Open Access Article
Open Access ArticleVirus adsorption and elution using cationic polymer brushes: potential applications for passive sampling in wastewater-based epidemiology†
Junya
Uchida
 a,
Miaomiao
Liu
a,
Miaomiao
Liu
 b,
Shizuka
Matsuyama
b,
Shizuka
Matsuyama
 a,
Hiroyuki
Katayama
*bc and
Takashi
Kato
a,
Hiroyuki
Katayama
*bc and
Takashi
Kato
 *acd
*acd
aDepartment of Chemistry and Biotechnology, School of Engineering, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8656, Japan. E-mail: kato@chiral.t.u-tokyo.ac.jp
bDepartment of Urban Engineering, School of Engineering, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8656, Japan. E-mail: katayama@env.t.u-tokyo.ac.jp
cResearch Center for Water Environment Technology, School of Engineering, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8656, Japan
dResearch Initiative for Supra-Materials, Shinshu University, 4-17-1 Wakasato, Nagano 380-8553, Japan
First published on 2nd April 2024
Abstract
Wastewater-based epidemiology (WBE) has been recognized as a promising approach for rapid monitoring of infectious diseases in local communities. Development of adsorption materials that efficiently capture viruses is important in WBE to provide precise information on the prevalence of viral infections. Herein, ionic polymer brushes are synthesized for the tuning of virus adsorption and elution. Quaternary ammonium-based cationic polymer brushes exhibit higher adsorption of enveloped and nonenveloped viruses than a low-molecular-weight amine adduct. Moreover, efficient and selective elution of Aichivirus from the polymer brushes is demonstrated. These cationic polymer brushes may be useful as materials for passive sampling of viruses from water.
Introduction
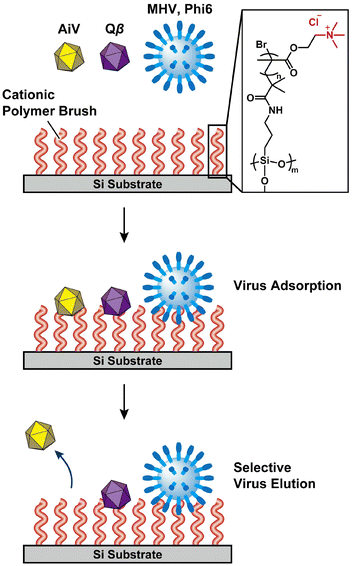
Wastewater-based epidemiology (WBE) has attracted much attention because it enables rapid monitoring of infectious diseases in local communities.1,2 Effective detection of viruses from sewage and environmental water is important to provide precise information on the prevalence of viral infections.3–5 For example, detection of Aichivirus (AiV), which is suggested as an appropriate indicator of viral contamination in the environment,6 may be useful to assess human faecal pollution.6–9 In WBE analysis, passive sampling is a virus collection method involving the placement of adsorption materials in water.10,11 Commercially available materials such as cotton gauze, glass beads, and polymer membranes have been used as adsorption materials to capture viruses.12–14 The tuning of the surface properties of these materials is considered to be a key to achieve efficient recovery of viruses from wastewater.14,15Our intention is to provide a new approach for tuning virus adsorption and elution for passive sampling of viruses from water (Fig. 1). We expected that ionic polymer brushes of grafting polymers may be useful in tuning virus adsorption and elution due to their electrostatic interactions with viruses. Grafting polymers on solid substrates have been widely studied to develop stimuli-responsive functional soft surfaces.16–23 For example, polyelectrolyte brushes were formed on a variety of surfaces to provide lubrication,24–26 antifouling,18,27,28 and antibacterial functions.29,30 Thermoresponsive surfaces based on poly(N-isopropylacrylamide) brushes have been developed for applications as substrates to control cell adhesion and detachment.29,31,32 We also reported thermoresponsive polymer brush matrices for tuning the morphologies of organic/inorganic hybrid thin films.33–35 Cationic polymer brushes may exhibit higher adsorption of viruses because most viruses are reported to be negatively charged at neutral pH.36 Cationic polymers and polymer hybrids have been studied for the development of virus adsorption,37 virus removal membranes,38–42 and antimicrobial materials.43–45 The use of polymer brushes for passive sampling of viruses from sewage has not been attempted, although purification of viral vectors for gene therapy by using thermoresponsive-anionic mixed polymer brushes has been reported.46
Here we report virus adsorption and elution using ionic polymer brushes grafted on the substrates (Fig. 1). The effects of the surface properties of the modified substrates on virus adsorption and elution were examined by using cocktail solutions containing different types of viruses.
Results and discussion
Materials design and synthesis
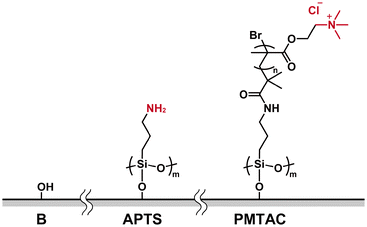
Ionic polymer brushes consisting of poly[2-(methacryloyloxy)ethyltrimethylammonium chloride] (PMTAC) (Fig. 2) were synthesized by surface-initiated atom transfer radical polymerization (ATRP) (Fig. S1†).47–49 An ATRP initiator, 2-bromo-2-methyl-N-(3-(triethoxysilyl)propyl)propanamide, was immobilized on a silicon substrate. This substrate was used for surface-initiated ATRP to obtain the PMTAC-grafted substrates. In addition, silicon substrates modified with 3-aminopropyltriethoxysilane (APTS) (Fig. 2) were prepared as a control material containing a low-molecular-weight amine branch. The PMTAC-grafted substrates (PMTAC) and APTS-functionalized substrates (APTS) as well as nonmodified bare silicon substrates (B) were used to examine virus adsorption and elution.Structures of the PMTAC-grafted surface
The formation of the PMTAC brushes grafted on the substrates was examined (Fig. 3, 4, Table 1, Fig. S2 and S3†). Fig. 3 shows a Fourier-transform infrared (FT-IR) spectrum of a silicon substrate that underwent surface-initiated ATRP. A strong absorption peak corresponding to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration of the PMTAC brushes was observed at 1723 cm−1.50,51 In addition, the surface exhibited an absorption band around 1481 cm−1. This can be assigned to a vibration of the quaternary ammonium group of the PMTAC brushes.50,51 These results show that the surface-initiated ATRP led to the preparation of the cationic polymer brushes consisting of PMTAC.
O stretching vibration of the PMTAC brushes was observed at 1723 cm−1.50,51 In addition, the surface exhibited an absorption band around 1481 cm−1. This can be assigned to a vibration of the quaternary ammonium group of the PMTAC brushes.50,51 These results show that the surface-initiated ATRP led to the preparation of the cationic polymer brushes consisting of PMTAC.
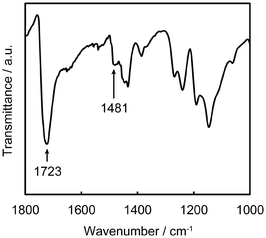 | ||
| Fig. 3 FT-IR spectrum of the poly[2-(methacryloyloxy)ethyltrimethylammonium chloride] (PMTAC) brush. | ||
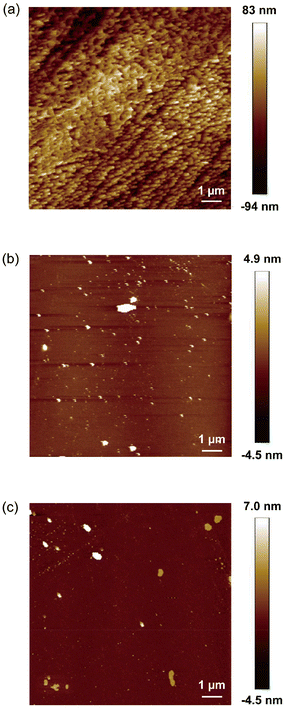 | ||
| Fig. 4 AFM images of (a) the poly[2-(methacryloyloxy)ethyltrimethylammonium chloride] (PMTAC)-grafted, (b) 3-aminopropyltriethoxysilane (APTS)-functionalized, and (c) nonmodified bare substrates. | ||
| Materialsa | Contact angle (degree) | Solvent-accessible cationsb (1015 cm−2) |
|---|---|---|
| a B: bare substrates; APTS: 3-aminopropyltriethoxysilane (APTS)-functionalized substrates; and PMTAC: poly[2-(methacryloyloxy)ethyltrimethylammonium chloride] (PMTAC)-grafted substrates. b The number of solvent-accessible cations was estimated with the fluorescein adsorption assay.53,54 | ||
| B | 36 ± 2.3 | 0.0 ± 0.0 |
| APTS | 50 ± 2.2 | 0.0 ± 0.0 |
| PMTAC | 69 ± 12 | 1.1 ± 0.4 |
The surface morphology of PMTAC was observed by atomic force microscopy (AFM) (Fig. 4a). The surface of PMTAC was sufficiently covered with the polymer brushes (Fig. 4a), while those of APTS (Fig. 4b), B (Fig. 4c), and the ATRP initiator-modified (Fig. S3†) substrates were smoother. Based on these observations, the PMTAC brushes are expected to efficiently interact with viruses to provide higher virus adsorption.
The surface wettability of PMTAC was examined by contact angle measurements (Table 1). The water contact angle of PMTAC was larger than those of APTS and B (Table 1). It was reported that surface wettability was affected by the nanostructures of the surface.52 The uneven surface of PMTAC (Fig. 4a) may result in the formation of a more hydrophobic surface than those of APTS and B. The substrates modified with the ATRP initiator also formed a hydrophobic surface with a contact angle of 86 ± 1.5°.
To gain further insights into the surface properties, the number of solvent-accessible cations on the surfaces was estimated (Table 1). A fluorescein adsorption assay53,54 was performed for PMTAC, APTS, and B. The charge density of PMTAC was estimated to be about 1015 cations per cm2 (Table 1). For APTS and B, no significant differences in the number of solvent-accessible cations were observed, although the contact angle of these substrates was different. The higher charge density of PMTAC may be useful for efficient adsorption of viruses.
Adsorption and elution of viruses
In the adsorption and elution experiments, four types of viruses including AiV, F-RNA-specific bacteriophage Qβ, Pseudomonas syringae phage Phi6, and murine hepatitis virus (MHV) were employed to represent different virus types in wastewater. AiV and Qβ are nonenveloped viruses, while MHV and Phi6 are enveloped viruses. The sizes of AiV and Qβ were reported to be around 30 nm,55–57 while those of MHV and Phi6 were reported to be approximately 80 nm.58,59 These nonenveloped and enveloped viruses were mixed in 100 mM phosphate buffered saline (PBS) to prepare feed solutions (referred to as Feed). To assess the virus adsorption, the feed solutions were placed on the surface-modified and bare silicon substrates for 5 h, followed by washing the substrates with PBS to collect the solution (referred to as Permeate). The elution of the viruses from the substrates was performed with sterile beef extract (3%, pH = 9), which is generally used for virus elution.15,60 After soaking the substrates in the beef extract for 1 h, the solution was collected as an eluate (referred to as Eluate). The concentrations of the four viruses in the Feed, Permeate, and Eluate were quantified with reverse transcription quantitative polymerase chain reaction (RT-qPCR) to estimate the virus adsorption and elution rates. Adsorption was quantified by calculating the logarithmic removal from the Feed to the Permeate (eqn (1)) whereas recovery (or elution) was determined by assessing the logarithmic change from the Feed to the Eluate (eqn (2)).| Adsorption rate (log10Ads) = \quad\quad log10(virus in Feed/virus in Permeate); | (1) |
| Elution rate (log10Loss) = log10(virus in Feed/virus in Eluate). | (2) |
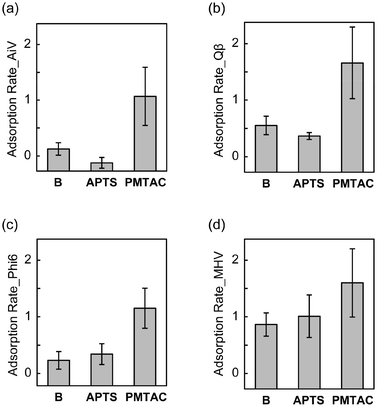
We found that the adsorption of the four tested viruses on the silicon substrates was enhanced by surface modification with the PMTAC brushes (Fig. 5). The adsorption rate of AiV by PMTAC increased to 1.07 ± 0.52 log10Ads (91.49% in percentage) (Fig. 5a), while those by B and APTS were 0.13 ± 0.11 log10Ads (25.11%) and −0.12 ± 0.10 log10Ads (−33.08%), respectively (Fig. 5a). Similar trends were observed for Qβ (Fig. 5b), Phi6 (Fig. 5c), and MHV (Fig. 5d). The increase in the virus adsorption by grafting the PMTAC brushes may be due to the electrostatic interactions between the cationic polymers and the viruses.61,62 It is noteworthy that the virus adsorption of PMTAC was higher than that of APTS (Fig. 5). The primary amino group of APTS is considered to be positively charged in the PBS solutions.63 These results suggest that the cationic PMTAC brushes grafted on the substrates efficiently interact with the viruses in the aqueous phase.
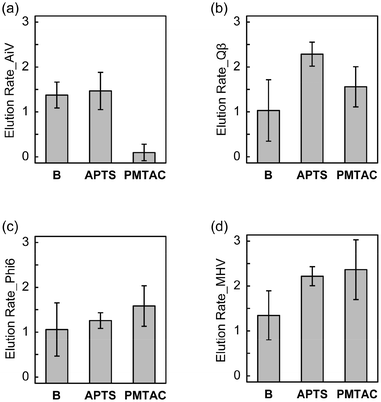
The elution rates of the nonenveloped and enveloped viruses from the surface-modified and bare substrates are shown in Fig. 6, where the values of elution rates closer to zero indicate higher recovery of viruses. PMTAC exhibited higher recovery (elution) of AiV (0.10 ± 0.18 log10Loss, 79.96% in percentage) than APTS (1.47 ± 0.41 log10Loss, 3.42% in percentage) and B (1.38 ± 0.29 log10Loss, 4.22% in percentage) (Fig. 6a). In contrast, the elution rates for Qβ (Fig. 6b), Phi6 (Fig. 6c), and MHV (Fig. 6d) rather decreased by the surface modification with the PMTAC brushes and APTS. Although the elution bahavior is complicated, a combination of electrostatic interactions and hydrophobic effects might result in the selective elution of AiV from the PMTAC brushes.64–66
Tables 2 and S1† summarize the average and significance of differences (p value) of adsorption and elution rates for the silicon-based surfaces. Statistically significant differences in virus adsorption were observed between PMTAC and B (p < 0.001 for AiV, Qβ, and Phi6, p < 0.01 for MHV). Moreover, PMTAC exhibited significantly higher elution of AiV compared to B (p < 0.001). Similar differences were observed between PMTAC and APTS in the virus adsorption (p < 0.001 for AiV, Qβ, and Phi6) and elution (p < 0.01 for AiV and Qβ) (Table S1†). Overall, the tuning of the virus adsorption and elution was demonstrated by using the PMTAC brushes (Fig. 1).
| Materialsa | Viruses | Adsorption | Elution | ||
|---|---|---|---|---|---|
| Averageb (log10Ads) | Averagec (Ads, %) | Averageb (log10Loss) | Averagec (Elu, %) | ||
| a B: bare substrates (n = 8); APTS: 3-aminopropyltriethoxysilane (APTS)-functionalized substrates (n = 4) and PMTAC: poly[2-(methacryloyloxy)ethyltrimethylammonium chloride] (PMTAC)-grafted substrates (n = 10). b The stars indicate the significance of differences (p value) between APTS and B, PMTAC and B. *: p < 0.05; **: p < 0.01; and ***: p < 0.001. c The average of adsorption rates (log10Ads) and elution rates (log10Loss) is calculated into the form of percentage. | |||||
| B | AiV | 0.13 | 25.11 | 1.38 | 4.22 |
| Qβ | 0.55 | 71.97 | 1.03 | 9.33 | |
| Phi6 | 0.23 | 41.33 | 1.06 | 8.72 | |
| MHV | 0.86 | 86.35 | 1.35 | 4.50 | |
| APTS | AiV | −0.12** | −33.08 | 1.47 | 3.42 |
| Qβ | 0.36* | 56.83 | 2.29*** | 0.52 | |
| Phi6 | 0.34 | 54.32 | 1.26 | 5.52 | |
| MHV | 1.01 | 90.23 | 2.22** | 0.61 | |
| PMTAC | AiV | 1.07*** | 91.49 | 0.10*** | 79.96 |
| Qβ | 1.66*** | 97.81 | 1.56 | 2.77 | |
| Phi6 | 1.15*** | 92.94 | 1.58 | 2.61 | |
| MHV | 1.60** | 97.50 | 2.36** | 0.43 | |
A plaque assay was performed to quantify Qβ as a representative infectious nonenveloped virus (Fig. 7). Comparison of the concentrations of Qβ measured by RT-qPCR and plaque assay provides insights into the inactivation of the phages.15 For the feed solutions, the differences in the concentrations of Qβ observed between RT-qPCR and plaque assay were around 2 log (Fig. 7). The bare and modified substrates also exhibited similar differences for both the Permeate (Fig. 7a) and Eluate (Fig. 7b). These results imply that the integrity of the virus particle is maintained in the adsorption and elution processes.
Conclusions
In conclusion, we have demonstrated that the cationic PMTAC brushes enhanced not only the adsorption of enveloped and nonenveloped viruses but also the elution of AiV. The surface modification with the cationic polymers can be applied to practical virus adsorption materials with high surface area such as gauze and glass fibers. Grafting the cationic polymer brushes may provide a new direction in controlling virus adsorption and elution for passive sampling of viruses from water. The efficient adsorption and elution of target viruses may lead to precise monitoring of infectious diseases in local communities.Experimental
Materials and methods
All the reagents for the preparation of the surface-modified substrates were purchased from Tokyo Chemical Industry (Tokyo, Japan), Sigma-Aldrich Japan (Tokyo, Japan), FUJIFILM Wako (Osaka, Japan), and Nacalai Tesque (Kyoto, Japan). These reagents were used as received unless otherwise stated. 2-Bromo-2-methyl-N-(3-(triethoxysilyl)propyl)propanamide was synthesized according to a procedure previously reported.67 The surface of silicone substrates was cleaned with a FEMTO SCIENCE plasma treatment chamber CIONE4 filled with argon (80 W, 1 min) before the surface modification.Fourier-transform infrared (FT-IR) spectra were obtained with a JASCO FT/IR-6100 Plus spectrometer and a JASCO IRT-5000 in the reflection mode. Atomic force microscopy (AFM) was performed using a Bruker MultiMode 8 atomic force microscope with a Nanoscope V controller. Contact angle measurements were performed using a Kyowa Interface Science DropMaster DMF-301 contact angle meter. The measurements were conducted at four different points for each substrate. UV-vis absorption spectroscopy was performed with a JASCO V-670 spectrophotometer.
Preparation of the surface-modified substrates
A solution of [2-(methacryloyloxy)ethyl]trimethylammonium chloride (75 wt% in H2O, 2.85 g, 10.3 mmol), methanol (2.0 mL), and deionized water (5.2 mL) was degassed by three freeze–pump–thaw cycles. N,N,N′,N′′,N′′-Pentamethyldiethylenetriamine (PMDETA) (64.6 mg, 0.37 mmol) and CuBr (12.6 mg, 0.088 mmol) were added to the deoxygenated solution under argon flow. The mixture was stirred for 5 min at 25 °C. To this mixture, the ATRP-initiator modified substrate was transferred. The mixture was stirred for 5 h at 25 °C. After removing the solution, the substrate was washed with methanol and deionized water and dried with a stream of argon to give a PMTAC-grafted substrate.
Estimation of the number of solvent-accessible cations
The number of solvent-accessible cations on the PMTAC-grafted, APTS-functionalized, and nonmodified bare substrates was estimated by a fluorescein adsorption assay according to procedures reported previously.53,54 The substrates with a total surface area of 2 cm2 were immersed in 1 wt% aqueous solution of sodium fluorescein (10 mL) for 20 min under gentle shaking. These substrates were washed with deionized water and placed in water (10 mL) with ultrasonication for 10 min. This washing process was repeated three times. To desorb the immobilized fluorescein, the substrates were immersed in a 0.1 wt% aqueous solution of cetyltrimethylammonium chloride (9 mL) for 30 min under gentle shaking. 0.1 M PBS buffer (pH 8.0) (1 mL) was added to the solution after the removal of the substrates. The absorbance of the solution at 501 nm was measured. The concentration of the desorbed fluorescein was estimated based on Beer–Lambert law with the extinction coefficient of fluorescein of 77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1, and an optical path length of 1 cm. The number of solvent-accessible cations on the substrates can be correlated with the concentration of fluorescein by assuming a 1
000 M−1 cm−1, and an optical path length of 1 cm. The number of solvent-accessible cations on the substrates can be correlated with the concentration of fluorescein by assuming a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexation of fluorescein with the cationic group. These measurements were performed three times for each substrate.
1 complexation of fluorescein with the cationic group. These measurements were performed three times for each substrate.
Virus preparation
Phi6 and MHV were used as surrogates to represent enveloped viruses, whereas Qβ and AiV were used for non-enveloped viruses. Bacteriophage Phi6 (NBRC 105899, National Institute of Technology and Evaluation, Tokyo, Japan) was propagated using P. syringae (NBRC14084, NITE) as the host bacterium according to the protocol. Bacteriophage Qβ (ATCC23631-B1, the American Type Culture Collection, Manassas, Virginia, USA) was propagated in the lab using Escherichia coli K12 A/λ F+ as the host bacteria.68 The AiV strain (strain A846/88 donated by Dr Yamashita from Shubun University, Aichi, Japan) and MHV A59 strain (ATCC VR-764) were propagated using buffalo green monkey kidney cells and DBT cells, respectively, as described before.69,70 Host cells were removed by centrifugation at 3000 rpm for 10 min followed by membrane filtration with a cellulose acetate filter (0.2 μm, DISMIC-25CS, Advantec, Tokyo, Japan). Then the viruses were further purified by gel filtration with an Illustra Microspin S-300 HR column (GE Healthcare, Tokyo, Japan).71,72 The purified viruses were quantified by RT-qPCR (MHV, AiV, Qβ and Phi6) and plaque assay (Qβ), aliquoted and stored at −80 °C until being used as intact virus stock. For each experiment, the 4 types of viruses were mixed into 1 mL of 100 mM PBS to make the feed solution (Feed) at the initial concentrations of 108 PFU mL−1 for Qβ, and 109 – 1010 copies per mL for AiV, Qβ, Phi6, and MHV.Virus adsorption and elution
A 5 μL feed solution was dropped onto a test substrate (1 cm × 1 cm) in a Petri dish. A cover glass was placed on top of the substrate. After incubation at 25 °C for 5 h, the substrate was washed with 100 mM PBS (2.5 mL). The solution was collected as the permeate (referred to as Permeate).The substrate used for virus adsorption was transferred to a 12-well cell culture plate and soaked in 2.5 mL of sterile beef extract (3%, pH = 9). After incubation at 25 °C for 1 h, the solution was collected as the eluate (referred to as Eluate). Throughout the experiments, the Feed, Permeate, and Eluate samples were stored at 4 °C until further analysis within 24 h. Successive treatment of adsorption and elution was performed for each test substrate.
Quantification of viruses
The concentrations of all the tested viruses in the Feed, Permeate, and Eluate were determined using reverse transcription quantitative polymerase chain reaction (RT-qPCR). Initially, RNA was extracted from 140 μL of each sample utilizing a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany). Subsequently, reverse transcription was performed using a High-Capacity cDNA Reverse Transcription Kit with an RNase Inhibitor (Applied Biosystems, USA). The resulting cDNA products were subjected to qPCR analysis. For the qPCR, a 25 μL reaction mixture was prepared, comprising 12.5 μL of TaqManTM Gene Expression Master Mix (Applied Biosystems), 1 μL of forward and reverse primers (10 μmol L−1), 0.5 μL of TaqMan probes (5 μmol L−1), and 5 μL of cDNA. The primers for Qβ,73 Phi6,74 AiV,75 and MHV76 were prepared according to the literature. RT-qPCR was carried out in duplicate on an ABI StepOnePlus thermocycler (Thermo Fisher, USA), following the recommended temperature conditions outlined in the previous studies, which varied depending on the virus type. For each 96-well RT-qPCR plate, negative controls were performed with sterile water. Standard curves were generated using 10-fold serial dilutions of pre-determined plasmid DNA containing the target gene sequence, covering target virus concentrations ranging from 106 to 100 copies per reaction. The amplification efficiencies exceeded 90% and the correlation factor (R2) was higher than 0.99. The assay limit of detection was 2 copy per μL (10 copy per reaction) for all the tested viruses. The process limit of detection was 1720 copies per mL sample.77In order to assess the adsorption and elution rates of infectious viruses in passive sampling, a plaque assay was conducted for the bacteriophage Qβ. To perform the assay, the Feed, Permeate, and Eluate samples were subjected to serial 10-fold dilution and plated onto LB agar plates pre-inoculated with host bacteria, specifically E. coli K12 F+ (A/λ). Duplicate plates were prepared for each sample at every dilution level. The plates were then incubated at 37 °C overnight to allow the formation of visible plaques, which were subsequently counted for analysis.
Author contributions
T. K. and H. K. conceived and designed the project. J. U., M. L., and S. M. performed the experiments and data analysis. J. U. and T. K. wrote the manuscript. All authors read and commented on the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by JST, CREST Grant Number JPMJCR20H3 and JSPS KAKENHI Grant Number JP19H05715 (Grant-in-Aid for Scientific Research on Innovative Area: Aquatic Functional Materials). We greatly appreciate Harumi Ishikawa, Terumi Seya, Miwako Nakagawa, and Dr Tippawan Singhopon for their valuable support in the experiments.References
- M. Kitajima, W. Ahmed, K. Bibby, A. Carducci, C. P. Gerba, K. A. Hamilton, E. Haramoto and J. B. Rose, Sci. Total Environ., 2020, 739, 139076 CrossRef CAS PubMed.
- A. C. Singer, J. R. Thompson, C. R. M. Filho, R. Street, X. Li, S. Castiglioni and K. V. Thomas, Nat. Water, 2023, 1, 408–415 CrossRef.
- K. M. S. de Melo Cassemiro, F. M. Burlandy, M. R. F. Barbosa, Q. Chen, J. Jorba, E. M. Hachich, M. I. Z. Sato, C. C. Burns and E. E. da Silva, PLoS One, 2016, 11, e0152251 CrossRef PubMed.
- J. Habtewold, D. McCarthy, E. McBean, I. Law, L. Goodridge, M. Habash and H. M. Murphy, Environ. Res., 2022, 204, 112058 CrossRef CAS PubMed.
- M. Kitajima, M. Murakami, R. Iwamoto, H. Katayama and S. Imoto, J. Travel Med., 2022, 29, taac004 CrossRef PubMed.
- M. Kitajima and C. P. Gerba, Pathogens, 2015, 4, 256–268 CrossRef PubMed.
- F. S. Le Guyader, J.-C. Le Saux, K. Ambert-Balay, J. Krol, O. Serais, S. Parnaudeau, H. Giraudon, G. Delmas, M. Pommepuy, P. Pothier and R. L. Atmar, J. Clin. Microbiol., 2008, 46, 4011–4017 CrossRef PubMed.
- K. Sdiri-Loulizi, M. Hassine, Z. Aouni, H. Gharbi-Khelifi, N. Sakly, S. Chouchane, M. Guediche, P. Pothier, M. Aouni and K. Ambert-Balay, Arch. Virol., 2010, 155, 1509–1513 CrossRef CAS PubMed.
- M. Kitajima, B. C. Iker, I. L. Pepper and C. P. Gerba, Sci. Total Environ., 2014, 488–489, 290–296 CrossRef CAS PubMed.
- G. Matrajt, B. Naughton, A. S. Bandyopadhyay and J. S. Meschke, Clin. Infect. Dis., 2018, 67, S90–S97 CrossRef CAS PubMed.
- C. Schang, N. D. Crosbie, M. Nolan, R. Poon, M. Wang, A. Jex, N. John, L. Baker, P. Scales, J. Schmidt, B. R. Thorley, K. Hill, A. Zamyadi, C.-W. Tseng, R. Henry, P. Kolotelo, J. Langeveld, R. Schilperoort, B. Shi, S. Einsiedel, M. Thomas, J. Black, S. Wilson and D. T. McCarthy, Environ. Sci. Technol., 2021, 55, 10432–10441 CrossRef CAS PubMed.
- B. Moore, Mon. Bull. Minist. Health Public Health Lab. Serv., 1948, 7, 241–248 Search PubMed.
- J. Voisin, B. Cournoyer and F. Mermillod-Blondin, Houille Blanche, 2015, 4, 52–57 CrossRef.
- F. Vincent-Hubert, B. Morga, T. Renault and F. Le Guyader, J. Appl. Microbiol., 2017, 122, 1039–1047 CrossRef CAS PubMed.
- M. Liu, J. Uchida, S. Matsuyama, T. Kato and H. Katayama, ACS ES&T Water, 2024, 4, 368–376 Search PubMed.
- J. O. Zoppe, N. C. Ataman, P. Mocny, J. Wang, J. Moraes and H.-A. Klok, Chem. Rev., 2017, 117, 1105–1318 CrossRef CAS PubMed.
- Y. Tsujii, K. Ohno, S. Yamamoto, A. Goto and T. Fukuda, Adv. Polym. Sci., 2006, 197, 1–45 CrossRef CAS.
- Y. Higaki, M. Kobayashi, D. Murakami and A. Takahara, Polym. J., 2016, 48, 325–331 CrossRef CAS.
- T. Masuda and M. Takai, J. Mater. Chem. B, 2022, 10, 1473–1485 RSC.
- M. Ejaz, S. Yamamoto, K. Ohno, Y. Tsujii and T. Fukuda, Macromolecules, 1998, 31, 5934–5936 CrossRef CAS.
- T. von Werne and T. E. Patten, J. Am. Chem. Soc., 1999, 121, 7409–7410 CrossRef CAS.
- M. Kohri, Polym. J., 2019, 51, 1127–1135 CrossRef CAS.
- Y. Takeoka, Polym. J., 2015, 47, 106–113 CrossRef CAS.
- H. Arafune, T. Kamijo, T. Morinaga, S. Honma, T. Sato and Y. Tsujii, Adv. Mater. Interfaces, 2015, 2, 1500187 CrossRef.
- K. Kitano, Y. Inoue, R. Matsuno, M. Takai and K. Ishihara, Colloids Surf., B, 2009, 74, 350–357 CrossRef CAS PubMed.
- U. Raviv, S. Giasson, N. Kampf, J.-F. Gohy, R. Jérôme and J. Klein, Nature, 2003, 425, 163–165 CrossRef CAS PubMed.
- M. Kobayashi, Y. Terayama, H. Yamaguchi, M. Terada, D. Murakami, K. Ishihara and A. Takahara, Langmuir, 2012, 28, 7212–7222 CrossRef CAS PubMed.
- J. B. Schlenoff, Langmuir, 2014, 30, 9625–9636 CrossRef CAS PubMed.
- M. Krishnamoorthy, S. Hakobyan, M. Ramstedt and J. E. Gautrot, Chem. Rev., 2014, 114, 10976–11026 CrossRef CAS PubMed.
- N. Hadjesfandiari, K. Yu, Y. Mei and J. N. Kizhakkedathu, J. Mater. Chem. B, 2014, 2, 4968–4978 RSC.
- T. Shimizu, M. Yamato, A. Kikuchi and T. Okano, Biomaterials, 2003, 24, 2309–2316 CrossRef CAS PubMed.
- Y. G. Takei, T. Aoki, K. Sanui, N. Ogata, Y. Sakurai and T. Okano, Macromolecules, 1994, 27, 6163–6166 CrossRef CAS.
- S. Kumar, T. Ito, Y. Yanagihara, Y. Oaki, T. Nishimura and T. Kato, CrystEngComm, 2010, 12, 2021–2024 RSC.
- Y. Han, T. Nishimura and T. Kato, Polym. J., 2014, 46, 499–504 CrossRef CAS.
- Y. Han, T. Nishimura and T. Kato, CrystEngComm, 2014, 16, 3540–3547 RSC.
- B. Michen and T. Graule, J. Appl. Microbiol., 2010, 109, 388–397 CrossRef CAS PubMed.
- M. D. Sobsey and B. L. Jones, Appl. Environ. Microbiol., 1979, 37, 588–595 CrossRef CAS PubMed.
- N. Marets, D. Kuo, J. R. Torrey, T. Sakamoto, M. Henmi, H. Katayama and T. Kato, Adv. Healthcare Mater., 2017, 6, 1700252 CrossRef PubMed.
- K. Hamaguchi, R. Ichikawa, S. Kajiyama, S. Torii, Y. Hayashi, J. Kumaki, H. Katayama and T. Kato, ACS Appl. Mater. Interfaces, 2021, 13, 20598–20605 CrossRef CAS PubMed.
- D. Kuo, M. Liu, K. R. S. Kumar, K. Hamaguchi, K. P. Gan, T. Sakamoto, T. Ogawa, R. Kato, N. Miyamoto, H. Nada, M. Kimura, M. Henmi, H. Katayama and T. Kato, Small, 2020, 16, 2001721 CrossRef CAS PubMed.
- D. Kuo, T. Sakamoto, S. Torii, M. Liu, H. Katayama and T. Kato, Polym. J., 2022, 54, 821–825 CrossRef CAS PubMed.
- T. Sakamoto, K. Asakura, N. Kang, R. Kato, M. Liu, T. Hayashi, H. Katayama and T. Kato, J. Mater. Chem. A, 2023, 11, 22178–22186 RSC.
- K. Fukushima, K. Kishi, K. Saito, K. Takakuwa, S. Hakozaki and S. Yano, Biomater. Sci., 2019, 7, 2288–2296 RSC.
- K. Lienkamp, A. E. Madkour, A. Musante, C. F. Nelson, K. Nüsslein and G. N. Tew, J. Am. Chem. Soc., 2008, 130(30), 9836–9843 CrossRef CAS PubMed.
- F. Nederberg, Y. Zhang, J. P. K. Tan, K. Xu, H. Wang, C. Yang, S. Gao, X. D. Guo, K. Fukushima, L. Li, J. L. Hedrick and Y.-Y. Yang, Nat. Chem., 2011, 3, 409–414 CrossRef CAS PubMed.
- K. Nagase, S. Kitazawa, T. Kogure, S. Yamada, K. Katayama and H. Kanazawa, Sep. Purif. Technol., 2022, 286, 120445 CrossRef CAS.
- K. Matyjaszewski and J. H. Xia, Chem. Rev., 2001, 101, 2921–2990 CrossRef CAS PubMed.
- J. Pyun and K. Matyjaszewski, Chem. Mater., 2001, 13, 3436–3448 CrossRef CAS.
- M. Kobayashi and A. Takahara, Chem. Rec., 2010, 10, 208–216 CrossRef CAS PubMed.
- K. Gao, Y. Su, L. Zhou, M. He, R. Zhang, Y. Liu and Z. Jiang, J. Membr. Sci., 2018, 548, 621–631 CrossRef CAS.
- S. L. Banerjee, T. Swift, R. Hoskins, S. Rimmer and N. K. Singha, J. Mater. Chem. B, 2019, 7, 1475–1493 RSC.
- K. Koch, B. Bhushan and W. Barthlott, Soft Matter, 2008, 4, 1943–1963 RSC.
- J. C. Tiller, C. J. Liao, K. Lewis and A. M. Klibanov, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 5981–5985 CrossRef CAS PubMed.
- E. Zorn, J. I. H. Knaack, N. Burmeister, N. Scharnagl, M. Rohnke, S. G. Wicha and W. Maison, Langmuir, 2023, 39, 11063–11072 CrossRef CAS PubMed.
- N. Shirasaki, T. Matsushita, Y. Matsui, T. Urasaki and K. Ohno, Water Res., 2009, 43, 605–612 CrossRef CAS PubMed.
- J. Langlet, L. Ogorzaly, J.-C. Schrotter, C. Machinal, F. Gaboriaud, J. F. L. Duval and C. Gantzer, J. Membr. Sci., 2009, 326, 111–116 CrossRef CAS.
- T. Yamashita, S. Kobayashi, K. Sakae, S. Nakata, S. Chiba, Y. Ishihara and S. Isomura, J. Infect. Dis., 1991, 164, 954–957 CrossRef CAS PubMed.
- M. Bárcena, G. T. Oostergetel, W. Bartelink, F. G. A. Faas, A. Verkleij, P. J. M. Rottier, A. J. Koster and B. J. Bosch, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 582–587 CrossRef PubMed.
- A. Katz, S. Peña, A. Alimova, P. Gottlieb, M. Xu and K. A. Block, Sci. Total Environ., 2018, 637–638, 104–111 CrossRef CAS PubMed.
- K. J. Schwab, R. D. Leon and M. D. Sobsey, Appl. Environ. Microbiol., 1995, 61, 531–537 CrossRef CAS PubMed.
- M. D. Sobsey and B. L. Jones, Appl. Environ. Microbiol., 1979, 37, 588–595 CrossRef CAS PubMed.
- R. Attinti, J. Wei, K. Kniel, J. T. Sims and Y. Jin, Environ. Sci. Technol., 2010, 44, 2426–2432 CrossRef CAS PubMed.
- J. Liu, H. Zhang, D. Xue, A. ul Ahmad, X. Xia, Y. Liu, H. Huang, W. Guo and H. Liang, RSC Adv., 2020, 10, 11393–11399 RSC.
- C. P. Gerba, Adv. Appl. Microbiol., 1984, 30, 133–168 CrossRef CAS PubMed.
- A. Armanious, M. Aeppli, R. Jacak, D. Refardt, T. Sigstam, T. Kohn and M. Sander, Environ. Sci. Technol., 2016, 50, 732–743 CrossRef CAS PubMed.
- J. Langlet, F. Gaboriaud, J. F. L. Duval and C. Gantzer, Water Res., 2008, 42, 2769–2777 CrossRef CAS PubMed.
- M. J. Mulvihill, B. L. Rupert, R. He, A. Hochbaum, J. Arnold and P. Yang, J. Am. Chem. Soc., 2005, 127, 16040–16041 CrossRef CAS PubMed.
- R. Kato, T. Asami, E. Utagawa, H. Furumai and H. Katayama, Water Res., 2018, 132, 61–70 CrossRef CAS PubMed.
- V. D. Canh, I. Kasuga, H. Furumai and H. Katayama, Food Environ. Virol., 2019, 11, 40–51 CrossRef CAS PubMed.
- V. D. Canh, S. Torii, H. Furumai and H. Katayama, Water Res., 2021, 189, 116674 CrossRef CAS PubMed.
- M. Kitajima, T. Oka, E. Haramoto, H. Katayama, N. Takeda, K. Katayama and S. Ohgaki, Appl. Environ. Microbiol., 2010, 76, 2461–2467 CrossRef CAS PubMed.
- M. Kitajima, T. Oka, E. Haramoto, N. Takeda, K. Katayama and H. Katayama, Environ. Sci. Technol., 2010, 44, 7116–7122 CrossRef CAS PubMed.
- S. Wolf, J. Hewitt and G. E. Greening, Appl. Environ. Microbiol., 2010, 76, 1388–1394 CrossRef CAS PubMed.
- L. Gendron, D. Verreault, M. Veillette, S. Moineau and C. Duchaine, Aerosol Sci. Technol., 2010, 44, 893–901 CrossRef CAS.
- M. Kitajima, A. Hata, T. Yamashita, E. Haramoto, H. Minagawa and H. Katayama, Appl. Environ. Microbiol., 2013, 79, 3952–3958 CrossRef CAS PubMed.
- G. D. Smith, P. J. Solenberg, M. C. Koenig, K. A. Brune and N. Fox, Comp. Med., 2002, 52, 456–460 CAS.
- W. Ahmed, A. Bivins, S. Metcalfe, W. J. Smith, M. E. Verbyla, E. M. Symonds and S. L. Simpson, Water Res., 2022, 213, 118132 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3lp00216k |
| This journal is © The Royal Society of Chemistry 2024 |