Open Access Article
Open Access ArticleInduced chiroptical properties of helical Eu(III) complex by electrostatic interaction with DNA†
Ziying
Li
 a,
Nana
Hitomi
a,
Hideyuki
Tanaka
b,
Hitomi
Ohmagari
b,
Kazuki
Nakamura
a,
Nana
Hitomi
a,
Hideyuki
Tanaka
b,
Hitomi
Ohmagari
b,
Kazuki
Nakamura
 a,
Miki
Hasegawa
a,
Miki
Hasegawa
 b and
Norihisa
Kobayashi
b and
Norihisa
Kobayashi
 *a
*a
aGraduate School of Engineering, Chiba University, 1-33 Yayoi-cho, Inage-ku, Chiba, 263-8522, Japan. E-mail: koban@faculty.chiba-u.jp
bCollege of Science and Engineering, Aoyama Gakuin University, 5-10-1 Fuchinobe, Chuo-ku, Sagamihara, Kanagawa 252-5258, Japan. E-mail: hasemiki@chem.aoyama.ac.jp
First published on 14th November 2023
Abstract
Circularly polarized luminescence (CPL) has significantly increased the interest in biological fields. In this research, a water-soluble Eu(III) complex with a helical complex structure, EuLCOOH, was incorporated in chiral DNA in aqueous solutions. The photoluminescence performance of this DNA/EuLCOOH hybrid system was investigated. Compared to EuLCOOH alone, emission intensity and emission lifetime were effectively improved in the presence of DNA. The major binding between EuLCOOH and DNA was proven to be the electrostatic interaction. Owing to this interaction, the chiral environment provided by DNA successfully induced CPL from EuLCOOH.
Introduction
The high prevalence and importance of chirality in biological fields have always been of great interest to scientists. In particular, the chiroptical property of circularly polarized luminescence (CPL) provides a rich resource of information on the chiral environment of molecules. CPL has received significant attraction in a wide range of biological applications such as bio-probes, biosensors and bio-imaging.1–7 CPL spectroscopy measures the luminescence difference between left and right circularly polarized lights. The degree of CPL is normally evaluated by the emission dissymmetry factor, glum = 2(IL − IR)/(IL + IR), where IL (IR) is the intensity of the left (right) CPL. Theoretically, glum is defined as glum = 4(|m|/|μ|) cosτ, where m and μ are magnetic and electric dipole transition moments, respectively, and τ is the angle between them.8 Therefore, a higher glum value prefers luminophores with magnetic dipole allowed and electric dipole forbidden transitions. Thus, lanthanide complexes have become the most potential candidates for CPL activity in the development of emission systems.3,9–11Notably, only the lanthanide ions located in a chiral environment are expected for CPL activity. This is because the dissymmetry in the ligand field of the chiral lanthanide complex guarantees their m and μ transitions are non-orthogonal (cosτ ≠ 0).11 One method to realize the chiral environment for the lanthanide ions in the complex is to coordinate them with chiral molecules. On the other hand, biopolymer-based materials have long attracted attention due to their highly ordered structure, environmentally friendly nature and potential applications as photo-functional materials.12–14 Natural chiral molecules such as DNA, which exhibits a characteristic helix structure, has the unique ability to bind various types of functional materials through electrostatic binding, intercalation and groove binding.15,16 In some DNA-based hybrid systems, DNA molecules were reported to transfer their natural chirality to their coordinated lumiphores and to induce the CPL activity in DNA/lumiphore hybrid systems.17,18 Moreover, DNA-based hybrid systems have been proven promising in significantly improving the photo-functional properties of metal complexes.19–21 DNA-based hybrid systems with high structural orders exhibit flexible responsive optical functions and can be applied as a soft crystal.13
Recently, some authors in this research have reported a novel water-soluble Eu(III) complex, EuLCOOH, in which two carboxyl groups are selected as hydrophilic skeletons and introduced into EuL. The water-solubility and excellent luminescence performances make EuLCOOH especially attractive in the biological field.22 In this study, a DNA/Eu(III) complex hybrid system based on the helical complex structure of EuLCOOH was fabricated and investigated in an aqueous solution. Compared with EuLCOOH alone, the photoluminescence performance of the Eu(III) compound was effectively improved, and structural chirality was endowed by the presence of DNA. The major binding mode between DNA and EuLCOOH was revealed to be electrostatic interaction. Based on this interaction, CPL was successfully induced from the helical Eu(III) complex through the chiral environment provided by DNA.
Experimental section
Materials
EuLCOOH was synthesized according to the reported procedure.22 The sodium salts of DNA (base pairs: ca. 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) were provided by Piotrek Co., Ltd. (Japan). These were marine-based salts that were first isolated from frozen salmon milt through a homogenization process followed by the removal of proteins and impurities.
000) were provided by Piotrek Co., Ltd. (Japan). These were marine-based salts that were first isolated from frozen salmon milt through a homogenization process followed by the removal of proteins and impurities.
Preparation of the DNA/EuLCOOH hybrid solutions
DNA/EuLCOOH hybrid solutions were prepared by mixing EuLCOOH and DNA in the aqueous solution. The concentration ratios of DNA and EuLCOOH ([DNA]:[EuLCOOH]) were 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 5
1, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 10
1, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 15
1, 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 20
1, 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 30
1 and 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The concentration of EuLCOOH was fixed at 0.02 mmol L−1.
1. The concentration of EuLCOOH was fixed at 0.02 mmol L−1.
Measurement and characterization
Oxygen dissolved in the DNA/EuLCOOH hybrid solutions was removed by bubbling nitrogen gas through the solution prior to carrying out the optical measurements. The absorption and CD (circular dichroism) spectra were obtained using a circular dichroism spectrometer (J-1100, JASCO Corporation, Japan). The photoluminescence spectra were acquired using a spectrofluorometer (FP-8500, JASCO Corporation, Japan). The emission lifetimes were determined using a time-resolved fluorescence spectrometer (Quantaurus-Tau C11367-21, Hamamatsu Photonics K. K., Japan). The CPL (circularly polarized luminescence) measurements were conducted using a previously reported system.23 This system consists of the following components: a 300 nm LED (M300L4, Thorlabs Japan Inc., Japan), an LED driver (DC2100, Thorlabs Japan Inc., Japan), a photoelastic modulator (PEM-90, Hinds Instruments Inc., United States), a photomultiplier tube (H7732-10, Hamamatsu Photonics K. K., Japan), a linearly polarized cubic prism (200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), a photomultiplier tube (H7732-10, Hamamatsu Photonics K. K., Japan), and a dual-phase DSP (digital signal processing) lock-in amplifier (7265, Signal Recovery Ltd., United Kingdom). The appropriate detection wavelengths of the monochromator and the PEM (photoelastic modulator) were controlled using a PC (Dell D11M). The molecular structure and coordination environment of EuLCOOH were reported to be extremely stable at pH = 4.0–9.7.22 In this research, all the experiments are carried out under solution conditions with a pH range of 5.0–8.0.
1), a photomultiplier tube (H7732-10, Hamamatsu Photonics K. K., Japan), and a dual-phase DSP (digital signal processing) lock-in amplifier (7265, Signal Recovery Ltd., United Kingdom). The appropriate detection wavelengths of the monochromator and the PEM (photoelastic modulator) were controlled using a PC (Dell D11M). The molecular structure and coordination environment of EuLCOOH were reported to be extremely stable at pH = 4.0–9.7.22 In this research, all the experiments are carried out under solution conditions with a pH range of 5.0–8.0.
Results and discussion
Photoluminescence performance of EuLCOOH in the presence of DNA
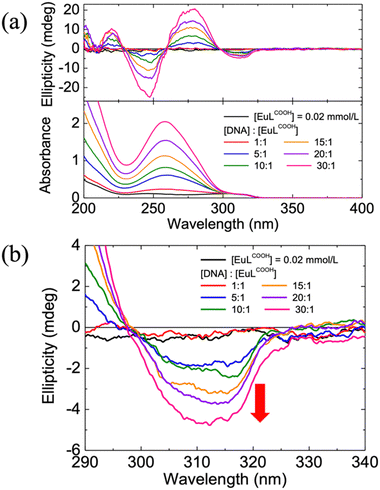
Circular dichroism (CD) spectroscopy originates from the interactions of chiral molecules with circularly polarized electromagnetic rays and exhibits high sensitivity and efficiency over the other conformational analysis. It suggests the difference between the absorption of right- and left-handed circularly polarized light and is quantified by ellipticity. Notably, CD spectroscopy is a highly sensitive diagnostic tool for determining the absolute configuration of chiral systems and monitoring their intermolecular interactions.24,25Fig. 1(a) shows the absorption and CD spectra of the EuLCOOH alone and the DNA/EuLCOOH hybrid solutions at various concentration ratios of DNA and EuLCOOH. DNA has a well-known axisymmetric helical structure and shows a characteristic absorption band near 260 nm, which is attributed to its nucleic acid bases, as illustrated in Fig. S1 (ESI†). For all solutions that DNA involved, the absorbance of DNA bases appeared and increased with the increasing DNA concentration. Moreover, an absorption band assigned to the π–π* transition of the ligands of EuLCOOH was observed around 305 nm.26 The unchanged absorption band from ligands of Eu(III) complex in all solutions indicates that the electronic transition of the ligands was not perturbed even when adding DNA. On the other hand, the CD behaviors of EuLCOOH obviously differed in the absence or presence of DNA. No CD signal was detected for EuLCOOH alone. The split Cotton effect with the first positive (275 nm) and second negative (250 nm) signals centered at the absorption peak of DNA was observed upon the addition of DNA because of its chiral structure. Furthermore, as the expanded CD spectra in Fig. 1(b) show, a negative ellipticity near 305 nm was induced, corresponding to the absorption of the Eu(III) complex. This suggests that DNA interacts with the Eu(III) complex and thus induces the chirality in its coordination structure without affecting the electronic transitions of the ligands. The ellipticity generated from the interaction between DNA and EuLCOOH was further amplified when additional DNA was added. Presumably, this phenomenon can be ascribed to the exciton coupling between multiple chromophores of the ligands of the Eu(III) complex upon interaction with DNA.27
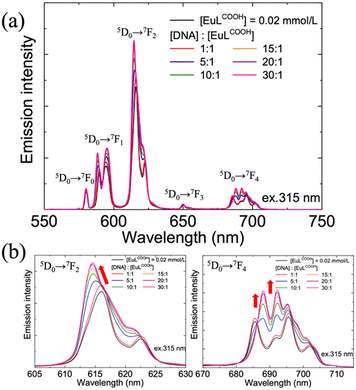
According to the induced chirality of EuLCOOH, the DNA in the second coordination sphere may have significantly distorted the first coordination sphere of EuLCOOH. High-resolution emission spectroscopy can well reflect even minor changes in the coordination sphere of the Eu(III) complex. Subsequently, emission measurements of EuLCOOH alone and the DNA/EuLCOOH hybrid solutions at various concentration ratios of DNA and EuLCOOH were shown in Fig. 2(a). Characteristic intense emission bands due to the 5D0 → 7FJ transition of the Eu(III) complex were observed at approximately 580 nm (J = 0), 590 nm (J = 1), 614 nm (J = 2), and 650 nm (J = 3). With the addition of DNA, the total emission intensity of EuLCOOH exhibits obvious enhancement. This can be explained by that the luminescence quenching caused by the vibration and rotation of the Eu(III) molecule is significantly suppressed. It is likely that the -OH oscillators of the water molecule are inhibited to some extent in DNA/EuLCOOH hybrid solutions. Therefore, the luminescence of EuLCOOH was effectively improved in the presence of DNA.
Fig. 2(b) shows the amplified spectra of the emission peaks corresponding to 5D0 → 7F2 (left) and 5D0 → 7F4 (right) transitions, respectively. These two transitions are known for their sensitivity to the structure of the matter around Eu3+ ions,28 and notable changes in their spectra patterns were observed in the presence of DNA. This indicates a perturbed ligand field of EuLCOOH owing to the interaction between DNA and EuLCOOH.
Combining the CD and luminescence spectroscopy results, it was concluded that DNA interacts with EuLCOOH and changes its coordination structure in aqueous solutions. On the other hand, the 5D0 → 7F1 transition is essentially independent of the chemical environment and acts as a reference for all transitions originating from the 5D0 excited state.29 The ratio (Irel) of the integrated intensity of insensitive 5D0 → 7F1 to hypersensitive 5D0 → 7F2 transitional probabilities is normally used to evaluate the site symmetry around the Eu3+ ions. A larger value of Irel indicates a lower symmetry occupied by the Eu3+ sites.30 For all EuLCOOH solutions, the values of Irel were almost constant at 2.25 with and without DNA (Fig. S2, ESI†). Although some variations in the coordination environment of EuLCOOH occurred reflected by changed luminescence and CD spectra, EuLCOOH maintained the site symmetry around the Eu3+ ions, even with the addition of DNA.
Time-resolved emission decay profiles were then obtained to further investigate the luminescence behaviour of the DNA/EuLCOOH hybrid solutions. Fig. 3 shows the emission decay profiles of EuLCOOH and the DNA/EuLCOOH hybrid solutions at various concentration ratios of DNA and EuLCOOH. The emission lifetime of the DNA/EuLCOOH hybrid solution became longer than that of EuLCOOH alone and further increased with increasing DNA concentration. Table 1 lists the values of the average emission lifetime (τave) and the contribution (%) of each exponential component (τ1 and τ2). The emission lifetime of EuLCOOH had only one exponential component (0.53 ms). This mono-exponential emission decay was attributed to the stable monolithic structure of EuLCOOH. Conversely, the DNA/EuLCOOH hybrid solutions exhibited multi-exponential emission decay with two components: τ1 and τ2. The short τ1 component (0.5 ms) was similar to that of the single component of EuLCOOH alone (0.53 ms). The longer τ2 component (1.1 ms) was derived from EuLCOOH with coordinating DNA. Therefore, the τ1 and τ2 components represent EuLCOOH without and with interacting DNA, respectively. With increasing concentrations of DNA, the contributions of the short lifetime τ1 component decreased and those of the long lifetime τ2 component increased. This indicates that more interaction between EuLCOOH and DNA occurred.
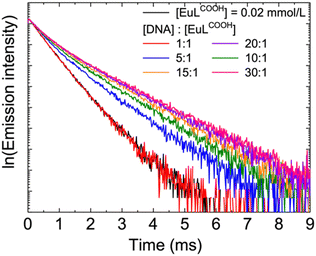 | ||
Fig. 3 Emission decay profiles of EuLCOOH and the DNA/EuLCOOH hybrid solutions. [DNA]:[EuLCOOH] were varied from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 30 1 to 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. 1. | ||
| [EuLCOOH] | τ 1 (ms) | ||
|---|---|---|---|
| τ ave (ms) | 0.53 | τ 2 (ms) | |
[DNA]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [EuLCOOH] [EuLCOOH] |
0.53 | 0.50 (%) | 1.10 (%) |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
0.54 | 94.2 | 5.8 |
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
0.77 | 54.5 | 45.5 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
0.88 | 36.7 | 63.3 |
15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
0.94 | 27.1 | 72.9 |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
0.97 | 22.4 | 77.6 |
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
0.99 | 17.8 | 82.3 |
Water molecules coordinated with Eu3+ ions are of great importance for understanding the surrounding environment and coordination sphere of Eu(III) complex in an aqueous solution. According to Horrocks’ equation, the coordinating water of the Eu(III) complex in aqueous solutions can be evaluated using its emission lifetime in H2O and D2O.31,32 The number of coordinating water molecules q is calculated as follows:
| q = 1.11[τH2O−1 − τD2O−1 − 0.31] |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 concentration ratio of DNA to EuLCOOH, the measured emission lifetimes in H2O and D2O were 0.77 ms and 2.04 ms, respectively. The value of q was thus calculated to be 0.55. This is similar to the contribution of the τ1 component (54.5%) for the same DNA/EuLCOOH hybrid solution ([DNA]:[EuLCOOH]= 5
1 concentration ratio of DNA to EuLCOOH, the measured emission lifetimes in H2O and D2O were 0.77 ms and 2.04 ms, respectively. The value of q was thus calculated to be 0.55. This is similar to the contribution of the τ1 component (54.5%) for the same DNA/EuLCOOH hybrid solution ([DNA]:[EuLCOOH]= 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). In this case, the value of q may be explained by the interaction ratio with DNA. The difference in q between EuLCOOH alone and the DNA/EuLCOOH hybrid solutions occurs because DNA competes for a portion of the complex coordination of Eu3+ ions from the water molecule. Water molecules in the second coordination sphere can shorten the emission lifetime of the 5D0 excited state, thereby extending the emission lifetime of the DNA/EuLCOOH hybrid solutions by being partially replaced to some extent by DNA.
1). In this case, the value of q may be explained by the interaction ratio with DNA. The difference in q between EuLCOOH alone and the DNA/EuLCOOH hybrid solutions occurs because DNA competes for a portion of the complex coordination of Eu3+ ions from the water molecule. Water molecules in the second coordination sphere can shorten the emission lifetime of the 5D0 excited state, thereby extending the emission lifetime of the DNA/EuLCOOH hybrid solutions by being partially replaced to some extent by DNA.
Interaction mode between EuLCOOH and DNA
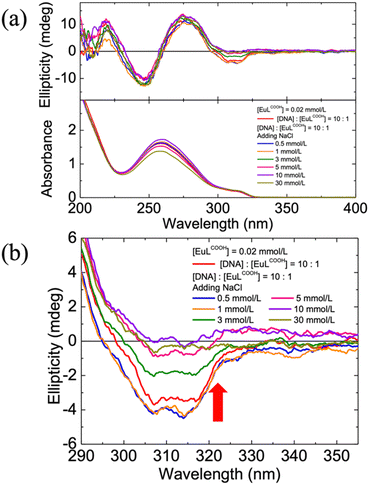
DNA is well known to be a negatively charged biological helical polymer. The electrostatic properties of its phosphate group and well-defined sites provided by its helical structure contributed to its characteristic counterion binding ability. Additionally, the Eu3+ ion possesses a high positive charge character owing to its high Lewis acidity from the well-shielded 4f orbitals.28,33,34 EuLCOOH complex acts as a stable cationic component with 2+ valence in an aqueous solution.22 Therefore, it is presumed that DNA interacts with EuLCOOH by mode of electrostatic binding.35,36 To clarify this hypothesis, an excess of Na+ ions (NaCl) was added to a DNA/EuLCOOH hybrid solution. The non-interference of Na+ ions with EuLCOOH alone in an aqueous solution was demonstrated by the unchanged emission spectra (Fig. S3(a), ESI†) and emission decay profiles (Fig. S3(b), ESI†) of EuLCOOH alone with and without a large amount of NaCl. Then, various concentrations of NaCl (0.5–30 mmol) were added sequentially to a DNA/EuLCOOH hybrid aqueous solution. The CD spectra, emission spectra, and emission lifetimes of these solutions were recorded to investigate the changes in the possible electrostatic interactions between DNA and EuLCOOH. As shown in Fig. 4, in addition to the excess Na+ ions, the induced negative CD signal derived from the Eu(III) complex interacting with DNA at approximately 310 nm, gradually tended towards zero. This phenomenon is caused by the cation exchange of Na+ and indicates the disassociation of the electrostatic interaction between the Eu3+ ion and DNA. Correspondingly, the changes that occurred in the emission spectra (Fig. S4, ESI†) and emission decay profiles (Fig. S5, ESI†) of the DNA/EuLCOOH hybrid solution due to the interaction with DNA also gradually disappeared and were finally approximated highly to EuLCOOH alone after adding excess Na+. As the emission lifetimes components analysis shows in Table S6 (ESI†), the contribution of τ1 component (0.5 ms) from EuLCOOH alone increases, and the contribution of τ2 component (1.1 ms) from EuLCOOH with coordinating DNA decrease with the gradual addition of Na+. This variation occurred in the ratio of the two emission exponential components, which revealed the process of disassociation of the electrostatic interaction between Eu3+ and DNA. These results do reinforce our hypothesis that EuLCOOH is mainly with the anionic DNA through electrostatic interaction.DNA melting is known as the process of heating DNA solutions to cause the tightly intertwined strands to cooperatively unravelling into single strands. This is an important biological process that can be applied to certain aspects of DNA.37–39 The effect of the DNA melting transition on the DNA/EuLCOOH hybrid solutions was investigated by evaluating the photoluminescence performance of the DNA/EuLCOOH hybrid solutions at temperatures from room temperature (25 °C) to 80 °C. Thermal quenching of the excited states of the Eu(III) complex sequentially occurred with the increasing temperature.28 Fig. S7 (ESI†) shows the emission spectra of the DNA/EuLCOOH hybrid solution ([DNA]: [EuLCOOH] = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) at various temperatures. The total emission intensity gradually decreased during the heating treatment. On the other hand, the shapes of the emission peaks from sensitive 5D0 → 7F2 and 5D0 → 7F4 transitions of EuLCOOH were retained over the entire temperature range. Therefore, the unaffected emission spectra suggest that the interaction between EuLCOOH and bases in DNA possess excellent thermal stability and this interaction is well kept at even under 80 °C. In the case of the emission lifetime of the DNA/EuLCOOH hybrid solution ([DNA]: [EuLCOOH] = 1
30) at various temperatures. The total emission intensity gradually decreased during the heating treatment. On the other hand, the shapes of the emission peaks from sensitive 5D0 → 7F2 and 5D0 → 7F4 transitions of EuLCOOH were retained over the entire temperature range. Therefore, the unaffected emission spectra suggest that the interaction between EuLCOOH and bases in DNA possess excellent thermal stability and this interaction is well kept at even under 80 °C. In the case of the emission lifetime of the DNA/EuLCOOH hybrid solution ([DNA]: [EuLCOOH] = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) through the heating process (Fig. S8, ESI†), the lifetime of EuLCOOH became progressively shorter owing to the thermal quenching. Interestingly, the emission lifetime of the DNA/EuLCOOH hybrid solution at 80 °C (0.60 ms) was still longer than that of EuLCOOH alone at room temperature of 25 °C (0.53 ms). Stable interactions, even at high temperatures, contribute to this excellent emission lifetime performance. Heating a DNA solution can result in the separation of strands, and the melting temperature of the DNA is normally lower than 80 °C, which was the highest temperature in this investigation. The melting process of a DNA solution is normally evaluated using absorption measurement.37 It is because the amount of UV light absorbed by DNA increases as the ratio of non-bonded base pairs increases during the melting process. Double helix stacking and a reduction in base parity are indicated by the changed electronic configuration of the bases during the melting process. Fig. S9 (ESI†) shows the absorption and CD spectra of the DNA/EuLCOOH hybrid solution ([DNA]: [EuLCOOH] = 1
30) through the heating process (Fig. S8, ESI†), the lifetime of EuLCOOH became progressively shorter owing to the thermal quenching. Interestingly, the emission lifetime of the DNA/EuLCOOH hybrid solution at 80 °C (0.60 ms) was still longer than that of EuLCOOH alone at room temperature of 25 °C (0.53 ms). Stable interactions, even at high temperatures, contribute to this excellent emission lifetime performance. Heating a DNA solution can result in the separation of strands, and the melting temperature of the DNA is normally lower than 80 °C, which was the highest temperature in this investigation. The melting process of a DNA solution is normally evaluated using absorption measurement.37 It is because the amount of UV light absorbed by DNA increases as the ratio of non-bonded base pairs increases during the melting process. Double helix stacking and a reduction in base parity are indicated by the changed electronic configuration of the bases during the melting process. Fig. S9 (ESI†) shows the absorption and CD spectra of the DNA/EuLCOOH hybrid solution ([DNA]: [EuLCOOH] = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) over the investigated temperature range. Absorbance peaks at approximately 257 nm were attributed to the electronic transition of DNA, and its relationship with temperature was shown in the insert of Fig. S9 (ESI†). The absorbance increased with the increasing temperature, suggesting the melting process of DNA in the DNA/EuLCOOH hybrid solution. Correspondingly, in the case of CD measurements, the second negative (250 nm) Cotton effect weakened with increasing temperature. Inside the DNA, bases gradually unstacked from neighbouring bases during heating, resulting in these variations including increased absorbance and changed CD signals.37 Moreover, the negative CD signal at approximately 315 nm from the EuLCOOH coordinating with DNA decreased and finally disappeared during heating. This indicates that EuLCOOH lost chirality which was obtained from its interaction with DNA at high temperatures owing to the denaturation of DNA. Although the interaction with DNA was not broken, as reflected in the emission spectra, EuLCOOH changed from binding to double-stranded DNA to single-stranded DNA binding in the case of high temperature.
30) over the investigated temperature range. Absorbance peaks at approximately 257 nm were attributed to the electronic transition of DNA, and its relationship with temperature was shown in the insert of Fig. S9 (ESI†). The absorbance increased with the increasing temperature, suggesting the melting process of DNA in the DNA/EuLCOOH hybrid solution. Correspondingly, in the case of CD measurements, the second negative (250 nm) Cotton effect weakened with increasing temperature. Inside the DNA, bases gradually unstacked from neighbouring bases during heating, resulting in these variations including increased absorbance and changed CD signals.37 Moreover, the negative CD signal at approximately 315 nm from the EuLCOOH coordinating with DNA decreased and finally disappeared during heating. This indicates that EuLCOOH lost chirality which was obtained from its interaction with DNA at high temperatures owing to the denaturation of DNA. Although the interaction with DNA was not broken, as reflected in the emission spectra, EuLCOOH changed from binding to double-stranded DNA to single-stranded DNA binding in the case of high temperature.
Induced CPL of EuLCOOH by interaction with DNA
As discussed in the CD measurement, the optical chirality of the helical EuLCOOH complex was due to electrostatic interactions with DNA. In general, CPL is considered a complementary tool for CD to investigate the optical chirality of a material. CPL measurements of EuLCOOH alone and the DNA/EuLCOOH hybrid solutions at various concentration ratios of DNA and EuLCOOH were performed, and their CPL spectra were shown in Fig. 5. As expected, no CPL signal was observed for EuLCOOH alone. In contrast, CPL signals appeared with the addition of DNA to the EuLCOOH solution and increased with an increasing concentration of DNA. A clear CPL signal assigned to the 5D0 → 7F4 transition was observed at approximately 655 nm. The luminescence dissymmetry, glum, was calculated to +0.22 when the concentration ratio of DNA to EuLCOOH was 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. This induced CPL performance of the helical Eu(III) complex can be attributed to the induced structural chirality, as reflected by CD measurement.
1. This induced CPL performance of the helical Eu(III) complex can be attributed to the induced structural chirality, as reflected by CD measurement.
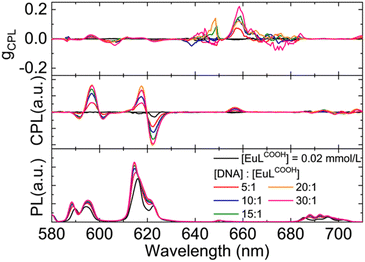 | ||
| Fig. 5 CPL spectra of EuLCOOH and the DNA/EuLCOOH hybrid solutions in various concentration ratios in water. The excitation wavelength was 300 nm. | ||
Conclusions
In this study, a water-soluble Eu(III) complex with helical ligands (EuLCOOH) was hybridized with DNA in an aqueous solution. The optical chirality of EuLCOOH was induced by its interaction with DNA. The photoluminescence performances of the DNA/EuLCOOH hybrid solutions were investigated using luminescence and luminescence lifetime measurements. It was found that EuLCOOH exhibited more brilliant luminescence with a longer lifetime in the addition of DNA. When an excess amount of Na+ ions was added to the DNA/EuLCOOH hybrid solution, the changes in the emission spectra and CD spectra resulting from the interaction between EuLCOOH and DNA disappeared. This result indicates that the major binding between DNA and EuLCOOH is an electrostatic interaction. Furthermore, this interaction contributes to the induced CPL signal of EuLCOOH due to the chirality of the DNA. This water-soluble Eu(III) material with excellent photo-luminescence performance and chiroptical properties is highly expected to be an effective bioanalytical tool in practical applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was partially supported by JSPS KAKENHI Grant Numbers 17H06377, 20K05641, 22H02154 and 23K04871. Z. L. received financial support from JST, the establishment of university fellowships towards the creation of science technology innovation, Grant Number JPMJFS2107. N. H. acknowledges the financial support from JPC (JAPAN PURE CHEMICAL) Scholarship Foundation, Kokudo Scholarship Foundation and Tomiyama Coltural Foundation.Notes and references
- C. P. Montgomery, B. S. Murray, E. J. New, R. Pal and D. Parker, Acc. Chem. Res., 2009, 42, 925–937 CrossRef CAS PubMed.
- G. Muller, Dalton Trans., 2009, 9692–9707 RSC.
- F. Zinna and L. Di Bari, Chirality, 2015, 27, 1–13 CrossRef CAS.
- Y. Dai, J. Chen, C. Zhao, L. Feng and X. Qu, Angew. Chem., Int. Ed., 2022, 61, e202211822 CrossRef CAS PubMed.
- E. Pershagen and K. E. Borbas, Angew. Chem., Int. Ed., 2015, 54, 1787–1790 CrossRef CAS.
- S. Mizukami, T. Yamamoto, A. Yoshimura, S. Watanabe and K. Kikuchi, Angew. Chem., Int. Ed., 2011, 50, 8750–8752 CrossRef CAS PubMed.
- A. Mohamadi and L. W. Miller, Bioconjugate Chem., 2016, 27, 2540–2548 CrossRef CAS.
- F. S. Richardson, Inorg. Chem., 1980, 19, 2806–2812 CrossRef CAS.
- Z. Li, H. Minami, K. Nakamura and N. Kobayashi, Chem. Phys. Chem., 2021, 22, 2511–2516 CrossRef CAS.
- Z. Li, K. Nakamura and N. Kobayashi, J. Mater. Chem. C, 2022, 11, 118–126 RSC.
- R. Carr, Doctoral dissertation, Durham University, 2014.
- R. P. Babu, K. O’Connor and R. Seeram, Prog. Biomater., 2013, 2, 8 CrossRef PubMed.
- M. Kato, H. Ito, M. Hasegawa and K. Ishii, Chem. – Eur. J., 2019, 25, 5105–5112 CrossRef CAS PubMed.
- R. J. B. Pinto, L. D. Carlos, P. A. A. P. Marques, A. J. D. Silvestre and C. S. R. Freire, J. App. Polym. Sci., 2014, 131, 41169 CrossRef.
- E. C. Long and J. K. Barton, Acc. Chem. Res., 1990, 23, 271–273 CrossRef CAS.
- C. V. Kumar, R. S. Turner and E. H. Asuncion, J. Photochem. Photobiol. A: Chem., 1993, 74, 231–238 CrossRef CAS.
- Q. Jiang, X. Xu, P.-A. Yin, K. Ma, Y. Zhen, P. Duan, Q. Peng, W.-Q. Chen and B. Ding, J. Am. Chem. Soc., 2019, 141, 9490–9494 CrossRef CAS PubMed.
- H. Minami, N. Itamoto, W. Watanabe, Z. Li, K. Nakamura and N. Kobayashi, Sci. Rep., 2020, 10, 18917 CrossRef CAS.
- S. Lin, L. Lu, T.-S. Kang, J.-L. Mergny, C.-H. Leung and D.-L. Ma, Anal. Chem., 2016, 88, 10290–10295 CrossRef CAS.
- K. Nakamura, H. Minami, A. Sagara, N. Itamoto and N. Kobayashi, J. Mater. Chem. C, 2018, 6, 4516–4522 RSC.
- B. J. Pages, D. L. Ang, E. P. Wright and J. R. Aldrich-Wright, Dalton Trans., 2015, 44, 3505–3526 RSC.
- S. Ogata, T. Shimizu, T. Ishibashi, Y. Ishiyone, M. Hanami, M. Ito, A. Ishii, S. Kawaguchi, K. Sugimoto and M. Hasegawa, New J. Chem., 2017, 41, 6385–6394 RSC.
- H. Tsumatori, T. Nakashima and T. Kawai, Org. Lett., 2010, 12, 2362–2365 CrossRef CAS PubMed.
- H. Tsukube and S. Shinoda, Chem. Rev., 2002, 102, 2389–2404 CrossRef CAS.
- S. R. Martin and M. J. Schilstra, Methods Cell Biol., 2008, 84, 263–293 CAS.
- H. Tsukube, A. Onimaru and S. Shinoda, Bull. Chem. Soc. Jpn., 2006, 79, 725–730 CrossRef CAS.
- M. Hasegawa, D. Iwasawa, T. Kawaguchi, H. Koike, A. Saso, S. Ogata, A. Ishii, H. Ohmagari, M. Iwamura and K. Nozaki, ChemPlusChem, 2020, 85, 294–300 CrossRef CAS PubMed.
- K. Binnemans, Coord. Chem. Rev., 2015, 295, 1–45 CrossRef CAS.
- C. Görller-Walrand, L. Fluyt, A. Ceulemans and W. T. Carnall, J. Chem. Phys., 1991, 95, 3099–3106 CrossRef.
- M. H. V. Werts, R. T. F. Jukes and J. W. Verhoeven, Phys. Chem. Chem. Phys., 2002, 4, 1542–1548 RSC.
- W. D. Horrocks Jr and D. R. Sudnick, J. Am. Chem. Soc., 1979, 101, 334–340 CrossRef.
- W. D. Horrocks Jr and D. R. Sudnick, Acc. Chem. Res., 1981, 14, 384–392 CrossRef.
- R. Díaz-Torres and S. Alvarez, Dalton Trans., 2011, 40, 10742 RSC.
- M. L. Aulsebrook, B. Graham, M. R. Grace and K. L. Tuck, Coord. Chem. Rev., 2018, 375, 191–220 CrossRef CAS.
- L. Su, D. Sen and H.-Z. Yu, Analyst, 2006, 131, 317–322 RSC.
- H. Minami, K. Nakamura and N. Kobayashi, J. Nanophotonics, 2018, 12, 1 Search PubMed.
- R. M. Wartell and A. S. Benight, Phys. Rep., 1985, 126, 67–107 CrossRef CAS.
- K. A. Heinrichs, J. M. Price, A. Raley, J. K. Berch, M. K. Murphy, H. Ka and P. Jm, Adv. Biochem. Biotechnol., 2017, 2, 118 Search PubMed.
- G. Khandelwal and J. Bhyravabhotla, PLoS One, 2010, 5, e12433 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ma00708a |
| This journal is © The Royal Society of Chemistry 2024 |