 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Microfluidic systems for infectious disease diagnostics
Thomas
Lehnert
 * and
Martin A. M.
Gijs
* and
Martin A. M.
Gijs

Laboratory of Microsystems, École Polytechnique Fédérale de Lausanne, Lausanne, CH-1015, Switzerland. E-mail: lehnert77@gmail.com
First published on 19th February 2024
Abstract
Microorganisms, encompassing both uni- and multicellular entities, exhibit remarkable diversity as omnipresent life forms in nature. They play a pivotal role by supplying essential components for sustaining biological processes across diverse ecosystems, including higher host organisms. The complex interactions within the human gut microbiota are crucial for metabolic functions, immune responses, and biochemical signalling, particularly through the gut–brain axis. Viruses also play important roles in biological processes, for example by increasing genetic diversity through horizontal gene transfer when replicating inside living cells. On the other hand, infection of the human body by microbiological agents may lead to severe physiological disorders and diseases. Infectious diseases pose a significant burden on global healthcare systems, characterized by substantial variations in the epidemiological landscape. Fast spreading antibiotic resistance or uncontrolled outbreaks of communicable diseases are major challenges at present. Furthermore, delivering field-proven point-of-care diagnostic tools to the most severely affected populations in low-resource settings is particularly important and challenging. New paradigms and technological approaches enabling rapid and informed disease management need to be implemented. In this respect, infectious disease diagnostics taking advantage of microfluidic systems combined with integrated biosensor-based pathogen detection offers a host of innovative and promising solutions. In this review, we aim to outline recent activities and progress in the development of microfluidic diagnostic tools. Our literature research mainly covers the last 5 years. We will follow a classification scheme based on the human body systems primarily involved at the clinical level or on specific pathogen transmission modes. Important diseases, such as tuberculosis and malaria, will be addressed more extensively.
1 Introduction
The burden of infectious diseases
Infectious diseases are undeniably linked to the fate of human society, be it on a regional or global scale. Even before the era of globalization, spreading of infectious pathogens by human migration caused substantial morbidity and mortality.1 Large parts of the native population of the Americas were devastated by smallpox and measles during the European conquest.2 More recently, the Spanish flu (1918–1920), one of the most severe pandemics in history, infected up to one third of the global population at that time with at least 50 million deaths.3,4 Over the last decades, a rise in human infectious disease outbreaks was observed on a global scale (time frame 1980–2013).5 An analysis based on disability-adjusted life years (DALY) indicated that in Europe (2009–2013) seasonal influenza burden was the highest, followed by tuberculosis, human immunodeficiency virus (HIV), and invasive pneumococcal disease (IPD).6 Another extensive study on the global burden of disease (1990–2019) also listed different infectious diseases, depending on the age category, among the top-ranked causes of DALYs.7 Increasing densification of populations in urban areas and global mobility fosters outbreaks of communicable diseases. This was dramatically demonstrated by the latest COVID-19 pandemic related to the air-borne SARS-CoV-2 coronavirus.8 Infectious diseases of poverty (including, among others, malaria, tuberculosis, AIDS and neglected tropical infections) disproportionately affect populations in developing countries.9–12 Endemic or epidemic outbreaks of tropical vector-borne diseases (e.g. dengue,13 Zika or chikungunya) or hemorrhagic fevers (e.g. Ebola14), as well as infections due to food- or water-borne pathogens (e.g. Salmonella infections15 or cholera) are recurring.16,17 Moreover, climate change affects regional vector and pathogen distributions, thereby playing an increasingly important role in the evolving global landscape of infectious diseases.18,19The current health system is facing growing challenges due to the fast and dynamic evolution of societal and environmental parameters that impact pathogen transmission, distribution and biological adaptation strategies.1 Precise epidemiological approaches to infectious diseases are needed to be prepared for future outbreaks, for instance by implementing technological advances enabling a broader application of pathogen genome sequencing.20 Vaccines are probably among one of the most important achievements of humanity, enabling the eradication (smallpox, poliomyelitis) or at least control (e.g. measles/mumps/rubella, hepatitis, influenza etc.) of several severe infectious diseases.21 For others, vaccines are currently under development or undergoing the WHO evaluation process.22 A prominent example is the recent approval of malaria vaccines, and the launch of large-scale vaccination campaigns.23,24 However, such protection does not yet exist against some major pathogens, such as AIDS/HIV, for instance. The importance of the development and fast implementation of new vaccine concepts in a context of emerging viral diseases and constantly arising genetic mutations became evident during the COVID-19 pandemic.25,26 Artificial intelligence is expected to facilitate vaccine or drug design and significantly support progress in the fight against infectious diseases in general.27
Emerging challenges of the global health system
One of the major upcoming threats to global health is antimicrobial resistance (AMR) to drugs, in particular for the ESKAPE pathogen species.28 Methicillin-resistant Staphylococcus aureus (MRSA) is only one example for a globally disseminated superbug.29 AMR is a leading cause of death around the world, with the highest burden in the developing world.30 Following the current trend, AMR is expected to cause more deaths than cancer in a few decades. New resistance mechanisms are emerging and spreading rapidly on a global scale, challenging our ability to treat common infectious diseases, due an increasingly limited availability of still efficient or new antibiotics. For instance, multidrug-resistant tuberculosis does not respond to the first-line drugs isoniazid and rifampicin, thus requiring extensive second-line treatments.31 Among major reasons for this situation are the empirical and often unnecessary prescription of (broad-spectrum) antibiotics in human disease management,32 but also abusive use in the agriculture/veterinary sector.33 New surveillance and antimicrobial stewardship strategies are therefore urgently needed.34,35 One of the keys is the development of rapid antimicrobial susceptibility testing (AST), including microfluidic and biosensor-based methods, for the appropriate choice of prescription at the point-of-care (POC).36–43Today diagnostics still relies on time-consuming pathogen culture-based methods and/or on techniques that are limited to central laboratory facilities. This is a particular problem for primary health care in low-resource settings, most likely facing severe constraints, due to a lack of infrastructure and related technical issues (e.g. hazardous electricity supply and refrigeration), health workers with insufficient qualification and limited accessibility in rural areas.44 As a consequence, the benefit and outcome of individual healthcare and disease management on a larger scale is very limited. To address these challenges, new paradigms for fast POC pathogen detection and identification, possibly combined with rapid AST, are required.45,46 Advanced microfluidic approaches demonstrate high potential in this regard.
Motivation and scope of the review
Our review offers an opportunity to explore recent research trends and emerging technologies in the field of microfluidic systems enabling rapid and sensitive detection of pathogens or biomarkers associated with infectious diseases. This topic encompasses various disciplines such as micro- and molecular biology, engineering, healthcare delivery, and public health. While existing reviews often focus on specific applications, pathogen types, or technologies, our aim is to provide a comprehensive resource for researchers and stakeholders seeking to advance infectious disease diagnostics. Throughout this review, we will highlight the latest advancements in sensitive biosensor technologies, innovative assay strategies, and advanced microfluidic integration, all of which show great promise in facilitating near-patient testing and enabling real POC diagnosis of infectious diseases. Furthermore, in a broader context, we aim to underscore the potential of microfluidics-based technology in addressing the increasing burden of infectious diseases on global healthcare.Our review begins by briefly introducing the scope and background of microfluidics and biosensors, along with a non-exhaustive summary of available review articles related to infectious disease diagnostics (section 2). Following this, we will elaborate on our discussion of recent microfluidic devices and platforms. We adopt a classification scheme primarily based on the human body systems involved at the clinical level. This classification includes infections of the respiratory tract (section 3), the urinary tract (section 4), the gastrointestinal tract (section 5) and the bloodstream (section 6). For other pathogens or diseases, categorization by specific transmission mode was more convenient, in particular for sexually transmitted infections (section 7) and vector-based infections (section 8). Additionally, based on the availability of recent developments in microfluidic systems, we will provide more in-depth discussions of key diseases, namely tuberculosis and malaria.
Our approach in this review is as follows: (i) in sections 3 to 8, we aim to provide a comprehensive overview of the state-of-the-art of microfluidic biosensor-based systems for infectious disease diagnostics, with an emphasis on microtechnological or microfluidic aspects. Recent devices for rapid AST will also be included. The timeframe covers the last 5 years (2018–2023, with a few exceptions). Corresponding tables outline the most relevant work, ordered by pathogens or analytical targets. (ii) Each section introduces the scope of the infection category, emphasizing microbiological, biomedical, or societal aspects. We believe this approach is crucial for our review, as it provides a concise insight into the complexity of each topic, especially for microfluidic system developers. This not only sets the framework that motivates advanced technological developments but may also help bridge the gap between research and clinical practice in this interdisciplinary field. To this end, we have included highly relevant articles related to each specific topic of our classification scheme. These articles do not focus on microfluidics but cover essential background information and may therefore have been published before 2018. (iii) Each section of our tutorial review also includes an overview of corresponding existing reviews, some of which were published before 2018. In general, these articles focus on specific applications, types of pathogens, or technologies. This approach allows the reader to explore a topic of particular interest more thoroughly.
2 Lab-on-a-chip devices for infectious disease diagnostics
2.1 Microfluidics and microfluidic devices
2.2 Biosensor technologies and analytical nucleic acid-based assays
Actual laboratory procedures, POC devices and commercial systems for clinical diagnosis of infectious diseases cannot necessarily meet the emerging needs of the global health system, such as the capability to respond rapidly and on a population-wide scale to the increasing risk of viral disease outbreaks, or to perform accurate informed diagnosis and screening campaigns of antimicrobial-resistant bacterial strains.87,88 Emerging diagnostic methods, based on microfluidic and biosensor integration are therefore being developed, aiming POC pathogen detection/identification with high sensitivity and specificity at early stages of infection.89,902.3 Microfluidic cartridge-based commercial systems
Commercial benchtop systems for infectious disease diagnostics aim to implement sample-to-answer strategies, often designed for fully integrated and automated NAAT methods for pathogen detection, requiring only minimal hands-on steps for sample preparation and assay protocol operations. Nevertheless, constraints like system or assays cost or the requirement of external power supplies may still limit the use in low resource settings. Wang et al. provide a comprehensive tabular comparison of microfluidic POC platforms for molecular diagnostics arranged by approval time.107 In a review on diagnostic tools for tackling febrile illness, Mitsakakis et al. proposed detailed descriptions of relevant microfluidic cartridge-based commercial platforms.108 Other authors focus on more specific applications, such as Nelson et al., who presented available POC tests and systems for respiratory viruses.109 Commercial assays for specific types of infections will be addressed in the corresponding sections of this review.The GeneXpert® (Cepheid, USA) is an example for a microfluidic cartridge-based RT-PCR system that returns test results in about an hour, including minimum sample preparation. Individual assay cartridges are generally designed for one or two pathogens.110 The BioFire® FilmArray® (BioFire Diagnostics/bioMérieux, USA) is a nested multiplex PCR system for panels of more than 20 targets and a throughput of up to 175 samples per day (unprocessed samples, results in about an hour). Reagents are stored in a pouch in freeze-dried format.111 Another chip-based approach is the VerePLEX™ Biosystem platform (Veredus Laboratories, Singapore) that offers chip panels for multiplex (more than 10) molecular testing of different pathogen families, including custom-designed applications (VereChip™). The cartridge comprises a microfluidic PCR unit and microarray modules for multiplexed DNA amplification and detection, respectively. Time to result is approximately 3.5 h.112 The Bosch Vivalytic Analyser (Bosch Healthcare Solutions, Germany) is an automated cartridge-based molecular diagnostics POC platform for rapid detection of multiple pathogens.113 The microfluidic cartridges have been developed by means of a rapid prototyping approach using generic polymer parts.114 A PCR test portfolio covering a wide range of pathogens is available.115 The LabDisk centrifugal microfluidic platform from Hahn-Schickard116 (Germany) and IMTEK (University of Freiburg, Germany) is a versatile technology that has been used for a variety of applications.108 The Rhonda player, a component of an in vitro diagnostic system based on the LabDisk technology, was successfully introduced to the market in 2020, in particular in combination with a SARS-CoV-2 RT-PCR test (Spindiag). The Rhonda player is now manufactured by Dialunox (Germany).117
2.4 Microfluidic-based diagnostics for infectious diseases – relevant review articles
Some review articles addressing the field of microfluidic-based infectious disease diagnostics form a broader perspective will be cited in the following (non-exhaustive list). X. Wang et al. discussed microfluidic strategies for molecular diagnostics of infectious diseases.107 Flores-Contreras et al. summarized microfluidic biosensing platforms for POC testing SARS-CoV-2 and seroprevalence.118 C. Wang et al. presented an extensive review on POC diagnostics for infectious diseases from the device/application perspective.119 Basiri et al. introduced microfluidic devices for detection of RNA viruses.120 Rezvani Jalal et al. was interested in magnetic nanomaterials in microfluidic sensors for virus detection, and applications were classified by the type of virus.121 Mitsakakis et al. approached the topic of infectious/tropical diseases by investigating diagnostic tools for febrile illness and enhancing patient management.108 Earlier relevant reviews have been proposed by Magro et al., who focused on NAAT combined with paper microfluidics for infectious diseases diagnosis,83 or Tay et al., who reviewed advances in microfluidics in combating infectious diseases.122 Damhorst et al. explored microfluidics and nanotechnology for detection of global infectious diseases, in particular for detection of HIV, malaria, and tuberculosis.123 A host of other review articles focusing on specific types of infections or diseases exists. We will summarize these articles in the corresponding sections of the present review. A large number of microfluidic biosensing platforms has also been designed for the detection of foodborne pathogens.124,1253 Respiratory tract infections
3.1 Scope and common pathogens
Respiratory tract infections (RTI) may be conveniently categorized as upper respiratory tract infection (URI) (nasal cavity, pharynx and larynx) or lower respiratory tract infection (LRI) affecting trachea, bronchi and the lung. Nevertheless, several pathogens, such as influenza viruses, may progressively infect the upper and lower parts likewise.126 Typically, respiratory pathogens may be detected in saliva, nasopharyngeal swabs or blood. Most of URIs (common cold, pharyngitis, sinusitis, etc.) are caused by viruses and are far less severe than LRIs, causing for instance whooping cough (pertussis) and or potentially life-threatening pneumonia.127,128 LRIs are among the leading causes of death, even before the COVID-19 pandemic outbreak. According to a WHO factsheet (2020), LRIs claimed 2.6 million lives in 2019, thus being the world's most deadly communicable disease category at that time.129 Superinfections, e.g. influenza virus-associated bacterial pneumonia, increase disease severity and mortality.130 Annual seasonal epidemics generated by influenza viruses type A or B are estimated to result in about 3 to 5 million cases of severe illness worldwide and hundreds of thousands respiratory deaths.131 A strain of the H1N1 influenza virus caused the extremely deadly Spanish flu pandemic outbreak in 1918.4 Among the multiple pathogenic conditions related to infections with coronaviruses, in particular the Middle East respiratory syndrome coronavirus (MERS-CoV) and the severe acute respiratory syndrome coronavirus (SARS-CoV), pneumonia-associated respiratory disorders are common.132 An ongoing WHO update indicated that the SARS-CoV-2/COVID-19 pandemic caused a cumulative number of nearly 7 million deaths until the end of 2023 worldwide.8 Other common viral pathogens possibly leading to serious respiratory illnesses include the respiratory syncytial virus (RSV), human parainfluenza viruses (HPIV), human adenoviruses (HAdV), human metapneumovirus (HMPV), human rhinovirus (HRV) or the human bocavirus (HBoV).109,133Streptococcus pneumoniae is the most prevalent bacterial microorganism pathogen in community-acquired pneumonia (CAP), with an increasing global burden related to drug-resistant strains.134,135 Among other CAP-causative pathogens are Klebsiella pneumoniae, Haemophilus influenzae, and Pseudomonas aeruginosa.136,137Mycoplasma pneumoniae generally causes mild infections, but which may evolve in more severe respiratory illness.138 Nosocomial pneumonia is the leading cause of mortality attributed to hospital-acquired infections and is significantly challenged by drug-resistance bacterial strains.139Tuberculosis, usually affecting the lungs, is caused by the bacillus Mycobacterium tuberculosis that spreads from person to person through the air. On a global scale, tuberculosis is still one of the leading causes of death due to an infectious agent and the second leading infectious killer after COVID-19 (2021).140 The scope of the disease and microfluidics-based tuberculosis diagnostics will be extensively discussed in a separate section.
3.2 Commercial platforms and reviews in the field
Nelson et al. analysed the landscape of current and future POC tests for common, emerging and novel respiratory viruses. In particular, this review provides extensive tabular overviews on available commercial devices for nucleic acid and antigen POC or near-POC tests, as well as links to corresponding datasheets, company websites or device evaluation studies. We refer to this review for more details on actual commercial systems.109 Huang et al. also evaluated the diagnostic accuracies of three multiplex PCR systems for the detection of viral respiratory infections.141 Examples among commercially available microfluidic molecular diagnostic systems are the GeneXpert® (Cepheid, USA) or the BioFire® FilmArray® (bioMérieux) which, for instance, a pneumonia panel test for 33 bacterial and viral clinically relevant RTI pathogens is available. The Rhonda system was used to screen the international biathlon season during the COVID-19 pandemic and a total of 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 182 tests were made during a 4 month period (2020–2021).142 The assay portfolio of the Bosch Vivalytic® also includes RTI tests, in particular a rapid SARS-CoV-2 test that can be performed within 39 min.143
182 tests were made during a 4 month period (2020–2021).142 The assay portfolio of the Bosch Vivalytic® also includes RTI tests, in particular a rapid SARS-CoV-2 test that can be performed within 39 min.143
Recent more general discussions on LOC-based methods for virus detection also include examples of respiratory pathogens.120,144 Fostered by the recent SARS-CoV-2 pandemic, a host of authors focused more specially on microfluidic and biosensor POC tools for respiratory virus detection. The non-exhaustive list of most recent articles includes a review by Breshears et al. on biosensor technology with a discussion on the background of airborne virus transmission,145 a contribution by Zhang et al. on advanced POC technologies for eight typical acute human respiratory viruses,146 a review by Tarim et al. on microfluidic virus detection methods for respiratory diseases,147 and discussions by Qin et al. of integrated micro- and nanosystems for COVID-19/viral infection diagnostics,148 or by Ribeiro et al. of RTI-related biosensor technologies, respectively.149 Flores-Contreras et al. explored emerging frontiers in POC testing SARS-CoV-2 and seroprevalence.118 In the review by Goud et al. on electrochemical diagnostics of infectious viral diseases, biosensors specifically designed for COVID-19/SARS-CoV-2 have been included.96 Zenhausern et al. explored microfluidic sample preparation for respiratory virus detection,150 whereas Krokhine et al., as well as Lee et al., discussed microfluidic sampling methods for airborne virus isolation and bioaerosol, respectively.151,152 Shabani et al. focused on laboratory detection methods for human coronaviruses, including RT-PCR, RT-LAMP, electrochemical and optical biosensors for RNA detection, and whole virus or viral proteins detection assays.153 Chen et al. also reviewed emerging detection technologies and auxiliary analysis for COVID-19.154 In the following, we discuss a selection of microfluidic/biosensor systems for respiratory virus and bacteria detection. Table 1 provides an overview of recent approaches for RTI pathogen detection.
| Pathogens | Device and assay principle | Performance indications | Ref. |
|---|---|---|---|
| Microfluidic platforms or devices based on NAAT assays | |||
| Up to 21 RTI viruses, SARS-CoV-2 variants | Multiplexed CRISPR-based droplet/microwell platform | 300–550 patient specimens in an 8 h working day | 155 |
| HAdV, HBoV, S. pneumonia | Hybridization chain reaction in an encoded particle platform | High multiplexing capability, low fM LOD values | 157 |
| SARS-CoV-2, RSV, influenza | Multiplexed chip-powered CRISPR/Cas12a system | Detection of co-infection in clinical swab samples | 159 |
| Up to 19 RTI pathogens | RT-PCR assays on a LoaD platform | Multiplex detection of pathogen panels in a single run within 200 min | 158 |
| RTI pathogens panels | Various (isothermal) NAAT-based LoaD systems | Simultaneous detection of several pathogens, typically within less than 1–2 h | 160–163 |
| Up to 21 RTI pathogens | Fully integrated RT-PCR array system | Process completed within 1.5 h. Tested with clinical samples. LOD ∼1 × 103 viral copies per mL | 164 |
| SARS-CoV-2 | Multifunctional micro-PCR droplet/microwell platform | Fast screening (running time 15 min). LOD of 10 nucleic acid copies per test | 165 |
| 11 RTI pathogens | Digital microfluidic RT-qPCR platform | LOD 12 to 150 copies per test, using positive plasmids samples | 166 |
| SARS-CoV-2, influenza, HPV | RT-PCR system with a gravity-driven microfluidic cartridge | qPCR in <30 min, up to 12 cartridges per test | 167 |
| 5 RTI pathogens | Quantitative multiplex digital PCR on a self-partitioning SlipChip | Melting curve analysis with a resolution of 1.5 K enabled amplicon classification | 179 |
| M. pneumoniae | qPCR on a 3D-printed device | Macrolide-resistant genes detection in PCR tubes fitted to the chip | 180 |
| B. pertussis | Paper/polymer hybrid microfluidic biochip integrated with LAMP | Tested with clinical samples. LOD 5 DNA copies, within 45 min | 182, 183 |
| Other microfluidic approaches | |||
| SARS-CoV-2 and H1N1 | Nanotemplating fluidic impedimetric assay with multiplexed readout | Parallel detection of viral load and specific antibodies in saliva or blood within 11 min | 168 |
| SARS-CoV-2 | 3D-printed LOC with multiplexed electrochemical outputs | Concurrent detection of SARS-CoV-2 RNA and anti-SARS-CoV-2 antibodies in saliva | 169 |
| SARS-CoV-2, influenza, HAdV, RSV | Immunoassays in microarray-format on a LoaD | Semi-automated analysis of 6 samples in 30 min (serum and nasopharyngeal samples) | 170 |
| SARS-CoV-2 | ELISA chip with a coil microreactor | Custom-developed antibodies and colorimetric read-out | 171 |
| Specific biosensing methods | |||
| SARS-CoV-2 and H1N1 | Aptamer-based detection on a rotational paper-based device | Aptamer attachment on a tetrahedral DNA framework improved assay performance | 174 |
| SARS-CoV-2 | Viral RNA sensing on ssDNA coated SiO2 slides | LOD 10 aM for viral RNA in saliva. Detection in <10 min | 175 |
| SARS-CoV-2 | Nucleic acid hybridization on a plasmonic biosensors | Thermoplasmonic heat generated on gold nanoislands improved performance | 172 |
| SARS-CoV-2 | Field-effect transistor based immunological assay | Graphene coating enhanced sensitivity. LOD 2.4 × 102 copies per mL with clinical samples | 173 |
3.3 Microfluidic systems for respiratory virus detection
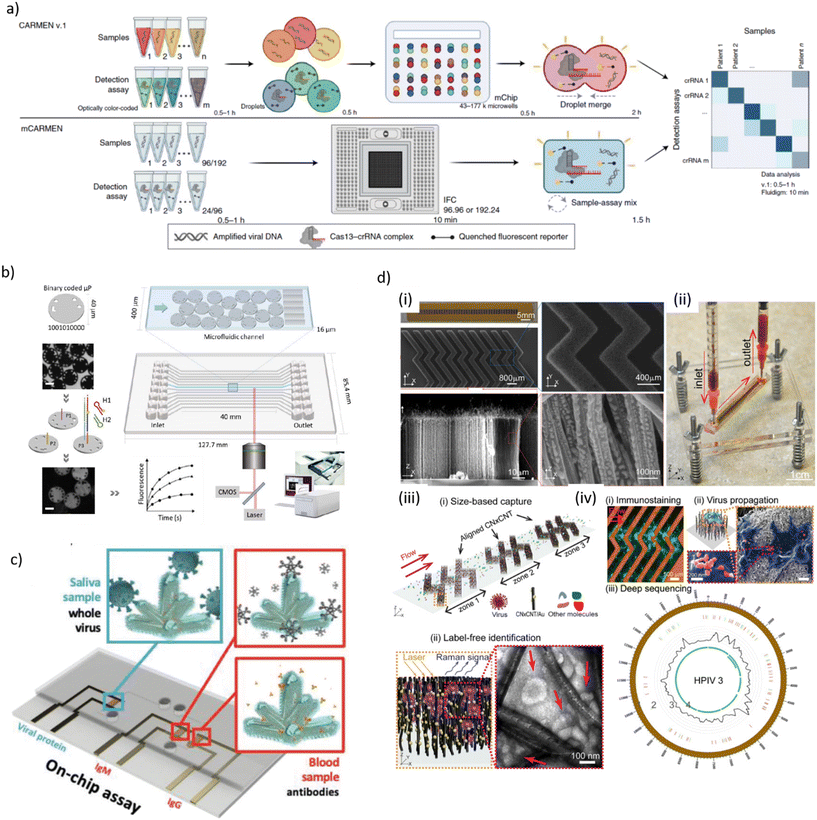 | ||
| Fig. 1 a) Schematic process flow of two virus and variant detection platforms, called combinatorial arrayed reactions for multiplexed evaluation of nucleic acids (CARMEN v.1, top) and its microfluidic version (mCARMEN, bottom), the latter combining CRISPR-based diagnostics and microfluidics. A mCARMEN respiratory virus panel allows testing for up to 21 viruses, including SARS-CoV-2, other coronaviruses and both influenza strains. b) Schematic representation of the processing of digitally barcoded microparticles using a microfluidic cartridge and an integrated instrument. Each microfluidic channel is embedded with encoded microparticles (P1, P2 and P3) that each serve as the substrate for the detection of a specific target. In presence of target, the hybridization chain reaction is initiated and the microparticles are identified during signal read-out. Scale bar is 20 μm. c) Schematic of an electrochemical microfluidic device for use with on-chip assays for the specific detection of whole viral particles in saliva and antibodies in blood using nanostructure-gold electrodes. d) Design and working principle of a microfluidic platform for effective virus capture and identification. (i) Photograph and SEM images of aligned carbon nanotubes (CNTs) exhibiting herringbone patterns decorated with gold nanoparticles for virus capture. (ii) Picture showing assembled device and processing of a blood sample. (iii) Illustration of size-based capture and in situ Raman spectroscopy for label-free optical virus identification. (iv) On-chip virus analysis and enrichment for next generation sequencing of human para-influenza virus type 3 (HPIV 3) [a) reproduced from ref. 155, ©2022, Creative Commons license, CC BY 4.0 (http://creativecommons.org/licenses/by/4.0/); b) reproduced from ref. 157, ©2023, CC BY 4.0; c) reproduced from ref. 168, ©2022, CC BY 4.0; d) reproduced from ref. 177, CC BY 4.0]. | ||
Rombach et al. introduced a LabDisk-format POC system named RespiDisk. This system enables multiplex RT-PCR detection of up to 19 viral and bacterial RTI pathogens from a single sample. Respiratory samples mimicking clinical conditions were loaded onto the disc for automated nucleic acid extraction (50 min), elution and target amplification (150 min).158 Liu et al. developed a multiplex analysis platform based on nested RPA and CRISPR/Cas12a-assisted virus identification for diagnosis of co-infections in the microfluidic format. The assays were designed for simultaneously detection of eight respiratory viral pathogen targets in nasopharyngeal samples, including SARS-CoV-2, RSV and influenza viruses/subtypes. The microfluidic chip was inserted in a centrifugal platform for running the assay steps, namely multiplex RT-RPA, subsequent RPA for separate amplification of each target gene and transfer into CRISPR/Cas12a detection chambers. LODs were 50–200 copies per mL depending on the assay with an on-chip protocol duration of 40 min.159 Other recent centrifugal microfluidic systems for RTI diagnostics (SARS-CoV-2, influenza A/B) implemented RT-LAMP/Cas12a detection or RT-qPCR,160,161 rapid differential diagnosis of seven human respiratory coronaviruses,162 or isothermal amplification for detection of 19 types of respiratory viruses.163
A fully automated microfluidic PCR-array platform, developed by Huang et al., could complete detection of 21 RTI pathogens (mainly viral and some bacterial) within 1.5 h with a LOD of 1.0 × 103 nucleic acid target copies per mL.164 Yin et al. designed a multifunctional rapid RT-PCR system for two different microfluidic chips, namely a microwell array chip of qualitative screening assays, or a droplet microfluidic chip for rapid quantification. Detection of SARS-CoV-2 virus sequences in serially diluted reference RNA samples was achieved within 15 min with a LOD of 10 copies per test.165 Another approach was a digital microfluidic (DMF) RT-qPCR platform for simultaneous detection of 11 viral and bacterial pathogens.166 Zai et al. operated a microfluidic test cartridge for multiplex RT-qPCR respiratory virus detection with passive gravity-driven fluid flow control. Assays were completed in 30 min with a LOD in the range of 200 RNA copies per mL (SARS-CoV-2, influenza A/B).167
Hydrophilic droplet surface energy traps served as virtual reaction chambers on a structure-free super-hydrophobic chip. Aptamer-coated magnetic microbeads provided the mobile substrates for the ELISA-like on-chip assay. A LOD of 0.032 hemagglutination units/reaction was reported for influenza A H1N1 detection.176 Yeh et al. used carbon nanotube (CNT) arrays with differential filtration porosity for virus enrichment combined with SERS identification in a microfluidic format (Fig. 1d). Au nanoparticle-decorated CNTs have been arranged in herringbone patterns for size-based capture and label-free detection. The device was validated with clinical samples from patients with rhinovirus, influenza A virus or HPIV infection. Viral detection was done in a few minutes with a 70-fold enrichment.177 Ramachandran et al. proposed a microfluidic assay for automated SARS-CoV-2 RNA detection using on-chip isotachophoresis (ITP) extraction of nucleic acids form clinical samples (5 min), followed by off-chip RT-LAMP preamplification (20–30 min) and on-chip ITP/CRISPR-based fluorescent target detection (SARS-CoV-2 N gene and E gene) (5 min). Electrokinetic protocols for ITP extraction and CRISPR/Cas12 enzymatic reactions were performed on a glass chip comprising two distinct cross-geometry channels. The LOD of the ITP/CRISPR method was found to be 10 copies per μL of viral RNA spiked into pooled nucleic acid extracts from negative clinical samples.178
3.4 Microfluidic systems for bacterial RTI pathogen panels
Yu et al. used the SlipChip technology to perform digital PCR for the detection of a panel of bacterial RTI-causative pathogens. As a proof of concept, the assay was designed for multiplex quantification of S. aureus, A. baumannii, S. pneumoniae, H. influenzae, and K. pneumoniae in a single test. The chip comprised two silanized glass microfluidic plates. The solution containing target nucleic acid templates and reactants was introduced into the chain-of-pearl channels of the top plate, which was subsequently slipped over the microwell array in the bottom plate. This operation resulted in surface tension-driven compartmentalization into a large number of reaction droplet partitions (2240 droplets with a volume of 4.5 nL). The chip was then placed on a thermal cycler for the PCR process. Differentiation of the target templates was performed by melting curve analysis of amplicons designed with different melting temperature Tm signatures and fluorescence detection by means of EvaGreen intercalation dye. Amplicons with Tm differences of 1.5 °C could be clearly separated.179A 3D-printed microfluidic device was developed for qPCR-based identification of M. pneumoniae mutant types with resistance to macrolide antibiotics. On-chip reservoirs contained sample solutions and PCR reactants, respectively, which were mixed and dispensed via pneumatic fluidic control into separately attached PCR tubes. The system was tested with plasmids containing a specific mutation, indicating a sensitivity 100 copies per reaction and a processing time of 80 min.180 Another device for POC detection of S. pneumoniae and M. pneumoniae took advantage of a polymer/paper microfluidic chip for genomic DNA extraction, performing LAMP in microchambers hosting chromatography paper substrates with pathogen-specific LAMP primers and calcein-mediated fluorescence detection. The analytical sensitivity of the LAMP microchamber reaction was 20 fg of target DNA.181 Dou et al. also proposed a hybrid microfluidic portable LAMP platform applied to the diagnosis of whooping cough (pertussis) in this case. The chip comprises six LAMP zones with paper disks for storage of DNA primers specific to B. pertussis. The assay reached a LOD of 5 DNA copies per LAMP zone (purified DNA samples) within 45 min. The clinical performance of the system, evaluated with lysates from B. pertussis spiked nasopharyngeal swabs and clinical samples from pediatric patients with signs of whooping cough, was comparable to real-time PCR tests.182,183 In an earlier approach, Huang et al. used a disc device with 24 test cells and pre-stored LAMP primers for multiplex identification of pathogens related to clinical pneumonia, in particular M. pneumoniae, S. aureus, and methicillin-resistant S. aureus. DNA samples and reactants were mixed off-chip prior to injection on the disc. The device had an analytical sensitivity of 10 nucleic acid copies. Assessment of with clinical samples demonstrated very good agreement with commercial real-time PCR systems.184
3.5 Tuberculosis
Acid-fast bacillus (AFB) smear microscopy performed from sputum is still widely used for initial TB diagnosis, even if this simple manual technique suffers from a lack of sensitivity (LOD ∼104 CFU mL−1).191Mtb culture is the gold standard for laboratory TB diagnosis and drug susceptibility testing, however, due to the slow Mtb growth rate, time-to-result may extend to several weeks.190,192 NAAT methods enable early detection and identification of mutations related to drug resistance. An update of WHO guidelines (2021) on rapid TB diagnostics outlines currently recommended technologies and products.193 WHO-endorsed TB diagnostics has been reviewed recently by Nandlal et al. and Hong et al., respectively.194,195
Commercial microfluidic PCR assays include Xpert® MTB/RIF (GeneXpert®, Cepheid, USA), a landmark development in TB diagnostics that detects MTBC bacteria and rifampicin (RIF) resistance within 2 h.196 Xpert® MTB/RIF Ultra has improved sensitivity (LOD 15.6 CFU mL−1 for MTBC) and Xpert® MTB/XDR was designed for detection of mutations resistant to 6 anti-TB drugs within 90 min.195 The VereMTB™ assay (VerePLEX™ Biosystem, Veredus Laboratories, Singapore) detects MTBC, several nontuberculous mycobacteria, as well as RIF and isoniazid (INH) resistance. Other WHO-endorsed on-chip RT-PCR assays are the Truenat® MTB, MTB Plus and MTB-RIF Dx assays (Molbio Diagnostics, India).197 Recently, Schlanderer et al. implemented a TB diagnostic workflow on the Rhonda player. MTBC detection including antibiotic resistance testing against the first-line antibiotics INH and RIF is performed on the disc from a single sputum sample. If the qPCR on-chip data indicates drug resistance, a detachable sample tube containing enriched MTBC DNA is available for subsequent comprehensive resistance profiling via targeted next generation sequencing (tNGS) in a centralized lab facility. This two-stage TB diagnostic can be completed within three days.198
Serological TB tests have insufficient diagnostic value.199 As a consequence, there is more focus on TB antigen detection, for instance lipoarabinomannan (LAM), a structural component of the outer cell wall of mycobacteria that may be released into urine.200 The LAM antigen can be detected on LFAs within minutes using unprocessed urine samples.201,202 Assays have also been developed for clinically relevant other TB antigens (such as MPT64, CPF-10 or ESAT-6).203–205 In a recent review on LFAs for detection of pathogenic bacteria, Sohrabi et al. also addressed assays specifically designed for TB diagnosis.80
Hong et al. discussed challenges in the development of rapid POC TB diagnosis and drug susceptibility testing, including some selected microfluidic and nanophotonic systems.194 Paul et al. discussed advanced integrative sensing technologies for detection of drug-resistant TB.190 Gupta et al. focused on developments in nano-biosensing technologies.206 Srivastava et al. also analysed biosensor-based detection.207 Earlier reviews on POC TB diagnosis, also discussing to some extend the potential of microfluidics, have been proposed by Mani et al., Wang et al., Dheda et al. and Niemz et al.208–211 In the following, we discuss a selection of microfluidic/biosensor systems for TB diagnostics. Table 2 provides an overview of recent approaches, including AST tools.
| Target | Device and assay principle | Performance indications | Ref. |
|---|---|---|---|
| Microfluidic devices based on NAAT assays | |||
| Mtb katG gene | LoaD for analysis of TB drug-resistance by mutation-specific PLP ligation and RCA | Mixing on-disc and real-time optomagnetic readout. Assay time of 2 h, LOD 5 pM | 212 |
| M. bovis BCG and Mtb (drug resistant strains) | LoaD for automated PCR analysis. Two separate DNA extracts are obtained | LOD of 10 CFU mL−1 in spiked sputum. Drug resistance testing with clinical samples | 214, 215 |
| Mtb gDNA | Modular LAMP paper-and-plastic POC device with dry-stored reagents | High analytical (10 copies of Mtb gDNA) and clinical sensitivity | 216 |
| Circulating cell-free Mtb-specific DNA | Droplet digital PCR using a commercial droplet generator | Absolute quantification of nucleic acid target sequences | 220, 221 |
| M. bovis BCG | Distinction of live/dead bacteria via PMA binding and on-chip PCR | Automated detection within 90 min | 223 |
| Mtb H37Ra bacilli | Modular sputum-to-genotype system with a lab-on-a-film gel element array | Multiplex detection of mutations. LOD 43 CFU mL−1 in raw sputum | 224, 225 |
| RIF-resistant Mtb (rpoB gene) | RT-PCR in on-chip reactors and high-resolution DNA melting-based TB test | Drug-resistance mutations were detected in clinical isolates. 20 PCR reactions per chip | 226 |
| Microfluidics for antimicrobial testing | |||
| M. smegmatis | Microfluidic chip for voltammetric detection of nucleic acid sequences | Antibiotic susceptibility apparent after 24 h through measuring 16SrRNA levels | 232 |
| M. smegmatis | On-chip cell trapping for visualization of growth and phenotypic alterations | Drug response assessed by real-time tracking for over 48 h at single-cell level | 233 |
| M. smegmatis (msm2570::Tn mutant) | Microfluidic-microscopy method to reveal antibiotic tolerance mechanisms | Antibiotic exposure of the msm2570::Tn mutant showed low number of lysed cells | 234 |
| M. smegmatis | Microfluidic acoustic trapping of live Mtb and Raman spectroscopy | Raman fingerprints change substantially upon INH exposure. Trapping for up to 8 h | 235 |
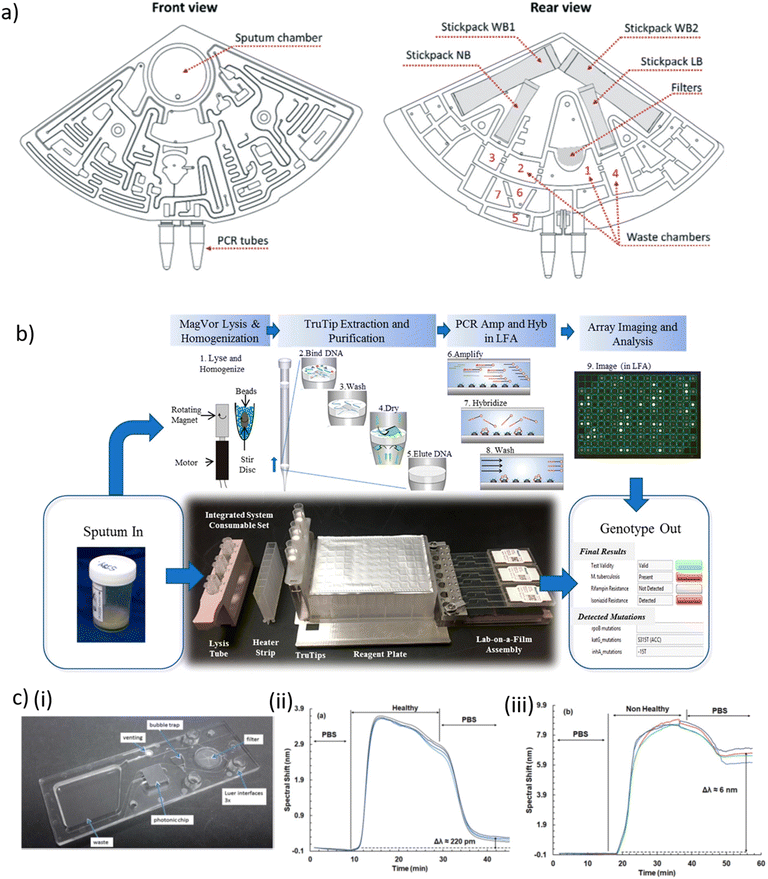 | ||
| Fig. 2 a) Design of a cartridge for Mtb diagnosis, three of which can be placed in a laboratory centrifuge in front view (left) and rear view (right), indicating the fluidic network with chambers and channels, the interfaces for sample input (sputum chamber) and product collection (PCR tubes), the filters, the waste collection chambers and 4 stickpacks for pre-storage of the reagents. Abbreviations: washing buffer 1 + 2 (WB1+2); lysis buffer (LB); neutralisation buffer (NB). b) Layout of the consumable in an automated Mtb sputum-to-genotype system for processing of six samples, in which the following steps occur. (1) Lysis and homogenization occur in lysis tubes, which include a magnetized stir disc and glass beads. (2) TruTip aspirates the sample mixed with a binding buffer, so that it flows through the pores of the matrix in the tip resulting in DNA bound to the matrix. (3) Porous matrix is washed to remove the impurities. (4) Matrix is dried with air. (5) Bound DNA is eluted into an elution buffer. (6) Purified DNA is amplified with an asymmetric PCR reaction. (7) Product (with fluorescent labels) hybridize to the gel elements. (8) Gel elements are washed to remove unbound product. (9) Image of the array is captured and analyzed. c) (i) Picture of a waveguide-based SiN nanophotonic chip with anti-LAM molecules covalently coupled to the SiN surface and assembled in a polymer cartridge. Spectral shift observed in the interferometric signal upon exposure of a waveguide to LAM, a biomarker for TB, in urine of (ii) a healthy and (iii) a non-healthy person. Presence of LAM in the sample leads to a long-term spectral shift [a) reproduced from ref. 214 with permission from the Royal Society of Chemistry; b) reprinted with permission from ref. 225, ©2020 American Chemical Society; c) reproduced from ref. 227 with permission from the Royal Society of Chemistry]. | ||
Droplet digital PCR (ddPCR) has been used in clinical applications thanks to its high accuracy and sensitivity for low-abundance DNA, and for absolute quantification of nucleic acid target sequences.217,218 Nyaruaba et al. recently reviewed the application of ddPCR as TB diagnostic tool.219 Several TB-related studies took advantage of the Bio-Rad QX200 droplet generator system (Bio-Rad Laboratories, USA) and ddPCR for detecting low levels of circulating Mtb-specific DNA, drug susceptibility testing and other applications.220–222 Based on a different approach, Wang et al. proposed a microfluidic system featuring 12 PCR reaction chambers enabling distinction of live/dead bacteria via photo-reactive propidium monoazide (PMA) binding. Selective covalent binding of PMA to DNA from dead bacteria inhibited PCR amplification. Heparin-binding haemagglutinin (HBHA) antibody-conjugated magnetic beads were used as capture probe against M. bovis Bacille Calmette–Guérin (BCG) and Mtb clinical isolates. Bacteria capture, thermolysis and DNA release, PMA treatment and rpoB gene PCR amplification was performed on-chip within 90 min and a reported LOD of 100 CFU.223 Kukhtin et al. developed a disposable lab-on-a-film that detects MDR-TB from sputum extracts.224 The device comprises a gel-based microarray printed onto a flexible polyester film. Target amplification and hybridization on the microarray was carried out within a single closed microfluidic flow-cell. Nucleic acid was purified off-chip using a pipet tip with an embedded matrix for nucleic acid isolation (TruTip, Akonni Biosystems). Initially a LOD of 32 CFU mL−1 for Mtb-spiked sputum was obtained. In a follow-up development improved sample homogenization and cell lysis was implemented in the workflow and a Mtb LOD of 43 CFU mL−1 in raw sputum was reported (Fig. 2b).225 Mbano et al. performed real-time PCR and subsequent high-resolution melting curve analysis on a microfluidic PDMS chip with 20 independent PCR chambers and fluorescence readout. RIF-resistant strains of Mtb were used to assess the performance of this method.226
4 Urinary tract infections
4.1 Scope and common uropathogens
The urinary tract system comprises the kidneys, the bladder, the ureters and the urethra. Measuring the abundance of specific biomarkers in urine is frequently used for non-invasive health monitoring.237,238 Paper-based devices, either conventional LFAs or more advanced designs, are well-suited for biochemical urine analysis in general.239–241 Urinary tract infection (UTI) occurs when uropathogens, such as bacteria from the vaginal area, the rectum or the skin, get into the urinary tract and move upwards into the bladder and eventually into the kidney.242–244 UTIs are among the most widespread community and hospital-acquired bacterial infections with hundreds of millions of people being affected annually worldwide, entailing a major global clinical and economic burden. Bladder infection (cystitis) is the most common type of UTI, whereas kidney infections (pyelonephritis) are less frequent but may have severe and even life-threatening consequences if pathogens spread into the bloodstream (urosepsis).245 Significant bacteriuria can be defined as a count of over 105 CFU of the same organism per mL of urine, but the threshold for UTI diagnosis may be set much lower in some cases (100–1000 CFU mL−1).246,247 By far the most common causative agent for both uncomplicated and complicated UTIs is uropathogenic E. coli (UPEC).248 Other bacterial species, with much lower prevalence and depending on the specific conditions, include K. pneumoniae, P. mirabilis, E. faecalis, P. aeruginosa, S. aureus and other species.246,247,249 Catheter-associated UTIs are strongly associated with complicated UTIs and are a common cause of secondary bloodstream infections.242,246,249 Also the prevalence of fungal Candida ssp UTIs is increasing.250At a first stage, UTI diagnosis is based on specific symptoms and prescription of broad-spectrum antibiotics is often the immediate choice for treatment. Urinalysis may include microscopic inspection to detect red or white blood cells in urine samples. Urinary dipsticks testing for nitrites and leukocyte esterase can be used, mainly to exclude the presence of infection.251 Urine culture remains the gold standard to confirm the presence of infection and for pathogen identification. This process usually takes 24 to 48 h.252 Subsequently, a second culture step may be necessary to determine the antibiotic resistance profile. The introduction of MALDI-TOF mass spectrometry has enabled fast identification of uropathogens, from cultures and possibly also directly from urine samples.253
4.2 Antimicrobial resistance of uropathogens and relevant reviews
Uncomplicate UTI occurring in healthy subjects normally can be easily treated with short-term antibiotic administration, however recurring UTIs due to persisting uropathogens and biofilm formation is an important health issue. Complicated UTI or kidney infections are associated with high AMR rates that require long-course antibiotic treatments, resulting in severe alteration of the normal microbiota and proliferation of resistant pathogen strains.242,249 UPEC bacteria have developed resistance against common antibiotics (e.g. ciprofloxacin, nitrofurantoin, ampicillin) with extremely high prevalence in some regions. Moreover, the emergence of UPEC strains possessing extendedspectrum βlactamases that confer resistance against thirdgeneration cephalosporins and monobactams, is observed in community-acquired UTIs.246,254,255 New technologies enabling early UTI diagnosis and fast POC pathogen identification combined with AST are required to improve therapeutic approaches. Due to the high prevalence of drug-resistant uropathogenic E. coli strains, but probably also because urine does not have a complicate sample matrix (as compared to blood for instance), a wide range of microfluidic fast AST methods, in particular enabled by single-cell analysis, has been developed with focus on UTI.UTI diagnostic methods for laboratory and hospital facilities have been recently reviewed by Santos et al. or Harris et al., for instance (including culture-based methods, microscopic urinalysis, flow cytometry, MALDI-TOF, PCR or FISH, etc.).256,257 Santos et al. also showed examples of innovative (culture-based) single-use POC tests that are currently available in the market for UTI diagnosis, as well as a summary of POC tests on paper for E. coli from various samples.256 Harris et al. more specifically focused on a discussion of currently available automated commercial UIT diagnostic systems.257 A host of recent and earlier reviews provided overviews on emerging technologies for POC urine analysis and/or UTI diagnosis, including discussions on AST methods and some microfluidic approaches.240,241,256–262 Paper devices with potential for UTI diagnosis have been recently reviewed by Hasandka et al. or Tai et al., for instance.241,262 Previously mentioned reviews discussing recent and emerging AST methods from a broader perspective are certainly also relevant for UTIs.42,43,59,263 In the following, we will first discuss innovative microfluidic and/or biosensor devices that have been specifically designed for UTI diagnostics. Recent LOC systems for fast AST of uropathogens will be presented in the second part of this section. Table 3 approaches for UTI diagnosis and fast AST on uropathogens.
| Pathogen | Device and assay principle | Performance indications | Ref. |
|---|---|---|---|
| Microfluidic devices for UTI diagnostics | |||
| E. coli (β-lactam antibiotics resistant) | Nanoelectrokinetic PAD with chromo-genic detection of β-lactamases | Label-free detection of drug-resistant bacteria within 7 min by cell phone. LOD 104 CFU mL−1 | 264 |
| E. coli | Pump-free immunomagnetic separation with colorimetric detection | Laminar flow control by paper pads. LOD 4.7 × 102 CFU mL−1 in urine | 265 |
| E. coli | Paper-based device for cultivating bacteria in situ and testing for nitrite | 104–107 CFU mL−1 quantified on a β-glucuronidase-specific substrate, within 6 h in urine | 266 |
| UTI bacterial pathogens | Enrichment on a herringbone chip and MALDI-TOF MS identification | Chaotic mixing enhanced bead/bacteria complex formation. Process takes 1.5 h | 267 |
| C. tropicalis | PNA-FISH protocol applied to pathogens in a microfluidic trap | Visual detection (1 × 105 cells per mL, within 6 h). Tested with spiked synthetic urine | 271 |
| E. coli | Bacteria enrichment on Si nanowires and MALDI-TOF MS identification | Detection of ∼103 CFU mL−1 in urine (after pre-culture for 4–6 h) | 272 |
| E. coli, P. putida, S. epidermidis | Plasmonic-assisted impedimetric detection on nanostructured surfaces | Hybrid 3D gold/graphene nanostructures enhanced sensitivity (LOD 20 CFU mL−1, 10 min) | 273 |
| Microfluidic chip-based devices for fast AST of uropathogens | |||
| E. coli (ATCC 25922) and UTI-positive urine | Bacteria growth monitoring on a droplet microfluidic platform | Single-cell AST in 90 min (first antibiotic) + 2 min for subsequent antibiotic conditions | 280 |
| UTI bacteria (16S rRNA gene) | Fluorogenic PNA probe-based hybridization assay on a droplet chip | Identification and single-cell AST from urine samples within 30 min. Clinical comparison study | 282, 283 |
| P. mirabilis, S. aureus, K. pneumoniae, E. coli | Pheno-molecular AST using PCR and digital high-resolution melt | Multiple bacterial species and susceptibility profiles identified in spiked urine within of ∼4 h | 284 |
| S. aureus, E. faecalis, E. coli, K. pneumoniae | Phenotypic analysis on parallelized droplet microfluidic platform | Simultaneous screening of 4 antibiotics per pathogens within 15–30 min | 285 |
| E. coli, other UTI pathogens | DMF for bacterial classification and AST using metabolic markers | Real-time bacterial metabolic monitoring. AST in <18 h, performed with resazurin dye | 287 |
| E. coli (ATCC 25922) and resistant strains | Combinatorial antibiotic screening with a nL-sized droplet SlipChip approach | MIC of E. coli against 4 antibiotics measured within 3 h on one chip | 288 |
| E. coli (ATCC 25922) | Multiplexed AST on a nL chamber array (resazurin indicator) | Antibiotic dilution on-chip. AST in 8–9 h. MIC determination required ∼2000 bacteria | 289 |
| S. epidermidis, M. bacteremicum, uropathogenic E. coli | Single-cell pathogen classification and AST in adaptable channels | Bacteria classification based on size and shape in urine. Single-cell AST within 30 min | 292 |
| E. coli, K. pneumoniae, S. saprophyticus | All-electrical AST of bacteria trapped/incubated in channel constrictions | Robust and sensitive resistance measurement of bacterial growth and AST within 2 h | 293 |
| E. coli, K. pneumoniae, P. aeruginosa, E. faecalis | Pneumatically-driven microfluidic chip for colorimetric AST assays | AST completed in 4.5–9 h in automated manner | 297 |
4.3 Microfluidics and biosensor for UTI diagnostics
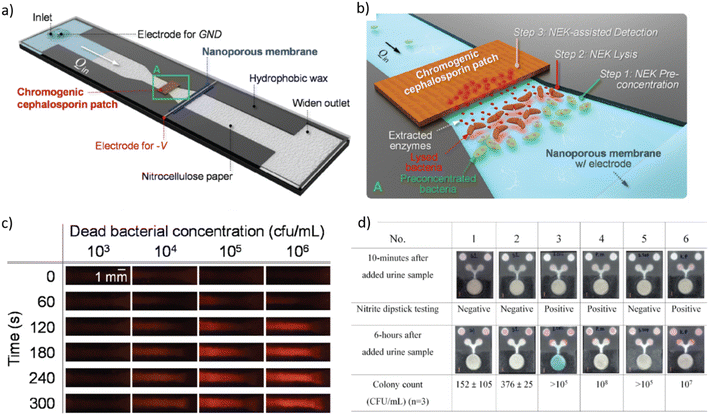 | ||
| Fig. 3 a) Schematic diagram of a nanoelectrokinetic (NEK) paper-based analytical device that is based on a constricted paper strip with a perm-selective nanoporous membrane placed perpendicular to the flow direction for inducing ion concentration polarization and inducing bacteria lysis, and with a chromogenic cephalosporin patch for detecting enzymes originating from the lysed bacteria. b) Zoom of rectangle A in a) with indication of the subsequent steps for bacterial detection. c) Color changes of a nitrocefin-coated patch over time for different bacterial concentrations. Color change from orange to red was detected via a cell phone-based read-out above bacterial concentrations of 104 CFU mL−1. d) In situ culture device for E. coli and testing for nitrite presence in urine. A colorimetric test for β-glucuronidase, a specific enzyme of E. coli, was implemented (blue color), while the pink color indicates presence of nitrite. The two circular zones on the top of the paper-based analytical device serve as a color control for the nitrite detection [a–c reprinted with permission from ref. 264, ©2022 Elsevier; d) reproduced from ref. 266, ©2019, Creative Commons license, CC BY 4.0 (http://creativecommons.org/licenses/by/4.0/). | ||
Shen et al. used a microfluidic chip featuring a herringbone mixer structure for pathogen enrichment via increased capture efficiency and subsequent detection by MALDI-TOF mass spectrometry. Vancomycin-modified magnetic beads were magnetically retained in the microfluidic channel for capturing the UTI pathogens (S. aureus, S. hominis, S. epidermidis and E. gallinarum). The process enabled pathogen identification directly from spiked human samples without bacterial culture (LOD 104–105 CFU mL−1, assay time 1.5 h).267 Chen et al. performed multiplex real-time RPA and pathogen detection (E. coli, S. aureus, S. typhimurium, P. mirabilis, and P. aeruginosa spiked into urine) on a centrifugal cartridge. Bacteria were concentrated/purified by means of a filter-pipette. Specific RPA primers and probes were preloaded in dedicated reaction chambers on the disc. The entire procedure, from bacterial enrichment to detection, was completed within 40 min (LOD in the range of 102 to 103 CFU mL−1).268 Olanrewaju et al. developed a modular system that incorporates an immunoaffinity column for rapid bacteria capture and fluorescence detection, as well as sequential retention burst valves and an on-chip capillary pump for autonomous liquid transport. The fluidic design allowed performing functional assay steps by pre-programmed and self-powered delivery of immunoassay reagents. Detection of E. coli was achieved in less than 7 min with a LOD of 1.2 × 102 CFU mL−1 (synthetic urine).269 Alves et al. implemented a quantitative E. coli fluorescence sandwich immunoassay in a microcapillary Teflon film strip array through which reagents were successively manually aspirated. By this means, large sample volumes could be passed through the capture antibody coated capillaries, resulting in a LOD of 240 CFU mL−1 (synthetic urine) in less than 25 min.270
4.4 Microfluidic devices for fast AST of uropathogens
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 pico-droplets containing single bacteria. In-line incubation was implemented and oil barrier plugs avoided cross-talk between adjacent pL-droplet groups. On-chip single-cell AST for four antibiotics was first characterized with a E. coli reference strain, and subsequently with clinical isolates. Clinically useful antibiograms with MIC values could be produced on a time scale of 90 min (mainly due to on-chip incubation) for the first antibiotic condition, plus 2 min for each subsequent condition. Furthermore, breakpoint testing with (filtered) uncomplicated UTI-positive clinical urine samples was performed.280 Mach et al. proposed a droplet microfluidic format for amplification-free detection and identification of single bacterial cells with fluorogenic PNA probes that target bacterial 16S rRNA.281 Subsequently, Kaushik et al. further implemented this approach for UTI diagnosis with single-cell resolution on a droplet platform. The platform was designed for both pathogen identification/classification (in particular E. coli) and AST from urine samples within the clinically relevant concentration range. Bacterial cells were encapsulated together with an antibiotic and multiple hybridization probes that target uropathogen-specific 16S rRNA sequences. A fluorescence two-color detection scheme allowed pathogen identification, whereas susceptibility to the antibiotic was revealed by signal intensities, corresponding to the relative amount of 16S rRNA per droplet produced by single cells exposed/not exposed to antibiotics. This pheno-molecular AST method was evaluated with 3 common antibiotics (gentamicin, ciprofloxacin, and ampicillin). The pL-droplet reaction volume enabled very short on-chip incubation times (10 min) and fast subsequent hybridization with the florescent probes (15 min), resulting in an overall assay time to result as short as 30 min. 50 human urine specimens were tested against ciprofloxacin to evaluate the clinical utility of the assay.282,283 This group also proposed another pheno-molecular AST platform implementing PCR and digital high-resolution melt (HRM) analysis to quantify bacterial DNA molecules. The core of the platform was a digital PCR nanowell array. Multiple bacterial uropathogens and corresponding susceptibility profiles were correctly identified within ∼4 h.284
000 pico-droplets containing single bacteria. In-line incubation was implemented and oil barrier plugs avoided cross-talk between adjacent pL-droplet groups. On-chip single-cell AST for four antibiotics was first characterized with a E. coli reference strain, and subsequently with clinical isolates. Clinically useful antibiograms with MIC values could be produced on a time scale of 90 min (mainly due to on-chip incubation) for the first antibiotic condition, plus 2 min for each subsequent condition. Furthermore, breakpoint testing with (filtered) uncomplicated UTI-positive clinical urine samples was performed.280 Mach et al. proposed a droplet microfluidic format for amplification-free detection and identification of single bacterial cells with fluorogenic PNA probes that target bacterial 16S rRNA.281 Subsequently, Kaushik et al. further implemented this approach for UTI diagnosis with single-cell resolution on a droplet platform. The platform was designed for both pathogen identification/classification (in particular E. coli) and AST from urine samples within the clinically relevant concentration range. Bacterial cells were encapsulated together with an antibiotic and multiple hybridization probes that target uropathogen-specific 16S rRNA sequences. A fluorescence two-color detection scheme allowed pathogen identification, whereas susceptibility to the antibiotic was revealed by signal intensities, corresponding to the relative amount of 16S rRNA per droplet produced by single cells exposed/not exposed to antibiotics. This pheno-molecular AST method was evaluated with 3 common antibiotics (gentamicin, ciprofloxacin, and ampicillin). The pL-droplet reaction volume enabled very short on-chip incubation times (10 min) and fast subsequent hybridization with the florescent probes (15 min), resulting in an overall assay time to result as short as 30 min. 50 human urine specimens were tested against ciprofloxacin to evaluate the clinical utility of the assay.282,283 This group also proposed another pheno-molecular AST platform implementing PCR and digital high-resolution melt (HRM) analysis to quantify bacterial DNA molecules. The core of the platform was a digital PCR nanowell array. Multiple bacterial uropathogens and corresponding susceptibility profiles were correctly identified within ∼4 h.284
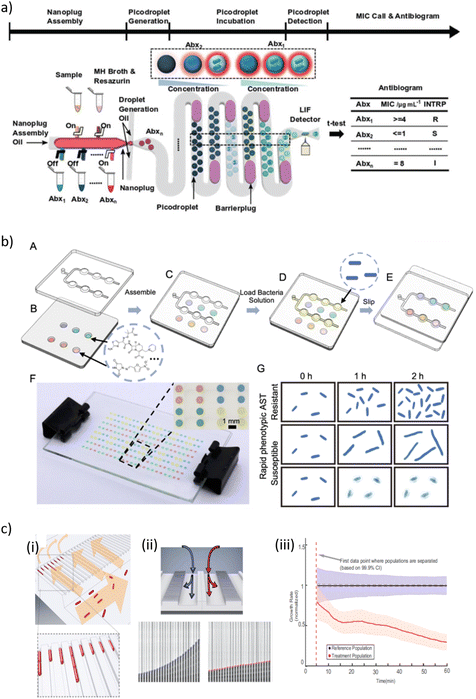 | ||
| Fig. 4 a) Overview of a so-called single-cell assembly line antibiotic susceptibility testing (SCALe-AST) device. It is an integrated droplet-based device with programmable microvalves to assemble bacteria sample, Mueller-Hinton II (MHMH) broth, resazurin, and antibiotics (denoted as Abx1, Abx2,…, Abxn) into nanoplugs and subsequently discretize the nanoplugs into groups of picodroplets encapsulating single bacteria. As each group of picodroplets flow through the built-in, 37 °C incubation channel, a barrier plug is introduced behind the picodroplets to keep them separated from adjacent groups of picodroplets, thus preventing cross-talk between different antibiotic conditions. The encapsulated single bacterium stops growing if it is susceptible to the applied antibiotic and the weakly fluorescent resazurin is reduced slowly, resulting in a weak fluorescence signal. In contrast, the bacterium proliferates if it is resistant to the applied antibiotic, and the weakly fluorescent resazurin is reduced to fluorescent resorufin quickly, resulting in a strong fluorescence signal within the picodroplet upon detection via a laser-induced-fluorescence (LIF) detector. By comparing picodroplet fluorescence intensity of different antibiotic concentrations, an antibiogram that provides the bacteria susceptibility categorization for multiple antibiotics with measured minimum inhibitor concentrations is constructed. b) Schematic presentations of the combinatorial screening (cs) SlipChip operation principle. (A) The top plate consists of a chain-of-pearls fluidic channel. (B) The bottom plate contains circular expansion microwells preloaded with different antibiotics. (C) The top and bottom plates are assembled in the initial loading position. (D) The bacterial solution (yellow) is introduced into the chain-of-pearls channel by pipetting. (E) The top plate is moved relative to the bottom plate to bring the chain-of-pearls channel into contact with the expansion channel by a manual slipping operation, and the aqueous solution containing the bacteria self-partitions into individual droplets that can be mixed with the preloaded antibiotics. (F) A bright-field photo of a cs-SlipChip loaded with an aqueous solution spiked with blue, red, yellow and green food dyes. (G) Schematic drawing of an antibiotic susceptibility/resistance profile obtained from the cs-SlipChip, as indicated by bacterial growth. c) Design and operation details of a microfluidic chip illustrating (i) the loading of rod-shaped bacterial cells (red) into cell traps. Arrows indicate flow direction during loading. (ii) Detection of growth rate effect of antibiotic. (ii, Top) Media with or without antibiotic are supplied to the two different rows of cell traps to test the effect of the antibiotic on the treatment population compared with the reference population. (ii, Bottom) The status of a single-cell trap from the reference population (left) and another single-cell trap from the treatment population (right) are shown every 2.5 min. The detected front-most cell pole position is given as a blue or red circle. (iii) The overlay of the two population's normalized growth rate distributions. The time of separation of the treatment population from the reference population occurs before the dashed magenta line, which indicates the first time point when different growth rates can be estimated [a) reprinted with permission from ref. 280, ©2021 John Wiley and Sons; b) reproduced from ref. 288 with permission from the Royal Society of Chemistry; c) reprinted with permission from ref. 291, Proceedings of the National Academy of Sciences]. | ||
Kang et al. developed an integrated quadruplex droplet device to screen several combinations of bacteria and/or antibiotics simultaneously. The device comprises four droplet generators and on-chip docking arrays (filled with >8000 droplets, droplet volume ∼110 pL) for incubation and observation. The performance of the system was tested with clinically relevant uropathogenic Gram-positive (S. aureus, E. faecalis) and Gram-negative (E. coli, K. pneumoniae) bacterial strains. Phenotypic AST was assessed for six concentrations of bactericidal and bacteriostatic antibiotics (oxacillin and tetracycline) at single cell resolution. Optical observation of bacteria proliferation in the droplets enabled MIC quantification for each bacteria/drug combination. Antibiotic susceptibility could be evaluated as fast as 15–30 min.285 Likewise, Sabhachandani et al. co-incapsulated bacteria and antibiotics on a droplet chip for phenotypic AST assessment (E. coli spiked human urine). Due to single-cell tracking, discriminatory readouts could be achieved within one 1 h of incubation in the on-chip droplet-docking array.286 Sklavounos et al. performed bacterial classification, breakpoint testing and phenotypic AST on a digital microfluidics platform (DMF). For on-chip AST, dilution series of antibiotics at different concentrations were generated by on-demand manipulation of μL-size droplets and mixed with bacteria-containing droplets (final concentration 5 × 105 CFU mL−1). For bacterial classification, droplets were mixed with different metabolic indicator droplets. The final droplet array was incubated for at least 16 h, resulting in a total of time-to-result of 18 h. AST was validated with two uropathogenic E. coli strains. Bacterial classification was performed independently or simultaneously with ciprofloxacin AST for E. coli, K. pneumoniae, P. mirabilis and S. aureus. As a proof-of-concept, multiplexed breakpoint testing was carried out with a multidrug resistant E. coli strain.287
5 Gastrointestinal tract infections
5.1 Scope and common pathogens
In most cases, gastrointestinal tract infection (GTI), in particular of the small intestine (enteritis), is acquired through fecal-contaminated food or drinking-water, by transmission from person-to-person or by contact with contaminated surfaces. Foodborne diseases represent a significant burden to public health.299 Ingested pathogens that escape the host defence in the upper gastrointestinal tract potentially may invade and multiply in the intestine, thus generating infection and disease, possibly progressing to infection of other body systems via the lymphatic system or the bloodstream (septic infection). Diarrhoeal disease is the most common outcome of GTI, with nearly 1.7 billion cases of childhood diarrhoeal disease every year. In the developing world diarrhoeal disease represents a major health problem, causing high morbidity and mortality rate, in particular through dehydration (WHO 2017).300 Many cases could be prevented by safe drinking-water and adequate sanitation. GTI may be caused by a host of bacteria, viruses or parasites. Pathogenic E. coli strains, such as Enterohemorrhagic E. coli (EHEC, O157:H7),301Salmonella,302Campylobacter,303Listeria monocytogenes,304Clostridium perfringens,305 or S. aureus306 are examples for important food- or waterborne bacterial pathogens.307 Ingestion of water or fish contaminated by pathogenic strains of Vibrio cholerae causes the potentially deadly acute diarrhoeal disease cholera. Cholera pandemic or endemic outbreaks regularly occur.308,309 Other examples for diseases caused by pathogenic enteric bacteria are shigellosis, a predominantly paediatric diarrhoeal disease with an exclusively human reservoir,310 or typhoid fever which is a life-threatening systemic infection caused by Salmonella Typhimurium.311 Viral gastroenteritis is also extremely widespread, with rotaviruses (children),312 noroviruses (adults)313 and hepatitis A virus314 being most prevalent pathogens transmitted by the fecal-oral route.315Helicobacter pylori, another common pathogen, causes infections of the stomach, which are often without symptoms but may generate gastritis or ulcers.3165.2 Diagnostic methods and relevant reviews
Conventional diagnosis protocols for viral or bacterial GTI infections include pathogen isolation from feces and culture, antigen or toxin detection (e.g. with rapid antigen immunochromatographic assays), MALDI-TOF and nucleic-acid tests.307,313 Emerging diagnostic tools and microfluidic assays that have been developed for other infectious pathogens and clinical samples may possibly also be adapted for GTI diagnosis.96,120 Xpert® Norovirus assay (Cepheid, USA) is an example for a commercial RT-PCR assays targeting an important GTI pathogen. The Bosch Vivalytic HSP test differs between the pathogens C. difficile, norovirus and rotavirus and delivers fast and precise results.115 Microfluidic and biosensor approaches dealing with GTI causative pathogens have been extensively reviewed in literature, in great majority with focus on pathogen detection from contaminated food- or waterborne samples. Very recently, Yin et al. discussed the current state-of-art and future perspectives of detection methods for foodborne viruses.317 Other recent review articles that include microfluidic devices have been worked out, for instance by Gao et al., who discussed advances in microfluidic devices for foodborne pathogen detection,318 by Shang et al. with focus on advances in nanomaterial-based microfluidic platforms,319 by Ranjbaran et al. on microfluidics at the interface of bacteria and fresh produce,320 or by Quintela et al. on advances and limitations of portable and rapid detection technologies for foodborne pathogens.321 Mi et al. summarized microfluidic biosensor tools for foodborne pathogenic bacteria and Su et al. investigated microfluidic nucleic acid tests of foodborne viruses.322,323 Other reviews focused on specific pathogens, such as POC methods for detection of norovirus by Zaczek-Moczydlowska et al. or POC diagnosis for E. coli O157:H7 in food and water by Rani et al.324,325 Shen et al. explored biosensor technologies for rapid detection of Salmonella in food.326 Wang et al. discussed microfluidic sampling and biosensing systems for foodborne E. coli and Salmonella.327 In the context of GTI diagnosis, only a few microfluidic devices have been designed for or tested directly with human samples, in particular fecal samples. Possible GTI diagnostic tools could also be derived from other applications, such as evolving technologies in clinical research in the gut microbiome era.328 On the other hand, enteric diseases are also a common problem in modern swine farming and POC devices developed for veterinary use could be suitable for adaptation to human samples and diagnostic approaches.329 In the following, considering the lack of literature on microfluidic devices specifically applied to POC clinical diagnosis of human GTI, we will extend our discussion to a selection of recent approaches covering pathogen detection from foodborne or non-human samples. Table 4 provides an overview of recent approaches for GTI pathogen detection.| Pathogen | Device and assay principle | Performance indications | Ref. |
|---|---|---|---|
| Systems for bacterial enteric pathogens | |||
| Gut microbiome, B. vulgatus | Droplet microfluidics applied to complex microbiome samples | Cultivation-free single-cell genetic assays and enrichment by sorting of positive droplets | 334 |
| Campylobacter spp. | On-chip chambers with chromogenic medium for identification and AST | Campylobacter spp. detected in milk and poultry meat. AST within 24 h | 335 |
| Enterohemorrhagic E. coli | Cartridges for PCR amplification and microarray hybridization | Fluidic operations with combined centrifugal/pneumatic actuation. Workflow of 2 h | 336 |
| Enterotoxins and enteric bacteria | Integrated LAMP and immunoassays on a LoaD platform | Different disc designs for single or dual assays. LOD 1.35–5.50 ng mL−1 (toxins), 1–30 cells (LAMP) | 337 |
| Foodborne pathogens | Microchip with paper pads for LAMP in multiple reaction chambers | LOD 0.013 ng μL−1 for purified E. coli DNA, LOD 12 CFU mL−1 for Salmonella spp. in milk | 338 |
| S. typhimurium | Colorimetric biosensor using bacteria-immune Au@Pt NP conjugates | Finger-driven mixing, LOD 168 CFU mL−1, within 25 min | 339 |
| E. coli O157:H7 | Integrated biosensor chip based on the RPA-CRISPR/Cas12a reaction | Finger-pressure actuation, LOD 10 CFU mL−1, within 2.5 h | 340 |
| Foodborne pathogens | Portable system and microfluidic cartridge with LAMP reaction wells | LOD 8 × 103 CFU mL (Shigella). Validated with artificially contaminated food samples | 341 |
| Systems for viral and parasitic enteric pathogens | |||
| Human norovirus | On-chip chamber digital RT-RAA for quantitative virus detection | Sample partition (10 min), amplification (20 min). LOD 1 cRNA copy per μL | 342 |
| Norovirus (capsids and intact viruses) | LFAs immunoassay based on dispersed particle aggregates | Virus detection at single copy level from water samples | 344 |
| Rotavirus A | Paper disc for nucleic acid extraction, LAMP and readout with the naked eye | LOD 1 × 103 virus copies per mL, within 30 min. Tested with clinical stool samples | 346 |
| Porcine enteric viruses | Multiplex colorimetric LAMP for visual detection of diarrhea-related viruses | Handheld operation of a fan-shaped chip. Testing <60 min, LOD of 100 DNA copies per μL | 347 |
| Porcine enteric viruses | RT-LAMP 3D-printed microfluidic device | LOD 101–102 RNA copies per reaction, appropriate for early-stage infection detection | 348 |
| Porcine enteric viruses | Multiplex RT-LAMP on LoaD | Detection of 3 viruses, LOD 101–102 RNA copies per μL, 1.5 h. Clinical fecal samples tested | 349 |
| Murine norovirus | Modular nucleic acid-based detection platform with colorimetric detection | LOD 10 PFU mg−1 within 30 min in fecal sample | 350 |
| Giardia | Giardia cysts purification using spiral inertial microfluidics | Recovery rates up to 75% from mouse feces with 0.75 mL min−1 throughput | 351 |
5.3 Microfluidic sample processing techniques
In the context of GTI diagnosis and gut microbiome studies, fecal sample processing and extraction of commensal or pathogenic bacteria colonizing the gastrointestinal tract is an important issue.330 Sample processing steps are critical aspects in the design of realistic diagnostic POC strategies. A challenge of GTI diagnostic workflows are specific problems related to the complex matrix of stool samples involving high variability of consistency, the presence of PCR inhibitors, and possibly low target of analyte concentrations. Three examples of microfluidic approaches emphasizing fecal samples processing are cited here. One on-chip method for liquefaction and homogenization of human stool samples was based on piezoelectric actuation for generating strong acoustic microvortex streaming and sample mixing in a PDMS microchannel. Sharp structures in the main channel enhanced the microstreaming effect, whereas an array of narrow parallel microchannels filtered large debris. The device could be operated in continuous manner with a throughput of 30 μL min−1.331 Kang et al. proposed a microfluidic cartridge for automated nucleic acids purification from fecal samples for POC diagnosis of gastroenteritis or gut microbiome analysis. The cartridge included a pre-treatment chamber for stool sample homogenization by electromagnetic actuation and subsequent filtering. An air pressure system controlled microvalve operation. The performance of the system was evaluated using fecal samples spiked with C. difficile or Enterovirus, as well as through metagenomics analysis of clinical fecal samples of diseased patients.332 Mosely et al. designed a sample introduction interface for on-chip nucleic acid analysis and applied the method to the detection of Helicobacter pylori from liquified stool samples. The multi-chamber DNA purification chip comprised a large chamber receiving liquid stool samples and a final small elution chamber. Extracted DNA was magnetically transported through interconnecting trapezoidal microfluidic conduits filled with an immiscible phase for filtration. DNA purification and 40-fold pre-concentration was achieved within 7 min from crude clinical stool samples.3335.4 Microfluidic devices for gastroenteric pathogen detection
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250.334 Ma et al. designed a microfluidic device for the identification of Campylobacter spp. and assessment of antimicrobial susceptibility profiles. The bacterial sample was distributed into 8 separated PDMS incubation chambers containing chromogenic agar for colorimetric growth detection. Campylobacter isolates from various agri-food food models were used in this study. C. jejuni was detected in raw milk (LOD 1 × 102 CFU mL−1, within 48 h), and Campylobacter spp. in chicken meat (LOD 1 × 104 CFU, after 60 h). For on-chip multiplexed AST and multidrug resistance testing, antibiotics were preloaded onto paper disks placed in each incubation chamber, followed by adding chromogenic agar and inoculation of bacterial suspension. C. jejuni susceptibility profiles were accurately determined for three types of antibiotics within 24 h.335
250.334 Ma et al. designed a microfluidic device for the identification of Campylobacter spp. and assessment of antimicrobial susceptibility profiles. The bacterial sample was distributed into 8 separated PDMS incubation chambers containing chromogenic agar for colorimetric growth detection. Campylobacter isolates from various agri-food food models were used in this study. C. jejuni was detected in raw milk (LOD 1 × 102 CFU mL−1, within 48 h), and Campylobacter spp. in chicken meat (LOD 1 × 104 CFU, after 60 h). For on-chip multiplexed AST and multidrug resistance testing, antibiotics were preloaded onto paper disks placed in each incubation chamber, followed by adding chromogenic agar and inoculation of bacterial suspension. C. jejuni susceptibility profiles were accurately determined for three types of antibiotics within 24 h.335
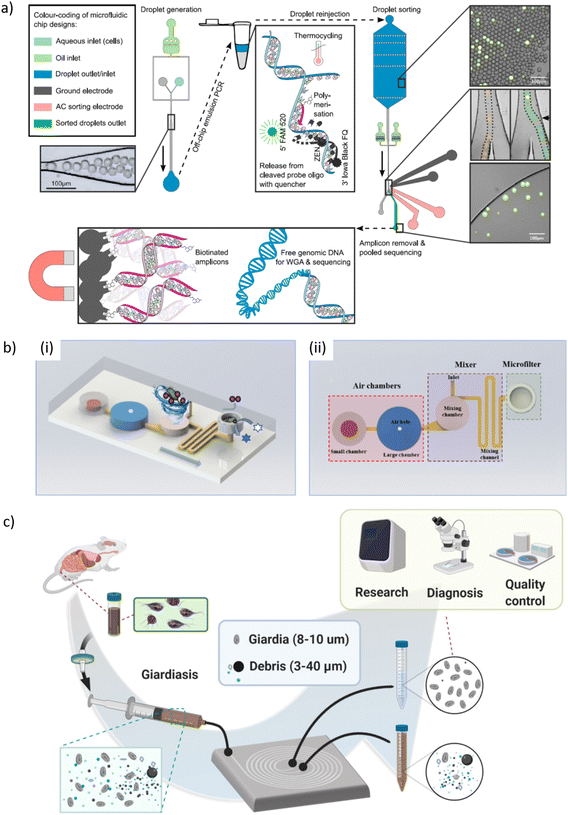 | ||
| Fig. 5 a) Microfluidic workflow, microfluidic chip designs, microscope images of their use, and schematics of molecular mechanisms of a droplet microfluidic platform for targeting individual cells from complex microbiome samples. (Top left) Droplet generation chip and photo of 39 μm-droplet generation with cells and reagents in oil. The arrow indicates the flow direction. Top center: Illustration of emulsion PCR in a thermocycler. If the target DNA sequence is present, it allows the binding of biotinylated primers and probes, thus the synthesis of corresponding amplicons. During strand extension, the TaqMan probes are cleaved, releasing fluorescent molecules. (Top right) Droplet sorting chip design and microscopy images. The microscopy pictures on the right show an emulsion before sorting and during sorting and the positive droplet enrichment post sorting. Here, fluorescent droplets indicate the presence of the gut bacterium B. vulgatus inside the droplet. Multiple microscopy images of the droplet sorting junction at different time points were overlaid and colored to demonstrate droplet flow-traces. In the right image, a small black arrow indicates the location of the nearby sorting electrode, and a bright green spot indicates the upstream location of fluorescence detection. The scale bars are 100 μm. (Bottom) Illustration of the removal of abundant amplicons with biotin-binding streptavidin beads (after pooling the sorted positive droplets) to purify genomic DNA for sequencing library preparation. Library preparation further involves whole-genome amplification (WGA) after binding adapter primers to the randomly broken genomic DNA fragments, followed by DNA fragment size selection. b) Schematic of the operation of a microfluidic biosensor chip for Salmonella detection. (i) The principle of this biosensor is based on the use of gold@platinum nanoparticles for specific bacterial labeling, a finger-driven mixer with two serial air chambers for efficient immunoreaction and a nuclear track membrane to be used as microfilter for bacterial isolation from the complex sample. (ii) Layout of the microfluidic chip. c) Schematic workflow, setup, and application of using an inertial microfluidic device to separate Giardia from a turbid fecal sample containing various sizes of contaminants [a) reprinted with permission from ref. 334, ©2021 Elsevier; b) reprinted with permission from ref. 339, ©2022 Elsevier; c) reprinted with permission from ref. 351, ©2022 AIP Publishing]. | ||
Geissler et al. designed a microfluidic cartridge installed in a centrifugal device, enabling thermal activation, internal pneumatic liquid pumping and valve control during rotation. Thanks to this dual actuation protocol, all fluidic operations for thermal lysis, PCR amplification and DNA microarray hybridization were performed in a fully-automated fashion. The system was validated for multiplexed detection of several enterohemorrhagic E. coli serotypes. The workflow took less than 2 h.336 Phaneuf et al. combined ultrasensitive immunoassays and isothermal amplification-based POC detection for both protein and nucleic acid targets on a portable centrifugal microfluidic platform. The panel of toxins and bacteria included three enterotoxins (cholera toxin, Staphylococcal enterotoxin B, and Shiga-like toxin 1) and three enteric bacteria (C. jejuni, E. coli, and S. typhimurium). All were detected with high sensitivity (LOD 1.35–5.50 ng mL−1 for immunoassays and 1–30 cells for isothermal amplification, <one hour). The system was capable of handling a complex sample matrix like stool.337 Zhang et al. performed LAMP of four foodborne pathogens (E. coli O157:H7, Salmonella spp., S. aureus, and V. parahaemolyticus) on a paper-embedded microchip. The device used centrifugal force for distributing the sample in radially arranged reaction chambers. Paper pads soaked by LAMP reagents were inserted in each chamber. The device was tested with milk samples spiked with Salmonella ssp. and on-chip DNA extraction on polydopamine-coated paper (LOD ∼12 CFU mL−1).338
Jin et al. developed a microfluidic biosensor for on-site detection of S. typhimurium from real pork samples (Fig. 5b). The device used finger-pressure air chambers for fluidic handing and a chamber for active on-chip mixing for forming Salmonella-immune Au@Pt nanoparticle conjugates. Sample concentration and colorimetric smartphone detection in H2O2-TMB substrate was carried out in a dedicated chamber. The device enabled Salmonella detection down to 168 CFU mL−1 in 25 min.339 Also Shang et al. developed a microfluidic cartridge with finger-pressure pneumatic actuation for fluidic manipulation. This device detected E. coli O157:H7 from 102 to 108 CFU mL−1 in 2.5 h with a LOD of 10 CFU mL−1. The workflow on the fully integrated device included immunomagnetic microbial separation, nucleic acid extraction and signal detection based on the RPA-CRISPR/Cas12a reaction. Spiked and real food samples were tested for system validation.340 A 10-well microfluidic chip was used by Cao et al. for the detection of multiple foodborne pathogens (Salmonella, S. aureus, E. coli O157:H7, and Shigell). Each reaction well was pre-loaded with lyophilized LAMP primers prior to multiplex on-chip LAMP with a LOD reaching reached 8 × 103 CFU mL−1 and an assay time of 45 min.341
Chung et al. proposed a paper-based norovirus POC assay combined with a smartphone fluorescence microscope. Norovirus solution and anti-norovirus fluorescent polystyrene particle suspension were successively added directly to the mainchannel of the μPAD from where they spread by capillary action and aggregated. The extent of particle immune-aggregation correlated to the norovirus concentration, enabling sensitive detection down to the single virus copy level, thus omitting the need of sample concentration or nucleic acid amplification steps. This protocol was also evaluated with field water samples. The LODs were 1 genome copy per μL in DI water and 10 copies per μL in reclaimed wastewater, respectively (assays time <20 min).344,345 Ye et al. used simple equipment-free glass fiber paper discs for rotavirus A DNA extraction, LAMP amplification and visual readout for POC diagnosis with the naked eye. The circular sample area of the paper disc was cut out and placed in a micro-well chip for the LAMP reaction and colorimetric detection. The time from sample to answer was <30 min with a LOD of 1 × 103 viral copies per mL. The device performance was evaluated with stool samples from pediatric diarrhea patients.346
A handheld PMMA microfluidic chip was designed for simultaneous colorimetric detection of four swine enteric viruses. DNA was extracted from the blood of the virus-infected swine and pipetted into the on-chip LAMP reaction chambers. To perform the LAMP reaction, the microfluidic chip was sealed and heated to 65 °C in a water bath for 60 min. The LOD of this simple POC device was 100 genomic viral DNA copies. The performance of the chip assay was comparable to commercial real-time PCR test kits.347 Another portable, 3D printed microfluidic device enabled multiplexed detection of porcine enteric viruses by real time RT-LAMP within 30 min and an analytical sensitivity of 10–100 genomic copies per reaction.348 Likewise, a microfluidic LAMP lab-on-a-disc enabled multiplex detection of a panel of viral porcine enteric pathogens from clinical fecal swine samples with analytical sensitivities in the range of 1 × 101 copies per μL to 1 × 102 RNA copies per μL within a total processing time of 1.5 h.349 Murine norovirus from fecal samples was detected using a foldable POC microfluidic chip module. Process steps included virus concentration by means of graphene oxide coated glass microbeads, lysis and RNA hybridization with colorimetric detection after an additional on-chip signal separation step. The detection sensitivity was 10 PFU mg−1 feces with an assay time of 30 min.350
Giardia is a small about 10 μm long parasite that causes diarrheal disease. Ding et al. designed a spiral microfluidic device to separate Giardia or other gastrointestinal pathogens from turbid samples, such as mouse fecal samples, by means of inertial microfluidics (Fig. 5c). The device provided a simple sample preparation method for bright-field microscopy diagnosis, achieving 75% Giardia recovery rate and 95% debris removal rate at a flow rate of 0.75 mL min−1.351 Another lab-on-a-disk device for parasite detection in stool sample was used for concentrating parasite eggs and removal of sample debris.352
6 Bloodstream infection and sepsis
6.1 Scope and common pathogens
Microbemia, or more specifically bacteremia, occurs when bacteria enter the bloodstream via the lymphatic system from sites of local infection in the body or via disrupted skin and mucous membrane. Bacteremia may be asymptomatic if viable bacteria can be rapidly removed by activation of immune response. However, if bacterial invasion persists and proliferates, e.g. due to high pathogenicity of the microbes or a weakened immune system, the bloodstream infection (BSI) leads to bacteria accumulation in other locations of the body, including artificial devices (such as implants or catheters), causing remote infections and possibly a serious systemic inflammatory response called sepsis. Sepsis is a major global health threat with a high incidence and mortality.353 A WHO report (released in 2020) indicates an estimated 49 million cases and 11 million sepsis-related deaths in 2017, accounting for approximately 20% of all-cause deaths globally.354 Sepsis can be defined as life-threatening organ dysfunction caused by a dysregulated host response to infection. Septic shock, triggered by dangerously low blood pressure, results in organ failure with a higher risk of mortality. Sepsis is characterized by significant alterations of clinical, laboratory and hemodynamic parameters due to circulatory and cellular/metabolic dysfunction.355–357 Sepsis can be considered as a biphasic syndrome, with the initial phase being characterized by overactivation of the inflammatory response (cytokine storm) followed by a second phase of persistent immuno-suppression where patients are strongly exposed to reoccurring primary or secondary infection with high risk of fatal outcome.357–359Pneumonia is the most common primary site of infection associated with sepsis, followed by bacteraemia without specific primary infection site location, UTIs and intraabdominal infection.355,356 Quantitative blood cultures indicate extremely low numbers of circulating bacteria in septic adults (typically a few tenths of bacteria per mL of blood), whereas bacterial counts may be significantly higher in neonates and children (up to 100–1000 CFU mL−1).360 Predominant bacterial species are staphylococci (S. aureus, S. epidermidis, S. pneumonia), Pseudomonas (P. aeruginosa), Enterobacter (E. coli), Klebsiella and Acinetobacter.355,356,361 Methicillin-resistant S. aureus (MRSA) was identified in about 10% of culture-positive patients in intensive care units.361 In the particular context of sepsis/sepsis shock fast antimicrobial treatment can be life-saving. For this reason, administration of broad-spectrum antibiotic as soon as possible, ideally within 1 h after the outbreak of symptoms, remains a strong recommendation in sepsis management.362
6.2 Methods and technologies for sepsis diagnosis and related reviews
G. Lippi introduced current strategies and their limitations for early sepsis diagnosis.363 The conventional culture-based workflow from incubation to pathogen identification is very time-consuming (typically in the range of 2 to 7 days) and additional subculture steps are needed for phenotypic AST.364 MALDI-TOF applied after positive BCs has significantly accelerated the pathogen identification process.365 Culture-independent molecular techniques using blood samples directly have been developed with an estimated reduction of the time to result to 6 from 48 h. Furthermore, array approaches enable multiplexing and identification of panels of relevant pathogen targets.364,366 Examples for commercial microfluidic systems include the Xpert® MRSA/SA Blood Culture assay (Cepheid, USA) that was designed for identification of methicillin-resistant S. aureus (MRSA) on a benchtop PCR system (GeneXpert®, Cepheid, USA). Time-to-result is 1 h after positive blood culture.367 The BioFire® FilmArray® Blood Culture Identification panels (BCID2) (BioFire® Diagnostics, USA) tests for 43 BSI targets (bacteria and antimicrobial resistance genes). Results are also available in 1 h from positive BC.365 The ePlex® system (GenMark Diagnostics, USA) takes advantage of an integrated assay based on droplet microfluidics, multiplex PCR amplification and electrochemical detection. The Sepsis Flow Chip® (Vitro/Master Diagnostica, Spain) detects a large panel of bacteria, fungus and antibiotic resistance genes. The multiplex PCR microarray is implemented in the 3D environment of a porous membrane enabling improved flow-through interaction of the DNA samples with immobilized probes.368 The abioSCOPE® POC platform detects an early protein biomarker of sepsis (pancreatic stone protein). The immunoassay-based system takes advantage of nanofluidic technology to accelerate molecular interactions, with test results obtained in a few minutes (Abionic SA, Epalinges, Switzerland).369A host of review articles covers the field of BSI diagnostics. Peker et al. provided an exhaustive overview of commercially available methods for the identification of microorganisms and detection of antimicrobial-resistant genes from positive blood cultures or directly from whole blood, respectively.364 Peri et al. reviewed culture-independent BSI detection systems.366 Sinha et al. also outlines the limitations of routine blood culture testing and discusses emerging molecular technologies.370 Jyoti et al. emphasized POC sensor-based methods for diagnosis of neonatal sepsis (including microfluidics).371 Zhang et al. more specifically focused on microfluidics for sepsis diagnosis.372 POC technologies were discussed by Oeschger et al., including pathogen removal devices and commercial systems for BSI detection.373 Other reviews on biosensors for BSI diagnostics have been elaborated by Kumar et al. or Tsounidi et al., respectively, including microfluidic approaches.374,375 Liu et al. reviewed detection of bacteria and sepsis-related biomarkers with aptamer-based biosensors.376 Pilecky et al. investigated pathogen enrichment from human whole blood,377 and also Burklund et al. looked into microfluidics tools for organism isolation from whole blood.378 In an earlier review, Li et al. discussed paper microfluidics for blood analysis from a more general perspective.379 In the following, we will explore different microfluidic approaches related to BSI and sepsis diagnosis. Table 5 provides an overview of selected recent systems.
| Analyte | Device and assay principle | Performance indications | Ref. |
|---|---|---|---|
| Detection and AST of sepsis-causing pathogens in whole blood | |||
| Sepsis-inducing bacteria | Micro-PCR chip with membrane-based filtration module | Rapid isolation of bacteria from blood by stirring-enhanced filtration and PCR within 4 h | 380 |
| Engineered E. coli strains, clinical isolates | Droplet digital PCR for rapid bacterial detection and AST in 10% whole blood | 3D droplet counter enabled high throughput (assay time 1 h, LOD 10 CFU mL−1). Applicable for a wide range of antibiotic resistance genes | 381 |
| E. coli, K. pneumoniae, E. faecalis, S. aureus | Culture-free detection and phenotypic AST by single-cell trapping | RBC depletion via dextran sedimentation. AST in <2 h. LOD 1–10 CFU mL−1 | 382 |
| Biomarker detection related to sepsis diagnosis | |||
| PCT, CRP, cytokines | Multiplexed electrochemical biosensor for rapid sepsis endotyping | ZnO/Au electrodes enhanced sensitivity. LOD of 1 pg mL−1, <5 min. Validation with clinical samples | 383–386 |
| PCT, CRP | Sensor microarray for nanoparticle-enhanced plasmonic detection | LOD 21 pg mL−1 for PCT and 36 pg mL−1 for CRP. Clinically relevant PCT levels in <15 min | 387 |
| CRP, NP | Immunoassays in microchannels with integrated optical detection | LOD 10 ng mL−1 for CRP and 2.1 ng mL−1 for spiked serum. Assay time 20 min | 388 |
| PCT | On-chip electrokinetically driven electrochemical immunoassay | LOD 0.04 ng mL−1 for PCT, time <20 min | 389 |
| CRP | ELISA on a sliding-strip 3D μPAD | Detection range 1–100 μg mL−1 in undiluted blood | 390 |
| IL-6 | Plasmonic immunosensor on paper | Portable device with smartphone detection. LOD 0.1 pg mL−1, within 17 min | 391 |
| IL-6 | Differential electrical counting of modified beads | LOD 127 pg mL−1, within 5 min | 392 |
| Cell-free DNA | Simple thread-based silicone tube device | cfDNA quantification (1–3 μg mL−1) spiked in plasma within 20 min | 393 |
| Sepsis diagnosis based on alterations of leucocyte properties | |||
| Neutrophils | Measurement of spontaneous motility in a channel array/maze chip | Machine-learning-based sepsis scoring validated with clinical samples | 394 |
| Leukocytes | Inertial cell sorting and isodielectric cell profiling | Continuous sepsis monitoring based on detecting activated/inactivated leukocytes | 395 |
| Leukocytes | Hydrodynamic interactions of immune cells with an on-chip pillar-array | Label-free immune profiling assay from unprocessed blood using shaped pillars, 15 min | 396 |
| Leukocytes | Microfluidic assay for combined CD64 and CD69 cell capture | Total analysis time 2 h. Assay validation with clinical samples | 397, 398 |
| Neutrophils | Quantification of CD64 expression by cell capture and differential counting | Multivariate regression using artificial neural networks. Validation with clinical samples | 399 |
6.3 Microfluidic devices for isolation and detection of sepsis-causing pathogens in whole blood
Blood is a complex matrix that often compromises direct application of molecular methods for pathogen detection, for instance due to the presence of PCR inhibitors or abundant interfering human DNA.400 Moreover, detection of extremely low pathogen concentrations encountered in clinical BSI/sepsis (as low as 1–10 bacteria per mL) in a background of blood cell concentrations that are orders of magnitude higher (RBCs ∼109 per mL; platelets ∼108 per mL; leukocytes ∼107 per mL) is extremely challenging.378 Sample preparation and pathogen enrichment therefore remains an important issue for implementing sepsis diagnosis strategies with high sensitivity.377,378 Kalyan et al. recently addressed inertial microfluidics enabling clinical research and blood cell/pathogen separation.401 In an earlier review, Pitt et al. considered various methods, including microfluidic approaches, for rapid separation of bacteria from blood, with emphasis on devices that are designed for processing mL-quantities enabling detection of extremely low pathogen concentrations.402 In the following, we present a selection of devices using different methods for bacteria isolation from blood cells for on-chip or off-chip detection.Fang et al. developed a PDMS microfluidic device for isolation and early detection of sepsis-causing bacteria directly from whole blood (Fig. 6a). The microfluidic chip assembly comprises different modules, namely (i) a pneumatically-actuated stirring-enhanced blood filtration module for extraction of bacteria-laden plasma, (ii) an adjacent chamber for bacteria capture on magnetic beads, and (iii) a micro-PCR module with four on-chip PCR chambers. Active on-chip micromixing prevented clogging of the filter membrane by the blood matrix and efficient bacteria isolation. Multiplex detection of relevant pathogens (E. coli, K. pneumoniae, P. aeruginosa, S. epidermidis and S. saprophyticus) was achieved within 4 h (LOD of 10 CFU mL−1 for the on-chip PCR reaction).380 Ohlsson et al. proposed a semi-integrated microfluidic method for bacteria detection from blood by preforming successive assay steps in connected devices. The process started with differential separation of bacteria from blood cells by on-chip acoustophoresis in a straight microchannel. The channel acted as ultrasonic resonant cavity in which blood cells migrate laterally and focus into a central stream, whereas live bacteria in plasma could be recovered from the off-center fluid fractions. The bacteria-containing plasma was then further processed in a glass capillary by acoustic-assisted bacteria trapping/enrichment on polystyrene particle clusters. Finally, bacteria were detected on a disposable PCR chip. A LOD of 1000 bacteria per mL was achieved within <2 h (P. putida spiked into whole blood).403 Dow et al. used microfluidic acoustophoretic separation and off-chip detection based on bacteriophage luminescence as optical reporter of the bacteria concentration (Fig. 6b). The system achieved a 40–60% recovery yield for concentrations from 102 to 106 CFU mL−1 (P. aeruginosa, E. coli, or S. aureus spiked in diluted whole blood). Acoustic purification resulted in a 33-fold LOD improvement of the bioluminescence assay.404 Microfluidic dielectrophoresis or elasto-inertial microfluidics are other techniques that have been explored earlier for label-free isolation of bacteria.
 | ||
| Fig. 6 a) (i) Exploded view of an integrated microfluidic chip for early detection of sepsis-inducing bacteria, featuring a stirring-enhanced filtration module, a bacteria-capturing module, and a micro-PCR module. (ii) The chip was equipped with microfluidic components (e.g. micro-stirrers, micro-pumps, micro-mixers, micro-valves, & microchambers). A magnet was placed underneath the micro-mixer for bead collection (not shown), and a thermo-electric cooler was placed under the four microchambers for PCR thermocycling (not shown). Microchambers for positive and negative PCR controls were also incorporated. EC, KP, PA, SS and SE stand for E. coli, K. pneumoniae, P. aeruginosa, S. saprophyticus, and S. epidermidis, respectively. (iii) Photograph of the integrated microfluidic chip. b) (i) Illustration of acoustic separation of bacteria from blood. A sample of bacteria and blood flows in the side inlets in laminar flow while a density-matched buffer flows in the center inlet. Acoustophoresis causes the blood cells (red and white blood cells) to migrate laterally toward the center of the channel and out the center outlet, while bacteria respond weakly to the acoustic field. This differential is the basis of isolation from blood cells. (ii) Illustration of the luminescent bacteriophage assay. Bacteriophage modified to incorporate a reporter luminescent protein are added to the blood sample and infect the target bacteria, using the host bacteria's machinery to amplify the reporter. Within one hour of incubation, a complementary substrate is added to induce the luminescent signal generation, which is recorded with a luminometer. c) Portable digital nanoparticle-enhanced plasmonic imager for biomarkers detection. (i) PCT and CRP, which are blood-circulating protein biomarkers secreted by the host body in response to systemic inflammation, are detected. A single-step bioassay directly in human serum enables rapid molecular results, critical for the early diagnosis of sepsis, by detecting individual Au nanoparticles (NPs) binding to the Au nanohole array (NHA). (ii) Prototype reader developed for highly sensitive and multiplexed detection of biomarkers. The device uses a CMOS camera and a narrow-band LED source to record the transmitted images from a nanoplasmonic chip. (iii) SEM image of an Au-NHA area after a bioassay showing the bound NPs. Inset shows a single nanoparticle bound inside a nanohole. (iv) Plasmonic image of the Au-NHA area with bound nanoparticles. The binding of Au-NPs on Au-NHAs causes local transmission suppression through distortion of plasmonic excitations in the Au-NHA and can be digitally detected using far-field imaging. The inset shows a normalized intensity contrast induced by a single nanoparticle trapped in a nanohole. d) Schematics of experiment and immune profiling workflow using deterministic lateral displacement (DLD) assays using L and L−1 pillar shapes for sorting. The whole blood DLD assay proceeds by loading the blood into the sample reservoirs of the PDMS DLD device which is used to simultaneously sort and measure the distribution of cells across the output region allowing size frequency distribution analysis. The device consists of two additional buffer reservoirs that sandwich the sample stream resulting in a precise injection of sample into the DLD region. The DLD region is composed of 21 DLD segments corresponding to 21 step measurement resolution ranging from size 6.0 to 16.0 μm in steps of 0.5 μm. The streams in input and output regions are in pseudo-color to show the differences between input and output. Scale bar is 200 μm [a) reproduced from ref. 380 with permission from the Royal Society of Chemistry; b) reproduced from ref. 404 with permission from the Royal Society of Chemistry; c) reprinted with permission from ref. 387, ©2020 John Wiley and Sons; d) reprinted with permission from ref. 396, ©2021 John Wiley and Sons]. | ||
6.4 Systems based on single-cell detection enabling fast AST in whole blood sample
Abram et al. implemented a digital one-step blood droplet PCR assay on a rapid diagnostic platform. Unprocessed whole blood samples spiked with target pathogens were mixed with reagents enabling inhibitor-resistant PCR and then compartmentalized on-chip. High-throughput droplet generation was achieved with four flow-focusing units in parallel. Subsequently, the blood droplets were collected for off-chip PCR and 3D volumetric fluorescence detection for digital target quantification. The assay achieved an analytical sensitivity of 10 CFU mL−1 within <1 h (10% blood spiked with an antibiotic resistant E. coli model strain containing the synthetic blaCTX-M-9 target gene). The applicability of the technology for rapid diagnosis of BSI was demonstrated with clinical isolates that have been interrogated for different molecular targets, including antibiotic resistance molecular markers.381 Forsyth reported a culture-free sample preparation and bacteria enrichment workflow for microbiological analysis of BSI and phenotypic AST by single-cell observation. Dextran sedimentation was used for efficient depletion of erythrocytes in whole blood spiked with clinically relevant bacteria (E. coli, K. pneumoniae, E. faecalis, and S. aureus). A centrifugation step was incorporated to enrich the sample. Subsequently, dextran-isolated bacteria were trapped in narrow microfluidic channels for single-cell detection and phenotypic AST based on bacteria growth dynamics upon exposure to different antibiotic concentrations. The workflow took less than 2 h and was performed with a clinically relevant pathogen concentration (10 CFU mL−1).3826.5 Biomarker detection for diagnosis of sepsis
A major challenge is to identify reliable biomarkers or combinations enabling rapid sepsis diagnosis and prognosis of outcome. As sepsis is an inflammatory syndrome involving numerous cellular processes most of the currently used biomarkers are not specific but may be indicative for other infectious conditions. The panel of relevant sepsis biomarkers has been reviewed by Pierrakos et al. and Biron et al., for instance.405,406 Reviews on biosensors also include discussions of biomarkers in sepsis diagnostics.371,375Two important sepsis-related biomarkers are C-reactive protein (CRP), a non-specific marker for acute immune response, and procalcitonin (PCT), a clinically relevant biomarker of sepsis diagnosis at early stages. Tanak et al. developed a dual-marker non-faradaic electrochemical impedimetric biosensor for the detection of sepsis. Zinc oxide coating of the interdigitated Au electrodes provided enhanced sensitivity. The choice of biomarkers, i.e. the early sepsis marker PCT and the late onset marker CRP, was expected to be useful for evaluating the severity of infection. Detection in whole blood with LODs of 0.10 ng mL−1 for PCT and 0.10 mg mL−1 for CRP, respectively, was reported.383 Further sensor array integration for monitoring five cytokines by means of a handheld electrochemical reader enhanced the capability of rapid POC sepsis endotyping (LOD 1 pg mL−1, results available in <5 min from a single drop of undiluted plasma).384 Subsequently, the system was validated by cohort studies with clinical samples.385,386 Belushkin et al. developed a digital nanoparticle-enhanced plasmonic imager for rapid and sensitive detection of CRP and PCT (Fig. 6c). A plasmonic gold nanohole array (Ti/Au film with 200 nm holes on a silica substrate) enabled detection of single Au nanoparticle (NP)-labeled molecules by means of a portable optical reader. The functionalized AuNP suspension mixed with blood serum was directly injected in the detection chamber of the microfluidic cartridge. Multiplexing was achieved by bioprinting different capture antibodies on the sensor array. The system showed very high sensitivity with LODs of 21 pg mL−1 for PCT and 36 pg mL−1 for CRP, respectively. Clinical validation with patient blood serum demonstrated that PCT levels typical for sepsis can be monitored in <15 min.387
Giannetti et al. implemented immunoassays for multiplex optical detection of CRP and neopterin (NP) on a multi-channel microfluidic flow cell in a sepsis diagnostic device. Combined measurement of both non-specific markers is expected to improve early diagnosis of systemic infection. A LOD of 10 ng mL−1 for CRP and 2.1 ng mL−1 for NP (spiked in human serum) was achieved, respectively, with a total assay time of 20 min.388 Molinero-Fernández et al. combined a PCT magneto-immunoassay format with flow-based electrochemical on-chip detection. The immunocomplex was injected in a cross-channel microfluidic chip and retained in the main channel while an enzymatic substrate was electrokinetically driven over the magnetic bead plug for downstream in-channel amperometric detection (LOD of 0.04 ng mL−1 for PCT standard solutions, analysis time <20 min).389 Verma et al. implemented a CRP ELISA assay in a microfluidic sliding-strip paper device. The paper strip, which contains the sensing area, was inserted in a functional dock, built up from multiple wax-patterned paper layers, defining the fluidic operations and for storage of dry reagents/detection antibody. The strip receives the blood sample at the initial position and is then moved further step-wise under successive reaction areas to perform the colorimetric ELISA assay. This simple device could be suitable for diagnosis of suspected neonatal sepsis, in particular in resource-limited setting.390 Kim et al. designed a microfluidic cartridge that incorporates an immuno-flow strip with colorimetric read-out for on-site sepsis diagnosis. The hybrid biosensor was designed for conducting a biochemical analysis (lactate assay) and immunoassays (PCT and CRT) simultaneously on the same membrane strip. In contrast to conventional LFAs, a 2D crossing flow protocol was implemented to perform the immunoassays.407
A plasmonic immunosensor for rapid detection of sepsis biomarkers in unprocessed blood, in this case the interleukin proinflammatory marker (IL-6), was proposed by Alba-Patino. Quantification of the colorimetric nanoprobe signal on a simple filter paper substrate was done in real time by means of a smartphone (LOD 0.1 pg mL−1, recombinant human IL-6 in buffer, within 17 min).391 Valera et al. used latex beads carrying the IL-6 biomarker that selectively binding to functionalized pillars in a capture chamber. On-chip coulter counting provided a measure for the IL-6 concentration (LOD 127 pg mL−1, within 5 min).392 A POC integrated magneto-electrochemical biosensor platform for sepsis diagnosis produced test results within 1 h from blood samples and detected IL-3 at a sensitivity of <10 pg mL−1.408
The content of circulating cell-free DNA (cfDNA) in plasma has been considered as an indicator for possibly sepsis fatality. Damodara et al. proposed a very simple microfluidic approach for low-cost POC detection of cfDNA. Sample and DNA-binding fluorescent dye were loaded into a silicone tube (length 1 cm) with twisted polyester threads and mixed by squeezing, prior to incubation and imaging. Quantification of cfDNA spiked in plasma in the range of 1–3 μg mL−1 was demonstrated, possible suitable to identify sepsis patients at high fatality risk.393
6.6 Sepsis diagnosis based on alterations of leucocyte properties
Microfluidic assays that analyse biochemical or biophysical properties of white blood cells have been developed as a tool for sepsis diagnosis. Neutrophils play an essential role in the response of the innate immune system.409 Babatunde et al. reviewed microfluidic devices that have been designed to investigate complex neutrophil migration behaviors.410Ellett et al. developed a microfluidic device for investigating sepsis-specific spontaneous migration signatures. The chip comprises RBC filters, an array of narrow neutrophil migration channels for measuring motility parameters and mazes allowing spontaneous binary changes of direction. Neutrophil track analysis and a machine-learning leveraged sepsis scoring system enabled accurate diagnosis of sepsis with clinical blood samples.394 Jeon et al. designed an integrated platform comprising two subsystems, namely (i) a multidimensional double-spiral microfluidic device for inertial isolation of leukocytes from peripheral blood, and (ii) an isodielectric separation chip for subsequent discrimination of activated/inactivated leukocytes based on dielectric cell properties. The authors demonstrated that leukocytes from septic and healthy human subjects could be discriminated.395 Zeming et al. proposed a microfluidic approach for monitoring rapidly changing host inflammatory response and severe immune response signatures, as occurs in acute septic conditions (Fig. 6d). For this purpose, an unprocessed blood sample was flushed through arrays of specifically designed pillars, enabling very rapid (15 min) profiling of immune response based on hydrodynamic interactions and sensing of biophysical signatures of white blood cells (size and deformability).396
A microfluidic chip developed by Zhou et al. took advantage of multiple affinity capture regions. Leukocytes with upregulated CD64 or CD69 expression levels due to inflammation or sepsis were retained in the flow channels by immunoaffinity on the functionalized sections. The interaction between cells and the substrate was increased via secondary flow patterns induced by herringbone structures in the channel.397,398 This assay builds on an earlier approach for capture of CD64+ cells only.411 Hassan et al. also proposed a microfluidic device for quantification of CD64 expression on neutrophils. The chip comprises two inlets for whole blood and RBC lysing buffer, respectively. Leukocytes were transported downstream through an anti-CD64 functionalized chamber enabling differential immunoaffinity counting of CD64+ cells.399,412
7 Sexually transmitted infections
7.1 Sexually transmitted pathogens and diseases
Sexually transmitted infections (STIs) are generally spread through unprotected sexual activity, whereas other transmission modes, for instance during pregnancy, child birth or through contact with infected body fluids, in particular blood, are possible. STIs have a profound impact on health and represent a major global burden for the public health system.413 More than 30 different pathogens can cause sexually transmitted diseases and over 1 million STIs are acquired every day worldwide (WHO STIs factsheet 2023).414 The four most common STIs are: chlamydia, a bacterial infection mainly caused by the species Chlamydia trachomatis (CT),415 gonorrhea with Neisseria gonorrhoeae (NG) as bacterial causative organism,416 trichomoniasis provoked by the protozoan parasite Trichomonas vaginalis (TV),417 and syphilis, which is due to infection with the bacterium Treponema pallidum (TP).418 These STIs are often asymptomatic or show non-specific symptoms, as a consequence most infected patients do not seek medical treatment. These diseases are actually curable with antimicrobial treatments, however, if untreated, serious impact on health may occur, including neurological and cardiovascular disease, infertility, stillbirths, and an increased risk of human immunodeficiency virus (HIV) infection.419 Rapidly increasing antimicrobial resistance is a growing threat for untreatable gonorrhea.420Most important currently incurable viral STIs or related causative pathogens are:414 hepatitis B virus (HBV),421 genital herpes simplex virus (HSV),422 HIV/AIDS423 and human papillomavirus (HPV).424 Hepatitis, in particular types B and C, leads to chronic disease such as liver cirrhosis and liver cancer. WHO estimates that 296 million people were living with chronic HBV infection in 2019, resulting in an estimated 820![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths, even though vaccines are available to prevent fatal outcome.425,426 Diagnostic devices focusing specifically on hepatitis C viruses (HCV) will be included in this section, even if transmission generally occurs by infected blood samples (e.g. by sharing syringes for drug injection) and not by sexual intercourse. Acquired immunodeficiency syndrome (AIDS) is a potentially life-threatening condition caused by HIV that continues to be a major global public health issue with an estimated 39.0 million people living with HIV infection at the end of 2022.414 HIV attacks the body's immune system, specifically white blood cells (CD4 cells), thus increasing the risk of morbidity through co-infections.419 HIV, as well as HSV, generate lifelong infections, but efficient antiviral therapies may suppress the viral load sufficiently to maintain manageable chronic conditions.427,428 Vaccines against HPV have been developed to prevent HPV-associated cancers, in particular cervical cancer.429
000 deaths, even though vaccines are available to prevent fatal outcome.425,426 Diagnostic devices focusing specifically on hepatitis C viruses (HCV) will be included in this section, even if transmission generally occurs by infected blood samples (e.g. by sharing syringes for drug injection) and not by sexual intercourse. Acquired immunodeficiency syndrome (AIDS) is a potentially life-threatening condition caused by HIV that continues to be a major global public health issue with an estimated 39.0 million people living with HIV infection at the end of 2022.414 HIV attacks the body's immune system, specifically white blood cells (CD4 cells), thus increasing the risk of morbidity through co-infections.419 HIV, as well as HSV, generate lifelong infections, but efficient antiviral therapies may suppress the viral load sufficiently to maintain manageable chronic conditions.427,428 Vaccines against HPV have been developed to prevent HPV-associated cancers, in particular cervical cancer.429
7.2 Current STI diagnostics landscape and recent reviews
Diagnostic procedures to detect STIs include both direct and indirect methods based on various samples, including blood, urine, and vaginal or endocervical swabs. Direct pathogen detection, like pathogen culture, antigen tests or nucleic acid tests can be used to examine localized and acute infections. On the other hand, antibody response to infection may become detectable only after weeks to months, thus are more suitable for diagnostic evaluation of chronic infection or the patient's actual immune status.430 Rapid LFA STI diagnostic tests for antibody/antigen detection are available, such as syphilis treponemal antibody tests (for instance Syphilis Health Check™, Diagnostics Direct, USA).431 These tests have limited sensitivity but deliver qualitative results in less than 30 min at the POC. NAAT-based commercial systems have been developed or adapted for POC STI diagnostics. As an example, Xpert® microfluidic PCR assays running on the GeneXpert® system have been designed for sexual health testing, including CT/NG and TV assays, as well as tests for viral STIs (including HIV, HBV/HCV and HPV). The Bosch Vivalytic STI PCR test can reliably analyse a patient sample for ten different pathogens at once. A urine sample or a urogenital swab is used as the sample type, results are available in 30–90 min.115 Another promising approach is the Visby Medical Sexual Health Test (Visby Medical, USA), which is a rapid (results within 30 min), handheld and easy-to-use PCR device with excellent sensitivity and specificity for CT, NG and TV detection.432Cristillo et al. reviewed the frame of POC STI diagnostics from a larger perspective, including current and emerging technologies, clinical and public health benefits and other aspects,433 whereas Wi et al. discussed challenges of STI diagnosis in resource-constrained settings.434 Caruso et al. compared current STI laboratory diagnostic tools with respect to performance, applicability, and adaptability to POC format.435 Toskin et al. looked at the state of STI POC testing technologies and the implications for health system integration.436 Discussions including commercial POC systems for STI testing, in particular for the four major curable STIs and HIV, have been provided by Adamson et al., Gaydos et al. or Hsieh et al., for instance.437–439 Hsieh et al. also investigated the gap of near-POC NAAT devices and the requirement to move to real POC applications. Thakur et al. recently reviewed high-performance biosensing systems for STI diagnostics,440 and Farokhzad et al. analyzed progress towards POC platforms from a materials chemistry-enabling perspective.441 Other reviews by Tharakan et al., Pai et al. or Eid et al. focused on novel technologies for HIV diagnostics, including nanotechnology and microfluidics.442–444 Xiao et al. provided an overview of POC tests for hepatitis B,445 other reviews addressed more specifically HCV diagnosis via microfluidics or biosensors for hepatitis diagnostics (HBV and others).446,447 Nath et al. discussed laboratory and POC techniques for diagnosis of HSV infections.448
Even if a wide range of STI diagnostic tools is currently available, high actual incidence rates of STI, emergence of antimicrobial drug resistance and the requirement of extended screening campaigns in low and middle-income countries motivate the development of new truly POC diagnostic devices with high sensitivity and short time-to-result. In the following we will discuss a selection of recent microfluidic and biosensor developments for STI diagnosis. Table 6 provides an outline.
| Analyte | Device and assay principle | Performance indications | Ref. |
|---|---|---|---|
| Microfluidic systems and assays for HIV/AIDS diagnostics | |||
| HIV viral particles | Filtration-assisted magnetofluidic cartridge qPCR platform | Uses a separate 3D-printed filter module, LOD 105 mL−1 HIV RNA (whole blood), 30 min | 449 |
| HIV-1 RNA | Fingerpick blood RT-LAMP test cartridge using magnetic transport | Viral load tests performed within 60 min in whole blood (threshold at 1000 copies per mL) | 450 |
| HIV-1 p24 antigen | Colorimetric ELISA assay on a 3D origami paper-based device | Semi-quantified results, LOD 0.03 ng mL−1 (spiked plasma), 10 min | 451 |
| HIV-1 RNA, HIV-1 virus | Cartridge for autonomous RT-LAMP with integrated LFA detection | LOD 3 × 105 HIV-1 viral particles per mL of whole blood, within 90 min | 452 |
| HIV-1 p24 antigen | HRP-linked immunoassay and Au nanorod based multicolor assay | Semi-quantitative analysis by naked eye in 1 h, LOD 0.5 ng mL−1 (buffer) | 453 |
| HIV-1 synthetic target | RCA combined with microfluidic affinity chromatography | Padlock probe-mediated detection, LOD <30 fM | 454 |
| HIV-1 plasmids | Wearable device for HIV-1 DNA RPA using human body heat | LOD 100 copies per mL (buffer), within 24 min | 455 |
| Microfluidic systems for HBV/HCV and HPV detection | |||
| HCV and HIV cDNA | Multiplexed digital droplet LAMP with scorpion-shaped probes | Quantification with LOD of 4 copies per reaction. Clinical plasma samples tested. | 456 |
| HCV RNA | LAMP assay cartridge with lateral flow detection | Result in <40 min. Tested with clinical samples | 457 |
| HBV, HCV, HIV nucleic acid targets | Self-driven microfluidic multiplex LAMP chip | Sample loading via on-chip vacuum. LOD 2 copies of target nucleic acid per μL | 458 |
| HPV | LAMP on LoaD with Chelex-100 based nucleic acid isolation | Multiplex detection of 5 high-risk HPV virus types, within 40 min | 459 |
| Up to 24 HPV genotypes | Reverse dot hybridization for HPV genotyping on a palm-sized cartridge | Automated operation with LOD of 103 copies per mL. Tested with clinical samples | 460 |
| HPV-16 and HPV-18 | SlipChip approach for viral load quantification with digital LAMP | On-chip self-partitioning of reaction droplets. Tested with clinical samples | 461 |
| Microfluidic systems for non-viral STIs | |||
| N. gonorrhoeae | Cartridge-based automated magneto-fluidic PCR platform | Detection in clinical samples with simultaneous antimicrobial resistance genotyping, <15 min | 462 |
| STI pathogens | Isothermal nucleic acid amplification on LoaD | Multiplex pathogen detection directly from genitourinary secretions, within 50 min | 463 |
| N. gonorrhoeae | Foldable paper platform based on thermophilic HDA and LFA detection | Clinically relevant LOD of 500 NG cells per device, run time 80 min | 464 |
7.3 Microfluidic systems and assays for HIV/AIDS diagnostics
In clinical settings, HIV/AIDS disease progression is monitored by CD4 counts,465 whereas p24 viral capsid protein antigen immunoassays or antigen/antibody combination assays are used for detecting acute HIV infection.466 NAAT-based RNA detection allows quantification of the viral load and early detection of infection. Trick et al. proposed a filtration-assisted magnetofluidic blood-to-PCR workflow for HIV RNA detection from blood (Fig. 7a).449 As described below, a similar approach was also used for other pathogens.462,467 In the present case, viral HIV particles were extracted from a droplet of whole blood by means of a filtration module and collected in a vial for lysis and RNA capture on magnetic beads. Subsequently, the filtered plasma mixture was loaded into the assay cartridge and inserted in the magnetofluidic qPCR platform. The system achieved a LOD of down to 1000 copies of HIV RNA per blood sample with a typical fingerprick volume of 10 μL (i.e. a LOD of 105 per mL) within in <30 min.449 The platform also enabled HCV RNA viral load quantitation from blood serum in approximately 1 h. The assay had a sensitivity of 45 IU per sample (corresponding to 4.500 IU mL−1) and may be suitable for screening patients with chronic HCV infections.468 Liu et al. developed a compact automated USB-interfaced analyzer aiming HIV self-testing. 100 μL of finger-prick blood would be collected and mixed in a tube containing lysis buffer and magnetic beads. In the present case, the device was tested with purified HIV RNA spiked into whole blood. The mixture is then manually transferred into a microfluidic cartridge and magnetically transported through a RNA binding chamber, a washing chamber and a RT-LAMP reaction chamber. Oil valve sections separate successive assay steps. Differentiating clinically relevant viral loads (threshold at 1000 copies per mL) was demonstrated with a turnaround time of 60 min. The RT-LAMP assay (validation in reaction tubes) had a LOD of 214 viral HIV RNA copies per mL for spiked whole blood.450 An immunoassay for HIV p24 antigen quantification was implemented by Li et al. in a centrifugal microchannel array chip format with a smartphone detection unit. Horseradish peroxidase (HRP)-catalyzed oxidation of tetramethylbenzidine (TMB) induced a color intensity change in the presence of the p24 antigen. LODs obtained were 0.17 ng mL−1 and 0.11 ng mL−1 for p24 antigen spiked in buffer or human serum, respectively.469 | ||
Fig. 7 a) Filtration-assisted magnetofluidic blood-to-PCR workflow. (i) To begin the test, (1) a droplet of blood is first deposited into the filter module and viral particles are rinsed through the filter membrane with phosphate buffered saline (PBS) using a syringe. The filter traps red blood cells (RBCs) while viral particles are small enough to pass through into a lysis and binding solution containing magnetic beads for capture of viral RNA. This entire filtered plasma mixture is (2) loaded into the assay cartridge, which is then (3) inserted into the instrument. (ii) The magnetic beads are transferred through an immiscible silicone oil layer into the cartridge's extruded wells containing preloaded reagent buffers using a 2-axis motorized magnet arm. (iii) Bead transfer between wells is conducted by lateral movement of the top permanent magnet with bead exchange into buffers using vertical translation of the magnet arm to attract beads into the well with the bottom magnet. The final transfer of beads into the PCR buffer allows direct elution of RNA due to the relatively alkaline pH, elevated temperature, and low salt conditions. b) Lateral flow detection of HCV LAMP assay. (i) The mechanism of the lateral flow strip. Two primers, FLP and BLP, are pre-labelled with biotin and FITC, respectively. The amplicon resulting from the LAMP reaction, contains both labelled primers as double-stranded DNA represented schematically by two gray lines. It is added onto the sample pad and moves towards the conjugate via capillary action. The streptavidin-labelled red particles bind with the biotin (from the FLP primer) on the amplicon and together move towards the test line. The anti-FITC antibody (Ab) on the strip captures the amplicon via its FITC label (from the BLP primer) at the test line forming a band. Any unbound red particles move towards the control line where they are captured via the biotin forming a second band. (ii) The assembly and interpretation of the lateral flow devices. The device consists of the water chamber (1), connecting channels (2), four LAMP reaction chambers (3), channels (4) and lateral flow strips (5). Following the incubation period, two bands indicate a positive reaction, one band indicates a negative and no bands indicate invalid results. P – positive, N – negative, 1 and 2 – sample in duplicate. (iii) Analytical sensitivity of the lateral flow method (top panel) compared to gel electrophoresis (middle panel) and fluorescence over time (bottom panel). Serial dilutions of plasmid JFH1 replicon were made based on copy number per reaction (log10). Black lines indicate median with interquartile range (n = 3 biologically independent experiments, each with three technical replicates). The different symbols are for each dilution, to ease visualisation (grey and white disc – 4.1, white lozenges – 3.8, inverse triangles – 3.5, black diamond – 3.2, grey circles – 2.9 and white squares – 2.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 copies per reaction, black disc is negative control – DI water). Statistical analysis for the fluorescence over time was performed using a parametric, one-way ANOVA. The F ratio = 55.56 and the degrees of freedom = 65. ****p ≤ 0.0001, Pos – positive HCV control, Neg – no template control, M – 100 bp NEB DNA ladder, C – control line, T – test line [a) reproduced from ref. 449 with permission from the Royal Society of Chemistry; b) reproduced from ref. 457, ©2021, Creative Commons license, CC BY 4.0 (http://creativecommons.org/licenses/by/4.0/)]. log10 copies per reaction, black disc is negative control – DI water). Statistical analysis for the fluorescence over time was performed using a parametric, one-way ANOVA. The F ratio = 55.56 and the degrees of freedom = 65. ****p ≤ 0.0001, Pos – positive HCV control, Neg – no template control, M – 100 bp NEB DNA ladder, C – control line, T – test line [a) reproduced from ref. 449 with permission from the Royal Society of Chemistry; b) reproduced from ref. 457, ©2021, Creative Commons license, CC BY 4.0 (http://creativecommons.org/licenses/by/4.0/)]. | ||
Paper-based devices open the way to real POC applications, based on electricity- and instrument-free infectious disease diagnosis. Chen et al. performed a colorimetric ELISA assay on a 3D origami μPAD sensor for HIV-1 p24 antigen detection. The μPAD contained wells with pre-dried detection antibodies and capture antibodies. Assay steps were sequentially performed by sliding a paper strip over the 3D stack inserted in a 3D-printed holder. Appropriate self-timing of the multistep assay was achieved by adjusting the buffer viscosity and the volume of the fluid path. Semi-quantitative evaluation of the signal visible to the naked eye on the μPAD was possible by comparison with a color chart diagram. The linear detection range for HIV-1 p24 antigen was 0.03 ng to 3 ng mL−1 with human plasma samples.451 Phillips et al. reported a fully-integrated autonomous POC analysis platform comprising a microfluidic μPAD installed in a small plastic housing with resistive heating elements, powered via a smartphone. The system was designed for automatically isolating HIV-1 RNA from whole blood, in situ RT-LAMP amplification, valve-controlled fluid transfer and detection on an internal LFA. The time-to-result was 90 min with a LOD of 3 × 105 HIV-1 viral particles (corresponding to 2.3 × 107 virus copies per mL of whole blood).452
A microfluidic immunosensor device for visual detection of HIV-1 p24 antigen with the naked eye was developed by Liu et al. The polymer cartridge for the multicolor immunosensor comprised an alternating alignment of circular reservoirs for assay reagents and elliptical reservoirs filled with mineral oil barriers. A HRP-linked magnetic bead-based immunoassay was implemented for detecting HIV-1 p24 antigens. Gold nanorods (AuNR) were used as chromogenic substrates. HRP-catalyzed TMB oxidation quantitatively mediated AuNR etching to produce a color change, which was visible within a HIV-1 p24 concentration range of 0–7 ng mL−1 (spiked serum samples). The assay enabled semiquantitative detection of HIV-1 p24 with a LOD of 0.5 ng mL−1 in buffer within 1 h. A significant color signal was displayed with 2 ng mL−1 HIV-1 p24 spiked in serum.453 Soares et al. used an agarose bead-based microfluidic device for affinity chromatographic capture and detection of padlock probe-mediated RCA products with a LOD below 30 fM for HIV-1 synthetic targets.454 Song et al. designed a microfluidic chip reactor for RT LAMP-based detection of HIV or HPV. Paraffin-encapsulated reagents were prestored in the reactor and a nucleic acid binding membrane decoupled the sample from the reaction volume. The device was tested with HPV-16 DNA spiked in saliva and human plasma laden with HIV (subtype C) virions.470 Kong et al. developed a wearable microfluidic device for rapid detection of HIV-1 DNA. The flexible chip, featuring a single fluidic 50 μL chamber containing RPA reagents and HIV-1 DNA, was immobilized by a wristband. The compatible temperature range for performing the RPA-based assay was simply provided by heat transfer from the human body. After incubation the chip was removed for fluorescence detection with a cellphone. HIV-1 DNA detection was achieved at 100 copies per mL in buffer within 24 min.455
A nanofluidic concentration device enabled highly efficient enrichment of nucleic acids and proteins directly from clinical samples. The approach, developed by Ouyang et al., took advantage of a hierarchical architecture with vertically stacked massively parallel microchannels in a first stage and downscaling to a single microchannel in the final stage of the device. The analytes were electrokinetically transported and concentrated through the nanochannel network construct comprising a cation-selective Nafion membrane held by plasma-bonded PDMS layers. The performance of the device was demonstrated by detecting the HIV p24 protein in a 3200-plex concentration device (0.6 mL sample volume). Fluorescence signals could be detected within 60 min for an initial target concentration of 10 to 100 aM (in diluted serum), representing a nearly 6 order-of-magnitude enhancement compared to the same assay without pre-concentration.471 Kadimisetty et al. discussed a self-powered flow-through membrane-based 3D-printed sample concentrator for highly sensitive molecular detection of HIV in whole blood at the POC.472
7.4 Microfluidic systems for HBV/HCV detection and other viral pathogens
Tan et al. adapted a droplet microfluidic technology for multiplexed LAMP and fluorescence detection of viral RNA. Multiplex detection in droplets was enabled by scorpion-shaped probes which, after activation by the LAMP reaction, generate a target specific color. Digital quantification of HCV and HIV cDNA with LOD as low as 4 copies was reported.456 Witkowska McConnell et al. proposed a low-cost pan-genotypic RT-LAMP assay for HCV detection (Fig. 7b). The cartridge-like assembly comprises four RT-LAMP reaction chambers, transfer channels and lateral flow strips for visual detection of amplification products in less than 40 min. Fluidic manipulation was done via finger pumps. Using optimized primers, the method was validated as part of a clinical study with samples from patients with a range of viral loads and genotypes.457 Li et al. designed a lab-on-a-disc for real-time PCR HBV DNA detection from whole blood. Versatility of the automated fluidic manipulation was enhanced by using a centrifugal device with two rotating shafts. On-chip wax valves were actuated by means of a laser diode. Detection of HBV in whole blood samples was feasible down to 102 copies per mL with a total assay time of about 48 min.469 LAMP-based multiplex detection of HBV, HCV and HIV was performed on a microfluidic chip designed by Xie et al. The PDMS chip comprises an array of 60 reaction chambers with target-specific primer solutions, fluidic channels and a vacuum chamber generating a negative pressure gradient for self-driven sample loading. An analytical sensitivity of 2 copies per μL of target viral nucleic acid after 50 min of isothermal amplification was reported.458A LAMP-based microfluidic lab-on-a-disc system designed by Zhao et al. provided automated diagnosis of five high-risk HPV types with high specificity within 40 min. The system integrated Chelex-100 based nucleic acid isolation and is capable of performing 40 detections simultaneously.459 A palm-sized PCR microfluidic cartridge, installed in a custom-made operating platform, enabled testing of 24 HPV genotypes. The whole cartridge comprised 3 functional areas for DNA extraction, for amplification in attached PCR tubes and a DNA microarray for reverse dot hybridization assay to identify multiple HPV genotypes, respectively. The LOD of the system for the detection of 24 HPV genotypes was 103 copies per mL.460 SlipChip technology for slip-induced self-partitioning and on-chip stationary droplet formation was applied to the quantification of HPV viral load by means of digital LAMP. The assay correctly identified clinical HPV-16 and HPV-18 positive samples with viral loads ranging from 7.0 × 102 copies per mL to 1.4 × 107 copies per mL.470 Goux et al. discussed the performance of nanophosphor lateral-flow assay for self-testing for HSV-2 seropositivity.473
7.5 Microfluidic systems for non-viral STIs
Shin et al. designed a stationary droplet NAAT magnetofluidic platform for CT screening. The core of the device is a thermoformed polymer cartridge with distinct fluidic compartments separated by a fluorinated oil layer. In the present case, the clinical sample matrix presented in the form of a vaginal swab was first expressed in a tube for cell lysis before being transferred into the cartridge. Pathogen DNA was captured on magnetic particles and magnetically transported through the cartridge for performing the assay protocol, i.e. successive rinsing steps, subsequent elution and LAMP amplification. The cartridge was installed in a compact processing unit for mechanical and thermal control, as well as for fluorescence signal acquisition. The duration of the automated workflow was 65 min with an assay sensitivity in the range of 102–103 target copies per sample. The performance of the platform was successfully tested in a clinical setting.467 More recently the platform has been used for detecting NG with simultaneous genotyping for resistance to ciprofloxacin. A clinical study was carried out with penile swab samples from sexual health clinics. In this case, rapid on-chip PCR (40 cycles were completed within 12 min) was implemented resulting in sample-to-answer diagnosis in less than 15 min. Initial manual operations were limited to swab elution and mixing with magnetic bead suspension. Depending on the amplified gene sequence an assay sensitivity in the range of 10 to 100 CFU was determined.462 The technology and assay principle were also adapted for HIV and HCV detection (see above).449,468Magnetic actuation was also implemented in another automated droplet-array platform with a 3D printed fluidic cartridge. The system was tested by microfluidic screening of STI pathogens directly from clinical urine samples.474 A microfluidic lab disc assay based on isothermal amplification was designed for processing eight clinical samples simultaneously targeting CT, NG, Mycoplasma hominis, and Ureaplasma urealyticum from genitourinary swabs. The turnaround time was 50 min with good detection limits and specificity.463 Horst et al. presented a POC paper-fluidic device for POC diagnosis of NG that is expected to bridge the gap between RDTs and laboratory NAATs. Patient samples derived from clinical urethral and vaginal swabs were mixed with lysis/DNA precipitation buffer and pipetted into the reaction chamber of the device for nucleic acid extraction and multiplexed thermophilic HDA, followed by visual lateral flow assay detection. The assay run time was 80 min from sample-to-result with a LOD of 500 genomic NG copies per reaction.464
8 Vector-borne infections
8.1 Scope of vector-borne infections
Vector-borne diseases are caused by parasites, bacteria or arboviruses that are transmitted mainly by bloodsucking infected arthropod insects, such as mosquitoes or ticks. The ingested disease-producing microorganisms reproduce in the animal (vector) and are (re-)transmitted to humans by insect bites. Vector-borne diseases account for more than 17% of all infectious diseases, causing more than 700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths annually.475 The burden is highest in tropical and subtropical areas, but vector-borne diseases are also widespread in Europe. The prevalence strongly depends on environmental conditions, such as climate factors, habit destruction or pesticide application, that impact the vector distribution. Vector control is a fundamental approach in WHO response to preventing disease.476,477 Global patterns are evolving with climate change.18,478,479 For instance, climate change enables the expansion of Lyme borreliosis, the most common bacterial (Borrelia burgdorferi) tick-borne disease in Europe.480,481 Invasive mosquito species, such as the Asian tiger mosquito (Aedes albopictus) are causing increasing concern in public health due to the possible transmission of tropical pathogens.482
000 deaths annually.475 The burden is highest in tropical and subtropical areas, but vector-borne diseases are also widespread in Europe. The prevalence strongly depends on environmental conditions, such as climate factors, habit destruction or pesticide application, that impact the vector distribution. Vector control is a fundamental approach in WHO response to preventing disease.476,477 Global patterns are evolving with climate change.18,478,479 For instance, climate change enables the expansion of Lyme borreliosis, the most common bacterial (Borrelia burgdorferi) tick-borne disease in Europe.480,481 Invasive mosquito species, such as the Asian tiger mosquito (Aedes albopictus) are causing increasing concern in public health due to the possible transmission of tropical pathogens.482
Dengue is the most prevalent arthropod-borne viral disease.483,484 The dengue virus (DENV) is transmitted by Aedes mosquitoes. About half of the world's population is at risk of dengue with an estimated 100–400 million infections occurring each year.485 Dengue currently is endemic in more than 100 countries with strongly growing incidence.486 Traditional methods for dengue virus detection are based on virus isolation in cell culture, serological assays, and molecular techniques.487 Emerging diagnostic biosensor technologies, including POC approaches, specifically focusing on dengue virus infection have been reviewed previously by Eivazzadeh-Keihan et al. or Darwish et al.488,489 Other viral mosquito-borne pathogens and diseases, mainly found in tropical and subtropical areas, include Lymphatic filariasis, Chikungunya virus (CHIKV), Zika virus (ZIKV), Yellow fever, Japanese encephalitis and West Nile viruses (listed according to global health significance).478 Review articles discussing microfluidic and biosensor devices for virus detection from a more general perspective generally also include tropical viruses such as DENV, ZIKV or CHIKV.96,120
Malaria, caused by the parasite Plasmodium, remains one of the most important vector-borne infectious diseases worldwide. This disease will therefore be discussed more extensively in a separate section. In the following, we first introduce systems for the detection of panels of fever-causing tropical pathogens and then discuss a selection of recent microfluidic or chip-based diagnostic systems for viral and bacterial vector-borne diagnostics. Table 7 provides an overview.
| Analyte | Device and assay principle | Performance indications | Ref. |
|---|---|---|---|
| Microfluidic platforms and devices for viral vector-borne diseases | |||
| 12-plex panel of fever-causing pathogens | LoaD platform for diagnosis of febrile illnesses | Tested with biobanked clinical samples. POC detection of coinfection within 2 h | 76 |
| 8 vector-borne viruses | Real-time or colorimetric LAMP assays on a self-powered microfluidic chip | LOD 50–500 plasmid copies per μL, 50 min | 490 |
| DENV, ZIKV, CHIKV and SARS-CoV-2 antibodies | Multiplex immunoassay platform incorporating a functionalized bead line array | Vacuum-driven fluid transfer. Chip with staggered herringbone micromixer. Test within 30 min | 491 |
| ZIKV RNA or virus | Modular design for sample pre-processing and LAMP. Spatial signal distribution readout | <32 min to distinguish pos/neg samples. LOD 2.7 × 102 RNA copies per μl (buffer) | 492 |
| ZIKV | LAMP assay chip with successive chambers and oil-filled valving sections | LOD 102 copies per mL (ZIKV spiked plasma), within 40 min | 493 |
| DENV | Paper origami/polymer device for a multiplex LAMP-based assay | Detection of 4 DENV serotypes, within 30 min (serum) | 494 |
| DENV, ZIKV, CHIKV | Lab-on-paper in LFA format for all-in-one molecular diagnostics | Multiplex detection of 3 viruses, within 60 min (serum) | 495 |
| CHIKV IgM | Paper-based device with CHIKV pseudo-particles for antibody capture | CHIKV IgM detected within <10 min (serum) | 496 |
| Microfluidic methods for Lyme disease diagnosis | |||
| Lyme-specific antibodies | Plasmonic biochip with antigen spot array for multiplex target screening | LOD in the fM range (clinical serum) | 497 |
| Lyme-specific antibodies | Paper-based multiplexed serodiagnostic test for early-stage Lyme disease | Sensing array with 7 Borrelia antigens. Machine learning based colorimetric detection | 498, 499 |
| Lyme-specific antibodies and biomarkers | Multiplexed serologic tests in microfluidic cassette functionalized with Borrelia proteins | Diagnosis of early and late Lyme disease using panels of clinical serum samples | 500 |
8.2 Microfluidic platforms enabling the detection of panels of fever-causing pathogens
Efficient management of febrile tropical illnesses, including patients presenting non-specific fever or fever with unknown origin, is a significant health care challenge.501,502 The availability of fast infection-specific true POC diagnostic tests, in particular for malaria and dengue, is indispensable to avoid presumptive therapy. Diagnostics of vector-borne infections and febrile illnesses in general, would greatly benefit from POC devices enabling the detection of wide panels of relevant pathogen simultaneously, by this reducing the important risk of misdiagnosis. Nonvector-borne emerging or reemerging tropical viruses, for instance the Ebola virus, could be included in the panel of fever-causing pathogens. Mitsakakis et al. elaborated a comprehensive review covering (commercial) diagnostic technologies and platforms for tackling febrile illness, including vector-borne diseases.108 The chip-based NAAT assay VereFever™ (VerePLEX™ Biosystem Veredus Laboratories, Singapore) is an example for a commercial approach that was designed for multiplex detection of a panel of viral pathogens (including DENV serotypes, ZIKV, CHIKV and others), as well as Plasmodium spp for malaria detection. Tan et al. evaluated the system performance and found a detection range from 250 to 4 × 107 PFU for CHIKV.503Hin et al. presented the FeverDisk platform, a LabDisk-format centrifugal device designed for differential diagnosis of tropical febrile illness with unknown origin.76 The platform enables simultaneous detection of a 12-plex panel of different infectious fever-causing pathogens, including several arboviruses collected during epidemics outbreaks (dengue, chikungunya), bacteria (e.g. Salmonella), and malaria parasite species. FeverDisk assays feature fully-automated integrated magnetic bead-based DNA purification and LAMP with fluorescence detection. Thanks to pre-stored reagents on the disc, only a single manual pipetting step for sample addition was required, providing fully integrated sample-to-answer operation. To validate POC capabilities of the device, two test series in African reference laboratories have been conducted with different types of biobanked or bacterial culture samples. Assays were performed in a total run time of less than two hours and in good agreement with reference method results. Malaria infections could be successfully confirmed in malaria-positive whole blood samples on a very short time scale. Likewise, the system confirmed different viral infections and co-infections. The analytical sensitivity of salmonella species was determined in a separate test series using cultured samples.76 The same authors also adapted this technology for vector monitoring (VectorDisk), i.e. for identifying mosquito and malaria parasite species, as well as insecticide resistance mechanisms.504
Yao et al. proposed a microfluidic RT-LAMP chip for multiplex detection of eight vector-borne viruses (DENV serotypes, CHIKV, yellow fever virus, Rift Valley fever, Japanese encephalitis virus). Rapid self-powered sample loading was achieved by PDMS-film vacuum pumping. Detection was either based on real-time LAMP or colorimetric read-out. The whole analysis could be completed in 50 min. The system was tested with human blood serum samples and infected mosquito samples.490 Moutallier et al. used a commercial microfluidic digital PCR system (Biomark™ HD, Standard BioTools, USA) for high-throughput detection of 64 mosquito-borne viruses in pools of mosquitoes from ZIKV endemic/epidemic areas.505
8.3 Microfluidic approaches for viral vector-borne human diseases
Ganguli et al. used a microfluidic diagnostics card with multiplex capability (ZIKV, DENV, CHIV) for detection of ZIKV in spiked whole human blood. The device comprises a sample PDMS preparation module and a second silicon chip module for the RT-LAMP assay. The integrated system allowed for hands-free sample processing and smartphone detection with a LOD corresponding to 1.56 × 105 PFU mL−1 for ZIKV within 35 min.508 Lee et al. designed a PDMS microfluidic chip inspired by conventional LFAs for multiplex one-step immunoassay detection (including DENV, ZIKV, CHIKV and SARS-CoV-2 biomarkers). Microfluidic on-chip components comprise a dried reagent storage chamber, a passive micromixer, and a vacuum-driven fluid transfer void. Self-assembled lines of functionalized polystyrene microbeads served as antigen or antibody capture/detection zones. Various biomarker combinations have been tested to demonstrate the versatility of the approach.491
An interesting assay for arbovirus detection was based isothermal amplification of ZIKV cDNA using padlock probes and RCA, combined with a bead-based microfluidic affinity chromatography enrichment platform.509 Jankelow et al. implemented LAMP assays for ZIKV detection from whole blood samples in a 3D printed microfluidic cartridge. The cartridge consists of two separate modules, involving several manual operations for mixing and other fluidic handling steps. A drop of unprocessed whole blood was introduced in the first module where viral RNA was released by chemical lysis and mixed with RT-LAMP reagents. The preprocessed sample solutions were then pipetted into the RT-LAMP reaction module, comprising parallel microfluidic compartments for spatial analysis of early fluorescent events of positive samples via smartphone real-time readout. A LOD of 2.7 × 102 viral RNA copies per μL (buffer) and 103 virus particles (in a 12.5 μl blood droplet) was determined. Discrimination of positive/negative samples was possible in less than 32 min.492 A LOC platform detected ZIKV within 40 min in spiked human plasma samples and a LOD of 102 copies per mL (spiked ZIKV nucleic acid plasma samples). The chip design featured successive reaction/washing and oil-filled valving chambers for automated sample transport by magnetic actuation, sequential on-chip assay steps, LAMP and colorimetric detection.493 In another approach, chitosan-modified SiO2 capillaries were embedded in a PDMS chip for on-chip sample lysis, ZIKV RNA enrichment and in situ RT-PCR amplification. A LOD of 50 TU (transduction units) per mL was reported with virus-like particles containing the ZIKV RNA sequences spiked in human raw saliva samples.510
 | ||
| Fig. 8 a) Schematic of the working principle of an origami-based paper device. The flow of the buffers is described with arrows. Layers 1–3 were made of Whatman 1 filter paper, except for the fact that the central region on layer 4 was constituted of a piece of a glass fiber filter membrane with a diameter of 5 mm for RNA extraction. Layer 5 consisted of a Y-shaped paper disc at the center and three pieces of 3 mm Whatman 1 filter paper circles in the periphery. These paper materials were sandwiched by a laminating film. Layer 6 was made of a double-sided adhesive tape, and six pieces of 3 mm Whatman 1 filter paper discs were sandwiched between the two layers of the adhesive film. Holes were created on two sides of the laminating film, and the upper layer of the adhesive film allowed the liquid to flow from layers 4 to 6. Layer 7 was made of PDMS to execute the LAMP reaction. Once the filter paper discs in layer 6 got wetted by the solution flowing from the above layers, the adhesive film got peeled off and pasted onto layer 7. An RT-LAMP reagent was added directly onto layer 7 for amplification and visualization of the result on top of the portable isothermal heating device. b) (i) Schematic representation of a single-step magneto-immunoassay operated using partial automation and based on a paper strip, in which magnetic particles washing, concentration and fluorescent detection are performed directly on-chip. (ii) Fluorescent reaction product catalyzed by the poly-HRP enzyme using a substrate (QuantaRed). (iii) Portable fluorimeter used for detection. c) Design of smartphone-based malaria detection. (i) The assembled device, showing the phone used to supply power, control the assay conditions (on/off, start/stop and temperature), collect results, communicate with the cloud, analyse data and provide geotagging. The diagnostic chip is shown, inserted into the heating element. The whole device, including the mobile phone, is lightweight (<500 g) and can be held in one hand, with the potential to enable diagnostics to be delivered anywhere. (ii) Open section view of the device and associated circuit. The numbered parts, respectively, are (1) the casing and main body of the device, (2) the aluminium band for receiving the diagnostic device and conducting heat for the nucleic acid amplification assay, (3) circuit components including a microcontroller, heater controller and power supply unit, and (4) the external port for thermal calibration. (iii) The plastic cartridge including a microfluidic circuit with chambers for the LAMP reaction and lateral flow strips for readout, as well as the QR code for traceability [a) reprinted with permission from ref. 494, ©2022 American Chemical Society; b) reprinted with permission from ref. 516, ©2022 Elsevier; c) reprinted with permission from ref. 517, ©2021 Springer Nature]. | ||
8.4 Microfluidic methods for Lyme disease diagnosis
The Sofia 2 Lyme FIA (QuidelOrtho, USA) is a commercially available immunofluorescence LFA for rapid detection of human IgM and IgG antibodies to B. burgdorferi. Flynn et al. recently discussed biosensors with focus on Lyme disease.518 Chou et al. presented a B. burgdorferi antigen microarray designed for the detection of serum antibodies in patients with active and convalescent Lyme disease. High sensitivity was achieved by plasmon-enhanced fluorescence signal detection. The approach has potential for screening of antibody targets predictive for disease status using microliter samples.497 Joung et al. developed a paper-based vertical flow immunodiagnostic assay comprising a spatially-multiplexed sensing membrane containing antigens against Lyme-specific antibodies in human sera. The assay was performed in 20 min with smartphone colorimetric readout and optimized by a deep-learning diagnostic algorithm.498,499 Arumugam et al. used a microfluidic platform for multiplexed serologic diagnosis of Lyme disease and screening of diagnostic biomarkers from clinical serum samples.500 The assay was improved by selecting a panel of 12 best-performing B. burgdorferi proteins. Diagnosis could differentiate different stages of the disease. The protocol uses microfluidic plastic cartridges featuring distinct channels with adsorbed Lyme antigens, enabling optical detection via gold-labeled detection antibodies and silver amplification reagents. Diagnostic tests can be performed in 15 min thus have potential for rapid POC diagnosis.519 Banović et al. took advantage of the fact that tick-borne pathogens may concentrated in the platelet fraction of blood.520 Differential detection of tick-borne pathogens was performed in human platelets, whole blood and ticks using a commercial digital PCR system. The assay allows detecting a large panel of relevant pathogens, including 27 bacterial species.8.5 Malaria
Three methods are commonly used for malaria diagnosis,528,529i.e. microscopy slide examination, NAATs, and immunochromatographic LFAs. Optical microscopic examination of patient blood smears remains the golden standard, allowing quantification of parasitaemia and identification of parasite species. However, these tests are often performed with suboptimal sensitivity, typically in the range a few 50–200 parasites per μL of blood.530 NAATs owing high sensitivity and specificity are suitable for detecting very low parasitaemia.531,532 The LOD for PCR diagnosis is in the range of 0.5–5 parasites per μL of blood. More recently, the potential of multiplex LAMP for POC malaria diagnosis has been assessed.533 Some NAAT-based LOC systems for malaria POC diagnostics are commercially available. The VereFever™ (VerePLEX™ Biosystem, Veredus Laboratories, Singapore) designed for multiplex detection of tropical viruses, also enables differentiation of Plasmodium species (200 to 4 × 105 for P. falciparum with spiked human blood).503 Truenat® Malaria Pv/Pf (Molbio Diagnostics, India), an on-chip real-time duplex microPCR test for P. vivax and P. falciparum, achieves a LOD comparable with PCR (<5 parasites per microlitre).534 Malaria rapid diagnostic tests are designed for detection of P. falciparum histidine-rich protein 2 (PfHRP2), P. falciparum lactase dehydrogenase (PfLDH), P. vivax lactate dehydrogenase (PvLDH), as well as for pan-pLDH and pan-aldolase antigen detection that are common to all human-infecting Plasmodium spp.535,536 Jimenez et al. evaluated the best performance of RTDs and determined a LOD of 0.4 to 1.6 ng mL−1 for PfHRP2 of 12.5 to 50 ng mL−1 for PvLDH ng mL−1 for samples derived from parasite culture or isolates.537 As an indication, Marquart et al. estimated an amount of 1.4 × 10−13 g of PfHRP2 produced per parasite per replication cycle.538
Current and emerging malaria diagnostics methods have been reviewed for instance by Mbanefo et al.529 Pham et al. also included LOC devices and specifications of market technologies.530 Thorne et al. more specifically focused on microfluidic devices for blood sample pre-concentration to facilitate malaria diagnosis.539 Kolluri et al. compared three different categories of LOC approaches, i.e. protein tests, nucleic acid tests and cell-based malaria assays.540 Ragavan et al. analysed biosensor technologies for malaria diagnosis.541 Mitsakakis et al. discussed the vision of a holistic approach in malaria management, comprising differential fever diagnosis and mosquito vector monitoring.542 Malaria diagnostics, in particular microfluidic approaches, may also probe biophysical properties for differentiating infected and non-infected red blood cells.540,543 in the following, we discuss different categories of microfluidic devices for malaria diagnosis. Table 8 provides an overview of selected recent approaches.
| Analyte | Device and assay principle | Performance indications | Ref. |
|---|---|---|---|
| Malaria antigen detection | |||
| PfLDH | Paper-based magneto-immunoassay with fluorescence detection | LOD 0.92 ng mL−1, <20 min | 516 |
| PfHRP2 | 3D paper immunoassay with on-chip mass spectrometry detection | Proof-of-concept, 4 test zones, LOD ≤5 nM, <10 min | 544 |
| PfLDH | Aptamer-based paper-based LFA with colorimetric enzymatic detection | Proof-of-concept, LOD in nM region | 545 |
| PfHRP2 | Microchannel capillary flow platform for chemiluminescence based ELISA | Autonomous sequential flow protocol LOD 8 ng mL−1, within 20 min | 546 |
| PfHRP2 | Immunoassay on capillary-driven chip with electroless Ag staining. | Ag film masks the fluorescent core of the Au labels. LOD <6 ng mL−1, within 20 min | 547 |
| PfLDH and PvLDH | Polymer film-based immuno-chromatographic device | LOD 50 ng mL−1 (PfLDH) and 100 ng mL−1 (PvLDH) | 548 |
| LAMP-based devices with different microfluidic formats | |||
| P. falciparum, pan-Plasmodium | Foldable multiplex LAMP paper device with lateral flow DNA detection | POC field-based diagnosis in <50 min, combined with deep learning for decision support | 549, 517 |
| Plasmodium spp., other pathogens | LoaD platform for diagnosis of febrile illnesses, including malaria | Tested with biobanked clinical samples. POC detection of coinfection within 2 h | 76 |
| P. falciparum, P. vivax | LoaD with quadruplex test capability | LOD ∼0.5 parasites per μl (blood), 50 min | 550 |
| P. falciparum (DNA amplicons) | Diffusion-based sensing of target amplicon/bead conjugates | 3 parasites per μL (blood) detectable within 45 min without DNA extraction | 551 |
Polymer-based diagnostic devices for malaria have been designed for a wide range assay formats and detection methods. Colbert et al. used a shallow sample well in a plastic chip for smartphone-enabled particle diffusometry. In this assay, LAMP amplicons of Plasmodium target genes bind on 400 nm polystyrene beads. Analysing the particle diffusivity allowed for detection of low concentrations of malaria DNA from unprocessed blood samples (3 parasites per μL were detectable within 45 min).551 Real-time fully-electronic DNA sensing of P. falciparum was achieved by electrochemically detecting pH changes during LAMP amplification on an ion-sensitive field-effect transistor (ISFET) sensor array incorporated in a LOC platform. Identification of drug-resistant mutations was.556
Conclusion
Infectious diseases represent a serious burden on global healthcare. This situation is particularly difficult in low-resource environments where infections are highly prevalent, with a broad spectrum of different pathogens, large variations in virulence and local distribution, as well as uncontrolled transmission routes. Nevertheless, the recent COVID-19 pandemic has underscored the fact that from the perspective of an infectious agent there is only one world without traceable boundaries. In developed countries, high-resolution diagnostics are available in centralized laboratories, albeit at a high cost due to specialized settings, instrumentation, and trained personnel. Unfortunately, these barriers limit the application of such solutions in the developing world. According to WHO recommendations, the ideal diagnostic test should satisfy the ASSURED criteria, i.e. being affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free, and deliverable. Microfluidics-based technology has the potential to meet these criteria, thereby significantly reducing healthcare diagnosis costs and opening up new diagnostic markets.Microfluidic point-of-care (POC) diagnostics has become increasingly valuable with respect to the management of infectious diseases. Typically, this routine involves conducting diagnostic tests outside of a centralized laboratory and in close proximity to the patient. Microfluidics technology serves as an integrating force by enabling the seamless conjunction of multiple assay steps on a single disposable device. These steps include sample pre-processing and purification, signal amplification, and detection, ensuring rapid and reliable test results. Integration also facilitates the development of portable, automated, and autonomous diagnostic systems. Accurate POC diagnostic testing already supports infection control and prevention strategies, primarily through the use of rapid lateral flow immunoassays. In a broader perspective, more advanced cheap and disposable paper-based microfluidic analytical devices are of particular interest for POC detection of infectious pathogens in real-world low-resource settings.
Meeting all ASSURED characteristics within a single device without compromises remains a challenging task. Currently existing immune-reaction-based and molecular assays integrated into microfluidic systems offer relatively sensitive pathogen detection but they often require prior knowledge of the infectious strains and specific assay designs. Furthermore, critical protocol steps such as patient sample pre-processing or pathogen culture and enrichment involve laboratory-based procedures, resulting in prolonged turnaround times. In fact, many of present-day microfluidic diagnostic tools may remain at a demonstrator stage, mainly due to the challenges of skilled operation, but also because of the absence of standardized microfluidic operations. Nevertheless, the number of microfluidic cartridge-based systems on the market is increasing. As an example, the GeneXpert® systems (Cepheid, USA), designed for PCR-based POC testing of a large panel of pathologies, are widely regarded as a valuable choice on the current market. However, the platform still incurs relatively high instrumentation and assay costs, and a trained technician is needed for sample pre-processing. An emerging alternative to PCR, which has gained increasing attention over the past years, is the isothermal LAMP technology for nucleic acid amplification. LAMP protocols allow for simpler instrumentation compared to thermal cycling for PCR amplification while retaining the essential features of gold standard nucleic acid-based testing. LAMP-based assays have been successfully performed on microfluidic devices but still need further development for effective implementation in clinical settings and POC testing.
Microfluidic approaches also offer promising capabilities and perspectives in phenotypic antibiotic susceptibility testing (AST). This is of particular importance with regard to the emerging major global health threat related to fast-spreading antibiotic resistance. Informed treatment, associated with the administration of the appropriate type and quantity of antibiotics, are crucial factors in this battle. AST protocols, which are based on differential bacterial growth in different antibiotic concentrations, reliably detect antibiotic resistance and allow for the determination of the minimum inhibitory concentration. Traditional AST methods involve time-consuming bacteria culturing with a time-to-result of up to several days, depending on the specific condition. This presents a serious bottleneck for timely diagnosis. Consequently, these methods often fail to guide medical treatment in the early stages of infection. While fast genotypic AST is possible, a detailed knowledge of the specific genetic antibiotic resistance marker to be tested is required. Shifting from conventional bacterial cultures to monitoring the growth of individual cells paves the way for rapid phenotypic AST. Microfluidic devices play a prominent role in this context by offering the possibility of confining single bacteria within microchannels or nanoliter droplets. Such devices enable the detection of changes in bacterial metabolism, morphology, or replication cycles in response to antibiotic stress with single-bacteria resolution. Single-cell technologies facilitate rapid AST protocols, with results obtained in less than 30 min. For the time being, there is a need to implement the capability to generate antibiograms within single-cell AST approaches. This can be achieved on-chip thanks to the possibility of precise fluidic control and the implementation of complex fluidic protocols. Scaling up this principle for fast multiplex phenotypic AST is an important area of current research. In general, diagnostic systems on the market provide varying levels of multiplexing capability, enabling the detection and differentiation of various infectious agents, as well as the quantification of their abundance in the mix. Additionally, the ability to process multiple patient samples simultaneously is an important feature. Unfortunately, there is usually a trade-off between high-throughput diagnostics and the extent of multiplexing, limiting the number of patient samples and/or pathogens that can be tested simultaneously. Highly integrated microfluidic systems can overcome these limitations by implementing parallel assay protocols strategies on a single chip or cartridge.
Next-generation diagnostic platforms will take advantage of optimized microfluidic designs, innovative detection schemes such as isothermal amplification or CRISPR-based assays, and potentially new functionalized nanomaterials. Screening campaigns are greatly facilitated by the use of inexpensive disposable test cartridges on portable devices, as well as large-scale POC monitoring of environmental samples for surveillance against potential health threats and infectious disease outbreaks. The convergence of wireless communication technologies with microfluidic sensors already enables instantaneous on-site monitoring, enhancing the real-time accessibility of diagnostic information. Smartphones may be used for optical detection of assay results at a remote location and seamless data transmission to centralized facilities. This integration of microfluidic and information technologies holds promise for enhancing the versatility, reliability, and widespread adoption of POC testing across diverse geographic regions. Recent advancements in medical informatics and data processing are expected to further support the efficiency and effectiveness of novel microfluidic diagnostic tools. Artificial intelligence algorithms will enable advanced analysis of large datasets, leading to diagnostics with improved prediction capabilities. Upcoming trends certainly also focus on personalized medicine initiatives, tailoring microfluidic systems to individual patient needs and contributing to precision medicine approaches. Additionally, the establishment of standardized protocols and quality control is necessary to facilitate regulatory approval and promote the implementation of microfluidic-based diagnostics in clinical settings. These steps are crucial for guiding the future development of microfluidics-based infectious disease diagnostics and harnessing the vast potential of microfluidics to address emerging healthcare challenges.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
Support to this work was provided by the École Polytechnique Fédérale de Lausanne.References
- R. E. Baker, A. S. Mahmud, I. F. Miller, M. Rajeev, F. Rasambainarivo, B. L. Rice, S. Takahashi, A. J. Tatem, C. E. Wagner, L.-F. Wang, A. Wesolowski and C. J. E. Metcalf, Nat. Rev. Microbiol., 2022, 20, 193–205 CrossRef CAS PubMed.
- K. B. Patterson and T. Runge, Am. J. Med. Sci., 2002, 323, 216–222 CrossRef.
- J. K. Taubenberger and D. M. Morens, Emerging Infect. Dis., 2006, 12, 15–22 CrossRef.
- D. Flecknoe, B. Charles Wakefield and A. Simmons, Med. Confl. Surviv., 2018, 34, 61–68 CrossRef PubMed.
- K. F. Smith, M. Goldberg, S. Rosenthal, L. Carlson, J. Chen, C. Chen and S. Ramachandran, J. R. Soc., Interface, 2014, 11, 20140950 CrossRef PubMed.
- A. Cassini, E. Colzani, A. Pini, M.-J. J. Mangen, D. Plass, S. A. McDonald, G. Maringhini, A. van Lier, J. A. Haagsma, A. H. Havelaar, P. Kramarz, M. E. Kretzschmar and on behalf of the Bc. Consortium, Eurosurveillance, 2018, 23, 17 CrossRef.
- GBD 2019 Diseases and Injuries Collaborators, Lancet, 2020, 396, 1204–1222 CrossRef.
- WHO Coronavirus (COVID-19) Dashboard, https://covid19.who.int, (accessed 5 January 2024).
- I. Bates, C. Fenton, J. Gruber, D. Lalloo, A. M. Lara, S. B. Squire, S. Theobald, R. Thomson and R. Tolhurst, Lancet Infect. Dis., 2004, 4, 267–277 CrossRef PubMed.
- I. Bates, C. Fenton, J. Gruber, D. Lalloo, A. M. Lara, S. B. Squire, S. Theobald, R. Thomson and R. Tolhurst, Lancet Infect. Dis., 2004, 4, 368–375 CrossRef PubMed.
- Z. A. Bhutta, J. Sommerfeld, Z. S. Lassi, R. A. Salam and J. K. Das, Infect. Dis. Poverty, 2014, 3, 21 CrossRef PubMed.
- H.-B. Weng, H.-X. Chen and M.-W. Wang, Infect. Dis. Poverty, 2018, 7, 67 CrossRef PubMed.
- C. Guo, Z. Zhou, Z. Wen, Y. Liu, C. Zeng, D. Xiao, M. Ou, Y. Han, S. Huang, D. Liu, X. Ye, X. Zou, J. Wu, H. Wang, E. Y. Zeng, C. Jing and G. Yang, Front. Cell. Infect. Microbiol., 2017, 7, 317 CrossRef PubMed.
- C. E. M. Coltart, B. Lindsey, I. Ghinai, A. M. Johnson and D. L. Heymann, Philos. Trans. R. Soc., B, 2017, 372, 20160297 CrossRef PubMed.
- R. Balasubramanian, J. Im, J.-S. Lee, H. J. Jeon, O. D. Mogeni, J. H. Kim, R. Rakotozandrindrainy, S. Baker and F. Marks, Hum. Vaccines Immunother., 2019, 15, 1421–1426 CrossRef PubMed.
- D. E. Bloom and D. Cadarette, Front. Immunol., 2019, 10, 549 CrossRef PubMed.
- WHO (ID 2023) Disease Outbreak News, https://www.who.int/emergencies/disease-outbreak-news, (accessed 5 January 2024).
- C. Caminade, K. M. McIntyre and A. E. Jones, Ann. N. Y. Acad. Sci., 2019, 1436, 157–173 CrossRef PubMed.
- N. E. Nnadi and D. A. Carter, PLoS Pathog., 2021, 17, e1009503 CrossRef CAS PubMed.
- J. T. Ladner, N. D. Grubaugh, O. G. Pybus and K. G. Andersen, Nat. Med., 2019, 25, 206–211 CrossRef CAS.
- S. Plotkin, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 12283–12287 CrossRef CAS PubMed.
- WHO Vaccines, https://www.who.int/teams/immunization-vaccines-and-biologicals/diseases, (accessed 3 February 2024).
- M. B. Laurens, Hum. Vaccines Immunother., 2020, 16, 480–489 CrossRef CAS PubMed.
- N. Aderinto, G. Olatunji, E. Kokori, S. Sikirullahi, J. E. Aboje and R. E. Ojabo, Malar. J., 2024, 23, 16 CrossRef PubMed.
- M. Brisse, S. M. Vrba, N. Kirk, Y. Liang and H. Ly, Front. Immunol., 2020, 11, 583077 CrossRef CAS.
- J.-L. Excler, M. Saville, S. Berkley and J. H. Kim, Nat. Med., 2021, 27, 591–600 CrossRef CAS PubMed.
- F. Wong, C. De La Fuente-Nunez and J. J. Collins, Science, 2023, 381, 164–170 CrossRef CAS PubMed.
- D. M. P. De Oliveira, B. M. Forde, T. J. Kidd, P. N. A. Harris, M. A. Schembri, S. A. Beatson, D. L. Paterson and M. J. Walker, Clin. Microbiol. Rev., 2020, 33, e00181-19 CrossRef PubMed.
- A. S. Lee, H. de Lencastre, J. Garau, J. Kluytmans, S. Malhotra-Kumar, A. Peschel and S. Harbarth, Nat. Rev. Dis. Primers, 2018, 4, 18033 CrossRef PubMed.
- Antimicrobial Resistance Collaborators, Lancet, 2022, 399, 629–655 CrossRef PubMed.
- C. Lange, R. E. Aarnoutse, J. W. C. Alffenaar, G. Bothamley, F. Brinkmann, J. Costa, D. Chesov, R. van Crevel, M. Dedicoat, J. Dominguez, R. Duarte, H. P. Grobbel, G. Günther, L. Guglielmetti, J. Heyckendorf, A. W. Kay, O. Kirakosyan, O. Kirk, R. A. Koczulla, G. G. Kudriashov, L. Kuksa, F. van Leth, C. Magis-Escurra, A. M. Mandalakas, B. Molina-Moya, C. A. Peloquin, M. Reimann, R. Rumetshofer, H. S. Schaaf, T. Schön, S. Tiberi, J. Valda, P. K. Yablonskii and K. Dheda, Int. J. Tuberc Lung Dis., 2019, 23, 645–662 CrossRef CAS PubMed.
- C. Llor, A. Moragas, C. Bayona, J. M. Cots, J. M. Molero, J. Ribas, J. F. Fóthy, I. Gutiérrez, C. Sánchez, J. Ortega, J. Arranz, J. Botanes and P. Robles, BMJ Open, 2017, 7, e015814 CrossRef PubMed.
- S. G. Llanos-Soto, N. Vezeau, M. Wemette, E. Bulut, A. Greiner Safi, P. Moroni, M. A. Shapiro and R. Ivanek, Prev. Vet. Med., 2021, 188, 105253 CrossRef PubMed.
- E. Tacconelli, F. Sifakis, S. Harbarth, R. Schrijver, M. van Mourik, A. Voss, M. Sharland, N. B. Rajendran, J. Rodríguez-Baño, J. Bielicki, M. de Kraker, S. Gandra, P. Gastmeier, K. Gilchrist, A. Gikas, B. P. Gladstone, H. Goossens, H. Jafri, G. Kahlmeter, F. Leus, C. Luxemburger, S. Malhotra-Kumar, G. Marasca, M. McCarthy, M. D. Navarro, M. Nuñez-Nuñez, A. Oualim, J. Price, J. Robert, H. Sommer, M. von Cube, C. Vuong, I. Wiegand, A. T. Witschi and M. Wolkewitz, Lancet Infect. Dis., 2018, 18, e99–e106 CrossRef PubMed.
- R. Laxminarayan, A. Duse, C. Wattal, A. K. M. Zaidi, H. F. L. Wertheim, N. Sumpradit, E. Vlieghe, G. L. Hara, I. M. Gould, H. Goossens, C. Greko, A. D. So, M. Bigdeli, G. Tomson, W. Woodhouse, E. Ombaka, A. Q. Peralta, F. N. Qamar, F. Mir, S. Kariuki, Z. A. Bhutta, A. Coates, R. Bergstrom, G. D. Wright, E. D. Brown and O. Cars, Lancet Infect. Dis., 2013, 13, 1057–1098 CrossRef PubMed.
- A. van Belkum and W. M. Dunne, J. Clin. Microbiol., 2013, 51, 2018–2024 CrossRef PubMed.
- A. van Belkum, T. T. Bachmann, G. Lüdke, J. G. Lisby, G. Kahlmeter, A. Mohess, K. Becker, J. P. Hays, N. Woodford, K. Mitsakakis, J. Moran-Gilad, J. Vila, H. Peter, J. H. Rex and W. M. Dunne, Nat. Rev. Microbiol., 2019, 17, 51–62 CrossRef CAS PubMed.
- S. Puttaswamy, S. K. Gupta, H. Regunath, L. P. Smith and S. Sengupta, Arch. Clin. Microbiol., 2018, 09(3), 83 Search PubMed.
- S. H. Needs, S. I. Donmez, S. P. Bull, C. McQuaid, H. M. I. Osborn and A. D. Edwards, Front. Mech. Eng., 2020, 6, 73 CrossRef.
- N. Qin, P. Zhao, E. A. Ho, G. Xin and C. L. Ren, ACS Sens., 2021, 6, 3–21 CrossRef CAS PubMed.
- B. Behera, G. K. Anil Vishnu, S. Chatterjee, V. S. N. Sitaramgupta V, N. Sreekumar, A. Nagabhushan, N. Rajendran, B. H. Prathik and H. J. Pandya, Biosens. Bioelectron., 2019, 142, 111552 CrossRef CAS PubMed.
- G. Maugeri, I. Lychko, R. Sobral and A. C. A. Roque, Biotechnol. J., 2019, 14, 1700750 CrossRef PubMed.
- A. J. Trotter, A. Aydin, M. J. Strinden and J. O'Grady, Curr. Opin. Microbiol., 2019, 51, 39–45 CrossRef CAS PubMed.
- P. Yager, T. Edwards, E. Fu, K. Helton, K. Nelson, M. R. Tam and B. H. Weigl, Nature, 2006, 442, 412–418 CrossRef CAS PubMed.
- N. P. Pai, C. Vadnais, C. Denkinger, N. Engel and M. Pai, PLoS Med., 2012, 9, e1001306 CrossRef PubMed.
- S. Ombelet, J.-B. Ronat, T. Walsh, C. P. Yansouni, J. Cox, E. Vlieghe, D. Martiny, M. Semret, O. Vandenberg, J. Jacobs, O. Lunguya, M.-F. Phoba, P. Lompo, T. Phe, S. Kariuki, P. N. Newton, D. A. B. Dance, C. Muvunyi, S. El Safi, B. Barbe, D. Falay, D. Affolabi, M. Page, C. Langendorf, Y. Gille, T. Leenstra, J. Stelling, T. Naas, T. Kesteman, D. Seifu, E. Delarocque-Astagneau, C. Schultsz, H. Schutt-Gerowitt, J. Letchford, H. Wertheim, G. Kahlmeter and A. Aidara Kane, Lancet Infect. Dis., 2018, 18, e248–e258 CrossRef PubMed.
- S. Bistafa, Rev. Bras. Ensino Fis., 2017, 40, e2603 Search PubMed.
- J. P. Brody, P. Yager, R. E. Goldstein and R. H. Austin, Biophys. J., 1996, 71, 3430–3441 CrossRef CAS PubMed.
- E. M. Purcell, Am. J. Phys., 1977, 45, 3–11 CrossRef.
- C.-Y. Lee and L.-M. Fu, Sens. Actuators, B, 2018, 259, 677–702 CrossRef CAS.
- A. Olanrewaju, M. Beaugrand, M. Yafia and D. Juncker, Lab Chip, 2018, 18, 2323–2347 RSC.
- T. Thorsen, R. W. Roberts, F. H. Arnold and S. R. Quake, Phys. Rev. Lett., 2001, 86, 4163–4166 CrossRef CAS PubMed.
- J. Zhang, S. Yan, D. Yuan, G. Alici, N.-T. Nguyen, M. Ebrahimi Warkiani and W. Li, Lab Chip, 2016, 16, 10–34 RSC.
- T. M. Squires and S. R. Quake, Rev. Mod. Phys., 2005, 77, 977–1026 CrossRef CAS.
- G. M. Whitesides, Nature, 2006, 442, 368–373 CrossRef CAS PubMed.
- M. A. Unger, H.-P. Chou, T. Thorsen, A. Scherer and S. R. Quake, Science, 2000, 288, 113–116 CrossRef CAS PubMed.
- T. Thorsen, S. J. Maerkl and S. R. Quake, Science, 2002, 298, 580–584 CrossRef CAS PubMed.
- J. Melin and S. R. Quake, Annu. Rev. Biophys. Biomol. Struct., 2007, 36, 213–231 CrossRef CAS PubMed.
- K. Zhang, S. Qin, S. Wu, Y. Liang and J. Li, Chem. Sci., 2020, 11, 6352–6361 RSC.
- A. Burmeister and A. Grünberger, Curr. Opin. Biotechnol., 2020, 62, 106–115 CrossRef CAS PubMed.
- F. J. H. Hol and C. Dekker, Science, 2014, 346, 1251821 CrossRef PubMed.
- M. Cornaglia, T. Lehnert and M. A. M. Gijs, Lab Chip, 2017, 17, 3736–3759 RSC.
- M. A. M. Gijs, F. Lacharme and U. Lehmann, Chem. Rev., 2010, 110, 1518–1563 CrossRef CAS PubMed.
- M. T. Guo, A. Rotem, J. A. Heyman and D. A. Weitz, Lab Chip, 2012, 12, 2146 RSC.
- L. Shang, Y. Cheng and Y. Zhao, Chem. Rev., 2017, 117, 7964–8040 CrossRef CAS PubMed.
- T. S. Kaminski and P. Garstecki, Chem. Soc. Rev., 2017, 46, 6210–6226 RSC.
- Y. Ding, P. D. Howes and A. J. deMello, Anal. Chem., 2020, 92, 132–149 CrossRef CAS PubMed.
- K. Matuła, F. Rivello and W. T. S. Huck, Adv. Biosyst., 2020, 4, 1900188 CrossRef PubMed.
- H. Song, M. R. Bringer, J. D. Tice, C. J. Gerdts and R. F. Ismagilov, Appl. Phys. Lett., 2003, 83, 4664–4666 CrossRef CAS PubMed.
- P. C. Gach, K. Iwai, P. W. Kim, N. J. Hillson and A. K. Singh, Lab Chip, 2017, 17, 3388–3400 RSC.
- J. Sánchez Barea, J. Lee and D.-K. Kang, Micromachines, 2019, 10, 412 CrossRef PubMed.
- O. Strohmeier, M. Keller, F. Schwemmer, S. Zehnle, D. Mark, F. von Stetten, R. Zengerle and N. Paust, Chem. Soc. Rev., 2015, 44, 6187–6229 RSC.
- J. Ducrée, Microsyst. Nanoeng., 2021, 7, 104 CrossRef PubMed.
- C. M. Miyazaki, E. Carthy and D. J. Kinahan, Processes, 2020, 8, 1360 CrossRef.
- V. Sunkara, S. Kumar, J. Sabaté Del Río, I. Kim and Y.-K. Cho, Acc. Chem. Res., 2021, 54, 3643–3655 CrossRef CAS PubMed.
- S. Hin, B. Lopez-Jimena, M. Bakheit, V. Klein, S. Stack, C. Fall, A. Sall, K. Enan, M. Mustafa, L. Gillies, V. Rusu, S. Goethel, N. Paust, R. Zengerle, S. Frischmann, M. Weidmann and K. Mitsakakis, PLoS Neglected Trop. Dis., 2021, 15, e0009177 CrossRef CAS PubMed.
- E. B. Bahadır and M. K. Sezgintürk, TrAC, Trends Anal. Chem., 2016, 82, 286–306 CrossRef.
- V.-T. Nguyen, S. Song, S. Park and C. Joo, Biosens. Bioelectron., 2020, 152, 112015 CrossRef CAS PubMed.
- Y. Liu, L. Zhan, Z. Qin, J. Sackrison and J. C. Bischof, ACS Nano, 2021, 15, 3593–3611 CrossRef CAS PubMed.
- H. Sohrabi, M. R. Majidi, M. Fakhraei, A. Jahanban-Esfahlan, M. Hejazi, F. Oroojalian, B. Baradaran, M. Tohidast, M. D. L. Guardia and A. Mokhtarzadeh, Talanta, 2022, 243, 123330 CrossRef CAS PubMed.
- E. Fu and C. Downs, Lab Chip, 2017, 17, 614–628 RSC.
- A. W. Martinez, S. T. Phillips, G. M. Whitesides and E. Carrilho, Anal. Chem., 2010, 82, 3–10 CrossRef CAS PubMed.
- L. Magro, C. Escadafal, P. Garneret, B. Jacquelin, A. Kwasiborski, J.-C. Manuguerra, F. Monti, A. Sakuntabhai, J. Vanhomwegen, P. Lafaye and P. Tabeling, Lab Chip, 2017, 17, 2347–2371 RSC.
- J. Ma, S. Yan, C. Miao, L. Li, W. Shi, X. Liu, Y. Luo, T. Liu, B. Lin, W. Wu and Y. Lu, Adv. Healthcare Mater., 2019, 8, 1801084 CrossRef PubMed.
- E. Noviana, T. Ozer, C. S. Carrell, J. S. Link, C. McMahon, I. Jang and C. S. Henry, Chem. Rev., 2021, 121, 11835–11885 CrossRef CAS PubMed.
- X. Qin, J. Liu, Z. Zhang, J. Li, L. Yuan, Z. Zhang and L. Chen, TrAC, Trends Anal. Chem., 2021, 143, 116371 CrossRef CAS.
- A. M. Caliendo, D. N. Gilbert, C. C. Ginocchio, K. E. Hanson, L. May, T. C. Quinn, F. C. Tenover, D. Alland, A. J. Blaschke, R. A. Bonomo, K. C. Carroll, M. J. Ferraro, L. R. Hirschhorn, W. P. Joseph, T. Karchmer, A. T. MacIntyre, L. B. Reller, A. F. Jackson and for the Infectious Diseases Society of America (IDSA), Clin. Infect. Dis., 2013, 57, S139–S170 CrossRef PubMed.
- S. Sachdeva, R. W. Davis and A. K. Saha, Front. Bioeng. Biotechnol., 2021, 8, 602659 CrossRef PubMed.
- A. Cassedy, A. Parle-McDermott and R. O'Kennedy, Front. Mol. Biosci., 2021, 8, 637559 CrossRef CAS PubMed.
- C. Deusenbery, Y. Wang and A. Shukla, ACS Infect. Dis., 2021, 7, 695–720 CrossRef CAS PubMed.
- B. Purohit, P. R. Vernekar, N. P. Shetti and P. Chandra, Sens. Int., 2020, 1, 100040 CrossRef.
- F. Huang, Y. Zhang, J. Lin and Y. Liu, Biosensors, 2021, 11, 190 CrossRef CAS PubMed.
- S. M. Yoo and S. Y. Lee, Trends Biotechnol., 2016, 34, 7–25 CrossRef CAS PubMed.
- O. Simoska and K. J. Stevenson, Analyst, 2019, 144, 6461–6478 RSC.
- E. Cesewski and B. N. Johnson, Biosens. Bioelectron., 2020, 159, 112214 CrossRef CAS PubMed.
- K. Y. Goud, K. K. Reddy, A. Khorshed, V. S. Kumar, R. K. Mishra, M. Oraby, A. H. Ibrahim, H. Kim and K. V. Gobi, Biosens. Bioelectron., 2021, 180, 113112 CrossRef CAS PubMed.
- D. Gao, X. Guo, Y. Yang, H. Shi, R. Hao, S. Wang, Z. J. Li, R. Zhao and H. Song, J. Biol. Eng., 2022, 16, 33 CrossRef CAS PubMed.
- M. De Falco, M. De Felice, F. Rota, D. Zappi, A. Antonacci and V. Scognamiglio, TrAC, Trends Anal. Chem., 2022, 148, 116538 CrossRef CAS.
- I. M. Artika, Y. P. Dewi, I. M. Nainggolan, J. E. Siregar and U. Antonjaya, Genes, 2022, 13, 2387 CrossRef CAS PubMed.
- L. Becherer, N. Borst, M. Bakheit, S. Frischmann, R. Zengerle and F. von Stetten, Anal. Methods, 2020, 12, 717–746 RSC.
- Y. Shang, J. Sun, Y. Ye, J. Zhang, Y. Zhang and X. Sun, Crit. Rev. Food Sci. Nutr., 2020, 60, 201–224 CrossRef CAS PubMed.
- Y. Bai, J. Ji, F. Ji, S. Wu, Y. Tian, B. Jin and Z. Li, Talanta, 2022, 240, 123209 CrossRef CAS PubMed.
- S. Barreda-García, R. Miranda-Castro, N. de-los-Santos-Álvarez, A. J. Miranda-Ordieres and M. J. Lobo-Castañón, Anal. Bioanal. Chem., 2018, 410, 679–693 CrossRef PubMed.
- L. Xu, J. Duan, J. Chen, S. Ding and W. Cheng, Anal. Chim. Acta, 2021, 1148, 238187 CrossRef CAS PubMed.
- M. M. Kaminski, O. O. Abudayyeh, J. S. Gootenberg, F. Zhang and J. J. Collins, Nat. Biomed. Eng., 2021, 5, 643–656 CrossRef CAS PubMed.
- J. de D. Habimana, R. Huang, B. Muhoza, Y. N. Kalisa, X. Han, W. Deng and Z. Li, Biosens. Bioelectron., 2022, 203, 114033 CrossRef CAS PubMed.
- X. Wang, X.-Z. Hong, Y.-W. Li, Y. Li, J. Wang, P. Chen and B.-F. Liu, Mil. Med. Res., 2022, 9, 11 Search PubMed.
- K. Mitsakakis, V. D'Acremont, S. Hin, F. von Stetten and R. Zengerle, Microelectron. Eng., 2018, 201, 26–59 CrossRef CAS PubMed.
- P. P. Nelson, B. A. Rath, P. C. Fragkou, E. Antalis, S. Tsiodras and C. Skevaki, Front. Cell. Infect. Microbiol., 2020, 10, 181 CrossRef CAS PubMed.
- Cepheid|GeneXpert System, www.cepheid.com, (accessed 8 January 2024).
- BioFire Diagnostics, https://www.biofiredx.com/, (accessed 8 January 2024).
- VerePLEXTM Biosystem, vereduslabs.com, (accessed 8 January 2024).
- Bosch Vivalytic Analyser, https://www.bosch-vivalytic.com/en/, (accessed 1 February 2024).
- D. Podbiel, L. Boecking, H. Bott, J. Kassel, D. Czurratis, F. Laermer, R. Zengerle and J. Hoffmann, J. Micromech. Microeng., 2020, 30, 115012 CrossRef CAS.
- Bosch Vivalytic tests, https://www.bosch-vivalytic.com/en/tests/, (accessed 1 February 2024).
- Hahn-Schickard, https://www.hahn-schickard.de/forschung-entwicklung/laborautomatisierung/labdisk-plattform, (accessed 8 January 2024).
- Dialunox, https://labdiskplayer.com/, (accessed 1 February 2024).
- E. A. Flores-Contreras, R. B. González-González, I. P. Rodríguez-Sánchez, J. F. Yee-De León, H. M. N. Iqbal and E. González-González, Biosensors, 2022, 12, 179 CrossRef CAS PubMed.
- C. Wang, M. Liu, Z. Wang, S. Li, Y. Deng and N. He, Nano Today, 2021, 37, 101092 CrossRef CAS PubMed.
- A. Basiri, A. Heidari, M. F. Nadi, M. T. P. Fallahy, S. S. Nezamabadi, M. Sedighi, A. Saghazadeh and N. Rezaei, Rev. Med. Virol., 2021, 31, e2154 CrossRef PubMed.
- N. Rezvani Jalal, P. Mehrbod, S. Shojaei, H. I. Labouta, P. Mokarram, A. Afkhami, T. Madrakian, M. J. Los, D. Schaafsma, M. Giersig, M. Ahmadi and S. Ghavami, ACS Appl. Nano Mater., 2021, 4, 4307–4328 CrossRef CAS.
- A. Tay, A. Pavesi, S. R. Yazdi, C. T. Lim and M. E. Warkiani, Biotechnol. Adv., 2016, 34, 404–421 CrossRef PubMed.
- G. L. Damhorst, M. Murtagh, W. R. Rodriguez and R. Bashir, Proc. IEEE, 2015, 103, 150–160 CAS.
- X. Fu, J. Sun, R. Liang, H. Guo, L. Wang and X. Sun, Trends Food Sci. Technol., 2021, 116, 115–129 CrossRef CAS.
- X. Weng, C. Zhang and H. Jiang, LWT, 2021, 151, 112172 CrossRef CAS.
- K. Subbarao and S. Mahanty, Immunity, 2020, 52, 905–909 CrossRef CAS PubMed.
- J. P. Mizgerd, N. Engl. J. Med., 2008, 358, 716–727 CrossRef CAS PubMed.
- N. Jain, R. Lodha and S. K. Kabra, Indian J. Pediatr., 2001, 68, 1135–1138 CrossRef CAS PubMed.
- The top 10 causes of death, https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death, (accessed 5 January 2024).
- K. M. Robinson, J. K. Kolls and J. F. Alcorn, Curr. Opin. Immunol., 2015, 34, 59–67 CrossRef CAS PubMed.
- Influenza (Seasonal), https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal), (accessed 5 January 2024).
- G. R. S. Budinger, A. V. Misharin, K. M. Ridge, B. D. Singer and R. G. Wunderink, J. Clin. Invest., 2021, 131, e149412 CrossRef CAS PubMed.
- D. Dandachi and M. C. Rodriguez-Barradas, J. Invest. Med., 2018, 66, 957–965 CrossRef PubMed.
- T. M. File, The American Journal of Medicine Supplements, 2004, 117, 39–50 CrossRef PubMed.
- S. Aliberti, G. S. Cook, B. L. Babu, L. F. Reyes, A. H. Rodriguez, F. Sanz, N. J. Soni, A. Anzueto, P. Faverio, R. F. Sadud, I. Muhammad, C. Prat, E. Vendrell, J. Neves, E. Kaimakamis, A. Feneley, R. Swarnakar, F. Franzetti, M. Carugati, M. Morosi, E. Monge and M. I. Restrepo, J. Infect., 2019, 79, 300–311 CrossRef PubMed.
- A. de Roux, S. Ewig, E. García, M. A. Marcos, J. Mensa, H. Lode and A. Torres, Eur. Respir. J., 2006, 27, 795–800 CrossRef CAS PubMed.
- S. B. A. Sattar and S. Sharma, Bacterial Pneumonia, 2022, https://www.ncbi.nlm.nih.gov/books/NBK513321 Search PubMed.
- K. B. Waites, L. Xiao, Y. Liu, M. F. Balish and T. P. Atkinson, Clin. Microbiol. Rev., 2017, 30, 747–809 CrossRef CAS PubMed.
- R. N. Jones, Clin. Infect. Dis., 2010, 51, S81–S87 CrossRef PubMed.
- WHO (2023) Tuberculosis (TB), https://www.who.int/news-room/fact-sheets/detail/tuberculosis, (accessed 5 January 2024).
- H.-S. Huang, C.-L. Tsai, J. Chang, T.-C. Hsu, S. Lin and C.-C. Lee, Clin. Microbiol. Infect., 2018, 24, 1055–1063 CrossRef CAS PubMed.
- K. Mjøsund, M. Ghaleb, L. Kolsrud, J. Carrabre, F. Kainzinger, D. Boehm, F. Bitterling and B. Wolfarth, Front. Sports Act. Living, 2023, 5, 1217463 CrossRef PubMed.
- L. A. Heger, N. Elsen, M. Rieder, N. Gauchel, U. Sommerwerck, C. Bode, D. Duerschmied, M. Oette and I. Ahrens, BMC Infect. Dis., 2022, 22, 486 CrossRef CAS PubMed.
- J. Zhuang, J. Yin, S. Lv, B. Wang and Y. Mu, Biosens. Bioelectron., 2020, 163, 112291 CrossRef CAS PubMed.
- L. E. Breshears, B. T. Nguyen, S. Mata Robles, L. Wu and J.-Y. Yoon, SLAS Technol., 2022, 27, 4–17 CrossRef CAS PubMed.
- Z. Zhang, P. Ma, R. Ahmed, J. Wang, D. Akin, F. Soto, B.-F. Liu, P. Li and U. Demirci, Adv. Mater., 2022, 34, 2103646 CrossRef CAS PubMed.
- E. A. Tarim, B. Karakuzu, C. Oksuz, O. Sarigil, M. Kizilkaya, M. K. A. A. Al-Ruweidi, H. C. Yalcin, E. Ozcivici and H. C. Tekin, Emergent Mater., 2021, 4, 143–168 CrossRef CAS PubMed.
- Z. Qin, R. Peng, I. K. Baravik and X. Liu, Matter, 2020, 3, 628–651 CrossRef PubMed.
- B. V. Ribeiro, T. A. R. Cordeiro, G. R. Oliveira e Freitas, L. F. Ferreira and D. L. Franco, Talanta Open, 2020, 2, 100007 CrossRef PubMed.
- R. Zenhausern, C.-H. Chen and J.-Y. Yoon, Biomicrofluidics, 2021, 15, 011503 CrossRef CAS PubMed.
- S. Krokhine, H. Torabi, A. Doostmohammadi and P. Rezai, Colloids Surf., B, 2021, 206, 111962 CrossRef CAS PubMed.
- I. Lee, E. Jeon and J. Lee, TrAC, Trends Anal. Chem., 2023, 158, 116880 CrossRef CAS PubMed.
- E. Shabani, S. Dowlatshahi and M. J. Abdekhodaie, Eur. J. Clin. Microbiol. Infect. Dis., 2021, 40, 225–246 CrossRef CAS PubMed.
- Y. Chen, S. Huang, L. Zhou, X. Wang, H. Yang and W. Li, J. Clin. Lab. Anal., 2022, 36, e24152 CrossRef CAS PubMed.
- N. L. Welch, M. Zhu, C. Hua, J. Weller, M. E. Mirhashemi, T. G. Nguyen, S. Mantena, M. R. Bauer, B. M. Shaw, C. M. Ackerman, S. G. Thakku, M. W. Tse, J. Kehe, M.-M. Uwera, J. S. Eversley, D. A. Bielwaski, G. McGrath, J. Braidt, J. Johnson, F. Cerrato, G. K. Moreno, L. A. Krasilnikova, B. A. Petros, G. L. Gionet, E. King, R. C. Huard, S. K. Jalbert, M. L. Cleary, N. A. Fitzgerald, S. B. Gabriel, G. R. Gallagher, S. C. Smole, L. C. Madoff, C. M. Brown, M. W. Keller, M. M. Wilson, M. K. Kirby, J. R. Barnes, D. J. Park, K. J. Siddle, C. T. Happi, D. T. Hung, M. Springer, B. L. MacInnis, J. E. Lemieux, E. Rosenberg, J. A. Branda, P. C. Blainey, P. C. Sabeti and C. Myhrvold, Nat. Med., 2022, 28, 1083–1094 CrossRef CAS PubMed.
- C. M. Ackerman, C. Myhrvold, S. G. Thakku, C. A. Freije, H. C. Metsky, D. K. Yang, S. H. Ye, C. K. Boehm, T.-S. F. Kosoko-Thoroddsen, J. Kehe, T. G. Nguyen, A. Carter, A. Kulesa, J. R. Barnes, V. G. Dugan, D. T. Hung, P. C. Blainey and P. C. Sabeti, Nature, 2020, 582, 277–282 CrossRef CAS PubMed.
- I. Rutten, D. Daems, K. Leirs and J. Lammertyn, Biosensors, 2023, 13, 100 CrossRef CAS PubMed.
- M. Rombach, S. Hin, M. Specht, B. Johannsen, J. Lüddecke, N. Paust, R. Zengerle, L. Roux, T. Sutcliffe, J. R. Peham, C. Herz, M. Panning, O. Donoso Mantke and K. Mitsakakis, Analyst, 2020, 145, 7040–7047 RSC.
- J. Liu, H. Wang, L. Zhang, Y. Lu, X. Wang, M. Shen, N. Li, L. Feng, J. Jing, B. Cao, X. Zou, J. Cheng and Y. Xu, Small, 2022, 18, 2200854 CrossRef CAS PubMed.
- H. Wang, J. Xu, S. Li, X. Wang, G. Liu, S. Yang, F. Zhao, Q. Liu, X. Chen, C. He and M. Li, Anal. Chim. Acta, 2023, 1242, 340812 CrossRef CAS PubMed.
- M. Ji, Y. Xia, F.-C. Loo, L. Li, H.-P. Ho, J. He and D. Gu, RSC Adv., 2020, 10, 34088–34098 RSC.
- H. Xiong, X. Ye, Y. Li, L. Wang, J. Zhang, X. Fang and J. Kong, Anal. Chem., 2020, 92, 14297–14302 CrossRef CAS PubMed.
- W. Xing, Y. Liu, H. Wang, S. Li, Y. Lin, L. Chen, Y. Zhao, S. Chao, X. Huang, S. Ge, T. Deng, T. Zhao, B. Li, H. Wang, L. Wang, Y. Song, R. Jin, J. He, X. Zhao, P. Liu, W. Li and J. Cheng, Engineering, 2020, 6, 1130–1140 CrossRef CAS PubMed.
- E. Huang, Y. Wang, N. Yang, B. Shu, G. Zhang and D. Liu, Anal. Bioanal. Chem., 2021, 413, 1787–1798 CrossRef CAS PubMed.
- H. Yin, Z. Tong, C. Shen, X. Xu, H. Ma, Z. Wu, Y. Qi and H. Mao, Lab Chip, 2022, 22, 2671–2681 RSC.
- H. Huang, K. Huang, Y. Sun, D. Luo, M. Wang, T. Chen, M. Li, J. Duan, L. Huang and C. Dong, Micromachines, 2022, 13, 1650 CrossRef PubMed.
- Y. Zai, C. Min, Z. Wang, Y. Ding, H. Zhao, E. Su and N. He, Lab Chip, 2022, 22, 3436–3452 RSC.
- R. Siavash Moakhar, C. del Real Mata, M. Jalali, H. Shafique, A. Sanati, J. de Vries, J. Strauss, T. AbdElFatah, F. Ghasemi, M. McLean, I. I. Hosseini, Y. Lu, S. G. Yedire, S. S. Mahshid, M. A. Tabatabaiefar, C. Liang and S. Mahshid, Adv. Sci., 2022, 9, 2204246 CrossRef CAS PubMed.
- D. Najjar, J. Rainbow, S. Sharma Timilsina, P. Jolly, H. de Puig, M. Yafia, N. Durr, H. Sallum, G. Alter, J. Z. Li, X. G. Yu, D. R. Walt, J. A. Paradiso, P. Estrela, J. J. Collins and D. E. Ingber, Nat. Biomed. Eng., 2022, 6, 968–978 CrossRef CAS PubMed.
- W. Teixeira, Y. Pallás-Tamarit, A. Juste-Dolz, A. Sena-Torralba, R. Gozalbo-Rovira, J. Rodríguez-Díaz, D. Navarro, J. Carrascosa, D. Gimenez-Romero, Á. Maquieira and S. Morais, Biosens. Bioelectron., 2022, 213, 114454 CrossRef CAS PubMed.
- J. Yang, V. M. Phan, C.-K. Heo, H. V. Nguyen, W.-H. Lim, E.-W. Cho, H. Poo and T. S. Seo, Sens. Actuators, B, 2023, 380, 133331 CrossRef CAS PubMed.
- G. Qiu, Z. Gai, Y. Tao, J. Schmitt, G. A. Kullak-Ublick and J. Wang, ACS Nano, 2020, 14, 5268–5277 CrossRef CAS PubMed.
- G. Seo, G. Lee, M. J. Kim, S.-H. Baek, M. Choi, K. B. Ku, C.-S. Lee, S. Jun, D. Park, H. G. Kim, S.-J. Kim, J.-O. Lee, B. T. Kim, E. C. Park and S. I. Kim, ACS Nano, 2020, 14, 5135–5142 CrossRef CAS PubMed.
- F. Li, J. Qi, Z. Ren, X. Hu, Y. Chen, B. Li and X. Fu, Microchem. J., 2023, 185, 108304 CrossRef CAS PubMed.
- P. Robin, L. Barnabei, S. Marocco, J. Pagnoncelli, D. Nicolis, C. Tarantelli, A. C. Tavilla, R. Robortella, L. Cascione, L. Mayoraz, C. M. A. Journot, M. Mensi, F. Bertoni, I. Stefanini and S. Gerber-Lemaire, Biosens. Bioelectron.: X, 2023, 13, 100302 CAS.
- P.-H. Lu, Y.-D. Ma, C.-Y. Fu and G.-B. Lee, Lab Chip, 2020, 20, 789–797 RSC.
- Y.-T. Yeh, K. Gulino, Y. Zhang, A. Sabestien, T.-W. Chou, B. Zhou, Z. Lin, I. Albert, H. Lu, V. Swaminathan, E. Ghedin and M. Terrones, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 895–901 CrossRef CAS PubMed.
- A. Ramachandran, D. A. Huyke, E. Sharma, M. K. Sahoo, C. Huang, N. Banaei, B. A. Pinsky and J. G. Santiago, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 29518–29525 CrossRef CAS PubMed.
- Y. Yu, Z. Yu, X. Pan, L. Xu, R. Guo, X. Qian and F. Shen, Analyst, 2022, 147, 625–633 RSC.
- A. Wang, Z. Wu, Y. Huang, H. Zhou, L. Wu, C. Jia, Q. Chen and J. Zhao, Biosensors, 2021, 11, 427 CrossRef CAS PubMed.
- H. Wang, Z. Ma, J. Qin, Z. Shen, Q. Liu, X. Chen, H. Wang, Z. An, W. Liu and M. Li, Biosens. Bioelectron., 2019, 126, 373–380 CrossRef CAS PubMed.
- M. Dou, J. Sanchez, H. Tavakoli, J. E. Gonzalez, J. Sun, J. Dien Bard and X. Li, Anal. Chim. Acta, 2019, 1065, 71–78 CrossRef CAS PubMed.
- M. Dou, N. Macias, F. Shen, J. Dien Bard, D. C. Domínguez and X. Li, EClinicalMedicine, 2019, 8, 72–77 CrossRef PubMed.
- G. Huang, Q. Huang, L. Xie, G. Xiang, L. Wang, H. Xu, L. Ma, X. Luo, J. Xin, X. Zhou, X. Jin and L. Zhang, Sci. Rep., 2017, 7, 6441 CrossRef PubMed.
- E. D. Carrol, J. E. Clark and A. J. Cant, Paediatr. Respir. Rev., 2001, 2, 113–119 CAS.
- B. A. Forbes, G. S. Hall, M. B. Miller, S. M. Novak, M.-C. Rowlinson, M. Salfinger, A. Somoskövi, D. M. Warshauer and M. L. Wilson, Clin. Microbiol. Rev., 2018, 31, e00038-17 CrossRef PubMed.
- C. J. Cambier, S. Falkow and L. Ramakrishnan, Cell, 2014, 159, 1497–1509 CrossRef CAS PubMed.
- B. B. Aldridge, M. Fernandez-Suarez, D. Heller, V. Ambravaneswaran, D. Irimia, M. Toner and S. M. Fortune, Science, 2012, 335, 100–104 CrossRef CAS PubMed.
- J. Millard, C. Ugarte-Gil and D. A. J. Moore, BMJ, 2015, 350, h882 CrossRef PubMed.
- A. Paul, N. Dutta, D. Moschou and G. Dutta, Sens. Int., 2020, 1, 100036 CrossRef.
- G. L. Hobby, A. P. Holman, M. D. Iseman and J. M. Jones, Antimicrob. Agents Chemother., 1973, 4, 94–104 CrossRef CAS PubMed.
- F. Rageade, N. Picot, A. Blanc-Michaud, S. Chatellier, C. Mirande, E. Fortin and A. van Belkum, Eur. J. Clin. Microbiol. Infect. Dis., 2014, 33, 867–870 CrossRef CAS PubMed.
- WHO consolidated guidelines on tuberculosis, https://www.who.int/publications-detail-redirect/9789240029415, (accessed 5 January 2024).
- J. M. Hong, H. Lee, N. V. Menon, C. T. Lim, L. P. Lee and C. W. M. Ong, Sci. Transl. Med., 2022, 14, eabj4124 CrossRef CAS PubMed.
- L. Nandlal, R. Perumal and K. Naidoo, Infect. Drug Resist., 2022, 15, 4971–4984 CrossRef PubMed.
- S. D. Lawn, P. Mwaba, M. Bates, A. Piatek, H. Alexander, B. J. Marais, L. E. Cuevas, T. D. McHugh, L. Zijenah, N. Kapata, I. Abubakar, R. McNerney, M. Hoelscher, Z. A. Memish, G. B. Migliori, P. Kim, M. Maeurer, M. Schito and A. Zumla, Lancet Infect. Dis., 2013, 13, 349–361 CrossRef CAS PubMed.
- C. Nikam, M. Kazi, C. Nair, M. Jaggannath, M. Manoj, R. Vinaya, A. Shetty and C. Rodrigues, Int. J. Mycobact., 2014, 3, 205–210 CrossRef PubMed.
- J. Schlanderer, H. Hoffmann, J. Lüddecke, A. Golubov, W. Grasse, E. V. Kindler, T. A. Kohl, M. Merker, C. Metzger, V. Mohr, S. Niemann, C. Pilloni, S. Plesnik, B. Raya, B. Shresta, C. Utpatel, R. Zengerle, M. Beutler and N. Paust, Lab Chip, 2024, 24, 74–84 RSC.
- K. R. Steingart, V. Ng, M. Henry, P. C. Hopewell, A. Ramsay, J. Cunningham, R. Urbanczik, M. D. Perkins, M. A. Aziz and M. Pai, Lancet Infect. Dis., 2006, 6, 664–674 CrossRef PubMed.
- J. Flores, J. C. Cancino and L. Chavez-Galan, Front. Microbiol., 2021, 12, 638047 CrossRef PubMed.
- S. D. Lawn, BMC Infect. Dis., 2012, 12, 103 CrossRef PubMed.
- M. A. Bulterys, B. Wagner, M. Redard-jacot, A. Suresh, N. R. Pollock, E. Moreau, C. M. Denkinger, P. K. Drain and T. Broger, J. Clin. Med., 2020, 9, 111 CrossRef CAS PubMed.
- X.-J. Cao, Y.-P. Li, J.-Y. Wang, J. Zhou and X.-G. Guo, BMC Infect. Dis., 2021, 21, 336 CrossRef CAS PubMed.
- D. Qiao, L. Li, J. Guo, S. Lao, X. Zhang, J. Zhang and C. Wu, Infect. Immun., 2011, 79, 3358–3365 CrossRef CAS PubMed.
- V. Boggaram, K. R. Gottipati, X. Wang and B. Samten, J. Biol. Chem., 2013, 288, 25500–25511 CrossRef CAS PubMed.
- S. Gupta and V. Kakkar, Biosens. Bioelectron., 2018, 115, 14–29 CrossRef CAS PubMed.
- S. K. Srivastava, C. J. M. van Rijn and M. A. Jongsma, RSC Adv., 2016, 6, 17759–17771 RSC.
- A. Niemz and D. S. Boyle, Expert Rev. Mol. Diagn., 2012, 12, 687–701 CrossRef CAS PubMed.
- V. Mani, S. Wang, F. Inci, G. De Libero, A. Singhal and U. Demirci, Adv. Drug Delivery Rev., 2014, 78, 105–117 CrossRef CAS PubMed.
- S. Wang, F. Inci, G. De Libero, A. Singhal and U. Demirci, Biotechnol. Adv., 2013, 31, 438–449 CrossRef PubMed.
- K. Dheda, M. Ruhwald, G. Theron, J. Peter and W. C. Yam, Respirology, 2013, 18, 217–232 CrossRef PubMed.
- G. A. S. Minero, M. Bagnasco, J. Fock, B. Tian, F. Garbarino and M. F. Hansen, Anal. Bioanal. Chem., 2020, 412, 2705–2710 CrossRef CAS PubMed.
- G. A. S. Minero, E. Tefiku, F. Garbarino and M. F. Hansen, IEEE Magn. Lett., 2020, 11, 3100105 CAS.
- A. R. Homann, L. Niebling, S. Zehnle, M. Beutler, L. Delamotte, M.-C. Rothmund, D. Czurratis, K.-D. Beller, R. Zengerle, H. Hoffmann and N. Paust, Lab Chip, 2021, 21, 1540–1548 RSC.
- M. Beutler, A. R. Homann, M. Mihalic, S. Plesnik, L. Niebling, M. Eckart, V. Allerheiligen, D. Czurratis, B. Maharjan, B. Shrestha, N. Parpieva, L. Turaev, Z. Sayfutdinov, S. Hofmann-Thiel, W. Grasse, C. Metzger-Boddien, N. Paust and H. Hoffmann, J. Mol. Diagn., 2021, 23, 643–650 CrossRef CAS PubMed.
- N. Kaur, J. S. Michael and B. J. Toley, Sci. Rep., 2019, 9, 15367 CrossRef PubMed.
- L. B. Pinheiro, V. A. Coleman, C. M. Hindson, J. Herrmann, B. J. Hindson, S. Bhat and K. R. Emslie, Anal. Chem., 2012, 84, 1003–1011 CrossRef CAS PubMed.
- H. Li, R. Bai, Z. Zhao, L. Tao, M. Ma, Z. Ji, M. Jian, Z. Ding, X. Dai, F. Bao and A. Liu, Biosci. Rep., 2018, 38, BSR20181170 CrossRef PubMed.
- R. Nyaruaba, C. Mwaliko, K. K. Kering and H. Wei, Tuberculosis, 2019, 117, 85–92 CrossRef CAS PubMed.
- N. Song, Y. Tan, L. Zhang, W. Luo, Q. Guan, M. Yan, R. Zuo, W. Liu, F. Luo and X.-L. Zhang, Emerging Microbes Infect., 2018, 7, 1–9 CAS.
- M. Yamamoto, R. Ushio, H. Watanabe, T. Tachibana, M. Tanaka, T. Yokose, J. Tsukiji, H. Nakajima and T. Kaneko, Int. J. Infect. Dis., 2018, 66, 80–82 CrossRef CAS PubMed.
- J. Luo, M. Luo, J. Li, J. Yu, H. Yang, X. Yi, Y. Chen and H. Wei, Int. J. Tuberc. Lung Dis., 2019, 23, 219–225 CrossRef CAS PubMed.
- C.-H. Wang, J.-R. Chang, S.-C. Hung, H.-Y. Dou and G.-B. Lee, Sens. Actuators, B, 2022, 365, 131968 CrossRef CAS.
- A. C. Kukhtin, T. Sebastian, J. Golova, A. Perov, C. Knickerbocker, Y. Linger, A. Bueno, P. Qu, M. Villanueva, R. C. Holmberg, D. P. Chandler and C. G. Cooney, Lab Chip, 2019, 19, 1217–1225 RSC.
- A. V. Kukhtin, R. Norville, A. Bueno, P. Qu, N. Parrish, M. Murray, D. P. Chandler, R. C. Holmberg and C. G. Cooney, Anal. Chem., 2020, 92, 5311–5318 CrossRef CAS PubMed.
- I. M. Mbano, T. Mandizvo, J. Rogich, T. T. R. Kunota, J. S. Mackenzie, M. Pillay and F. K. Balagaddé, J. Appl. Lab. Med., 2020, 5, 440–453 CrossRef PubMed.
- D. Martens, P. Ramirez-Priego, M. S. Murib, A. A. Elamin, A. B. Gonzalez-Guerrero, M. Stehr, F. Jonas, B. Anton, N. Hlawatsch, P. Soetaert, R. Vos, A. Stassen, S. Severi, W. V. Roy, R. Bockstaele, H. Becker, M. Singh, L. M. Lechuga and P. Bienstman, Anal. Methods, 2018, 10, 3066–3073 RSC.
- P. Ramirez-Priego, D. Martens, A. A. Elamin, P. Soetaert, W. Van Roy, R. Vos, B. Anton, R. Bockstaele, H. Becker, M. Singh, P. Bienstman and L. M. Lechuga, ACS Sens., 2018, 3, 2079–2086 CrossRef CAS PubMed.
- N. Singh, B. Dahiya, V. S. Radhakrishnan, T. Prasad and P. K. Mehta, Int. J. Nanomed., 2018, 13, 8523–8535 CrossRef CAS PubMed.
- S. Gupta, P. Bhatter and V. Kakkar, Tuberculosis, 2021, 127, 102055 CrossRef CAS PubMed.
- N. Ariffin, N. A. Yusof, J. Abdullah, S. F. Abd Rahman, N. H. Ahmad Raston, N. Kusnin and S. Suraiya, J. Sens., 2020, 2020, 1–10 CrossRef.
- H. Ghorbanpoor, A. N. Dizaji, I. Akcakoca, E. O. Blair, Y. Ozturk, P. Hoskisson, T. Kocagoz, H. Avci, D. K. Corrigan and F. D. Guzel, Sens. Actuators, A, 2022, 339, 113515 CrossRef CAS.
- H. Wang, G. M. Conover, S.-I. Han, J. C. Sacchettini and A. Han, Microsyst. Nanoeng., 2021, 7, 37 CrossRef CAS PubMed.
- M. Elitas, N. Dhar and J. D. McKinney, Antibiotics, 2021, 10, 794 CrossRef CAS PubMed.
- V. O. Baron, M. Chen, B. Hammarstrom, R. J. H. Hammond, P. Glynne-Jones, S. H. Gillespie and K. Dholakia, Commun. Biol., 2020, 3, 236 CrossRef CAS PubMed.
- R. J. H. Hammond, V. O. Baron, K. Oravcova, S. Lipworth and S. H. Gillespie, J. Antimicrob. Chemother., 2015, 70, 2823–2827 CrossRef CAS PubMed.
- A. Masajtis-Zagajewska and M. Nowicki, Clin. Chim. Acta, 2017, 471, 286–291 CrossRef CAS PubMed.
- S. Aitekenov, A. Gaipov and R. Bukasov, Talanta, 2021, 223, 121718 CrossRef CAS PubMed.
- E. Lepowsky, F. Ghaderinezhad, S. Knowlton and S. Tasoglu, Biomicrofluidics, 2017, 11, 051501 CrossRef PubMed.
- E. Mahoney, J. Kun, M. Smieja and Q. Fang, J. Electrochem. Soc., 2020, 167, 037518 CrossRef CAS.
- W.-C. Tai, Y.-C. Chang, D. Chou and L.-M. Fu, Biosensors, 2021, 11, 260 CrossRef CAS PubMed.
- A. L. Flores-Mireles, J. N. Walker, M. Caparon and S. J. Hultgren, Nat. Rev. Microbiol., 2015, 13, 269–284 CrossRef CAS PubMed.
- M. Medina and E. Castillo-Pino, Ther. Adv. Urol., 2019, 11, 1756287219832172 Search PubMed.
- R. D. Klein and S. J. Hultgren, Nat. Rev. Microbiol., 2020, 18, 211–226 CrossRef CAS PubMed.
- R. Öztürk and A. Murt, World J. Urol., 2020, 38, 2669–2679 CrossRef PubMed.
- B. Foxman, Nat. Rev. Urol., 2010, 7, 653–660 CrossRef PubMed.
- C. Walsh and T. Collyns, Surgery, 2020, 38, 191–196 Search PubMed.
- T. J. Wiles, R. R. Kulesus and M. A. Mulvey, Exp. Mol. Pathol., 2008, 85, 11–19 CrossRef CAS PubMed.
- F. M. E. Wagenlehner, T. E. Bjerklund Johansen, T. Cai, B. Koves, J. Kranz, A. Pilatz and Z. Tandogdu, Nat. Rev. Urol., 2020, 17, 586–600 CrossRef PubMed.
- M. Gajdács, I. Dóczi, M. Ábrók, A. Lázár and K. Burián, Cent. Eur. J. Urol., 2019, 72, 209–214 Search PubMed.
- E. Paydaş Hataysal, B. Saraçlıgil, H. Türk Dağı and H. Vatansev, Eur. Respir. J., 2019, 5, 613–617 CrossRef.
- D. Fallon, J. Clin. Pathol., 2003, 56, 608–612 CrossRef CAS PubMed.
- M. Oyaert and J. Delanghe, Ann. Lab. Med., 2018, 39, 15–22 CrossRef PubMed.
- M.-D. Phan, K. M. Peters, S. Sarkar, S. W. Lukowski, L. P. Allsopp, D. G. Moriel, M. E. S. Achard, M. Totsika, V. M. Marshall, M. Upton, S. A. Beatson and M. A. Schembri, PLoS Genet., 2013, 9, e1003834 CrossRef PubMed.
- P. Bajaj, N. S. Singh and J. S. Virdi, Front. Microbiol., 2016, 7, 417 Search PubMed.
- M. Santos, M. Mariz, I. Tiago, J. Martins, S. Alarico and P. Ferreira, J. Pharm. Biomed. Anal., 2022, 219, 114889 CrossRef CAS PubMed.
- M. Harris and T. Fasolino, J. Lab. Med., 2022, 46, 3–15 CrossRef CAS.
- M. S. Kumar, S. Ghosh, S. Nayak and A. P. Das, Biosens. Bioelectron., 2016, 80, 497–510 CrossRef CAS PubMed.
- M. Davenport, K. E. Mach, L. M. D. Shortliffe, N. Banaei, T.-H. Wang and J. C. Liao, Nat. Rev. Urol., 2017, 14, 298–310 Search PubMed.
- M. S. Kumar and A. P. Das, Adv. Colloid Interface Sci., 2017, 249, 53–65 CrossRef CAS PubMed.
- R. Lei, R. Huo and C. Mohan, Expert Rev. Mol. Diagn., 2020, 20, 69–84 CrossRef CAS PubMed.
- A. Hasandka, A. R. Singh, A. Prabhu, H. R. Singhal, M. S. G. Nandagopal and N. K. Mani, Anal. Bioanal. Chem., 2022, 414, 847–865 CrossRef CAS PubMed.
- D. J. Shin, N. Andini, K. Hsieh, S. Yang and T.-H. Wang, Annu. Rev. Anal. Chem., 2019, 12, 41–67 CrossRef CAS PubMed.
- W. Kim, J. S. Park, D. Lee, J. Seo, L. P. Lee and S. J. Kim, Biosens. Bioelectron., 2022, 213, 114350 CrossRef CAS PubMed.
- Z. D. Call, I. Jang, B. J. Geiss, D. S. Dandy and C. S. Henry, Anal. Chem., 2022, 94, 7545–7550 CrossRef CAS PubMed.
- J. Noiphung and W. Laiwattanapaisal, Sci. Rep., 2019, 9, 1555 CrossRef PubMed.
- Y. Shen, J. Yi, M. Song, D. Li, Y. Wu, Y.-J. Liu, M. Yang and L. Qiao, Analyst, 2021, 146, 4146–4153 RSC.
- J. Chen, Y. Xu, H. Yan, Y. Zhu, L. Wang, Y. Zhang, Y. Lu and W. Xing, Lab Chip, 2018, 18, 2441–2452 RSC.
- A. O. Olanrewaju, A. Ng, P. Decorwin-Martin, A. Robillard and D. Juncker, Anal. Chem., 2017, 89, 6846–6853 CrossRef CAS PubMed.
- I. P. Alves and N. M. Reis, Biosens. Bioelectron., 2019, 145, 111624 CrossRef CAS PubMed.
- V. B. Barbosa, C. F. Rodrigues, L. Cerqueira, J. M. Miranda and N. F. Azevedo, Front. Bioeng. Biotechnol., 2022, 10, 987669 CrossRef PubMed.
- Y. Li, T. Wang and J. Wu, Analyst, 2021, 146, 1151–1156 RSC.
- R. Moakhar, T. AbdelFatah, A. Sanati, M. Jalali, S. E. Flynn, S. S. Mahshid and S. Mahshid, ACS Appl. Mater. Interfaces, 2020, 12, 23298–23310 CrossRef PubMed.
- M. Basak, S. Mitra, M. Gogoi, S. Sinha, H. B. Nemade and D. Bandyopadhyay, ACS Appl. Bio Mater., 2022, 5, 5321–5332 CrossRef CAS PubMed.
- R. Pandey, Y. Lu, E. Osman, S. Saxena, Z. Zhang, S. Qian, A. Pollinzi, M. Smieja, Y. Li, L. Soleymani and T. Hoare, ACS Sens., 2022, 7, 985–994 CrossRef CAS PubMed.
- D. Yang, H. Zhou, N. E. Dina and C. Haisch, R. Soc. Open Sci., 2018, 5, 180955 CrossRef PubMed.
- H. Ilhan, B. Guven, U. Dogan, H. Torul, S. Evran, D. Çetin, Z. Suludere, N. Saglam, İ. H. Boyaci and U. Tamer, Talanta, 2019, 201, 245–252 CrossRef CAS PubMed.
- Y. Li, G. Xie, J. Qiu, D. Zhou, D. Gou, Y. Tao, Y. Li and H. Chen, Sens. Actuators, B, 2018, 258, 803–812 CrossRef CAS.
- S. Reyes, N. Le, M. D. Fuentes, J. Upegui, E. Dikici, D. Broyles, E. Quinto, S. Daunert and S. K. Deo, Int. J. Mol. Sci., 2020, 21, 5015 CrossRef CAS PubMed.
- P. Zhang, A. M. Kaushik, K. Hsieh, S. Li, S. Lewis, K. E. Mach, J. C. Liao, K. C. Carroll and T.-H. Wang, Small Methods, 2022, 6, 2101254 CrossRef CAS PubMed.
- K. E. Mach, A. M. Kaushik, K. Hsieh, P. K. Wong, T.-H. Wang and J. C. Liao, Analyst, 2019, 144, 1565–1574 RSC.
- A. M. Kaushik, K. Hsieh, K. E. Mach, S. Lewis, C. M. Puleo, K. C. Carroll, J. C. Liao and T.-H. Wang, Adv. Sci., 2021, 8, 200341 Search PubMed.
- K. Hsieh, K. E. Mach, P. Zhang, J. C. Liao and T.-H. Wang, Acc. Chem. Res., 2022, 55, 123–133 CrossRef CAS PubMed.
- P. Athamanolap, K. Hsieh, C. M. O'Keefe, Y. Zhang, S. Yang and T.-H. Wang, Anal. Chem., 2019, 91, 12784–12792 CrossRef CAS PubMed.
- W. Kang, S. Sarkar, Z. S. Lin, S. McKenney and T. Konry, Anal. Chem., 2019, 91, 6242–6249 CrossRef CAS PubMed.
- P. Sabhachandani, S. Sarkar, P. C. Zucchi, B. A. Whitfield, J. E. Kirby, E. B. Hirsch and T. Konry, Microchim. Acta, 2017, 184, 4619–4628 CrossRef CAS.
- A. A. Sklavounos, C. R. Nemr, S. O. Kelley and A. R. Wheeler, Lab Chip, 2021, 21, 4208–4222 RSC.
- X. Li, X. Liu, Z. Yu, Y. Luo, Q. Hu, Z. Xu, J. Dai, N. Wu and F. Shen, Lab Chip, 2022, 22, 3952–3960 RSC.
- M. Osaid, Y.-S. Chen, C.-H. Wang, A. Sinha, W.-B. Lee, P. Gopinathan, H.-B. Wu and G.-B. Lee, Lab Chip, 2021, 21, 2223–2231 RSC.
- J. Avesar, D. Rosenfeld, M. Truman-Rosentsvit, T. Ben-Arye, Y. Geffen, M. Bercovici and S. Levenberg, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E5787–E5795 CrossRef CAS PubMed.
- Ö. Baltekin, A. Boucharin, E. Tano, D. I. Andersson and J. Elf, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 9170–9175 CrossRef PubMed.
- H. Li, P. Torab, K. E. Mach, C. Surrette, M. R. England, D. W. Craft, N. J. Thomas, J. C. Liao, C. Puleo and P. K. Wong, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 10270–10279 CrossRef CAS PubMed.
- Y. Yang, K. Gupta and K. L. Ekinci, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 10639–10644 CrossRef PubMed.
- V. Kara, C. Duan, K. Gupta, S. Kurosawa, D. J. Stearns-Kurosawa and K. L. Ekinci, Lab Chip, 2018, 18, 743–753 RSC.
- V. Shumeiko, G. Hidas, C. Nowogrodski, Y. Pinto, O. Gofrit, M. Duvdevani and O. Shoseyov, Sensors, 2021, 21, 5902 CrossRef PubMed.
- S. H. Needs, H. M. I. Osborn and A. D. Edwards, J. Microbiol. Methods, 2021, 187, 106199 CrossRef CAS PubMed.
- K.-W. Hsu, W.-B. Lee, H.-L. You, M. S. Lee and G.-B. Lee, Lab Chip, 2021, 21, 755–763 RSC.
- J. Gao, H. Li, P. Torab, K. E. Mach, D. W. Craft, N. J. Thomas, C. M. Puleo, J. C. Liao, T.-H. Wang and P. K. Wong, Nanomedicine, 2019, 17, 246–253 CrossRef CAS PubMed.
- S. M. Pires, B. N. Desta, L. Mughini-Gras, B. T. Mmbaga, O. E. Fayemi, E. M. Salvador, T. Gobena, S. E. Majowicz, T. Hald, P. S. Hoejskov, Y. Minato and B. Devleesschauwer, Curr. Opin. Food Sci., 2021, 39, 152–159 CrossRef PubMed.
- WHO 2017 Diarrhoeal disease, https://www.who.int/news-room/fact-sheets/detail/diarrhoeal-disease, (accessed 6 January 2024).
- Y. Nguyen and V. Sperandio, Front. Cell. Infect. Microbiol., 2012, 2, 90 CAS.
- L. A. Knodler and J. R. Elfenbein, Trends Microbiol., 2019, 27, 964–965 CrossRef CAS PubMed.
- D. Costa and G. Iraola, Clin. Microbiol. Rev., 2019, 32, e00072-18 CrossRef PubMed.
- M. M. Koopmans, M. C. Brouwer, J. A. Vázquez-Boland and D. van de Beek, Clin. Microbiol. Rev., 2022, e00060-19 Search PubMed.
- I. Mehdizadeh Gohari, M. A. Navarro, J. Li, A. Shrestha, F. Uzal and B. A. McClane, Virulence, 2021, 12, 723–753 CrossRef CAS PubMed.
- S. M. E. Toubar, A. A. Elbialy, M. M. M. Zaky and A. S. El-Shafey, AASCIT Journal of Health, 2018, 5, 34–38 Search PubMed.
- R. M. Humphries and A. J. Linscott, Clin. Microbiol. Rev., 2015, 28, 3–31 CrossRef PubMed.
- WHO Cholera – Global situation, https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON426, (accessed 6 January 2024).
- J. Deen, M. A. Mengel and J. D. Clemens, Vaccine, 2020, 38, A31–A40 CrossRef PubMed.
- S. Baker and H. C. The, Curr. Opin. Infect. Dis., 2018, 31, 449–454 CrossRef PubMed.
- M. M. Gibani, C. Britto and A. J. Pollard, Curr. Opin. Infect. Dis., 2018, 31, 440–448 CrossRef PubMed.
- S. E. Crawford, S. Ramani, J. E. Tate, U. D. Parashar, L. Svensson, M. Hagbom, M. A. Franco, H. B. Greenberg, M. O'Ryan, G. Kang, U. Desselberger and M. K. Estes, Nat. Rev. Dis. Primers, 2017, 3, 17083 CrossRef PubMed.
- E. Robilotti, S. Deresinski and B. A. Pinsky, Clin. Microbiol. Rev., 2015, 28, 134–164 CrossRef CAS PubMed.
- G. Di Cola, A. C. Fantilli, M. B. Pisano and V. E. Ré, Int. J. Food Microbiol., 2021, 338, 108986 CrossRef CAS PubMed.
- K. Bányai, M. K. Estes, V. Martella and U. D. Parashar, Lancet, 2018, 392, 175–186 CrossRef PubMed.
- S. E. Crowe, N. Engl. J. Med., 2019, 380, 1158–1165 CrossRef PubMed.
- L. Yin, Y. Li, W. Zhang, X. Han, Q. Wu, Y. Xie, J. Fan and L. Ma, J. Agric. Food Chem., 2023, 71, 3551–3563 CrossRef CAS PubMed.
- D. Gao, Z. Ma and Y. Jiang, TrAC, Trends Anal. Chem., 2022, 157, 116788 CrossRef CAS.
- Y. Shang, X. Xiang, Q. Ye, Q. Wu, J. Zhang and J.-M. Lin, TrAC, Trends Anal. Chem., 2022, 147, 116509 CrossRef CAS.
- M. Ranjbaran and M. S. Verma, Trends Food Sci. Technol., 2022, 128, 102–117 CrossRef CAS.
- I. A. Quintela, T. Vasse, C.-S. Lin and V. C. H. Wu, Front. Microbiol., 2022, 13, 1054782 CrossRef PubMed.
- W. Su, D. Liang and M. Tan, Trends Food Sci. Technol., 2021, 113, 97–109 CrossRef CAS.
- F. Mi, C. Hu, Y. Wang, L. Wang, F. Peng, P. Geng and M. Guan, Anal. Bioanal. Chem., 2022, 414, 2883–2902 CrossRef CAS PubMed.
- A. Rani, V. B. Ravindran, A. Surapaneni, N. Mantri and A. S. Ball, Int. J. Food Microbiol., 2021, 349, 109233 CrossRef CAS PubMed.
- M. A. Zaczek-Moczydlowska, A. Beizaei, M. Dillon and K. Campbell, Trends Food Sci. Technol., 2021, 114, 684–695 CrossRef CAS.
- Y. Shen, L. Xu and Y. Li, Compr. Rev. Food Sci. Food Saf., 2021, 20, 149–197 CrossRef PubMed.
- B. Wang and B. Park, Foodborne Pathog. Dis., 2022, 19, 359–375 CrossRef CAS PubMed.
- A. Ajayi, T. Jolaiya and S. Smith, J. Clin. Med., 2020, 9, 2565 CrossRef CAS PubMed.
- R. Li, X. Tian, J. Pang, L. Li, J. Yuan, Z. Tian and Z. Wang, Viruses, 2022, 14, 1355 CrossRef PubMed.
- W.-K. Wu, C.-C. Chen, S. Panyod, R.-A. Chen, M.-S. Wu, L.-Y. Sheen and S.-C. Chang, J. Formosan Med. Assoc., 2019, 118, 545–555 CrossRef PubMed.
- S. Zhao, W. He, Z. Ma, P. Liu, P.-H. Huang, H. Bachman, L. Wang, S. Yang, Z. Tian, Z. Wang, Y. Gu, Z. Xie and T. Jun Huang, Lab Chip, 2019, 19, 941–947 RSC.
- J. Kang, C. Park, J. Lee, J. Namkung, S. Y. Hwang and Y. S. Kim, BioChip J., 2017, 11, 76–84 CrossRef CAS.
- O. Mosley, L. Melling, M. D. Tarn, C. Kemp, M. M. N. Esfahani, N. Pamme and K. J. Shaw, Lab Chip, 2016, 16, 2108–2115 RSC.
- A. Pryszlak, T. Wenzel, K. W. Seitz, F. Hildebrand, E. Kartal, M. R. Cosenza, V. Benes, P. Bork and C. A. Merten, Cells Rep. Methods, 2022, 2, 100137 CrossRef CAS PubMed.
- L. Ma, M. Petersen and X. Lu, Appl. Environ. Microbiol., 2020, 86, e00096-20 CrossRef PubMed.
- M. Geissler, D. Brassard, L. Clime, A. V. C. Pilar, L. Malic, J. Daoud, V. Barrère, C. Luebbert, B. W. Blais, N. Corneau and T. Veres, Analyst, 2020, 145, 6831–6845 RSC.
- C. R. Phaneuf, B. Mangadu, H. M. Tran, Y. K. Light, A. Sinha, F. W. Charbonier, T. P. Eckles, A. K. Singh and C.-Y. Koh, Biosens. Bioelectron., 2018, 120, 93–101 CrossRef CAS PubMed.
- M. Zhang, J. Liu, Z. Shen, Y. Liu, Y. Song, Y. Liang, Z. Li, L. Nie, Y. Fang and Y. Zhao, BMC Microbiol., 2021, 21, 197 CrossRef CAS PubMed.
- N. Jin, L. Xue, Y. Ding, Y. Liu, F. Jiang, M. Liao, Y. Li and J. Lin, Biosens. Bioelectron., 2023, 220, 114844 CrossRef CAS PubMed.
- Y. Shang, G. Xing, X. Liu, H. Lin and J.-M. Lin, Anal. Chem., 2022, 94, 16787–16795 CrossRef CAS PubMed.
- Y. Cao, C. Ye, C. Zhang, G. Zhang, H. Hu, Z. Zhang, H. Fang, J. Zheng and H. Liu, Food Control, 2022, 134, 108694 CrossRef CAS.
- Z. Qin, X. Xiang, L. Xue, W. Cai, J. Gao, J. Yang, Y. Liang, L. Wang, M. Chen, R. Pang, Y. Li, J. Zhang, Y. Hu and Q. Wu, Microchem. J., 2021, 164, 106050 CrossRef CAS.
- R. Chand and S. Neethirajan, Biosens. Bioelectron., 2017, 98, 47–53 CrossRef CAS PubMed.
- S. Chung, L. E. Breshears, S. Perea, C. M. Morrison, W. Q. Betancourt, K. A. Reynolds and J.-Y. Yoon, ACS Omega, 2019, 4, 11180–11188 CrossRef CAS PubMed.
- S. Chung, L. E. Breshears, A. Gonzales, C. M. Jennings, C. M. Morrison, W. Q. Betancourt, K. A. Reynolds and J.-Y. Yoon, Nat. Protoc., 2021, 16, 1452–1475 CrossRef CAS PubMed.
- X. Ye, J. Xu, L. Lu, X. Li, X. Fang and J. Kong, Anal. Chim. Acta, 2018, 1018, 78–85 CrossRef CAS PubMed.
- S. Wen, J. Zhang, R. Zhao, J. Gao, N. Wang, T. Lu, R. Xie, X. Sun, B. Xiao, Z. Duan and A. Chen, ACS Agric. Sci. Technol., 2022, 2, 805–812 CrossRef CAS.
- M. El-Tholoth, H. Bai, M. G. Mauk, L. Saif and H. H. Bau, Lab Chip, 2021, 21, 1118–1130 RSC.
- L. Zhou, Y. Chen, X. Fang, Y. Liu, M. Du, X. Lu, Q. Li, Y. Sun, J. Ma and T. Lan, Anal. Chim. Acta, 2020, 1125, 57–65 CrossRef CAS PubMed.
- W. Y. Cui, H. J. Yoo, Y. G. Li, C. Baek and J. Min, Biosens. Bioelectron., 2022, 199, 113878 CrossRef CAS PubMed.
- L. Ding, S. Razavi Bazaz, T. Hall, G. Vesey and M. Ebrahimi Warkiani, Biomicrofluidics, 2022, 16, 014105 CrossRef CAS.
- S. Sukas, B. van Dorst, A. Kryj, O. Lagatie, W. De Malsche and L. J. Stuyver, Micromachines, 2019, 10, 852 CrossRef PubMed.
- K. E. Rudd, S. C. Johnson, K. M. Agesa, K. A. Shackelford, D. Tsoi, D. R. Kievlan, D. V. Colombara, K. S. Ikuta, N. Kissoon, S. Finfer, C. Fleischmann-Struzek, F. R. Machado, K. K. Reinhart, K. Rowan, C. W. Seymour, R. S. Watson, T. E. West, F. Marinho, S. I. Hay, R. Lozano, A. D. Lopez, D. C. Angus, C. J. L. Murray and M. Naghavi, Lancet, 2020, 395, 200–211 CrossRef PubMed.
- World Health Organization, Global report on the epidemiology and burden of sepsis: current evidence, identifying gaps and future directions, World Health Organization, Geneva, 2020 Search PubMed.
- D. C. Angus and T. van der Poll, N. Engl. J. Med., 2013, 369, 840–851 CrossRef CAS PubMed.
- F. B. Mayr, S. Yende and D. C. Angus, Virulence, 2014, 5, 4–11 CrossRef PubMed.
- C. Nedeva, J. Menassa and H. Puthalakath, Front. Cell Dev. Biol., 2019, 7, 108 CrossRef PubMed.
- R. S. Hotchkiss, G. Monneret and D. Payen, Nat. Rev. Immunol., 2013, 13, 862–874 CrossRef CAS PubMed.
- F. Venet and G. Monneret, Nat. Rev. Nephrol., 2018, 14, 121–137 CrossRef CAS PubMed.
- P. Yagupsky and F. S. Nolte, Clin. Microbiol. Rev., 1990, 3, 11 CrossRef PubMed.
- J.-L. Vincent, J. Rello, J. Marshall, E. Silva, A. Anzueto, C. D. Martin, R. Moreno, J. Lipman, C. Gomersall, Y. Sakr, K. Reinhart and for the EPIC II Group of Investigators, JAMA, J. Am. Med. Assoc., 2009, 302, 2323–2329 CrossRef CAS PubMed.
- L. Evans, A. Rhodes, W. Alhazzani, M. Antonelli, C. M. Coopersmith, C. French, F. R. Machado, L. Mcintyre, M. Ostermann, H. C. Prescott, C. Schorr, S. Simpson, W. J. Wiersinga, F. Alshamsi, D. C. Angus, Y. Arabi, L. Azevedo, R. Beale, G. Beilman, E. Belley-Cote, L. Burry, M. Cecconi, J. Centofanti, A. Coz Yataco, J. De Waele, R. P. Dellinger, K. Doi, B. Du, E. Estenssoro, R. Ferrer, C. Gomersall, C. Hodgson, M. H. Møller, T. Iwashyna, S. Jacob, R. Kleinpell, M. Klompas, Y. Koh, A. Kumar, A. Kwizera, S. Lobo, H. Masur, S. McGloughlin, S. Mehta, Y. Mehta, M. Mer, M. Nunnally, S. Oczkowski, T. Osborn, E. Papathanassoglou, A. Perner, M. Puskarich, J. Roberts, W. Schweickert, M. Seckel, J. Sevransky, C. L. Sprung, T. Welte, J. Zimmerman and M. Levy, Intensive Care Med., 2021, 47, 1181–1247 CrossRef PubMed.
- G. Lippi, Clin. Chem. Lab. Med., 2019, 57, 1281–1283 CrossRef CAS PubMed.
- N. Peker, N. Couto, B. Sinha and J. W. Rossen, Clin. Microbiol. Infect., 2018, 24, 944–955 CrossRef CAS PubMed.
- B. Fiori, T. D'Inzeo, A. Giaquinto, G. Menchinelli, F. M. Liotti, F. de Maio, G. De Angelis, G. Quaranta, D. Nagel, M. Tumbarello, B. Posteraro, M. Sanguinetti and T. Spanu, J. Clin. Microbiol., 2016, 54, 576–584 CrossRef CAS PubMed.
- A. M. Peri, P. N. A. Harris and D. L. Paterson, Clin. Microbiol. Infect., 2022, 28, 195–201 CrossRef PubMed.
- M. P. McHugh, B. J. Parcell, F. M. MacKenzie, K. E. Templeton and Scottish Microbiology and Virology Network (SMVN) Molecular Diagnostics Evaluation Group, J. Med. Microbiol., 2020, 69, 552–557 CrossRef CAS PubMed.
- A. Galiana, J. Coy, A. Gimeno, N. M. Guzman, F. Rosales, E. Merino, G. Royo and J. C. Rodríguez, PLoS One, 2017, 12, e0177627 CrossRef PubMed.
- Abionic SA, www.abionic.com, (accessed 09 February 2024).
- M. Sinha, J. Jupe, H. Mack, T. P. Coleman, S. M. Lawrence and S. I. Fraley, Clin. Microbiol. Rev., 2018, 31, e00089-17 CrossRef PubMed.
- A. Jyoti, S. Kumar, V. Kumar Srivastava, S. Kaushik and S. Govind Singh, Clin. Chim. Acta, 2021, 521, 45–58 CrossRef CAS PubMed.
- Y. Zhang, Y. Zhou, Y. Yang and D. Pappas, Analyst, 2021, 146, 2110–2125 RSC.
- T. Oeschger, D. McCloskey, V. Kopparthy, A. Singh and D. Erickson, Lab Chip, 2019, 19, 728–737 RSC.
- S. Kumar, S. Tripathy, A. Jyoti and S. G. Singh, Biosens. Bioelectron., 2019, 124–125, 205–215 CrossRef CAS PubMed.
- D. Tsounidi, P. S. Petrou and I. Raptis, IEEE Sens. J., 2021, 21, 12840–12855 CAS.
- L. Liu, Z. Han, F. An, X. Gong, C. Zhao, W. Zheng, L. Mei and Q. Zhou, J. Nanobiotechnol., 2021, 19, 216 CrossRef PubMed.
- M. Pilecky, A. Schildberger, D. Orth-Höller and V. Weber, Diagn. Microbiol. Infect. Dis., 2019, 94, 7–14 CrossRef PubMed.
- A. Burklund and J. X. J. Zhang, Ann. Biomed. Eng., 2019, 47, 1657–1674 CrossRef PubMed.
- H. Li and A. J. Steckl, Anal. Chem., 2019, 91, 352–371 CrossRef CAS PubMed.
- Y.-L. Fang, C.-H. Wang, Y.-S. Chen, C.-C. Chien, F.-C. Kuo, H.-L. You, M. S. Lee and G.-B. Lee, Lab Chip, 2021, 21, 113–121 RSC.
- T. J. Abram, H. Cherukury, C.-Y. Ou, T. Vu, M. Toledano, Y. Li, J. T. Grunwald, M. N. Toosky, D. F. Tifrea, A. Slepenkin, J. Chong, L. Kong, D. V. Del Pozo, K. T. La, L. Labanieh, J. Zimak, B. Shen, S. S. Huang, E. Gratton, E. M. Peterson and W. Zhao, Lab Chip, 2020, 20, 477–489 RSC.
- B. Forsyth, P. Torab, J.-H. Lee, T. Malcom, T.-H. Wang, J. C. Liao, S. Yang, E. Kvam, C. Puleo and P. K. Wong, Biosensors, 2021, 11, 288 CrossRef CAS PubMed.
- A. S. Tanak, B. Jagannath, Y. Tamrakar, S. Muthukumar and S. Prasad, Anal. Chim. Acta: X, 2019, 3, 100029 CAS.
- A. S. Tanak, S. Muthukumar, S. Krishnan, K. L. Schully, D. V. Clark and S. Prasad, Biosens. Bioelectron., 2021, 171, 112726 CrossRef CAS PubMed.
- A. S. Tanak, A. Sardesai, S. Muthukumar, S. Krishnan, D. A. Striegel, K. L. Schully, D. V. Clark and S. Prasad, Biosens. Bioelectron.: X, 2022, 10, 100144 CAS.
- A. S. Tanak, A. Sardesai, S. Muthukumar and S. Prasad, Bioeng. Transl. Med., 2022, 7, e10310 CrossRef CAS PubMed.
- A. Belushkin, F. Yesilkoy, J. J. González-López, J. C. Ruiz-Rodríguez, R. Ferrer, A. Fàbrega and H. Altug, Small, 2020, 16, 1906108 CrossRef CAS PubMed.
- A. Giannetti, C. Trono, G. Porro, C. Domenici, M. Puntoni and F. Baldini, Chemosensors, 2020, 8, 12 CrossRef CAS.
- A. Molinero-Fernández, M. A. López and A. Escarpa, Analyst, 2020, 145, 5004–5010 RSC.
- M. S. Verma, M.-N. Tsaloglou, T. Sisley, D. Christodouleas, A. Chen, J. Milette and G. M. Whitesides, Biosens. Bioelectron., 2018, 99, 77–84 CrossRef CAS PubMed.
- A. Alba-Patiño, S. M. Russell, M. Borges, N. Pazos-Pérez, R. A. Álvarez-Puebla and R. De La Rica, Nanoscale Adv., 2020, 2, 1253–1260 RSC.
- E. Valera, J. Berger, U. Hassan, T. Ghonge, J. Liu, M. Rappleye, J. Winter, D. Abboud, Z. Haidry, R. Healey, N.-T. Hung, N. Leung, N. Mansury, A. Hasnain, C. Lannon, Z. Price, K. White and R. Bashir, Lab Chip, 2018, 18, 1461–1470 RSC.
- S. Damodara, J. Arora, P. C. Liaw, A. E. Fox-Robichaud and P. R. Selvaganapathy, Microchim. Acta, 2022, 189, 146 CrossRef CAS PubMed.
- F. Ellett, J. Jorgensen, A. L. Marand, Y. M. Liu, M. M. Martinez, V. Sein, K. L. Butler, J. Lee and D. Irimia, Nat. Biomed. Eng., 2018, 2, 207–214 CrossRef CAS PubMed.
- H. Jeon, D.-H. Lee, B. Jundi, M. Pinilla-Vera, R. M. Baron, B. D. Levy, J. Voldman and J. Han, ACS Sens., 2021, 6, 2747–2756 CrossRef CAS PubMed.
- K. K. Zeming, R. Vernekar, M. T. Chua, K. Y. Quek, G. Sutton, T. Krüger, W. S. Kuan and J. Han, Small, 2021, 17, 2006123 CrossRef CAS PubMed.
- Y. Zhang, Y. Zhou, W. Li, V. Lyons, A. Johnson, A. Venable, J. Griswold and D. Pappas, Anal. Chem., 2018, 90, 7204–7211 CrossRef CAS PubMed.
- Y. Zhou, Y. Zhang, A. Johnson, A. Venable, J. Griswold and D. Pappas, Anal. Chim. Acta, 2019, 1062, 110–117 CrossRef CAS PubMed.
- U. Hassan, R. Zhu and R. Bashir, Lab Chip, 2018, 18, 1231–1240 RSC.
- N. Mancini, S. Carletti, N. Ghidoli, P. Cichero, R. Burioni and M. Clementi, Clin. Microbiol. Rev., 2010, 23, 235–251 CrossRef CAS PubMed.
- S. Kalyan, C. Torabi, H. Khoo, H. W. Sung, S.-E. Choi, W. Wang, B. Treutler, D. Kim and S. C. Hur, Micromachines, 2021, 12, 257 CrossRef PubMed.
- W. G. Pitt, M. Alizadeh, G. A. Husseini, D. S. McClellan, C. M. Buchanan, C. G. Bledsoe, R. A. Robison, R. Blanco, B. L. Roeder, M. Melville and A. K. Hunter, Biotechnol. Prog., 2016, 32, 823–839 CrossRef CAS PubMed.
- P. Ohlsson, M. Evander, K. Petersson, L. Mellhammar, A. Lehmusvuori, U. Karhunen, M. Soikkeli, T. Seppä, E. Tuunainen, A. Spangar, P. von Lode, K. Rantakokko-Jalava, G. Otto, S. Scheding, T. Soukka, S. Wittfooth and T. Laurell, Anal. Chem., 2016, 88, 9403–9411 CrossRef CAS PubMed.
- P. Dow, K. Kotz, S. Gruszka, J. Holder and J. Fiering, Lab Chip, 2018, 18, 923–932 RSC.
- B. M. Biron, A. Ayala and J. L. Lomas-Neira, Biomarker Insights, 2015, 10s4, BMI.S29519 CrossRef PubMed.
- C. Pierrakos and J.-L. Vincent, Crit. Care, 2010, 14, R15 CrossRef PubMed.
- S.-W. Kim, I.-H. Cho, G.-S. Lim, G.-N. Park and S.-H. Paek, Biosens. Bioelectron., 2017, 98, 7–14 CrossRef CAS PubMed.
- J. Min, M. Nothing, B. Coble, H. Zheng, J. Park, H. Im, G. F. Weber, C. M. Castro, F. K. Swirski, R. Weissleder and H. Lee, ACS Nano, 2018, 12, 3378–3384 CrossRef CAS PubMed.
- C. Rosales, Front. Physiol., 2018, 9, 113 CrossRef PubMed.
- K. A. Babatunde, J. M. Ayuso, S. C. Kerr, A. Huttenlocher and D. J. Beebe, Front. Immunol., 2021, 12, 781535 CrossRef CAS PubMed.
- Y. Zhang, W. Li, Y. Zhou, A. Johnson, A. Venable, A. Hassan, J. Griswold and D. Pappas, Analyst, 2018, 143, 241–249 RSC.
- U. Hassan, T. Ghonge, B. Reddy, M. Patel, M. Rappleye, I. Taneja, A. Tanna, R. Healey, N. Manusry, Z. Price, T. Jensen, J. Berger, A. Hasnain, E. Flaugher, S. Liu, B. Davis, J. Kumar, K. White and R. Bashir, Nat. Commun., 2017, 8, 15949 CrossRef CAS PubMed.
- Y. Zheng, Q. Yu, Y. Lin, Y. Zhou, L. Lan, S. Yang and J. Wu, Lancet Infect. Dis., 2022, 22, 541–551 CrossRef PubMed.
- WHO STI factsheet 2023, https://www.who.int/news-room/fact-sheets/detail/sexually-transmitted-infections-(stis), (accessed 7 January 2024).
- C. Elwell, K. Mirrashidi and J. Engel, Nat. Rev. Microbiol., 2016, 14, 385–400 CrossRef CAS PubMed.
- M. K. Morgan and C. F. Decker, Disease-a-Month, 2016, 62, 260–268 CrossRef PubMed.
- F. Mercer and P. J. Johnson, Trends Parasitol., 2018, 34, 683–693 CrossRef PubMed.
- K. G. Ghanem, S. Ram and P. A. Rice, N. Engl. J. Med., 2020, 382, 845–854 CrossRef PubMed.
- S. R. Galvin and M. S. Cohen, Nat. Rev. Microbiol., 2004, 2, 33–42 CrossRef CAS PubMed.
- B. Mlynarczyk-Bonikowska, C. Kowalewski, A. Krolak-Ulinska and W. Marusza, Int. J. Mol. Sci., 2022, 23, 10499 CrossRef CAS PubMed.
- M. Iannacone and L. G. Guidotti, Nat. Rev. Immunol., 2022, 22, 19–32 CrossRef CAS PubMed.
- K. Madavaraju, R. Koganti, I. Volety, T. Yadavalli and D. Shukla, Front. Cell. Infect. Microbiol., 2021, 10, 617578 CrossRef PubMed.
- J. O. Kahn and B. D. Walker, N. Engl. J. Med., 1998, 339, 33–39 CrossRef CAS PubMed.
- M. E. Sabatini and S. Chiocca, Br. J. Cancer, 2020, 122, 306–314 CrossRef PubMed.
- WHO Hepatitis B factsheet 2023, https://www.who.int/news-room/fact-sheets/detail/hepatitis-b, (accessed 7 January 2024).
- J. K.-T. Ho, B. Jeevan-Raj and H.-J. Netter, Viruses, 2020, 12, 126 CrossRef CAS PubMed.
- K. Yoshimura, J. Infect. Chemother., 2017, 23, 12–16 CrossRef PubMed.
- L. A. Sadowski, R. Upadhyay, Z. W. Greeley and B. J. Margulies, Viruses, 2021, 13, 1228 CrossRef CAS PubMed.
- L. Cheng, Y. Wang and J. Du, Vaccines, 2020, 8, 391 CrossRef CAS PubMed.
- T. Meyer, Microorganisms, 2016, 4, 25 CrossRef PubMed.
- C. C. Bristow, J. D. Klausner and A. Tran, Clin. Infect. Dis., 2020, 71, S52–S57 CrossRef CAS PubMed.
- S. R. Morris, C. C. Bristow, M. R. Wierzbicki, M. Sarno, L. Asbel, A. French, C. A. Gaydos, L. Hazan, L. Mena, P. Madhivanan, S. Philip, S. Schwartz, C. Brown, D. Styers, T. Waymer and J. D. Klausner, Lancet Infect. Dis., 2021, 21, 668–676 CrossRef CAS PubMed.
- A. D. Cristillo, C. C. Bristow, R. Peeling, B. Van Der Pol, S. H. de Cortina, I. K. Dimov, N. P. Pai, D. Jin Shin, R. Y. T. Chiu, C. Klapperich, P. Madhivanan, S. R. Morris and J. D. Klausner, Sex. Transm. Dis., 2017, 44, 211–218 CrossRef PubMed.
- T. E. Wi, F. J. Ndowa, C. Ferreyra, C. Kelly-Cirino, M. M. Taylor, I. Toskin, J. Kiarie, N. Santesso and M. Unemo, J. Int. AIDS Soc., 2019, 22, e25343 CrossRef PubMed.
- G. Caruso, A. Giammanco, R. Virruso and T. Fasciana, Int. J. Environ. Res. Public Health, 2021, 18, 1038 CrossRef CAS PubMed.
- I. Toskin, V. Govender, K. Blondeel, M. Murtagh, M. Unemo, C. Zemouri, R. W. Peeling and J. Kiarie, Sex. Transm. Infect., 2020, 96, 342–347 CrossRef PubMed.
- K. Hsieh, J. H. Melendez, C. A. Gaydos and T.-H. Wang, Lab Chip, 2022, 22, 476–511 RSC.
- P. C. Adamson, M. J. Loeffelholz and J. D. Klausner, Arch. Pathol. Lab. Med., 2020, 144, 1344–1351 CrossRef PubMed.
- C. A. Gaydos, Y. C. Manabe and J. H. Melendez, Sex. Transm. Dis., 2021, 48, S71–S77 CrossRef PubMed.
- D. Thakur, T. Fatima, P. Sharma, M. R. Hasan, N. Malhotra, M. Khanuja, S. K. Shukla and J. Narang, Process Biochem., 2023, 126, 223–237 CrossRef CAS.
- N. Farokhzad and W. Tao, Trends Chem., 2021, 3, 589–602 CrossRef CAS.
- J. Eid, M. Mougel and M. Socol, Viruses, 2020, 12, 982 CrossRef CAS PubMed.
- S. Tharakan, O. Faqah, W. Asghar and A. Ilyas, Biosensors, 2022, 12, 949 CrossRef CAS PubMed.
- N. P. Pai, A. Karellis, J. Kim and T. Peter, Lancet HIV, 2020, 7, e574–e581 CrossRef PubMed.
- Y. Xiao, A. J. Thompson and J. Howell, Cells, 2020, 9, 2233 CrossRef CAS PubMed.
- V. Narayanamurthy, Z. E. Jeroish, K. S. Bhuvaneshwari and F. Samsuri, Anal. Methods, 2021, 13, 740–763 RSC.
- S. Hassanpour, B. Baradaran, M. de la Guardia, A. Baghbanzadeh, J. Mosafer, M. Hejazi, A. Mokhtarzadeh and M. Hasanzadeh, Microchim. Acta, 2018, 185, 568 CrossRef PubMed.
- P. Nath, M. A. Kabir, S. K. Doust and A. Ray, Infect. Dis. Rep., 2021, 13, 518–539 CrossRef PubMed.
- A. Y. Trick, H. T. Ngo, A. H. Nambiar, M. M. Morakis, F.-E. Chen, L. Chen, K. Hsieh and T.-H. Wang, Lab Chip, 2022, 22, 945–953 RSC.
- T. Liu, G. Choi, Z. Tang, A. Kshirsagar, A. J. Politza and W. Guan, Biosens. Bioelectron., 2022, 209, 114255 CrossRef CAS PubMed.
- C.-A. Chen, H. Yuan, C.-W. Chen, Y.-S. Chien, W.-H. Sheng and C.-F. Chen, Lab Chip, 2021, 21, 1908–1915 RSC.
- E. A. Phillips, T. J. Moehling, K. F. K. Ejendal, O. S. Hoilett, K. M. Byers, L. A. Basing, L. A. Jankowski, J. B. Bennett, L.-K. Lin, L. A. Stanciu and J. C. Linnes, Lab Chip, 2019, 19, 3375–3386 RSC.
- D. Liu, Y. Zhang, M. Zhu, Z. Yu, X. Ma, Y. Song, S. Zhou and C. Yang, Anal. Chem., 2020, 92, 11826–11833 CrossRef CAS PubMed.
- R. R. G. Soares, J. C. Varela, U. Neogi, S. Ciftci, M. Ashokkumar, I. F. Pinto, M. Nilsson, N. Madaboosi and A. Russom, Biosens. Bioelectron., 2020, 166, 112442 CrossRef CAS PubMed.
- M. Kong, Z. Li, J. Wu, J. Hu, Y. Sheng, D. Wu, Y. Lin, M. Li, X. Wang and S. Wang, Talanta, 2019, 205, 120155 CrossRef CAS PubMed.
- Y.-L. Tan, A.-Q. Huang, L.-J. Tang and J.-H. Jiang, Chem. Sci., 2021, 12, 8445–8451 RSC.
- W. Witkowska McConnell, C. Davis, S. R. Sabir, A. Garrett, A. Bradley-Stewart, P. Jajesniak, J. Reboud, G. Xu, Z. Yang, R. Gunson, E. C. Thomson and J. M. Cooper, Nat. Commun., 2021, 12, 6994 CrossRef CAS PubMed.
- C. Xie, S. Chen, L. Zhang, X. He, Y. Ma, H. Wu, B. Zou and G. Zhou, Anal. Bioanal. Chem., 2021, 413, 2923–2931 CrossRef CAS PubMed.
- X. Zhao, X. Li, W. Yang, J. Peng, J. Huang and S. Mi, Analyst, 2021, 146, 5102–5114 RSC.
- R. Wang, J. Wu, X. He, P. Zhou and Z. Shen, Micromachines, 2021, 12, 263 CrossRef PubMed.
- Z. Yu, W. Lyu, M. Yu, Q. Wang, H. Qu, R. F. Ismagilov, X. Han, D. Lai and F. Shen, Biosens. Bioelectron., 2020, 155, 112107 CrossRef CAS PubMed.
- A. Y. Trick, J. H. Melendez, F.-E. Chen, L. Chen, A. Onzia, A. Zawedde, E. Nakku-Joloba, P. Kyambadde, E. Mande, J. Matovu, M. Atuheirwe, R. Kwizera, E. A. Gilliams, Y.-H. Hsieh, C. A. Gaydos, Y. C. Manabe, M. M. Hamill and T.-H. Wang, Sci. Transl. Med., 2021, 13, eabf6356 CrossRef CAS PubMed.
- X. Ye, Y. Li, L. Wang, X. Fang and J. Kong, Talanta, 2021, 221, 121462 CrossRef CAS PubMed.
- A. L. Horst, J. M. Rosenbohm, N. Kolluri, J. Hardick, C. A. Gaydos, M. Cabodi, C. M. Klapperich and J. C. Linnes, Biomed. Microdevices, 2018, 20, 35 CrossRef PubMed.
- J. Hoffman, J. van Griensven, R. Colebunders and M. McKellar, HIV Ther., 2010, 4, 27–39 CrossRef.
- M. Stone, J. Bainbridge, A. M. Sanchez, S. M. Keating, A. Pappas, W. Rountree, C. Todd, S. Bakkour, M. Manak, S. A. Peel, R. W. Coombs, E. M. Ramos, M. K. Shriver, P. Contestable, S. V. Nair, D. H. Wilson, M. Stengelin, G. Murphy, I. Hewlett, T. N. Denny and M. P. Busch, J. Clin. Microbiol., 2018, 56, e02045-17 CrossRef PubMed.
- D. J. Shin, P. Athamanolap, L. Chen, J. Hardick, M. Lewis, Y. H. Hsieh, R. E. Rothman, C. A. Gaydos and T. H. Wang, Sci. Rep., 2017, 7, 4495 CrossRef CAS PubMed.
- D. J. Shin, A. Y. Trick, Y.-H. Hsieh, D. L. Thomas and T.-H. Wang, Sci. Rep., 2018, 8, 9793 CrossRef PubMed.
- F. Li, Y. Zheng, J. Wu, L. Zhao, L. Shui, Q. Pu and S. Liu, Talanta, 2019, 203, 83–89 CrossRef CAS PubMed.
- J. Song, C. Liu, M. G. Mauk, J. Peng, T. Schoenfeld and H. H. Bau, Anal. Chem., 2018, 90, 1209–1216 CrossRef CAS PubMed.
- W. Ouyang and J. Han, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 16240–16249 CrossRef CAS PubMed.
- K. Kadimisetty, K. Yin, A. M. Roche, Y. Yi, F. D. Bushman, R. G. Collman, R. Gross, L. Feng and C. Liu, Analyst, 2021, 146, 3234–3241 RSC.
- H. J. Goux, B. Raja, K. Kourentzi, J. R. C. Trabuco, B. V. Vu, A. S. Paterson, A. Kirkpatrick, B. Townsend, M. Lee, V. T. T. Truong, C. Pedroza and R. C. Willson, PLoS One, 2019, 14, e0225365 CrossRef CAS PubMed.
- B. Shu, L. Lin, B. Wu, E. Huang, Y. Wang, Z. Li, H. He, X. Lei, B. Xu and D. Liu, Biosens. Bioelectron., 2021, 181, 113145 CrossRef CAS PubMed.
- WHO Vector-borne diseases, https://www.who.int/news-room/fact-sheets/detail/vector-borne-diseases, (accessed 7 January 2024).
- WHO Global vector control response 2017–2030, https://www.who.int/publications-detail-redirect/9789241512978, (accessed 7 January 2024).
- A. L. Wilson, O. Courtenay, L. A. Kelly-Hope, T. W. Scott, W. Takken, S. J. Torr and S. W. Lindsay, PLoS Neglected Trop. Dis., 2020, 14, e0007831 CrossRef CAS PubMed.
- L. H. V. Franklinos, K. E. Jones, D. W. Redding and I. Abubakar, Lancet Infect. Dis., 2019, 19, e302–e312 CrossRef PubMed.
- J. Rocklöv and R. Dubrow, Nat. Immunol., 2020, 21, 479–483 CrossRef PubMed.
- A. C. Steere, F. Strle, G. P. Wormser, L. T. Hu, J. A. Branda, J. W. R. Hovius, X. Li and P. S. Mead, Nat. Rev. Dis. Primers, 2016, 2, 16090 CrossRef PubMed.
- S. Li, L. Gilbert, S. O. Vanwambeke, J. Yu, B. V. Purse and P. A. Harrison, Environ. Health Perspect., 2019, 127, 067010 CrossRef PubMed.
- R. Bellini, A. Michaelakis, D. Petrić, F. Schaffner, B. Alten, P. Angelini, C. Aranda, N. Becker, M. Carrieri, M. Di Luca, E. Fălcuţă, E. Flacio, A. Klobučar, C. Lagneau, E. Merdić, O. Mikov, I. Pajovic, D. Papachristos, C. A. Sousa, A. Stroo, L. Toma, M. I. Vasquez, E. Velo, C. Venturelli and M. Zgomba, Travel Med. Infect. Dis., 2020, 35, 101691 CrossRef PubMed.
- M. G. Guzman and E. Harris, Lancet, 2015, 385, 453–465 CrossRef.
- A. Wilder-Smith, E.-E. Ooi, O. Horstick and B. Wills, Lancet, 2019, 393, 350–363 CrossRef.
- WHO Dengue and severe dengue, https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue, (accessed 7 January 2024).
- X. Yang, M. B. M. Quam, T. Zhang and S. Sang, J. Travel Med., 2021, 28, taab146 CrossRef PubMed.
- M. G. Guzmán and G. Kourí, Int. J. Infect. Dis., 2004, 8, 69–80 CrossRef PubMed.
- R. Eivazzadeh-Keihan, P. Pashazadeh-Panahi, T. Mahmoudi, K. K. Chenab, B. Baradaran, M. Hashemzaei, F. Radinekiyan, A. Mokhtarzadeh and A. Maleki, Microchim. Acta, 2019, 186, 329 CrossRef PubMed.
- N. T. Darwish, S. D. Sekaran and S. M. Khor, Sens. Actuators, B, 2018, 255, 3316–3331 CrossRef CAS.
- Y. Yao, N. Zhao, W. Jing, Q. Liu, H. Lu, W. Zhao, W. Zhao, Z. Yuan, H. Xia and G. Sui, Sens. Actuators, B, 2021, 333, 129521 CrossRef CAS.
- W. Lee, H. Kim, P. K. Bae, S. Lee, S. Yang and J. Kim, Biosens. Bioelectron., 2021, 190, 113388 CrossRef CAS PubMed.
- A. M. Jankelow, H. Lee, W. Wang, T.-H. Hoang, A. Bacon, F. Sun, S. Chae, V. Kindratenko, K. Koprowski, R. A. Stavins, D. D. Ceriani, Z. W. Engelder, W. P. King, M. N. Do, R. Bashir, E. Valera and B. T. Cunningham, Analyst, 2022, 147, 3838–3853 RSC.
- S. Sharma, M. A. Kabir and W. Asghar, Arch. Pathol. Lab. Med., 2020, 144, 1335–1343 CrossRef CAS PubMed.
- P. Biswas, G. N. Mukunthan Sulochana, T. N. Banuprasad, P. Goyal, D. Modak, A. K. Ghosh and S. Chakraborty, ACS Sens., 2022, 7, 3720–3729 CrossRef CAS PubMed.
- Y. Seok, B. S. Batule and M.-G. Kim, Biosens. Bioelectron., 2020, 165, 112400 CrossRef CAS PubMed.
- G. Theillet, G. Grard, M. Galla, C. Maisse, M. Enguehard, M. Cresson, P. Dalbon, I. L. Leparc-Goffart and F. Bedin, J. Med. Virol., 2019, 91, 899–910 CrossRef CAS PubMed.
- E. Chou, E. Lasek-Nesselquist, B. Taubner, A. Pilar, E. Guignon, W. Page, Y.-P. Lin and N. C. Cady, PLoS One, 2020, 15, e0228772 CrossRef CAS PubMed.
- H.-A. Joung, Z. S. Ballard, A. Ma, D. K. Tseng, H. Teshome, S. Burakowski, O. B. Garner, D. D. Carlo and A. Ozcan, Lab Chip, 2019, 19, 1027–1034 RSC.
- H.-A. Joung, Z. S. Ballard, J. Wu, D. K. Tseng, H. Teshome, L. Zhang, E. J. Horn, P. M. Arnaboldi, R. J. Dattwyler, O. B. Garner, D. Di Carlo and A. Ozcan, ACS Nano, 2020, 14, 229–240 CrossRef CAS PubMed.
- S. Arumugam, S. Nayak, T. Williams, F. S. D. S. Maria, M. S. Guedes, R. C. Chaves, V. Linder, A. R. Marques, E. J. Horn, S. J. Wong, S. K. Sia and M. Gomes-Solecki, J. Clin. Microbiol., 2019, 57, e01142-19 CrossRef PubMed.
- W. F. Wright and P. G. Auwaerter, Open Forum Infect. Dis., 2020, 7, ofaa132 CrossRef PubMed.
- J. Elven, P. Dahal, E. A. Ashley, N. V. Thomas, P. Shrestha, K. Stepniewska, J. A. Crump, P. N. Newton, D. Bell, H. Reyburn, H. Hopkins and P. J. Guérin, BMC Med., 2020, 18, 279 CrossRef PubMed.
- J. J. L. Tan, M. Capozzoli, M. Sato, W. Watthanaworawit, C. L. Ling, M. Mauduit, B. Malleret, A.-C. Grüner, R. Tan, F. H. Nosten, G. Snounou, L. Rénia and L. F. P. Ng, PLoS Neglected Trop. Dis., 2014, 8, e3043 CrossRef PubMed.
- S. Hin, D. Baumgartner, M. Specht, J. Lüddecke, E. M. Arjmand, B. Johannsen, L. Schiedel, M. Rombach, N. Paust, F. von Stetten, R. Zengerle, N. Wipf, P. Müller, K. Mavridis, J. Vontas and K. Mitsakakis, Processes, 2020, 8, 1677 CrossRef CAS.
- S. Moutailler, L. Yousfi, L. Mousson, E. Devillers, M. Vazeille, A. Vega-Rúa, Y. Perrin, F. Jourdain, F. Chandre, A. Cannet, S. Chantilly, J. Restrepo, A. Guidez, I. Dusfour, F. V. S. De Abreu, T. P. Dos Santos, D. Jiolle, T. M. Visser, C. J. M. Koenraadt, M. Wongsokarijo, M. Diallo, D. Diallo, A. Gaye, S. Boyer, V. Duong, G. Piorkowski, C. Paupy, R. L. De Oliveira, X. De Lamballerie and A.-B. Failloux, Viruses, 2019, 11, 904 CrossRef CAS PubMed.
- T. Liao, X. Wang, M. Donolato, E. Harris, M. M. Cruz, A. Balmaseda and R. Y. L. Wang, Diagnostics, 2020, 10, 372 CrossRef CAS PubMed.
- I. Alejo-Cancho, J. Navero-Castillejos, A. Peiró-Mestres, R. Albarracín, J. Barrachina, A. Navarro, V. Gonzalo, V. Pastor, J. Muñoz and M. J. Martínez, PLoS Neglected Trop. Dis., 2020, 14, e0008082 CrossRef PubMed.
- A. Ganguli, A. Ornob, H. Yu, G. L. Damhorst, W. Chen, F. Sun, A. Bhuiya, B. T. Cunningham and R. Bashir, Biomed. Microdevices, 2017, 19, 73 CrossRef CAS PubMed.
- R. R. G. Soares, A. Pettke, A. Robles-Remacho, S. Zeebaree, S. Ciftci, M. Tampere, A. Russom, M.-R. Puumalainen, M. Nilsson and N. Madaboosi, Sens. Actuators, B, 2021, 336, 129723 CrossRef CAS.
- X. Zhu, J. Zhao, A. Hu, J. Pan, G. Deng, C. Hua, C. Zhu, Y. Liu, K. Yang and L. Zhu, Micromachines, 2020, 11, 186 CrossRef PubMed.
- S. Pal, A. L. Dauner, I. Mitra, B. M. Forshey, P. Garcia, A. C. Morrison, E. S. Halsey, T. J. Kochel and S.-J. L. Wu, PLoS One, 2014, 9, e113411 CrossRef PubMed.
- R. Luo, N. Fongwen, C. Kelly-Cirino, E. Harris, A. Wilder-Smith and R. W. Peeling, Clin. Microbiol. Infect., 2019, 25, 659–666 CrossRef CAS PubMed.
- M. Kikuti, J. S. Cruz, M. S. Rodrigues, A. S. Tavares, I. A. D. Paploski, M. M. O. Silva, P. M. Santana, L. B. Tauro, G. A. O. F. Silva, G. S. Campos, J. M. G. Araújo, U. Kitron, M. G. Reis and G. S. Ribeiro, PLoS One, 2019, 14, e0213301 CrossRef PubMed.
- K. Kaarj, P. Akarapipad and J.-Y. Yoon, Sci. Rep., 2018, 8, 12438 CrossRef PubMed.
- M. Shehata Draz, M. Venkataramani, H. Lakshminarayanan, E. Saygili, M. Moazeni, A. Vasan, Y. Li, X. Sun, S. Hua, X. G. Yu and H. Shafiee, Nanoscale, 2018, 10, 11841–11849 RSC.
- K. Arias-Alpízar, A. Sánchez-Cano, J. Prat-Trunas, E. de la Serna Serna, O. Alonso, E. Sulleiro, A. Sánchez-Montalvá, A. Diéguez and E. Baldrich, Biosens. Bioelectron., 2022, 215, 114513 CrossRef PubMed.
- X. Guo, M. A. Khalid, I. Domingos, A. L. Michala, M. Adriko, C. Rowel, D. Ajambo, A. Garrett, S. Kar, X. Yan, J. Reboud, E. M. Tukahebwa and J. M. Cooper, Nat. Electron., 2021, 4, 615–624 CrossRef CAS.
- C. Flynn and A. Ignaszak, Biosensors, 2020, 10, 137 CrossRef CAS PubMed.
- S. Nayak, A. Sridhara, R. Melo, L. Richer, N. H. Chee, J. Kim, V. Linder, D. Steinmiller, S. K. Sia and M. Gomes-Solecki, Sci. Rep., 2016, 6, 35069 CrossRef CAS PubMed.
- P. Banović, E. Piloto-Sardiñas, D. Mijatović, A. Foucault-Simonin, V. Simin, I. Bogdan, D. Obregón, L. Mateos-Hernández, S. Moutailler and A. Cabezas-Cruz, Acta Trop., 2023, 238, 106756 CrossRef PubMed.
- R. Amino, S. Thiberge, B. Martin, S. Celli, S. Shorte, F. Frischknecht and R. Ménard, Nat. Med., 2006, 12, 220–224 CrossRef CAS PubMed.
- M. Prudêncio, A. Rodriguez and M. M. Mota, Nat. Rev. Microbiol., 2006, 4, 849–856 CrossRef PubMed.
- T. F. de Koning-Ward, M. W. A. Dixon, L. Tilley and P. R. Gilson, Nat. Rev. Microbiol., 2016, 14, 494–507 CrossRef CAS PubMed.
- R. N. Price, R. J. Commons, K. E. Battle, K. Thriemer and K. Mendis, Trends Parasitol., 2020, 36, 560–570 CrossRef.
- World malaria report 2022, https://www.who.int/publications-detail-redirect/9789240064898, (accessed 7 January 2024).
- T. Bousema, L. Okell, I. Felger and C. Drakeley, Nat. Rev. Microbiol., 2014, 12, 833–840 CrossRef CAS.
- K. Elfving, D. Shakely, M. Andersson, K. Baltzell, A. S. Ali, M. Bachelard, K. I. Falk, A. Ljung, M. I. Msellem, R. S. Omar, P. Parola, W. Xu, M. Petzold, B. Trollfors, A. Björkman, M. Lindh and A. Mårtensson, PLoS One, 2016, 11, e0146054 CrossRef PubMed.
- B. A. Mathison and B. S. Pritt, J. Clin. Microbiol., 2017, 55, 2009–2017 CrossRef CAS.
- A. Mbanefo and N. Kumar, Trop. Med. Infect. Dis., 2020, 5, 102 CrossRef PubMed.
- N. M. Pham, W. Karlen, H.-P. Beck and E. Delamarche, Malar. J., 2018, 17, 260 CrossRef PubMed.
- M. S. Cordray and R. R. Richards-Kortum, Am. J. Trop. Med. Hyg., 2012, 87, 223–230 CrossRef CAS PubMed.
- S. Britton, Q. Cheng and J. S. McCarthy, Malar. J., 2016, 15, 88 CrossRef.
- S. Sharma, J. Singh, A. Sen and A. R. Anvikar, J. Vector Borne Dis., 2022, 59, 29–36 CrossRef CAS PubMed.
- C. B. Nair, J. Manjula, P. A. Subramani, P. B. Nagendrappa, M. N. Manoj, S. Malpani, P. K. Pullela, P. V. Subbarao, S. Ramamoorthy and S. K. Ghosh, PLoS One, 2016, 11, e0146961 CrossRef PubMed.
- J. C. Mouatcho and J. P. D. Goldring, J. Med. Microbiol., 2013, 62, 1491–1505 CrossRef CAS PubMed.
- A. N. Mukkala, J. Kwan, R. Lau, D. Harris, D. Kain and A. K. Boggild, Curr. Infect. Dis. Rep., 2018, 20, 49 CrossRef PubMed.
- A. Jimenez, R. R. Rees-Channer, R. Perera, D. Gamboa, P. L. Chiodini, I. J. González, A. Mayor and X. C. Ding, Malar. J., 2017, 16, 128 CrossRef PubMed.
- L. Marquart, A. Butterworth, J. S. McCarthy and M. L. Gatton, Malar. J., 2012, 11, 74 CrossRef CAS PubMed.
- N. Thorne, L. Flores-Olazo, R. Egoávil-Espejo, E. A. Vela, J. Noel, J. Valdivia-Silva and D. van Noort, Micromachines, 2021, 12, 1245 CrossRef PubMed.
- N. Kolluri, C. M. Klapperich and M. Cabodi, Lab Chip, 2018, 18, 75–94 RSC.
- K. V. Ragavan, S. Kumar, S. Swaraj and S. Neethirajan, Biosens. Bioelectron., 2018, 105, 188–210 CrossRef CAS PubMed.
- K. Mitsakakis, S. Hin, P. Müller, N. Wipf, E. Thomsen, M. Coleman, R. Zengerle, J. Vontas and K. Mavridis, Int. J. Environ. Res. Public Health, 2018, 15, 259 CrossRef PubMed.
- M. Depond, B. Henry, P. Buffet and P. A. Ndour, Front. Physiol., 2020, 10, 1613 CrossRef PubMed.
- S. Jackson, S. Lee and A. K. Badu-Tawiah, Anal. Chem., 2022, 94, 5132–5139 CrossRef CAS PubMed.
- A. M. Ogunmolasuyi, R. Fogel, H. Hoppe, D. Goldring and J. Limson, Malar. J., 2022, 21, 174 CrossRef CAS PubMed.
- S. Ghosh and C. H. Ahn, Analyst, 2019, 144, 2109–2119 RSC.
- N. M. Pham, S. Rusch, Y. Temiz, H.-P. Beck, W. Karlen and E. Delamarche, Biomed. Microdevices, 2019, 21, 24 CrossRef PubMed.
- J. Choi, S.-J. Cho, Y. T. Kim and H. Shin, Biomed. Microdevices, 2019, 21, 86 CrossRef CAS PubMed.
- J. Reboud, G. Xu, A. Garrett, M. Adriko, Z. Yang, E. M. Tukahebwa, C. Rowell and J. M. Cooper, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 4834–4842 CrossRef CAS PubMed.
- G. Choi, T. Prince, J. Miao, L. Cui and W. Guan, Biosens. Bioelectron., 2018, 115, 83–90 CrossRef CAS PubMed.
- A. J. Colbert, K. Co, G. Lima-Cooper, D. H. Lee, K. N. Clayton, S. T. Wereley, C. C. John, J. C. Linnes and T. L. Kinzer-Ursem, Malar. J., 2021, 20, 380 CrossRef CAS PubMed.
- G. Ruiz-Vega, K. Arias-Alpízar, E. de la Serna, L. N. Borgheti-Cardoso, E. Sulleiro, I. Molina, X. Fernàndez-Busquets, A. Sánchez-Montalvá, F. J. del Campo and E. Baldrich, Biosens. Bioelectron., 2020, 150, 111925 CrossRef CAS PubMed.
- N. K. Singh, P. Jain, S. Das and P. Goswami, Anal. Chem., 2019, 91, 4213–4221 CrossRef CAS PubMed.
- S. Ghosh, K. Aggarwal, T. U. Vinitha, T. Nguyen, J. Han and C. H. Ahn, Microsyst. Nanoeng., 2020, 6, 5 CrossRef CAS PubMed.
- R. Clément, A. Bienvenu, A. Lavoignat, G. Bonnot, B. Doumèche and S. Picot, Talanta, 2023, 252, 123839 CrossRef PubMed.
- K. Malpartida-Cardenas, N. Miscourides, J. Rodriguez-Manzano, L.-S. Yu, N. Moser, J. Baum and P. Georgiou, Biosens. Bioelectron., 2019, 145, 111678 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |


