin alone, it was observed that doxorubicin showed very little cytotoxicity at low concentrations (10, 100, 500 nM) with cell viability of 100 ± 4% to 80 ± 3%. This confirms that NAD has better cytotoxicity with IC50 value of 6 ± 2 nM compared to free drugs. We also tested NAD cytotoxic effect on MCF-7 at different time period of 48 h and 72 h (Fig. SI-1†). We observed that the cell viability decreased at 48 and 72 h compared to 24 h. The cell viability at 24 h at 2, 4, 6, 8 and 10 nM concentration deceased from 99 ± 4 to 90 ± 3 and 80 ± 3%, 98 ± 5 to 75 ± 4 and 60 ± 5%, 50 ± 3 to 40 ± 4 and 30 ± 2%, 42 ± 3 to 31 ± 3 and 25 ± 2%, 31 ± 2.2 to 18 ± 1.5 and 11 ± 1% at 48 and 72 h. No significant differences in the cell viability were observed for cells treated with NAD at the concentration less than or equal to 1 nM for all three time point. Next, we studied the cell morphological changes using optical microscopy. The viable untreated cancer cells expressed similar normal cancer cell structures and intact nuclear structures (Fig. 5C(i)). Doxorubicin-treated (100 nM) MCF-7 cells (Fig. 5C(ii)) showed no noticeable morphological changes. On the other hand, the NAD-treated (100 nM) MCF-7 cells showed characteristic attributes of cellular death with loss of cell membrane integrity (Fig. 5C(iii)), cell detachment, nuclear disintegration, and cell shrinkage compared to untreated cancer cells. From Fig. 5A, we observed that NAD reduced the cell viability by less than 50%, even at 10 nM concentration. The cell viability decreased from 31 ± 3% to 10 ± 3% with an increase in NAD concentration from 10–1000 nM. The half maximal (50%) inhibitor concentration (IC50) value for NAD was measured to be less than 10 nM. By comparing the NAD cytotoxicity of doxorubicin alone, it was observed that doxorubicin showed very little cytotoxicity at low concentrations (10, 100, 500 nM) with cell viability of 100 ± 4% to 80 ± 3%. This confirms that NAD has better cytotoxicity even at the nanomolar range compared to free drugs. Next, we studied the cell morphological changes using optical microscopy. The viable untreated cancer cells expressed similar normal cancer cell structures and intact nuclear structures (Fig. 5C(i)). Doxorubicin-treated (100 nM) MCF-7 cells (Fig. 5C(ii)) showed no noticeable morphological changes. On the other hand, the NAD-treated (100 nM) MCF-7 cells showed the characteristic attributes of cellular death with loss of cell membrane integrity (Fig. 5C(iii)), cell detachment, nuclear disintegration, and cell shrinkage compared to untreated cancer cells.Subsequently, for further confirmation, we performed AO/EB staining to differentiate between viable, apoptotic, and necrotic cells. The stained cells were discriminated into viable (green fluorescence), early apoptotic (bright green fluorescence and condensed chromatin), late apoptotic (yellow/orange fluorescence), and nonviable cells (red fluorescence).41 We observed untreated cancer cells demonstrating the normal green fluorescent nuclear structure and an intact configuration without any noticeable cellular death (Fig. 5C(iv)). Doxorubicin-treated cells (100 nM) showed a much less apoptotic effect (Fig. 5C(v)). But NAD-treated (100 nM) MCF-7 cells showed increased necrotic and apoptotic attributes of cellular death in MCF-7, resulting in increased orange and red fluorescence (Fig. 5C(vi)). No fluorescence interference from doxorubicin at 100 nM without any staining was observed (Fig. SI-2†). This finding suggests that NAD induces significant apoptotic cellular death in breast cancer cells without inducing toxicity in normal cells. So, NAD could be an alternative promising therapeutic agent compared to free drugs.
Doxorubicin has been used as a cytostatic drug for breast cancer treatment, and studies reported that doxorubicin causes serious side effects during cancer treatment.42,43 Numerous research efforts have been dedicated to developing effective targeted delivery of doxorubicin drugs.44–46 However, the acidic and hypoxic tumor microenvironment hinders their therapeutic potential. Earlier reports have shown that doxorubicin liposomes induced cytotoxicity in breast and other cancer cell lines at a concentration range between 0.6 and 2.6 μM.47,48 But in our system, the synthesized NAD exhibits excellent anticancer efficacy even at a nanomolar concentration.
Studies reported that nanoparticles with smaller sizes less than 200 nm can more easily penetrate cellular barriers and tissues enhancing their ability to reach target sites that might be inaccessible to larger particles or conventional drugs.49,50 This improved cellular uptake can lead to better bioavailability of the drug payload within the cells, allowing for more efficient drug delivery.51 Several studies reported that the most optimal size for enhanced permeability effect is in the range between 100 and 200 nm.52–55 Thus, we speculate, NAD with size less than 100 nm could have excellent anti-cancer efficacy at nano molar concentration due to their high bioavailability and EPR effect. The results are in accordance with Grebinyk et al., where they have studied the cytotoxic effect doxorubicin loaded C60 fullerene nanocomplex against different leukemic cancer cells at various nanomolar concentrations (1 to 1000 nM) and found that drug loaded fullerene nanocomplexes exhibited higher cytotoxic potential in comparison with the free doxorubicin.56 In another study, Susa et al., reported that doxorubicin loaded lipid modified dextran based polymeric nano system showed tenfold higher activity than free drug alone with IC50 value of 30 nM against U2OS cancer cell line which also aligns with our results.57
3.6 FACS analysis of NAD-induced apoptosis in MCF-7 cells
Next, we performed a flow cytometry analysis to evaluate whether NAD induces apoptosis or necrosis in MCF-7 cells using the Annexin V-APC/PI discrimination assay. A fluorescently labelled Annexin V calcium-binding protein detects the phosphatidyl serine located on the cell membrane of apoptotic cells. To distinguish apoptotic and necrotic cells, co-staining of cells with propidium iodide was performed, as previously described.58In our assay, proper gating strategies were employed and the homogeneous population of MCF-7 cells was identified using forward and side scatter plots (Fig. 6A). The doxorubicin intensity spill over to the PI channel in meagre amount is taken care by background subtraction and fluorochrome compensation (Fig. SI-3†). Fig. 6B depicts the Annexin V-APC/PI-stained control cells with 93% of viable cells while NAD-treated (100 nM) MCF-7 cells of 92% of cell population had undergone late apoptosis and the remaining 8% of cell population experienced necrosis (Fig. 6C). We observed that MCF-7 experienced major cell death through apoptosis with an increased apoptotic index by NAD than the control cells. Fig. 6D shows the clear peak shift in the fluorescence intensity of the control and NAD-treated cells undergoing apoptosis with increased uptake of Annexin V dye. Similarly, we also observed that the amount of PI uptake by the necrotic cells upon NAD treatment was higher when compared with the control cells (Fig. 6E). These results are in concordance with the in vitro cell cytotoxicity and AO/EB staining assays.
Kucuksayan et al., studied the cytotoxic efficacy of doxorubicin-loaded poly (lactic-co-glycolic acid) (PLGA) nanoparticles (NP).59 They observed that doxorubicin-loaded PLGA-NP induces apoptosis in MCF-7 cancer cells in 22.7% of the population. In another study, Li et al., showed that doxorubicin-loaded gold nanoparticles induced apoptosis in 43% of the HEPG-2 cell population.60 Membrane fusogenic liposomes loaded with doxorubicin also exhibited an apoptosis rate of 13.2% in MCF-7 cells.61 Compared to the existing data, our results with NAD have shown a multi-fold increase in apoptosis rate.
3.7 Effect of NAD on cell cycle
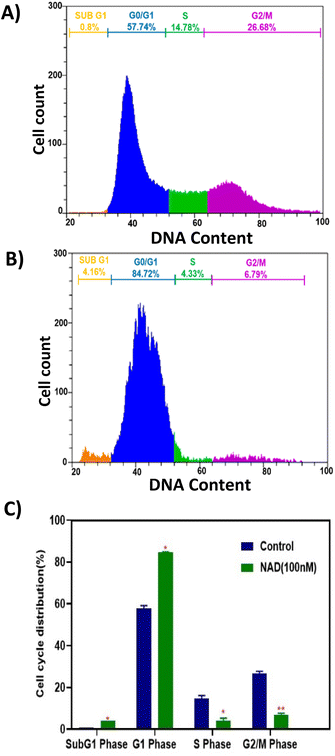
Flow cytometry analysis was performed to evaluate NAD's effect on the cell cycle progression of MCF-7 cell lines. Fig. 7A shows the cell cycle progression for control cells based on the DNA content in each stage. The MCF-7 cells treated with NAD (100 nM) efficiently induced the cells to accumulate in the sub G1, G0/G1 phase with a drastic reduction in the S and G2/M phases (Fig. 7B) compared to the control (Fig. 7A). The mean percentage of the cells in the sub G1, G0/G1 phase, S phase, and G2/M phase of the cell cycle for control was 0.8%, 57.74 ± 1%, 14.78 ± 1.44%, and 26.68 ± 1.14%, respectively, and for NAD-treated samples were 4.16 ± 0.6%, 84.72 ± 0.13%, 4.33 ± 1.02%, and 6.79 ± 0.89% respectively. G2/M phase of the cell cycle for control was 35.6%, 13.6%, and 18.94%, respectively, and for NAD-treated samples were 41.58%, 9.32%, and 3.34% respectively.Fig. 7C represents the statistics of the cell cycle distribution between control and treated cells. From the paired t-tests (p < 0.05) it is observed that there is a significant difference between the control and NAD-treated cells in cell cycle distribution. NAD-treated cells have shown nearly a five-fold increase in the subG1 phase arrest and prominent cell cycle arrest in G0/G1 phase when compared to control as an indication of late apoptosis. In addition, G2/M phase also showed a distinct six-fold reduction in the NAD treated samples compared to the control. The results indicated that cells were impeded in their cycle progression at an initial stage of the interphase. Similar results have been reported before, which concord with our results on arresting the cells at the G0/G1 phase with the subsequent induction of cellular apoptosis in MCF-7 cells but at a higher drug concentration level.62,63
4. Conclusion
Although several nanocarriers have been formulated in the past decade, such strategies were unsuccessful due to the concerns associated with their biocompatibility, colloidal stability, and side effects due to higher concentration.64,65 In this study, we have demonstrated NA with excellent long-term stability at various conditions. These nanocarriers have also shown sustained release properties, which can reduce the risks of sudden drug release and increasing patient compliance. The present study provides evidence for the biocompatible nature of NA in normal cells and its ability to cause cytotoxicity in cancer cells when loaded with anticancer drugs by predominantly inducing apoptosis followed by cell cycle arrest at the G0/G1 phase, even at a low nanomolar concentration. All these findings indicate that the NA could function as an efficient next-generation drug carrier system in vitro to improve the therapeutic efficacy of drugs.Author contributions
Swathi Sudhakar has designed the project. Swathi Sudhakar, Poornima Budime Santhosh, Kaviya Vijayalakshmi Babunagappan, Abirami Seetharaman, and Subastri Ariraman carried out experiments. Swathi Sudhakar, Poornima Budime Santhosh, Kaviya Vijayalakshmi Babunagappan, Abirami Seetharaman, and Subastri Ariraman analyzed the results. Swathi Sudhakar, Poornima Budime Santhosh, Kaviya Vijayalakshmi Babunagappan, and Abirami Seetharaman wrote the manuscript. Swathi Sudhakar, Julia Genova, and Natasa Poklar Ulrih finalized the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful for the financial support from IIT Madras through the Institutions of Eminence (IoE) Scheme (Grant No. SP22231226CPETWOCSMHOC), (Grant No. SP22231226CPETWOHCTHOC) and RF22230395AMNFIG009000 from the Ministry of Education, Government of India. We also gratefully acknowledge the financial support from the project KP-06-DB8/01.12.2020 with the Bulgarian National Science Fund under the National Scientific Program P. Beron 2020. We would like to thank Prof. Pijyush Gosh for providing access to FTIR studies in his lab. We would like to thank Prof. Ramakrishnan S. for providing us with a Multimode Plate Reader. We would also like to thank Dr Nathiya Muthalagu for providing us with MCF-7 cell lines for our study.References
- H. Nsairat, D. Khater, U. Sayed, F. Odeh, A. A. Bawab and W. Alshaer, Heliyon, 2022, 8, e09394 CrossRef CAS PubMed.
- L. Sercombe, T. Veerati, F. Moheimani, S. Y. Wu, A. K. Sood and S. Hua, Front. Pharmacol, 2015, 6, 286 CrossRef PubMed.
- T. O. B. Olusanya, R. R. H. Ahmad, D. M. Ibegbu, J. R. Smith and A. A. Elkordy, Molecules, 2018, 23, 907 CrossRef PubMed.
- P. Nakhaei, R. Margiana, D. O. Bokov, W. K. Abdelbasset, M. A. J. Kouhbanani, R. S. Varma, F. Marofi, M. Jarahian and N. Beheshtkhoo, Front. Bioeng. Biotechnol., 2021, 9, 705886 CrossRef PubMed.
- D. Lombardo and M. A. Kiselev, Pharmaceutics, 2022, 14, 543 CrossRef CAS.
- A. Ota, D. Gmajner, M. Šentjurc and N. P. Ulrih, Archaea, 2012, 1–9 CrossRef PubMed.
- M. Á. Cabrera and J. M. Blamey, Biol. Res., 2018, 5, 37 CrossRef.
- S. Jain, A. Caforio and A. J. Driessen, Front. Microbiol., 2014, 5, 641 CrossRef PubMed.
- P. B. Santhosh and J. Genova, ACS Omega, 2022, 8, 1–9 CrossRef PubMed.
- L. Krishnan and G. D. Sprott, J. Drug Target., 2003, 11, 515–524 CrossRef CAS PubMed.
- B. Hashemi, A. Akbarzadeh and M. Arjmand, Biomed. Pharmacol. J., 2016, 9, 215–228 CrossRef.
- G. B. Patel and G. D. Sprott, Crit. Rev. Biotechnol., 1999, 19, 317–357 CrossRef CAS.
- M. J. Altube, M. M. B. Martínez, B. Malheiros, P. C. Maffía, L. R. S. Barbosa, M. J. Morilla and E. L. Romero, Mol. Pharm., 2019, 17, 70–83 CrossRef.
- N. Charó, H. Jerez, S. Tatti, E. L. Romero and M. Schattner, Pharmaceutics, 2022, 14(4), 736 CrossRef.
- H. E. Jerez, M. J. Altube, Y. B. Gándola, L. González, M. C. Gonzalez, M. J. Morilla and E. L. Romero, Eur. J. Pharm. Biopharm., 2021, 160, 42–54 CrossRef CAS.
- F. L. Parra, A. T. Caimi, M. J. Altube, D. E. Cargnelutti, M. E. Vermeulen, M. A. De Farias, R. V. Portugal, M. J. Morilla and E. L. Romero, Front. Bioeng. Biotechnol., 2018, 6, 163 CrossRef.
- S. E. Alavi, H. Mansouri, M. K. M. Esfahani, F. Movahedi, A. Akbarzadeh and M. Chiani, Indian J. Clin. Biochem., 2014, 29, 150–153 CrossRef CAS PubMed.
- I. Milek, B. Cigić, M. Skrt, G. Kaletunç and N. P. Ulrih, Can. J. Microbiol., 2005, 51, 805–809 CrossRef CAS PubMed.
- D. Gmajner, A. Ota, M. Šentjurc and N. P. Ulrih, Chem. Phys. Lipids, 2011, 164, 236–245 CrossRef CAS.
- P. B. Santhosh, B. Drašler, D. Drobne, M. E. Kreft, S. Kralj, D. Makovec and N. P. Ulrih, Int. J. Nanomed., 2015, 10, 6089–6104 CAS.
- R. Vakili-Ghartavol, S. M. Rezayat, R. Faridi-Majidi, K. Sadri and M. R. Jaafari, Sci. Rep., 2020, 10, 5569 CrossRef CAS.
- A. G. Gomez, S. Syed, K. Marshall and Z. Hosseinidoust, ACS Omega, 2019, 4(6), 10866–10876 CrossRef.
- R. Domínguez-Ríos, D. R. Sánchez-Ramírez, K. Ruiz-Saray, P. E. Oceguera-Basurto, M. Almada, J. Juárez, A. Zepeda-Moreno, A. del Toro-Arreola, A. Topete and A. Daneri-Navarro, Colloids Surf., B, 2019, 178, 199–207 CrossRef PubMed.
- J. Barbeau, S. Cammas-Marion, P. Auvray and T. Benvegnu, J. Drug Delivery, 2011, 396068 Search PubMed.
- N. P. Ulrih, G. Dejan and R. Peter, Appl. Microbiol. Biotechnol., 2009, 84, 249–260 CrossRef CAS.
- A. A. Khafoor, A. S. Karim and S. M. Sajadi, Results in Surfaces and Interfaces, 2023, 11, 100124 CrossRef.
- M. D. Fulton and W. Najahi-Missaoui, Int. J. Mol. Sci., 2023, 24, 6615 CrossRef CAS PubMed.
- L. Šturm and N. P. Ulrih, Int. J. Mol. Sci., 2021, 22, 6547 CrossRef PubMed.
- S. Behzadi, V. Serpooshan, W. Tao, M. A. Hamaly, M. Y. Alkawareek, E. C. Dreaden, D. Brown, A. M. Alkilany, O. C. Farokhzad and M. Mahmoudi, Chem. Soc. Rev., 2017, 46, 4218–4244 RSC.
- F. Villanueva-Flores, A. Castro-Lugo, O. T. Ramírez and L. A. Palomares, Nanotechnology, 2020, 31, 132002 CrossRef.
- A. Soni, M. P. Bhandari, G. K. Tripathi, P. Bundela, P. K. Khiriya, P. S. Khare, M. K. Kashyap, A. Dey, B. Vellingiri, S. Sundaramurthy, A. Suresh and J. M. Pérez de la Lastra, J. Cell. Mol. Med., 2023, 27, 737–762 CrossRef CAS.
- M. J. Nirmala, U. Kizhuveetil, A. Johnson, G. Balaji, R. Nagarajan and V. Muthuvijayan, RSC Adv., 2023, 13, 8606–8629 RSC.
- K. Yoncheva, B. Tzankov, Y. Yordanov, I. Spassova, D. Kovacheva, M. Frosini, M. Valoti and V. Tzankova, J. Drug Delivery Sci. Technol., 2020, 59, 101870 CrossRef CAS.
- S. Bandak, A. Ramu, Y. Barenholz and A. Gabizo, Pharm. Res., 1999, 16, 841–846 CrossRef CAS.
- M. E. O'Brien, N. Wigler, M. Inbar, R. Rosso, E. Grischke, A. Santoro, R. Catane, D. G. Kieback, P. Tomczak, S. P. Ackland, F. Orlandi, L. Mellars, L. Alland and C. Tendler, Ann. Oncol., 2004, 15(3), 440–449 CrossRef.
- H. Onafuye, S. Pieper, D. Mulac, J. Cinatl Jr, M. N. Wass, K. Langer and M. Michaelis, Beilstein J. Nanotechnol., 2019, 10, 1707–1715 CrossRef CAS.
- A. U. Rehman, Z. Omran, H. Anton, Y. Mely, S. Akram, T. F. Vandamme and N. Anton, Eur. J. Pharm. Biopharm., 2018, 133, 331–338 CrossRef CAS PubMed.
- R. Teranishi, R. Matsuki, E. Yuba, A. Harada and K. Kono, Pharmaceutics, 2016, 9, 4 CrossRef.
- M. V. Liberti and J. W. Locasale, Trends Biochem. Sci., 2016, 41, 211–218 CrossRef CAS.
- P. B. Santhosh, T. Tenev, L. Šturm, N. P. Ulrih and J. Genova, Int. J. Mol. Sci., 2023, 24, 10226 CrossRef CAS PubMed.
- A. Subastri, V. Arun, P. Sharma, A. Suyavaran, S. Nithyananthan, G. M. Alshammari, B. Aristatile, V. Dharuman and C. Thirunavukkarasu, Chem. Biol. Interact., 2018, 295, 73–83 CrossRef CAS.
- C. J. Lovitt, T. B. Shelper and V. M. Avery, BMC Cancer, 2018, 18, 1–11 CrossRef PubMed.
- D. Zheng, Y. Zhang, M. Zheng, T. Cao, G. Wang, L. Zhang, R. Ni, J. Brockman, H. Zhong and G.-C. Fan, Clin. Sci., 2019, 133, 1505–1521 CrossRef CAS PubMed.
- E. Bagheri, M. Alibolandi, K. Abnous, S. M. Taghdisi and M. Ramezani, J. Mater. Chem. B, 2021, 9, 1351–1363 RSC.
- Y. Zhao, S. Tang, J. Guo, M. Alahdal, S. Cao, Z. Yang, F. Zhang, Y. Shen, M. Sun and R. Mo, Sci. Rep., 2017, 7, 44758 CrossRef CAS PubMed.
- Y. Hazeri, A. Samie, M. Ramezani, M. Alibolandi, E. Yaghoobi, S. Dehghani, R. Zolfaghari, F. Khatami, T. Zavvar and M. A. Nameghi, J. Drug Deliv. Sci. Technol., 2022, 71, 103285 CrossRef CAS.
- L. Wang, W. G. Wang, Z. Rui and D. S. Zhou, Drug Deliv., 2016, 23, 3200–3208 CrossRef CAS PubMed.
- N. Chowdhury, S. Chaudhry, N. Hall, G. Olverson, Q.-J. Zhang, T. Mandal, S. Dash and A. Kundu, AAPS PharmSciTech, 2020, 21, 1–12 CrossRef.
- P. Shashi, A. Geeta, C. Ramesh, A. Bahar and N. Surendra, Artif. Cells, Nanomed., Biotechnol., 2016, 44(1), 83–91 CrossRef.
- E. A. G. Stephanie, A. R. Patricia, D. P. Patrick, J. C. Luft, J. M. Victoria, E. N. Mary and M. D. Joseph, Proc. Natl. Acad. Sci. U. S. A., 2008, 105(33), 11613–11618 CrossRef PubMed.
- G. Gregoriadis and S. U. Paul, J. Pharmaceut. Sci., 1989, 78(8), 693 Search PubMed.
- W. Jun, J. Pers. Med., 2021, 11, 771 CrossRef PubMed.
- N. Yuko, M. Ai, L. C. Peter and K. Hisataka, Bioconjugate Chem., 2016, 17, 2225–2238 Search PubMed.
- K. Jinseong, C. Hanhee, L. Dong-Kwon, K. J. Min and K. Kwangmeyung, Int. J. Mol. Sci., 2023, 24, 10082 CrossRef.
- L. Stephano, J. Pers. Med., 2022, 12, 1259 CrossRef.
- A. Grebinyk, S. Prylutska, O. Chepurna, S. Grebinyk, Y. Prylutskyy, U. Ritter, T. Y. Ohulchanskyy, O. Matyshevska, T. Dandekar and M. Frohme, Nanoscale Res. Lett., 2019, 14, 61 CrossRef.
- M. Susa, A. K. Iyer, K. Ryu, F. J. Hornicek, H. Mankin, M. M. Amiji and Z. Duan, BMC Cancer, 2009, 9, 399 CrossRef.
- L. C. Crowley, B. J. Marfell, A. P. Scott and N. J. Waterhouse, Cold Spring Harb. Protoc., 2016, 2016, 953–957 Search PubMed.
- E. Kucuksayan, F. Bozkurt, M. T. Yilmaz, A. Sircan-Kucuksayan, A. Hanikoglu and T. Ozben, Sci. Rep., 2021, 11, 13027 CrossRef CAS.
- L.-S. Li, B. Ren, X. Yang, Z.-C. Cai, X.-J. Zhao and M.-X. Zhao, Pharmaceuticals, 2021, 14, 101 CrossRef CAS.
- H. Deng, K. Song, X. Zhao, Y. Li, F. Wang, J. Zhang, A. Dong and Z. Qin, ACS Appl. Mater. Interfaces, 2017, 9, 9315–9326 CrossRef CAS.
- Z. Yuan, H. Meng-lei, L. Miao, C. Ying, S. Yang, C. Han, L. Dao-zhou, M. Qi-bing and Z. Si-yuan, Sci. Rep., 2016, 6, 35267 CrossRef PubMed.
- T. Katerina, P. Katerina, P. Klara, B. Svatopluk, H. Marketa, K. Mary, K. Eva, Z. Jana, M. Lukas, H. Jana, M. Jakub, K. Argiris, B. Aristides, K. Hana and Z. Radek, Int. J. Nanomed., 2015, 10, 949–961 Search PubMed.
- R. Foldbjerg, D. A. Dang and H. Autrup, Arch. Toxicol., 2011, 85, 743–750 CrossRef CAS PubMed.
- N. Ganot, S. Meker, L. Reytman, A. Tzubery and E. Y. Tshuva, J. Visualized Exp., 2013, 81, e50767 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3na00953j |
| This journal is © The Royal Society of Chemistry 2024 |