Open Access Article
Open Access ArticleSynergy between the clavanins as a weapon against multidrug-resistant Enterobacter cloacae†
Marvin D.
Naing
 a,
Samuel A.
Juliano
a and
Alfredo M.
Angeles-Boza
a,
Samuel A.
Juliano
a and
Alfredo M.
Angeles-Boza
 *ab
*ab
aDepartment of Chemistry, University of Connecticut, Storrs 06269, USA. E-mail: alfredo.angeles-boza@uconn.edu
bInstitute of Materials Science, University of Connecticut, Storrs 06269, USA
First published on 16th May 2024
Abstract
Finding new antibiotics that can act synergistically with each other offers many benefits such as lower dosages used for each drug, improved pathogen clearance, and ability to act against multi-drug resistant strains. In this study, six peptides isolated from the tunicate Styela clava were evaluated for their synergistic interaction using the checkerboard assay and the time kill kinetics assay. Using two different tests, we report synergy between clavanin D and clavaspirin in both tests and synergy between clavanin A and B only in the checkerboard test when used against the multidrug resistant E. cloacae 0136. This work demonstrates the possible cooperativity between homologous AMPs from a single organism and the advantage of using two susceptibility tests instead of one when testing synergistic combinations.
As predicted by Alexander Fleming, we have misused antibiotics and we now find ourselves facing an increasing prevalence of antibiotic-resistant bacteria. This social dilemma, known as antibiotic resistance, is a global threat.1,2 To overcome this problem, we and many other groups are developing new antimicrobials and therapies that will eventually replace our current arsenal to better fight off superbugs in the future.3–6
The recent emergence of Enterobacter cloacae strains resistant to broad-spectrum antibiotics, including the last-resort carbapenems, has caused increased interest in finding therapies against this group of microorganisms. The E. cloacae complex contains common human pathogens that cause a wide variety of infections. Further, being a Gram-negative pathogen, the double membrane structure with the outer membrane containing lipopolysaccharide and lipid A makes it harder for many antimicrobial agents to penetrate.7
Antimicrobial peptides (AMPs) continue to be a promising avenue for antimicrobial drug discovery and development because of their broad-spectrum activity encompassing bacteria, fungi, viruses, and unicellular protozoa.8,9 Various organisms produce multiple AMPs with high homology as part of their innate immune defense; however, the benefits of just a few mutations in their sequence are unknown. The clavanin family of AMPs, found in the hemocytes of the tunicate Styela clava,10 consists of six peptides (Table 1). Five of them have ca. 80% sequence similarity whereas the sixth member only possess ∼30% sequence similarity to ClavA, the archetypical member of this family.11 Since their discovery, the clavanins, particularly ClavA, have been shown to possess potent antimicrobial activity against several Gram-positive and Gram-negative bacteria as well as some fungi.10,12–19 It is generally assumed that the other clavanins would follow the same mechanism of action (MoA) as ClavA but this doesn't rationalize why the tunicate would spend resources to produce multiple similar peptides with the same MoA. We hypothesize that one of the reasons for the production of these multiple homologues is a synergistic activity among the peptides produced. More importantly, we could take advantage of this synergistic behavior to develop therapies to combat nosocomial pathogens.
| Peptide | Abbr. | Sequence | MIC, μM (μg mL−1) | % helicity20 | |
|---|---|---|---|---|---|
| pH 5.5 | pH 7.4 | ||||
| Y′ = DOPA residue.14 | |||||
| Clavanin A | ClavA | VFQFL GKIIH HVGNF VHGFS HVF-NH2 | >128 (341) | >128 (341) | 50.1 |
| Clavanin B | ClavB | VFQFL GRIIH HVGNF VHGFS HVF-NH2 | 128 (344) | >128 (344) | 45.1 |
| Clavanin C | ClavC | VFHLL GKIIH HVGNF VY′GFS HVF-NH2 | 16 (46) | >128 (372) | 36.3 |
| Clavanin D | ClavD | AFKLL GRIIH HVGNF VY′GFS HVF-NH2 | 8 (23) | >128 (371) | 21.6 |
| Clavanin E | ClavE | LFKLL GKIIH HVGNF VHGFS HVF-NH2 | 8 (23) | >128 (368) | 49.0 |
| Clavaspirin | ClavS | FLRFI GSVIH GIGHL VHHIG VAL-NH2 | 4 (11) | 128 (348) | 39.8 |
| Magainin 2 | Mag2 | GIGKF LHSAK KFGKA FVGEI MNS-NH2 | 4 (4.9) | 4 (4.9) | — |
In this study, we identify synergistic pairs among the clavanins that are active against the Gram-negative ESKAPE pathogen, E. cloacae (CDC AR isolate bank #0136).21 We employed the checkerboard assay to spot the synergistic pairs that can combat E. cloacae under acidic conditions. The time-kill kinetics assay is then used to further confirm the synergy among the pairs. Having both tests agree with each other means that there is undeniable synergy among the pair.22
All members of the clavanin family were synthesized using solid-phase peptide synthesis, purified and characterized according to published protocols (Fig. S1–S6†). The sequences of the peptides are shown in Table 1. These peptides are cationic, α-helical (Fig. 1), amphipathic, and are rich in histidine and phenylalanine residues. Clavanin A and B are the most similar pair, with only a K to R mutation at position 7. Clavanin C and D are the only ones that contain a L-DOPA residue in place of Y15.14 Using a standard broth microdilution assay in MHB media, the minimum inhibitory concentration (MIC) of the clavanins against E. cloacae 0136, were determined at two different pH values: 5.5 and 7.4. This bacterial strain is resistant to a number of antibiotic classes such as aminoglycosides, β-lactams, sulfonamides, and trimethoprim.21Table 1 summarizes the results with Magainin2, a pore-forming cationic AMP, used as a control.13 We tested all the peptides at a maximum concentration of 128 μM because higher concentrations lead to precipitation of the peptides when mixed in MHB media. To our knowledge, this is the first report on the activity of the clavanins against E. cloacae.
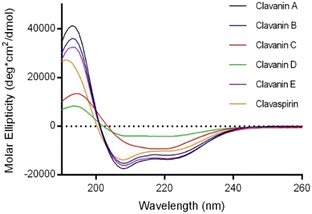 | ||
| Fig. 1 CD spectroscopy data for all the clavanins taken at 50 μM concentration in 50% TFE solution showing the typical α-helix signals. | ||
From the above data, the clavanins have higher activity at pH 5.5 than at pH 7.4, confirming previous reports on ClavA and ClavS.11,13 This is attributed to the multiple histidine residues in their sequence that get protonated at lower pH, making it more cationic and thus increasing the affinity towards the negatively charged microbial membrane. In S. clava, the vacuoles of phagocytic hemocytes, where the clavanins are found, undergo acidification as part of the process to eliminate the phagocytosed organisms.12 Recent reports suggest that E. cloacae can survive inside the acidic lysosomal environment of macrophages after disrupting normal phagocytic trafficking.23 Succeeding experiments were performed at pH 5.5 based on these results.
We used two methodologies to determine synergy among clavanins. The first method is the checkerboard assay which involves the calculation of the fractional inhibitory concentration (FIC, ESI† eqn (S2)) from the observed MICs alone and in combination.24 One downside of this method is that it is not capable of testing more than two drugs at one time and it only measures one endpoint with either a growth or no growth reading within the experimental time frame.25 The way the FIC is calculated is similar to the Loewe additivity model (ESI† eqn (S3)) of synergy and the isobologram analysis, both of which operate under the assumption that a compound cannot interact with itself and two different compounds that have the same effect are equivalent or additive.26,27 The second method is the time-kill kinetics assay which measures changes in bacterial titer over time.25 Both methods are based on the Clinical and Laboratory Standards Institute (CLSI) guidelines.25,28 To offer a more pragmatic way of testing, the concentrations that had the lowest FIC from the checkerboard assay was the one used in the time-kill assay. This assay also has a different criterion for pronouncing synergy. A deviation of ≥2 log decrease in CFU mL−1 is considered synergistic.22 This is similar to what the Bliss independence model implies (ESI† eqn (S5)). There is strong synergy if the combination is synergistic in both tests and a weak synergy if only synergistic in one test. Having two methodologies that operate under two different synergy models would allow us to gauge the strength of the synergy observed between the clavanins as well as attain a more consistent synergy result.
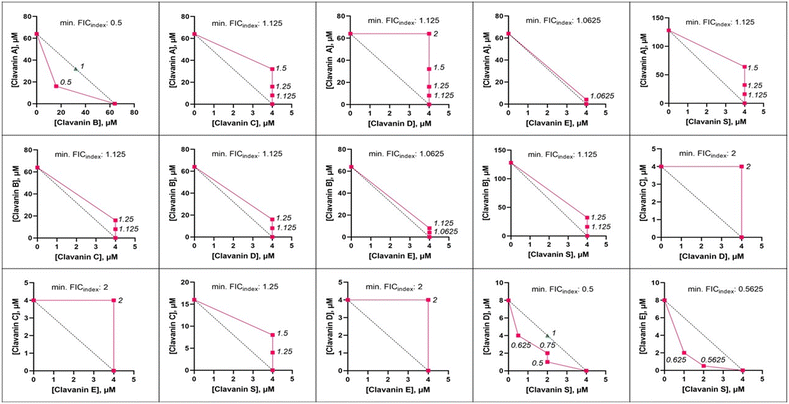
Despite the similarities in the amino acid sequence and activity of the clavanins, there is marked difference in terms of their pair-wise activity (Fig. 2, ESI† Table S1). The results from the checkerboard assay and the isobologram analysis show that only two pairs are synergistic against E. cloacae 0136, namely, ClavA + ClavB and ClavD + ClavS. One pair, ClavE + ClavS, has an additive interaction. The rest of the combinations have no effect or are indifferent from each other. No pair resulted in an antagonistic interaction.
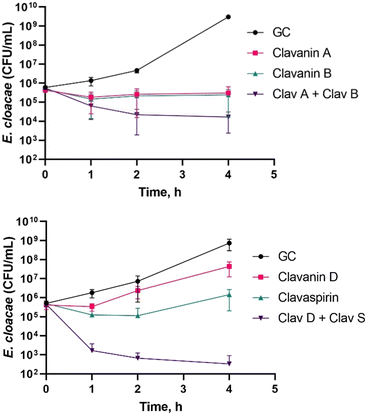
We then performed the time-kill kinetics assay on the two synergistic pairs reported above. The combination of ClavD and ClavS is synergistic (Fig. 3) based on the criteria mentioned above. The ClavA and B pair, which shows a bacteriostatic response, is not deemed synergistic using this assay since the difference between the response of the most active component and the actual response of the combination is only about 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log decrease in CFU. Nonetheless, the deviation implies that the two peptides are interacting albeit a weaker response. The high sequence similarity for ClavA and B, which differ in only one amino acid, could be the reason for this weak synergy. Both amino acids, K and R, are basic and would carry a positive charge under these conditions. This may be the reason why they behave similarly at the concentration tested, both individually and in combination. The difference between ClavA and ClavB could lie in their ability to penetrate the membrane because Arg residues are known to insert themselves in lipid membranes better than Lys residues resulting to an increased cellular penetration.29
log decrease in CFU. Nonetheless, the deviation implies that the two peptides are interacting albeit a weaker response. The high sequence similarity for ClavA and B, which differ in only one amino acid, could be the reason for this weak synergy. Both amino acids, K and R, are basic and would carry a positive charge under these conditions. This may be the reason why they behave similarly at the concentration tested, both individually and in combination. The difference between ClavA and ClavB could lie in their ability to penetrate the membrane because Arg residues are known to insert themselves in lipid membranes better than Lys residues resulting to an increased cellular penetration.29
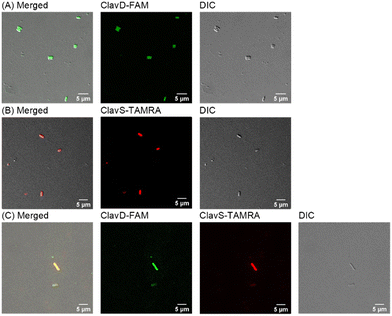
The results of the checkerboard assay and the time-kill assay confirmed synergy between ClavD and ClavS warranting further mechanistic investigation. We performed confocal microscopy studies on ClavD and ClavS to get insight on their behavior and interaction in the presence of cells. Both ClavD and ClavS were tagged with either 5(6)-carboxytetramethylrhodamine (TAMRA) or 5(6)-carboxyfluorescein (FAM) by substituting a phenylalanine residue (F2 and F4 in ClavD and ClavS, respectively) with a lysine protected by a 4-methyltrityl (Mtt) group as shown in Scheme S1.† The replacement of a hydrophobic residue with a hydrophobic dye would have the least impact on the activity of the peptides, an approach we have followed in the past.30,31 After tagging the peptides, their MICs were assessed to make sure that the dye did not affect their antimicrobial activity. The susceptibility assay showed that the MICs of the tagged peptides were the same as those of the untagged peptides. A checkerboard assay confirmed that the ClavD-FAM and ClavS-TAMRA are synergistic and this verifies a previous report stating that common fluorescent probes have minimal to no effect on the antimicrobial activity of AMPs.32 When E. cloacae cells were incubated for 30 min with either ClavD-FAM and ClavS-TAMRA, we observed full translocation of the peptides, suggesting that there is an internal target for the peptides (Fig. 4A and B). In the combined treatment (Fig. 4C), the majority of the bacterial cells have both peptides inside the cells whereas only a small number have ClavS-TAMRA localized on the cell membrane. This is in contrast to the cells treated with ClavS-TAMRA only (vide supra). In all cases, there was no evidence of membrane damage.
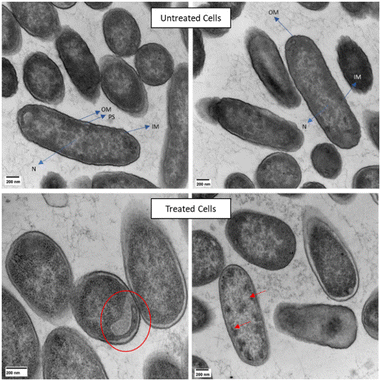
Lack of membrane damage was also observed in transmission electron microscopy (TEM) experiments done on E. cloacae 0136 treated with ClavD + ClavS (Fig. 5). Cells treated with the ClavD + ClavS combination show internal damage while keeping the membrane intact (no blisters or protruding bubbles/blebs). Regions of low electron density on some of the cells can be observed which accounts for intracellular damage similar to what was reported for rhizobium strains treated with polymyxin B.33 Some cells contained black spots (very electron-dense areas) which can be evidence for aggregated intracellular components as a result of the exposure to both peptides. This is also similar to what was observed with colistin-treated E. coli.34
Our evidence indicates that the synergistic pair does not act primarily on membrane disruption but on an intracellular target. To our knowledge, this is the first reported synergistic interaction between two AMPs of this type. There are several examples in the literature on synergy between AMPs but there is no universal agreement on the molecular characteristics of peptides that lead to synergy.35 For example, abaecin, an AMP that is suspected of targeting the molecular chaperone DnaK, synergizes with hymenoptaecin, a pore-forming AMP.9 In this case, hymenoptaecin opens up pores in the bacterial membrane which facilitate easier entry for abaecin and provide easy access to internal targets. Similarly, the less active AMPs from the winter flounder, such as WF3, are able to control the membrane interactions of the more active winter flounder AMPs, such as pleurocidin, resulting to a potentiated activity at much lower concentrations.36 Combinations that don't have this type of cooperativity are also possible. For example, magainin and PGLa demonstrated synergy by forming defined supramolecular structures along the membrane surface that leads to increased permeabilization and eventually, cell death.37,38
In summary, the criteria used in this study utilizing both checkerboard and time-kill kinetics assays for two different synergy models, identified two new synergistic AMP pairs that are active against the MDR strain E. cloacae 0136. This indicates cooperativity between AMPs produced by a single organism. ClavA and ClavB, together, are bacteriostatic against E. cloacae. Against this problematic pathogen, the combination of ClavD and ClavS is bactericidal and provides a promising avenue for the development of novel therapies, although we still need to identify the specific target(s) of such a synergistic pair. Confocal microscopy and TEM results suggest that both ClavD and ClavS can translocate inside the cells without damaging the membrane (ESI† video). This suggests that the MOA is intracellular and that these peptides can interfere with processes inside the bacterial cell. Future studies will focus on establishing the targets of this synergistic pair.
Author contributions
S. A. J.: conceptualization, investigation, and data curation. M. D. N.: conceptualization, data curation, investigation, writing – review and editing. A. M. A.-B., conceptualization, funding acquisition, supervision, writing – review and editing. This work was supported by the National Science Foundation through MCB1715494 to A. M. A.-B.Conflicts of interest
There are no conflicts to declare.Notes and references
- S. S. Kadri, Crit. Care Med., 2020, 48, 939 CrossRef PubMed.
- Centers for Disease Control and Prevention (U.S.), Antibiotic resistance threats in the United States, 2019, Centers for Disease Control and Prevention (U.S.), 2019 Search PubMed.
- J. Portelinha and A. M. Angeles-Boza, ChemBioChem, 2021, 22, 1646–1655 CrossRef CAS PubMed.
- J. Portelinha, S. S. Duay, S. I. Yu, K. Heilemann, M. D. J. Libardo, S. A. Juliano, J. L. Klassen and A. M. Angeles-Boza, Chem. Rev., 2021, 121, 2648–2712 CrossRef CAS PubMed.
- T. Misawa, C. Goto, N. Shibata, M. Hirano, Y. Kikuchi, M. Naito and Y. Demizu, MedChemComm, 2019, 10, 896–900 RSC.
- D. Ben Hur, G. Kapach, N. A. Wani, E. Kiper, M. Ashkenazi, G. Smollan, N. Keller, O. Efrati and Y. Shai, J. Med. Chem., 2022, 65, 9050–9062 CrossRef CAS PubMed.
- R. Jasim, M.-L. Han, Y. Zhu, X. Hu, M. Hussein, Y.-W. Lin, Q. Zhou, C. Dong, J. Li and T. Velkov, Int. J. Mol. Sci., 2018, 19, 2356 CrossRef PubMed.
- M. Mahlapuu, J. Håkansson, L. Ringstad and C. Björn, Front. Cell. Infect. Microbiol., 2016, 6, 194 Search PubMed.
- L. Duong, S. P. Gross and A. Siryaporn, Front. Med. Technol., 2021, 3, 640981 CrossRef PubMed.
- C. Zhao, L. Liaw, I. Hee Lee and R. I. Lehrer, FEBS Lett., 1997, 410, 490–492 CrossRef CAS PubMed.
- I.-H. In, C. Zhao, T. Nguyen, L. Menzel, A. J. Waring, R. I. Lehrer and M. A. Sherman, J. Pept. Res., 2001, 58, 445–456 CrossRef.
- S. A. Juliano, S. Pierce, J. A. deMayo, M. J. Balunas and A. M. Angeles-Boza, Biochemistry, 2017, 56, 1403–1414 CrossRef CAS PubMed.
- I. H. Lee, Y. Cho and R. I. Lehrer, Infect. Immun., 1997, 65, 2898–2903 CrossRef CAS PubMed.
- R. I. Lehrer, Integr. Comp. Biol., 2003, 43, 313–322 CrossRef CAS PubMed.
- E. J. M. van Kan, R. A. Demel, A. van der Bent and B. de Kruijff, Biochim. Biophys. Acta, Biomembr., 2003, 1615, 84–92 CrossRef CAS PubMed.
- E. J. M. van Kan, R. A. Demel, E. Breukink, A. van der Bent and B. de Kruijff, Biochemistry, 2002, 41, 7529–7539 CrossRef CAS PubMed.
- E. J. M. van Kan, D. N. Ganchev, M. M. E. Snel, V. Chupin, A. van der Bent and B. de Kruijff, Biochemistry, 2003, 42, 11366–11372 CrossRef CAS PubMed.
- S. A. Juliano, L. F. Serafim, S. S. Duay, M. Heredia Chavez, G. Sharma, M. Rooney, F. Comert, S. Pierce, A. Radulescu, M. L. Cotten, M. Mihailescu, E. R. May, A. I. Greenwood, R. Prabhakar and A. M. Angeles-Boza, ACS Infect. Dis., 2020, 6, 1250–1263 CrossRef CAS PubMed.
- O. N. Silva, E. S. F. Alves, C. de la Fuente-Núñez, S. M. Ribeiro, S. M. Mandal, D. Gaspar, A. S. Veiga, M. A. R. B. Castanho, C. A. S. Andrade, J. M. Nascimento, I. C. M. Fensterseifer, W. F. Porto, J. R. Correa, R. E. W. Hancock, S. Korpole, A. L. Oliveira, L. M. Liao and O. L. Franco, Sci. Rep., 2016, 6, 27128 CrossRef CAS PubMed.
- M. M. Juban, M. M. Javadpour and M. D. Barkley, in Antibacterial Peptide Protocols, Humana Press, New Jersey, 1997, vol. 78, pp. 73–78 Search PubMed.
- J. D. Lutgring, M.-J. Machado, F. H. Benahmed, P. Conville, R. M. Shawar, J. Patel and A. C. Brown, J. Clin. Microbiol., 2018, 56, e01415–e01417 Search PubMed.
- J. Tang, K. Wennerberg and T. Aittokallio, Front. Pharmacol., 2015, 6, 181 Search PubMed.
- G. Parau, F. Bisaro, H. Marshall and M. A. Valvano, Access Microbiol., 2019, 1, 227 Search PubMed.
- C. D. Doern, J. Clin. Microbiol., 2014, 52, 4124–4128 CrossRef PubMed.
- Antimicrobial Susceptibility Testing Protocols, ed. R. Schwalbe, L. Steele-Moore and A. C. Goodwin, CRC Press, Boca Raton, 2007 Search PubMed.
- S. Lederer, T. M. H. Dijkstra and T. Heskes, Front. Pharmacol., 2018, 9, 31 CrossRef PubMed.
- R. Huang, L. Pei, Q. Liu, S. Chen, H. Dou, G. Shu, Z. Yuan, J. Lin, G. Peng, W. Zhang and H. Fu, Front. Pharmacol., 2019 DOI:10.3389/fphar.2019.01222.
- M07: Dilution AST for Aerobically Grown Bacteria - CLSI, https://clsi.org/standards/products/microbiology/documents/m07/, (accessed February 20, 2024).
- L. Li, I. Vorobyov and T. W. Allen, J. Phys. Chem. B, 2013, 117, 11906–11920 CrossRef CAS PubMed.
- M. D. J. Libardo, C. de la Fuente-Nuñez, K. Anand, G. Krishnamoorthy, P. Kaiser, S. C. Pringle, C. Dietz, S. Pierce, M. B. Smith, A. Barczak, S. H. E. Kaufmann, A. Singh and A. M. Angeles-Boza, ACS Infect. Dis., 2018, 4, 1623–1634 CrossRef CAS PubMed.
- M. D. J. Libardo, V. Y. Gorbatyuk and A. M. Angeles-Boza, ACS Infect. Dis., 2016, 2, 71–81 CrossRef CAS PubMed.
- C. Zhao, A. Fernandez, N. Avlonitis, G. Vande Velde, M. Bradley, N. D. Read and M. Vendrell, ACS Comb. Sci., 2016, 18, 689–696 CrossRef CAS PubMed.
- I. Komaniecka, K. Zamłyńska, R. Zan, M. Staszczak, J. Pawelec, I. Seta and A. Choma, Acta Biochim. Pol., 2016, 63, 517–525 CrossRef CAS PubMed.
- N. Hazime, Y. Belguesmia, I. Kempf, A. Barras, D. Drider and R. Boukherroub, Pharmaceuticals, 2022, 15, 682 CrossRef CAS PubMed.
- J. He, C. G. Starr and W. C. Wimley, Biochim. Biophys. Acta, Biomembr., 2015, 1848, 8–15 CrossRef CAS PubMed.
- M. Clarke, C. K. Hind, P. M. Ferguson, G. Manzo, B. Mistry, B. Yue, J. Romanopulos, M. Clifford, T. T. Bui, A. F. Drake, C. D. Lorenz, J. M. Sutton and A. J. Mason, npj Antimicrob. Resist., 2023, 1, 1–16 CrossRef.
- E. Han and H. Lee, RSC Adv., 2015, 5, 2047–2055 RSC.
- C. Aisenbrey, M. Amaro, P. Pospíšil, M. Hof and B. Bechinger, Sci. Rep., 2020, 10, 11652 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4md00070f |
| This journal is © The Royal Society of Chemistry 2024 |