Open Access Article
Open Access ArticleMicrofluidic approach to correlate C. elegans neuronal functional aging and underlying changes of gene expression in mechanosensation†
Jason
Wan‡
 a,
Jimmy L.
Ding‡
b and
Hang
Lu
a,
Jimmy L.
Ding‡
b and
Hang
Lu
 *abc
*abc
aWallace H. Coulter Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, GA 30332, USA
bPetit Institute for Bioengineering and Bioscience, Interdisciplinary BioEngineering Program, Georgia Institute of Technology, Atlanta, GA 30332, USA. E-mail: hang.lu@gatech.edu
cSchool of Chemical & Biomolecular Engineering, Georgia Institute of Technology, Atlanta, GA 30332, USA
First published on 23rd April 2024
Abstract
The aging process has broad physiological impacts, including a significant decline in sensory function, which threatens both physical health and quality of life. One ideal model to study aging, neuronal function, and gene expression is the nematode Caenorhabditis elegans, which has a short lifespan and relatively simple, thoroughly mapped nervous system and genome. Previous works have identified that mechanosensory neuronal structure changes with age, but importantly, the actual age-related changes in the function and health of neurons, as well as the underlying genetic mechanisms responsible for these declines, are not fully understood. While advanced techniques such as single-cell RNA-sequencing have been developed to quantify gene expression, it is difficult to relate this information to functional changes in aging due to a lack of tools available. To address these limitations, we present a platform capable of measuring both physiological function and its associated gene expression throughout the aging process in individuals. Using our pipeline, we investigate the age-related changes in function of the mechanosensing ALM neuron in C. elegans, as well as some relevant gene expression patterns (mec-4 and mec-10). Using a series of devices for animals of different ages, we examined subtle changes in neuronal function and found that while the magnitude of neuronal response to a large stimulus declines with age, sensory capability does not significantly decline with age; further, gene expression is well maintained throughout aging. Additionally, we examine PVD, a harsh-touch mechanosensory neuron, and find that it exhibits a similar age-related decline in magnitude of neuronal response. Together, our data demonstrate that our strategy is useful for identifying genetic factors involved in the decline in neuronal health. We envision that this framework could be applied to other systems as a useful tool for discovering new biology.
1. Introduction
Age-related declines in sensorimotor control are one of the most prominent hallmarks of aging.1–3 In humans, progressive hearing loss, impaired balance, and an increase in falls have been shown to be age-related and pose major health concerns for the elderly population.4–7 There are treatments and strategies to address many of these aging-related symptoms, but the underlying reasons for the decline in sensorimotor control remain unclear. This is perhaps due to the large number of factors contributing to the aging-dependent phenotypes and thus, a lack of comprehensive tools to study and quantify functional aging (i.e., how functions, such as a neuron's ability to sense environmental cues, change with age). It is difficult to characterize aging because, unlike many other diseases, it cannot be traced to a single factor. Due to the complexity and difficulty of conducting lifelong human studies, researchers often turn towards more well-defined model organisms such as mice, flies, C. elegans, and yeast. C. elegans, a microscopic roundworm, is an ideal model for aging research due to its short natural lifespan (∼3 weeks), self-fertilization for easy culturing of large, isogenic populations, significant genetic homology to humans, including especially well-conserved aging pathways,8,9 and broad genetic toolbox for gene expression-based studies. C. elegans are simultaneously a useful model for studying neurological function due to their simple and compact nervous system, well-defined physiology on molecular and functional levels,10,11 fully mapped connectome,12,13 and well-characterized behaviors.14,15 Since these roundworms are optically transparent, they are also easily adaptable for high-throughput in vivo and in situ fluorescent imaging-based assays.16 For example, several microfluidic platforms have been developed for C. elegans to image,17–19 screen and sort individuals,17,20 deliver reagents more efficiently,21,22 study complex behaviors,23–26 and perform long-term animal culture with single-animal resolution.27While traditional methods have established a basic understanding of sensorimotor control, these approaches are limited as they either rely on indirect methods to suggest changes in function or require manual manipulation and expertise. For example, the classical mechanosensation assay involves manual delivery of mechanical stimulus with an eyebrow hair or metal pick and visual scoring of the behavior (i.e., how the animal responds).28,29 While it enabled important findings, this assay requires expertise and can vary between experimenters, both in the application of the stimulus and the scoring. Significant advancements, through technologies such as force probes and microfluidic platforms,23,30–34 have allowed for more accurate consistent delivery of stimuli. In addition, these technologies have been combined with genetic tools to improve the depth of quantifiable information. For instance, fluorescence imaging of transgenic strains of C. elegans allows researchers to visualize and quantify how touch receptor neurons – the most upstream component of the sensorimotor pathway – accumulate physical defects with age.1,35,36 By measuring these trends across different genetic backgrounds, researchers also found that gene expression and regulation of specific genes are essential to neuronal health.37 For example, loss-of-function mutations in mec-4 and mec-10, two genes that encode a protein complex that forms the transmembrane channel essential for mechanotransduction, result in an acceleration of age-dependent, physical defects in neurons.1,38 While these morphological changes, such as blebbing and branching of neurites, may suggest declines in function, they are only a proxy and require further, direct investigation to confirm whether function is actually affected.
To quantify neuronal cellular functional output, the most commonly used technique is patch-clamp electrophysiology.39,40 While this technique allows for the highest precision, such as recording single-channel currents, it requires dissection of an animal as well as expertise in locating a specific neuron of interest.41 This limits the experimental throughput, does not allow for recovery of the animal for further experimentation, and both the gluing process and dissection are likely to alter the neuronal or circuit response and add to experimental variability. To address these shortcomings, microfluidics devices designed specifically to deliver mechanical stimuli and measure the in vivo calcium response of C. elegans, using the genetically encoded calcium indicator GCaMP, have been developed to quantify mechanosensation.30,32 These devices reduce the amount of manual labor needed and improve the consistency of the assay by delivering well-controlled mechanical stimuli using deformable polydimethylsiloxane (PDMS) membranes. In addition, these devices can deliver stimuli with different magnitudes, durations, and frequencies.30–32 While these devices are foundational to improving mechanosensation assays, importantly, these findings do not demonstrate whether differences in calcium responses can attributed to neuronal health or function (i.e., a larger response may not necessarily indicate healthier function of a neuron). Additionally, these devices were designed for specific sizes of animals and do not extend their applications beyond measuring a single time point, restricting their utility for measuring functional aging. To be able to attribute measured differences in function to age and not experimental variability, it is essential to keep the stimulus constant between age groups and individuals. This poses another significant challenge as the animals' size and other physical properties, such as cuticle thickness and elasticity, change with age.33,42
In this work, we aim to synthesize previous designs and improve these assays to quantify functional aging. We do so by adapting the microfluidic stimulus-delivery strategy to differently aged animals, and importantly, linking functional changes to gene expression patterns with single-animal precision. Using a computational model, we first design a microfluidic-based strategy to deliver a consistent, high-frequency gentle touch stimulus to differently aged animals. Next, to ensure our platform can measure the subtle changes of functional aging in vivo, we validate our ability to quantify declines in neuronal function with a pharmacological perturbation. We then apply this microfluidic pipeline to measure the changes in aging. Together, our platform enables us to correlate age-related changes in functional responses and gene expression with single-animal, sub-cellular resolution. This work created a new opportunity to quantify neuron function with age and possibly link the change in function to an underlying genetic reason. This platform was designed to effectively study aging-related changes, which was previously not possible. Surprisingly, we found that age-related mechanosensation decline is not monotonous. Our study also reveals that the temporal expression of certain genes related to mechanosensation is non-monotonous and we find that the change in gene expression pattern and neuron function occur at similar time points.
2. Results and discussion
2.1 Integrated microfluidic pipeline to measure neuronal functional aging and gene expression
To deliver precise stimuli to individual animals and quantify their stimulus-evoked calcium response (i.e., functional neuronal activity) and gene expression profiles in vivo, we created an automated microfluidic-based pipeline (Fig. 1). Using this pipeline, we specifically examine the ALM neuron, a gentle touch mechanosensory neuron. In the gentle touch neurons in C. elegans, it is known that degenerin/epithelial sodium channels (DEG/ENaC) are responsible for mechanotransduction (Fig. 1a and b).43,44 While there are other components to this larger family of genes and proteins, here we focused on MEC-4 and MEC-10 since these proteins form the transmembrane channel essential to the neurons' mechanosensation function (Fig. 1b). To further characterize age-dependent neuronal changes, we use the activities and channel-expressions in the ALM touch receptor neuron as a model system.Previous fluorescence-based studies have established platforms to measure gene expression but relating that information to a biological function is difficult with existing technologies. Similarly, studies on neuronal function are limited as they would require large perturbations, such as genetic mutations, to measure how gene expression and regulation influence function. Further, understanding the effects of aging on neuronal function requires complex methods and tools designed to correlate function and gene expression in aging populations of animals. We designed a new pipeline to address these limitations (Fig. 1c and d). Two parallel populations were allowed to age following standard plate-based protocols, sampled at different ages, and tested in the microfluidic systems. Performing longitudinal observations on the same animals at each age would greatly increase the required complexity of the platform, reduce throughput, and risk losing or injuring animals during recovery and re-loading. Instead, we subsampled from the populations for each age group. Because the population of the animals are well-controlled and isogenic, each sub-sample should be representative of the population as a whole. To assess neuronal functions, one population carrying a genetically encoded calcium reporter GCaMP was used; the second population of animals carrying a translational reporter was imaged for gene expression quantification. By measuring reporter trends in each population during aging with our platform, we could correlate neuronal function to gene expression in the context of aging, enabling us to ask how age-related changes of gene expression may influence neuronal function.
The genes we focused on are mec-4 and mec-10. We used our microfluidic platform to enable high-throughput imaging of fluorescent translational reporters (Fig. 1e), a common technique that can provide a proxy readout of a gene's expression.45–47 Here, we could image single neurons and quantify their expression profiles. This same platform could also serially load individual animals with GCaMP, apply a touch stimulus, monitor the functional response of the mechanosensory neuron, and then empty the channel for the next animal (Fig. 1f). While single touches could evoke a neuronal response, we found that delivering multiple mechanical stimuli at a high frequency (2.7 ± 0.1 Hz) resulted in a larger, more robust neuronal response.32
2.2 Computational modeling and empirical validation to accommodate different aged populations of C. elegans
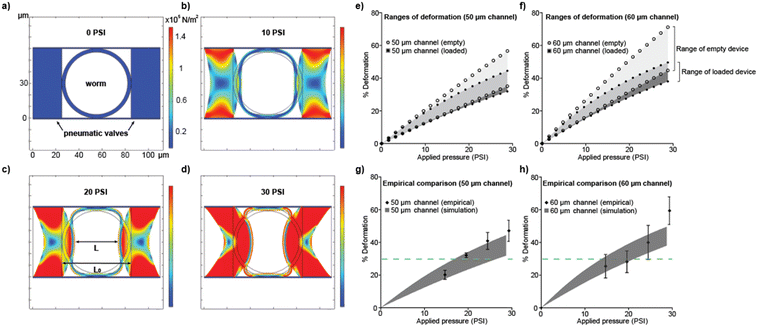
Delivering consistent stimuli with the pneumatic actuators across different ages and devices is key to measuring functional aging, as it helps to ensure that changes detected in neuronal function are indeed due to aging and not due to experimental variation. This is made challenging primarily due to a combination of variable properties from both the device and the animal. C. elegans increase, and then decrease, in size appreciably as they age and also exhibit changes in other physical properties, such as the thickness of the cuticle and elasticity of the body.48 PDMS, which is used for the microfluidic device, also has a relatively wide range of stiffness due to raw material variability and device fabrication factors such as curing time and temperature. This complexity is perhaps an important contributing factor to the limited tools available to study functional aging. To address these challenges, we designed devices for each age of the animal by scaling the imaging channel of the device to different widths and heights while carefully considering the stimulus each device design could deliver to an animal. We used a computational model to estimate the stimulus capability of each device design and aid in ensuring consistent stimulus delivery.While there may be different metrics to compare between the devices, percent deformation was the most physically relevant detail49 in our investigation, and we aimed to keep this value consistent between all individuals loaded into the microfluidic pipeline. To ensure the consistency of the stimulus, regardless of age or size, we use our model to examine the effects of varying relevant parameters. The goal of the model was to first verify this type of stimulus delivery was possible and identify the range of values (i.e., the amount of pressure) needed to achieve the same stimulus. To achieve a gentle touch stimulus that the neuron of interest was sensitive to, we aimed for ∼30% deformation based on previous literature.30 The model was explained in detail in the Material and methods section. Briefly, using our model (a 2D cross-section of the stimulation/imaging channel), we varied the pressure applied and calculated the resulting percent deformation (Fig. 2a–d). The percent deformation is calculated by dividing the shortest distance between the actuators at a given time, L, by the distance between them at t = 0, L0.
We used the model to consider several experimental variables and their impact on the resulting deformation, including the size of the animal, the geometry of the device, and a range of possible material properties of both the devices and animals. The model generated plots of the resulting deformation per an applied pressure depending on these various factors (Fig. 2e and f). To account for batch-to-batch variation in the devices, we considered a range of PDMS properties for both sizes of devices. After calculating an upper and lower range of possible PDMS stiffnesses, both were modeled and plotted (Fig. 2e), with the grey shaded area representing possible PDMS material values between these two limits.
We found that for any given deformation percentage, the potential variability, represented by the difference in pressure required for the upper and lower material limits, was manageable and could be adjusted for with a relatively small change in input pressure (<10 PSI). In addition, we considered the effect of the presence of an animal in the loading chamber on the upper and lower bounds of deformation, and found that removing the animal from the channel significantly shifts the curves (Fig. 2e). This suggests that simply calibrating the device with an empty channel to achieve 30% deformation was not accurate to fully deliver 30% deformation to the animal (Fig. 2e). We next considered the animal's changing physical properties, such as cuticle thickness and elasticity, generating upper and lower bounds using data from the literature. We found that these effects are negligible compared to the range of PDMS properties (Fig. S1, ESI†). These findings demonstrated that, in our application, the device properties play a more significant role than those of the biological sample. This suggests that our platform is less sensitive to the physical properties of C. elegans than expected, and therefore can be used for studying mechanosensation in most if not all C. elegans strains, even mutants with altered physical properties, e.g. muscular or cuticle mutants.
To address the differences in sizes of animals at different ages, we modeled two devices with 50 μm and 60 μm wide imaging channels. These imaging channel dimensions were found to be the most relevant based on experimental results, with days 2 and 15 animals best fitting in a 50 μm wide device while days 5 and 10 animals best fit in a 60 μm wide device. Alongside the 50 μm device that was discussed above, the 60 μm device model was also used to generate a range of potential deformation values (Fig. 2f). Studying these device dimensions using the model, we demonstrated that both devices were capable of delivering similar stimuli in our target range of ∼30% deformation (Fig. 2e and f). Taken together, we found, using our computational model, that a 30% deformation (a relatively gentle stimulus) was achievable with all channel widths and for a range of fabrication temperatures.
With a calibration curve provided by our computational model, we could estimate the range of values needed to deliver the target stimulus we wanted consistently delivered across all devices. We found exact pressure values through empirical testing of each batch of devices used by measuring the percent deformation while applying different amounts of pressure. The empirical results collected are compared against the ranges generated by the model; we establish that our empirical results closely fall within the range of computational model, especially around the desired deformation for our use case (Fig. 2g and h). At high pressure, beyond the target deformation, the empirical results begin to deviate from predicted values. This suggests that the model is overestimating the resistance of the material to deformation at high stress and that highly crosslinked PDMS is more hyperelastic than expected by the model. Another major contributing factor may be that the model assumes an idealized geometry, whereas the fabrication process leads to actual asymmetry of thickness, with the membrane being thinner than specified at y = 0, where it is bonded to glass at the bottom. These empirical results validate our model and stimulus delivery strategy at the target deformation, demonstrating the utility of the model during the device design. Ultimately, we validate that, regardless of the age of the animal, we could deliver consistent mechanical stimuli to evaluate functional aging.
2.3 Validating the microfluidic-based strategy to evaluate neuronal health
While GCaMP has been used as a proxy for neuronal activity, its use in measuring function has been typically limited to measuring only whether a neuron is activated or not. This resolution was enough for qualitative studies, such as finding which neurons respond to stimuli of a certain magnitude or applied to a specific region of the animal, but its use as a measurement for neuronal health is not yet well established. In other words, it is unclear whether a larger stimulus-evoked calcium response is due to the neurons being healthy. Because we are interested in the graded response, we first wanted to ensure that our platform was capable of yielding quantifiable calcium dynamics, and that varying calcium dynamics reflect relevant biological changes, such as those possibly found in the aging process. To accomplish this, we first validated our system in a pharmacological application.Amiloride specifically inhibits DEG/ENaC channels,50,51 and when applied to C. elegans, we were able to measure its influence on the neuronal response dynamics (Fig. 3a). We compared two populations: a control, untreated group and a group treated with 3 mM amiloride. Briefly, each animal was loaded into the microfluidic chip, and a stimulus of 8 gentle touch presses at 2.7 ± 0.1 Hz was applied. While a single press is sufficient to produce a significant neuronal response,30 we found that high-frequency presses resulted in even stronger responses (Fig. 3a) and thus a larger dynamic range of data.32 By quantifying the peaks of each animal's stimulus-evoked response, we found that the drug treatment, as expected, significantly decreases the neuronal response (Fig. 3b). Previous studies showed similar trends using the patch-clamping technique,40,52 reinforcing our demonstration that this calcium-sensitive indicator was similarly capable of measuring neuronal function without requiring dissection of the animal. This indicated that our platform can not only measure whether a neuron responds or not but also the strength and general health of its functional response. In addition, these findings also showed that with simple modifications our platform may enable functional in vivo drug screening.
2.4 Age-related declines of neuronal function in a mechanosensory neuron
Mechanosensation is one of the primary modes of sensation, which allows for animals to successfully respond to their environment and perform critical functions, such as avoiding threats or navigating to food sources. Specifically, it is responsible for touch, hearing, and balance – all of which are known functions that decline with age in humans and contribute to significant health risks.4–7 These behaviors require healthy, functioning neurons, but current methods to study mechanosensation, especially in the context of aging, are limited as they typically require either large perturbations to see broad phenotypes or labor-intensive and invasive methods to measure neuronal response and function.We engineered a microfluidic-based functional aging assay that addressed these shortcomings and was applied to differently aged populations of animals to measure functional aging and answer how neuronal health and function were changing with age. Animals were cultured and sub-sampled at different time points (days 2, 5, 10, and 15 of adulthood) during the lifespan (∼20 days) (Fig. 1c). Each sampled group was then loaded into our pipeline to measure both gene expression and neuronal activity. Briefly, each animal was loaded into the microfluidic chip, which applied stimulus pulses comprised of 8 gentle touch presses at 2.7 ± 0.1 Hz for 3 seconds with 40 second gaps between each pulse. A single pulse was sufficient to produce a consistent neuronal response, and we extended the stimulus protocol to allow for the measurement of additional features of neuronal function. In total, each animal was stimulated 9 times with this brief pulse, followed by a longer 30 second stimulus also at 2.7 ± 0.1 Hz (Fig. 4a–d). The repeated pulses acted as a measure of the consistency of the response to the same stimulus as well as an assay for habituation, a form of learning that can imply that neuronal plasticity is functioning properly. The longer stimulus at the end produced the maximum amplitude that the worm's neuronal response is capable of, a measurement of its total dynamic range. We performed this assay in three independent trials. The average response of all individuals across the three trials is shown by plotting the normalized GCaMP signal intensity over time (Fig. 4a). Comparing the average neuronal responses of each age group revealed notable differences and trends in aging. Along with the peaks of the neuronal responses, aging also influenced the dynamics (e.g., decay rate) of the neuronal traces (Fig. S2, ESI†).
The differences between age groups are more accentuated when comparing individual peaks. The first peak (Fig. 4b) demonstrated a stereotypical neuronal response from a single applied stimulus pulse. When comparing the magnitude of response at the population level, there is no significant difference between age groups (Fig. 4b and e). This means that for this initial stimulus, mechanosensory function is surprisingly robust to age-related changes. Subsequent stimulus pulses were shown to consistently produce similar response peaks, with no statistically distinguishable difference between the first and ninth peaks in nearly all cases (Fig. 4c and S3†).
The final 30 second stimulus delivery (Fig. 4d and f) produced a different trend to those found in the other measurements: there was a decline in response as populations aged, with a notably large difference between day 2 and day 10. While the magnitude response of some age groups for some trials did not demonstrate a statistically significant difference, linear regression demonstrated that there was a significantly non-zero decline for all trials from day 2 to day 10 (Fig. S4†). The larger dynamic range demonstrated by younger animals implies that while older animals remain capable of sensing mechanical stimuli, younger animals may be better at differentiating stimuli of differing magnitudes, whereas older animals respond similarly to stimuli regardless of magnitude. In comparison to the declining trend seen from day 2 to day 10, the day 15 animals exhibited a higher average magnitude than the younger day 10 group. This departure from the declining trend may be explained by the fact that, due to attrition from aging and subsampling the population, only a small fraction of animals from the starting population remained in the day 15 group. This means that compared to younger age groups, the day 15 age group may be over-represented by animals with above-average healthspan, whereas some low responders observed in day 10 have died by the next observed time point. This may also explain why the day 15 group exhibited the largest variance between individuals, as there may be two groups being sampled with a relatively small sample size – one group being abnormally healthy and the other following the previous trend of decline in function. This observation suggests that mechanosensory neuronal output may be correlated with health and that decline in the amplitude of a mechanosensory response to a strong stimulus could be a predictor of decline in health and death. While the three replicate trials produced overall similar trends, variability of biological and environmental factors and relatively low sample size led to some observed variance (Fig. S3†).
These results together suggest that age-related declines in mechanosensation are complex, and may depend on the exact nature of the stimuli. Currently dissecting this complexity is still difficult due to the high variability observed among individuals within each age group. However, the data suggest that some aspects of mechanosensation, such as the sensing of short, gentle pulses, appear to be relatively robust to aging-related changes, while other aspects, such as capability to differentiate the magnitude of a stimulus, may decline with age. There may be an ecological reason for younger animals to be sensitive to different magnitudes of stimuli: this ability may be important for escaping from danger prior to reaching and during the reproductive period; post reproduction, there may be a reduced need. Motor neurons have been observed to follow a similar trend;53 to the best of our knowledge, observations as reported in this study on mechanosensory neurons have not been reported before. Taken together, this suggests that there may be a global mechanism regulating functional aging common to multiple neuron types.
2.5 Age-related changes of gene expression in mec-4 and mec-10
To link the functional output measured (Fig. 4) to molecular mechanisms, we next looked to measure gene expression. While we know from the literature that genetic mutations impair biological function (e.g., mutating genes that form the mechanotransduction channel and measuring how the neuron responds to mechanical stimuli), expressions/maintenance/changes over age of these channels have not been characterized. Our platform, in contrast, enabled us to measure gene expression and couple this information to the trends in functional aging to identify how the genes mec-4 and mec-10 may be behind these age-related changes. We imaged fluorescent translational reporters where the fluorescence intensity levels may suggest trends in gene expression pattern.45 It should be noted that the accuracy of this method is limited as changes in rates of protein degradation and turnover may vary during aging. These experiments were done in parallel with the functional imaging described above and the data from the replicate trials, which are similar to the example trial shown below, can be found in the supplemental section (Fig. S5†). Looking at population-level data, there was an increase in expression of both mec-4 and mec-10 with age, followed by a decrease at the final time point (Fig. 5a). Since our platform enabled single-animal resolution, we were able to inspect expression levels and compare them within individual animals; here each point corresponds to a single animal and its mec-4 and mec-10 expression levels (Fig. 5b–e). The Pearson correlation coefficient, R, was calculated to determine the relationship between the expression levels. The expression levels of the two genes across the different ages indicated a strong positive correlation, perhaps with the tightest correlation at around early-to-mid-life (Fig. 5c and d). These results suggested that the regulatory elements of mec-4 and mec-10 seem to be maintained with age, where animals with high expression of mec-4 also had high expression of mec-10. This was expected as MEC-4 and MEC-10 physically form a complex together, suggesting that they may be regulated by similar mechanisms. Surprisingly, we found that the day 10 increase in both gene expressions was primarily driven by a group of high-expressing animals rather than the whole population (Fig. 5d), and that this population of high expressers were not present in the day 15 group (Fig. 5e and S6, ESI†). This may imply that high expressing animals may be more short-lived. This observation mirrors the previously mentioned mechanosensory differences of the day 15 age group and further supports the possibility that day 15 is sampling from a population of differing distribution compared to the other age groups. Because the microfluidic platform we developed is capable of measuring both functional responses and gene expression patterns across different ages and therefore allows correlation between the two.Several trends in functional response and gene expression observed in our assay potentially parallel each other, pointing to areas of interest for future research. For example, the continued regulation mec-4 and mec-10 ratio throughout the lifespan of animals may be connected to the robustness of mechanosensory response. As mec-4 and mec-10 expression increase from day 2 to day 10, the mechanosensory neuronal maximum amplitude declines, matching trends seen in previous literature that link overexpression of these genes with mechanosensory neuronal death (Fig. 4 and 5). Similarly, the increased expression levels, which may be connected to lower magnitude response and may negatively impact lifespan, as seen by the lack of high expressing animals at day 15, parallels the smaller fraction of low responding animals at this same time point (Fig. 4f, 5b and S6, ESI†). We propose a potential explanation where aging animals experience steadily increasing channel proteins (mec-4 and mec-10) that eventually progresses beyond a healthy threshold, leading to neuronal functional degradation in day 10 animals, which perish before day 15. This is further supported in literature as gain-of-function mutations in mec-4 are detrimental to mechanosensory neuronal health.1,36,44 Furthermore, even in isogenic populations of C. elegans grown on the same conditions, there is animal-to-animal variability in lifespan. Since day 15 is towards the end of the lifespan, it is reasonable to assume that perhaps the animals measured here are the longer-lived, and perhaps healthier, portion of the population. In other words, healthier, longer-lived animals are more likely to survive to day 15 and be sampled and observed to have lower expression levels and higher amplitude responses. This would further support the notion that relatively well-controlled, low expression of mec-4 and mec-10 contributes to healthier aging and prevents earlier death.
2.6 A different class of mechanosensory neuron demonstrates similar age-related trends
While this pipeline was optimized for the purposes of delivering gentle touch stimulus to differently aged populations and measuring the resulting neuronal response, the general operational strategy is readily adaptable to address a variety of biological questions by changing the stimulus delivered or the genes examined. For example, the pneumatic actuator is capable of applying a variety of different mechanical stimuli and could be used to examine age-related changes to sensitivity to specific parameters, such as harsh versus gentle touch and high and low frequencies. Furthermore, other classes of mechanosensory neurons such as proprioceptive, nociceptive PVD, and the dopaminergic head neurons ADE and CEP, can be examined as well, by using different transgenes. Beyond mechanosensation, this pipeline could be modified to examine other sensory modes coupled to relevant gene expression and aging, such as the use of chemical stimuli (e.g., different drug compounds or food pheromones).We utilize the adaptability of this platform to assay PVD, a mechanosensory neuron of a different class, and determine whether the age-related changes in function of ALM are unique or a general phenomenon experienced by other sensory neurons. PVD is a multimodal nociceptor with complex branching dendrites that is sensitive to harsh touch as well as temperature.54 It does not express MEC-4 but does express MEC-10.43,54 We applied the same microfluidic strategy using a different transgenic strain with a different promoter, and changing the stimulus protocol to a single harsh press (40% deformation). This generated a set of similar response curves to the long stimulus applied to ALM, where the average day 10 response was notably lower than the other populations (Fig. 6a). When plotting the maximum amplitude of each individual, for each age group, it is seen that the peak height of day 2 and day 5 individuals are significantly higher than day 10 (Fig. 6b). It is also seen that the majority of day 10 and 15 individuals demonstrate a low functional response. Despite a few outliers with the largest peak heights of all individuals, the day 15 age group is not significantly different than day 10. Similar to ALM, the day 15 outliers and resulting higher average functional response may be due to a survivor bias in which healthier animals are more likely to survive to this time point, whereas the very low responders seen in day 10 may have died by this point. Studying age-related changes in function in PVD reveals a similar trend compared to ALM, implying that this trend may be a general phenomenon common to aging-related changes in sensory neurons. We note that at present, direct comparisons are inappropriate due to the different stimuli applied, as well as variability among populations and the sampling frequency. Nevertheless, our platform measured age-related functional changes in two separate sensory neuronal classes. We find that similar to the gentle touch neuron ALM, PVD also demonstrates an overall age-related decline in maximum amplitude of the neuronal response.
3. Conclusion
A major bottleneck in aging studies is the lack of tools available to measure both functional aging and its associated underlying molecular mechanisms. While there are platforms to measure function, including those that deliver mechanical stimuli and measure the stimulus-evoked neuronal response, they have not been adapted for aging studies.30,32 Further, these functional assays generally cannot identify or evaluate which genes are relevant to the function without large genetic perturbations, such as mutations. Similarly, while there have been advancements to measure gene expression in whole animals, such as microfluidic tools for high-throughput fluorescence-based imaging17,21,55 or even scRNA-seq, it is difficult to tie together gene expression with a meaningful biological function without expansive prior knowledge of the gene's role. Without the development of new, more comprehensive tools, the field is limited to assays measuring either a biological function or gene expression at single timepoints.This work demonstrates that adapting high-throughput, high-content microfluidic platforms to measure both functional and gene expression data can yield meaningful findings and correlations in aging. This strategy should be easily applied to other platforms. For example, microfluidic-based lifespan platforms have already been developed.18,27,56 While they were created to study behavior or physical structures, perhaps incorporating gene expression quantification can further elucidate underlying mechanisms of aging. A well-studied phenotype in C. elegans is a decline in movement with age where young animals move much more than old animals.24,27,57–59 By measuring how other sensorimotor-related genes change with age, it may be possible to identify which genes need to be maintained to prevent the decline in movement and other healthspan metrics. While we adapted our current pipeline to investigate age-related decline in mechanosensation, we hope to establish this work as a framework for future functional aging studies.
4. Material and methods
4.1 Device design and fabrication
The device design is adapted from a previous design30 to interrogate differently aged animals. As C. elegans age, they increase in size and then decrease at the end of their life. To accommodate for this, we designed several variants of the device with different imaging channel widths ranging from 50–75 μm. We found that the day 2 and day 15 animals fit best in the 50 μm device, and day 5 and day 10 animals fit best in the 60 μm device. Since the worms were not immobilized using drugs or cooling, the animals' head or tail can move in the imaging channel of the microfluidic chip. This movement sometimes blurs images. To reduce the movement of the head or tail part of the worms, a three-step vertical tapering of the imaging channel was used. The thickness of the first layer was 20 μm, and the second and third layers were 15 μm for the 50 μm deep imaging channel. The design of the width of the actuated membrane is 150 μm, the distance between the first and second sets of membranes is 200 μm, and the distance between the second and third sets of membranes is 250 μm. The placement of the actuator membrane, combined with the pneumatic valve which controls the animals head position, is designed to deliver a stimulus to the neuronal process directly above the neuron body, allowing for a consistent stimulus and simultaneous imaging of the neuron. The width of the actuator is large relative to potential variability in animal size and neuron position to mitigate the effects of morphological heterogeneity. Additional considerations to actuator positioning are recommended to accommodate for animals with significant changes to morphology.Microfluidic device fabrication has been previously described in greater detail.30 Briefly, the master mold of the microfluidic device was fabricated with SU-8 2015 (Microchem), a negative photoresist using standard UV photolithography. All microfluidic master wafers were then treated with tridecafluoro-1,1,2,2-tetrahydrooctyl-1-trichlorosilane vapor (Sigma-Aldrich). The microfluidic devices were fabricated in polydimethylsiloxane (PDMS, Dow Corning Sylgard 184) by traditional soft lithography. To create the actuated PDMS structure to touch and trap worms, a multi-layer soft lithography process was used. For the bottom flow layer of features, PDMS in a 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio between the elastomer and curing agent was deposited via spin coating to create a thin layer. For the top control layer, 10
1 ratio between the elastomer and curing agent was deposited via spin coating to create a thin layer. For the top control layer, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PDMS was directly poured onto a blank master to create a thick and mechanically rigid handle layer. Both layers were then baked at 70 °C for 30 minutes until the control layer was still underbaked and slightly sticky. The layers were manually aligned and additional 10
1 PDMS was directly poured onto a blank master to create a thick and mechanically rigid handle layer. Both layers were then baked at 70 °C for 30 minutes until the control layer was still underbaked and slightly sticky. The layers were manually aligned and additional 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PDMS was poured and cured for several hours.
1 PDMS was poured and cured for several hours.
4.2 Computational deformation modeling
The COMSOL stationary solver was used to analyze the effect of varying pressure on the deformation of the stimulus actuator membrane. The values for the material properties of the PDMS and C. elegans, such as Young's modulus and density, were taken from several sources and a range of potential PDMS material properties based on a 20% variation in cross-linker ratio and temperature were estimated.60–63Using these literature sources, the range of material properties used was estimated as follows:
To accommodate the extreme deformations produced, both the PDMS and cuticle were modeled as hyperplastic materials. The hyperelastic material model is commonly applied to biological tissues and rubbers and has been previously used for both PDMS and C. elegans.65–68 The model chosen was simplified to follow a non-compressible Neo–Hookean model, where the Lamé parameter was defined as μ = E/3, as the compression of the PDMS membrane was not the point of interest of the study.
A 2D cross sectional view of the imaging channel was used in the study, for both a 50 μm and 60 μm channel width and matching worm diameter. The bottom and top edges of the device were constrained to zero deformation to simulate their attachment to glass and the massive amount of significantly stiffer PDMS surrounding the membrane. The outside faces of the actuators were assigned a boundary pressure, whereas the insides and the entire outside of the worm were paired into contact pairs. The worm was modeled as a ring of hyperelastic material based on the properties of the cuticle. An internal pressure was defined within the ring, increasing in magnitude as the area of the ring decreased according to
| Pint = (0.1 MPa) × (Areat=0/Areaenclosed)1.4 − 1, |
Because compressibility of the worm has been observed in ref. 60, and furthermore the model does not consider the sections of the worm before and beyond the actuator, about 90% of the total length of the animal, which would experience the large majority of the resulting forces and deformation produced by the compression, the interior of the worm is considered to be a compressible fluid rather than an incompressible liquid (i.e. water). The initial pressure value implies that at approximately 40% deformation of the modeled section, about 4% of the entire animal, the internal pressure is 0.1 MPa, which roughly agrees with the observed results in prior literature.60
The pressure applied to the deformable membrane was parametrically increased and contact between the PDMS membrane and the animal was solved for nonlinearly. A cut-line drawn through the center of each membrane was used to extract the distance between the two membranes during deformation and estimate percent deformation.
4.3 Device operation and imaging
All imaging experiments were performed using a Leica DMIRB inverted microscope with a 40× air objective (NA 0.75). Video sequences were captured using a Hamamatsu EM-CCD camera with 100 ms exposure time. Simultaneous two-color imaging was performed using a DV2 beamsplitter (Photometrics) containing a GFP/RFP filter set. Excitation light for fluorescence imaging was delivered through a projector system69 and was kept on during the entire experiment to prevent any light response during stimulus recording. Animals that were not oriented head-first were unloaded without imaging, and only head-first animals were observed. A pneumatic valve at the exit of the imaging area is used to ensure the head of each loaded animal is fixed, allowing for stimulus to be delivered at a consistent position relative to the target neuron. Animals were allowed to rest in the channel for 90s after loading to acclimate to any mechanical stimulus and response to loading and return to an inactive state. Stimuli were delivered 10 seconds after recording the baseline activity of neurons using the pneumatically actuated actuators on the microfluidic device. The operational pressure was calibrated per device and per day to produce a 30% deformation. This magnitude reflects previously used values in existing literature.30For strain validation and qualitative imaging (Fig. 1e), we used a spinning disk confocal microscope (PerkinElmer UltraVIEW VoX) equipped with a Hamamatsu FLASH 4 sCMOS camera and a 60x oil immersion objective.
4.4 Image processing, analysis, and statistics
Fluorescence intensities for each frame were obtained using customized neuron-tracking MATLAB scripts.30 Fluorescence values were computed by subtracting the background intensity (F = IG_ROI − IG_Back). GCaMP intensities were measured as the mean pixel intensity of the 100 brightest pixels of a circular region of interest (ROI) of 20-pixel radius. Background intensities were measured as the mean pixel intensity of an ring surrounding the ROI and were subsequently subtracted. Calcium traces were computed as the change in F, . Baseline values were computed as the mean of F prior to stimulus delivery.
. Baseline values were computed as the mean of F prior to stimulus delivery.
All statistical tests and graphs were performed and plotted in GraphPad Prism. For the amiloride drug screen comparison between treated and untreated animals, we used a one-tailed, unpaired t-test (Fig. 3). When analyzing the differences between peaks with age and gene expression changes (Fig. 4 and 5), we used a Kruskal–Wallis test followed by Dunn's multiple comparison test. To quantify the dynamics of the calcium imaging, one-phase association and one-phase dissociation non-linear curves were fit using GraphPad Prism (Fig. S2, ESI†).
4.5 Animal culture
Worms were grown following standard protocols on NGM agar plates and fed on Escherichia coli (E. coli) OP50 lawns and maintained at 20 °C for at least two generations. For aging studies, synchronized L1 larvae of the F2 generation animals were obtained from a 2 hour hatch-off of each strain and grown on NGM agar plates until L4 stage on separate plates. At L4, animals were transferred to NGM plates supplemented with 5 μM C22, a compound that interrupts eggshell formation and prevents viable progeny without impacting the development or longevity of adults.27,70 We found that the C22 treatment required multiple days before complete interruption of eggshell formation. We therefore designated day 2 adults as the earliest time point for data collection to ensure that the populations were correctly age-controlled. Following this, days 5, 10, and 15 were selected as linearly distributed time points. The smaller gap between day 2 and day 5 was chosen to improve time resolution at earlier ages where changes were expected to occur at a faster rate. Animals were transferred every day for the first 5 days of adulthood (when the majority of eggs are laid) and then transferred every other day until day 10 adults on NGM plates with C22.When treated with amiloride, animals were exposed to a 3 mM amiloride solution. A single dosage of amiloride was applied to the animals directly before experimentation.
4.6 C. elegans strains
The C. elegans strains used in this work were GT372 aSi31[lox2272 Cbr-unc-119(+) lox2272 + pmec-7::jGCaMP7F::ras-2CAAX:SL2:mScarlet-I::ras-2CAAX] II; unc-119(ed3) III, GT406 unc-119(ed3) III; aEx48[pmec-4::mec-4::mNeonGreen (50 ng μl−1) + pmec-10::mec-10::mScarlet-I (50 ng μl−1) + pDSP2 (Cbr-unc-119(+)) (50 ng μl−1)], and GT367 unc-119(ed3) III; aEx45[ser-2p3b::jGCaMP7F::ras-2CAAX::SL2::mScarlet-I::ras-2CAAX (50 ng μl−1) + pDSP2 (Cbr-unc-119(+)) (50 ng μl−1)]. For the MEC-4::mScarlet and MEC-10::mNeonGreen dual reporter strain GT406, the constructs for pmec-4::mec-4::mNeonGreen and pmec-10::mec-10::mScarlet-I were generated using previously published methods.46,47 Briefly, pmec-4::mec-4::mScarlet-I, pmec-10::mec-10::mNeonGreen, and a positive selection marker that rescues unc-119(−) were co-injected at 50 ng μL−1 into QL74 ttTi5605 II; unc-119(ed3) III. For the GCaMP calcium reporter strains GT372 (ALM) and GT367 (PVD), the development of GT367 followed a similar procedure to GT406, except in this case, membrane-localized ser-2p3b::jGCaMP7F and mScarlet-I dual reporter construct was injected at 50 ng μL−1 into QL74 and integrated using71 to develop the single-copy integrant strain GT372.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank Dr. Dhaval S. Patel for technical guidance and mentorship in strain construction, Hyun Jee Lee for assistance in strain injections and screening, and Dr. Gongchen Sun for discussions on the manuscript. This work was supported by NIH R01AG056436 and R01NS096581 to HL, NSF 1764406 to HL, and an NSF Graduate Research Fellowship to JW. HL thanks C. Luthio for moral support.References
- C.-L. Pan, C.-Y. Peng, C.-H. Chen and S. McIntire, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(22), 9274, DOI:10.1073/pnas.1011711108.
- S. K. Hunter, H. M. Pereira and K. G. Keenan, J. Appl. Physiol., 2016, 121(4), 982–995, DOI:10.1152/japplphysiol.00475.2016.
- H. Degens and M. T. Korhonen, Maturitas, 2012, 73(3), 197–201, DOI:10.1016/j.maturitas.2012.07.015.
- B. E. Maki and W. E. McIlroy, Age Ageing, 2006, 35(suppl_2), ii12–ii18, DOI:10.1093/ageing/afl078.
- M. D. Seidman, N. Ahmad and U. Bai, Ageing Res. Rev., 2002, 1(3), 331–343, DOI:10.1016/S1568-1637(02)00004-1.
- A. J. Campbell, J. Reinken, B. C. Allan and G. S. Martinez, Age Ageing, 1981, 10(4), 264–270, DOI:10.1093/ageing/10.4.264.
- T. Yamasoba, F. R. Lin, S. Someya, A. Kashio, T. Sakamoto and K. Kondo, Hear. Res., 2013, 303, 30–38, DOI:10.1016/j.heares.2013.01.021.
- C. Kenyon, J. Chang, E. Gensch, A. Rudner and R. Tabtiang, Nature, 1993, 366(6454), 461–464, DOI:10.1038/366461a0.
- C. J. Kenyon, Nature, 2010, 464(7288), 504–512, DOI:10.1038/nature08980.
- S. R. Wicks and C. H. Rankin, J. Neurosci., 1995, 15(3), 2434, DOI:10.1523/JNEUROSCI.15-03-02434.1995.
- M. Chalfie, J. E. Sulston, J. G. White, E. Southgate, J. N. Thomson and S. Brenner, J. Neurosci., 1985, 5(4), 956, DOI:10.1523/JNEUROSCI.05-04-00956.1985.
- J. G. White, E. Southgate, J. N. Thomson and S. Brenner, Philos. Trans. R. Soc., B, 1986, 314(1165), 1–340, DOI:10.1098/rstb.1986.0056.
- L. R. Varshney, B. L. Chen, E. Paniagua, D. H. Hall and D. B. Chklovskii, PLoS Comput. Biol., 2011, 7(2), e1001066, DOI:10.1371/journal.pcbi.1001066.
- M. Liu, A. K. Sharma, J. W. Shaevitz and A. M. Leifer, eLife, 2018, 7, e36419, DOI:10.7554/eLife.36419.
- D. A. Porto, J. Giblin, Y. Zhao and H. Lu, Sci. Rep., 2019, 9(1), 5182, DOI:10.1038/s41598-019-41349-0.
- J. Wan and H. Lu, Lab Chip, 2020, 20(24), 4528–4538, 10.1039/D0LC00881H.
- K. Chung, M. M. Crane and H. Lu, Nat. Methods, 2008, 5(7), 637–643, DOI:10.1038/nmeth.1227.
- S. Saberi-Bosari, J. Huayta and A. San-Miguel, Lab Chip, 2018, 18(20), 3090–3100, 10.1039/C8LC00655E.
- H. Lee, S. A. Kim, S. Coakley, P. Mugno, M. Hammarlund, M. A. Hilliard and H. Lu, Lab Chip, 2014, 14(23), 4513–4522, 10.1039/C4LC00789A.
- M. M. Crane, K. Chung, J. Stirman and H. Lu, Lab Chip, 2010, 10(12), 1509–1517, 10.1039/B927258E.
- J. Wan, G. Sun, J. Dicent, D. S. Patel and H. Lu, Lab Chip, 2020, 20(2), 266–273, 10.1039/C9LC00896A.
- G. Sun, J. Wan and H. Lu, Biomicrofluidics, 2019, 13(6), 064101, DOI:10.1063/1.5124827.
- P. D. McClanahan, J. H. Xu and C. Fang-Yen, Integr. Biol., 2017, 9(10), 800–809, 10.1039/c7ib00120g.
- M. A. Churgin, S.-K. Jung, C.-C. Yu, X. Chen, D. M. Raizen and C. Fang-Yen, eLife, 2017, 6, e26652, DOI:10.7554/eLife.26652.
- N. Chronis, M. Zimmer and C. I. Bargmann, Nat. Methods, 2007, 4(9), 727–731, DOI:10.1038/nmeth1075.
- M. Rahman, H. Edwards, N. Birze, R. Gabrilska, K. P. Rumbaugh, J. Blawzdziewicz, N. J. Szewczyk, M. Driscoll and S. A. Vanapalli, Sci. Rep., 2020, 10(1), 16190, DOI:10.1038/s41598-020-73002-6.
- K. N. Le, M. Zhan, Y. Cho, J. Wan, D. S. Patel and H. Lu, Commun. Biol., 2020, 3(1), 297, DOI:10.1038/s42003-020-1013-2.
- M. Chalfie and J. Sulston, Dev. Biol., 1981, 82(2), 358–370, DOI:10.1016/0012-1606(81)90459-0.
- O. Hobert, D. G. Moerman, K. A. Clark, M. C. Beckerle and G. Ruvkun, J. Cell Biol., 1999, 144(1), 45–57, DOI:10.1083/jcb.144.1.45.
- Y. Cho, D. A. Porto, H. Hwang, L. J. Grundy, W. R. Schafer and H. Lu, Lab Chip, 2017, 17(15), 2609–2618, 10.1039/C7LC00465F.
- Y. Cho, D. N. Oakland, S. A. Lee, W. R. Schafer and H. Lu, Lab Chip, 2018, 18(4), 601–609, 10.1039/c7lc01201b.
- A. L. Nekimken, H. Fehlauer, A. A. Kim, S. N. Manosalvas-Kjono, P. Ladpli, F. Memon, D. Gopisetty, V. Sanchez, M. B. Goodman, B. L. Pruitt and M. Krieg, Lab Chip, 2017, 17(6), 1116–1127, 10.1039/C6LC01165A.
- M. Backholm, W. S. Ryu and K. Dalnoki-Veress, Proc. Natl. Acad. Sci. U. S. A., 2013, 110(12), 4528, DOI:10.1073/pnas.1219965110.
- K. Hong, I. Mano and M. Driscoll, J. Neurosci., 2000, 20(7), 2575–2588, DOI:10.1523/JNEUROSCI.20-07-02575.2000.
- L. A. Herndon, P. J. Schmeissner, J. M. Dudaronek, P. A. Brown, K. M. Listner, Y. Sakano, M. C. Paupard, D. H. Hall and M. Driscoll, Nature, 2002, 419(6909), 808–814, DOI:10.1038/nature01135.
- Y. Chen, S. Bharill, R. O'Hagan, E. Y. Isacoff and M. Chalfie, G3: Genes, Genomes, Genet., 2016, 6(4), 1121–1130, DOI:10.1534/g3.115.023507.
- M. L. Toth, I. Melentijevic, L. Shah, A. Bhatia, K. Lu, A. Talwar, H. Naji, C. Ibanez-Ventoso, P. Ghose, A. Jevince, J. Xue, L. A. Herndon, G. Bhanot, C. Rongo, D. H. Hall and M. Driscoll, J. Neurosci., 2012, 32(26), 8778, DOI:10.1523/JNEUROSCI.1494-11.2012.
- K. Hong and M. Driscoll, Nature, 1994, 367(6462), 470–473, DOI:10.1038/367470a0.
- M. B. Goodman, D. H. Hall, L. Avery and S. R. Lockery, Neuron, 1998, 20(4), 763–772, DOI:10.1016/s0896-6273(00)81014-4.
- R. O'Hagan, M. Chalfie and M. B. Goodman, Nat. Neurosci., 2005, 8(1), 43–50, DOI:10.1038/nn1362.
- M. B. Goodman, T. H. Lindsay, S. R. Lockery and J. E. Richmond, Methods Cell Biol., 2012, 107, 409–436, DOI:10.1016/B978-0-12-394620-1.00014-X.
- A. Reina, A. B. Subramaniam, A. Laromaine, A. D. T. Samuel and G. M. Whitesides, PLoS One, 2013, 8(7), e69651, DOI:10.1371/journal.pone.0069651.
- M. Huang and M. Chalfie, Nature, 1994, 367(6462), 467–470, DOI:10.1038/367467a0.
- M. Driscoll and M. Chalfie, Nature, 1991, 349(6310), 588–593, DOI:10.1038/349588a0.
- T. Boulin, J. F. Etchberger and O. Hobert, Reporter gene fusions, WormBook: The C. elegans Research Community, 2006 Search PubMed.
- Y. Chen, S. Bharill, E. Y. Isacoff and M. Chalfie, Proc. Natl. Acad. Sci. U. S. A., 2015, 112(37), 11690, DOI:10.1073/pnas.1515968112.
- M. Chatzigeorgiou, L. Grundy, K. S. Kindt, W.-H. Lee, M. Driscoll and W. R. Schafer, J. Neurophysiol., 2010, 104(6), 3334–3344, DOI:10.1152/jn.00330.2010.
- M. Rahimi, S. Sohrabi and C. T. Murphy, Biophys. J., 2022, 121(4), 515–524, DOI:10.1016/j.bpj.2022.01.013.
- A. L. Eastwood, A. Sanzeni, B. C. Petzold, S.-J. Park, M. Vergassola, B. L. Pruitt and M. B. Goodman, Proc. Natl. Acad. Sci. U. S. A., 2015, 112(50), E6955, DOI:10.1073/pnas.1514138112.
- M. B. Goodman, G. G. Ernstrom, D. S. Chelur, R. O'Hagan, C. A. Yao and M. Chalfie, Nature, 2002, 415(6875), 1039–1042, DOI:10.1038/4151039a.
- D. S. Chelur, G. G. Ernstrom, M. B. Goodman, C. A. Yao, L. Chen, R. O'Hagan and M. Chalfie, Nature, 2002, 420(6916), 669–673, DOI:10.1038/nature01205.
- S. L. Geffeney, J. G. Cueva, D. A. Glauser, J. C. Doll, T. H.-C. Lee, M. Montoya, S. Karania, A. M. Garakani, B. L. Pruitt and M. B. Goodman, Neuron, 2011, 71(5), 845–857, DOI:10.1016/j.neuron.2011.06.038.
- J. Liu, B. Zhang, H. Lei, Z. Feng, J. Liu, A.-L. Hsu and X. Z. S. Xu, Cell Metab., 2013, 18(3), 392–402, DOI:10.1016/j.cmet.2013.08.007.
- M. Chatzigeorgiou, S. Yoo, J. D. Watson, W.-H. Lee, W. C. Spencer, K. S. Kindt, S. W. Hwang, D. M. Miller 3rd, M. Treinin, M. Driscoll and W. R. Schafer, Nat. Neurosci., 2010, 13(7), 861–868, DOI:10.1038/nn.2581.
- E. V. Entchev, D. S. Patel, M. Zhan, A. J. Steele, H. Lu and Q. Ch'ng, eLife, 2015, 4, e06259, DOI:10.7554/eLife.06259.
- S. E. Hulme, S. S. Shevkoplyas, A. P. McGuigan, J. Apfeld, W. Fontana and G. M. Whitesides, Lab Chip, 2010, 10(5), 589–597, 10.1039/B919265D.
- C. Huang, C. Xiong and K. Kornfeld, Proc. Natl. Acad. Sci. U. S. A., 2004, 101(21), 8084, DOI:10.1073/pnas.0400848101.
- B. L. Newell Stamper, J. R. Cypser, K. Kechris, D. A. Kitzenberg, P. M. Tedesco and T. E. Johnson, Aging Cell, 2018, 17(1), e12704, DOI:10.1111/acel.12704.
- C. F. Glenn, D. K. Chow, L. David, C. A. Cooke, M. S. Gami, W. B. Iser, K. B. Hanselman, I. G. Goldberg and C. A. Wolkow, J. Gerontol., Ser. A, 2004, 59(12), 1251–1260, DOI:10.1093/gerona/59.12.1251.
- W. Gilpin, S. Uppaluri and C. P. Brangwynne, Biophys. J., 2015, 108(8), 1887–1898, DOI:10.1016/j.bpj.2015.03.020.
- I. D. Johnston, D. K. McCluskey, C. K. L. Tan and M. C. Tracey, J. Micromech. Microeng., 2014, 24(3), 035017, DOI:10.1088/0960-1317/24/3/035017.
- Z. Wang, A. A. Volinsky and N. D. Gallant, J. Appl. Polym. Sci., 2014, 131(22) DOI:10.1002/app.41050.
- C. L. Essmann, D. Martinez-Martinez, R. Pryor, K.-Y. Leung, K. B. Krishnan, P. P. Lui, N. D. E. Greene, A. E. X. Brown, V. M. Pawar, M. A. Srinivasan and F. Cabreiro, Nat. Commun., 2020, 11(1), 1043, DOI:10.1038/s41467-020-14785-0.
- J. Mark, Polymer Data Handbook, Oxford University Press, 1999 Search PubMed.
- L. C. S. Nunes, Mater. Sci. Eng., A, 2011, 528(3), 1799–1804, DOI:10.1016/j.msea.2010.11.025.
- C. Cardoso, C. S. Fernandes, R. Lima and J. Ribeiro, Mech. Res. Commun., 2018, 90, 26–33, DOI:10.1016/j.mechrescom.2018.04.007.
- D. Z. Tansel, J. Brenneman, G. K. Fedder and R. Panat, J. Micromech. Microeng., 2020, 30(6), 067001, DOI:10.1088/1361-6439/ab82f4.
- P. Ciarletta, M. Ben Amar and M. Labouesse, Philos. Trans. R. Soc., A, 2009, 367(1902), 3379–3400, DOI:10.1098/rsta.2009.0088.
- J. N. Stirman, M. M. Crane, S. J. Husson, S. Wabnig, C. Schultheis, A. Gottschalk and H. Lu, Nat. Methods, 2011, 8(2), 153–158, DOI:10.1038/nmeth.1555.
- S. E. Weicksel, A. Mahadav, M. Moyle, P. G. Cipriani, M. Kudron, Z. Pincus, S. Bahmanyar, L. Abriola, J. Merkel, M. Gutwein, A. G. Fernandez, F. Piano, K. C. Gunsalus and V. Reinke, Development, 2016, 143(19), 3540–3548, DOI:10.1242/dev.140046.
- J. Ding, L. Peng, S. Moon, H. J. Lee, D. S. Patel and H. Lu, G3: Genes, Genomes, Genet., 2023, 13(10) DOI:10.1093/g3journal/jkad183.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3lc01080e |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |