Open Access Article
Open Access ArticleSynthesis, characterization, and application of a biocompatible gene delivery nanocarrier constructed from gold nanostars and a chitosan–cyclodextrin–poly(ethylene imine) graft polymer†
Farbod
Tabesh
ab,
Golnaz
Haghverdi
ab,
Kireeti Phani
Devarakonda
 ab,
Tarik F
Massoud
ab,
Tarik F
Massoud
 ab and
Ramasamy
Paulmurugan
ab and
Ramasamy
Paulmurugan
 *ab
*ab
aMolecular Imaging Program at Stanford (MIPS), Department of Radiology, Stanford University School of Medicine, Stanford, CA, USA. E-mail: paulmur8@stanford.edu
bCanary Center for Cancer Early Detection, Department of Radiology, Stanford University School of Medicine, Palo Alto, CA, USA
First published on 10th July 2024
Abstract
Gene therapy can be an efficient method to treat genetic diseases, including cancer. However, the lack of biocompatible gene delivery systems with minimal toxicity limits the clinical success of this approach. We report the synthesis of an effective gene carrier using gold nanostars (AuNSs) in combination with chitosan (CS), β-cyclodextrin (CD), and branched-poly(ethylene imine) (bPEI) as a bionanocomposite (BNC) with minimal toxicity for gene delivery applications. We synthesized and characterized the AuNS@CS–CD–bPEI (AuNS@CCP) BNC using various analytical methods. Ultraviolet-visible spectroscopy confirmed the successful synthesis of AuNSs from Au nanoseeds. Nuclear magnetic resonance proved the conjugation of CS and CD upon its multistep synthesis. Transmission electron microscopy images confirmed the median size of AuNSs as 53 nm, which increased to 70 nm for the AuNS@CCP BNC. Energy dispersive X-ray (EDX) analysis further confirmed the presence of a polymeric layer of CS–CD–bPEI on AuNSs. We also measured the zeta potential of AuNS@CCP using dynamic light scattering to check its ability to bind to nucleic acids, which was +28.2 mV. The applicability of the BNC in nucleic acid transfection was evaluated in various mammalian cell lines (HEK293T, LN308, MDA-MB-231, and CT-26) using firefly luciferase-zetagreen (FLuc-ZsGreen) reporter genes by optical imaging, and synthetic mRNA coding for SARS-CoV-2 spike protein by immunoblot analysis to prove the ability of the BNC to transfect both DNA and RNA. We also characterized the N/P ratio of the BNC with the nucleic acids to optimize cell transfection efficiency. The cytotoxicity study (MTT assay) of the BNC showed no significant toxicity when used at the optimal N/P ratio. Overall, we show that the synthesized BNC has a high potential for use as a gene delivery agent for in vivo applications, potentially using various delivery routes such as intranasal, intramuscular, intrathecal, and intraperitoneal in treating genetic diseases and for vaccines in infectious diseases.
Introduction
Gene delivery is defined as the transfer of genes into cells to achieve gene expression to accomplish a cure against genetic disorders.1 Viral-mediated gene transduction and nonviral gene transfection using various carriers are well-established delivery methods in gene therapy applications.2 Viral-mediated gene transduction is highly efficient; however, safety issues, including toxicity, viral-induced immune responses, and the limited cargo capacity, are all downsides of this method.1,3 On the other hand, the nonviral gene delivery methods have some advantages, including lower immunogenicity and tumorgenicity, simplicity, biocompatibility, high loading efficacy, and repeated deliveries to achieve sustained gene expression in vivo.2–5In many nonviral gene delivery methods, the nucleic acid could simply be complexed with the carrier for transfection. Carriers such as nanostructures, liposomes, cationic lipids, and polymers, to name a few, have been reported for nonviral gene delivery methods.6–12 Gold nanoparticles (AuNPs) are one of the widely used nanostructures for nonviral gene delivery in biomedical research.13 Gold (Au) nanostructures are nontoxic and highly biocompatible compared to other nanomaterials;14 they also benefit from their ease of synthesis, optical properties, and surface plasmon resonance properties, which allows for tracking nanostructure delivery and biodistribution in vivo using optical and CT imaging.15 Accordingly, Kim et al.16 prepared Au nanoparticles (NPs) conjugated with a DNA aptamer to detect matrix metalloproteinase-9 (MMP-9) activity in the tumor microenvironment. They also showed that the as-prepared AuNPs did not show cytotoxicity but a strong binding affinity to MMP-9 in vitro and in vivo. Paul et al.17 used AuNPs to enhance the DNA adsorption rate onto graphene oxide. They found that adding AuNPs increases the DNA adsorption rate on graphene oxide by 100-fold. Xu et al.18 produced AuNPs and conjugated them with a peptide sequence to improve DNA binding capacity for gene delivery. Functionalized AuNPs were synthesized in situ by reducing AuHCl4 with NaBH4 in the presence of the peptide. Their DNA release results showed that the complex had low toxicity and high delivery efficiency. However, the functionalized AuNPs had a strong binding affinity to plasmid DNA, thus affecting the DNA release capacity for gene expression. Therefore, despite the advantages of AuNPs, their transfection efficiency is poor. It is also reported that nanocomposites containing AuNPs can show higher transfection efficiency than AuNPs.19,20
Therapeutic genes can be delivered to target tissues using various routes, such as intravascular, intravenous, intraarterial, intraperitoneal, intramuscular, intrathecal, and intranasal.21–27 While each technique has merits and demerits, e.g., intranasal delivery is more efficient for respiratory diseases, including vaccine delivery in infectious diseases and suicide gene therapy in lung cancer, since it targets both upper and lower respiratory tissues.28,29 However, AuNPs are effective in DNA delivery, and it is essential to use polymers or polymeric nanocomposites with mucoadhesive properties to achieve high transfection efficiency upon intranasal delivery. In addition, mucoadhesive polymers induce mucosal immunity when transfection is used for vaccine applications in respiratory diseases.30 An optimal method would be to use natural polymers such as polysaccharides owing to their biocompatibility, nontoxicity, cost-effectiveness, availability, high nucleic acid-carrying capacity, and mucoadhesivity.31–33
Since nucleic acids are negatively charged, the chosen carrier must have a positive surface charge to bind to nucleic acids.1,4,34 Therefore, cationic polymers are more suitable for nucleic acid delivery applications. Chitosan (CS) is one of the widely used cationic polymers to make gene carriers.3,35,36 CS is a hydrolysis product of chitin (Ch) that contains D-glucosamine and N-acetyl-D-glucosamine; therefore, hydrolysis of chitin results in deacetylated D-glucosamine.37 The higher the degree of deacetylation, the more the –NH2 groups are present, resulting in a more positive surface charge. The positive surface charge of the polymer (from –NH3+) can bind to negatively charged DNA.38 In addition to the cationic properties of the natural polymers, chitosan has the benefit of biocompatibility and favorable mucoadhesivity resulting from the hydration of chitosan chains with mucus,39–42 which facilitates the use of chitosan for lung-specific gene delivery applications. Despite these advantages, CS has poor transfection efficiency, specifically for primary cells. To overcome this problem, we have established a method to conjugate CS with a highly efficient cationic poly(ethylene imine) (PEI). Branched-PEI (bPEI) is an amorphous cationic polymer with an amine backbone synthesized by a ring-opening reaction of aziridine and has had a wide range of applications in gene delivery.43 PEI has become a gold-standard transfection agent in recent years.44 However, excess PEI can cause cell toxicity by inducing apoptotic or necrotic cell death.45,46 We hypothesize that the conjugation of CS and bPEI may improve the transfection efficiency of CS while avoiding the toxicity associated with free bPEI and achieving desirable mucoadhesivity and biocompatibility for intranasal delivery applications.
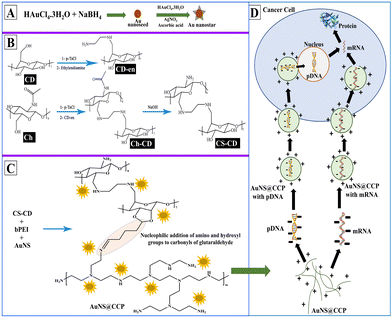
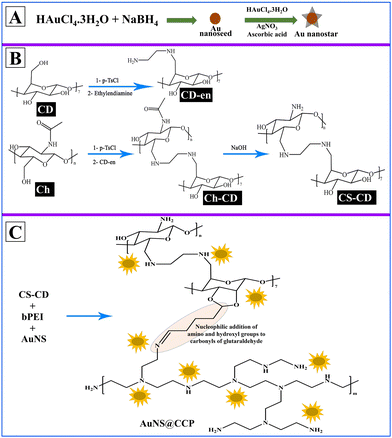
We aimed to prepare a biocompatible gene delivery system with high transfection efficiency for DNA and RNA, which could be used with different delivery methods. We also conjugated CS with β-cyclodextrin (β-CD) to facilitate the addition of targeting peptides/proteins/aptamers for targeted delivery applications in vivo. Since β-CD is a cyclic heptasaccharide, it provides a ring suitable for interacting with small cyclic compounds such as adamantane (host–guest chemistry). Thus, we included β-CD in the formulation to create more options for making the bionanocomposite (BNC) selective for targeted deliveries.47 We prepared the AuNS@CS–CD–bPEI (CCP) BNC using a multistep synthesis process (Scheme 1A–C). Using various characterization methods, we confirmed the successful synthesis of BNCs with solid binding properties to DNA and RNA (Scheme 1D). By transfection in various cells, we also confirmed the ability of AuNS@CCP to deliver the loaded nucleic acids inside cells for functional expression of delivered genes with no nonspecific toxicity. The novelties of this study are that the developed BNC can be used in various gene delivery applications selectively to the target cells by reacting the CD portion of the BNC with adamantane-functionalized target ligands (host–guest chemistry) with high efficiency. In addition, this BNC is biocompatible with no apparent toxicity. It is worth mentioning that it can be used to deliver nucleic acids through various delivery methods such as intranasal, intravenous, intramuscular, subcutaneous, etc. Moreover, because of the greater surface-enhanced Raman scattering properties of Au nanostars (AuNSs) over AuNPs, one can use this feature to image AuNSs upon delivery in vivo to the target tissues using various imaging modalities such as CT, photoacoustic, and near-infrared imaging.48,49
Results and discussion
Synthesis and characterization of AuNSs
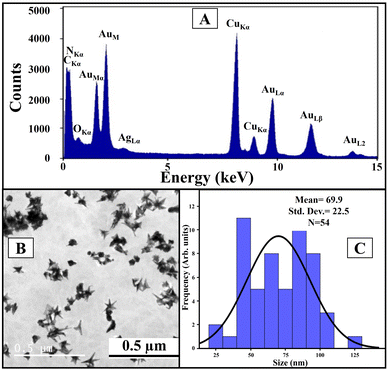
We used Au nanoseed-assisted synthesis of AuNSs (Scheme 1A). We acquired the ultraviolet-visible (UV-vis) spectra of Au nanoseeds and AuNSs to confirm the synthesis. The Au nanoseed spectrum showed a descending trend beyond approximately 500 nm, while the AuNS peak at approximately 630 nm indicated the star structure (Fig. 1A).50–52 This absorbance is also derived from the localized surface plasmon resonance of the AuNSs.53 We then conducted the elemental analysis of AuNSs using energy dispersive X-ray (EDX) spectroscopy (Fig. 1B) to find the presence of substantial elements. We observed representative peaks of Au and Ag from AuNSs and C and O from the surfactant. The morphology and size of AuNSs were determined using transmission electron microscopy (TEM), as depicted in Fig. 1C. As expected, a star structure was observed for AuNSs with a uniform shape and distribution. Based on the size histogram (Fig. 1D) obtained from the TEM image, AuNSs had a mean size of 53 nm, while their hydrodynamic diameter measured with the DLS device was ∼155 nm. This could be due to the interaction between water and the surfactant layer around AuNSs (which can be seen as a pale layer around the spikes in Fig. S1, ESI†) that absorbs water. Therefore, it causes aggregation owing to hydrogen bonding and attraction forces between hydrophilic functional groups in the surfactant and water. The hydrodynamic size distribution is provided in Fig. S2 (ESI†).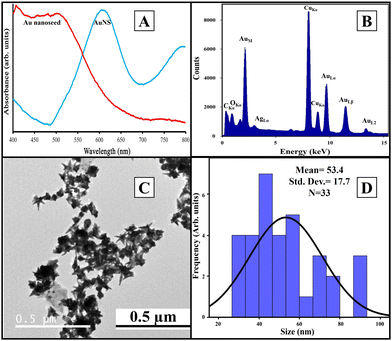 | ||
| Fig. 1 UV-vis spectra of Au nanoseeds and AuNSs (A), EDX spectrum of AuNSs (B), TEM image of AuNSs (C), and physical size histogram of AuNSs obtained from the TEM image (D). | ||
Synthesis and characterization of CS–CD
We synthesized CS–CD using a multistep process, as shown in Scheme 1B, which we adopted from a previous study.54 We confirmed each step of the CS–CD synthesis process by proton nuclear magnetic resonance (1H NMR) spectroscopy (Fig. 2). We started the synthesis by adding ethylenediamine (en) to β-CD, which showed the appearance of new peaks at around δ = 1.3, 2.3, and 3.2 ppm (–CH2, –NH2, and –NH, respectively) confirming the addition of en to β-CD. Meanwhile, the tosylation of Ch was confirmed by the existence of the peaks of aromatic hydrogens of the tosyl group at δ = 7.4–7.7 ppm. The disappearance of the tosyl (δ = 7.4–7.7 ppm) confirmed the attachment of β-CD to Ch. We then hydrolyzed Ch–CD to CS–CD, which was subsequently confirmed by vanishing of the Ch amide peak (δ = 8 ppm) and the appearance of deacylated nitrogen of CS (δ = 1.8 ppm). We also recorded the 1H NMR spectra of bPEI and AuNS@CCP (Fig. S3, ESI†). Hydrogens of methylene and various amino groups of bPEI appeared at δ ∼ 2.5 and 5–6.5 ppm. Due to the presence of numerous amino groups, interactions among them make their spectrum complicated (Fig. S3A, ESI†). On the other hand, for AuNS@CCP, the important peaks of CS and bPEI overlapped, and bPEI peaks appeared predominantly owing to their higher ratio in the polymeric structure (Fig. S3B, ESI†). The resemblance of these two spectra confirmed the presence of bPEI in the polymeric matrix.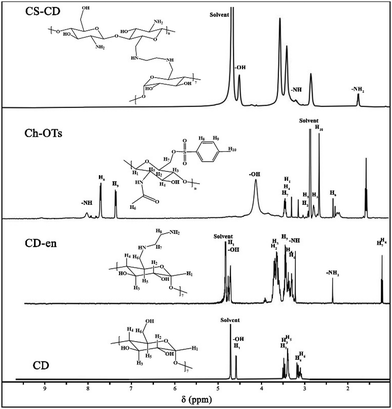 | ||
| Fig. 2 1H NMR spectra of CD, CD–en, Ch–OTs (tosylated chitin), and CS–CD. 5–8 mg of each sample was dissolved in 2% v/v DCl in D2O to acquire TEM spectra. | ||
Synthesis and characterization of AuNS@CCP
After incorporating AuNSs into CS–CD–bPEI graft polymer (AuNS@CCP) (Scheme 1C), we performed EDX spectroscopy to confirm the coexistence of AuNSs and the graft polymer (Fig. 3A). The AuNS@CCP spectrum showed peaks of Au, Ag, C, N, and O with different energy levels. Also, the intensities of C and N peaks were significantly increased in the EDX spectrum of AuNS@CCP (Fig. 3A), confirming the co-existence of AuNSs and the polymeric matrix. We acquired the TEM image of AuNS@CCP (Fig. 3B) and its size histogram (Fig. 3C). The TEM image showed AuNSs with a median size of 70 nm in the polymeric matrix with a reasonable and uniform dispersion and distribution. The size of AuNSs increased from 53 to 70 nm in the polymeric matrix (AuNS@CCP), indicating the functional coverage and interaction of AuNSs with the graft polymer. A very pale layer around the AuNSs can be seen in higher magnification TEM images in Fig. S4 (ESI†). Since the polymeric matrix is hydrophilic, the hydrophilic functional groups tend to absorb water. Therefore, the hydrodynamic diameter of AuNS@CCP should be larger than its particle diameter. Its hydrodynamic diameter was 668 nm, measured with the DLS device. The hydrodynamic size distribution is provided in Fig. S5 (ESI†). Based on the TEM images, we concluded that the chemical and physical conditions of the synthesized particles were intact while also confirming that the high pressure of 30k psi in the microfluidic system helped to achieve uniformly distributed particles without affecting the morphologies of the AuNSs and AuNS@CCP.Gel retardation assay (GRA) for AuNS@CS–CD, AuNS@bPEI, AuNS@CCP, and CCP
A component can bind with nucleic acid with a positive surface charge. Thus, we studied the surface charge of different components to investigate their potential DNA binding. Zeta potentials for CD, CS–CD, and AuNS@CCP were 6.74 ± 0.67, 19.0 ± 7.26, and 28.2 ± 2.35 mV, respectively. Next, we used a GRA to confirm the DNA binding ability of AuNS@CS–CD, AuNS@bPEI, AuNS@CCP, and CCP. As shown in Fig. 4A, we observed minimal binding for all the conditions used for AuNS@CS–CD, while AuNS@bPEI showed a significant level of DNA binding under all the conditions used for the study. We observed similar results for AuNS@CCP and CCP, indicating that adding AuNSs to the system did not affect the DNA binding efficiency of CCP. We measured the zeta potential of the AuNS@CCP/pcDNA-FLuc-ZsGreen plasmid at different polymer/plasmid DNA (N/P) ratios (Fig. 4B) and found that at an N/P ratio of 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, the zeta potential was 28.8 ± 4.0 mV, and at 14
1, the zeta potential was 28.8 ± 4.0 mV, and at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, it was 7.6 ± 4.3 mV.
1, it was 7.6 ± 4.3 mV.
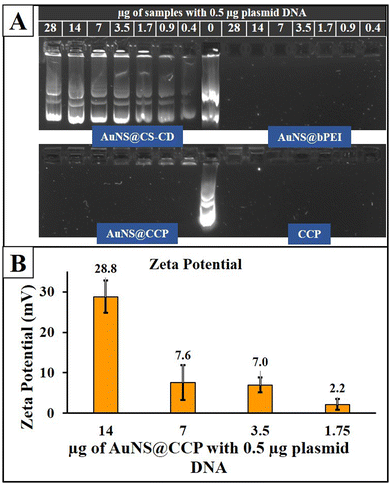 | ||
| Fig. 4 GRA results for AuNS@CS–CD, AuNS@bPEI, AuNS@CCP, and CCP (A) and zeta potential outcomes of the AuNS@CCP/plasmid DNA polyplex at different N/P ratios (B). | ||
Transfection and cytotoxicity evaluation of different BNCs in mammalian cells
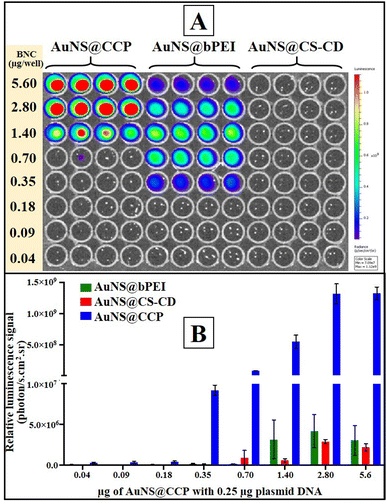
After characterizing BNCs for their physicochemical properties, we assessed the transfection efficiency of different BNCs in mammalian cells using the pcDNA-FLuc-ZsGreen reporter plasmid. We first tested the transfection efficiency of CS, CS–CD, and AuNS@CCP in HEK293T cells to observe the influence of each component on the other one, and the results are shown in Fig. S6 (ESI†). We did not observe acceptable transfection from CS, as expected. In addition, CS–CD did not show a much stronger transfection profile than CS. On the other hand, as expected, AuNS@CCP showed higher transfection efficiency than CS and CS–CD, which could be due to the availability of abundant –NH2 groups of bPEI for binding with DNA. We also investigated the transfection efficiency of AuNS@CS–CD, AuNS@bPEI, and AuNS@CCP. We observed a very low level of transfection when we used AuNS@CS–CD, which agreed with the DNA binding ability results (Fig. 5A and B). In contrast, AuNS@bPEI and AuNS@CCP showed strong luciferase signals as measured using an IVIS optical imaging system. Notably, both AuNS@bPEI and AuNS@CCP had their highest transfection efficiencies at an N/P ratio of 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The transfection efficiency of AuNS@CCP is ∼2500-fold higher than that of AuNS@bPEI (Fig. 5B).
1. The transfection efficiency of AuNS@CCP is ∼2500-fold higher than that of AuNS@bPEI (Fig. 5B).
We also studied the transfection efficiency of the FLuc-ZsGreen plasmid in two vector backbones (pcDNA and pVAX1) in HEK293T cells using AuNS@CCP. As depicted in Fig. 6A, the transfection efficiency increased after an increase in the amount of AuNS@CCP, but after reaching its maximum levels, the transfection efficiency began to decrease. The maximum level of transfection was observed with 3.5 μg AuNS@CCP for the pcDNA-FLuc-ZsGreen plasmid (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 N/P). In comparison, it was 7.0 μg AuNS@CCP for pVAX1-FLuc-ZsGreen (28
1 N/P). In comparison, it was 7.0 μg AuNS@CCP for pVAX1-FLuc-ZsGreen (28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 N/P). We also measured the cytotoxicity of AuNS@CCP using an MTT assay in HEK293T cells by transfecting pcDNA-FLuc-ZsGreen and pVAX1-FLuc-ZsGreen plasmids. According to ISO 10993-5, a viability lower than 70% is considered cytotoxic, and based on the cytotoxicity results (Fig. 6B), no toxicity was observed for both samples, confirming AuNS@CCP as a biocompatible gene carrier. These results indicated that AuNS@CCP is not toxic to cells at its highest transfection efficiency dose.
1 N/P). We also measured the cytotoxicity of AuNS@CCP using an MTT assay in HEK293T cells by transfecting pcDNA-FLuc-ZsGreen and pVAX1-FLuc-ZsGreen plasmids. According to ISO 10993-5, a viability lower than 70% is considered cytotoxic, and based on the cytotoxicity results (Fig. 6B), no toxicity was observed for both samples, confirming AuNS@CCP as a biocompatible gene carrier. These results indicated that AuNS@CCP is not toxic to cells at its highest transfection efficiency dose.
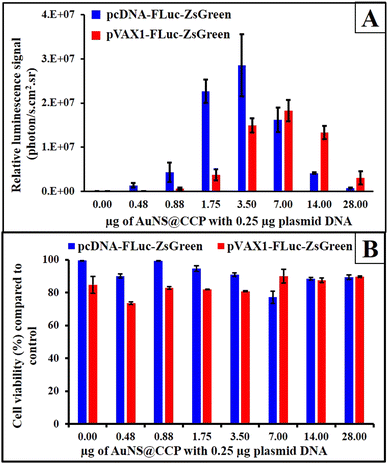 | ||
| Fig. 6 Transfection of 0.25 μg pcDNA-FLuc-ZsGreen and pVAX1-FLuc-ZsGreen by different quantities of AuNS@CCP (A) and MTT assay results for studying the cytotoxicity of AuNS@CCP (B). | ||
We further studied the cytotoxicity of AuNSs and AuNS@CCP without complexing with plasmid DNA to observe their natural toxicity in CT-26 and HEK293T cells (Fig. S7, ESI†). As expected, AuNSs showed no toxicity to normal and cancer cells. In comparison, AuNS@CCP showed slight toxicity in higher amounts (above 10 μg for CT-26 and 40 μg for HEK293T cells). The mild toxicity of AuNS@CCP at higher amounts could be due to an increase in the amount of bPEI, which might increase the positive charge and toxicity to the cells. Therefore, we concluded that a combination of CS, bPEI, and AuNSs could increase the biocompatibility of the hybrid system.
We also studied the transfection efficiency of pcDNA-FLuc-ZsGreen in different cell lines (HEK293T, LN-308, MDA-MB-231, and CT-26) using AuNS@CCP (Fig. 7A and B). The results, as expected, showed the highest transfection efficiency with HEK293T cells, followed by LN-308 and MDA-MB-231 cells. In addition, the transfection efficiency was also reduced by decreasing the amount of AuNS@CCP. However, it increased in the case of CT-26 cells, for which the lower amount of AuNS@CCP had a better transfection efficiency; this could be because of the growth rate, phagocytic properties, and transcriptional activity of the CT-26 cells.55
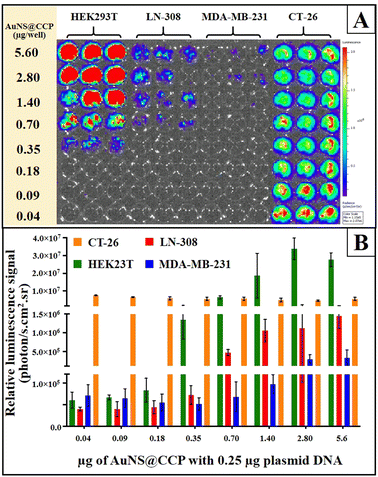 | ||
| Fig. 7 Transfection of 0.25 μg of pcDNA-FLuc-ZsGreen to HEK293T, LN-308, MDA-MB-231, and CT-26 cell lines by different amounts of AuNS@CCP (A) and its results (B). | ||
Evaluation of AuNS@CCP for its potential in RNA transfection using mRNA coding for SARS-CoV-2 spike as a protein model
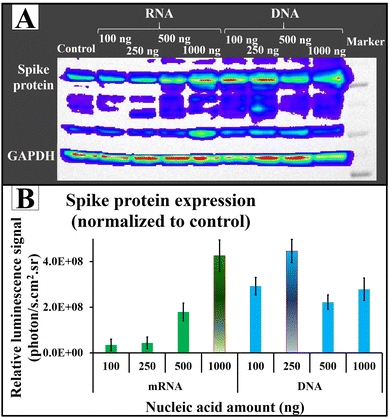
After validating the transfection efficiency of AuNS@CCP using plasmid DNA coding FLuc-ZsGreen in different cell lines, we also tested its potential application for RNA transfection using mRNA coding for SARS-CoV-2 spike protein. We used plasmid DNA (pcDNA-Spike) coding for spike protein for comparison. We found an increasing trend in the spike protein expression by increasing the amount of nucleic acid (Fig. 8A). Of note, the quantities of expressed spike protein were almost the same for both RNA and DNA (Fig. 8B). Since a DNA cargo must cross the cell wall (first barrier) and then penetrate the nucleus (second barrier) to release the DNA for the transcription and translation processes, and the mRNA cargo faces only one barrier (cell wall) to release the mRNA to be translated into proteins, we expected less spike protein after DNA transfection compared with mRNA transfection. Surprisingly, AuNS@CCP showed strong carrier properties because it penetrated two barriers to deliver the nucleic acid content, which could be why we observed almost similar amounts of spike protein. Indeed, this could be a considerable advantage for a gene carrier carrying either RNA or DNA for various applications, including DNA- and RNA-based vaccines. Additionally, 250 ng of DNA showed the same protein expression level as 1000 ng mRNA. We assumed that increasing the DNA amount makes the total surface charge unsuitable to penetrate the nucleus membrane, which is less permeable.Experimental
Materials
We purchased HAuCl4·3H2O, NaBH4, ascorbic acid, Triton™ X-100, chitin (Art No. C7170), β-CD, p-toluenesulfonyl chloride (p-TsCl), triethylamine (Et3N), lithium chloride (LiCl), ammonium chloride (NH4Cl), ethylenediamine (en), boric acid, dimethyl sulfoxide (DMSO) and glutaraldehyde (GA, 50% w/v) from Sigma-Aldrich, USA. Similarly, we used Dulbecco's modified eagle's medium (DMEM, cell growth medium), phosphate-buffered saline, 0.5 M ethylenediaminetetraacetic acid (EDTA), trypsin–EDTA (0.05%), fetal bovine serum (FBS), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), AgNO3, acetonitrile, and NaOH purchased from Thermo Fisher Scientific, USA. We procured anhydrous N,N-dimethylacetamide (DMAc), bPEI (MW: 2000 g mol−1), and glacial acetic acid (AcOH) from Chem-Impex Int'l. Inc., Polysciences, Inc., and Merck Co., respectively. Ethanol (EtOH, 100%) was purchased from Gold Shield Dist., Inc. Agarose was purchased from IBI Scientific, USA. Tris base was purchased from Bio-Rad Laboratories, Inc., USA. We constructed eukaryotic plasmid vectors expressing the Luciferase-ZsGreen reporter gene and spike protein under the CMV promoter using the gene constructs provided by BEI-NIAID, USA.Devices
We used a Milli-Q® (IQ 7000, France) water purifier as a pure water source for our various synthesis processes and for preparing different reagents. We used a Branson CPX2800H bath sonicator (40 kHz) and a Branson SLPe probe sonicator (40 kHz, 150 W) as homogenizing devices and a Sorvall™ WX+ Ultracentrifuge Series (Thermo Scientific™) to centrifuge the samples. We used the in vivo Imaging System (IVIS) (IVIS® Lumina II, Caliper Lifesciences, USA) to optically image the luciferase plasmid transfected in cells using different transfection agents developed in this study.Methods
Characterization of AuNSs and AuNS@CCP
We used transmission electron microscopy (TEM) and energy dispersive X-ray spectroscopy (FEI Tecnai G2 F20 X-TWIN transmission electron microscope 200 kV, USA) to study the morphology and size of the AuNS@CCP samples and to elementally characterize them. We diluted 5 μL of the sample with 5 μL water, then dropped it on a copper 400 mesh (Electron Microscopy Sciences, LC400-Cu-100) and allowed it to dry for 24 h before using it for TEM imaging. We used dynamic light scattering (DLS, Zetasizer Nano ZS90, UK) to measure the surface charge of the samples. We diluted 10 μL of each sample with 1 mL of Milli-Q® water and then transferred it to a cuvette for the measurement. To record the ultraviolet-visible (UV-vis) spectra of the samples, we added 20 μL of each sample to a 96-well plate and analyzed it using an Infinite® M1000 microplate reader (USA) and then recorded the spectra in the wavelength range of 400–800 nm. We applied nuclear magnetic resonance (NMR) (Agilent 400-MR NMR spectrometer, USA) to study the chemical bonding between the functional groups. We dissolved 10 mg of each sample in 2 mL of the NMR solvent (1.96 mL D2O and 0.04 mL DCl) to acquire 1H NMR spectra of the samples.Gel retardation assay (GRA)
We used GRA to study the DNA binding efficiency of AuNS@CCP. We prepared agarose gel (0.7% w/v) in 0.5× TBE buffer (Tris base, boric acid, EDTA 0.5 M). We prepared the DNA with polymer complex in various ratios by adding different amounts of AuNS@CCP and plasmid DNA. We incubated the samples at room temperature for 15 min before resolving them in agarose gel by running at 50 V for 45–60 min.Cell culture
We cultured HEK293T (human embryonic kidney cells), LN-308 (p53-deficient glioblastoma cells), MDA-MB-231 (triple-negative breast cancer), and CT-26 (colon cancer) cell lines in DMEM supplemented with 10% FBS, 100 units per mL penicillin and 100 μg per mL streptomycin and incubating them at 37 °C, 5% CO2, and 95% air. We passaged the cells when their confluency reached about 80% by trypsinizing them. For transfection, we counted the cells and plated them either in 48-well (50k cells per well) or 24-well (75k cells per well) plates and incubated them for 24 h before use for transfection.Transfection efficiency by bioluminescence imaging
We first studied the transfection efficiency of CS, CS–CD, bPEI, and AuNS@CCP for delivering plasmid DNA in HEK293T cells. We then tested the transfection efficiency of AuNS@CCP under various conditions, such as different AuNS@CCP/plasmid ratios (N/P ratios), different plasmid amounts, and various cell lines. Typically, we diluted the samples in 10% w/v dextrose and diluted the plasmid in serum-free Opti-MEM before mixing them. We incubated the component/plasmid complex at RT for about 25–30 min before transferring them to cell culture plates. We incubated the cells for 24–48 h at 37 °C, 5% CO2, and 95% air, then imaged them for bioluminescence signals using the IVIS optical imaging system by adding D-luciferin (Dluc, 150 μg mL−1). We acquired the signals using the bioluminescence imaging mode and quantified them by drawing regions of interest (ROI) over the wells.Cytotoxicity evaluation
We conducted the MTT assays to study the cytotoxicity of AuNSs and AuNS@CCP with and without complexing with plasmid DNA. We seeded 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in 96-well plates, and after 24 h, we added AuNS@CCP complexed with 0.25 μg plasmid equivalent per well. We aspirated the medium 24 h after transfection and added 12.5 μM of MTT solution to each well, and incubated it for 2 h at 37 °C, 5% CO2, and 95% air. We carefully aspirated the liquid, added 0.1 mL of DMSO to each well, and incubated it for 30 min at 37 °C. We spectroscopically measured the samples at 570 nm and plotted the graph by normalizing the results using the control samples as a reference (relative growth rate by control as 100%). The same procedure was carried out for AuNSs and AuNS@CCP without complexing with plasmid DNA.
000 cells per well in 96-well plates, and after 24 h, we added AuNS@CCP complexed with 0.25 μg plasmid equivalent per well. We aspirated the medium 24 h after transfection and added 12.5 μM of MTT solution to each well, and incubated it for 2 h at 37 °C, 5% CO2, and 95% air. We carefully aspirated the liquid, added 0.1 mL of DMSO to each well, and incubated it for 30 min at 37 °C. We spectroscopically measured the samples at 570 nm and plotted the graph by normalizing the results using the control samples as a reference (relative growth rate by control as 100%). The same procedure was carried out for AuNSs and AuNS@CCP without complexing with plasmid DNA.
AuNS@CCP mediated transfection of mRNA and plasmid DNA coding for SARS-CoV-2 spike protein
We evaluated the transfection efficiency of AuNS@CCP for both RNA and DNA to show its capability of delivering nucleic acids as a vaccine. We used mRNA and plasmid DNA coding for SARS-CoV-2 spike protein in different nucleic concentrations at a constant nucleic acid/polymer ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 for transfection in HEK293T cells. We collected the cells 24 h after transfection, lysed the cells, and used the total for western blotting analysis using an anti-spike protein antibody as a detection antibody. After resolving the proteins in an acrylamide gel and transferring them onto a cellulose membrane, we detected the expressed spike protein by adding the anti-spike antibody (rabbit polyclonal antibody from BEI-NIAID, USA) followed by an HRP-conjugated anti-rabbit secondary antibody. We imaged the membrane by adding a chemiluminescent HRP-substrate using an IVIS optical CCD camera. We quantified the amounts of expressed spike protein by ROI measurement, and the results were normalized to control wells. We used GAPDH as a housekeeping protein to normalize the protein loading.
10 for transfection in HEK293T cells. We collected the cells 24 h after transfection, lysed the cells, and used the total for western blotting analysis using an anti-spike protein antibody as a detection antibody. After resolving the proteins in an acrylamide gel and transferring them onto a cellulose membrane, we detected the expressed spike protein by adding the anti-spike antibody (rabbit polyclonal antibody from BEI-NIAID, USA) followed by an HRP-conjugated anti-rabbit secondary antibody. We imaged the membrane by adding a chemiluminescent HRP-substrate using an IVIS optical CCD camera. We quantified the amounts of expressed spike protein by ROI measurement, and the results were normalized to control wells. We used GAPDH as a housekeeping protein to normalize the protein loading.
Conclusions
We prepared a simple gene nanocarrier based on AuNSs, CS, β-CD, and bPEI for gene delivery applications. We demonstrated the efficiency of the as-prepared BNC by delivering the pcDNA-FLuc-ZsGreen reporter plasmid, pcDNA-Spike plasmid, and synthetic mRNA coding for SARS-CoV-2 spike protein in various cell lines. We used different methods, such as NMR, UV-vis, TEM, EDX, and DLS, to characterize and study the properties of the prepared nanocomposites. The TEM images and UV-vis spectra of Au nanoseeds and AuNSs confirmed the formation of gold star-like structures with a size of approximately 53 nm for AuNSs and 70 nm for AuNS@CCP. We also prepared AuNS@CS–CD and AuNS@bPEI to compare with and study the effect of each component on the transfection efficiency of AuNS@CCP. The zeta potential for AuNS@CCP was +28.8 ± 4.0 mV. We studied the DNA binding ability of the nanocomposite using a gel retardation assay, in which we observed an excellent binding profile for AuNS@bPEI and AuNS@CCP while finding a very low binding for AuNS@CS–CD. The transfection study revealed that AuNS@CCP had the highest transfection efficiency and the best polymer/plasmid ratio of 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Further, the MTT assay results showed no cytotoxicity for AuNS@CCP at this particular ratio. Western blotting results indicated the same amount of spike protein expression by transfecting both mRNA and plasmid DNA, demonstrating the high capability of AuNS@CCP as a nanocarrier for both DNA and RNA in potential vaccine applications. Further validation in vivo is necessary to demonstrate its future use in various gene therapy applications. In conclusion, this biocompatible gene carrier BNC can be used as an efficient and potent gene carrier, with potential for various applications such as targeted cancer therapy and vaccines.
1. Further, the MTT assay results showed no cytotoxicity for AuNS@CCP at this particular ratio. Western blotting results indicated the same amount of spike protein expression by transfecting both mRNA and plasmid DNA, demonstrating the high capability of AuNS@CCP as a nanocarrier for both DNA and RNA in potential vaccine applications. Further validation in vivo is necessary to demonstrate its future use in various gene therapy applications. In conclusion, this biocompatible gene carrier BNC can be used as an efficient and potent gene carrier, with potential for various applications such as targeted cancer therapy and vaccines.
Author contributions
Farbod Tabesh: conceptualization, methodology, software, validation, formal analysis, data curation, writing – original draft, writing – review & editing, project administration. Golnaz Haghverdi: technical assistance and data curation. Kireeti Phani Devarakonda: technical assistance and data curation. Tarik F Massoud: conceptualization, review & editing. Ramasamy Paulmurugan: conceptualization, methodology, software, validation, formal analysis, investigation, data curation, writing – original draft, review & editing, supervision, project administration, funding acquisition, resources. All authors reviewed and approved the manuscript.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors do not have any competing interests to declare.Acknowledgements
General: The authors would like to thank the Canary Center at Stanford, Department of Radiology, for providing the facility and resources to conduct this research. The authors would also like to thank Stanford Animal Histology Services for preparing histology. They would like to thank Mallesh Pandrala, Sourabh Mehta, and Suresh Thangudu for their discussion and assistance during this work. Funding: The Gary Glazer-GE Fund (Department of Radiology, Stanford University) and Anges Inc. partly supported this work. NIH S10OD023518-01A1 Award for the Celigo S Imaging Cytometer (200-BFFL-S) to RP is acknowledged.References
- J. Malina, H. Kostrhunova, V. Novohradsky, P. Scott and V. Brabec, Nucleic Acids Res., 2022, 50, 674–683 CrossRef CAS PubMed.
- Z. Krut, D. Gazit, Z. Gazit and G. Pelled, Bioengineering, 2022, 9, 190 CAS.
- Y. Wang, Y. Li, Z. Huang, B. Yang, N. Mu, Z. Yang, M. Deng, X. Liao, G. Yin and Y. Nie, Carbohydr. Polym., 2022, 290, 119499 CrossRef CAS.
- B. Cheng, H.-H. Ahn, H. Nam, Z. Jiang, F. J. Gao, I. Minn and M. G. Pomper, Pharmaceutics, 2022, 14, 373 CrossRef CAS.
- Y. Cao, Y. F. Tan, Y. S. Wong, M. W. J. Liew and S. Venkatraman, Mar. Drugs, 2019, 17, 381 CrossRef CAS PubMed.
- Y. Li, B. Qiu, Z. Li, X. Wang, Z. He, D. M. Sandoval, R. Song, A. Sigen, C. Zhao and M. Johnson, JCR, 2024, 367, 327–338 CAS.
- C. Guo, Y. Liu, Z. Zhao, Y. Wu, Q. Kong and Y. Wang, JCR, 2024, 365, 1004–1018 CAS.
- N. Song, Z. Sun, B. Wang, X. Liu, B. Hu, N. Chen, S. Zhang and Z. Yu, Acta Biomater., 2023, 250–261 Search PubMed.
- H. Liu, X. Zhu, Y. Wei, C. Song and Y. Wang, Biomed. Pharmacother., 2023, 157, 114065 CrossRef CAS.
- E. Demirel and Y. Y. Durmaz, Eur. Polym. J., 2023, 186, 111841 CrossRef CAS.
- M. Brard, C. Lainé, G. Réthoré, I. Laurent, C. Neveu, L. Lemiegre and T. Benvegnu, JOC, 2007, 72, 8267–8279 CrossRef CAS.
- D. Sahoo, E. N. Atochina-Vasserman, D. S. Maurya, M. Arshad, S. S. Chenna, N. Ona, J. A. Vasserman, H. Ni, D. Weissman and V. Percec, J. Am. Chem. Soc., 2024, 146(6), 3627–3634 CrossRef CAS.
- D. A. Giljohann, D. S. Seferos, W. L. Daniel, M. D. Massich, P. C. Patel and C. A. Mirkin, in Spherical Nucleic Acids, ed. C. A. Mirkin, Taylor & Francis, 2020, vol. 1, pp. 55–90 Search PubMed.
- M. A. Malik, A. A. Hashmi, A. S. Al-Bogami and M. Y. Wani, J. Mater. Chem. B, 2024, 12, 552–576 RSC.
- W.-C. Ko, S.-J. Wang, C.-Y. Hsiao, C.-T. Hung, Y.-J. Hsu, D.-C. Chang and C.-F. Hung, Molecules, 2022, 27, 1551 CrossRef CAS PubMed.
- J. Kim, M. Y. Anthony, K. P. Kubelick and S. Y. Emelianov, Photoacoustics, 2022, 25, 100307 CrossRef PubMed.
- T. Paul, P. Mohapatra and P. P. Mishra, Appl. Surf. Sci., 2022, 577, 151696 CrossRef CAS.
- X. Xu, Y. Liu, Y. Yang, J. Wu, M. Cao and L. Sun, Colloids Surf., A, 2022, 640, 128491 CrossRef CAS.
- J. Darmadi and A. Anwar, E3S Web Conf., 2024, DOI:10.1051/e3sconf/202448803020.
- A. Anil, J. Chaskar, A. B. Pawar, A. Tiwari and A. C. Chaskar, J. Biotechnol., 2024, 382, 8–20 CrossRef CAS PubMed.
- E. C. Greig, R. Rabiee and A. R. Afshar, Am. J. Ophthalmol. Case Rep., 2024, 33, 101987 CrossRef PubMed.
- J. Li, Z. Lu, L. Xu, J. Wang, S. Qian, Q. Hu and Y. Ge, ACS Appl. Bio Mater., 2024, 7, 1081–1094 CrossRef CAS PubMed.
- S. Qian, J. Chen, Y. Zhao, X. Zhu, D. Dai, L. Qin, J. Hong, Y. Xu, Z. Yang and Y. Li, Cytotherapy, 2024, 26, 113–125 CrossRef CAS.
- A. E. Ryckman, N. M. Deschenes, B. M. Quinville, K. J. Osmon, M. Mitchell, Z. Chen, S. J. Gray and J. S. Walia, MTMCD, 2024, 32 Search PubMed.
- T. Wan, J. Zhong, Q. Pan, T. Zhou, Y. Ping and X. Liu, Sci. Adv., 2022, 8, eabp9435 CrossRef CAS PubMed.
- J. Yao, S. Atasheva, N. Wagner, N. C. Di Paolo, P. L. Stewart and D. M. Shayakhmetov, Mol. Ther., 2024, 32, 103–123 CrossRef CAS.
- H. Zhang, Z. Liu, H. Lihe, L. Lu, Z. Zhang, S. Yang, N. Meng, Y. Xiong, X. Fan and Z. Chen, Adv. Healthcare Mater., 2024, 13, 2303261 CrossRef CAS PubMed.
- X. Lin, Y. Sheng, X. Zhang, Z. Li, Y. Yang, J. Wu, Z. Su, G. Ma and S. Zhang, J. Controlled Release, 2022, 346, 380–391 CrossRef CAS PubMed.
- V. Muripiti, A. Velidandia, Y. P. Sharma, R. Gondru, C. Arya and J. Banothu, J. Drug Delivery Sci. Technol., 2024, 93, 105457 CrossRef CAS.
- M. W. Russell, Z. Moldoveanu, P. L. Ogra and J. Mestecky, Front. Immunol., 2020, 11, 3221 Search PubMed.
- C. Zhang, J. Chen, Y. Song, J. Luo, P. Jin, X. Wang, L. Xin, F. Qiu, J. Yao and G. Wang, ACS Appl. Mater. Interfaces, 2022, 14, 2587–2596 CrossRef CAS.
- S. Mallakpour and F. Tabesh, in Handbook of polymer nanocomposites for industrial applications, ed. C. M. Hussain, Elsevier, 2021, pp. 503–528 Search PubMed.
- S. Mallakpour, V. Behranvand and F. Tabesh, in Natural polymers-based green adsorbents for water treatment, ed. S. Kalia, Elsevier, 2021, pp. 159–193 Search PubMed.
- B. Maiti and S. Bhattacharya, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2022, 14, e1759 CAS.
- H. Tong, Q. Shi, J. C. Fernandes, L. Liu, K. Dai and X. Zhang, Curr. Gene Ther., 2009, 9, 495–502 CrossRef CAS.
- R. Sarvari, M. Nouri, S. Agbolaghi, L. Roshangar, A. Sadrhaghighi, A. M. Seifalian and P. Keyhanvar, Int. J. Polym. Mater., 2022, 71, 246–265 CrossRef CAS.
- L. Bastiaens, L. Soetemans, E. D'Hondt and K. Elst, in Chitin and Chitosan: Properties and Applications, ed. C. G. B. Lambertus and A. M. van den Broek, Wiley, 2019, pp. 1–34 Search PubMed.
- G. Lin, J. Huang, M. Zhang, S. Chen and M. Zhang, Nanomaterials, 2022, 12, 584 CrossRef CAS PubMed.
- C. Peptu, A. C. Humelnicu, R. Rotaru, M. E. Fortuna, X. Patras, M. Teodorescu, B. I. Tamba and V. Harabagiu, in Chitin and Chitosan: Properties and Applications, ed. C. G. B. Lambertus and A. M. van den Broek, Wiley, 2019, pp. 259–289 Search PubMed.
- M. Safarzadeh, S. Mohammadi-Yeganeh, F. Ghorbani-Bidkorbeh and M. H. M. Hoseini, Life Sci., 2022, 297, 120459 CrossRef CAS.
- Y. Gao and Y. Wu, Int. J. Biol. Macromol., 2022, 203, 379–388 CrossRef CAS PubMed.
- T. Pol, W. Chonkaew, L. Hocharoen, N. Niamnont, N. Butkhot, Y. M. Roshorm, S. Kiatkamjornwong, V. P. Hoven and K. Pratumyot, ACS Omega, 2022, 7, 10056–10068 CrossRef CAS PubMed.
- H. Mousazadeh, E. Bonabi and N. Zarghami, Carbohydr. Polym., 2022, 276, 118747 CrossRef CAS PubMed.
- V. K. Mishra and M. Ahmed, in Gold Nanoparticles for Drug Delivery, ed. P. Kesharwani, Elsevier, 2024, ch. 15, pp. 393–417 Search PubMed.
- N. Fattahi, L. Gorgannezhad, S. F. Masoule, N. Babanejad, A. Ramazani, M. Raoufi, E. Sharifikoloue, A. Foroumadi and M. Khoobi, Adv. Colloid Interface Sci., 2024, 325, 103119 CrossRef CAS.
- M. Wang, H. Huang, Y. Sun, M. Wang, Z. Yang, Y. Shi and L. Liu, Int. J. Biol. Macromol., 2024, 255, 128354 CrossRef CAS PubMed.
- B. Chabaud, H. Bonnet, R. Lartia, A. Van Der Heyden, R. Auzély-Velty, D. Boturyn, L. Coche-Guérente and G. V. Dubacheva, Langmuir, 2024, 40, 4646–4660 CrossRef CAS.
- I. B. Becerril-Castro, I. Calderon, N. Pazos-Perez, L. Guerrini, F. Schulz, N. Feliu, I. Chakraborty, V. Giannini, W. J. Parak and R. A. Alvarez-Puebla, Analysis Sensing, 2022, 2, e202200005 CrossRef CAS.
- Y. C. Dong, M. Hajfathalian, P. S. Maidment, J. C. Hsu, P. C. Naha, S. Si-Mohamed, M. Breuilly, J. Kim, P. Chhour and P. Douek, Sci. Rep., 2019, 9, 14912 CrossRef PubMed.
- M. M. Vega, A. Bonifacio, V. Lughi, S. Marsi, S. Carrato and V. Sergo, J. Nanopart. Res., 2014, 16, 1–6 CrossRef CAS.
- M. Wang, X. Cao, W. Lu, L. Tao, H. Zhao, Y. Wang, M. Guo, J. Dong and W. Qian, RSC Adv., 2014, 4, 64225–64234 RSC.
- F. Liebig, R. Henning, R. M. Sarhan, C. Prietzel, C. N. Schmitt, M. Bargheer and J. Koetz, RSC Adv., 2019, 9, 23633–23641 RSC.
- N. Cennamo, G. D'Agostino, A. Donà, G. Dacarro, P. Pallavicini, M. Pesavento and L. Zeni, Sensors, 2013, 13, 14676–14686 CrossRef PubMed.
- U. K. Sukumar, R. J. Bose, M. Malhotra, H. A. Babikir, R. Afjei, E. Robinson, Y. Zeng, E. Chang, F. Habte and R. Sinclair, Biomaterials, 2019, 218, 119342 CrossRef CAS PubMed.
- Y.-J. Chen, W.-M. Chang, Y.-W. Liu, C.-Y. Lee, Y.-H. Jang, C.-D. Kuo and H.-F. Liao, Chem. – Biol. Interact., 2009, 181, 440–446 CrossRef CAS PubMed.
- U. S. Kumar, R. Afjei, K. Ferrara, T. F. Massoud and R. Paulmurugan, ACS Nano, 2021, 15, 17582–17601 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ma00433g |
| This journal is © The Royal Society of Chemistry 2024 |