Open Access Article
Open Access ArticleLive cell screening to identify RNA-binding small molecule inhibitors of the pre-let-7–Lin28 RNA–protein interaction†
Sydney L.
Rosenblum
 a,
Dalia M.
Soueid
b,
George
Giambasu
a,
Dalia M.
Soueid
b,
George
Giambasu
 c,
Steve
Vander Roest
d,
Alexander
Pasternak
e,
Erin F.
DiMauro
e,
Vladimir
Simov
c,
Steve
Vander Roest
d,
Alexander
Pasternak
e,
Erin F.
DiMauro
e,
Vladimir
Simov
 *e and
Amanda L.
Garner
*e and
Amanda L.
Garner
 *ab
*ab
aProgram in Chemical Biology, University of Michigan, 210 Washtenaw Avenue, Ann Arbor, MI 48109, USA. E-mail: algarner@umich.edu
bDepartment of Medicinal Chemistry, College of Pharmacy, University of Michigan, 1600 Huron Parkway, NCRC B520, Ann Arbor, MI 48109, USA
cComputational Chemistry, Merck & Co., Inc., Boston, MA 02115, USA. E-mail: george.giambasu@merck.com
dCenter for Chemical Genomics, Life Sciences Institute, University of Michigan, 210 Washtenaw Avenue, Ann Arbor, MI 48109, USA
eDiscovery Chemistry, Merck & Co., Inc., Boston, MA 02115, USA. E-mail: alexander_pasternak@merck.com; erin.dimauro@merck.com; vladimir_simov@merck.com
First published on 19th March 2024
Abstract
Dysregulation of the networking of RNA-binding proteins (RBPs) and RNAs drives many human diseases, including cancers, and the targeting of RNA–protein interactions (RPIs) has emerged as an exciting area of RNA-targeted drug discovery. Accordingly, methods that enable the discovery of cell-active small molecule modulators of RPIs are needed to propel this emerging field forward. Herein, we describe the application of live-cell assay technology, RNA interaction with protein-mediated complementation assay (RiPCA), for high-throughput screening to identify small molecule inhibitors of the pre-let-7d–Lin28A RPI. Utilizing a combination of RNA-biased small molecules and virtual screening hits, we discovered an RNA-binding small molecule that can disrupt the pre-let-7–Lin28 interaction demonstrating the potential of RiPCA for advancing RPI-targeted drug discovery.
Introduction
It is well understood the paramount role that RNAs play in maintaining human health. No longer thought of as solely playing an intermediary role in the central dogma, RNAs have been demonstrated to perform multiple functions in the cell, including non-canonical tasks in the regulation of transcription, splicing, and translation, RNA function through RNA–RNA interactions, as well as catalysis. These functions are carried out by a pantheon of RNA molecules ranging in size and structure, from small ∼22 nucleotide (nt) microRNAs (miRNAs) to >200 nt messenger RNAs (mRNAs), ribosomal RNAs (rRNAs) and long non-coding RNAs (lncRNAs) to highly structured RNA catalysts, or ribozymes. In combination with discoveries connecting aberrant RNA biology with human diseases, a new generation of RNA drug hunters has been called to arms with the goal of unleashing small molecule chemistry for manipulating the cellular activities of RNAs.1An important consideration in the field of RNA-targeted drug discovery is the extensive networking of RNAs with RNA-binding proteins (RBPs).2,3 Indeed, RNAs are invariably bound to and often modified by RBPs.4–7 These RBPs, in turn, regulate many aspects of coding and non-coding RNA biology, including RNA processing, nuclear export, function, localization, and stability.4–7 In-line with the significance of these RBP functions, dysregulation of RNA–RBP networks has been linked to many human diseases, including cancers and neurodegeneration.8–11 As such, targeting of RNA–protein interactions (RPIs) has emerged as a strategy for uncovering small molecules capable of modulating the function of cellular RNAs.12
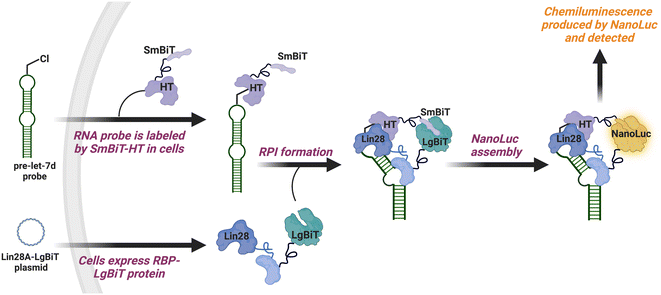
To catalyze hit discovery efforts in this nascent field of RNA–protein interaction manipulation, our laboratory developed live-cell assay technology for the detection of RPIs, RNA interaction with protein-mediated complementation assay (RiPCA) (Fig. 1).13,14 In RiPCA, cells are first engineered to stably express the small subunit of split NanoLuciferase (NanoLuc®) (SmBiT) fused to HaloTag® (HT) (SmBiT–HT).15,16 These cells are then transiently co-transfected with a plasmid encoding an RBP fused to the large subunit of NanoLuc® (LgBiT) and an RNA substrate that is chemically modified to contain a chloroalkane HT ligand. Following uptake, the RNA becomes covalently labeled by SmBiT–HT to enable NanoLuc®-mediated chemiluminescence signal generation upon binding to the LgBiT-tagged RBP and complementation of the SmBiT and LgBiT tags to reassemble functional NanoLuc®. Towards our goal of using RiPCA in high-throughput screening (HTS), we recently reported an optimized RiPCA protocol, RiPCA 2.0, which we subsequently used to detect a number of RBP interactions with precursor miRNAs (pre-miRNAs) and messenger RNAs (mRNAs).17,18 Building upon these successful optimization studies, we were eager to demonstrate RiPCA's applicability for HTS. Herein we disclose our first RiPCA-based screening campaign focused on identifying small molecule inhibitors of the interaction of Lin28A, an RBP, with the pre-miRNA, pre-let-7d.
Results and discussion
Adaptation of RiPCA for HTS
In our previously reported assay development efforts, RiPCA was performed in 96-well format; however, for HTS, higher throughput was required. Using our previously developed RiPCA 2.0 assay for the pre-let-7d–Lin28A interaction (Fig. 1),17 we miniaturized the assay to 384-well format to reduce reagent consumption and enable HTS (Fig. S1†). In addition to adaptation to liquid handling, several points of optimization were needed for miniaturization, including the number of cells per well, procedure for media removal prior to NanoLuc® substrate addition, and transfection scale-up (Fig. S2 and S3†). As an additional assay characterization step, we performed a pilot screen of 320 known drugs from the LOPAC library (Fig. S4†). From these optimization studies, which demonstrated that RiPCA could be performed in high throughput with suitable assay statistics (signal-to-background (S/B): 22–32; Z′ factor: 0.3–0.68; Fig. S3 and S4†), we were encouraged that RiPCA would be amenable to HTS and subsequently employed this protocol for screening small molecule libraries.Screening campaign
Lin28 is a highly conserved oncofetal RBP that plays crucial roles in development, pluripotency, cell cycle, organismal and cellular growth, metabolism, and immunity.19–23 The most well-characterized RNA target of Lin28 is the tumor suppressor miRNA family, let-7, where Lin28 functions as a specific inhibitor of let-7 maturation by binding to a hairpin loop found within the pri- and pre-let-7 intermediates.24,25 Although Lin28 is largely not expressed in adult somatic tissues (Fig. S5†) and restricted to embryonic development in mammals;23,26 in cancer, it becomes transcriptionally activated resulting in the development of poorly differentiated tumors.19,20,23 Loss of let-7 through Lin28 reactivation has been observed in at least 15% of all human cancers, including lung, breast, liver, esophageal, stomach, ovarian, prostate and colon cancers, and chronic lymphocytic leukemia.23,27–29 Relatedly, reduced let-7 levels have been found to correlate with high grade tumors, poor patient prognosis, and drug resistance to existing therapies.20,23,28 Dysregulation of the let-7/Lin28 pathway drives nearly all of the hallmarks of cancer, including regulation of cellular proliferation and metabolism, mediating cell death resistance, metastasis, angiogenesis and immune evasion, and promoting tumor-associated inflammation,23,30–32 as the let-7 family plays an important role in down-regulating cellular oncogenes including RAS and its mutant isoforms, MYCs and HMGA2 among others.33 Accordingly, disruption of the let-7/Lin28 interaction has emerged as a promising anti-cancer therapeutic strategy,23 and we were excited to deploy RiPCA to meet the goal of discovering drug-like and cell-active small molecule inhibitors of the pre-let-7d–Lin28A interaction.As chemical libraries, we initially assembled a comprehensive set of 13.5 K compounds (i.e., RNA ![[K with combining low line]](https://www.rsc.org/images/entities/char_004b_0332.gif) nowledge
nowledge ![[B with combining low line]](https://www.rsc.org/images/entities/char_0042_0332.gif) ased
ased ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) et) that was designed to encompass various direct mechanisms of RNA function modulation, including published and proprietary RNA and RBP binders, as well as splicing modulators. To build the 13.5 K RNA KBS, the preliminary set of RNA tools subsequently underwent similarity (SIM) expansion by leveraging in silico tools that focused on shape similarity searches and established pharmacophore interactions with different types of RNA secondary structures, as well as synthesis of additional libraries and singleton analogues for added chemical and structural diversity. In addition to the RNA KBS collection, we also created a Virtual Library set of 4.3 K compounds selected using an iterative virtual screening approach based on large-scale pharmacophore-based docking34–36 and rescoring using Glide scoring functions.37–40 Virtual screening was based on the Merck & Co., Inc., Rahway, NJ, USA compound collection against published structures of Lin28 with the pre-element of pre-let-7 and other small RNA fragments (PDB IDs: 5UDZ, 3TS0, 3TRZ, 3TS2)41,42 and apo-form structures (PDB IDs: 4ALP, 4A4I, 4A75, 4A76, 3ULJ),43 mainly focusing on the cold shock domain (CSD) and zinc knuckle domains (ZKD). Lastly, small molecule similarity screening based on previously reported let-7–Lin28 disrupters, TUTase inhibitors, and pre-miRNA hairpin binding molecules was used to complete the Virtual Library.44–52
et) that was designed to encompass various direct mechanisms of RNA function modulation, including published and proprietary RNA and RBP binders, as well as splicing modulators. To build the 13.5 K RNA KBS, the preliminary set of RNA tools subsequently underwent similarity (SIM) expansion by leveraging in silico tools that focused on shape similarity searches and established pharmacophore interactions with different types of RNA secondary structures, as well as synthesis of additional libraries and singleton analogues for added chemical and structural diversity. In addition to the RNA KBS collection, we also created a Virtual Library set of 4.3 K compounds selected using an iterative virtual screening approach based on large-scale pharmacophore-based docking34–36 and rescoring using Glide scoring functions.37–40 Virtual screening was based on the Merck & Co., Inc., Rahway, NJ, USA compound collection against published structures of Lin28 with the pre-element of pre-let-7 and other small RNA fragments (PDB IDs: 5UDZ, 3TS0, 3TRZ, 3TS2)41,42 and apo-form structures (PDB IDs: 4ALP, 4A4I, 4A75, 4A76, 3ULJ),43 mainly focusing on the cold shock domain (CSD) and zinc knuckle domains (ZKD). Lastly, small molecule similarity screening based on previously reported let-7–Lin28 disrupters, TUTase inhibitors, and pre-miRNA hairpin binding molecules was used to complete the Virtual Library.44–52
Using our optimized HTS workflow (Fig. S1†), the combined collection of 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 797 small molecules (RNA KBS and Virtual Library) was screened at 10 μM final concentration in RiPCA (Fig. 2A; plate layout can be found in Fig. S6†). For each plate, a pre-miR-21 probe was used as a positive control and DMSO was used as a negative control. Throughout our screening campaign, the assay performed well with an average plate Z′ factor of 0.37 and average S/B of 14.7. Our screening funnel can be found in Fig. 2B. Using a hit criterion of >55% inhibition, 992 compounds were identified as primary hits (5.6% hit rate); 758 hits came from the RNA KBS and 234 hits came from the Virtual Library. Because we anticipated that some hits may be false positives due to cytotoxicity, we subsequently performed a counterscreen using the CellTiter-Glo® (CTG) cell viability assay. By eliminating compounds that exhibited toxicity of >20% at 10 μM in at least two of three replicates, 840 compounds (85% of the primary hits) were then taken on to triplicate analysis using RiPCA. Of the 840 compounds tested, 240 hits (28.6% of the primary hits) demonstrated inhibitory activity of >55% in two of the three replicates and were tested in dose–response assays. About one-third of tested molecules displayed dose-dependent inhibition, which was defined as pAC50 value >5 (78 compounds; 7.9% of the primary hits).
797 small molecules (RNA KBS and Virtual Library) was screened at 10 μM final concentration in RiPCA (Fig. 2A; plate layout can be found in Fig. S6†). For each plate, a pre-miR-21 probe was used as a positive control and DMSO was used as a negative control. Throughout our screening campaign, the assay performed well with an average plate Z′ factor of 0.37 and average S/B of 14.7. Our screening funnel can be found in Fig. 2B. Using a hit criterion of >55% inhibition, 992 compounds were identified as primary hits (5.6% hit rate); 758 hits came from the RNA KBS and 234 hits came from the Virtual Library. Because we anticipated that some hits may be false positives due to cytotoxicity, we subsequently performed a counterscreen using the CellTiter-Glo® (CTG) cell viability assay. By eliminating compounds that exhibited toxicity of >20% at 10 μM in at least two of three replicates, 840 compounds (85% of the primary hits) were then taken on to triplicate analysis using RiPCA. Of the 840 compounds tested, 240 hits (28.6% of the primary hits) demonstrated inhibitory activity of >55% in two of the three replicates and were tested in dose–response assays. About one-third of tested molecules displayed dose-dependent inhibition, which was defined as pAC50 value >5 (78 compounds; 7.9% of the primary hits).
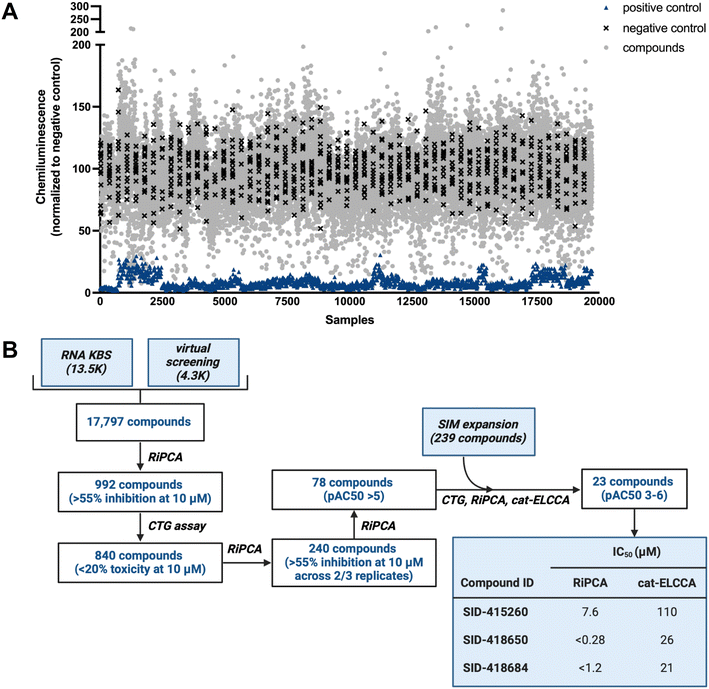 | ||
| Fig. 2 RiPCA HTS. (A) Campaign view of screening data. (B) HTS funnel and tabulated IC50 values for the 3 most active hits in RiPCA and cat-ELCCA. | ||
Hit validation
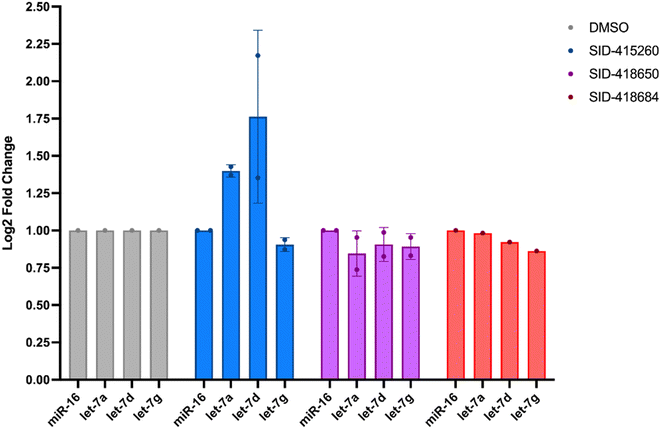
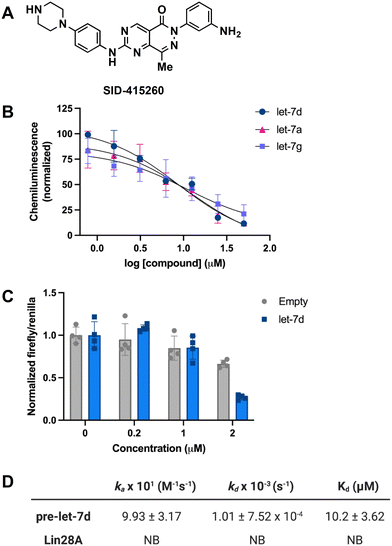
In addition to testing confirmed hits from our RiPCA primary screen, we also performed structure–activity relationship (SAR) studies in subsequent characterization assays through hit expansion to afford an additional 239 compounds with 2D and 3D structural similarity to the 78 hits. Compounds were first analyzed in the CTG assay (10 μM in triplicate), and those that exhibited an average toxicity of >40% were removed. The remaining 264 compounds were then analyzed for dose-dependent cellular inhibitory activity in RiPCA, as well as biochemical inhibitory activity using our laboratory's catalytic enzyme-linked click chemistry assay (cat-ELCCA) assay technology53 that we previously used to screen against the pre-let-7–Lin28 RPI.44 In cat-ELCCA, only 8.7% of tested compounds exhibited concentration-dependent inhibition. Representative data from RiPCA, cat-ELCCA, and CTG for the 3 most active compounds from RiPCA possessing a common pyrimido[4,5-d]pyridazine-5(6H)-ones pharmacophore can be found in Fig. S7.† Interestingly, all 3 of these hits were more active in RiPCA than in cat-ELCCA (Fig. 2B and S7†).While it is uncommon, examples of molecules with greater potency in cells have been described, including several examples of small molecules targeting RNA–protein complexes.54 Thus, to provide further evidence of inhibitory activity against the pre-let-7–Lin28 RPI, we determined how these hits impacted the production of mature let-7 in cells. In brief, Lin28A-expressing T-47D breast cancer cells55 were treated with compounds (5 μM for 48 h), and let-7 levels were quantified from extracted RNA via qRT-PCR. In addition to let-7d, we also examined two other let-7s, let-7a-1 and let-7g. As shown in Fig. 3, excitingly, one of the hits, SID-415260 (structure in Fig. 4A), showed increased levels of two let-7s, let-7d and let-7a-1, which were previously shown to be regulated by Lin28A in this cell line.55 Of note, treatment with higher compound concentrations was limited by toxicity in this cell line (data not shown). We additionally tested the inhibitory activity of SID-415260 in RiPCAs developed for pre-let-7a-1 and -7g.17 Notably, RPI disruption was similar across the let-7s with measured IC50 values of 10.7 μM, 7.4 μM, and 12.5 μM, respectively, for pre-let-7a, -7d, and -7g (Fig. 4B). As let-7g levels were not impacted by compound treatment in our qRT-PCR studies, this could point to additional levels of regulation of the maturation of this let-7 isoform in this cell line.
To provide further evidence that SID-415260 restored production of let-7, we tested the activity of this compound using the pmiRGLO dual-luciferase miRNA reporter assay.56 NTERA-2 cells which express Lin28A and Lin28B were transiently transfected with a pmiRGLO reporter containing a let-7d target site in the 3′ untranslated region of the firefly luciferase gene56,57 and then treated with varying concentrations of compound for 48 h. Excitingly, in-line with our qRT-PCR results, we observed a decrease in luminescence signal produced by firefly luciferase over an empty vector control indicating enhanced let-7 activity (Fig. 4C).
SID-415260 belongs to a known class of pyrimido[4,5-d]pyridazine-5(6H)-ones reported as selective Wee1 kinase inhibitors, with the aminopyrimidine moiety making a key hinge interaction with Cys379 in the Wee1 ATP binding pocket.58 The carbonyl group in the bicyclic pyrimido-pyridazinone has also been shown to engage with the Asn376 residue of the Wee1 gatekeeper region, further contributing to kinase activity and selectivity.58 In our hands, SID-415260 inhibited Wee1 with an IC50 value of 35 nM across a panel of 264 kinases with an additional 18 kinases having measured IC50 values between 298 nM and 1 μM (Table S4†). The promiscuity of this compound was anticipated based on the presence of the aminopyrimidine core which is a common hinge-binding motif found in many multi-kinase inhibitors,59,60 including Wee1 kinase inhibitors of varying selectivity.61–64 Of note, we confirmed that SID-415260 does not affect Lin28A phosphorylation (Fig. S8†), as this post-translational modification has been previously shown to impact Lin28A function.65 Structurally, SID-415260 shows high 2D and 3D similarity relative to known RNA binders and modulators, including risdiplam, SMN-C3, and SMN-C5.66 It contains a central bicyclic core with an aryl group at one end and a basic piperazine cycloalkyl group at the other end of the molecule. Based on this structural similarity, as well as precedence for kinase inhibitors as RNA binding ligands,67–69 it was included as part of the RNA KBS.
To decipher the mechanism of inhibitory action, we performed biophysical binding assays using surface plasmon resonance (SPR) spectroscopy. Biotinylated pre-let-7d and Lin28A were immobilized on high-sensitivity biotin–streptavidin sensors and SID-415260 was flowed over the immobilized biomolecules. The association (ka) and dissociation (kd) rates and binding constant (Kd) were obtained by fitting to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 interaction model based on duplicate data sets. As shown in Fig. 4D, in-line with its prediction as a potential RNA-binding ligand, the compound was found to bind exclusively to the RNA with a Kd value of 10.2 μM (sensorgrams can be found in Fig. S9†). To the best of our knowledge, this is the first reported direct pre-let-7-binding disruptor of the let-7/Lin28 interaction, as previously discovered compounds directly bind to Lin28 (ref. 44, 46–49 and 70) or function upstream at the level of Lin28 expression in cells.45,71
1 interaction model based on duplicate data sets. As shown in Fig. 4D, in-line with its prediction as a potential RNA-binding ligand, the compound was found to bind exclusively to the RNA with a Kd value of 10.2 μM (sensorgrams can be found in Fig. S9†). To the best of our knowledge, this is the first reported direct pre-let-7-binding disruptor of the let-7/Lin28 interaction, as previously discovered compounds directly bind to Lin28 (ref. 44, 46–49 and 70) or function upstream at the level of Lin28 expression in cells.45,71
Conclusions
As the field of small molecule RNA-targeted drug discovery expands, it necessitates the development of new technologies for enabling lead finding with functional cellular activity. Herein we disclose how RiPCA can be employed for HTS to discover small molecule inhibitors of cellular RPIs. Using the pre-let-7–Lin28 RPI as a model, we successfully miniaturized and automated RiPCA and screened biased libraries of small molecules to identify inhibitors of this cancer-relevant RPI. Excitingly, these efforts resulted in the uncovering of a Wee1 kinase inhibitor that binds to pre-let-7 with micromolar affinity and exhibits promising bioactivity in Lin28-expressing cancer cell lines. Our results demonstrate how RiPCA can be used to identify drug-like hits for targeting RPIs and future work will focus on optimizing affinity and specificity for pre-let-7 and applying this technology to additional RNA/RPI systems of relevance to human health and disease.Author contributions
Conceptualization: S. L. R., A. P., E. F. D., V. S., A. L. G.; data curation: G. G.; formal analysis: S. L. R., D. M. S., G. G., S. V. R., A. L. G.; funding acquisition: A. L. G.; investigation: S. L. R., D. M. S., G. G.; methodology: S. L. R., D. M. S., G. G., S. V. R.; project administration: V. S., A. L. G.; supervision: V. S., A. L. G.; visualization: S. L. R., D. M. S., A. L. G.; writing – original draft: S. L. R., V. S., A. L. G.; writing – review and editing: S. L. R., D. M. S., V. S., A. L. G.Conflicts of interest
A. L. G. is an advisor to and holds equity in Skyhawk Therapeutics.Acknowledgements
This work was supported by the NIH (R01 GM135252 to A. L. G. and T32 GM14530 to D. M. S) and Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, USA. We would like to thank Dr. Aaron Robida for advice during HTS optimization, as well as Andy Liaw, Kiersten Tovar, Serena (Guangyun) Xu, and Mary Jo Wildey for their help with screening data analysis and visualization. Fig. 1, 2B, 4D, and S1† were created using Biorender.References
- A. L. Garner, Contemporary progress and opportunities in RNA-targeted drug discovery, ACS Med. Chem. Lett., 2023, 14, 251–259 CrossRef CAS PubMed.
- S. Gerstberger, M. Hafner and T. Tuschl, A census of human RNA-binding proteins, Nat. Rev. Genet., 2014, 15, 829–845 CrossRef CAS PubMed.
- M. W. Hentze, A. Castello, T. Schwarzl and T. Preiss, A brave new world of RNA-binding proteins, Nat. Rev. Mol. Cell Biol., 2018, 19, 327–341 CrossRef CAS PubMed.
- E. Jankowsky and M. E. Harris, Specificity and non-specificity in RNA-protein interactions, Nat. Rev. Mol. Cell Biol., 2015, 16, 533–544 CrossRef CAS PubMed.
- E. L. Van Nostrand, P. Freese, G. A. Pratt, X. Wang, X. Wei, R. Xiao, S. M. Blue, J.-Y. Chen, N. A. L. Cody, D. Dominguez, S. Olson, B. Sundararaman, L. Zhan, C. Bazile, L. P. B. Bouvrette, J. Bergalet, M. O. Duff, K. E. Garcia, C. Gelboin-Burkhart, M. Hochman, N. J. Lambert, H. Li, M. P. McGurk, T. B. Nguyen, T. Palden, I. Rabano, S. Sathe, R. Stanton, A. Su, R. Wang, B. A. Yee, B. Zhou, A. L. Louie, S. Algner, X.-D. Fu, E. Lecuyer, C. B. Burge, B. R. Graveley and G. W. Yeo, A large-scale binding and functional map of human RNA-binding proteins, Nature, 2020, 583, 711–719 CrossRef CAS PubMed.
- B. S. Zhao, I. A. Roundtree and C. He, Post-transcriptional gene regulation by mRNA modifications, Nat. Rev. Mol. Cell Biol., 2017, 18, 31–42 CrossRef CAS PubMed.
- G. Singh, G. Pratt, G. W. Yeo and M. J. Moore, The clothes make the mRNA: past and present trends in mRNP fashion, Annu. Rev. Biochem., 2015, 84, 325–354 CrossRef CAS PubMed.
- B. Pereira, M. Billaud and R. Almeida, RNA-binding proteins in cancer: old players and new actors, Trends Cancer, 2017, 3, 506–528 CrossRef CAS PubMed.
- P. S. Choi and A. Thomas-Tikhonenko, RNA-binding proteins of COSMIC importance in cancer, J. Clin. Invest., 2021, 131, e151627 CrossRef CAS PubMed.
- E. G. Conlon and J. L. Manley, RNA-binding proteins in neurodegeneration: mechanisms in aggregate, Genes Dev., 2017, 31, 1509–1528 CrossRef CAS PubMed.
- A. H. Corbett, Post-transcriptional regulation of gene expression and human disease, Curr. Opin. Cell Biol., 2018, 52, 96–104 CrossRef CAS PubMed.
- P. Wu, Inhibition of RNA-binding proteins with small molecules, Nat. Rev. Chem., 2020, 4, 441–458 CrossRef CAS PubMed.
- S. R. Rosenblum, D. A. Lorenz and A. L. Garner, A Live-Cell Assay for the Detection of pre-microRNA-Protein Interactions, RSC Chem. Biol., 2021, 2, 241–247 RSC.
- S. R. Rosenblum and A. L. Garner, RiPCA: an assay for the detection of RNA-protein interactions in live cells, Curr. Protoc., 2022, 2, e358 CrossRef CAS PubMed.
- A. S. Dixon, M. K. Schwinn, M. P. Hall, K. Zimmerman, P. Otto, T. H. Lubben, B. L. Butler, B. F. Binkowski, T. Machleidt, T. A. Kirkland, M. G. Wood, C. T. Eggers, L. P. Encell and K. V. Wood, NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells, ACS Chem. Biol., 2016, 11, 400–408 CrossRef CAS PubMed.
- G. V. Los, L. P. Encell, M. G. McDougall, D. D. Hartzell, N. Karassina, C. Zimprich, M. G. Wood, R. Learish, R. F. Ohana, M. Urh, D. Simpson, J. Mendez, K. Zimmerman, P. Otto, G. Vidugiris, J. Zhu, A. Darzins, D. H. Klaubert, R. F. Bulleit and K. V. Wood, HaloTag: a novel protein labeling technology for cell imaging and protein analysis, ACS Chem. Biol., 2008, 3, 373–382 CrossRef CAS PubMed.
- S. R. Rosenblum and A. L. Garner, Optimization of RiPCA for the live-cell detection of pre-microRNA-protein interactions, ChemBioChem, 2022, 23, e202200508 CrossRef CAS PubMed.
- D. M. Soueid and A. L. Garner, Adaptation of RiPCA for the live-cell detection of mRNA-protein interactions, Biochemistry, 2023, 62, 3323–3336 CrossRef CAS PubMed.
- S. R. Viswanathan and G. Q. Daley, Lin28: a microRNA regulator with a macro role, Cell, 2010, 140, 445–449 CrossRef CAS PubMed.
- J. E. Thornton and R. I. Gregory, How does Lin28 let-7 control development and disease?, Trends Cell Biol., 2012, 22, 474–482 CrossRef CAS PubMed.
- N. Shyh-Chang and G. Q. Daley, Lin28: primal regulator of growth and metabolism in stem cells, Cell Stem Cell, 2013, 12, 395–406 CrossRef PubMed.
- S. Jiang and D. Baltimore, RNA-binding protein Lin28 in cancer and immunity, Cancer Lett., 2016, 375, 108–113 CrossRef CAS PubMed.
- J. Balzeau, M. R. Menezes, S. Cao and J. P. Hagan, The LIN28/let-7 pathway in cancer, Front. Genet., 2017, 3, 31 Search PubMed.
- S. R. Viswanathan, G. Q. Daley and R. I. Gregory, Selective blockade of microRNA processing by Lin28, Science, 2008, 320, 97–100 CrossRef CAS PubMed.
- M. A. Newman, J. M. Thomson and S. M. Hammond, Lin-28 interaction with the let-7 precursor loop mediates regulated microRNA processing, RNA, 2008, 14, 1539–1549 CrossRef CAS PubMed.
- D. H. Yang and E. G. Moss, Temporally regulated expression of Lin-28 in diverse tissues of the developing mouse, Gene Expression Patterns, 2003, 3, 719–726 CrossRef CAS PubMed.
- H. Ling, M. Fabbri and G. A. Calin, MicroRNAs and other non-coding RNAs as targets for anticancer drug development, Nat. Rev. Drug Discovery, 2013, 12, 847–865 CrossRef CAS PubMed.
- S. R. Viswanathan, J. T. Powers, W. Einhorn, Y. Hoshida, T. L. Ng, S. Toffanin, M. O'Sullivan, J. Lu, L. A. Phillips, V. L. Lockhart, S. P. Shah, P. S. Tanwar, C. H. Mermel, R. Beroukhim, M. Azam, J. Teixeira, M. Meyerson, T. P. Hughes, J. M. Llovet, J. Radich, C. G. Mullighan, T. R. Golub, P. H. Sorensen and G. Q. Daley, Lin28 promotes transformation and is associated with advanced human malignancies, Nat. Genet., 2009, 41, 843–848 CrossRef CAS PubMed.
- S. Lin and R. I. Gregory, MicroRNA biogenesis pathways in cancer, Nat. Rev. Cancer, 2015, 15, 321–333 CrossRef CAS PubMed.
- T. Wang, G. Wang, D. Hao, X. Liu, D. Wang, N. Ning and X. Li, Aberrant regulation of the LIN28A/LIN28B and let-7 loop in human malignant tumors and its effects on the hallmarks of cancer, Mol. Cancer, 2015, 14, 125 CrossRef PubMed.
- H. Xiong, W.-X. Zhao, J. Wang, B. J. Seifer, C. Ye, Y. Chen, Y. Jia, C. Chen, J. Shen, L. Wang, X. Sui and J. Zhou, Oncogenic mechanisms of Lin28 in breast cancer: new functions and therapeutic opportunities, Onco Targets Ther, 2017, 8, 25721–25735 Search PubMed.
- J. W. Franses, J. Philipp, P. Missios, I. Bhan, A. Liu, C. Yashaswini, E. Tai, H. Zhu, M. Ligorio, B. Nicholson, E. M. Tassoni, N. Desai, A. S. Kulkarni, A. Szabolcs, T. S. Hong, A. S. Liss, C. Fernandez-del Castillo, D. P. Ryan, S. Maheswaran, D. A. Haber, G. Q. Daley and D. T. Ting, Pancreatic circulating tumor cell profiling identifies LIN28B as a metastasis driver and drug target, Nat. Commun., 2020, 11, 3303 CrossRef CAS PubMed.
- S. Roush and F. J. Slack, The let-7 family of microRNAs, Trends Cell Biol., 2008, 18, 505–516 CrossRef CAS PubMed.
- D. Joseph-McCarthy, B. E. Thomas 4th, M. Belmarsh, D. Moustakas and J. C. Alvarez, Pharmacophore-based molecular docking to account for ligand flexibility, Proteins, 2003, 51, 172–188 CrossRef CAS PubMed.
- T. Halgren, New method for fast and accurate binding-site identification and analysis, Chem. Biol. Drug Des., 2007, 69, 146–148 CrossRef CAS PubMed.
- T. A. Halgren, Identifying and characterizing binding sites and assessing druggability, J. Chem. Inf. Model., 2009, 49, 377–389 CrossRef CAS PubMed.
- R. A. Friesner, J. L. Banks, R. B. Murphy, T. A. Halgren, J. J. Klicic, D. T. Mainz, M. P. Repasky, E. H. Knoll, M. Shelley, J. K. Perry, D. E. Shaw, P. Francis and P. S. Shenkin, Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy, J. Med. Chem., 2004, 47, 1739–1749 CrossRef CAS PubMed.
- T. A. Halgren, R. B. Murphy, R. A. Friesner, H. S. Beard, L. L. Frye, W. T. Pollard and J. L. Banks, Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening, J. Med. Chem., 2004, 47, 1750–1759 CrossRef CAS PubMed.
- R. A. Friesner, R. B. Murphy, M. P. Repasky, L. L. Frye, J. R. Greenwood, T. A. Halgren, P. C. Sanschagrin and D. T. Mainz, Extra precision Glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes, J. Med. Chem., 2006, 49, 6177–6196 CrossRef CAS PubMed.
- Y. Yang, K. Yao, M. P. Repasky, K. Leswing, R. Abel, B. K. Shoichet and S. V. Jerome, Efficient exploration of chemical space with docking and deep learning, J. Chem. Theory Comput., 2021, 17, 7106–7119 CrossRef CAS PubMed.
- L. Wang, Y. Nam, A. K. Lee, C. Yu, K. Roth, C. Chen, E. M. Ransey and P. Sliz, LIN28 zinc knuckle domain is required and sufficient to induce let-7 oligouridylation, Cell Rep., 2017, 17, 2664–2675 CrossRef PubMed.
- Y. Nam, C. Chen, R. I. Gregory, J. J. Chou and P. Sliz, Molecular basis for interaction of let-7 microRNAs with Lin28, Cell, 2011, 147, 1080–1091 CrossRef CAS PubMed.
- F. Mayr, A. Schutz, N. Doge and U. Heinemann, The Lin28 cold-shock domain remodels pre-let-7 microRNA, Nucleic Acids Res., 2012, 40, 7492–7506 CrossRef CAS PubMed.
- D. A. Lorenz, T. Kaur, S. A. Kerk, E. E. Gallagher, J. Sandoval and A. L. Garner, Expansion of cat-ELCCA for the discovery of small molecule inhibitors of the pre-let-7-Lin28 RNA-protein interaction, ACS Med. Chem. Lett., 2018, 9, 517–521 CrossRef CAS PubMed.
- M. Roos, U. Pradere, R. P. Ngondo, A. Behera, S. Allegrini, G. Civenni, J. A. Zagalak, J.-R. Marchand, M. Menzi, H. Towbin, J. Scheuermann, D. Neri, A. Caflisch, C. V. Catapano, C. Claudo and J. Hall, A small-molecule inhibitor of Lin28, ACS Chem. Biol., 2016, 11, 2773–2781 CrossRef CAS PubMed.
- D. Lim, W. G. Byun, J. Y. Koo, H. Park and S. B. Park, Discovery of a small-molecule inhibitor of protein-microRNA interaction using binding assay with a site-specifically labeled Lin28, J. Am. Chem. Soc., 2016, 138, 13630–13638 CrossRef CAS PubMed.
- D. Lim, W. G. Byun and S. B. Park, Restoring let-7 microRNA biogenesis using a small-molecule inhibitor of the protein-RNA interaction, ACS Med. Chem. Lett., 2018, 9, 1181–1185 CrossRef CAS PubMed.
- H. L. Lightfoot, E. A. Miska and S. Balasubramanian, Identification of small molecule inhibitors of the Lin28-mediated blockage of pre-let-7g processing, Org. Biomol. Chem., 2016, 14, 10208–10216 RSC.
- L. Wang, R. G. Rowe, A. Jaimes, C. Yu, Y. Nam, D. S. Pearson, J. Zhang, X. Xie, W. Marion, G. J. Heffron, G. Q. Daley and P. Sliz, Small-molecule inhibitors disrupt let-7 oligouridylation and release the selective blockage of let-7 processing by LIN28, Cell Rep., 2018, 23, 3091–3101 CrossRef CAS PubMed.
- W. G. Byun, D. Lim and S. B. Park, Discovery of small-molecule modulators of protein-RNA interactions by fluorescence intensity-based binding assay, ChemBioChem, 2020, 21, 818–824 CrossRef CAS PubMed.
- S. Lin and R. I. Gregory, Identification of small molecule inhibitors of Zcchc11 TUTase activity, RNA Biol., 2015, 12, 792–800 CrossRef PubMed.
- A. Cording, M. Gormally, P. J. Bond, M. Carrington, S. Balasubramanian, E. A. Miska and B. Thomas, Selective inhibitors of trypanosomal uridylyl transferase RET1 establish druggability of RNA post-transcriptional modifications, RNA Biol., 2017, 14, 611–619 CrossRef PubMed.
- A. L. Garner, cat-ELCCA: catalyzing drug discovery through click chemistry, Chem. Commun., 2018, 54, 6531–6539 RSC.
- A. G. Schwald and I. Cornella-Taracido, Causes and significance of increased compound potency in cellular or physiological contexts, J. Med. Chem., 2018, 61, 1767–1773 CrossRef PubMed.
- E. Piskounova, C. Polytarchou, J. E. Thornton, R. J. LaPierre, C. Pothoulakis, J. P. Hagan, D. Iliopoulos and R. I. Gregory, Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms, Cell, 2011, 147, 1066–1079 CrossRef CAS PubMed.
- K. Gumireddy, D. D. Young, X. Xiong, J. B. Hogenesch, Q. Huang and A. Deiters, Small-molecule inhibitors of microRNA miR-21 function, Angew. Chem., Int. Ed., 2008, 47, 7482–7484 CrossRef CAS PubMed.
- A. W. Robertson, J. Sandoval, O. G. Mohamed, Y. Zhuang, E. E. Gallagher, J. J. Schmidt, L. Caratelli, A. Menon, P. J. Schultz, R. M. Torrez, C. L. Hay, B. A. Bell, P. A. Price, A. L. Garner and A. Tripathi, Discovery of surfactins as inhibitors of microRNA processing using cat-ELCCA, ACS Med. Chem. Lett., 2021, 12, 878–886 CrossRef CAS PubMed.
- V. J. Alli, P. Yadav, V. Suresh and S. S. Jadav, Synthetic and medicinal chemistry approaches toward WEE1 kinase inhibitors and its degraders, ACS Omega, 2023, 8, 20196–20233 CrossRef CAS PubMed.
- A. Spallarossa, B. Tasso, E. Russo, C. Villa and C. Brullo, The development of FAK inhibitors: a five-year update, Int. J. Mol. Sci., 2022, 23, 6381 CrossRef CAS PubMed.
- M. H. Keylor, A. Gulati, S. D. Kattar, R. E. Johnson, R. W. Chau, K. A. Margrey, M. J. Ardolino, C. Zarate, K. E. Poremba, V. Simov, G. J. Morriello, J. J. Acton, B. Pio, X. Yan, R. L. Palte, S. E. McMinn, L. Nogle, C. A. Lesburg, D. Adpressa, S. Lin, S. Neelamkavil, P. Liu, J. Su, L. G. Hegde, J. D. Woodhouse, R. Faltus, T. Xiong, P. J. Ciaccio, J. Piesvaux, K. M. Otte, H. B. Wood, M. E. Kennedy, D. J. Bennett, E. F. DiMauro, M. J. Fell and P. H. Fuller, Structure-guided discovery of aminoquinazolines as brain-penetrant and selective LRRK2 inhibitors, J. Med. Chem., 2022, 65, 838–856 CrossRef CAS PubMed.
- H. Hirai, Y. Iwasawa, M. Okada, T. Arai, T. Nishibata, M. Kobayashi, T. Kimura, N. Kaneko, J. Ohtani, K. Yamanaka, H. Itadani, I. Takahashi-Suzuki, K. Fukasawa, H. Oki, T. Nambu, J. Jiang, T. Sakai, H. Arakawa, T. Sakamoto, T. Sagara, T. Yoshizumi, S. Mizuarai and H. Kotani, Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents, Mol. Cancer Ther., 2009, 8, 2992–3000 CrossRef CAS PubMed.
- B. D. Palmer, J. B. Smaill, G. W. Rewcastle, E. M. Dobrusin, A. Kraker, C. W. Moore, R. W. Steinkampf and W. A. Denny, Structure–activity relationships for 2-anilino-6-phenylpyrido[2,3-d]pyrimidin-7(8H)-ones as inhibitors of the cellular checkpoint kinase Wee1, Bioorg. Med. Chem. Lett., 2005, 15, 1931–1935 CrossRef CAS PubMed.
- Y. Tong, M. Torrent, A. S. Florjancic, K. D. Bromberg, F. G. Buchanan, D. C. Ferguson, E. F. Johnson, L. M. Lasko, D. Maag, P. J. Merta, A. M. Olson, D. J. Osterling, N. Soni, A. R. Shoemaker and T. D. Penning, Pyrimidine-based tricyclic molecules as potent and orally efficacious inhibitors of Wee1 kinase, ACS Med. Chem. Lett., 2015, 6, 58–62 CrossRef CAS PubMed.
- A. Mastracchio, C. Lai, M. Torrent, K. Bromberg, F. G. Buchanan, D. Ferguson, V. Bontcheva, E. F. Johnson, L. Lasko, D. Maag, A. R. Shoemaker and T. D. Penning, Investigation of biaryl heterocycles as inhibitors of Wee1 kinase, Bioorg. Med. Chem. Lett., 2019, 29, 1481–1486 CrossRef CAS PubMed.
- K. M. Tsanov, D. S. Pearson, Z. Wu, A. Han, R. Triboulet, M. T. Seligson, J. T. Powers, J. K. Osborne, S. Kane, S. P. Gygi, R. I. Gregory and G. Q. Daley, LIN28 phosphorylation by MAPK/ERK couples signalling to the post-transcriptional control of pluripotency, Nat. Cell Biol., 2017, 19, 60–67 CrossRef CAS PubMed.
- H. Ratni, R. S. Scalco and A. H. Stephan, Risdiplam, the first approved small molecule splicing modifier drug as a blueprint for future transformative medicines, ACS Med. Chem. Lett., 2021, 12, 874–877 CrossRef CAS PubMed.
- P. Zhang, X. Liu, D. Abegg, T. Tanaka, Y. Tong, R. I. Benhamou, J. Baisden, G. Crynen, S. M. Meyer, M. D. Cameron, A. K. Chatterjee, A. Adibekian, J. L. Childs-Disney and M. D. Disney, Reprogramming of protein-targeted small-molecule medicines to RNA by ribonuclease recruitment, J. Am. Chem. Soc., 2021, 143, 13044–13055 CrossRef CAS PubMed.
- M. D. Shortridge, V. Vidalala and G. Varani, The kinase inhibitor palbociclib is a potent and specific RNA-binding molecule, bioRxiv, 2022, preprint, DOI:10.1101/2022.01.20.477126.
- M. D. Shortridge, B. Chaubey, H. J. Zhang, T. Pavelitz, V. Vidadala, C. Tang, G. L. Olsen, G. A. Calin and G. Varani, Drug-like small molecules that inhibit expression of the oncogenic microRNA-21, ACS Chem. Biol., 2023, 18, 237–250 CrossRef CAS PubMed.
- L. Borgelt, F. Li, P. Hommen, P. Lampe, J. Hwang, G. L. Goebel, S. Sievers and P. Wu, Trisubstituted pyrrolinones as small-molecule inhibitors disrupting the protein–RNA Interaction of LIN28 and Let-7, ACS Med. Chem. Lett., 2021, 12, 893–898 CrossRef CAS PubMed.
- P. Sin-Chan, I. Mumal, T. Suwal, B. Ho, X. Fan, I. Singh, Y. Du, M. Lu, N. Patel, J. Torchia, D. Popovski, M. Fouladi, P. Guilhamon, J. R. Hansford, S. Leary, L. M. Hoffman, J. M. Mulcahy Levy, A. Lassaletta, P. Solano-Paez, E. Rivas, A. Reddy, G. Y. Gillespie, N. Gupta, T. E. Van Meter, H. Nakamura, T. T. Wong, Y. S. Ra, S. K. Kim, L. Massimi, R. G. Grundy, J. Fangusaro, D. Johnston, J. Chan, L. Lafay-Cousin, E. I. Hwang, Y. Wang, D. Catchpoole, J. Michaud, B. Ellezam, R. Ramanujachar, H. Lindsay, M. D. Taylor, C. E. Hawkins, E. Bouffet, N. Jabado, S. K. Singh, C. L. Kleinman, D. Barsyte-Lovejoy, X. N. Li, P. B. Dirks, C. Y. Lin, S. C. Mack, J. N. Rich and A. Huang, A C19MC-LIN28A-MYCN Oncogenic Circuit Driven by Hijacked Super-enhancers Is a Distinct Therapeutic Vulnerability in ETMRs: A Lethal Brain Tumor, Cancer Cell, 2019, 36, 51–67 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Material and methods, RiPCA HTS protocol, other assay protocols, supplemental figures and tables. See DOI: https://doi.org/10.1039/d4md00123k |
| This journal is © The Royal Society of Chemistry 2024 |