Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Photodynamic treatment of Staphylococcus aureus with non-iron hemin analogs in the presence of hydrogen peroxide†
Badhu Prashanthika
Sivasubramaniam
a,
Benjamin M.
Washer
 a,
Yuichiro
Watanabe‡
a,
Yuichiro
Watanabe‡
 a,
Kathryn E.
Ragheb
b,
J. Paul
Robinson
b and
Alexander
Wei
a,
Kathryn E.
Ragheb
b,
J. Paul
Robinson
b and
Alexander
Wei
 *a
*a
aDepartment of Chemistry, Purdue University, 560 Oval Drive, West Lafayette, IN 47907, USA. E-mail: alexwei@purdue.edu
bCollege of Veterinary Medicine, Purdue University, 625 Harrison Street, West Lafayette, IN 47907, USA
First published on 3rd May 2024
Abstract
Bacteria subjected to antiseptic or antibiotic stress often develop tolerance, a trait that can lead to permanent resistance. To determine whether photodynamic agents could be used to counter tolerance, we evaluated three non-iron hemin analogs (M-PpIX; M = Al, Ga, In) as targeted photosensitizers for antimicrobial photodynamic inactivation (aPDI) following exposure to sublethal H2O2. Al-PpIX is an active producer of ROS whereas Ga- and In-PpIX are more efficient at generating singlet oxygen. Al- and Ga-PpIX are highly potent aPDI agents against S. aureus and methicillin-resistant strains (MRSA) with antimicrobial activity (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction in colony-forming units) at nanomolar concentrations. The aPDI activities of Al- and Ga-PpIX against S. aureus were tested in the presence of 1 mM H2O2 added at different stages of growth. Bacteria exposed to H2O2 during log-phase growth were less susceptible to aPDI but bacteria treated with H2O2 in their postgrowth phase exhibited aPDI hypersensitivity, with no detectable colony growth after treatment with 15 nM Ga-PpIX.
log reduction in colony-forming units) at nanomolar concentrations. The aPDI activities of Al- and Ga-PpIX against S. aureus were tested in the presence of 1 mM H2O2 added at different stages of growth. Bacteria exposed to H2O2 during log-phase growth were less susceptible to aPDI but bacteria treated with H2O2 in their postgrowth phase exhibited aPDI hypersensitivity, with no detectable colony growth after treatment with 15 nM Ga-PpIX.
Introduction
Antimicrobial resistance is a global health threat that continues to grow in severity.1–3 The rise of bacterial pathogens with multi-drug resistance, coupled with the sluggish development of novel antibiotics,4,5 is driving the need for innovative approaches to combat nosocomial contamination and infections.6 One possibility is to develop treatments that can override tolerance, an important but under-appreciated forerunner to more permanent forms of antimicrobial resistance.7 Bacterial subpopulations can develop tolerance if treatment times are insufficient; surviving cells can activate numerous repair genes that also increase the rate of mutations which lead to permanent resistance.8To find ways of overcoming stress adaptation and tolerance, we considered antimicrobial photodynamic inactivation (aPDI), a topical, non-invasive treatment modality which has shown strong potential for combating drug-resistant strains of bacterial pathogens.9,10 Some reports suggest that aPDI agents have intriguing potential to increase microbial susceptibility to antibiotics, possibly due to higher membrane permeability.11,12 aPDI agents can also be used as light-activated antiseptics for decolonizing bacteria on dental implants, food packaging, and other surfaces that require sterilization.13–15 aPDI uses a photosensitizer (PS) to produce singlet oxygen (1O2) or reactive oxygen species (ROS) in a localized manner and offers a multimodal mechanism for killing bacteria, as opposed to antibiotics that act on a specific target. Importantly, the delivery of aPDI agents to bacterial colonies may be sufficient to produce a localized killing effect with limited adverse response from host cells and tissues.9
Many synthetic and natural dyes have been studied as photosensitizers.16 Tetrapyrrole-based macrocycles such as porphyrins, chlorins, and phthalocyanines are used extensively as aPDI agents, many of which are either clinically approved or currently under clinical trials for topical treatments such as cancer and acne.17,18 In particular, protoporphyrin IX (PpIX) and hematoporphyrin derivatives (HpD) have demonstrated good aPDI activity in vivo,19,20 and the polycation tetrakis(1-methyl-4-pyridinium)porphyrin (TMPyP) is highly active against multi-drug resistant bacteria as well as fungal species.21 However, many lack target specificity which contributes toward high PS loadings and subsequent phototoxicity.19 Current efforts to address the latter challenge include chemical modifications for targeted PS delivery, liposomal encapsulation, and conjugation to nanoparticles.22 Metal ions can also influence the PS properties of porphyrins and related tetrapyrroles by modulating intersystem crossing and 1O2 quantum yield.23
We24,25 and others26,27 have been investigating strategies for targeted aPDI based on the innate affinity of bacterial pathogens for hemin (Fe-PpIX), the oxidized form of heme. Many bacteria have hemin acquisition systems for the purpose of harvesting iron, an essential mineral for virulence and growth.28 Some express cell-surface receptors enabling the direct acquisition of hemin,29,30 whereas others deploy a more sophisticated system based on the release and diffusion of hemin-harvesting proteins (hemophores).28 Non-iron hemin analogs are thus promising candidates for targeted aPDI, as they can be delivered through a Trojan-horse mechanism.31,32 For example, replacing the Fe(III) core with isoelectronic Ga(III) results in a fluorescent derivative that can be delivered to a variety of bacteria via their hemin acquisition systems.24 Ga-PpIX is also a strong photosensitizer and can mediate aPDI of planktonic S. aureus and MRSA at nanomolar levels after just a few seconds of exposure to 405 nm light from an LED source. In contrast, Ga-PpIX has very low cytotoxicity with keratinocytes (HaCaT) and kidney cells (HEK293) maintaining 85–90% viability at 20 μM, the highest concentration tested.24
Multiple studies have shown that aPDI potency can be augmented in the presence of or pretreatment with H2O2, albeit with relatively high PS loadings and H2O2 levels.33–40 There are several postulates for the potentiation of aPDI by H2O2: (i) greater permeability of cell walls or membranes after H2O2 exposure,37,41 (ii) increased production of ROS and 1O2,37 and (iii) increased O2 levels from H2O2 disproportionation, possibly mediated by catalase-like activity.40,42,43 The latter effect may be helpful to combat bacterial infections in hypoxic or anaerobic environments.
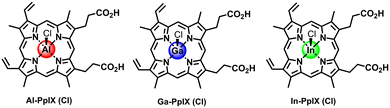
In this work we compare the PS and aPDI activities of non-iron hemin analogs (M-PpIX) with group 13 metal ions, namely Al(III), Ga(III), and In(III) (Fig. 1), and their potential for synergy with sublethal H2O2 (1 mM) to enhance antiseptic action against S. aureus and drug-resistant variants. The synergy can be remarkable under the right circumstances: H2O2-challenged strains of S. aureus can be reduced below the limit of detection at loadings as low as 15 nM M-PpIX, depending on the PS type as well as conditioning methods used.
Results and discussion
Chemical and photophysical characterization of M-PpIX
Characterization of Ga-PpIX(Cl) has been described previously24 and is included for analysis of periodic trends. The absorption spectra of all M-PpIX(Cl) salts (8 μM in DMSO) exhibit intense Soret bands at 405 nm, similar to PpIX but with narrower linewidths, and lack two peaks in the Q band region at 500 and 625 nm, characteristic of porphyrin–metal ion complexes (Fig. 2).44 Photoluminescence spectroscopy reveals a large blueshift in M-PpIX emission (45–55 nm relative to PpIX), with a primary emission band at 575–585 nm and a secondary band at 628–635 nm (Table 1). The fluorescence quantum yields (ΦFL) for 405 nm excitation of Al-PpIX, Ga-PpIX and In-PpIX are 12%, 6.3% and 1% respectively, the latter diminished by the faster intersystem crossing rate due to spin–orbit coupling (heavy atom effect).45 On the other hand, 1O2 quantum yields for M-PpIX are enhanced by the heavy-atom effect: ΦSO for Ga- and In-PpIX are 0.45–0.46, whereas that for Al-PpIX is only 0.12.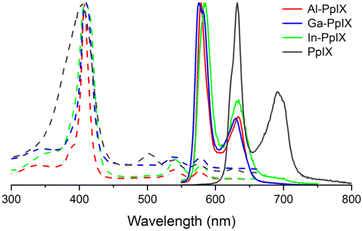 | ||
| Fig. 2 Normalized absorption (dashed curves) and luminescence spectra (solid curves) of M-PpIX(Cl) (M = Al, Ga, and In) and PpIX in 8 μM in DMSO. | ||
Flow cytometry analysis of M-PpIX uptake by S. aureus
Our lab has determined previously that active cultures of S. aureus acquire Ga-PpIX rapidly (within seconds) and with high affinity,24 which we attribute to the expression of cell-surface hemin receptors such as Isd proteins.31,46 The fluorescence of Ga-PpIX (and lack of fluorescence from Fe-PpIX) enabled us to establish CSHR specificity by a hemin competition experiment.24 With regard to the possible identity of CSHRs, X-ray crystal structures have been reported of Ga- and In-PpIX bound to the NEAT3 domain of IsdH receptors expressed by S. aureus.47,48The strong fluorescence of Al- and Ga-PpIX also make them ideal candidates for using flow cytometry (FC) to study their time-dependent uptake. Bacterial suspensions of S. aureus cultured in iron-deficient conditions were treated with Al- or Ga-PpIX diluted in PBS (final concentration 10 μM) and incubated at room temperature between 60 s and 60 min, then fixed with 4% paraformaldehyde and subjected directly to FC. In both cases, fluorescence signals were already at their maximum intensity from the initial injection and remained little unchanged over time (Fig. 3 and S1, ESI†). This indicates M-PpIX uptake to be a diffusion-controlled process with low susceptibility to efflux pump activity, which is known to play a role in heme homeostasis.49 We also performed a competitive uptake assay using a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of M-PpIX and hemin, which strongly impacted signal intensity and confirmed the results of our previous study by fluorescence imaging.24 We note that S. aureus cultured under iron-replete conditions responded less avidly to M-PpIX, implying that CSHR expression is upregulated upon iron deprivation.
1 ratio of M-PpIX and hemin, which strongly impacted signal intensity and confirmed the results of our previous study by fluorescence imaging.24 We note that S. aureus cultured under iron-replete conditions responded less avidly to M-PpIX, implying that CSHR expression is upregulated upon iron deprivation.
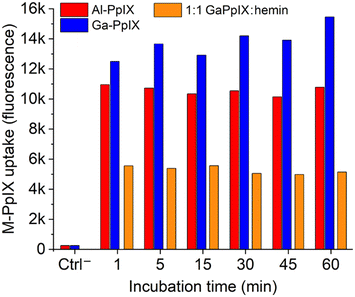 | ||
Fig. 3 Histogram of flow cytometry (FC) data for S. aureus (PCI 1203) cultured in Fe-deficient media, following treatment with 10 μM M-PpIX, where M = Al (red) or Ga (blue). Specificity for M-PpIX uptake by S. aureus established by competition assay using a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Ga-PpIX 1 Ga-PpIX![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) hemin mixture (orange). All bacteria were fixed after M-PpIX treatment and before FC injection; see ESI† for original FC data. hemin mixture (orange). All bacteria were fixed after M-PpIX treatment and before FC injection; see ESI† for original FC data. | ||
The diffusion-controlled uptake of M-PpIX led us to postulate that CHSR expression alone should be sufficient for targeted delivery. To test this, S. aureus cultures were fixed prior to treatment with M-PpIX and subjected to FC analysis at various time points (Fig. S2, ESI†). Remarkably, no apparent differences in fluorescence intensity or uptake over time were observed, with results essentially identical to those shown in Fig. 3. We thus consider M-PpIX uptake to be an energy-independent process mediated purely by cognate CSHRs presented on the bacterial surface.
Antibacterial and aPDI activity of M-PpIX
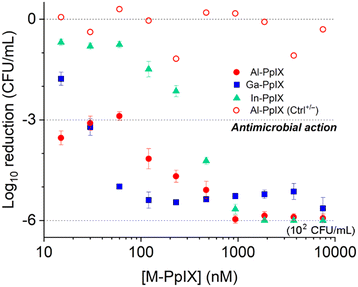
Antimicrobial activities of M-PpIX derivatives were evaluated using planktonic S. aureus plus four clinical isolates of MRSA cultured in iron-deficient media (Fig. 4 and Table 2). In the absence of light, the minimum inhibitory concentration (MIC) values for all combinations of M-PpIX and S. aureus was 30 μM; with 30 seconds of 405 nm irradiation, the MIC values were lowered to 15 μM. The full potency of aPDI was established by quantifying colony log reduction following irradiation, with 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reductions in the nanomolar range. Al- and Ga-PpIX were most active against S. aureus with 3
log reductions in the nanomolar range. Al- and Ga-PpIX were most active against S. aureus with 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction values as low as 0.015 μM and 0.03 μM respectively; aPDI with In-PpIX was much less potent (0.24 μM) although still comparable to TMPyP, a leading PS.24 Little to no reduction in S. aureus populations was observed in the absence of 405 nm light (Ctrl+/−) or M-PpIX (Ctrl−/+). Interestingly, Al-PpIX was less potent against three of the four MRSA strains whereas the aPDI activity of Ga-PpIX was essentially constant. This indicates that (i) the less susceptible MRSA strains have active mechanisms for quenching ROS, and (ii) none of the bacteria have defense mechanisms to counter 1O2, which has an aqueous lifetime of just a few microseconds.50
log reduction values as low as 0.015 μM and 0.03 μM respectively; aPDI with In-PpIX was much less potent (0.24 μM) although still comparable to TMPyP, a leading PS.24 Little to no reduction in S. aureus populations was observed in the absence of 405 nm light (Ctrl+/−) or M-PpIX (Ctrl−/+). Interestingly, Al-PpIX was less potent against three of the four MRSA strains whereas the aPDI activity of Ga-PpIX was essentially constant. This indicates that (i) the less susceptible MRSA strains have active mechanisms for quenching ROS, and (ii) none of the bacteria have defense mechanisms to counter 1O2, which has an aqueous lifetime of just a few microseconds.50
| Bacterial strain | Activity | Al-PpIX (μM) | Ga-PpIX (μM) | In-PpIX (μM) |
|---|---|---|---|---|
a Standard conditions: 108 CFU mL−1 in Fe-deficient media prior to dilution with PBS.
b MIC: data obtained 16 h after M-PpIX treatment.
c aPDI: 30 s irrad. with 405 nm LED source (4.2 J cm−2).
d Minimum concentrations for 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction. log reduction.
|
||||
| S. aureus (PCI 1203) | MIC (dark)b | 30 | 30 | 30 |
| MIC (irrad)b,c | 15 | 15 | 15 | |
| aPDId | 0.015 | 0.03 | 0.23 | |
| MRSA, USA 300 | aPDI | 0.015 | 0.03 | 0.03 |
| (NRS 383) | ||||
| MRSA, USA 300 | aPDI | 0.06 | 0.03 | 0.12 |
| (NRS 385) | ||||
| MRSA, USA 300 | aPDI | 0.23 | 0.015 | 0.23 |
| (NRS 386) | ||||
| MRSA, USA 300 | aPDI | 0.06 | 0.03 | 0.46 |
| (NRS 387) | ||||
aPDI activity of M-PpIX against H2O2-challenged S. aureus
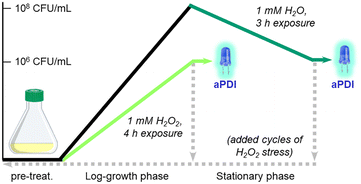
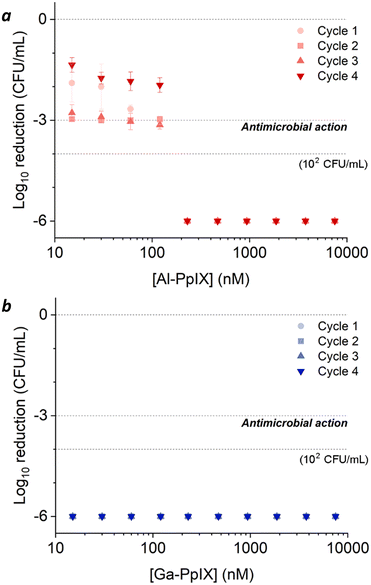
The prevalence of S. aureus and MRSA in hospital-associated infections is supported by their ability to adapt and survive adverse conditions during routine sterilizations. For example, S. aureus employs a variety of defense mechanisms to combat acute oxidative stress such as biofilm formation and the overexpression of catalase, superoxide dismutase, and staphyloxanthin.51 For longer periods of stress, bacteria can transform into small-colony variants (SCVs) with high tolerance to antibiotics and H2O2.52,53 In this work, we investigated the aPDI efficacy of Al- and Ga-PpIX against S. aureus following their adaptation to sublethal levels of H2O2.The experimental design for this study is summarized in Scheme 1. S. aureus (PCI 1203) cultured in Fe-deficient media were challenged with 1 mM H2O2 at either an early stage (4 h during log-growth phase) or a late stage (3 h during stationary phase) followed by aPDI with Al- or Ga-PpIX (cycle 1); the initial population in both cases was 106 CFU mL−1. In addition, cultures of H2O2-challenged bacteria were harvested then resubjected to 1 mM H2O2 during early- or late-stage growth (cycles 2–4) to determine adaptation to oxidative stress over time.
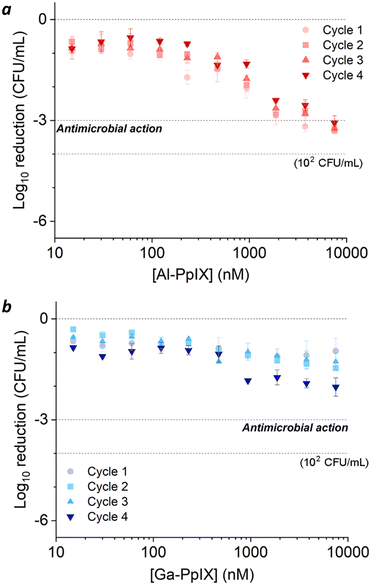
Bacteria challenged with 1 mM H2O2 during early-stage growth had low sensitivity to aPDI. A 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction could only be achieved with Al-PpIX above 4 μM, a tolerance that persisted over several growth cycles (Fig. 5a). Low sensitivity to ROS is expected as antioxidant enzymes are upregulated in response to oxidative stress;54 for example, aPDI of E. coli and E. faecalis can induce the upregulation of oxidative stress genes such as dps, hypR, and soxRS55,56 as well as general stress markers such as σ-factors,55,57 and aPDI treatment of S. aureus with PpIX can increase the expression of superoxide dismutase.58
log reduction could only be achieved with Al-PpIX above 4 μM, a tolerance that persisted over several growth cycles (Fig. 5a). Low sensitivity to ROS is expected as antioxidant enzymes are upregulated in response to oxidative stress;54 for example, aPDI of E. coli and E. faecalis can induce the upregulation of oxidative stress genes such as dps, hypR, and soxRS55,56 as well as general stress markers such as σ-factors,55,57 and aPDI treatment of S. aureus with PpIX can increase the expression of superoxide dismutase.58
Surprisingly, H2O2-challenged bacteria were also highly tolerant to Ga-PpIX-mediated aPDI, which did not produce a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction even at 10 μM (Fig. 5b). To our knowledge, there are no reports linking known oxidative stress responses with protection against 1O2. To explain the low sensitivity to 1O2 we considered the possibility that H2O2 exposure lowered CSHR expression and M-PpIX avidity, however FC analysis of H2O2-treated S. aureus with Ga-PpIX did not reveal any differences in time-dependent uptake (Fig. S3, ESI†). Further studies are needed to elucidate the apparently low susceptibility of these cultures to Ga-PpIX-mediated aPDI.
log reduction even at 10 μM (Fig. 5b). To our knowledge, there are no reports linking known oxidative stress responses with protection against 1O2. To explain the low sensitivity to 1O2 we considered the possibility that H2O2 exposure lowered CSHR expression and M-PpIX avidity, however FC analysis of H2O2-treated S. aureus with Ga-PpIX did not reveal any differences in time-dependent uptake (Fig. S3, ESI†). Further studies are needed to elucidate the apparently low susceptibility of these cultures to Ga-PpIX-mediated aPDI.
The effects of aPDI were strikingly different for S. aureus cultures challenged with H2O2 after their log-growth phase (Fig. 6). A 3 h exposure to 1 mM H2O2 reduced the bacterial population from 108 to 106 CFU mL−1, which was insufficient for antimicrobial activity by itself but caused surviving populations to become hypersensitive to aPDI. Cultures experienced a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction when treated with 0.015 μM Al-PpIX and below the limit of detection (∼6
log reduction when treated with 0.015 μM Al-PpIX and below the limit of detection (∼6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction) was achieved at 0.24 μM, several fold lower than that for naïve S. aureus (Table 3). Most impressively, Ga-PpIX had outstanding potency with reduction below the limit of detection achieved at 0.015 μM, the lowest PS concentration tested in this study.
log reduction) was achieved at 0.24 μM, several fold lower than that for naïve S. aureus (Table 3). Most impressively, Ga-PpIX had outstanding potency with reduction below the limit of detection achieved at 0.015 μM, the lowest PS concentration tested in this study.
| Condition | aPDIa | Al-PpIX (μM) | Ga-PpIX (μM) |
|---|---|---|---|
a Reduction in CFU mL−1 from initial population.
b Standard conditions: 7 h in Fe-deficient media (4 h growth, 3 h lag); 108 CFU mL−1 prior to dilution with PBS. aPDI: 30 s irrad. with 405 nm LED source (4.2 J cm−2).
c 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction in CFU mL−1.
d Initial 2 log reduction in CFU mL−1.
d Initial 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction not included in aPDI.
e Cycle 1.
f Below 100 CFU mL−1.
g Cycle 2. n/o = not observed. log reduction not included in aPDI.
e Cycle 1.
f Below 100 CFU mL−1.
g Cycle 2. n/o = not observed.
|
|||
| Standardb (no H2O2 added) | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log |
0.015 | 0.03 |
6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log |
0.90 | 0.24c | |
| 1 mM H2O2, growth phased | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log |
3.76e | n/o |
6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) logf logf |
n/o | n/o | |
| 1 mM H2O2, stationary phased | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log |
0.015g | <0.015 |
6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) logf logf |
0.24 | ≤0.015 | |
The dramatically higher aPDI potency following post-growth H2O2 exposure may be due to an increase in either catalase activity or cell wall permeability. The latter effect has been noted in both microbial and mammalian cells and is thought to be responsible for increased drug or PS uptake.12,36,40,41 Moreover, bacterial cultures are less adept at responding to stress in the stationary phase compared with the log-growth phase, when survival genes can be actively selected.59,60 With regard to hypersensitivity to Ga-PpIX, we postulate that H2O2 induces the upregulation of catalase which increases the local concentration of O2. Photodynamic inactivation is much more potent in hyperoxic environments, which can be triggered by a variety of catalysts in the presence of H2O2.42,43,61
The relative susceptibility of H2O2-challenged S. aureus can evolve with the number of exposures and growth cycles. For cultures treated with 1 mM H2O2 during log-phase growth, tolerance to aPDI remains high but a gradual increase in sensitivity to Ga-PpIX-mediated aPDI can be observed (Fig. 5b). Repeated H2O2 exposures may be selecting for subpopulations that produce high levels of catalase, resulting in higher local O2 concentrations and greater sensitivity to 1O2. On the other hand, cultures treated with 1 mM H2O2 postgrowth exhibit decreased sensitivity to Al-PpIX-mediated aPDI at later cycles (Fig. 6a), which can be ascribed to an adaptation against ROS stress. H2O2 is also known to trigger a bacterial SOS response that activates the upregulation of DNA repair enzymes,53,62 and this may also contribute toward aPDI tolerance over time. We note that while Al-PpIX loses its efficacy at low concentrations after repeated H2O2 exposures, its rate of ROS production above 0.1 μM is sufficient to overcome tolerance and eradicate H2O2-adapted bacteria.
Conclusions
Non-iron hemin analogs with group 13 atoms (Al-, Ga-, and In-PpIX) are excellent photosensitizers for targeted aPDI against S. aureus and MRSA strains. Fluorescent Al-PpIX produces mostly ROS whereas Ga- and InPpIX are more efficient at producing singlet oxygen. Al- and Ga-PpIX are captured by S. aureus within seconds and can mediate aPDI at 405 nm excitation with 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reductions at 15–30 nM, but their potencies are greatly reduced against bacteria grown in the presence of 1 mM H2O2. On the other hand, S. aureus exposed to H2O2 in their postgrowth phase are hypersensitized to aPDI, with 6
log reductions at 15–30 nM, but their potencies are greatly reduced against bacteria grown in the presence of 1 mM H2O2. On the other hand, S. aureus exposed to H2O2 in their postgrowth phase are hypersensitized to aPDI, with 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction at 240 nM for Al-PpIX and 15 nM for Ga-PpIX. The stark contrast in the aPDI sensitivity of S. aureus challenged by H2O2 at different stages in its lifecycle underscores a complex relationship between stress response and adaptation, revealing opportunities to exploit potential weaknesses in bacterial defense mechanisms.
log reduction at 240 nM for Al-PpIX and 15 nM for Ga-PpIX. The stark contrast in the aPDI sensitivity of S. aureus challenged by H2O2 at different stages in its lifecycle underscores a complex relationship between stress response and adaptation, revealing opportunities to exploit potential weaknesses in bacterial defense mechanisms.
Experimental
General
Hemin chloride and all reagents were obtained from commercial sources and used as received. The preparation of PpIX and the Cl salts of Al-PpIX, Ga-PpIX, and In-PpIX have been described in the literature previously and were reproduced with little modification.24,63–65 Absorption spectra of M-PpIX were collected on a Varian Cary50 spectrometer using a 1 cm quartz cuvette; luminescence spectra and fluorescence quantum yields (ΦFL) were measured using an Edinburgh Instruments FLS 980 spectrometer with an integrating sphere accessory. EPR studies were performed using a Bruker EMX X-band spectrometer operating at 9.5 GHz and 5.02 mW with a field modulation amplitude of 5 g at 100 kHz and used to estimate ΦSO with 405 nm excitation and TMPyP as a reference compound, based on the EPR method reported by Nakamura et al.66 EPR data analysis is provided in ESI† (Fig. S4).Microbiological cultures
S. aureus PC 1203 was obtained from the American Type Culture Collection (ATCC 10537) and cultured at 37 °C in standard tryptic soy (TS) broth (Fe-replete conditions), and also in media containing 3 mM 2,2′-bipyridine (Fe-deficient conditions). Clinical isolates of MRSA were cultured in a similar fashion.24,25 Cultures were typically incubated for 16 h then diluted to achieve an optical density of 0.5 at 600 nm (108 CFU mL−1). Bacterial counts were determined by plating serial dilutions on agar dishes and incubating at 37 °C for 16 h.Bacterial uptake assays using flow cytometry
FC studies were performed using a Cytoflex instrument (λex = 488 nm; 585/42 nm emission filter). Stock solutions of Al- or Ga-PpIX were prepared in DMSO (1 mM) and diluted in phosphate buffered saline (PBS, pH 7.4) just prior to use. Bacterial suspensions (108 CFU in 0.5 mL) were incubated with M-PpIX (10 μM) for specified periods between 10 s and 30 min, then fixed with 0.5 mL of 4% paraformaldehyde and subjected to FC analysis without further washing at a flow rate of 10 μL min−1. Bacterial cells (N = 105) were gated by defining a region of interest (ROI) based on forward and side scattering (FSC and SSC) parameters (Fig. S1, ESI†). For experiments involving labeling by inactivated samples, fixed bacteria were collected by centrifugation and redispersed in PBS before treatment with Ga-PpIX. Samples were incubated between 0.5 and 30 min, then analyzed by flow cytometry without further washing.Antimicrobial activities
MIC values were obtained using the broth microdilution method.67 Bacterial suspensions (final concentration 5 × 106 CFU mL−1) were diluted with one volume of M-PpIX solution with twofold serial dilution in microwells and incubated at 37 °C for 16 h. MIC values were determined by visual turbidity and confirmed in some cases by plating surviving bacteria on agar plates.aPDI assays were performed on planktonic S. aureus and MRSA (106–108 CFU mL−1) using 96-well plates irradiated by a 405 nm LED array (Rainbow Technology Systems, 140 mW cm−2). In a typical study, bacteria were cultured in standard TS broth then sub-cultured under Fe-deficient conditions prior to serial dilution in microtiter plates (0.2 mL per well). Bacterial suspensions were treated with 100 μL of M-PpIX with final concentrations ranging from 0.015 to 60 μM, followed immediately with a 30 s exposure to 405 nm light (4.2 mJ cm−2). Irradiated bacteria were then plated onto agar plates using serial tenfold dilutions; controls included bacteria irradiated without M-PpIX (Ctrl−/+) and bacteria with M-PpIX but no irradiation (Ctrl+/−). Bacterial counts were determined by the drop-plate method using TS-agar plates;68 aPDI susceptibilities were quantified by subtracting final log counts from initial values (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 or log
8 or log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6).
6).
aPDI studies with H2O2-treated bacteria
H2O2-tolerant (aPDI-insensitive) cultures were induced by cultivating S. aureus in standard media at 37 °C for 16 h. Optical density was adjusted to 0.5 followed by 100-fold dilution in Fe-deficient media containing 1 mM H2O2 and incubation at 37 °C for 4 h (log-growth phase; Scheme 1). This typically yielded bacterial densities close to 106 CFU mL−1; aPDI assays were then performed using the procedure described above. Repeat growth cycles were performed by transferring 50 μL of the H2O2-treated culture into 5 mL of fresh Fe-replete media and incubating at 37 °C for 16 h. Optical density was adjusted to 0.5 followed by 100-fold dilution in Fe-deficient media with 1 mM H2O2 as above.H2O2-challenged (aPDI-sensitive) cultures were induced by first cultivating S. aureus in Fe-deficient media until 108 CFU mL−1 was achieved, followed by post-growth treatment with sublethal H2O2 (final concentration 1 mM) and incubation at 37 °C for 4 h (stationary phase, Scheme 1) with final bacterial densities close to 106 CFU mL−1. aPDI assays and repeat growth cycles were performed using the procedures described above.
Author contributions
B. P. S. conducted all microbiological and aPDI studies; B. M. W. and B. P. S. synthesized M-PpIX derivatives and performed EPR analysis of ΦSO; Y. W. collected optical properties and ΦFL; B. P. S., K. E. R. and J. P. R. conducted FC analysis; B. P. S. and A. W. designed the research study and wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge financial support from the U.S. National Science Foundation (CHE-2204206), the National Institutes of Health (T32-GM125620, P30-CA023168), the Purdue Drug Discovery Institute (fellowship to B. P. S.), the Japan Society for the Promotion of Science (fellowship to Y. W.), and the Center for Food Safety Engineering at Purdue University, funded by the U.S. Department of Agriculture (59-8072-1-002, 58-8042-0-061). Clinical isolates of MRSA were kindly provided by Mohammed Seleem. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the U.S. Department of Agriculture.References
- Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report, World Health Organization, 2021.
- E. Tacconelli, et al. , Lancet Infect. Dis., 2018, 18, 318–327 CrossRef PubMed; T. T. Yoshikawa, J. Am. Geriatr. Soc., 2002, 50, 226–229 CrossRef PubMed.
- Antibiotic Resistance Threats in the United States, Centers for Disease Control and Prevention (U.S.), 2019.
- T. T. Yoshikawa, J. Am. Geriatr. Soc., 2002, 50, 226–229 CrossRef PubMed.
- S. Ashkenazi, J. Paediatr. Child Health, 2013, 49, E179–E182 CrossRef PubMed.
- R. I. Aminov, Front. Microbiol., 2010, 1, 1 Search PubMed.
- A. Brauner, O. Fridman, O. Gefen and N. Q. Balaban, Nat. Rev. Microbiol., 2016, 14, 320–330 CrossRef CAS PubMed.
- E. M. Windels, J. E. Michiels, B. V. D. Bergh, M. Fauvart and J. Michiels, mBio, 2019, 10, 5–19 CrossRef PubMed.
- M. Wainwright, T. Maisch, S. Nonell, K. Plaetzer, A. Almeida, G. P. Tegos and M. R. Hamblin, Lancet Infect. Dis., 2017, 17, e49–e55 CrossRef PubMed.
- M. S. Mulani, E. E. Kamble, S. N. Kumkar, M. S. Tawre and K. R. Pardesi, Front. Microbiol., 2019, 10, 539 CrossRef PubMed.
- A. Rapacka-Zdonczyk, A. Wozniak, M. Pieranski, A. Woziwodzka, K. P. Bielawski and M. Grinholc, Sci. Rep., 2019, 9, 9423 CrossRef PubMed.
- A. Woźniak, N. Burzyńska, I. Zybała, J. Empel and M. Grinholc, J. Photochem. Photobiol., B, 2022, 235, 112554 CrossRef PubMed.
- D. F. Silva, J. L. Toledo Neto, M. F. Machado, J. R. Bochnia, A. S. Garcez and A. A. Foggiato, Photodiagn. Photodyn. Ther., 2019, 27, 345–353 CrossRef CAS PubMed.
- I. Buchovec, A. Gricajeva, L. Kalėdienė and P. Vitta, Int. J. Mol. Sci., 2020, 21, 6932 CrossRef CAS PubMed.
- V. S. Ghate, W. Zhou and H.-G. Yuk, Compr. Rev. Food Sci. Food Saf., 2019, 18, 402–424 CrossRef PubMed.
- F. Cieplik, D. Deng, W. Crielaard, W. Buchalla, E. Hellwig, A. Al-Ahmad and T. Maisch, Crit. Rev. Microbiol., 2018, 44, 571–589 CrossRef CAS PubMed.
- J. F. Lovell, T. W. B. Liu, J. Chen and G. Zheng, Chem. Rev., 2010, 110, 2839–2857 CrossRef CAS PubMed.
- X. Li, J. F. Lovell, J. Yoon and X. Chen, Nat. Rev. Clin. Oncol., 2020, 17, 657–674 CrossRef PubMed.
- A.-G. Niculescu and A. M. Grumezescu, Appl. Sci., 2021, 11, 3626 CrossRef CAS.
- E. F. G. Dickson, J. C. Kennedy and R. H. Pottier, in Photodynamic Therapy, ed. T. Patrice, Royal Society of Chemistry, 2003, pp. 81–104 Search PubMed.
- A. Garcia-Sampedro, A. Tabero, I. Mahamed and P. Acedo, J. Porphyrins Phthalocyanines, 2019, 23, 11–27 CrossRef CAS.
- M. Klausen, M. Ucuncu and M. Bradley, Molecules, 2020, 25, 5239 CrossRef CAS PubMed.
- L. B. Josefsen and R. W. Boyle, Met.-Based Drugs, 2008, 276109 Search PubMed.
- A. V. Morales-de-Echegaray, T. R. Maltais, L. Lin, W. Younis, N. R. Kadasala, M. N. Seleem and A. Wei, ACS Infect. Dis., 2018, 4, 1564–1573 CrossRef CAS PubMed.
- A. V. Morales-de-Echegaray, L. Lin, B. Sivasubramaniam, A. Yermembetova, Q. Wang, N. S. Abutaleb, M. N. Seleem and A. Wei, Nanoscale, 2020, 12, 21734–21742 RSC.
- K. Michalska, M. Rychłowski, M. Krupińska, G. Szewczyk, T. Sarna and J. Nakonieczna, Mol. Pharmaceutics, 2022, 19, 1434–1448 CrossRef CAS PubMed.
- H. Zhang, Q. Li, X. Qi, Y. Li, H. Ma, M. Grinholc, J. Nakonieczna, B. Yu, X. Wang and L. Zhang, Chem. Eng. J., 2023, 451, 138261 CrossRef CAS.
- H. Contreras, N. Chim, A. Credali and C. Goulding, Curr. Opin. Chem. Biol., 2014, 19, 34–41 CrossRef CAS PubMed.
- J. E. Choby and E. P. Skaar, J. Mol. Biol., 2016, 428, 3408–3428 CrossRef CAS PubMed.
- N. D. Hammer and E. P. Skaar, Annu. Rev. Microbiol., 2011, 65, 129–147 CrossRef CAS PubMed.
- I. Stojiljkovic, B. D. Evavold and V. Kumar, Expert Opin. Invest. Drugs, 2001, 10, 309–320 CrossRef CAS PubMed.
- N. T. Vu, Y. Moriwaki, J. M. M. Caaveiro, T. Terada, H. Tsutsumi, I. Hamachi, K. Shimizu and K. Tsumoto, Protein Sci., 2013, 22, 942–953 CrossRef CAS PubMed.
- D. Kunz, J. Wirth, A. Sculean and S. Eick, Photodiagn. Photodyn. Ther., 2019, 26, 306–312 CrossRef CAS PubMed.
- Z. Cai, Y. Li, Y. Wang, S. Chen, S. Jiang, H. Ge, L. Lei and X. Huang, Photodiagn. Photodyn. Ther., 2019, 25, 382–388 CrossRef CAS PubMed.
- E.-M. Decker, V. Bartha, A. Kopunic and C. von Ohle, J. Periodontal Res., 2017, 52, 162 CrossRef CAS PubMed.
- A. S. Garcez, S. C. Núñez, M. S. Baptista, N. A. Daghastanli, R. Itri, M. R. Hamblin and M. S. Ribeiro, Photochem. Photobiol. Sci., 2011, 10, 483–490 CrossRef CAS PubMed.
- C. McCullagh and P. K. J. Robertson, Environ. Sci. Technol., 2006, 40, 2421–2425 CrossRef CAS PubMed.
- S. M. Yang, D. W. Lee, H. J. Park, M. H. Kwak, J. M. Park and M.-G. Choi, Photochem. Photobiol., 2019, 95, 833–838 CrossRef CAS PubMed.
- M. Nie, R. C. E. Silva, K. T. de Oliveira, V. S. Bagnato, A. N. de Souza Rastelli, W. Crielaard, J. Yang and D. M. Deng, Biofouling, 2021, 37, 656–665 CrossRef CAS PubMed.
- Y. Li, J. Du, S. Huang, S. Wang, Y. Wang, Z. Cai, L. Lei and X. Huang, Photodiagn. Photodyn. Ther., 2022, 37, 102691 CrossRef CAS PubMed.
- R. S. Funk and J. P. Krise, Mol. Pharmaceutics, 2007, 4, 154–159 CrossRef CAS PubMed.
- X. Sun, J. Sun, Y. Sun, C. Li, J. Fang, T. Zhang, Y. Wan, L. Xu, Y. Zhou, L. Wang and B. Dong, Adv. Funct. Mater., 2021, 31, 2101040 CrossRef CAS.
- B. Wang, L. Zhou, Y. Guo, H. Guo, Y. Zhong, X. Huang, Y. Ge, Q. Wang, X. Chu, Y. Jin, K. Lan, M. Yang and J. Qu, Bioact. Mater., 2022, 12, 314–326 CAS.
- Y. Hu, P. Geissinger and J. C. Woehl, J. Lumin., 2011, 131, 477–481 CrossRef CAS.
- Y. Shimizu and T. Azumi, J. Phys. Chem., 1982, 86, 22–26 CrossRef CAS.
- A. W. Maresso, G. Garufi and O. Schneewind, PLoS Pathog., 2008, 4, e1000132 CrossRef PubMed.
- Y. Moriwaki, J. M. M. Caaveiro, Y. Tanaka, H. Tsutsumi, I. Hamachi and K. Tsumoto, Biochemistry, 2011, 50, 7311–7320 CrossRef CAS PubMed.
- N. T. Vu, Y. Moriwaki, J. M. M. Caaveiro, T. Terada, H. Tsutsumi, I. Hamachi, K. Shimizu and K. Tsumoto, Protein Sci., 2013, 22, 942–953 CrossRef CAS PubMed.
- C. A. Wakeman, D. L. Stauff, Y. Zhang and E. P. Skaar, J. Bacteriol., 2014, 196, 1335–1342 CrossRef PubMed.
- B. A. Lindig, M. A. J. Rodgers and A. P. Schaap, J. Am. Chem. Soc., 1980, 102, 5590–5593 CrossRef CAS.
- D. Frees and H. Ingmer, Stress Responses in Staphylococcus aureus, in Staphylococcus: Genetics and Physiology, ed. G. A. Somerville, Caister Academic Press, 2016, pp. 221–248 Search PubMed.
- R. A. Proctor, C. von Eiff, B. C. Kahl, K. Becker, P. McNamara, M. Herrmann and G. Peters, Nat. Rev. Microbiol., 2006, 4, 295–305 CrossRef CAS PubMed.
- K. L. Painter, E. Strange, J. Parkhill, K. B. Bamford, D. Armstrong-James and A. M. Edwards, Infect. Immun., 2015, 83, 1830–1844 CrossRef CAS PubMed.
- C. Staerck, A. Gastebois, P. Vandeputte, A. Calenda, G. Larcher, L. Gillmann, N. Papon, J.-P. Bouchara and M. J. J. Fleury, Microb. Pathog., 2017, 110, 56–65 CrossRef CAS PubMed.
- D. Manoil, A. Parga, C. Hellesen, A. Khawaji, M. Brundin, S. Durual, V. Özenci, H. Fang and G. N. Belibasakis, J. Photochem. Photobiol., B, 2022, 234, 1011–1344 CrossRef PubMed.
- L. Agnez-Lima, P. Mascio, B. Demple and C. Menck, Biol. Chem., 2001, 382, 1071–1075 CAS.
- J. R. Anthony, K. L. Warczak and T. J. Donohue, Proc. Natl. Acad. Sci. U. S. A., 2005, 18, 6502–6507 CrossRef PubMed.
- J. Nakonieczna, E. Michta, M. Rybicka, M. Grinholc, A. Gwizdek-Wiśniewska and K. P. Bielawski, BMC Microbiol., 2010, 10, 323 CrossRef CAS PubMed.
- A. Rodríguez-Rojas, J. J. Kim, P. R. Johnston, O. Makarova, M. Eravci, C. Weise and J. Rolff, PLoS Genet., 2020, 16, 3 Search PubMed.
- V. N. Fritsch, V. V. Loi, T. Busche, Q. Tung, R. Lill, P. Horvatek, C. Wolz, J. Kalinowski and H. Antelmann, Free Radical Biol. Med., 2020, 161, 351–364 CrossRef CAS PubMed.
- W. Xiu, L. Wan, K. Yang, X. Li, L. Yuwen, H. Dong, Y. Mou, D. Yang and L. Wang, Nat. Commun., 2022, 13, 3875 CrossRef CAS PubMed.
- M. Grinholc, A. Rodziewicz, K. Forys, A. Rapacka-Zdonczyk, A. Kawiak, A. Domachowska, G. Golunski, C. Wolz, L. Mesak, K. Becker and K. P. Bielawski, Appl. Microbiol. Biotechnol., 2015, 99, 9161–9176 CrossRef CAS PubMed.
- E. L. Dodd and D. Bohle, Polyhedron, 2016, 108, 36–42 CrossRef CAS.
- D. R. Davydov, G. V. Ponomarev, E. Bobrovnikova-Marjon, D. C. Haines and J. A. Peterson, Biotechnol. Appl. Biochem., 2013, 64, 41–51 CrossRef PubMed.
- S. B. Kahl, J. J. Schaeck and M. S. Koo, J. Org. Chem., 1997, 62, 1875–1880 CrossRef CAS.
- K. Nakamura, K. Ishiyama, H. Ikai, T. Kanno, K. Sasaki, Y. Niwano and M. Kohno, J. Clin. Biochem. Nutr., 2011, 49, 87–95 CrossRef CAS PubMed.
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID), Clin. Microbiol. Infect., 2003, 9, ix–xv CrossRef.
- B. Herigstad, M. Hamilton and J. Heersink, J. Microbiol. Methods, 2001, 44, 121–129 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Synthesis of Al-PpIX; additional flow cytometry data; EPR data for quantum yields of photogenerated 1O2. See DOI: https://doi.org/10.1039/d4md00148f |
| ‡ Current address: Department of Polymer Chemistry, Kyoto University, Nishikyo-ku, Kyoto, 615-8510 Japan. |
| This journal is © The Royal Society of Chemistry 2024 |