 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
SeParate: multiway fluorescence-activated droplet sorting based on integration of serial and parallel triaging concepts†
Wannes
Verbist
 a,
Jolien
Breukers
a,
Jolien
Breukers
 a,
Sapna
Sharma
b,
Iene
Rutten
a,
Hans
Gerstmans
a,
Sapna
Sharma
b,
Iene
Rutten
a,
Hans
Gerstmans
 a,
Lotte
Coelmont
b,
Francesco
Dal Dosso
a,
Kai
Dallmeier
a,
Lotte
Coelmont
b,
Francesco
Dal Dosso
a,
Kai
Dallmeier
 b and
Jeroen
Lammertyn
b and
Jeroen
Lammertyn
 *a
*a
aDepartment of Biosystems – Biosensors Group, KU Leuven, Willem de Croylaan 42, Box 2428, 3001 Leuven, Belgium. E-mail: jeroen.lammertyn@kuleuven.be
bDepartment of Microbiology, Immunology and Transplantation, Rega Institute, Laboratory of Virology and Chemotherapy, Molecular Vaccinology and Vaccine Discovery, KU Leuven, 3000 Leuven, Belgium
First published on 20th February 2024
Abstract
Fluorescence-activated droplet sorting (FADS) has emerged as a versatile high-throughput sorting tool that is, unlike most fluorescence-activated cell sorting (FACS) platforms, capable of sorting droplet-compartmentalized cells, cell secretions, entire enzymatic reactions and more. Recently, multiplex FADS platforms have been developed for the sorting of multi-fluorophore populations towards different outlets in addition to the standard, more commonly used, 2-way FADS platform. These multiplex FADS platforms consist of either multiple 2-way junctions one after the other (i.e. serial sorters) or of one junction sorting droplets in more than 2 outlets (i.e. parallel sorters). In this work, we present SeParate, a novel platform based on integrating ![[s with combining low line]](https://www.rsc.org/images/entities/char_0073_0332.gif)
![[e with combining low line]](https://www.rsc.org/images/entities/char_0065_0332.gif) rial and
rial and ![[p with combining low line]](https://www.rsc.org/images/entities/char_0070_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[r with combining low line]](https://www.rsc.org/images/entities/char_0072_0332.gif) allel sorting principles for accur
allel sorting principles for accur![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[e with combining low line]](https://www.rsc.org/images/entities/char_0065_0332.gif) multiplex droplet sorting that is able to mitigate limitations of current multiplex sorters. We show the SeParate platform and its capability in highly accurate 4-way sorting of a multi-fluorophore population into four subpopulations with the potential to expand to more. More specifically, the SeParate platform was thoroughly validated using mixed populations of fluorescent beads and picoinjected droplets, yielding sorting accuracies up to 100% and 99.9%, respectively. Finally, transfected HEK-293T cells were sorted employing two different optical setups, resulting in an accuracy up to 99.5%. SeParate's high accuracy for a diverse set of samples, including highly variable biological specimens, together with its scalability beyond the demonstrated 4-way sorting, warrants a broad applicability for multi-fluorophore studies in life sciences, environmental sciences and others.
multiplex droplet sorting that is able to mitigate limitations of current multiplex sorters. We show the SeParate platform and its capability in highly accurate 4-way sorting of a multi-fluorophore population into four subpopulations with the potential to expand to more. More specifically, the SeParate platform was thoroughly validated using mixed populations of fluorescent beads and picoinjected droplets, yielding sorting accuracies up to 100% and 99.9%, respectively. Finally, transfected HEK-293T cells were sorted employing two different optical setups, resulting in an accuracy up to 99.5%. SeParate's high accuracy for a diverse set of samples, including highly variable biological specimens, together with its scalability beyond the demonstrated 4-way sorting, warrants a broad applicability for multi-fluorophore studies in life sciences, environmental sciences and others.
1 Introduction
Fluorescence-activated cell sorting (FACS) has become an indispensable tool for high-throughput fluorescence-based cell studies in various fields including, but not limited to, cancer research,1 immunology,2 stem cell research3 and environmental research.4 Despite its widespread use, FACS has intrinsic limitations as it does not allow the sorting of cells based on their secretion products,5 needs sufficiently large cell samples (>105)6 and is limited in its applicability for the enrichment of rare cells.7 Furthermore, due to the stress exerted on the cells, the FACS process might lead to activation of stress pathways and/or cell death8,9 and hence the loss of original biological information. To address these limitations, in the last few decades, fluorescence-activated droplet sorting (FADS) emerged as a technology enabling high-throughput sorting of microfluidic droplets encapsulating (single) entities.5,10,11 Because FADS platforms manipulate and sort intact droplets, they allow for the sorting of cells based on their secretion products and products of enzymatic reactions in addition to fluorescent cells.10 Since every droplet acts as an isolated reaction chamber, FADS enables sorting in highly parallelized experiments where it has been employed either as the sole droplet manipulation technique10,12 or integrated with other complex droplet manipulation techniques such as picoinjection,13,14 droplet merging,15 incubation16,17 and droplet dispensing.18,19 Moreover, in FADS platforms, a high-speed camera enables the visualization of each droplet sorting event which is only recently made possible for FACS with the more expensive FACSDiscover™.20 Due to its versatility, high-throughput and usability in parallelized experiments, FADS is multidisciplinary applicable as shown by its use in the study of directed evolution,21,22 screening of drugs,23,24 diagnostics25,26 and more.FADS platforms are categorized based on droplet deflection principles: acoustic,27,28 magnetic,29,30 pneumatic31,32 and electric33–35 control. Electric control, and more specifically dielectrophoresis (DEP), is the most commonly used method applicable to a wide range of droplet sizes (diameters of a few microns up to 337 μm16,34,36) ultimately allowing for 2-way sorting (for instance one population of interest and one waste population) at throughputs ranging from 4 Hz16 to 30 kHz,33 depending on chip design and droplet size.34 While 2-way sorting may be sufficient for sorting out one population of interest, it does not provide the versatility of a multiway FACS capable of sorting cells into parallel tubes (up to six) and well plates (up to 384 wells).20,37 Currently, most FADS platforms cannot handle samples with more than one population of interest, as required in several applications such as multiplexed DNA detection or multiplexed quantification of pathogens.38–41 To address this need, multiplex high-throughput FADS platforms were developed. These platforms can be subdivided into serial and parallel sorters based on their microfluidic designs. More specifically, serial sorters consist of consecutive 2-way junctions while parallel sorters consist of one junction splitting into multiple outlets (Fig. 1a and b). Serial sorters, using four42 or five43 consecutive 2-way junctions have been described, capable of sorting 100 μm diameter droplets at 3 Hz or 40 μm diameter droplets at a throughput of 700 Hz, respectively. Typically, serial approaches are prone to errors due to differences in the timing of droplet arrival at the sorting junction due to polydispersity or small inaccuracies of the pump setup.6 This is the consequence of the distance between the point of detection and the sorting junction of interest. Alternatively, several parallel sorters have been described, achieving up to 3-way44,45 or 5-way6,46 sorting. In the platform of Caen et al.,6 different DEP forces were implemented to sort droplets (i.e. 45 μm diameter at 200 Hz) to the five different outlets. In the platform of Isozaki et al.,46 a sequentially addressable dielectrophoretic array consisting of a specialized series of electrodes was introduced. This platform employed small DEP forces per electrode, leading to a stepwise displacement. In doing so, the risk of droplet breakage was reduced, avoiding lowered sorting accuracies and altered droplet sizes. Additionally, by allowing multiple sequential droplets to be attracted simultaneously, the electrode array enabled higher throughput sorting for 50 μm droplets (473 Hz) compared to more standard electrode systems, where consecutive droplet attraction is limited due to the previous droplet passing by the whole electrode. Typically, parallel sorting approaches either require a more complex experimental setup (i.e. more complex electronics setup and manual calibration steps),46 or the number of outlets is limited by the amount of deflection achievable by an electrode without risking the electrosplitting of droplets.6,44,45
 | ||
| Fig. 1 Different design strategies for multiplex FADS. The microfluidic channels are represented in light-grey, the working electrodes in red and the ground electrodes in dark-grey. The different sorting outlets used in the respective studies are indicated with numbers. (a) A serial sorting design, adapted from Vyawahare et al.,43 that consists of consecutive 2-way junctions. (b) The design principle of a parallel droplet sorting platform, consisting of one sorting junction splitting into five channels adapted from Caen et al.6 (c) The design of the SeParate platform presented in this work, consisting of serialized 3-way junctions. | ||
Next to custom-made FADS platforms presented in literature, a small number of commercial FADS platforms are available. The Pico-Mine®,47 Cyto-Mine®48 (both from Sphere Fluidics, UK) and CelliGo™ (Nexcelom, USA)49 allow for droplet sorting at a frequency of respectively 300 Hz, 200 Hz and 10 kHz but only support 2-way sorting. The Styx (Atrandi Biosciences, Lithuania)50 and the Modaflow™ (LiveDrop, Belgium)51 respectively allow sorting up to 30 kHz and 2.5 kHz and may in principle be used in a multiway sorting configuration, though not yet documented.
In this work, we propose a microfluidic chip-based multiplex sorting platform by integrating ![[s with combining low line]](https://www.rsc.org/images/entities/char_0073_0332.gif)
![[e with combining low line]](https://www.rsc.org/images/entities/char_0065_0332.gif) rial and
rial and ![[p with combining low line]](https://www.rsc.org/images/entities/char_0070_0332.gif)
![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[r with combining low line]](https://www.rsc.org/images/entities/char_0072_0332.gif) allel sorting principles for accur
allel sorting principles for accur![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif)
![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif)
![[e with combining low line]](https://www.rsc.org/images/entities/char_0065_0332.gif) droplet sorting: SeParate (Fig. 1c), mitigating some limitations of both parallel and serial sorters and providing an alternative methodology to facilitate 4-way sorting to existing platforms (see section 3.5 and Table 1 for an extensive comparison). The SeParate platform is validated in a 4-way configuration for droplets with a diameter of 60 μm at a throughput of approximately 80 Hz. The sorting performance of the SeParate platform is, for a first time, characterized thoroughly using three different model systems with increasing complexity and intra-subpopulation variation in fluorescence intensities; i.e. encapsulated beads, picoinjected droplets and encapsulated living cells. Using these model systems, we illustrate the robustness, flexibility and usability of SeParate for life-science applications. Apart from showing the sorting performance in terms of accuracy, the influence of the sorting threshold and optical system on the performance are evaluated.
droplet sorting: SeParate (Fig. 1c), mitigating some limitations of both parallel and serial sorters and providing an alternative methodology to facilitate 4-way sorting to existing platforms (see section 3.5 and Table 1 for an extensive comparison). The SeParate platform is validated in a 4-way configuration for droplets with a diameter of 60 μm at a throughput of approximately 80 Hz. The sorting performance of the SeParate platform is, for a first time, characterized thoroughly using three different model systems with increasing complexity and intra-subpopulation variation in fluorescence intensities; i.e. encapsulated beads, picoinjected droplets and encapsulated living cells. Using these model systems, we illustrate the robustness, flexibility and usability of SeParate for life-science applications. Apart from showing the sorting performance in terms of accuracy, the influence of the sorting threshold and optical system on the performance are evaluated.
| Serial sorters | Parallel sorters | SeParate | |||||
|---|---|---|---|---|---|---|---|
| Frenzel et al.42 | Vyawahare et al.43 | Girault et al.44 | Blaha et al.45 | Caen et al.6 | Isozaki et al.46 | ||
| a The works of Caen et al.6 and Isozaki et al.46 report sorting accuracy in terms of sorting actuation efficiency, a method based on comparing the droplet's fluorescent signal to the generated on-chip pulse, potentially resulting in an overestimation of the sorting performance. Other works reported the sorting accuracy by checking the sorted droplet populations after sorting using imaging or by reculturing cells. b This number is given by the amount of used outlets, including the channel (often referred to as waste channel) for negative droplets. | |||||||
| Throughput (Hz) | 3 | 700 | 10 | 100 | 200 | 473 | 80 |
| Sorting accuracy | Up to 97.4% for droplets with fluorescent dye | Up to 98.1% for beads | Up to 91% for cells | Up to 97% for cells | Up to 98.4% for droplets with fluorescent dyea | Up to 100% droplets with fluorescent dyea | Up to 100% for beads |
| Up to 95% for cells | Up to 100% for cellsa | Up to 99.9% for picoinjected droplets | |||||
| Up to 99.5% for cells | |||||||
| Degree of multiway sorting presentedb | 4 | 6 | 3 | 3 | 5 | 5 | 4 |
| Scaling methodology | Adding 2-way junctions | Adding parallel channels and using stronger DEP forces | Adding parallel channels and adjusting dSADA (more electrodes…) | Adding 3-way junctions | |||
| Consideration when scaling | Differences in the timing of droplet arrival | Electrosplitting | Electrosplitting, complexity of chip fabrication | Differences in the timing of droplet arrival | |||
| Setup complexity | Medium | High | Medium | Low | Low | High | Medium |
| Standard electronics setup | Custom electronics setup | Custom electronics setup | Standard electronics setup | Standard electronics setup | Custom electronics setup | Standard electronics setup | |
| Manual calibration of delays | Manual calibration of delays | No manual calibration reported | Minimal manual calibration | No manual calibration reported | Extensive manual calibration of delays, pulse time and pulse strength | Manual calibration of delays | |
2 Material and methods
2.1 Reagents and materials
SU-8 2075 photoresist was purchased from Chimie Tech Services (France) and 3-inch silicon wafers from Microchemicals GmbH (Germany). DC Sylgard 184 elastomer was purchased from Farnell (Belgium) for polydimethylsiloxane (PDMS) production. From Sigma Aldrich (Belgium), propylene glycol methyl ether acetate (PGMEA), phosphate-buffered saline (PBS) and trichloro(1H,1H,2H,2H-perfluorooctyl)silane were bought. ThermoFisher Scientific (USA) supplied Alexa Fluor 488 (AF 488), Alexa Fluor 568 (AF 568), fetal bovine serum (FBS) and microfluidic tubing with inner diameters of 0.56 and 0.30 mm with respective outer diameters of 1.07 and 0.75 mm. HEK-293T (human embryonic kidney; CRL-3216) cells were obtained from ATCC (USA). Yellow (i.e. green fluorescent) and Nile Red (i.e. red fluorescent) beads were purchased from Gentaur (Belgium) and rainbow (i.e. green + red fluorescent) beads from PolyAn (Germany). HFE-7500 oil from 3M (Belgium) was used to dilute 008-FluoroSurfactant from RAN biotechnologies (USA) to 2% (w/w). Fast read cell counting chambers with counting grids were obtained from VWR (Belgium). Calcium soda glass slides with a thickness of 1 and 0.13–0.16 mm (thickness no. 1) and Sterican needles with 0.60 mm external diameter were obtained from Carl Roth Gmbh (Germany). Biopsy punches of 0.75 and 1 mm in diameter were acquired from BAP Medical B.V. (Netherlands).2.2 Microfluidic chip fabrication
The microfluidic chips were fabricated following a standard soft-lithography protocol.52 In short, a 3 inch wafer was cleaned using acetone and dried, after which it was spin-coated with SU-8 2075 for 35 s at 4000 rpm to result in a layer of 60 μm height. Subsequently, the wafer was baked at 65 °C for 3 min followed by 7 min and 20 s at 95 °C. UV exposure at 180 mJ cm−2 was performed while a chromium-on-glass photomask was positioned on top of the wafer. Finally, the wafer was baked at 65 °C for 3 min, followed by 6 min and 30 s at 95 °C and developed for 6 min in PGMEA. For the 40 μm high chips, the cleaned 3 inch wafer was spin-coated with SU-8 2025 for 35 s at 2000 rpm and baked at 65 °C for 3 min followed by 6 min at 95 °C. The chromium-on-glass photomask was positioned on top of the wafer and exposed at 160 mJ cm−2. The final bake was performed at 65 °C for 1 min, followed by 6 min at 95 °C, followed by 5 min of PGMEA development.Then, PDMS was mixed with curing agent in a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio and desiccated for 20 min. After casting the desiccated PDMS on the mold, the PDMS was baked at 65 °C for at least 3 h. After hardening, the PDMS slab was removed from the mold and holes were punched at the location of inlets (using a 1 mm puncher) and outlets (using a 0.75 mm puncher). Finally, the PDMS slab and glass slide were activated in a plasma oven (Blackhole Lab, France) for 2 min at high power, followed by application of the PDMS slab on the glass and baking for at least 2 h at 65 °C. This method was used for all the microfluidic chip designs in combination with 1 mm glass slides except for sorting chips used in the cell sorting experiments where 0.13–0.16 mm glass slides were used to be compatible with the short working distance objective.
1 ratio and desiccated for 20 min. After casting the desiccated PDMS on the mold, the PDMS was baked at 65 °C for at least 3 h. After hardening, the PDMS slab was removed from the mold and holes were punched at the location of inlets (using a 1 mm puncher) and outlets (using a 0.75 mm puncher). Finally, the PDMS slab and glass slide were activated in a plasma oven (Blackhole Lab, France) for 2 min at high power, followed by application of the PDMS slab on the glass and baking for at least 2 h at 65 °C. This method was used for all the microfluidic chip designs in combination with 1 mm glass slides except for sorting chips used in the cell sorting experiments where 0.13–0.16 mm glass slides were used to be compatible with the short working distance objective.
2.3 Generation of different droplet populations
A population of approximately 2 × 106 droplets was generated and temporarily stored in an oil-filled reinjection vessel that consisted of an upside-down 1.5 mL Eppendorf tube (VWR, Belgium) glued to a 1 mm glass slide with two openings, as previously reported.53 One opening served as the inlet for droplets during production and outlet during reinjection, while the other side opening was connected to a flow sensor for flow control in the storage vessel.
Droplet production and droplet picoinjection were performed on the same chip at a channel height of 40 μm (detailed chip design is represented in Fig. S2†), with droplets produced at 300 Hz using flow rates of 2 and 20 μL min−1 for the water and oil phase, respectively, aiming at a droplet population with a diameter of 60 μm. After production, droplets passed 3 injectors which were, from top to bottom, respectively filled with AF 488 (i.e. green fluorescent dye), AF 568 (i.e. red fluorescent dye) and PBS (Fig. S4a†). In one cycle of 42 minutes, 3 sweeps between 0.6 and 1.4 μL min−1 of 2 minutes each were performed to produce droplets with varying concentrations and combinations of green, red and green + red fluorescence (Fig. S4b†). To end up with the same injected volume per droplet, the sum of the injector flow rates was kept constant at 2 μL min−1. More specifically for green or red droplet generation, the injector containing respectively green or red fluorescent dye was swept between 0.6 and 1.4 μL min−1 while injector 3, containing PBS, was swept inversely from 1.4 to 0.6 μL min−1. For the green + red droplets, one of the two dyes was swept from 0.6 to 1.4 μL min−1 while the other one was swept from 1.4 to 0.6 μL min−1. In between sweeps, only PBS was injected for 12 minutes at a flow rate of 2 μL min−1. To stop injecting, the equilibrium pressure of the injector, pre-calibrated as described by Breukers et al.,53 was set and the corresponding electrode was switched off. A population of approximately 2 × 106 picoinjected droplets was stored in the storage vessel before being reinjected in a sorting chip, as described in section 2.3.1.
2.4 Droplet sorting
After positioning the sorting chip on the stage of an inverted Olympus IX-73 microscope (Japan), 0.75 mm tubing was connected to each sorting outlet and put in a separate 1.5 mL Eppendorf for droplet capture after sorting. The earlier prepared droplet population (see section 2.3) was reinjected from the storage vessel into the sorting chip at a flow rate of 0.5 μL min−1. During reinjection, droplets were equally spaced by an oil phase flown at 15 μL min−1. The region of LED excitation (CoolLED, pE-300white, UK) was tailored by an adjustable field-stop towards the size of one droplet (i.e. the interrogation zone), eliminating crosstalk between droplets (Fig. 2a). Fig. 2b presents an overview of the optical setup. LED excitation and emitted fluorescence from droplets passing by were guided through a triple bandpass filter (380/55–470/30–557/35 nm, Chroma Technology, USA). The emitted fluorescence was then separated by a dichroic mirror (590 nm, Chroma Technology) towards the 2 photon multiplier tubes (PMTs) (PMM02, Thorlabs, USA), equipped with emission filters for detection of green (525/50 nm, Chroma Technology) and red (590 nm long pass (LP) or 638 nm LP, Chroma Technology) fluorescence for PMT 1 and PMT 2, respectively. During the bead and picoinjected droplet sorting experiments, a 20× (numerical aperture (NA) 0.45, working distance (WD) 6.60–7.80 mm, Olympus) objective was used in combination with the 638 nm LP filter in front of PMT 2. During cell sorting, a 40× (NA 0.95, WD 0.18 mm, Olympus) objective was used, and either the 638 nm or 590 nm LP filter was used. | ||
| Fig. 2 The sorting setup used. (a) Droplets are reinjected from the left and spaced by the oil phase. After reaching the interrogation zone, indicated with a black circle, droplets with green fluorescent content are attracted by electrode 1 at sorting junction 1 and attracted again at junction 2 by electrode 2. Droplets with red fluorescent content are attracted by electrode 1 at junction 1 and droplets with green + red fluorescent content by electrode 3. In these experiment, electrode 4 was not used, but would theoretically enable up to 5-way sorting. Droplets that were never attracted ended up in the waste channel. (b) Schematic representation of the optical setup. Adapted from Breukers et al.53 (c) Overview of the equipment. Black arrows illustrate the electronic connections linking the different modules of the setup. Created with https://BioRender.com. | ||
During sorting, the output signal of both PMTs was fed to an Arduino (UNO Wifi) which ran a custom-made program to compare the incoming voltage with a user-set threshold (Fig. 2c). In case the signal exceeded the threshold, the Arduino triggered the pulse generator (TGP110, AIM-TTi, UK) after which the pulse was amplified by a voltage amplifier (TREK 2220-ce, Acal bfi, Belgium). This resulted in a 30 kHz squared pulse of 5 ms at 700 Vpp. The pulse was passed on to the on-chip electrode channels (filled with 5 M NaCl) via alligator clamps connected to a needle and syringe for DEP-based attraction. Apart from generating the triggers, the Arduino was programmed to open and close 3 relays (PLA 171, RS components, Belgium) which were positioned in the path between the high-voltage amplifier and their respective on-chip electrodes (electrodes 1–3, Fig. 2a and c) to separately control electrode actuation, resulting in the ability to perform 4-way sorting. This way, a high-voltage pulse was directed to electrode 1 in the case of a droplet with green fluorescent content, followed by a delayed pulse of electrode 2, guiding these droplets to the green channel (Fig. 2a). This time delay was determined once for every chip before the start of an experiment by visual confirmation of droplet arrival at sorting junction 2 using a high-speed camera (Phantom Micro C110, Vision Research, USA). For sorting of droplets with red fluorescent content to the red channel, only electrode 1 was triggered and for droplets with green + red fluorescent content, electrode 3 was triggered. The detailed design of the sorting chip is depicted in Fig. S3 of ESI.† PMT output data was recorded using a data acquisition system (DAQ, NI USB-6002), running an in-house developed LabVIEW (National Instruments, USA) script and analyzed in Matlab (Mathworks, USA).
2.5 Droplet imaging and statistical analysis
The analysis of droplets from the population before sorting was performed by imaging the content of the reinjection vessel and the post-sorting analysis of droplets by imaging droplets from the tubing connected to the outlet as droplets accumulated there instead of going into the Eppendorf. Droplets were loaded on the counting chamber and imaged on an inverted microscope (Nikon Ti-Eclipse, Japan) using a 10× (NA 0.25, WD 10.5 mm, Nikon) objective with 1× or 1.5× internal magnification. Imaging was performed using a CoolLED (pE-300white) and FITC and TRITC filter cubes. To accurately determine the fluorescence intensity, the shading correction tool of NIS elements software was used (considering as reference a sample with uniform fluorescence at a similar intensity of the sample). The fluorescence intensity was measured using ImageJ 2.9.0 (National Institutes of Health, USA), and processed in Matlab and Excel (Microsoft, USA).Droplet diameters were determined by analysis of bright field images in Matlab after filtering out droplets deviating 10% from the average diameter due to droplet merging and/or breaking in the post-processing. Sorting performance was assessed based on the sorting accuracy, defined as the fraction of correctly sorted droplets over the total amount of droplets. For the accuracy assessment, doublets (i.e. droplets containing two beads or cells), were classified as droplets containing one bead or cell with a fluorescent signal of both beads or cells combined. For the determination of the sorting accuracy of the picoinjected droplets, droplets deviating 10% from the average droplet area were filtered out to ensure potential picoinjection inconsistencies would not influence the sorting performance. Significant differences between conditions were determined using JMP Pro 16 (UK) by running a generalized linear model with logit link function on the weighted average of the repeated measures as the categorical response variable is binomially distributed.57
3 Results and discussion
3.1 Sorting chip features and operation
We developed the SeParate platform, a chip-based FADS platform that allows the sorting of droplets encapsulating particles or dyes based on two fluorescent labels into four subpopulations (green only, red only, green + red and no fluorescence). Our approach employs a design that is based on serially splitting one channel into three (Fig. 3a). The design was made as such that when a droplet reaches a junction and is not attracted by the electrodes, it will flow towards the centered channel (i.e. waste channel at junction 1, and the red or green + red channel at junction 2) due to the lower resistance in that channel as a consequence of the larger dimensions. By splitting into three channels, we aimed to: (1) balance the pressures, ensuring there is no preference for non-attracted droplets to deviate from the centered channel, and (2) sort the droplets in either off-centered channel (i.e. towards junction 2 and the green channel at junction 2) using similar pulse amplitudes. As shown in Caen et al.,6 sorting with one pulse amplitude is not possible with more than three parallel channels as a higher pulse amplitude is required to deviate droplets further from the channel center.Droplet spacing by the spacer oil (Fig. 2a) ensured the arrival of one droplet at a time at the interrogation zone (indicated with a black circle in Fig. 3a–c) at a throughput of approximately 80 Hz. As the delay between droplet detection and attraction at junction 1 was observed to be negligible (<1 ms), the interrogation zone was positioned in correspondence with the tip of electrodes 1 and 3, aiming for efficient attraction from the beginning of the sorting junction onwards. Once attracted at junction 1 (Fig. 3b and c, Movie S1 and S2†), droplets can be attracted again at sorting junction 2 towards the green channel by electrode 2 (Fig. 3d, Movie S3 and S4†). Due to the distance between the interrogation zone and junction 2 (i.e. 1024 μm), the time delay (40.4 ± 2.7 ms, average of 5 different chips) was measured and adjusted in the Arduino software for every chip at the start of the experiment. While this chip design theoretically allows up to 5-way sorting by using electrode 4, the following experiments were performed using 4-way sorting as the samples consisted of four distinct subpopulations (i.e. showing green, red, green + red of no fluorescence).
3.2 Sorting of a mixed bead population
For the first characterization of the SeParate platform, three types of fluorescent beads (green, red and green + red) were encapsulated to produce a droplet population showing highly intense fluorescent signals with little intra-subpopulation variation in fluorescence intensity (see Section S3 of ESI†). Droplets with an average diameter of 56.9 ± 1.2 μm were generated and subsequently reinjected and sorted. The population consisted of 4.1% loaded droplets with one of the three bead types, a lower loading efficiency than the estimated 13.5% using Poisson statistics.58 This is a likely consequence of diluting the beads in the microfluidic loading reservoir over time by constant delivery of fresh PBS.Sorting of this mixed bead population was performed using thresholds of 2 and 1.6 V for green and red signals, respectively, which was well above background fluorescence (for details see Fig. S7a†). Fig. 4a illustrates a typical outcome of such a sorting experiment and Fig. 4b the weighted average sorting accuracy with correctly sorted droplets indicated as ‘success’ while any other droplet was indicated as ‘fail’. In general, sorting resulted in subpopulations with high accuracies of 96.5 ± 0.4% for the green channel, 96.4 ± 0.4% for the red channel, 100% for the green + red channel and 99.8 ± 0.1% for the waste channel (more details in Table S1†), which is more accurate than reported before in for instance Frenzel et al.42 (84.5–97.4%) and Vyawahare et al.43 (91.3–98.1%) for 3-way fluorescently-dyed droplet sorting and 5-way mixed bead sorting, respectively. This comparison considers only studies with similar methods of determining sorting accuracy (i.e. by studying the sorted droplet populations after sorting) in contrast to Caen et al.6 and Isozaki et al.,46 where accuracies were determined by the sorting actuation accuracy (i.e. comparing the droplet's fluorescent signal to the generated on-chip pulse), which could result in an overestimation of the sorting performance as long-term effects affecting the sorting (e.g. dirt on-chip, inconsistency with droplet reinjection) might be overlooked.42
 | ||
| Fig. 4 Outcome of sorting of a droplet population with 3 encapsulated bead types. (a) Overlays of bright- and widefield images of droplets retrieved from the indicated microfluidic channels (i.e. whereto they were sorted). Green + red fluorescent beads appear as yellow due to the overlaying. Scale bar = 100 μm. (b) Droplet sorting accuracy obtained from imaging droplets retrieved from their respective microfluidic channels. For every channel more than 355 droplets were analyzed (Table S1†). The error bars indicate one standard error of the mean (n = 3). | ||
3.3 Sorting of a population of picoinjected droplets with time-variant settings
To show the versatility of the SeParate platform and the capability to sort samples with higher intra-subpopulation variation in fluorescence intensity (see Section S3 of ESI†), droplets with varying contents of fluorescent dye(s) were generated. For this purpose, we used a recently described serial picoinjector technology53 that enables the generation of highly monodisperse droplets with various fluorescent dye combinations and concentrations using a single chip in a semi-automated way.After picoinjection, the resulting droplet population had an average diameter of 61.1 ± 1.5 μm and consisted of droplets showing varying green fluorescence (5.4%), droplets showing varying red fluorescence (4.4%), droplets showing varying green + red fluorescence (4.1%) and 86.1% non-fluorescent droplets (Fig. 5a). This was in line with the targeted populations (see section 2.3.2). Fig. 5b shows the green and red fluorescence intensities of such a sample of the population where every dot represents one droplet. Four subpopulations can be distinguished: a non-fluorescent subpopulation consisting of droplets injected with only PBS, a green subpopulation, a red subpopulation and a subpopulation of droplets containing both green and red dye. The latter shows a linear relationship between green and red fluorescence intensity, which is to be expected as the injection flow rate of the two dyes were inversely proportional related. The picoinjected droplet population was sorted at different thresholds to show the effect of the threshold on the sorting performance for a sample with increased intra-subpopulation variation in fluorescence intensity. Three different thresholds were evaluated: 4, 2.5 and 1 V for both the green and red PMT signals as is shown in Fig. S5, S6† and 5. From these experiments, it was clear that lowering the threshold improved the accuracy significantly with 1 V being the best condition at sorting accuracies of 99.1 ± 0.2% for the green channel, 98.6 ± 0.2% for the red channel, 99.9 ± 0.1% for the green + red channel and 99.9 ± 0.1% for the waste channel (Fig. 5c).
 | ||
| Fig. 5 The droplet population created by three time-variant picoinjectors before and after sorting. (a) Wide- and brightfield images of a sample of the picoinjected droplet population before sorting, visualizing differences in intensity between droplets. In the overlay image, green + red fluorescent droplets appear as yellow. (b) The fluorescence intensities of every imaged droplet which shows the effect of time-variant picoinjection as there are four distinct populations present: a green, a red and a green + red subpopulation and one non-fluorescent population, of which the first three are spread between low and high fluorescence intensities. Dot colors represent the channel from which a droplet was retrieved after sorting with a threshold of 1 V. The dotted lines correspond to the fluorescence intensity value below which 99% of the droplets from the green and waste channel (for the horizontal line) or the red and waste channel (for the vertical line) are located. (c) Sorting accuracy for all 4 channels a threshold of 1 V. For every channel, at least 2682 droplets were analyzed (Table S3†). Error bars represent one standard error of the mean (n = 3). | ||
The inverse correlation between an increased accuracy and a decreasing threshold is clear from Fig. S5a, b† and 5. A too high threshold results in a lack of detection of droplets of lower fluorescence intensity and thus their wrong identification, whereas a too low threshold (below 1 V) would result in every droplet being sorted towards the green + red channel. The accuracies for a threshold of 1 V are similar (or even slightly better) to the ones presented in section 3.2 indicating that an increasing intra-subpopulation variation in fluorescence intensity, introduced by time-variant injection of fluorescent dyes, does not influence the performance of the sorter when an optimized threshold is used.
3.4 Sorting of transfected cells
Finally, the SeParate platform was used to sort encapsulated plasmid-transfected HEK-293T cells. The thus studied cell population showed a high heterogeneity. Successfully transfected cells, expressing the viral construct, displayed both green and red fluorescence. Cells showing only green fluorescence resulted from the uptake of plasmid, but no expression of the viral construct. Cells with only red fluorescence are the result of an infection by viral particles produced and released in the culture medium by other cells. Whereas the majority of cells appeared colorless, i.e. neither transfected nor infected, respective fluorescence intensities depend on multiple factors such as (1) plasmid uptake in the cells, (2) permissiveness of individual cells in expressing the viral construct, or (3) timing and susceptibility of individual cells in the sample to be infected. This resulted, on top of the four possible combinations of fluorescent signals, in a high intra-subpopulation variation in fluorescence intensities (see Section S3 of ESI†) with the corresponding PMT signals ranging from just above the background until PMT saturation.Cell encapsulation resulted in a population of 57.9 ± 1.0 μm droplets of which 0.4% were loaded with green fluorescent cells, 2.1% with red fluorescent cells and 0.4% with green + red fluorescent cells. This is in line with what was observed in a cell sample before encapsulation, where red fluorescent cells were 4.3 times more present than green fluorescent cells and 3.5 times more than green + red cells (Fig. S9†).
As in general the fluorescence intensity of the cells was low, cell sorting was performed using the 40× short working distance objective allowing for more sensitive detection. Further, the threshold for these experiments was set right above the background signal of the PMT, aiming for the detection of as many cells as possible. Cell sorting was performed with a 638 nm LP filter in front of PMT 2 (Fig. 2b) using thresholds between 3.6 and 4 V (adjusted if needed due to small changes in background fluorescence) for green fluorescent and 2 V for red fluorescent signal. Sorting results are illustrated in Fig. 6a, showing accuracies of 69.9 ± 1.2% for the green channel, 99.1 ± 0.2% for the red channel, 97.1 ± 0.4% for the green + red channel and 98.4 ± 0.2% for the waste channel. The lower accuracy in the green channel compared to the others is explained by the strict filtering at PMT 2, resulting in only the more intense red fluorescent cells reaching above the background and thus the sorting threshold (Fig. S10†). This ultimately resulted in a fraction of green + red cells, namely those with low red fluorescence intensity, getting sorted to the green channel (Fig. 6b–d), and some low intense red cells ending up in the waste channel. Moreover, in general, a higher number of empty droplets were wrongly sorted (Table S4†) compared to the sorting experiments presented in sections 3.2 and 3.3. This is explained by the thresholding strategy, as setting the threshold just above the background might lead to wrong sort events due to small disturbances (e.g. electrical noise on the Arduino).
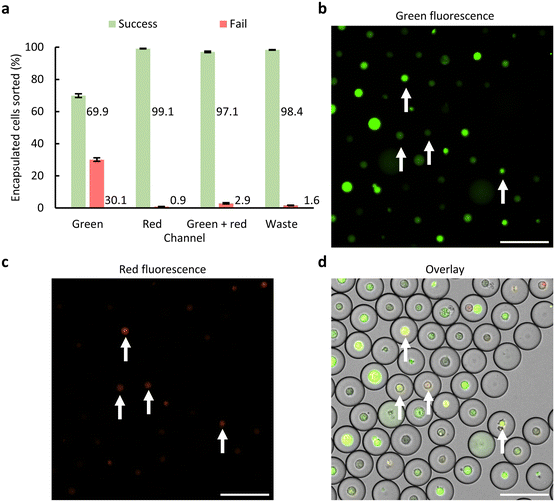 | ||
| Fig. 6 Sorting performance of the encapsulated cell population using the 638 nm LP setup. (a) Sorting accuracy for the 4 channels determined by analyzing at least 1446 droplets per channel (Table S4†). Error bars indicate one standard error of the mean (n = 3). (b and c) Widefield images of droplets retrieved from the green channel, white arrows indicate green + red fluorescent cells that were sorted wrongly due to the more strict filtering of PMT 2, resulting in weak fluorescent red signal not to be picked up. (d) Overlay of the widefield images and the brightfield image where green + red cells appear as yellow. Scale bar = 100 μm. | ||
To reduce the sorting errors in the green and waste channels, the optical setup was adjusted by placing a 590 nm LP filter in front of PMT 2, allowing for more red fluorescent signal to pass to PMT 2. Thresholds between 2.8 V and 3.8 V for green and between 3 V and 5.6 V for red fluorescent signals were used. This setup resulted in an increased accuracy for the green (92.8 ± 0.7%) and waste channels (99.5 ± 0.1%), while the red channel showed a similar accuracy as before, as high as 96.6 ± 0.3% (Fig. 7a). However, the green + red channel showed a decreased sorting accuracy of 84.8 ± 0.8%, a consequence of wrongly sorted green fluorescent cells with high intensities (Fig. 7b–d). Their presence is explained by the bleed-through of the fluorescent signal of highly intense green cells in PMT 2 due to overlapping emission spectra59 in combination with the low thresholds used, allowing for this signal to be picked up in PMT 2 (details in Fig. S10 and S11†). Similar to the sorting experiment with 638 nm LP, there were in general more empty droplets sorted due to the threshold being close to the background of the PMTs. Note that cell viability, although crucial for certain types of downstream processing, was not measured as the focus of these experiments was the performance characterization of the sorting platform.
 | ||
| Fig. 7 Sorting performance of the encapsulated cell population for the setup using 590 nm LP. (a) Sorting accuracy for the 4 channels, where each one is represented by at least 1316 droplets (Table S5†). Error bars indicate one standard error of the mean (n = 3). (b and c) Widefield images of droplets retrieved from the green + red channel, white arrows indicate bright green fluorescent cells that were sorted wrongly due to bleed-through. (d) Overlay of the widefield images and the brightfield image of the droplets with green + red fluorescent cells appearing as yellow. Scale bar = 100 μm. | ||
As a consequence of the challenging characteristics of this cell population in terms of optical detection (i.e. overlap of emission spectra of fluorophores), both tested optical setups (590 nm and 638 nm LP) did not result in high accuracies for respectively the green + red and green channels. As this is an issue linked to the model system and setup optics, yet not to the SeParate platform itself, potential solutions entail: (1) using different fluorophores with more separated emission spectra, (2) using sequential excitation,43,60 (3) applying spectral compensation to correct for this60,61 or (4) by implementing modulated excitation sources, synchronized with the acquisition setup.62 Another solution, that does not require a change in model system nor optical setup, is (5) running the sample on both 590 nm and 638 nm LP setups, which would result in proper separation as well since three out of four channels showed high accuracy.
3.5 Positioning in the field of multiplex sorting
In this section, we compare the SeParate platform to other multiplex droplet sorting platforms. An overview of multiple key parameters can be found in Table 1. We want to note that, while sorting throughput and accuracy are major characteristics of a sorting platform, comparing platforms on only these parameters can be misleading due to a number of reasons. First, the droplet size, which is different between the presented studies, has a major impact on the throughput that can be achieved.34 Larger droplets namely require stronger DEP forces to allow enough displacement from their original path and since stronger DEP forces potentially lead to electrosplitting of droplets, lower throughputs (and thus longer droplet actuation) are often implemented to avoid this. Second, the readout system and the accompanying characteristics might influence throughputs and accurate signal capture (e.g. microprocessors with higher sample rates allow faster and more accurate signal capture and processing). Third, in some studies the sorting accuracy is calculated based on the on-chip droplet actuation after signal detection, instead of capturing all droplets and characterizing the sorted populations (e.g. via imaging). We believe that the latter approach, which was used in this work, is most suitable to characterize the actual sorter's performance, and that the former approach might lead to overestimation of droplet sorting accuracies as long-term effects affecting the sorting (e.g. dirt on-chip, inconsistency with droplet reinjection) are overlooked. Finally, the used sample might influence the performance (e.g. dyed droplets compared to encapsulated cells or beads are generally easier to detect due to a more uniform and wider signal).The SeParate platform, presented in this work, enabled highly accurate droplet sorting (slightly higher than presented in other works) and in a 4-way configuration, which is a degree of multiway sorting that is in line with earlier presented platforms, for three different samples ranging from low to high intra-subpopulation variation and from a low to a high biological relevance. This in-depth study of the sorting performance using model systems with, well-studied, different properties shows SeParate's reliability, reproducibility (experiments were performed in triplicate) and broad applicability. In the presented work, the sorting platform was characterized more thoroughly than most earlier presented works which were shown for the sorting of just one6,42,44,45 or maximally two43,46 samples.
Typically, serial sorters allow scaling beyond the presented degree of multiway sorting by adding additional 2-way junctions.43 As a result, the manual calibration will be more extensive and the susceptibility to differences in droplet arrival timing (e.g. due to polydispersity, inaccuracies of the pump setup…) might increase, resulting in lower sorting accuracies. Parallel sorters, on the other hand, are expected to have limited scalability. To allow upscaling of the platforms of Girault et al.,44 Blaha et al.45 and Caen et al.,6 stronger DEP forces would be required to attract droplets, introducing the risk of droplet electrosplitting. The dSADA46 potentially allows for upscaling without electrosplitting, however this will require significant changes to the chip design and/or the electrode design which are yet to be shown. An additional downside of the latter is the required complex setup (handling) which might prove difficult to implement in environments less-specialized in electronics.
The SeParate platform consists of serialized 3-way junctions and is as such able to combine some advantages of both serial and parallel platforms. SeParate is scalable beyond the presented degree of multiway sorting by adding additional 3-way junctions. Similar to serial sorters, this will result in a more extensive calibration and potentially more issues with differences in droplet arrival timing. However, as every junction is a 3-way junction, this will be less pronounced compared to conventional serial sorters since the amount of junctions will be significantly lower for the same amount of outlets (e.g. SeParate would require 3 sequential junctions for 19 outlets, while other serial sorters would require 18 junctions to get to 19 outlets). This scaling can be done, in contrast to most parallel systems, without introducing electrosplitting as no stronger DEP forces are required to add outlets, and without complexifying the setup and/or chip design drastically.
To summarize, while the presented platform is able to mitigate some limitations of serial and parallel sorters, it is not resolving all of them (e.g. susceptibility to differences in droplet arrival). Therefore we believe that depending on the characteristics of the sample and the setup, researchers should choose a platform based on the relevant characteristics (scalability, number of outlets, setup complexity…) as presented in Table 1.
4 Conclusions
To show the robustness of the SeParate platform, it was, for a first time, extensively characterized using three model (bio)systems of increasing complexity due to an increasing intra-subpopulation variation within the to be sorted specimen. Initial characterization of the SeParate platform was performed using a population consisting of three types of bead with well-defined high and homogenous fluorescence intensities. Here, we reached sorting accuracies of above 96.4%. In the next step, the platform was challenged using a sample with a higher level of intra-subpopulation variation regarding fluorescence intensities as droplets were created using picoinjection of varying amounts and combinations of fluorescent dyes while sweeping. Different sorting thresholds (4, 2.5 and 1 V) were tested, leading to accuracies above 98.6% for all channels for the optimal threshold of 1 V.Finally, the SeParate platform was characterized for plasmid-transfected HEK-293T cells characterized by an overall relatively low fluorescence and high biological heterogeneity requiring the use of a more sensitive 40× objective and low sorting thresholds. Particularly challenging was the fact that the population showed a high variation in fluorescence intensity ranging from just above the background until PMT saturation. Two optical setups were tested, resulting in accuracies ranging from 92.8% to 99.5% for the channels not affected by the optical limitations this setup and model system posed. As for these fluorophores, none of the tested optical setups resulted in high accuracies for all channels, different strategies to improve the sorting accuracy even further were discussed, such as using different fluorophores, using sequential excitation sources, applying spectral compensation or by implementing modulated excitation sources, synchronized with the acquisition setup. The main requirement for a strategy to be viable is that the operation and/or calculation speed is fast enough to detect, process and sort every droplet of interest. With this detailed study, showing highly accurate sorting capabilities for different sample types, we hope to inspire other researchers in the field to thoroughly characterize novel droplet sorters, which will facilitate more objective comparisons between different sorting platforms in the future.
Apart from allowing single-cell studies in a multiplexed configuration,38,63 we anticipate broad applicability in other fields such as directed evolution and enzymatic reaction studies.64 Further, we show flexibility in the thresholding, allowing sorting based on the level of intensity in addition to sorting based on a fixed fluorescent signal, a potentially interesting additional feature in multiplex sorting studies e.g. for fluorescence intensity-based enzyme studies.10 Finally, the capacity of droplet microfluidics to integrate with other modules was exploited by coupling droplet sorting with reagent addition via picoinjection. While in this study we used different chips per manipulation, their integration on one chip, reducing the amount of handling, is a potential path for future research. This integration is not limited to picoinjection as other manipulations, like droplet dispensing18,19 and incubation,16,17 have proven to be powerful modules to couple to FADS platforms as well. Additional future work includes adding electrodes to our current 3-way junctions to further scale the multiway sorting, enabling the SeParate for even more multiway sorting by adding a third junction and accompanying electrodes. As this might be challenging due to the multitude of channels, stacking of channels and/or electrodes might be needed using for example 3D printing technologies.46 Finally, upgrading our setup by switching our Arduino for an FPGA will lead to increased sample rates, more accurate detection and potentially increased throughputs of the platform.
Author contributions
W. V.: conceptualization, planning and execution of experiments, data analysis and writing of the manuscript. J. B.: contribution to hardware and software for experimental setups, manuscript revision and editing. S. S.: cell line work and transfection. I. R.: contribution to software for experimental setups, manuscript revision and editing. H. G.: manuscript revision. L. C.: manuscript revision. F. D. D.: conceptualization, experimental design, data analysis, manuscript revision and editing. K. D.: conceptualization, manuscript revision and editing, supervision, project fundings. J. L.: conceptualization, experimental design, data analysis, manuscript revision and editing, supervision, project fundings.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
The authors gratefully acknowledge the financial support from KU Leuven (IDN/20/011, C3/19/059, Special research Fund iBOF/23/005, Lab of Excellence), the Flemish Research Foundation (FWO) Excellence of Science (EOS) program (30981113, VirEOS project and 40007527, VirEOS2), the European Union's Horizon 2020 research and innovation program (733176, RABYD-VAX consortium) and the Bill and Melinda Gates Foundation (OPP1195179).References
- X. Jin, Z. Demere, K. Nair, A. Ali, G. B. Ferraro and T. Natoli, et al., A metastasis map of human cancer cell lines, Nature, 2020, 588(7837), 331–336 CrossRef CAS PubMed.
- S. Varchetta, D. Mele, B. Oliviero, S. Mantovani, S. Ludovisi and A. Cerino, et al., Unique immunological profile in patients with COVID-19, Cell. Mol. Immunol., 2021, 18(3), 604–612 CrossRef CAS PubMed.
- K. Tiklová, S. Nolbrant, A. Fiorenzano, Å. K. Björklund, Y. Sharma and A. Heuer, et al., Single cell transcriptomics identifies stem cell-derived graft composition in a model of Parkinson's disease, Nat. Commun., 2020, 11(1), 2434 CrossRef PubMed.
- H. Liu, R. Xue, Y. Wang, E. Stirling, S. Ye and J. Xu, et al., FACS-iChip: a high-efficiency iChip system for microbial ‘dark matter’ mining, Mar. Life Sci. Technol., 2021, 3(2), 162–168 CrossRef CAS PubMed.
- L. Mazutis, J. Gilbert, W. L. Ung, D. A. Weitz, A. D. Griffiths and J. A. Heyman, Single-cell analysis and sorting using droplet-based microfluidics, Nat. Protoc., 2013, 8(5), 870–891 CrossRef CAS PubMed.
- O. Caen, S. Schütz, M. S. S. Jammalamadaka, J. Vrignon, P. Nizard and T. M. Schneider, et al., High-throughput multiplexed fluorescence-activated droplet sorting, Microsyst. Nanoeng., 2018, 4(1), 33 CrossRef PubMed.
- K. Cai, S. Mankar, T. Ajiri, K. Shirai and T. Yotoriyama, An integrated high-throughput microfluidic circulatory fluorescence-activated cell sorting system (μ-CFACS) for the enrichment of rare cells, Lab Chip, 2021, 21(16), 3112–3127 RSC.
- E. M. Llufrio, L. Wang, F. J. Naser and G. J. Patti, Sorting cells alters their redox state and cellular metabolome, Redox Biol., 2018, 16, 381 CrossRef CAS PubMed.
- I. Andrä, H. Ulrich, S. Dürr, D. Soll, L. Henkel and C. Angerpointner, et al., An Evaluation of T-Cell Functionality After Flow Cytometry Sorting Revealed p38 MAPK Activation, Cytometry, Part A, 2020, 97(2), 171–183 CrossRef PubMed.
- J. C. Baret, O. J. Miller, V. Taly, M. Ryckelynck, A. El-Harrak and L. Frenz, et al., Fluorescence-activated droplet sorting (FADS): Efficient microfluidic cell sorting based on enzymatic activity, Lab Chip, 2009, 9(13), 1850–1858 RSC.
- J. Panwar, R. Utharala, L. Fennelly, D. Frenzel and C. A. Merten, iSort enables automated complex microfluidic droplet sorting in an effort to democratize technology, Cells Rep. Methods, 2023, 3(5), 100478 CrossRef CAS PubMed.
- D. Vallejo, A. Nikoomanzar, B. M. Paegel and J. C. Chaput, Fluorescence-Activated Droplet Sorting for Single-Cell Directed Evolution, ACS Synth. Biol., 2019, 8(6), 1430–1440 CrossRef CAS PubMed.
- J. De Jonghe, T. S. Kaminski, D. B. Morse, M. Tabaka, A. L. Ellermann and T. N. Kohler, et al., spinDrop: a droplet microfluidic platform to maximise single-cell sequencing information content, Nat. Commun., 2023, 14(1), 4788 CrossRef CAS PubMed.
- Y. Liu, S. Wang, M. Lyu, R. Xie, W. Guo and Y. He, et al., Droplet Microfluidics Enables Tracing of Target Cells at the Single-Cell Transcriptome Resolution, Bioengineering, 2022, 9(11), 674 CrossRef CAS PubMed.
- L. Nan, T. Mao and H. C. Shum, Self-synchronization of reinjected droplets for high-efficiency droplet pairing and merging, Microsyst. Nanoeng., 2023, 9(1), 24 CrossRef CAS PubMed.
- T. Beneyton, I. P. M. Wijaya, P. Postros, M. Najah, P. Leblond and A. Couvent, et al., High-throughput screening of filamentous fungi using nanoliter-range droplet-based microfluidics, Sci. Rep., 2016, 6(1), 1–10 CrossRef PubMed.
- Y. Zhou, Z. Yu, M. Wu, Y. Lan, C. Jia and J. Zhao, Single-cell sorting using integrated pneumatic valve droplet microfluidic chip, Talanta, 2023, 253, 124044 CrossRef CAS.
- R. H. Cole, S. Y. Tang, C. A. Siltanen, P. Shahi, J. Q. Zhang and S. Poust, et al., Printed droplet microfluidics for on demand dispensing of picoliter droplets and cells, Proc. Natl. Acad. Sci. U. S. A., 2017, 114(33), 8728–8733 CrossRef CAS PubMed.
- L. Nan, M. Y. A. Lai, M. Y. H. Tang, Y. K. Chan, L. L. M. Poon and H. C. Shum, On-Demand Droplet Collection for Capturing Single Cells, Small, 2020, 16(9), 1902889 CrossRef CAS PubMed.
- BD FACSDiscover™ S8 Cell Sorter [Internet].
- A. Autour and M. Ryckelynck, Ultrahigh-Throughput Improvement and Discovery of Enzymes Using Droplet-Based Microfluidic Screening, Micromachines, 2017, 8(4), 128 CrossRef.
- L. Weng and J. E. Spoonamore, Droplet microfluidics-enabled high-throughput screening for protein engineering, Micromachines, 2019, 10(11), 734 CrossRef PubMed.
- Y. Wang, F. Bian, L. Shang, Z. Chen, K. Zhu and Y. Zhao, Advances of droplet-based microfluidics in drug discovery, Expert Opin. Drug Discovery, 2020, 969–979 CrossRef PubMed.
- N. Shembekar, C. Chaipan, R. Utharala and C. A. Merten, Droplet-based microfluidics in drug discovery, transcriptomics and high-throughput molecular genetics, Lab Chip, 2016, 16(8), 1314–1331 RSC.
- J. Chung, H. Shao, T. Reiner, D. Issadore, R. Weissleder and H. Lee, et al., Microfluidic Cell Sorter (μFCS) for On-chip Capture and Analysis of Single cells, Adv. Healthcare Mater., 2012, 1(4), 432–436 CrossRef CAS PubMed.
- H. Tavakoli, W. Zhou, L. Ma, S. Perez, A. Ibarra and F. Xu, et al., Recent advances in microfluidic platforms for single-cell analysis in cancer biology, diagnosis and therapy, TrAC, Trends Anal. Chem., 2019, 117, 13–26 CrossRef CAS PubMed.
- L. Schmid, D. A. Weitz and T. Franke, Sorting drops and cells with acoustics: acoustic microfluidic fluorescence-activated cell sorter, Lab Chip, 2014, 14(19), 3710–3718 RSC.
- S. Li, X. Ding, F. Guo, Y. Chen, M. I. Lapsley and S. C. S. Lin, et al., An On-Chip, Multichannel Droplet Sorter Using Standing Surface Acoustic Waves, Anal. Chem., 2013, 5468–5474 CrossRef CAS PubMed.
- B. Teste, N. Jamond, D. Ferraro, J. L. Viovy and L. Malaquin, Selective handling of droplets in a microfluidic device using magnetic rails, Microfluid. Nanofluid., 2015, 19, 141–153 CrossRef CAS.
- K. Zhang, Q. Liang, S. Ma, X. Mu, P. Hu and Y. Wang, et al., On-chip manipulation of continuous picoliter-volume superparamagnetic droplets using a magnetic force, Lab Chip, 2009, 2992–2999 RSC.
- Z. Cao, F. Chen, N. Bao, H. He, P. Xu and S. Jana, et al., Droplet sorting based on the number of encapsulated particles using a solenoid valve, Lab Chip, 2013, 13, 171 RSC.
- L. Wu, P. Chen, Y. Dong, X. Feng and B. F. Liu, Encapsulation of single cells on a microfluidic device integrating droplet generation with fluorescence-activated droplet sorting, Biomed. Microdevices, 2013, 553–560 CrossRef PubMed.
- A. Sciambi and A. R. Abate, Accurate microfluidic sorting of droplets at 30 kHz, Lab Chip, 2014, 15(1), 47–51 RSC.
- A. Isozaki, Y. Nakagawa, M. H. Loo, Y. Shibata, N. Tanaka and D. L. Setyaningrum, et al., Sequentially addressable dielectrophoretic array for high-throughput sorting of large-volume biological compartments, Sci. Adv., 2020, 6(22), 1–12 Search PubMed.
- K. Ahn, C. Kerbage, T. P. Hunt, R. M. Westervelt, D. R. Link and D. A. Weitz, Dielectrophoretic manipulation of drops for high-speed microfluidic sorting devices, Appl. Phys. Lett., 2006, 88(2), 1–3 CrossRef.
- M. Leman, F. Abouakil, A. D. Griffiths and P. Tabeling, Droplet-based microfluidics at the femtolitre scale, Lab Chip, 2015, 15(3), 753–765 RSC.
- MA900 Multi-Application Cell Sorter - Adaptable Sorting - Sony Biotechnology [Internet].
- P. O. Krutzik and G. P. Nolan, Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling, Nat. Methods, 2006, 3(5), 361–368 CrossRef CAS PubMed.
- Y. Li, Y. Thi, H. Cu and D. Luo, Multiplexed detection of pathogen DNA with DNA-based fluorescence nanobarcodes, Nat. Biotechnol., 2005, 885–889 CrossRef CAS PubMed.
- H. W. Yu, I. S. Kim, R. Niessner and D. Knopp, Multiplex competitive microbead-based flow cytometric immunoassay using quantum dot fluorescent labels, Anal. Chim. Acta, 2012, 750, 191–198 CrossRef CAS PubMed.
- Z. Wang, Q. Xu, S. Liu, Y. Liu, Y. Gao and M. Wang, et al., Rapid and multiplexed quantification of Salmonella, Escherichia coli O157:H7, and Shigella flexneri in ground beef using flow cytometry, Talanta, 2022, 238, 123005 CrossRef CAS PubMed.
- D. Frenzel and C. A. Merten, Microfluidic train station: highly robust and multiplexable sorting of droplets on electric rails, Lab Chip, 2017, 17(6), 1024–1030 RSC.
- S. Vyawahare, M. Brundage, A. Kijac, M. Gutierrez, M. De Geus and S. Sinha, et al., Sorting droplets into many outlets, Lab Chip, 2021, 21(21), 4262–4273 RSC.
- M. Girault, H. Kim, H. Arakawa, K. Matsuura, M. Odaka and A. Hattori, et al., An on-chip imaging droplet-sorting system: a real-time shape recognition method to screen target cells in droplets with single cell resolution, Sci. Rep., 2017, 7(1), 1–10 CrossRef PubMed.
- M. E. Blaha, S. Hasan, C. Dusny and D. Belder, Fluorescence lifetime activated droplet sorting (FLADS) for label-free sorting of Synechocystis sp. PCC6803, Lab Chip, 2022, 22(8), 1604–1614 RSC.
- A. Isozaki, D. Huang, Y. Nakagawa and K. Goda, Dual sequentially addressable dielectrophoretic array for high-throughput, scalable, multiplexed droplet sorting, Microfluid. Nanofluid., 2021, 25(4), 1–10 CrossRef.
- Picodroplet technology platform|Pico-Mine®|Sphere Fluidics [Internet].
- D. Josephides, S. Davoli, W. Whitley, R. Ruis, R. Salter and S. Gokkaya, et al., Cyto-Mine: An Integrated, Picodroplet System for High-Throughput Single-Cell Analysis, Sorting, Dispensing, and Monoclonality Assurance, SLAS Technol., 2020, 25(2), 177–189 CrossRef PubMed.
- A. Gérard, A. Woolfe, G. Mottet, M. Reichen, C. Castrillon and V. Menrath, et al., High-throughput single-cell activity-based screening and sequencing of antibodies using droplet microfluidics, Nat. Biotechnol., 2020, 38(6), 715–721 CrossRef PubMed.
- Technical note your ground control in high-throughput biology high-throughput screening powered by microfluidics-based droplet sorting.
- Modaflow|Flow cytometer|Fluorescence activated cell sorting [Internet].
- A. S. Hansen, N. Hao and E. K. OShea, High-throughput microfluidics to control and measure signaling dynamics in single yeast cells, Nat. Protoc., 2015, 10(8), 1181–1197 CrossRef CAS PubMed.
- J. Breukers, H. Op de Beeck, I. Rutten, M. López Fernández, S. Eyckerman and J. Lammertyn, Highly flexible and accurate serial picoinjection in droplets by combined pressure and flow rate control, Lab Chip, 2022, 22(18), 3475–3488 RSC.
- S. Sharma, M. A. Schmid, L. Sanchez Felipe, J. Grenelle, S. J. F. Kaptein and L. Coelmont, et al., Small-molecule inhibitors of TBK1 serve as an adjuvant for a plasmid-launched live-attenuated yellow fever vaccine, Hum. Vaccines Immunother., 2020, 16(9), 2196–2203 CrossRef CAS PubMed.
- D. B. Kum, N. Mishra, R. Boudewijns, I. Gladwyn-Ng, C. Alfano and J. Ma, et al., A yellow fever-Zika chimeric virus vaccine candidate protects against Zika infection and congenital malformations in mice, npj Vaccines, 2018, 3, 56 CrossRef CAS PubMed.
- M. Rasulova, T. Vercruysse, J. Paulissen, C. Coun, V. Suin and L. Heyndrickx, et al., A High-Throughput Yellow Fever Neutralization Assay, Microbiol. Spectrum, 2022, 10(3), 21 Search PubMed.
- A. Agresti, Categorical data analysis, John Wiley & Sons, 2012 Search PubMed.
- D. J. Collins, A. Neild, A. deMello, A. Q. Liu and Y. Ai, The Poisson distribution and beyond: Methods for microfluidic droplet production and single cell encapsulation, Lab Chip, 2015, 15(17), 3439–3459 RSC.
- S. Padilla-Parra, N. Audugé, M. Coppey-Moisan and M. Tramier, Dual-color fluorescence lifetime correlation spectroscopy to quantify protein–protein interactions in live cell, Microsc. Res. Tech., 2011, 74(8), 788–793 CrossRef CAS PubMed.
- T. Zimmermann, Spectral imaging and linear unmixing in light microscopy, Adv. Biochem. Eng./Biotechnol., 2005, 95, 245–265 CrossRef PubMed.
- T. Schmit, M. Klomp and M. N. Khan, in An Overview of Flow Cytometry: Its Principles and Applications in Allergic Disease Research BT - Animal Models of Allergic Disease: Methods and Protocols, ed. K. Nagamoto-Combs, New York, NY, 2021, pp. 169–182 Search PubMed.
- J. Panwar and C. A. Merten, Fluorescence crosstalk reduction by modulated excitation-synchronous acquisition for multispectral analysis in high-throughput droplet microfluidics, Lab Chip, 2023, 23(11), 2514–2520 RSC.
- T. Maetzig, J. Ruschmann, C. K. Lai, M. Ngom, S. Imren and P. Rosten, et al., A Lentiviral Fluorescent Genetic Barcoding System for Flow Cytometry-Based Multiplex Tracking, Mol. Ther., 2017, 25(3), 606–620 CrossRef CAS PubMed.
- X. Fu, Y. Zhang, Q. Xu, X. Sun and F. Meng, Recent Advances on Sorting Methods of High-Throughput Droplet-Based Microfluidics in Enzyme Directed Evolution, Front. Chem., 2021, 9, 1–9 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3lc01075a |
| This journal is © The Royal Society of Chemistry 2024 |

