Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
One-pot synthesis of black biopolymeric eumelanin pigment by indigenous salt-tolerant Pseudomonas stutzeri SGM-1†
Swapnil G.
Mahajan
 *,
Vinod S.
Nandre
,
Kisan M.
Kodam
and
Mohan V.
Kulkarni
*,
Vinod S.
Nandre
,
Kisan M.
Kodam
and
Mohan V.
Kulkarni
Division of Biochemistry, Department of Chemistry, Savitribai Phule Pune University, Pune-411007, Maharashtra, India. E-mail: swapnilmahajan87@gmail.com; drmvkulkarni@gmail.com
First published on 4th January 2024
Abstract
For the first time, a soil-isolated indigenous salt-tolerant Pseudomonas stutzeri SGM-1 mediated one-pot synthesis of extracellular eumelanin is reported using only L-DOPA as the substrate. Biophysical characterizations of the eumelanin were performed using UV-Vis, FTIR, XRD, TGA, FESEM-EDS, DLS, and zeta potential analyses, thus confirming the colloidal microparticles as eumelanin. The synthesized eumelanin was acid-resistant, alkali-soluble, and insoluble in distilled water and many organic solvents. This one-pot synthesis method can reduce the use of hazardous chemicals and harsh extraction methods in the manufacturing and purification of eumelanin.
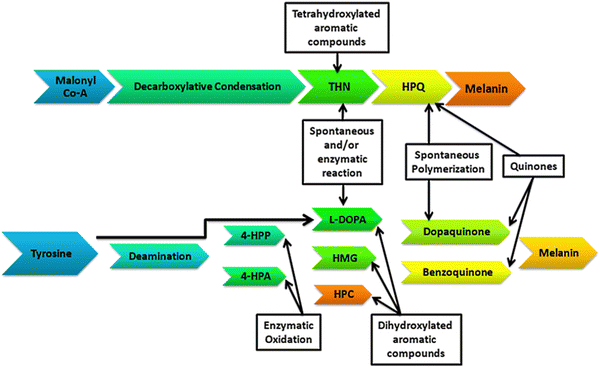
The generic term ‘melanin’ refers to an ancient pigment found in nature that is negatively charged, hydrophobic, ubiquitous, heterogeneous, and resistant. As per evolutionary history, melanin appeared very early in most living kingdoms on the Earth. This early presence of melanin has been proven by its occurrence in old fossils of dinosaurs, early-bird species, and non-avian species, like cephalopods and theropods, dating long back to the Jurassic period.1–3,35 The name melanin is derived from an ancient Greek word ‘melanos’ meaning ‘dark’. Structurally, melanins are diverse biopolymeric complex pigments having relatively diverse structures, which to date have not been identified completely. Many attempts have been made in the last more than 50 years to define the complete structures of this pigment but all those afforded either partial or incomplete definitions. This was because of their wide diversity in occurrences, compositions, colors, sizes, and functions, and this is obvious as melanins can be found in all kingdoms of life. Solano (2014) proposed a widespread and simple definition to include all types of melanin as heterogeneous polymers derived by the oxidation of phenols and subsequent polymerization of intermediate phenols and their resulting quinones.4 The synthesis and distribution of melanin pigments (comprising eumelanin, a brownish-black pigment, and pheomelanin, a yellowish-orange type of pigment), known as melanogenesis, occurs by a process in specialized cells called melanocytes in the epidermis of the skin.5 This pigmentation originating from melanogenesis is long-lasting, unlike the pigmentation arising from the oxidation of already-existing melanin pigment, generally referred to as ‘tanning’.
The skin, hair, and eye colors of organisms are determined by the presence of the melanin pigments, and more precisely depends on the ratio between the two types of melanin pigments in the mixture.6,36 The amount of the pigments present offers skin the color from white (vitiligo, which is a condition of a lack of melanin pigmentation) to brown–black (increased melanin density), more specifically the ratio of eumelanin–pheomelanin determines these differences in human skin pigmentation.7 In addition to defining the important human phenotype trait, melanin pigments serve a critical role in photoprotection by the property of ultraviolet radiation (UV) absorption.7,8 If the epidermal melanocytes produce a quantity of pheomelanin that is higher compared to eumelanin, the skin is of lighter color and has a higher susceptibility to sunburn, leading to a higher quantity of reactive oxygen species and ultimately resulting in the carcinogenesis process.9,10 Raper and Mason first established the melanin biosynthetic pathway in 1930 and 1940.11 Melanosomes, which are subcellular lysosome-like organelles, are present in melanocyte cells, in which melanin pigments are synthesized and stored.12 Melanin pigment synthesis is a cascading reaction that is catalyzed by different enzymes in a step-by-step process. The brownish- to black-colored pigment eumelanin is formed through the oxidative polymerization of 5,6-dihydroxyindole (DHI). L-Tyrosine is the main precursor for melanogenesis that is hydroxylated to L-DOPA enzymatically by tyrosinase and followed by melanin synthesis by a series of proteins related to tyrosinase.13 However, the yellowish-orange-colored pheomelanin, which is a soluble biopolymer in alkaline solution, results from oxidative polymerization of the cysteinyldopa (formed through the condensation of cysteine amino acid with dopaquinone). The tyrosinase enzyme is an important enzyme for the melanogenesis process, and is located only in the membrane of the melanosomes, where homeostasis of the melanogenic system is controlled by the concentration of L-tyrosine and that of L-DOPA, whereby ultimately the melanocytes coordinate with this system for melanogenesis.14 In eukaryotes, melanogenesis involves multiple enzymes, in addition to tyrosinase, for catalyzing L-tyrosine, leading to the synthesis of different forms of melanin pigments. However, in prokaryotes, this process of melanin synthesis comprises a series of reactions involving a single melanogenic enzyme.15
Depending on the geographical conditions on the Earth, organisms are adapted for the synthesis of melanin pigments for photoprotection. The process of melanogenesis occurs in almost every taxon of living organisms. The study of the synthesis, functioning, and characterization of melanin pigments is broadly focused on higher animals and humans, but little is known about the pathways and functioning of the melanin pigments in microorganisms.16–18,38 Regarding the bacterial world, tyrosine metabolism and pigment synthesis were discovered more than a century ago.19,20 Microorganisms synthesize their melanin pigments enzymatically through a single melanogenic enzyme (Fig. 1). Tyrosinase (EC 1.14.18.1) is an important metalloenzyme containing copper that catalyzes the rate-limiting step in melanin biosynthesis by converting the tyrosine to L-DOPA. Tyrosinase is widely distributed phylogenetically in different life forms. Tyrosinase catalyzes melanin synthesis through the hydroxylation of o-monophenols to o-diphenols and subsequently to reactive o-quinones.
Pseudomonas stutzeri is a marine bacterium often associated with rice plants and was reported by several researchers to be interesting to study for its melanin synthesis potential. A Pseudomonas stutzeri epiphytic strain HMGM-7 isolated from a red seaweed Hypnea musciformis from the Gulf of Mannar, India, was reported to produce up to 6.7 g L−1 melanin when grown in a seawater medium (Table 1). The black-colored, polymeric melanin, extracellularly produced by the HMGM-7 strain, was reported to be insoluble in distilled water and organic solvents except for phenol, readily soluble in alkali, and precipitated in acidic solutions. Furthermore, the researchers reported that the produced melanin was non-toxic when tested for cytotoxicity for two human cancer cell lines, namely A549 and HeLa.21 The authors reported this as the first report on a marine Pseudomonas stutzeri strain producing melanin without L-tyrosine supplementation in a seawater production medium. Another isolate, the Pseudomonas stutzeri strain BTCZ-10 from 97 m-depth marine sediments from the Arabian sea, was reported to produce 47.47 μg ml−1 melanin in a chemically defined minimal medium serving L-tyrosine (2 g L−1) as the sole carbon and nitrogen source (Table 1).22 However, in addition to the P. stutzeri strains HMGM-7 and BTCZ-10, there have been no other P. stutzeri strains reported for melanin synthesis from the marine origin and none from soil origin to date.
| S. no. | Pseudomonas stutzeri strain | Growth medium | Yield | Ref. |
|---|---|---|---|---|
| 1 | HMGM-7 | Seawater medium | 6.7 g L−1 | Kumar et al. 2013 |
| 2 | BTCZ-10 | Chemically defined minimal medium with L-tyrosine (2 g L−1) | 47.47 μg ml−1 | Kurian et al. 2014 |
| 3 | SGM-1 | L-DOPA (200 mg L−1) in DI-H2O | 126 mg L−1 | This study |
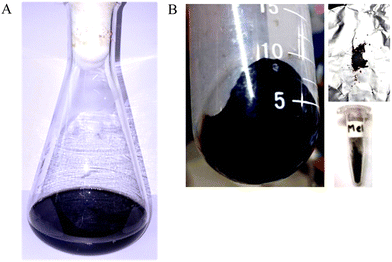
The isolate Pseudomonas stutzeri SGM-1 (GenBank Accn. No: KJ769179 & KJ769180), which was earlier reported for its salt and other abiotic stress tolerance features by our lab, was screened for its eumelanin synthesis potential from an L-DOPA-containing liquid medium.23 Briefly, the inoculum of Pseudomonas stutzeri SGM-1 was prepared by overnight incubation in a nutrient broth medium at 37 °C; the culture was centrifuged three times (REMI, India) and then the pellet was aseptically washed with sterile ultrapure water, to remove all traces of the medium. Next, this pellet was added to sterile ultrapure water containing 200 mg L−1L-DOPA and incubated at 37 °C and 150 rpm for 24 h. Post-incubation, the colorless L-DOPA solution attained a dark-black color, qualitatively confirming eumelanin synthesis. Next, the cells were separated from the supernatant containing extracellular colloidal eumelanin by centrifugation at 6000 rpm at 4 °C for 10 min. The typical dark-black-colored extracellular eumelanin synthesized by P. stutzeri SGM-1 was observed (Fig. 2A). The synthesized eumelanin was extracellular; therefore, there was no requirement for physicochemical or enzymatic rupture of the cells to yield the synthesized eumelanin pigment. The collected colloidal eumelanin was transferred to new sterile tubes and then lyophilized (Labconco Freezone 2.5 plus, USA) to obtain eumelanin powder, which was then stored in a tightly closed vial (Fig. 2B). Overall, for the 200 mg L−1 of L-DOPA used, the total yield of the dry weight of eumelanin pigment liberated was 126 mg L−1. The pigment yield could then be maximized by increasing the substrate concentration and to prepare the biomass used for pigment production.
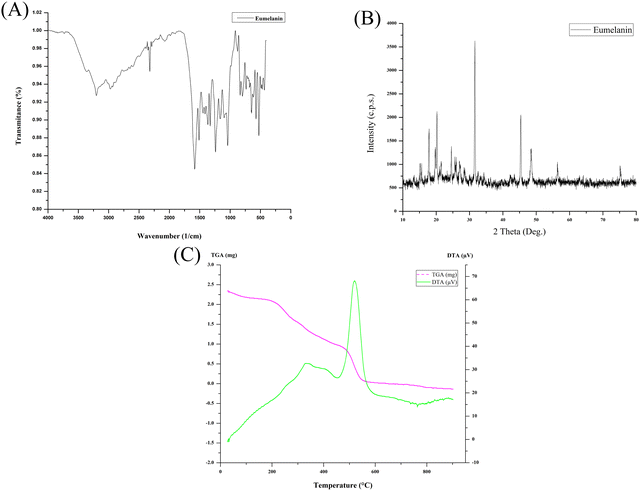
The solubility of the lyophilized powder of eumelanin was tested in the following sequence with distilled water, 1 N HCl, 1 M NaOH, ethanol, methanol, n-hexane, chloroform, acetone, DMSO, benzene, acetic acid, ethyl acetate, phenol, and petroleum ether, and stirred for 30 min. The produced eumelanin demonstrated the reported common features, like precipitation in acidic solutions, insolubility in water and other organic solvents tested, and could be dispersed in strong alkali to form a colloidal solution. UV-Vis spectral measurements (Shimadzu UV-1800, Japan) were taken to confirm the ultraviolet light-absorption property of the produced eumelanin, which monotonically increased the absorption toward the UV range (Fig. S1, ESI†). The lyophilized eumelanin powder was analyzed directly on an FT-IR-ATR spectrometer (Bruker-ALPHA-II, Bruker, USA) in the range of 4000–400 cm−1 with a resolution of 1 cm−1 (Fig. 3A). The FTIR results highlighted the presence of the carboxylic group and its ion form (–COO–), hydroxylic, phenolic groups, and aromatic –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C– stretching. These stretching vibrations obtained for the above-mentioned functional groups were related to the base constituents and were confirmatory for the eumelanin pigment molecule. The produced eumelanin was scanned using an X-ray diffractometer (PANalytical X’PERT PRO), and it was found that the XRD spectrum lacked the structured diffraction pattern corresponding to any crystallinity (Fig. 3B). The overall spectrum was dominated by a non-Bragg diffraction pattern. The observation of broad features, also known as non-Bragg features, is because of the absence of coherent scattering structures in a compound, attributed to the amorphous nature of the compound analyzed.24,25 The XRD spectrum of eumelanin demonstrated non-Bragg features attributable to the amorphous nature of the powdered eumelanin. From the position of the peaks shown in Fig. 3B, the overall eumelanin crystallographic pattern at 2θ displayed its amorphous nature, which was also in accordance with the previous reports.25–27 Melanins are amorphous, insoluble, and heterogeneous and their structures are uncertain, and here the X-ray diffraction of P. stutzeri SGM-1 eumelanin highlighted the same.
C– stretching. These stretching vibrations obtained for the above-mentioned functional groups were related to the base constituents and were confirmatory for the eumelanin pigment molecule. The produced eumelanin was scanned using an X-ray diffractometer (PANalytical X’PERT PRO), and it was found that the XRD spectrum lacked the structured diffraction pattern corresponding to any crystallinity (Fig. 3B). The overall spectrum was dominated by a non-Bragg diffraction pattern. The observation of broad features, also known as non-Bragg features, is because of the absence of coherent scattering structures in a compound, attributed to the amorphous nature of the compound analyzed.24,25 The XRD spectrum of eumelanin demonstrated non-Bragg features attributable to the amorphous nature of the powdered eumelanin. From the position of the peaks shown in Fig. 3B, the overall eumelanin crystallographic pattern at 2θ displayed its amorphous nature, which was also in accordance with the previous reports.25–27 Melanins are amorphous, insoluble, and heterogeneous and their structures are uncertain, and here the X-ray diffraction of P. stutzeri SGM-1 eumelanin highlighted the same.
Thermogravimetric (TG) data for the lyophilized powder of the eumelanin sample was recorded using a Discovery-TGA-5500 thermal analyzer (TA Instruments, USA) under an inert atmosphere of nitrogen gas. An alumina crucible was used for this measurement at a heating rate of 1 °C min−1 in the temperature range of 10–600 °C. The eumelanin produced here was a natural biopolymer synthesized by P. stutzeri SGM-1 from L-DOPA obtained extracellularly by centrifugation and it did not require processing involving complex and harsh purification steps. The eumelanin demonstrated high thermal stability (Fig. 3C) with a mass loss of up to 220 °C caused by the weight of the water, suggesting that the initial 20% mass of the eumelanin was strongly bound to water molecules. A weight loss of ∼40% was observed between 200 °C and 400 °C, attributable to melanin degradation because of the non-attachment of the eumelanin to the cellular molecules. The gradual loss of eumelanin up to 800 °C indicated the complete combustion of the produced pigment molecule. The differential thermal analysis of the eumelanin demonstrated a slower endothermic decomposition, while rapid exothermic decomposition was observed by a sharp peak at 500 °C. The thermogravimetric pattern of this P. stutzeri SGM-1 eumelanin was in line with the previously reported TGA results for melanin.25
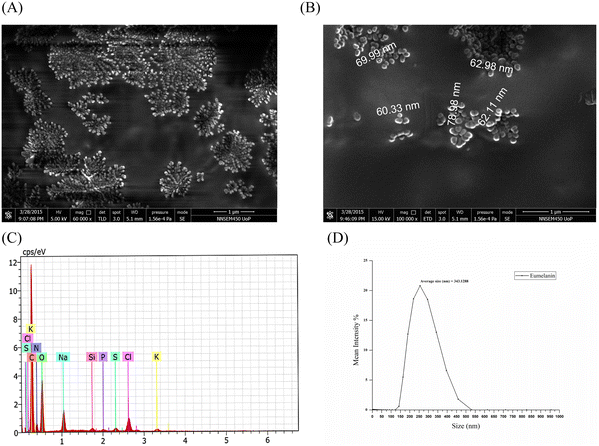
The lyophilized eumelanin powder was resuspended in 100% ethanol solution and was loaded on to a 0.5 cm × 0.5 cm glass slide, which, after drying, was coated with a platinum sputter coater (Quorum 150ES). The images were taken by field-emission scanning electron microscopy (FESEM: Inspect F50, FEI Europe BV, The Netherlands; FP 2031/12, SE Detector R580) at different magnifications, which demonstrated that spherical eumelanin nano- and microparticles had been synthesized by the isolate (Fig. 4A and B). The spherical structures of oligomers bound together to form a granular structure of melanin granules with a higher size (μm). These multi-μm-sized eumelanin granules formed the aggregated amorphous structure of the pigment. This aggregation of the pigment granules was dependent on the surface tension of the water/air/substrate system.28 However, the morphology and size of the melanin were dependent on the procedure followed for the extraction and purification of the pigment. The spherical granular pigments obtained from P. stutzeri SGM-1 demonstrated an aggregated pattern under FESEM and formed a colloidal suspension when added to water.
Energy-dispersive X-ray spectroscopy of the eumelanin microparticles (Brookhaven; NY; USA, NanoBrook series, equipped with an SVE-175 small volume electrode) demonstrated the elemental composition, in order from higher to lower, as C, O, N, Na, Cl, K, Si, S, and P, respectively. This result highlighted that the majority of elements were C, O, and N, corresponding to the carboxylic (–COOH), phenolic/hydroxylic (–OH), and amino (–NH) groups present in eumelanin's composition. Fig. 4C shows the weight % of each element constituting the eumelanin pigment which were similar to in the previously reported melanin pigment elemental composition.27,29 The particle size of the eumelanin particles was analyzed by dynamic light scattering (DLS). DLS analyzes the fluctuations in the scattered light intensity with time owing to Brownian motion of the scattering particles in a solution and provides information about the hydrodynamic diameter of those particles. Since the eumelanin particles synthesized were not perfect spheres and without smooth surfaces, the DLS could indicate an apparent size of the dynamic hydrated particle in solution. From Fig. 4D, it could be found that the particle size distribution was from 125 to 525 nm with an average of 343 nm; thus, the synthesized eumelanin was considered to be micro-particulate.30 However, as per Fig. S2 (ESI†), the surface charge analysis by zeta potential analysis showed a value of –41.7 mV with a zeta deviation of 14.8 mV and conductivity of 1.29 mS cm−1; thus, the eumelanin microparticles were considered stable and negatively charged.
There are earlier reports about melanin pigment synthesis by Pseudomonas stutzeri isolates from a marine origin, namely, P. stutzeri strain HMGM-7 and P. stutzeri strain BTCZ-10.21,22 However, in addition to these strains (HMGM-7 and BTCZ-10), there are no other P. stutzeri strains reported for melanin synthesis from a marine origin and none from a soil origin. With research advancements in melanin pigments, there has been a substantial increase in the application of melanin in pharmaceutical, medicine, cosmetics, and agriculture fields, but achieving a cost-effective high yield is the prime concern for its production. Furthermore, nowadays the semiconductor feature of this biopolymer has seen its application in many biosensor products.31,32,37 Currently, obtaining melanin from animal and plant sources involves several steps and the use of harsh chemical and physical methods. Moreover, there are few microbial (fungal and bacterial) sources reported for the green synthesis of melanin, but those methods use intracellular enzymatic synthesis and involve the enzymatic digestion of the melanosomes, which is time-consuming and costly when considered for the industrial scale.33,34 Hence, there is a requirement for a cost-effective, green, and simple downstream method for obtaining melanin. In the present study, the melanin synthesized from L-DOPA in sterile ultrapure water by the P. stutzeri strain SGM-1 was extracellular; therefore, there was no need for physicochemical or enzymatic rupture of the cells to yield the synthesized melanin pigment. Hence, the downstream process to yield melanin becomes fast, involving just a few simple steps and a cost-effective process.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The author thanks the Department of Chemistry, Savitribai Phule Pune University, Pune-07 for infrastructure and analytical facilities.Notes and references
- F. Zhang, S. L. Kearns, P. J. Orr, M. J. Benton, Z. Zhou, D. Johnson, X. Xu and X. Wang, Nature, 2010, 463(7284), 1075–1078 CrossRef CAS PubMed.
- R. A. Wogelius, P. L. Manning, H. E. Barden, N. P. Edwards, S. M. Webb, W. I. Sellers and K. G. Taylor, et al. , Science, 2011, 333(6049), 1622–1626 CrossRef CAS PubMed.
- K. Glass, S. Ito, P. R. Wilby, T. Sota, A. Nakamura, C. R. Bowers and J. D. Simon, et al. , Proc. Natl. Acad. Sci. U. S. A., 2012, 109(26), 10218–10223 CrossRef CAS PubMed.
- F. Solano, New J. Sci., 2014, 1–28 CAS.
- J. Bonaventure, M. J. Domingues and L. Larue, Pigm. Cell Melanoma Res., 2013, 26(3), 316–325 CrossRef CAS PubMed.
- M. Maranduca, D. Branisteanu, D. Serban, D. Branisteanu, G. Stoleriu, N. Manolache and I. Serban, Oncol. Lett., 2019, 17(5), 4183–4187 CAS.
- J. Y. Lin and D. E. Fisher, Nature, 2007, 445(7130), 843–850 CrossRef CAS PubMed.
- G.-E. Costin and V. J. Hearing, FASEB J., 2007, 21(4), 976–994 CrossRef CAS PubMed.
- S. Wu, J. Han, F. Laden and A. A. Qureshi, Cancer Epidemiol. Prev. Biomarkers, 2014, 23(6), 1080–1089 CrossRef PubMed.
- S. Okazaki, Y. Funasaka, K. Wakamatsu, S. Kawana and H. Saeki, J. Dermatol., 2015, 42(4), 382–390 CrossRef CAS PubMed.
- M. Sugumaran, Int. J. Mol. Sci., 2016, 17(9), 1576 CrossRef PubMed.
- M. S. Marks and M. C. Seabra, Nat. Rev. Mol. Cell Biol., 2001, 2(10), 738–748 CrossRef CAS PubMed.
- C. Niu and H. A. Aisa, Molecules, 2017, 22(8), 1303 CrossRef PubMed.
- A. Slominski, M. A. Zmijewski and J. Pawelek, Pigm. Cell Melanoma Res., 2011, 25(1), 14–27 CrossRef PubMed.
- J. Borovansky and P. A. Riley, Melanins and Melanosomes, John Wiley & Sons, 2011 Search PubMed.
- L. Zecca, D. Tampellini, M. Gerlach, P. Riederer, R. G. Fariello and D. Sulzer, Mol. Pathol., 2001, 54(6), 414–418 CAS.
- R. Halaban, Pigm. Cell Res., 2000, 13(1), 4–14 CrossRef CAS PubMed.
- A. Slominski, J. Wortsman, P. M. Plonka, K. U. Schallreuter, R. Paus and D. J. Tobin, J. Invest. Dermatol., 2005, 124(1), 13–21 CrossRef CAS PubMed.
- M. W. Beijerinck, KNAW, Proc., 1911, 13(II), 1066–1077 CAS.
- C. E. Skinner, J. Bacteriol., 1938, 35(4), 415–424 CrossRef CAS PubMed.
- C. Ganesh Kumar, N. Sahu, G. Narender Reddy, R. B. N. Prasad, N. Nagesh and A. Kamal, Lett. Appl. Microbiol., 2013, 57(4), 295–302 CrossRef CAS PubMed.
- N. K. Kurian, N. Harisree and B. Sarita, Int. J. Curr. Biotechnol., 2014, 2(5), 6–11 CAS.
- S. G. Mahajan, V. S. Nandre, R. C. Salunkhe, Y. S. Shouche and M. V. Kulkarni, Biocatal. Agric. Biotechnol., 2020, 27, 101652 CrossRef.
- A. Casadevall, PLoS Pathog., 2012, 8(8), e1002808 CrossRef CAS PubMed.
- S. S. A. O. Sajjan, G. B. Kulkarni, A. S. Nayak, S. B. Mashetty and T. B. Karegoudar, Korean J. Microbiol. Biotechnol., 2013, 41(1), 60–69 CrossRef CAS.
- S. Subianto, PhD thesis, Inorganic Material Research Program, the Queensland University of Technology, 2006.
- A. Mbonyiryivuze, Z. Y. Nuru, L. Kotsedi, B. Mwakikunga, S. M. Dhlamini, E. Park and M. Maaza, Mater. Today: Proc., 2015, 2(7), 3988–3997 Search PubMed.
- Y. Liu, V. R. Kempf, J. Brian Nofsinger, E. E. Weinert, M. Rudnicki and K. Wakamatsu, et al. , Pigm. Cell Res., 2003, 16(4), 355–365 CrossRef CAS PubMed.
- V. Manirethan, N. Gupta, R. M. Balakrishnan and K. Raval, Environ. Sci. Pollut. Res., 2020, 27(20), 24723–24737 CrossRef CAS PubMed.
- I. Calle de la, D. Soto-Gómez, P. Pérez-Rodríguez and J. E. López-Periago, Food Anal. Methods, 2019, 12(5), 1140–1151 CrossRef.
- E. Bronze-Uhle, M. Silva, J. Paulin, C. Battocchio and C. Graeff, Synthesis of water-soluble melanin, 2015 Search PubMed.
- E. Vahidzadeh, A. P. Kalra and K. Shankar, Biosens. Bioelectron., 2018, 122, 127–139 CrossRef CAS PubMed.
- S. R. Pombeiro-Sponchiado, G. S. Sousa, J. C. Andrade, H. F. Lisboa and R. C. Gonçalves, Melanin, 2017, 47–75 CAS.
- K. Y. Choi, Front. Bioeng. Biotechnol., 2021, 9, 765110 CrossRef PubMed.
- R. G. De La Garza, P. Sjövall, R. Hauff and J. Lindgren, Palaeontology, 2023, 66(4), e12668 CrossRef.
- D. I. Schlessinger, M. Anoruo and J. Schlessinger, Biochemistry, melanin, StatPearls, 2022 Search PubMed.
- A. Menichetti, A. Mavridi-Printezi, D. Mordini and M. Montalti, Biosensors, 2023, 13(11), 956 CrossRef CAS PubMed.
- K. Wakamatsu and S. Ito, Int. J. Mol. Sci., 2023, 24(9), 8305 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ma01097f |
| This journal is © The Royal Society of Chemistry 2024 |