Open Access Article
Open Access ArticleCell membrane fusion induced by surface modification with cell-penetrating peptide–lipid conjugates that facilitates close contact between distinct membranes†
Yuya
Sato
 ab,
Teruhiko
Baba
ab,
Teruhiko
Baba
 b,
Takeyuki
Uchida
c,
Ung-il
Chung
a and
Yuji
Teramura
b,
Takeyuki
Uchida
c,
Ung-il
Chung
a and
Yuji
Teramura
 *bde
*bde
aDepartment of Bioengineering, Graduate School of Engineering, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo, 113-8656, Japan
bCellular and Molecular Biotechnology Research Institute (CMB), National Institute of Advanced Industrial Science and Technology (AIST), AIST Tsukuba Central 5, 1-1-1 Higashi, Tsukuba, Ibaraki 305-8565, Japan. E-mail: y.teramura@aist.go.jp; Tel: +81(0)29-861-6582
cResearch Institute of Electrochemical Energy, Department of Energy and Environment, National Institute of Advanced Industrial Science and Technology (AIST), 1-8-31 Midorigaoka, Ikeda, Osaka 563-8577, Japan
dDepartment of Immunology, Genetics and Pathology (IGP), Uppsala University, Dag Hammarskjölds väg 20, SE-751 85, Uppsala, Sweden
eMaster's/Doctoral Program in Life Science Innovation (T-LSI), University of Tsukuba, 1-1-1 Tennodai, Tsukuba, Ibaraki 305-8577, Japan
First published on 13th May 2024
Abstract
Cell fusion has several biotechnological applications. We previously reported cell surface modification with interactive ligands to induce cell–cell attachment, resulting in higher cell fusion efficiency when combined with a polyethylene glycol (PEG)-induced method. However, spontaneous cell fusion never occurred, even though cell–cell contact was strongly induced by ligand modification. The interactive ligands placed between the two membranes might obstruct spontaneous cell fusion during lipid exchange between the two cellular membranes. We hypothesized that two distinct cell membranes must be brought together to realize efficient membrane fusion by temporal ligand modification. Here, we aimed to induce spontaneous membrane fusion with single-polymer conjugates using liposomes and cells. We synthesized poly(ethylene glycol)-lipids (PEG-lipids) carrying the Tat peptide (Tat-PEG-lipid), a representative cell-penetrating peptide, and used it for cell–cell anchoring. We found that Tat-PEG-lipid interacted with the bilayer membrane via the Tat peptide and lipid domain and induced membrane fusion when the lipid was nonanoyl (C9) or dodecanoyl (C12) and the PEG chain was 5 kDa, as assessed by quartz crystal microbalance with dissipation monitoring, dynamic light scattering, and fluorescence resonance energy transfer measurements, transmission electron microscopy, and confocal microscopy. It was found that Tat-PEG-lipid could induce membrane attachment and fusion of cells as well as liposomes, but it did not remain stable on the membrane. Thus, membrane fusion could be effectively promoted by the attachment of lipid membranes induced by transient surface modification with a Tat-PEG-lipid containing a nonanoyl or dodecanoyl lipid chain and a 5 kDa PEG chain.
Introduction
Cell fusion is often observed in several biological processes in which two distinct lipid bilayer membranes are fused into the same membrane to form heterogeneous and homogeneous cells. As new cells can be fabricated by cell fusion, this technique has been applied to several biotechnologies. For example, monoclonal antibodies are produced from hybridomas, which are fused cells of B cells and myeloma cells,1 and breeding different plant species has been attempted by protoplast fusion.2 Studies have reported that stem cell3 and therapeutic cells4–6 can be produced using cell fusion techniques. Reprogramming of somatic cell nuclei can be induced by cell fusion with embryonic stem cells, which can produce pluripotent stem cells with a major histocompatibility complex (MHC) similar to the MHC of the donor. This is a promising technique for cell sources in allogeneic transplantation owing to the reduced immune rejection.3 According to another rat study, long-term graft survival was improved by allogeneic transplantation of pancreatic islet cells fused with mesenchymal stem cells (MSCs). Nuclear reprogramming occurs in fused islet cells, promotes angiogenesis and regulates inflammation by enhancing the properties of MSCs.4In vitro cell fusion was achieved using a virus,7 electricity,8 and poly(ethylene glycol) (PEG).9,10 Although cell fusion has been studied, fusion efficiency varies depending on operator skill and the cell line used, these three methods remain inefficient. The poor efficiency in vitro is due to the lack of cell–cell contact. In cells, the soluble N-ethylmaleimide-sensitive-factor attachment protein receptor (SNARE) family of membrane proteins is involved in various cell fusion processes to form an α-helix bundle that brings the cell membranes of two separate cells together and promotes cell fusion.11 In fact, vesicle fusion never occurred, but rather the cells aggregated when SNARE function was inhibited.12 Therefore, the current approach to address this issue is to induce cell–cell attachment by combining three conventional methods for a high-efficiency cell fusion.13–15 For example, two cells can be optically arranged in close contact with each other to induce cell fusion using optical tweezers.16 Cell fusion has been achieved by laser irradiation of gold nanoparticles placed between two attached cells. Thus, this study showed the proof of concept that cell–cell attachment is important for cell fusion.
Recently, cell surface modification has also been used to induce cell–cell attachment, in which cell surfaces are modified with various functional ligands, such as single-stranded DNA and oligopeptides, to make them interact with each other.13,17–20 For example, we selected two types of oligopeptides and three repeated units of EIAALEK and KIAALKE sequences, which tend to interact.21 Cell–cell attachment can be achieved through a specific interaction between two peptides by cell surface modification with peptide-conjugated PEG-lipids. We then improved the cell fusion efficiency by combining it with the conventional PEG-induced method.14 Another report demonstrated that the cell–cell attachment efficiency was improved by cell surface modification with DNA-conjugated cholesterol combined with a PEG-induced method.13 Thus, close contact between separate cell membranes is an important factor for improving cell fusion efficiency. However, spontaneous cell fusion never occurred, although cell–cell contact was strongly induced by cell surface modification. The use of interactive ligands might be an obstacle to spontaneous cell fusion because the molecules are placed between the two membranes and may act as a barrier inhibiting lipid exchange between the two cellular membranes.
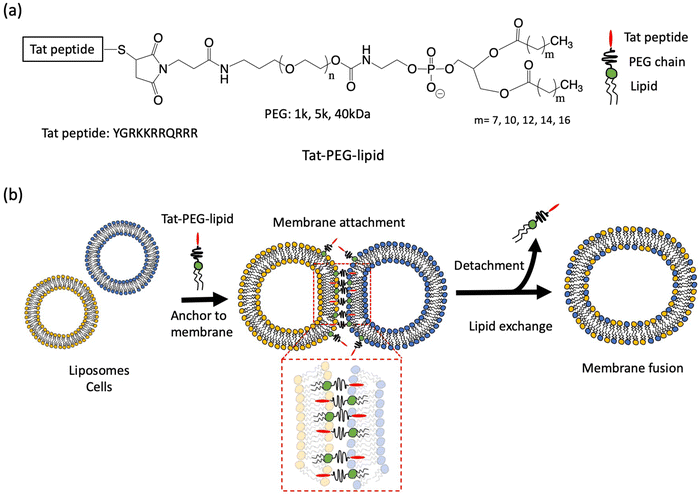
To address this issue, we designed a cell-penetrating peptide (CPP)-conjugated PEG-lipid to induce the cell–cell attachment of two cells, in which the single polymer conjugate can bring distinct cellular membranes together. Here, the cationic Tat peptide (YGRKKRRQRRR), a representative CPP,22 was conjugated to the end of the PEG chain of the PEG-lipid (Tat-PEG-lipid). The cell surface can be modified with Tat-PEG-lipids, where the Tat peptide can be displayed on the cell surface.18,23,24 Because the Tat peptide is membrane-permeable, Tat-PEG-lipids inserted into a cellular membrane can anchor to the neighboring cellular membrane through the interaction between the Tat peptide and another cellular membrane. We assumed that the cell–cell attachment by this Tat-PEG-lipid could induce spontaneous cell fusion because it is a single polymer and can achieve closer contact between two cells compared to ligand-PEG-lipids, where the two polymers interact with each other from both cellular membranes. In this study, we examined the membrane fusion of liposomes and cells (CCRF-CEM and adipose-derived mesenchymal stem cells (ADSCs)) using various Tat-PEG-lipids with different PEG chains and lipid lengths. Since the liposome has been used as the cell membrane model, we considered that it was possible to study the mechanism of membrane fusion by various analytical methods. Finally, we examined the membrane fusion of heterologous cells using two different cells (CCRF-CEM and ADSCs).
Materials and methods
Materials
1,2-Dinonanoyl-sn-glycero-3-phosphocholine (DC9PC) was purchased from Avanti Polar Lipids (Alabaster, AL, USA). 1,2-Dilauroyl-sn-glycero-3-phosphoethanolamine (lipid(C12)) was purchased from Bachem (Bubendorf, Switzerland). 1,2-Dimyristoyl-sn-glycero-3-phosphoethanolamine (lipid(C14)), 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (lipid(C16)), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine (lipid(C18)), α-3-[(3-maleimido-1-oxopropyl)aminopropyl-ω-(succinimidyloxy carboxy)]polyethylene glycol (Mal-PEG (1 kDa)-NHS, MW 1000 Da), α-3-[(3-maleimido-1-oxopropyl)aminopropyl-ω-(succinimidyloxy carboxy)]polyethylene glycol (Mal-PEG (5 kDa)-NHS, MW 5000 Da), α-3-[(3-maleimido-1-oxopropyl)aminopropyl-ω-(succinimidyloxy carboxy)]polyethylene glycol (Mal-PEG (40 kDa)-NHS, MW 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da), MPC polymer, and 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) were purchased from NOF (Tokyo, Japan). 5(6)-Carboxyfluorescein (CF), rhodamine 6G (R6G), triethylamine (TEA), TLC plate silica gel, 1,6-diphenyl-1,3,5-hexatriene (DPH), bovine serum albumin (BSA), and CPP (Tat: YGRKKRRQRRRC and FITC-Tat: FITC-YGRKKRRRQRRRC) were purchased from Sigma-Aldrich (St. Louis, MO, USA). DiI, DiO, 4′,6-diamidino-2-phenylindole (DAPI), and spin columns were purchased from Thermo Fisher Scientific (Waltham, MA, USA). L-Cysteine, 2-Aminoethanol (ethanolamine), iodine, phosphotungstic acid hydrate, diethyl ether, chloroform, methanol, chloroform-d, methanol-d4, acetone, dichloromethane (super dehydrated), cholesterol, sodium dodecyl sulfate (SDS), tetrahydrofuran (THF), sodium chloride (NaCl), sodium hydroxide (NaOH), dimethyl sulfoxide (DMSO), ethanol, Dulbecco's phosphate-buffered saline (PBS; pH 7.4), 1-dodecanethiol, hydrochloric acid (HCl), 4% Paraformaldehyde Phosphate Buffer Solution, and a cholesterol kit (Cholesterol E-Test Wako) were purchased from Fujifilm Wako Pure Chemical (Osaka, Japan). Phospholipase D extracted from Streptomyces sp. (sPLD, T-39) was purchased from Asahi Kasei Pharma (Tokyo, Japan). Glass-bottomed dishes were purchased from Matsunami Glass Industries (Osaka, Japan). A 50 mL round-bottom flask and 96-well plate were purchased from Iwaki (Tokyo, Japan). Extrusion filters (1.0, 0.4, 0.2, and 0.1 μm) (Nuclepore Track-Etch Membrane) were purchased from Whatman (Maidstone, UK). The CCRF-CEM cell line, established from acute lymphoblastic leukemia T cells, was purchased from the Health Science Research Resources Bank (Tokyo, Japan). RPMI 1640 medium, fetal bovine serum (FBS), penicillin, streptomycin, and trypan blue solution were purchased from Invitrogen (Carlsbad, CA, USA). Human-adipose derived stem cells (ADSCs), and adipose-derived stem cells basal medium with supplements were purchased from Lonza (Walkersville, MD, USA). Cell Counting Kit-8 was purchased from Dojindo Laboratories (Kumamoto, Japan).
000 Da), MPC polymer, and 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) were purchased from NOF (Tokyo, Japan). 5(6)-Carboxyfluorescein (CF), rhodamine 6G (R6G), triethylamine (TEA), TLC plate silica gel, 1,6-diphenyl-1,3,5-hexatriene (DPH), bovine serum albumin (BSA), and CPP (Tat: YGRKKRRQRRRC and FITC-Tat: FITC-YGRKKRRRQRRRC) were purchased from Sigma-Aldrich (St. Louis, MO, USA). DiI, DiO, 4′,6-diamidino-2-phenylindole (DAPI), and spin columns were purchased from Thermo Fisher Scientific (Waltham, MA, USA). L-Cysteine, 2-Aminoethanol (ethanolamine), iodine, phosphotungstic acid hydrate, diethyl ether, chloroform, methanol, chloroform-d, methanol-d4, acetone, dichloromethane (super dehydrated), cholesterol, sodium dodecyl sulfate (SDS), tetrahydrofuran (THF), sodium chloride (NaCl), sodium hydroxide (NaOH), dimethyl sulfoxide (DMSO), ethanol, Dulbecco's phosphate-buffered saline (PBS; pH 7.4), 1-dodecanethiol, hydrochloric acid (HCl), 4% Paraformaldehyde Phosphate Buffer Solution, and a cholesterol kit (Cholesterol E-Test Wako) were purchased from Fujifilm Wako Pure Chemical (Osaka, Japan). Phospholipase D extracted from Streptomyces sp. (sPLD, T-39) was purchased from Asahi Kasei Pharma (Tokyo, Japan). Glass-bottomed dishes were purchased from Matsunami Glass Industries (Osaka, Japan). A 50 mL round-bottom flask and 96-well plate were purchased from Iwaki (Tokyo, Japan). Extrusion filters (1.0, 0.4, 0.2, and 0.1 μm) (Nuclepore Track-Etch Membrane) were purchased from Whatman (Maidstone, UK). The CCRF-CEM cell line, established from acute lymphoblastic leukemia T cells, was purchased from the Health Science Research Resources Bank (Tokyo, Japan). RPMI 1640 medium, fetal bovine serum (FBS), penicillin, streptomycin, and trypan blue solution were purchased from Invitrogen (Carlsbad, CA, USA). Human-adipose derived stem cells (ADSCs), and adipose-derived stem cells basal medium with supplements were purchased from Lonza (Walkersville, MD, USA). Cell Counting Kit-8 was purchased from Dojindo Laboratories (Kumamoto, Japan).
Coating of containers with MPC polymers
Because the Tat peptide is positively charged due to the arginine and lysine residues contained in the sequence, we were concerned that non-specific adsorption could occur on the plastic and glass surfaces. Therefore, we used the MPC polymer to coat all plastic and glass surfaces used in all experiments to prevent non-specific adsorption.25,26 Briefly, the glass-bottom dishes, plastic tubes, tips, and culture dishes were treated with MPC polymer solution (0.5% in ethanol) for overnight, then the excess solution was removed, followed by air drying.Synthesis of DC9PE
DC9PC (28 mg) and diethyl ether (10 mL) were mixed in a glass centrifuge tube with a screw cap (Iwaki, capacity: 30 mL). To this solution, ethanolamine-HCl aqueous solution (2 mL, 250 mM, pH 4.2) and sPLD aqueous solution (1 mL, 30 U mL−1) were added, and the tube was sealed tightly. After shaking the tube with a high-speed shaking machine (As One ASCM-1, 1000 rpm, 90 min) in a low-temperature incubator (Yamato Scientific IJ200, 35 °C), diethyl ether was volatilized with a stream of nitrogen. Ultrapure water (0.2 mL) was added to the remaining aqueous phase, followed by chloroform (4 mL) and methanol (8 mL), which were mixed to form a homogeneous solution. Chloroform (1 mL) and ultrapure water (1 mL) were added to form an inhomogeneous solution, followed by centrifugation (As One C-12B centrifuge, 2500 rpm (650g) for 15 min at room temperature) to obtain two clear phases. The lower chloroform phase was evaporated and dried to obtain DC9PE (lipid(C9)). DC9PC and DC9PE were analyzed by a mass spectrometer (The MStation JMS-700, JEOL, Tokyo, Japan). DC9PC is 538.4 m/z and DC9PE is 496.3 m/z.Synthesis of Mal-PEG-lipid
Maleimidyl poly(ethylene glycol)-conjugated phospholipids (Mal-PEG(1k, 5k, 40k)-lipid) were synthesized as previously reported.18 Briefly, lipid(C9), lipid(C12), lipid(C14), lipid(C16), or lipid(C18) (9.8 mg, 11 mg, 13 mg, 14 mg, 15 mg respectively, 1 eq.), Mal-PEG(5k, 40k)-NHS (5k: 90 mg, 90 mg, 90 mg, 90 mg, 90 mg, 40k; 7.2 × 102 mg, 7.2 × 102 mg, 7.2 × 102 mg, 7.2 × 102 mg, 7.2 × 102 mg respectively, 1.1 eq.) and triethylamine (5.0 μL) were dissolved in dichloromethane (15 mL) and stirred for 2 d at room temperature. The reaction solution was reprecipitated with diethyl ether (3.0 L) to obtain Mal-PEG(5k, 40k)-lipids (yield: Mal-PEG(5k)-lipid(C9, C12, C14, C16, or C18): 85, 82, 80, 83, 86% and Mal-PEG(40k)-lipid(C9, C12, C14, C16, or C18): 79, 82, 85, 90, 85%). Also, lipid(C9), lipid(C12) or lipid(C16) (8.2 mg, 9.6 mg, 11 mg respectively), Mal-PEG(1k)-NHS (21 mg, 21 mg, 21 mg respectively) and triethylamine (5.0 μL) were dissolved in dichloromethane (15 mL) and stirred for 2 d at room temperature. The reaction solution was reprecipitated with diethyl ether (3.0 L) to obtain Mal-PEG(1k)-lipids as a white powder (yield: Mal-PEG(1k)-lipid(lipid(C9), lipid(C12), or lipid(C16): 78, 67, 49%)). All samples were analyzed by 1H-NMR spectroscopy (AscendTM 600; Bruker, Billerica, Massachusetts, USA).
1H-NMR (CDCl3, 600 MHz, ™ppm) and Rf value of silica TLC (chloroform: methanol = 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 by volume) Mal-PEG(1k)-lipid(C9): 0.88 (t, 6H, –CH3), 1.25 (br, 20H, –CH2–), 3.64 (br, 89H, PEG), 6.71 (s, 1.5H, –HC
1 by volume) Mal-PEG(1k)-lipid(C9): 0.88 (t, 6H, –CH3), 1.25 (br, 20H, –CH2–), 3.64 (br, 89H, PEG), 6.71 (s, 1.5H, –HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–, maleimide) and Rf value: 0.083. Mal-PEG(1k)-lipid(C12): 0.88 (t, 6H, –CH3), 1.25 (br, 33H, –CH2–), 3.64 (br, 91H, PEG), 6.71 (s, 1.5H, –HC
CH–, maleimide) and Rf value: 0.083. Mal-PEG(1k)-lipid(C12): 0.88 (t, 6H, –CH3), 1.25 (br, 33H, –CH2–), 3.64 (br, 91H, PEG), 6.71 (s, 1.5H, –HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–, maleimide), and Rf value: 0.081. Mal-PEG(1k)-lipid(C16): 0.88 (t, 6H, –CH3), 1.25 (br, 49H, –CH2–), 3.64 (br, 90H, PEG), 6.71 (s, 1.5H, –HC
CH–, maleimide), and Rf value: 0.081. Mal-PEG(1k)-lipid(C16): 0.88 (t, 6H, –CH3), 1.25 (br, 49H, –CH2–), 3.64 (br, 90H, PEG), 6.71 (s, 1.5H, –HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–, maleimide), and Rf value: 0.13. Mal-PEG(5k)-lipid(C9): 0.88 (t, 6H, –CH3), 1.25 (br, 22H, –CH2–), 3.64 (br, 459H, PEG), 6.71 (s, 1.6H, –HC
CH–, maleimide), and Rf value: 0.13. Mal-PEG(5k)-lipid(C9): 0.88 (t, 6H, –CH3), 1.25 (br, 22H, –CH2–), 3.64 (br, 459H, PEG), 6.71 (s, 1.6H, –HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–, maleimide), and Rf value: 0.25. Mal-PEG(5k)-lipid(C12): 0.88 (t, 6H, –CH3), 1.25 (br, 33H, –CH2–), 3.64 (br, 493H, PEG), 6.71 (s, 1.5H, –HC
CH–, maleimide), and Rf value: 0.25. Mal-PEG(5k)-lipid(C12): 0.88 (t, 6H, –CH3), 1.25 (br, 33H, –CH2–), 3.64 (br, 493H, PEG), 6.71 (s, 1.5H, –HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–, maleimide), and Rf value: 0.24. Mal-PEG(5k)-lipid(C14): 0.88 (t, 6H, –CH3), 1.25 (br, 40H, –CH2–), 3.64 (br, 495H, PEG), 6.71 (s, 1.5H, –HC
CH–, maleimide), and Rf value: 0.24. Mal-PEG(5k)-lipid(C14): 0.88 (t, 6H, –CH3), 1.25 (br, 40H, –CH2–), 3.64 (br, 495H, PEG), 6.71 (s, 1.5H, –HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–, maleimide), and Rf value: 0.23. Mal-PEG(5k)-lipid(C16): 0.88 (t, 6H, –CH3), 1.25 (br, 48H, –CH2–), 3.64 (br, 480H, PEG), 6.70 (s, 1.6H, –HC
CH–, maleimide), and Rf value: 0.23. Mal-PEG(5k)-lipid(C16): 0.88 (t, 6H, –CH3), 1.25 (br, 48H, –CH2–), 3.64 (br, 480H, PEG), 6.70 (s, 1.6H, –HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–, maleimide), and Rf value: 0.24. Mal-PEG(5k)-lipid(C18): 0.88 (t, 6H, –CH3), 1.25 (br, 57H, –CH2–), 3.64 (br, 450H, PEG), 6.71 (s, 1.5H, –HC
CH–, maleimide), and Rf value: 0.24. Mal-PEG(5k)-lipid(C18): 0.88 (t, 6H, –CH3), 1.25 (br, 57H, –CH2–), 3.64 (br, 450H, PEG), 6.71 (s, 1.5H, –HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–, maleimide), and Rf value: 0.29. Mal-PEG(40k)-lipid(C9) 0.88 (t, 6H, –CH3), 1.25 (br, 20H, –CH2–), 3.64 (br, 3800H, PEG), 6.71 (s, 1.5H, –HC
CH–, maleimide), and Rf value: 0.29. Mal-PEG(40k)-lipid(C9) 0.88 (t, 6H, –CH3), 1.25 (br, 20H, –CH2–), 3.64 (br, 3800H, PEG), 6.71 (s, 1.5H, –HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–, maleimide), and Rf value: 0.25. Mal-PEG(40k)-lipid(C12): 0.88 (t, 6H, –CH3), 1.25 (br, 32H, –CH2–), 3.64 (br, 3800H, PEG), 6.71 (s, 1.6H, –HC
CH–, maleimide), and Rf value: 0.25. Mal-PEG(40k)-lipid(C12): 0.88 (t, 6H, –CH3), 1.25 (br, 32H, –CH2–), 3.64 (br, 3800H, PEG), 6.71 (s, 1.6H, –HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–, maleimide), and Rf value: 0.27. Mal-PEG(40k)-lipid(C14): 0.88 (t, 6H, –CH3), 1.25 (br, 40H, –CH2–), 3.64 (br, 3700H, PEG), 6.71 (s, 1.4H, –HC
CH–, maleimide), and Rf value: 0.27. Mal-PEG(40k)-lipid(C14): 0.88 (t, 6H, –CH3), 1.25 (br, 40H, –CH2–), 3.64 (br, 3700H, PEG), 6.71 (s, 1.4H, –HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–, maleimide), and Rf value: 0.26. Mal-PEG(40k)-lipid(C16): 0.88 (t, 6H, –CH3), 1.25 (br, 48H, –CH2–), 3.64 (br, 3600H, PEG), 6.71 (s, 1.5H, –HC
CH–, maleimide), and Rf value: 0.26. Mal-PEG(40k)-lipid(C16): 0.88 (t, 6H, –CH3), 1.25 (br, 48H, –CH2–), 3.64 (br, 3600H, PEG), 6.71 (s, 1.5H, –HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–, maleimide), and Rf value: 0.33. Mal-PEG(40k)-lipid(C18): 0.88 (t, 6H, –CH3), 1.25 (br, 55H, –CH2–), 3.64 (br, 4300H, PEG), 6.71 (s, 1.6H, –HC
CH–, maleimide), and Rf value: 0.33. Mal-PEG(40k)-lipid(C18): 0.88 (t, 6H, –CH3), 1.25 (br, 55H, –CH2–), 3.64 (br, 4300H, PEG), 6.71 (s, 1.6H, –HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–, maleimide), and Rf value: 0.23.
CH–, maleimide), and Rf value: 0.23.
To inactivate maleimide groups in Mal-PEG-lipids, cysteine solution (100 μL, 1 mg mL−1, in PBS) was added to Mal-PEG-lipids (1 mg) to obtain control PEG-lipids (10 mg mL−1, in PBS).
Synthesis of Tat-PEG-lipids
Tat-PEG lipids were synthesized as previously described.18 Briefly, the Tat peptide (Tat: YGRKKRRQRRRC or FITC-Tat: FITC-YGRKKRRQRRRC) and Mal-PEG-lipid were conjugated via a thiol-maleimide reaction between the maleimide group of Mal-PEG-lipid and the cysteine residue at the C-terminus of the peptides. The following were mixed and stirred at room temperature for 1 d: Mal-PEG(1k)-lipids (100 μL, 10 mg mL−1 in DMSO, 1 eq), Mal-PEG(5k)-lipids (100 μL, 10 mg mL−1 in PBS, 1 eq.) or Mal-PEG(40k)-lipids (100 μL, 10 mg mL−1 in PBS, 1 eq.) and Tat peptide solution (Tat: 122, 116, 108 μL) for Mal-PEG(1k)-lipids(C9, C12, C16) respectively, Tat: 33, 33, 32, 32, 32 μL, FITC-Tat: 40, 40, 39, 39, 39 μL for Mal-PEG(5k)-lipids(C9, C12, C14, C16, C18) respectively, Tat: 4.4, 4.4, 4.4, 4.4, 4.4 μL for Mal-PEG(40k)-lipids(C9, C12, C14, C16, C18) respectively, 10 mg mL−1 in (DMSO, 1.1 eq). Thus, we obtained Tat-PEG lipids with different PEG chains (1k, 5k and 40 kDa) and lipid lengths (C9, C12, C14, C16, and C18).Critical micelle concentration (CMC) of Tat-PEG(5k)-lipids
DPH was used to determine the CMC of the Tat-PEG-lipids (lipid(C9), lipid(C12), lipid(C14), lipid(C16), lipid(C18)). Tat-PEG-lipid solution (18 μL, 10.0, 1.0, 1.0 × 10−1, 1.0 × 10−2, 1.0 × 10−3, 1.0 × 10−4, 1.0 × 10−5 mg mL−1 in PBS) and DPH solution (2.0 μL, 30 μM in THF) were mixed. After incubation for 1 h at 37 °C, PBS (480 μL) was added for dilution. The fluorescence intensity of the resulting solution was measured using a spectrofluorometer (FP-6500; JASCO Co., Tokyo, Japan) (excitation wavelength: 357 nm, emission wavelength: 430 nm). The CMC of Tat-PEG-lipids (lipid(C9), lipid(C12), lipid(C14), lipid(C16), lipid(C18)) were 500, 104, 75, 52, and 26 μM, respectively.Liposome preparation
Liposomes were prepared as previously described.24 DPPC solution (1 mL, 10 mg mL−1, in ethanol) and cholesterol solution (530 μL, 10 mg mL−1, in ethanol) were mixed (DPPC: cholesterol = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 by mol) and evaporated using a rotary evaporator (37 °C, 80 rpm, 10 min; RV 10, digital V, IKA, Staufen im Breisgau, Germany) to form a lipid film. After overnight vacuum drying in a desiccator (Vacuum Desiccator Model VS, As One, Osaka, Japan), the lipid was hydrated with PBS (1.5 mL) or CF solution (1.5 mL, 0.1 mM or 50 mM in water, pH 7.4, for fluorescent labeling) or R6G solution (1.5 mL, 0.2 mM in water, for fluorescent labeling) then vigorously stirred for 2 h to prepare liposome suspension at room temperature. For the fluorescence labeled liposome with CF or R6G, the liposome suspension was centrifuged (MX301, Tomy Seikop Co., Ltd, Tokyo, Japan, 70 min, 20
1 by mol) and evaporated using a rotary evaporator (37 °C, 80 rpm, 10 min; RV 10, digital V, IKA, Staufen im Breisgau, Germany) to form a lipid film. After overnight vacuum drying in a desiccator (Vacuum Desiccator Model VS, As One, Osaka, Japan), the lipid was hydrated with PBS (1.5 mL) or CF solution (1.5 mL, 0.1 mM or 50 mM in water, pH 7.4, for fluorescent labeling) or R6G solution (1.5 mL, 0.2 mM in water, for fluorescent labeling) then vigorously stirred for 2 h to prepare liposome suspension at room temperature. For the fluorescence labeled liposome with CF or R6G, the liposome suspension was centrifuged (MX301, Tomy Seikop Co., Ltd, Tokyo, Japan, 70 min, 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 4 °C) to remove free CF or R6G. This procedure was repeated thrice. Finally, the liposome pellet was resuspended in PBS (1.5 mL). The resulting liposome suspension was extruded by an extruder (Avanti Polar Lipids, Inc., Birmingham, AL, USA) into membrane filters (pore sizes: 1.0, 0.4, 0.2, and 0.1 μm) to obtain liposomes of uniform size. The solution was then passed through each extrusion filter 21 times. Then, the liposome suspension was centrifuged once (70 min, 20
000g, 4 °C) to remove free CF or R6G. This procedure was repeated thrice. Finally, the liposome pellet was resuspended in PBS (1.5 mL). The resulting liposome suspension was extruded by an extruder (Avanti Polar Lipids, Inc., Birmingham, AL, USA) into membrane filters (pore sizes: 1.0, 0.4, 0.2, and 0.1 μm) to obtain liposomes of uniform size. The solution was then passed through each extrusion filter 21 times. Then, the liposome suspension was centrifuged once (70 min, 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 4 °C) to completely remove free CF or R6G again. Liposomes and liposomes encapsulating CF or R6G (liposomes(CF) and liposomes(R6G), respectively) were prepared as described above.
000g, 4 °C) to completely remove free CF or R6G again. Liposomes and liposomes encapsulating CF or R6G (liposomes(CF) and liposomes(R6G), respectively) were prepared as described above.
Determination of cholesterol concentration in liposomes
The cholesterol concentration in the liposomes was quantified using a cholesterol kit according to the manufacturer's instructions. The liposome suspension (20 μL, in PBS) was mixed with SDS solution (20 μL, SDS aq, 1 mg mL−1, in pure water) and incubated for 30 min at 37 °C to solubilize the liposomes. Cholesterol standards (cholesterol concentrations of 0, 100, 200, 400, and 600 mg dL−1) were prepared using cholesterol standard solutions. Then, chromogenic reagent (3 mL), and the liposome suspension (40 μL) or the standard samples (40 μL) were mixed and incubated for 5 min at 37 °C. The absorbance was measured at 600 nm using a plate reader (AD 200 Plate Reader; Beckman Coulter Inc., Miami, FL, USA). Therefore, we calculated the cholesterol concentration in the liposome suspension.Diameter and surface charge of liposomes
Liposome suspension ([cholesterol] = 4.6 mM, in PBS) was diluted in PBS (liposome suspension: PBS = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 [5 μL
9 [5 μL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 μL]), placed in a particle size measurement cell (50 μL, High Precision Cell [ZEN2112]), and the intensity averaged size distribution and polydispersity (PDI) were measured by dynamic light scattering (DLS) using a Zetasizer Nano ZS (Malvern Instruments Co., Malvern, UK). Liposome suspensions ([cholesterol] = 4.6 mM, in PBS) were also diluted in sodium chloride solution (NaCl aq, 1.0 mM, in pure water) (liposome suspension: NaCl aq = 1
45 μL]), placed in a particle size measurement cell (50 μL, High Precision Cell [ZEN2112]), and the intensity averaged size distribution and polydispersity (PDI) were measured by dynamic light scattering (DLS) using a Zetasizer Nano ZS (Malvern Instruments Co., Malvern, UK). Liposome suspensions ([cholesterol] = 4.6 mM, in PBS) were also diluted in sodium chloride solution (NaCl aq, 1.0 mM, in pure water) (liposome suspension: NaCl aq = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 49 [20 μL
49 [20 μL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 980 μL]) and the surface potential was measured using a Zetasizer Nano ZS.
980 μL]) and the surface potential was measured using a Zetasizer Nano ZS.
Analysis of interaction between Tat peptide and liposomes by a quartz crystal microbalance with dissipation monitoring (QCM-D)
Q-senseE4 (Biolin Scientific AB, Stockholm, Sweden) was used to conduct the QCM-D measurements. When any substance is adsorbed onto the sensor surface, the oscillator frequency (Δf) decreases. The change in the oscillator frequency can be used to calculate the amount of adsorbed material on a specific surface. In this study, liposome adsorption was investigated using QCM-D to study the interactions between liposomes and Tat.The gold sensor chip was sonicated with ethanol, methanol, and acetone for 5 min each and irradiated with UV/O3 for 15 min using an oxygen plasma machine (SKB401Y-02; Sun Energy Corporation, Osaka, Japan) to clean the surface of the sensor chips. The sensor chips were immersed overnight in a 1-dodecanethiol solution (1 mM in ethanol) to form a C12-self-assembled monolayer (SAM) of terminal CH3 on the substrate surface. The sensor chips were washed with ethanol to remove unreacted 1-dodecanethiol and then air dried. The sensor chip was placed in the QCM-D chamber and exposed to PBS (pH 7.4) until a stable baseline signal was obtained. Solutions of PEG(5k)-lipid(C16), Tat-PEG(5k), or Tat-PEG(5k)-lipid(C16) (50 μg mL−1 in PBS) were injected for 18 min, followed by washing with PBS for 8 min. Next, a solution of BSA (1 mg mL−1 in PBS) was injected for 8 min to block non-specific binding, followed by another PBS wash step. Liposome suspension ([cholesterol] = 1.8 mM, in PBS) was then injected, followed by washing with PBS. All measurements were performed at room temperature and the results are expressed as the average of at least three independent experiments. The adsorption capacity of each material was used from the resonance frequency change (Δf) and dissipation (ΔD) at the 7th overtone.27
Analysis of interaction of PEG-lipids with different lipid lengths by QCM-D
We studied the interaction of different PEG-lipids with different lipid lengths (lipid(C9), lipid(C12), lipid(C14), lipid(C16), and lipid(C18)) on the hydrophobic C12-SAM surface using QCM-D. A solution of PEG(5k)-lipids (lipid(C9), (C12), (C14), (C16), or (C18)) (50 μg mL−1 in PBS) was injected for 20 min. Then, PBS was allowed to flow for 30 min to observe the detachment of the PEG-lipids from the surface to evaluate stability. All measurements were performed at room temperature and the results are expressed as the average of at least three independent experiments.Analysis of liposomes modified with Tat-PEG-lipids
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 70 min at 4 °C, and the supernatant was completely removed. Then, a solution of Tat-PEG-lipid (50 μL, 1 mg mL−1, in PBS) was mixed with the liposome pellet and incubated for 1, 3, and 5 h at 37 °C. For comparison with each different concentration of Tat-PEG-lipids, a solution of Tat-PEG-lipid (50 μL, 0.2, 1.0, and 5.0 mg mL−1, in PBS) was mixed with the liposome pellet and incubated for 5 h at 37 °C. The diameter and zeta potential of Tat-PEG-lipid-treated liposomes were measured as described above. A mixed solution of Tat-PEG-lipid-treated liposomes was analyzed using a Zetasizer Nano ZS. Visible precipitate was present in liposome suspension when treated with Tat-PEG(5k)-lipids(C9,12). The intensity averaged size distribution and PDI data were obtained from liposome suspension including the visible aggregates.
000g for 70 min at 4 °C, and the supernatant was completely removed. Then, a solution of Tat-PEG-lipid (50 μL, 1 mg mL−1, in PBS) was mixed with the liposome pellet and incubated for 1, 3, and 5 h at 37 °C. For comparison with each different concentration of Tat-PEG-lipids, a solution of Tat-PEG-lipid (50 μL, 0.2, 1.0, and 5.0 mg mL−1, in PBS) was mixed with the liposome pellet and incubated for 5 h at 37 °C. The diameter and zeta potential of Tat-PEG-lipid-treated liposomes were measured as described above. A mixed solution of Tat-PEG-lipid-treated liposomes was analyzed using a Zetasizer Nano ZS. Visible precipitate was present in liposome suspension when treated with Tat-PEG(5k)-lipids(C9,12). The intensity averaged size distribution and PDI data were obtained from liposome suspension including the visible aggregates.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 70 min at 4 °C, and the supernatant was completely removed. Then, a solution of FITC-Tat-PEG-lipid (50 μL, 1 mg mL−1, in PBS) was mixed with the liposome pellet and incubated for 1 h at 37 °C. The liposome suspension was centrifuged (20
000g for 70 min at 4 °C, and the supernatant was completely removed. Then, a solution of FITC-Tat-PEG-lipid (50 μL, 1 mg mL−1, in PBS) was mixed with the liposome pellet and incubated for 1 h at 37 °C. The liposome suspension was centrifuged (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 70 min, 4 °C) and washed with PBS (1.0 mL), then the supernatant was taken out to remove free FITC-Tat-PEG(5k)-lipids and obtain the liposome pellet, which was then resuspended in PBS (80 μL). The resulting liposome suspension (40 μL) was diluted with PBS (3.0 mL). The fluorescence intensity of the liposome suspension was measured using a spectrofluorometer (FP-6500; JASCO Corp., Tokyo, Japan) (Ex: 488 nm, emission: 526 nm). We then calculated the number of Tat-PEG (5k)-lipid molecules incorporated into the liposomes from calibration using FITC-Tat.
000g, 70 min, 4 °C) and washed with PBS (1.0 mL), then the supernatant was taken out to remove free FITC-Tat-PEG(5k)-lipids and obtain the liposome pellet, which was then resuspended in PBS (80 μL). The resulting liposome suspension (40 μL) was diluted with PBS (3.0 mL). The fluorescence intensity of the liposome suspension was measured using a spectrofluorometer (FP-6500; JASCO Corp., Tokyo, Japan) (Ex: 488 nm, emission: 526 nm). We then calculated the number of Tat-PEG (5k)-lipid molecules incorporated into the liposomes from calibration using FITC-Tat.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 70 min at 4 °C, and the supernatant was completely removed. Then, a solution of Tat-PEG-lipid (50 μL, 1 mg mL−1, in PBS) was mixed with the liposome pellet and incubated for 1 h at 37 °C. The mixture was diluted with PBS (150 μL) for further experiments. The modified liposomes(CF) (200 μL, in PBS) were seeded into glass-bottom dishes and incubated for 30 min at room temperature. The dish was washed five times with PBS (1.0 mL) to remove unbound liposomes(CF) and the glass substrate surface was observed using a confocal laser scanning microscope (LSM700; Carl Zeiss Microscopy Co. Ltd, Jena, Germany). The acquired fluorescence images (five different pictures per sample for each experiment) were analyzed using image processing software (ImageJ version 1.52o, NIH, Bethesda, MD, USA). The average value of five images was calculated as the fluorescence intensity for each sample. Results are presented as the mean ± SD. In this study, we used glass-bottom dishes for observation under a confocal laser scanning microscope.
000g for 70 min at 4 °C, and the supernatant was completely removed. Then, a solution of Tat-PEG-lipid (50 μL, 1 mg mL−1, in PBS) was mixed with the liposome pellet and incubated for 1 h at 37 °C. The mixture was diluted with PBS (150 μL) for further experiments. The modified liposomes(CF) (200 μL, in PBS) were seeded into glass-bottom dishes and incubated for 30 min at room temperature. The dish was washed five times with PBS (1.0 mL) to remove unbound liposomes(CF) and the glass substrate surface was observed using a confocal laser scanning microscope (LSM700; Carl Zeiss Microscopy Co. Ltd, Jena, Germany). The acquired fluorescence images (five different pictures per sample for each experiment) were analyzed using image processing software (ImageJ version 1.52o, NIH, Bethesda, MD, USA). The average value of five images was calculated as the fluorescence intensity for each sample. Results are presented as the mean ± SD. In this study, we used glass-bottom dishes for observation under a confocal laser scanning microscope.
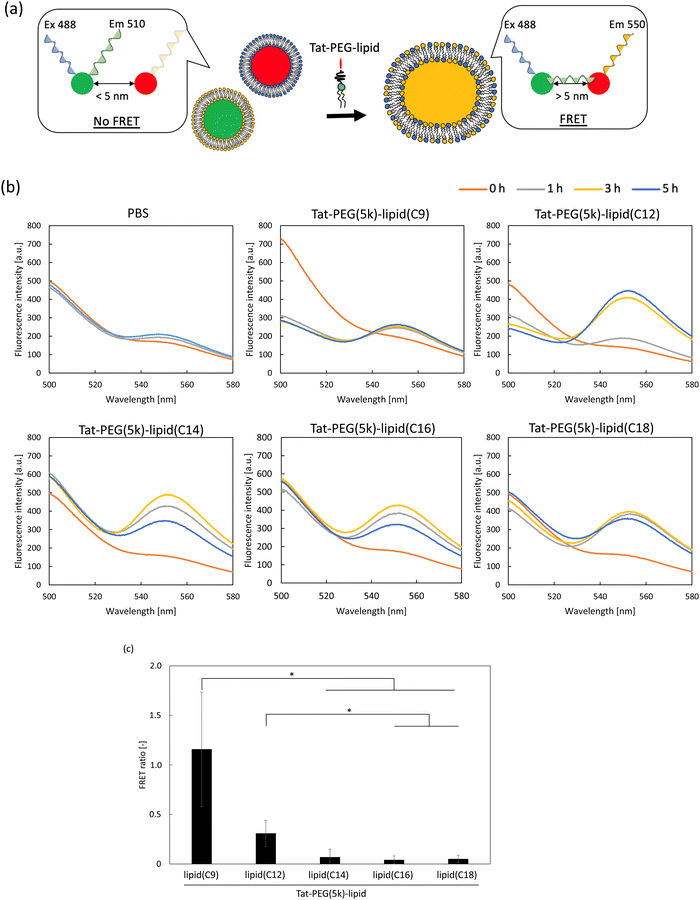
Fluorescence resonance energy transfer (FRET) measurement of liposomes treated with Tat-PEG(5k)-lipids
Liposome (CF) suspension (10 μL, [cholesterol] = 4.6 mM, in PBS) and liposome (R6G) suspension (10 μL, [cholesterol] = 4.6 mM, in PBS) were mixed and centrifuged (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 70 min, 4 °C), then the supernatant was removed to obtain a mixed liposome (CF) and liposome (R6G) pellet. A solution of Tat-PEG(5k)-lipid(lipid(C9), lipid(C12), lipid(C14), lipid(C16), lipid(C18), 50 μL, in PBS, 1.0 mg mL−1) was added into the pellet and incubated for 0, 1, 3, 5 h at 37 °C. The liposome suspension was diluted in PBS (450 μL). Fluorescence spectra (Em range: 500–580 nm) were measured by excited at 488 nm on a FP-6500 spectrofluorometer.
000g for 70 min, 4 °C), then the supernatant was removed to obtain a mixed liposome (CF) and liposome (R6G) pellet. A solution of Tat-PEG(5k)-lipid(lipid(C9), lipid(C12), lipid(C14), lipid(C16), lipid(C18), 50 μL, in PBS, 1.0 mg mL−1) was added into the pellet and incubated for 0, 1, 3, 5 h at 37 °C. The liposome suspension was diluted in PBS (450 μL). Fluorescence spectra (Em range: 500–580 nm) were measured by excited at 488 nm on a FP-6500 spectrofluorometer.
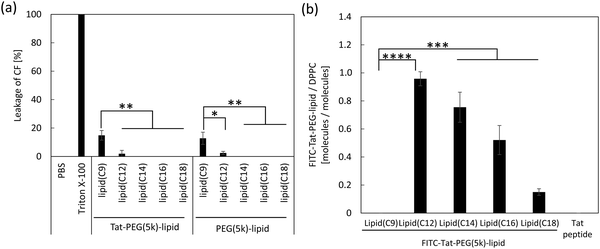
Measurement of released CF from liposomes treated with Tat-PEG-lipids
The CF released from the liposomes was analyzed before and after treatment with Tat-PEG(5k)-lipids to evaluate the liposome fusion.28 Liposomes encapsulating 50 mM CF (20 μL, [cholesterol] = 4.6 mM, in PBS) were centrifuged (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 70 min, 4 °C) to obtain a liposome(CF) pellet. The liposome(CF) pellet and Tat-PEG(5k)-lipid(lipid(C9), lipid(C12), lipid(C14), lipid(C16), lipid(C18)) solution (50 μL, in PBS, 1.0 mg mL−1) or Triton X-100 (for control) were mixed and incubated for 1 h at 37 °C. After incubation, the solution was diluted with PBS (950 μL). The fluorescence intensity of the solution was measured using a spectrofluorometer (excitation wavelength: 488 nm; emission wavelength: 517 nm).
000g for 70 min, 4 °C) to obtain a liposome(CF) pellet. The liposome(CF) pellet and Tat-PEG(5k)-lipid(lipid(C9), lipid(C12), lipid(C14), lipid(C16), lipid(C18)) solution (50 μL, in PBS, 1.0 mg mL−1) or Triton X-100 (for control) were mixed and incubated for 1 h at 37 °C. After incubation, the solution was diluted with PBS (950 μL). The fluorescence intensity of the solution was measured using a spectrofluorometer (excitation wavelength: 488 nm; emission wavelength: 517 nm).
Measurement of detached Tat-PEG-lipids form liposomes
Liposome suspension (20 mL, [cholesterol] = 4.6 mM, in PBS) was centrifuged at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 70 min at 4 °C, and the supernatant was completely removed. Then, a solution of FITC-Tat-PEG-lipid (50 μL, 1 mg mL−1, in PBS) was mixed with the liposome pellet and incubated for 1 h at 37 °C. The fluorescence intensity of the solution was measured using a spectrofluorometer (excitation wavelength: 488 nm; emission wavelength: 517 nm).
000g for 70 min at 4 °C, and the supernatant was completely removed. Then, a solution of FITC-Tat-PEG-lipid (50 μL, 1 mg mL−1, in PBS) was mixed with the liposome pellet and incubated for 1 h at 37 °C. The fluorescence intensity of the solution was measured using a spectrofluorometer (excitation wavelength: 488 nm; emission wavelength: 517 nm).
Cell culture
CCRF-CEM were cultured in RPMI-1640 medium supplemented with 10% FBS, 50 U mL−1 penicillin, and 50 μg mL−1 streptomycin at 37 °C in an atmosphere of 5% CO2 and 95% air. ADSCs were cultured in the complete medium containing FBS, at 37 °C in an atmosphere of 5% CO2 and 95% air.Cell fusion of CCRF-CEMs and ADSCs with Tat-PEG-lipids
A cell suspension of CCRF-CEM (1 mL, 1.0 × 105 cells) and DiO solution (5 μL) were mixed and then incubated at 37 °C for 15 min for the labeling. The resulting cells were washed with culture medium (9 mL) by centrifugation (180 × g, 3 min) to remove free DiO. We also labeled ADSCs with DiI in the same way. Then, we collected those cell pellets and well mixed the labeled CCRF-CEM and ADSCs. A solution of Tat-PEG(5k)-lipid (lipid(C9), lipid(C12), 50 μL, in PBS, 1.0 mg mL−1) or PBS (as a control) was added into the mixed cell pellet and then incubated for 6 h at 37 °C. The cells were observed by confocal laser scanning microscopy at 0, 3, and 6h. For observation of adherent fused cells at 24 h by confocal laser scanning microscopy, 1.5 mL of culture medium was added to the cell suspension and then seeded on the glass-bottom dish, and then incubated for 18 h at 37 °C. After 18 h seeding, the adhered fusion cells were washed with PBS to remove unfused cells. Adhered fusion cells were fixed in 4% paraformaldehyde for 30 min at 37 °C, followed by two washes with PBS. The fixed cells were treated with DAPI (7 mL, in PBS, 1 μg mL−1) for 30 min at 37 °C, followed by washing with PBS. The cells were observed under a confocal microscope.Cell viability assay
Cytotoxicity was evaluated by the MTT assay using a Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer's instructions. The cell suspension of CCRF-CEM and ADSCs was seeded on 96-well plates at a density of 2 × 105 cells per well and added a solution of Tat-PEG(5k)-lipid(lipid(C9), lipid(C12)) with each different concentration to each final concentration (100 μL, in PBS, 0.1, 0.5, and 1.0 mg mL−1) and then incubated for 6 h at 37 °C. After 6 h incubation, Cell Counting Kit-8 solution was added, and the cells were incubated for 2 h at 37 °C. The absorbance at 450 nm was measured by a plate reader.Statistical analysis
All results are repeated at least three times and are presented as means ± standard deviations. For statistical calculations, we used one-way ANOVA, followed by Dunnett's multiple comparison test, performed using GraphPad Prism 8 for MacOS version 8.4.2 (GraphPad Software, La Jolla, CA, USA). The cutoff for statistical difference was p < 0.05, with levels of significance indicated as follows: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.Results and discussion
Analysis of interaction between Tat peptide and liposomes by QCM-D
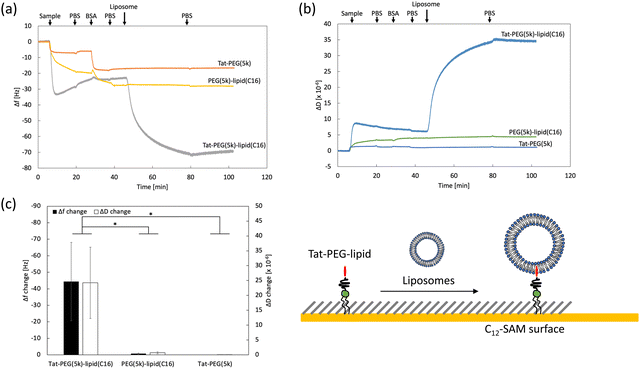
The Tat peptide, a representative CPP, was covalently conjugated to maleimide-PEG-lipids with different PEG chains (1k, 5k, and 40 kDa) and lipid lengths (C9, C12, C14, C16, and C18); these conjugates were used for this study (Scheme 1).We previously evaluated the interactions of PEG-lipid derivatives with ligands by QCM-D using a hydrophobic C12-self-assembled monolayer (SAM) surface.29 Here, we studied the interaction between Tat-PEG-lipid and liposomes by measuring Δf and ΔD, where Tat-PEG-lipid(C16) was selected because the lipid (C16) was stably anchored on the SAM (Fig. S1, ESI†). First, we treated the sensor surface with Tat-PEG-lipids, followed by blocking with BSA and liposomes. As controls, we also used PEG-lipids without the Tat peptide and Tat-PEG without lipids (Fig. 1 and Fig. S6, S7, ESI†). When we flowed the Tat-PEG-lipid onto the hydrophobic SAM surface, there was rapid and large binding of the Tat-PEG-lipid by hydrophobic interactions with the lipid domain and Tat peptide (Fig. 1a). We also detected substantial binding of liposomes onto immobilized Tat-PEG-lipid, in which the liposome was presumably bound by the interaction between the Tat peptide and lipid membrane. A large change in ΔD during liposome binding indicated soft and highly viscoelastic properties, which also suggests liposome binding (Fig. 1b and c). Because the Tat peptide is membrane-permeable, liposomes can be anchored to the immobilized Tat peptides.
When we used PEG-lipid without the Tat peptide as a control, a similar binding of PEG-lipid was observed. However, the initial binding rate was slower than that of Tat-PEG-lipid (Fig. 1a). Because there was almost no difference in the binding amount between them, the hydrophobic interaction between the lipid domain and the SAM surface was mainly involved, but not the Tat peptide. No liposome binding was detected on the PEG-lipid-treated surface, indicating that there was no interaction between the liposomes and the PEG chain. When we used Tat-PEG without the lipid domain as another control, less binding between Tat-PEG than both Tat-PEG-lipid and PEG-lipid, and no binding of liposomes to the treated surface was observed (Fig. 1a). This indicates that the Tat peptide did not interact with the SAM surface. Thus, we found that the Tat peptide interacted with the lipid membrane and could be anchored to liposomes. Therefore, Tat-PEG-lipid can interact with the lipid membrane via both the lipid domain and the Tat peptide, where Tat-PEG-lipid can act as a bridge between liposomes.
Analysis of interaction of Tat-PEG-lipid with liposome by DLS and zeta potential measurement
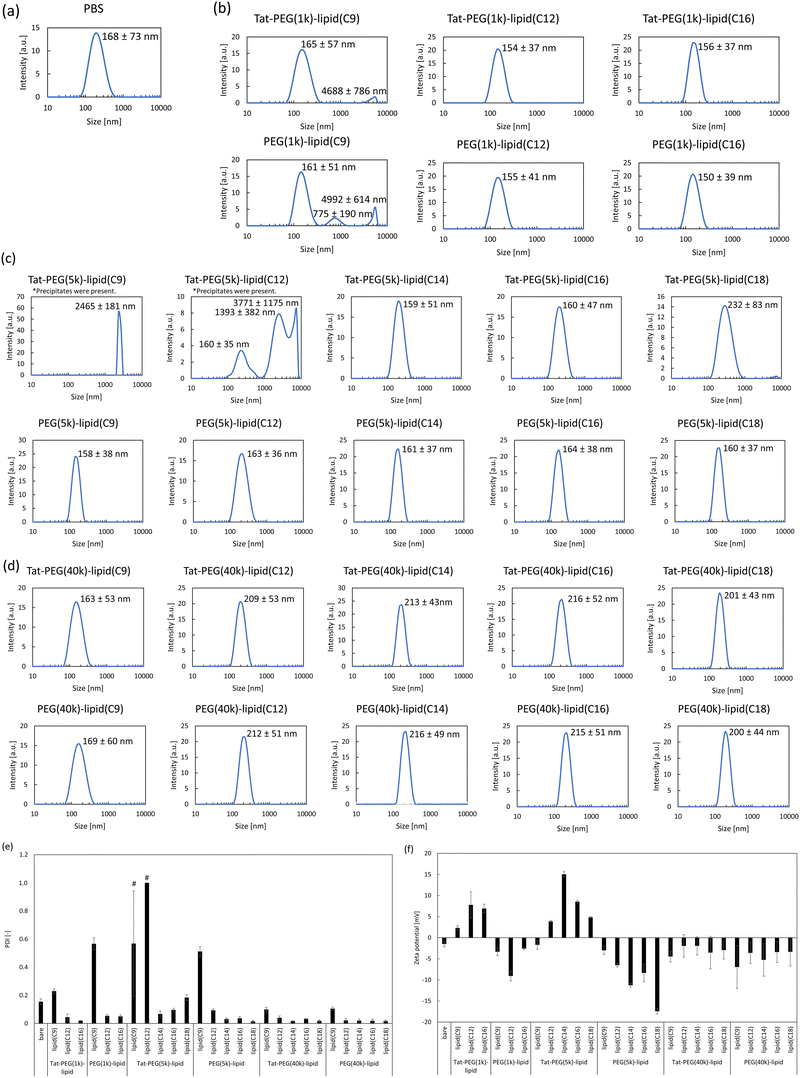
Next, we mixed Tat-PEG-lipids with the liposomes and measured their intensity averaged size distribution, PDI, and zeta potential (Fig. 2 and Fig. S4, ESI†). Here, we used Tat-PEG-lipids with different PEG chains (1k, 5k, and 40 kDa) and lipid lengths (C9, C12, C14, C16, and C18). We incubated the Tat-PEG-lipids with liposomes (DPPC/cholesterol = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, by molar ratio) for 1 h at 37 °C, and also used PEG-lipids without Tat peptide as a control. When we incubated liposomes with the Tat-PEG(5k)-lipid(C9) (0.2 mg mL−1), no change was detected in the liposome size. On the other hand, when we incubated liposomes with the Tat-PEG(5k)-lipid(C9) with 1.0 and 5.0 mg mL−1, the liposome size increased although there was no significant difference between those concentrations (Fig. S4, ESI†). Therefore, we decided to use 1.0 mg mL−1 for further experiments. No change was detected in the liposome size during incubation with Tat-PEG-lipids and PEG-lipids, except for Tat-PEG(5k)-lipid(C9), Tat-PEG(5k)-lipid(C12), and PEG(5k)-lipid(C9) (Fig. 2a–d). For Tat-PEG(1k, 40k)-lipid, the size of the modified liposome could not increase because the spontaneous liposome attachment hardly occurred due to the inappropriate spacer length of the 1kDa and 40 kDa PEG. However, when we incubated liposomes with the Tat-PEG(5k)-lipid(C9), Tat-PEG(5k)-lipid(C12), and PEG(5k)-lipid(C9), the liposome size greatly increased; in particular, the size of liposome treated with Tat-PEG(5k)-lipid(C9) showed a dramatic increase up to several micron meter levels. We also observed the treated liposomes(CF) using confocal laser scanning microscopy. Large liposomes were observed for Tat-PEG(5k)-lipid(C9) and Tat-PEG(5k)-lipid(C12), whereas hardly any large liposomes were detected in the other groups (Fig. S2, ESI†). A similar tendency was observed for PDI, in which liposomes treated with Tat-PEG(5k)-lipid(C9), Tat-PEG(5k)-lipid(C12), and PEG(5k)-lipid(C9) showed an increase in PDI, although some other lipids also showed an increase (Fig. 2e). These results suggest the aggregation and/or fusion of liposomes upon the addition of Tat-PEG(5k)-lipid(C9), Tat-PEG(5k)-lipid(C12), and PEG(5k)-lipid(C9).
1, by molar ratio) for 1 h at 37 °C, and also used PEG-lipids without Tat peptide as a control. When we incubated liposomes with the Tat-PEG(5k)-lipid(C9) (0.2 mg mL−1), no change was detected in the liposome size. On the other hand, when we incubated liposomes with the Tat-PEG(5k)-lipid(C9) with 1.0 and 5.0 mg mL−1, the liposome size increased although there was no significant difference between those concentrations (Fig. S4, ESI†). Therefore, we decided to use 1.0 mg mL−1 for further experiments. No change was detected in the liposome size during incubation with Tat-PEG-lipids and PEG-lipids, except for Tat-PEG(5k)-lipid(C9), Tat-PEG(5k)-lipid(C12), and PEG(5k)-lipid(C9) (Fig. 2a–d). For Tat-PEG(1k, 40k)-lipid, the size of the modified liposome could not increase because the spontaneous liposome attachment hardly occurred due to the inappropriate spacer length of the 1kDa and 40 kDa PEG. However, when we incubated liposomes with the Tat-PEG(5k)-lipid(C9), Tat-PEG(5k)-lipid(C12), and PEG(5k)-lipid(C9), the liposome size greatly increased; in particular, the size of liposome treated with Tat-PEG(5k)-lipid(C9) showed a dramatic increase up to several micron meter levels. We also observed the treated liposomes(CF) using confocal laser scanning microscopy. Large liposomes were observed for Tat-PEG(5k)-lipid(C9) and Tat-PEG(5k)-lipid(C12), whereas hardly any large liposomes were detected in the other groups (Fig. S2, ESI†). A similar tendency was observed for PDI, in which liposomes treated with Tat-PEG(5k)-lipid(C9), Tat-PEG(5k)-lipid(C12), and PEG(5k)-lipid(C9) showed an increase in PDI, although some other lipids also showed an increase (Fig. 2e). These results suggest the aggregation and/or fusion of liposomes upon the addition of Tat-PEG(5k)-lipid(C9), Tat-PEG(5k)-lipid(C12), and PEG(5k)-lipid(C9).
In addition, we studied the time-course change of liposome size during the fusion process by Tat-PEG-lipids (Fig. S3, S8 and S9, ESI†). Apparently, the size increase was detected till 3 h and no further reaction occurred till 5 h for the case with Tat-PEG(5k)-lipid(C9), Tat-PEG(5k)-lipid(C12). We considered that while more liposomes fused with time, the probability of collision frequency of liposomes gradually decreased, and then finally the reaction stopped.
Because the Tat peptide (YGRKKRRQRRR) has cationic arginine residues in its sequence, the modified liposome should be positively charged. In contrast, bare and PEG-lipid-modified liposomes are negatively charged.24 When we measured the zeta potential of liposomes mixed with control PEG(1k, 5k, 40k)-lipids, a negative charge was observed, indicating the PEGylation of liposomes (Fig. 2f). In contrast, the zeta potential of the liposomes was positively charged in the case of Tat-PEG(1k, 5k)-lipids, except for Tat-PEG(5k)-lipid(C9) and Tat-PEG(5k)-lipid(C12). We assumed that the zeta potential was almost zero (or neutral) because of the rapid detachment of Tat-PEG(5k)-lipid(C9) and Tat-PEG(5k)-lipid(C12) from the liposome surface, whereas other Tat-PEG(5k)-lipids remained on the liposome surface and showed a positive charge. For Tat-PEG(1k)-lipids, the zeta potential of the modified liposomes was positive (Fig. 2f), indicating that the liposome surface was modified with Tat-PEG(1k)-lipids. However, no change was detected in liposome size treated with Tat-PEG(1k)-lipids. Therefore, we considered that the PEG(1k) spacer was too short to reach and interact with liposomes. For Tat-PEG(40k)-lipids, the zeta potential of the modified liposomes was negative (Fig. 2f). This was because the spontaneous incorporation of Tat-PEG(40k)-lipids hardly occurred due to the large extended volume effect of the 40 kDa PEG. Therefore, Tat-PEG(40k)-lipids could not induce the aggregation and fusion of liposomes.
Thus, we assumed that the aggregation and/or fusion of liposomes was induced by mixing Tat-PEG(5k)-lipid(C9) and Tat-PEG(5k)-lipid(C12) with liposomes, in which Tat-PEG(5k)-lipid(C9) and Tat-PEG(5k)-lipid(C12) could interact but rapidly detach from the liposome surface. In fact, when we studied the detachment of PEG(5k)-lipids with different lipid lengths (C9, C12, C14, C16, and C18) from the hydrophobic SAM surface using QCM-D, Tat-PEG(5k)-lipid(C9) and Tat-PEG(5k)-lipid(C12) showed rapid detachment, which supported the detachment of these two Tat-PEG-lipids from the liposome surface (Fig. S1, ESI†).
Observation of liposome morphology by TEM
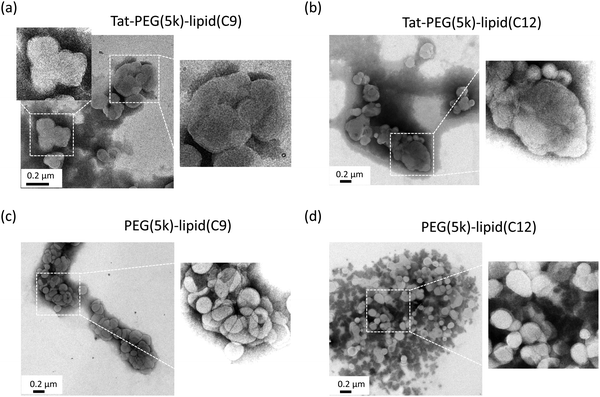
Next, we observed the modified liposomes using TEM to analyze whether they were aggregated or fused after treatment with Tat-PEG(5k)-lipid(C9) and Tat-PEG(5k)-lipid(C12) (Fig. 3). In this study, PEG (5k)-lipid (C9) and PEG (5k)-lipid (C12) without the Tat peptide were used as controls. When the liposomes were treated with the control PEG(5k)-lipid(C9) and PEG(5k)-lipid(C12), liposomes with a diameter of approximately 100 nm were clearly observed. However, there were some aggregations due to artifacts during sample preparation for TEM observation. Because the liposomes were PEGylated by the spontaneous insertion of PEG(5k)-lipid(C9) and PEG(5k)-lipid(C12), they were still intact. This result is consistent with that of the DLS analysis (Fig. 2). In contrast, liposomes larger than 100 nm were observed for both Tat-PEG(5k)-lipid(C9) and Tat-PEG(5k)-lipid(C12), which seemed to be fused but not aggregated. The border between each liposome in the large aggregates could not be observed, but they were apparently larger single liposomes. Some smaller liposomes were observed, which were closely located with large liposomes. Therefore, we assumed that the large liposomes were the result of small liposomes fusing. This result was also consistent with the size distribution date of the DLS analysis (Fig. 2). Therefore, we decided to fuse the liposome membrane by adding Tat-PEG(5k)-lipid(C9) and Tat-PEG(5k)-lipid(C12) to the liposomes.Analysis of membrane fusion by FRET
To analyze membrane fusion, we used FRET (Fig. 4). Here, we used two types of liposomes encapsulating different fluorescent dyes, 5(6)-carboxyfluorescein (CF) and rhodamine 6G (R6G), and studied the FRET during the process by adding Tat-PEG(5k)-lipids (Fig. 4a and b). When we added PBS to a suspension of the two labeled liposomes as a control, a small increase in fluorescence was observed at approximately 550 nm. Although each fluorescent dye was separately encapsulated within the liposomes, the fluorescence emitted from CF could excite R6G inside another liposome because the lipid membrane thickness was approximately 5 nm. However, this was not considered as an energy transfer, since there was no direct interaction between the two fluorescent dyes. In contrast, when Tat-PEG(5k)-lipid(C9) and Tat-PEG(5k)-lipid(C12) were added to the liposomal suspension, a rapid increase in fluorescence at approximately 550 nm and a decrease in fluorescence at approximately 500 nm were observed. We assumed that this was mainly due to FRET between CF and R6G inside the fused liposomes. In the case of other Tat-PEG-lipids, such as Tat-PEG(5k)-lipid(C14), Tat-PEG(5k)-lipid(C16), and Tat-PEG(5k)-lipid(C18), only an increase in fluorescence was observed at approximately 550 nm, whereas no decrease in fluorescence was observed at approximately 500 nm, which was more pronounced than in the PBS control group. We assumed that the fluorescence emitted from CF could excite R6G inside another liposome, which was in closer contact with each other owing to the Tat-PEG-lipid, resulting in greater fluorescence detection. However, there was no direct interaction between the two fluorescent dyes, because no decrease in fluorescence was observed at approximately 500 nm. When we calculated the FRET efficiency (Fig. 4c), a higher ratio was observed for the liposomes treated with Tat-PEG(5k)-lipid(C9) and Tat-PEG(5k)-lipid(C12) than for the others. Based on these results, we hypothesized that membrane fusion was induced by Tat-PEG(5k)-lipid(C9) and Tat-PEG(5k)-lipid(C12).Evaluation of membrane leakage of liposome and stability by Tat-PEG-lipid
To evaluate membrane leakage, we directly measured the fluorescence intensity of CF released from liposome(CF) after the addition of Tat-PEG-lipids to the liposome suspension. Because the fluorescence from CF was quenched inside the liposome owing to its high concentration, no fluorescence was detected (PBS in Fig. 5a). In contrast, when we lysed the liposome(CF) with Triton X-100, CF was released and emitted fluorescence (Triton X-100 in Fig. 5a). When Tat-PEG(5k)-lipid(C9) and Tat-PEG(5k)-lipid(C12) were added to liposome(CF), some CF leakage was detected in both groups. In particular, highly increased leakage was detected with the addition of Tat-PEG(5k)-lipid(C9). In contrast, no leakage was observed in the other Tat-PEG-lipid groups. The same tendency was observed for the PEG-lipid groups without the Tat peptide, in which PEG(5k)-lipid(C9) and PEG(5k)-lipid(C12) showed a higher leakage of CF from the liposome(CF). This result indicated that shorter lipid domains (C9 and C12) influenced membrane integrity and destroyed the membrane structure by spontaneous insertion into liposomes. Thus, membrane destruction by Tat-PEG-lipids may involve membrane fusion.We also measured the fluorescence intensity of liposomes after treatment with FITC-labeled Tat-PEG(5k)-lipids to analyze their stability on the liposome membrane (Fig. 5b). We mixed a solution of FITC-Tat-PEG-lipids with the liposome suspension for 1 h and measured the fluorescence intensity after washing by centrifugation. The amount of FITC-Tat-PEG(5k)-lipids incorporated into liposomes increased with shorter acyl chain lengths of the lipids, except for FITC-Tat-PEG(5k)-lipid(C9) (Fig. 5b). However, almost no fluorescence was detected in liposomes treated with Tat-PEG(5k)-lipids (C9). We also measured the CMC of Tat-PEG(5k)-lipids because it is strongly involved in spontaneous incorporation into liposomes, as previously reported.24 The increase in the amount of FITC-Tat-PEG(5k)-lipids could be due to an increase in CMC, in which more monomeric FITC-Tat-PEG(5k)-lipids could interact with the liposome surface, resulting in greater incorporation of Tat-PEG(5k)-lipids. However, FITC-Tat-PEG(5k)-lipid(C9) was not detected in the treated liposomes, although its CMC was the highest among the Tat-PEG(5k)-lipids. When we consider the dissociation of Tat-PEG(5k)-lipids from the liposome surface, the detachment rate should increase with an increase in CMC because fewer hydrophobic interactions occur with the lipid membrane. Therefore, FITC-Tat-PEG(5k)-lipid(C9) could be more rapidly incorporated into the liposome surface, but at the same time, it also quickly detached from the surface, whereas FITC-Tat-PEG(5k)-lipid(C9) was anchored to the liposomes via the Tat peptide and lipid domain to induce membrane fusion.
Thus, we found that Tat-PEG(5k)-lipid(C9) induced membrane fusion of liposomes more efficiently than the other Tat-PEG(5k)-lipids. Liposomes are brought together via Tat-PEG(5k)-lipids, in which the Tat peptide and lipid domains are anchored to the membranes. Once the two membranes are in close contact, lipid exchange can occur between the two membranes to induce membrane fusion, while the Tat-PEG(5k)-lipid detaches from the surface. Therefore, lipid exchange was not inhibited in the presence of Tat-PEG(5k)-lipid anchored between the lipid membranes.
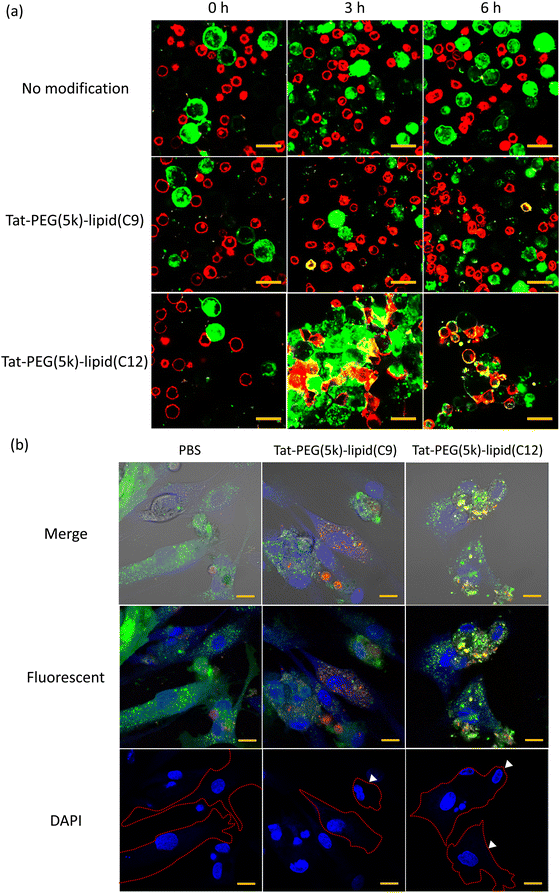
Cell fusion of ADSCs and CCRF-CEM by Tat-PEG-lipid
To study membrane fusion of living cells by Tat-PEG-lipid, we used two different cells (ADSCs and CCRF-CEM) (Fig. 6). When Tat-PEG(5k)-lipid(C12) was added to mixed cell suspension, the cell–cell attachment started to occur immediately (Fig. 6a), and the cell aggregation was observed during 6 h. Also, we could observe yellow areas at the interface between two cells at 3 h, which indicated the membrane fusion of ADSCs (green) and CCRF-CEM (red). This membrane fusion was more pronounced 6 h after Tat-PEG(5k)-lipid(C12) was added. On the other hand, no cell–cell attachment nor membrane fusion was observed in the control group, in which PBS was added into the mixed cell suspension of ADSCs and CCRF-CEM. In the case of adding Tat-PEG(5k)-lipid(C9) into mixed cell suspension, membrane fusion hardly occurred, which was a different result from the liposome study. Also, we stained the nucleic acids with DAPI (blue) to observe the adhered fusion cells (Fig. 6b). While no fusion of cells was observed for adherent cells treated with PBS, the adherent fusion cells were observed when treatment with Tat-PEG(5k) lipid(C9, 12), and there were multiple nuclei observed in some cells (arrows in Fig. 6b). The cell morphology after treatment with Tat-PEG(5k)-lipid(C9, 12) differed from the control cells which were treated with PBS. The cells treated with Tat-PEG(5k)-lipid(C12) adhered but did not spread well, keeping round on the substrate, indicating the influence of cell fusion with floating cells, CCRF-CEM. The cell viability was influenced by interaction with Tat-PEG-lipids during the cell fusion (Fig. S5, ESI†). The cell viability decreased with an increase of Tat-PEG(5k)-lipid(C9, C12) concentration. This was because the hydrophobic lipid part destroyed the cellular membrane integrity. On the other hand, the interaction between Tat-PEG(5k)-lipid(C9, C12) and cellular membrane could also induce cell fusion. Therefore, this is a tradeoff relationship.Thus, we found that Tat-PEG(5k)-lipid(C12) could induce the strong cell–cell attachment and subsequent membrane fusion between ADSCs and CCRF-CEM more efficiently.
Conclusion
Tat-PEG(5k)-lipid(C9) and Tat-PEG(5k)-lipid(C12) were spontaneously incorporated into bilayer membrane, where the Tat peptide and lipid domain interacted with the lipid membrane to anchor the neighboring bilayer membranes. Lipid exchange easily occurs to induce membrane fusion during close contact between lipid membranes and the detachment of Tat-PEG(5k)-lipid(C9) and Tat-PEG(5k)-lipid(C12). The detachment of Tat-PEG(5k)-lipid(C9) and Tat-PEG(5k)-lipid(C12) by weak hydrophobic interactions with the lipid membrane could result in the spontaneous membrane fusion.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This research was supported in part by a Grant-in-Aid for Scientific Research (B) (18H03528, 22H03966) and a Grant-in-Aid for JSPS Fellows (24KJ0722) from the JSPS KAKENHI, a Grant-in-Aid for Scientific Research for Fostering Joint International Research (18KK0305) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan, and JST SPRING (JPMJSP2108). We would like to thank Ms Fumiko Hosaka for cell fusion experiments.References
- G. Galfre and C. Milstein, Preparation of monoclonal antibodies: strategies and procedures, Methods Enzymol., 1981, 73, 3–46 CrossRef CAS PubMed.
- P. S. Carlson, H. H. Smith and R. D. Dearing, Parasexual interspecific plant hybridization, Proc. Natl. Acad. Sci. U. S. A., 1972, 69, 2292–2294 CrossRef CAS PubMed.
- M. Tada, Y. Takahama, K. Abe, N. Nakatsuji and T. Tada, Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells, Curr. Biol., 2001, 11, 1553–1558 CrossRef CAS PubMed.
- G. Yanai, T. Hayashi, Q. Zhi, K. C. Yang, Y. Shirouzu, T. Shimabukuro, A. Hiura, K. Inoue and S. Sumi, Electrofusion of mesenchymal stem cells and islet cells for diabetes therapy: a rat model, PLoS One, 2013, 8, e64499 CrossRef CAS PubMed.
- X. Wang, H. Willenbring, Y. Akkari, Y. Torimaru, M. Foster, M. Al-Dhalimy, E. Lagasse, M. Finegold, S. Olson and M. Grompe, Cell fusion is the principal source of bone-marrow-derived hepatocytes, Nature, 2003, 422, 897–901 CrossRef CAS PubMed.
- Q. L. Ying, J. Nichols, E. P. Evans and A. G. Smith, Changing potency by spontaneous fusion, Nature, 2002, 416, 545–548 CrossRef CAS PubMed.
- Y. Okada, Sendai virus-induced cell fusion, Methods Enzymol., 1993, 221, 18–41 CAS.
- M. M. Lo, T. Y. Tsong, M. K. Conrad, S. M. Strittmatter, L. D. Hester and S. H. Snyder, Monoclonal antibody production by receptor-mediated electrically induced cell fusion, Nature, 1984, 310, 792–794 CrossRef CAS PubMed.
- Q. F. Ahkong, D. Fisher, W. Tampion and J. A. Lucy, Mechanisms of cell fusion, Nature, 1975, 253, 194–195 CrossRef CAS PubMed.
- K. Honda, Y. Maeda, S. Sasakawa, H. Ohno and E. Tsuchida, The components contained in polyethylene glycol of commercial grade (PEG-6,000) as cell fusogen, Biochem. Biophys. Res. Commun., 1981, 101, 165–171 CrossRef CAS PubMed.
- G. Stenbeck, Soluble NSF-attachment proteins, Int. J. Biochem. Cell Biol., 1998, 30, 573–577 CrossRef CAS PubMed.
- J. E. Rothman, Mechanisms of intracellular protein transport, Nature, 1994, 372, 55–63 CrossRef CAS PubMed.
- F. Gao, D. Yang, F. Xu, X. Ma and P. Wang, Promoting Cell Fusion by Polyvalent DNA Ligands, Nano Lett., 2022, 22, 3018–3025 CrossRef CAS PubMed.
- A. Yoshihara, S. Watanabe, I. Goel, K. Ishihara, K. N. Ekdahl, B. Nilsson and Y. Teramura, Promotion of cell membrane fusion by cell-cell attachment through cell surface modification with functional peptide-PEG-lipids, Biomaterials, 2020, 253, 120113 CrossRef CAS PubMed.
- C. Y. Chang, J. A. Tai, Y. Sakaguchi, T. Nishikawa, Y. Hirayama and K. Yamashita, Enhancement of polyethylene glycol-cell fusion efficiency by novel application of transient pressure using a jet injector, FEBS Open Bio., 2023, 13, 478–489 CrossRef CAS PubMed.
- A. Bahadori, L. B. Oddershede and P. M. Bendix, Hot-nanoparticle-mediated fusion of selected cells, Nano Res., 2017, 10, 2034–2045 CrossRef CAS.
- S. Asif, K. N. Ekdahl, K. Fromell, E. Gustafson, A. Barbu, K. Le Blanc, B. Nilsson and Y. Teramura, Heparinization of cell surfaces with short peptide-conjugated PEG-lipid regulates thromboinflammation in transplantation of human MSCs and hepatocytes, Acta Biomater., 2016, 35, 194–205 CrossRef CAS PubMed.
- Y. Teramura, S. Asif, K. N. Ekdahl, E. Gustafson and B. Nilsson, Cell Adhesion Induced Using Surface Modification with Cell-Penetrating Peptide-Conjugated Poly(ethylene glycol)-Lipid: A New Cell Glue for 3D Cell-Based Structures, ACS Appl. Mater. Interfaces, 2017, 9, 244–254 CrossRef CAS PubMed.
- Y. Teramura and H. Iwata, Cell surface modification with polymers for biomedical studies, Soft Matter, 2010, 6, 1081–1091 RSC.
- M. Noiri, K. Asawa, N. Okada, T. Kodama, Y. Murayama, Y. Inoue, K. Ishihara, K. N. Ekdahl, B. Nilsson and Y. Teramura, Modification of human MSC surface with oligopeptide-PEG-lipids for selective binding to activated endothelium, J. Biomed. Mater. Res. A, 2019, 107, 1779–1792 CrossRef CAS PubMed.
- J. R. Litowski and R. S. Hodges, Designing heterodimeric two-stranded alpha-helical coiled-coils: the effect of chain length on protein folding, stability and specificity, J. Pept. Res., 2001, 58, 477–492 CrossRef CAS PubMed.
- S. Futaki, Membrane-permeable arginine-rich peptides and the translocation mechanisms, Adv. Drug Delivery Rev., 2005, 57, 547–558 CrossRef CAS PubMed.
- M. Noiri, Y. Goto, Y. Sato, N. Nakamura, K. Ishihara and Y. Teramura, Exogenous Cell Surface Modification with Cell Penetrating Peptide-Conjugated Lipids Causes Spontaneous Cell Adhesion, ACS Appl. Bio Mater., 2021, 4, 4598–4606 CrossRef CAS PubMed.
- Y. Sato, K. Asawa, T. Huang, M. Noiri, N. Nakamura, K. N. Ekdahl, B. Nilsson, K. Ishihara and Y. Teramura, Induction of Spontaneous Liposome Adsorption by Exogenous Surface Modification with Cell-Penetrating Peptide-Conjugated Lipids, Langmuir, 2021, 37, 9711–9723 CrossRef CAS PubMed.
- S. Asif, K. Asawa, Y. Inoue, K. Ishihara, B. Lindell, R. Holmgren, B. Nilsson, A. Ryden, M. Jensen-Waern, Y. Teramura and K. N. Ekdahl, Validation of an MPC Polymer Coating to Attenuate Surface-Induced Crosstalk between the Complement and Coagulation Systems in Whole Blood in In Vitro and In Vivo Models, Macromol. Biosci., 2019, 19, e1800485 CrossRef PubMed.
- K. Ishihara, Revolutionary advances in 2-methacryloyloxyethyl phosphorylcholine polymers as biomaterials, J. Biomed. Mater. Res., Part A, 2019, 933–943 CrossRef CAS PubMed.
- Y. Teramura and M. Takai, Quartz Crystal Microbalance, The Surface Science Society of Japan, Compendium of Surface and Interface Analysis, Springer Singapore, Singapore, 2018, pp. 509–520 Search PubMed.
- S. Takeoka, N. Kimura, H. Ohno and E. Tsuchida, Polymerization of Liposomes Evaluated from Molecular Weight Distribution of Diene-Type Phospholipid Polymers, Polym. J., 1990, 22, 867–874 CrossRef CAS.
- Y. Teramura, K. Kuroyama and M. Takai, Influence of molecular weight of PEG chain on interaction between streptavidin and biotin-PEG-conjugated phospholipids studied with QCM-D, Acta Biomater., 2016, 30, 135–143 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ma00193a |
| This journal is © The Royal Society of Chemistry 2024 |