Recent advances in micro-physiological systems for investigating tumor metastasis and organotropism
Heejeong
Yoon
 a,
Jonathan
Sabaté del Río
a,
Jonathan
Sabaté del Río
 b,
Seung Woo
Cho
a and
Tae-Eun
Park
b,
Seung Woo
Cho
a and
Tae-Eun
Park
 *a
*a
aDepartment of Biomedical Engineering, College of Information and Biotechnology, Ulsan National Institute of Science and Technology (UNIST), Ulsan 44919, Republic of Korea. E-mail: tepark@unist.ac.kr
bCenter for Algorithmic and Robotized Synthesis (CARS), Institute for Basic Science (IBS), Ulsan 44919, Republic of Korea
First published on 23rd January 2024
Abstract
Tumor metastasis involves complex processes that traditional 2D cultures and animal models struggle to fully replicate. Metastatic tumors undergo a multitude of transformations, including genetic diversification, adaptation to diverse microenvironments, and modified drug responses, contributing significantly to cancer-related mortality. Micro-physiological systems (MPS) technology emerges as a promising approach to emulate the metastatic process by integrating critical biochemical, biomechanical, and geometrical cues at a microscale. These systems are particularly advantageous simulating metastasis organotropism, the phenomenon where tumors exhibit a preference for metastasizing to particular organs. Organotropism is influenced by various factors, such as tumor cell characteristics, unique organ microenvironments, and organ-specific vascular conditions, all of which can be effectively examined using MPS. This review surveys the recent developments in MPS research from the past five years, with a specific focus on their applications in replicating tumor metastasis and organotropism. Furthermore, we discuss the current limitations in MPS-based studies of organotropism and propose strategies for more accurately replicating and analyzing the intricate aspects of organ-specific metastasis, which is pivotal in the development of targeted therapeutic approaches against metastatic cancers.
1. Introduction
Tumors pose a significant challenge due to their complexity and heterogeneity, with tumor metastasis being a crucial factor contributing to elevated patient mortality rates.1,2 Metastasis is the process by which cancer cells from a primary tumor spread to other parts of the body, where they can form secondary tumors.3 Metastatic tumors undergo a variety of changes, including 1) genetic diversity caused by cellular mutations, 2) phenotypic changes in tumor cells adapting to different microenvironments, and 3) alterations in the pharmacokinetic/pharmacodynamic properties of drugs due to infiltration into different organs.4–6 These changes make the treatment of metastatic tumors more challenging.Conventional tumor models, including the two-dimensional (2D) in vitro culture system and in vivo animal models, have yielded valuable insights into metastasis over many years.7,8 However, to effectively investigate metastasis and develop preventive or therapeutic strategies for cancer patients, many researchers concur on the need for advanced metastasis models that transcend the limitations of traditional approaches.7,8 While 2D cultures of cell lines or primary cells derived from actual tumors do share some biological characteristics with their parent tumors, they often exhibit altered behaviors when adapted to the 2D in vitro environment, lacking 3D structural context.9–11 This can result in differences in cell proliferation, migration, and responses to treatments, potentially yielding misleading results. Moreover, the limited complexity of the microenvironment in 2D culture systems, including the absence of key elements like blood vessels, various neighboring cell types, and extracellular components, makes it challenging to replicate the intricate interactions between cancer cells and their microenvironment during the metastasis process.12,13
Of paramount importance, metastasis involves a multistep process encompassing invasion, intravasation, circulation, extravasation, and colonization,3 which traditional 2D cultures cannot fully reproduce, thereby constraining the comprehensive study of the entire metastatic cascade. Animal tumor models offer a more physiologically relevant environment compared to 2D cultures, enabling the investigation of microenvironmental factors. However, it is crucial to acknowledge that animal models do not entirely mirror human biology due to genetic, immune response, and metabolic differences.14 Consequently, these limitations pose a significant barrier in global drug development against metastatic cancers, particularly with the high threshold of clinical trials.
Micro-physiological system (MPS), also known as organ-on-a-chip technology, is currently under active research as potential alternatives to, or reductions of, animal experiments.15 MPS aims to replicate the structure and function of specific human organs or tissues by creating microscale models that integrate living cells and reproduce key physiological features.16 These systems offer a more physiologically relevant environment compared to a traditional 2D cell culture, potentially yielding more advanced results even at the cellular experimentation stage. In the context of tumor metastasis, fluid flow, such as blood or lymph plays a crucial role in the spread of tumor cells from the primary site to distant organs or tissues.17 Incorporating fluid flow into tumor MPS models enables researchers to simulate the metastatic process more accurately. These advancements are expected to improve the accuracy of preclinical testing of anti-cancer drugs, in line with growing awareness of the limitations in previous preclinical methodologies.18–21 A recent example includes a study demonstrating that an MPS integrating 3D bone Ewing sarcoma and heart muscle tissues could accurately reflect the clinical impact of linsitinib, an anti-cancer drug that failed in phase 2 clinical trials due to cardiotoxicity, despite its initial promise in cancer treatment.18 The increasing evidence supporting the superior predictive accuracy of MPS over traditional cell-based assays is fueling the expansion of the MPS market in the pharmaceutical industry.22
The capability of MPS to replicate the structure, function, and blood flow of tissues offers valuable insights into the study of metastatic organotropism.23–26 Organotropism is known as the phenomenon where a tumor selectively metastasizes following a non-random distribution among distant organs.27 For example, breast cancer can metastasize to different sites, including bone, lung, liver, and brain. However, the luminal sub-type has a greater propensity to metastasize to the bone, whereas triple-negative breast cancer tends to favor visceral organs.28 The organotropism feature of tumor is determined by tumor-intrinsic factor, the circulation pattern of tumor cells, the organ-specific environment, and the interaction between tumor cells and cell or matrix in the specific organ.27 Specific surface proteins of tumor cells and their interaction with organ-specific signal molecules also play a role in organotropism. MPS technology enables the investigation of these interactions by faithfully mimicking the tissue-specific microenvironment.28–31 Such insights hold promise for predicting metastasis into specific organs from primary tumors, thereby fostering the development of organotropism biomarkers and preventive strategies against metastasis.
This review is designed to assist researchers in selecting the optimal MPS platform for their studies on tumor metastasis and organotropism. Initially, we aim to equip researchers with a comprehensive knowledge of the essential biological factors that should be considered when employing MPS for these studies. Following this, we examine various MPS platforms that have been specifically tailored for investigating tumor metastasis. Additionally, we delve into recent MPS research that concentrates on organotrophic metastasis, and offer strategies to enhance the study of organotropism using MPS technologies. This approach aims to provide a detailed roadmap for researchers navigating the complex landscape of tumor metastasis and organ-specific cancer research using MPS.
2. Key steps of metastasis
2.1. Epithelial–mesenchymal transition (EMT)
The epithelial–mesenchymal transition (EMT) plays a pivotal role in the spread of tumor.32 This process transforms epithelial tumor cells, which are typically cohesive and adherent, into more mobile and invasive mesenchymal cells.32 These transformed cells have the capability to detach from the primary tumor, penetrate nearby tissues, and migrate into the bloodstream. EMT in tumors can be initiated by a variety of elements within the tumor environment, including low oxygen levels, fluid dynamics, inflammatory signals, and growth factors like TGF-β and EGF, along with components of the extracellular matrix (ECM).33 Therefore, recapitulation of these intricate environmental factors can be helpful for simulating the EMT process or developing therapeutic approaches in MPS. In in vitro systems, the state of EMT can be assessed using established markers that span a range of characteristics, from indicators of cellular stemness to markers of morphological alterations and transcriptional regulators.34 For instance, the analysis of E-cadherin (an epithelial marker) and vimentin (a mesenchymal marker) at the transcriptional level or through immunofluorescent microscopy has been extensively utilized in traditional 2D platforms,34 as well as in MPSs.35–372.2. Intravasation and extravasation
The dynamic nature of tumor metastasis, fundamentally involving cell movement, renders microfluidic systems highly effective tools, particularly for studying intravasation and extravasation. These processes, which entail the movement of tumor cells through blood vessel endothelium, are key applications making MPS technology valuable.38Intravasation, the process of tumor cells entering the circulation, encompasses following steps: 1) the endothelial basement membrane underlying the endothelium is degraded by enzymes such as matrix metalloproteinases, 2) tumor cells migrate towards blood vessels, 3) they adhere to the endothelium, and penetration into blood vessels via induced changes in endothelial cells.39 In the context of intravasation, the features of highly vascularized tumor areas, such as disorganized and permeable blood vessels, facilitate the entry of tumor cells into the bloodstream.40 Tumor vasculatures, being narrower and less orderly than normal vessels, provide an easier path for tumor cell movement.40 Moreover, they are more prone to thrombus formation and exhibit greater inflammation than normal vessels.41 Therefore, understanding and managing these tumor vessels is critical in cancer treatment and improving patient outcomes.42 Hence, it is crucial to precisely mimic the angiogenesis process, stimulated by tumor cells, and the property of resulting tumor vasculature within the MPS. This accurate replication is essential for an accurate representation of tumor–endothelial interactions during intravasation.
Extravasation, the process of tumor cells migrating from the bloodstream into secondary tissues, involves several steps: 1) circulating tumor cells (CTCs) either get physically trapped in small capillaries or come into proximity with endothelial cells lining blood vessels, 2) CTCs adhere to endothelial cells via interactions between surface proteins, 3) they migrate across the endothelial layer, a process that includes breaking down endothelial junctions and the basement membrane, and 4) they invade the surrounding tissue.39 This process is influenced by factors such as the expression of adhesion molecules on both tumor and endothelial cells, and the dynamics of blood flow, which affect CTC adhesion to vessel walls.39 These aspects can be effectively simulated in MPS. Increased vascular permeability, potentially caused by inflammatory signals and exosomes from the primary tumor,43 also facilitates CTCs to escape from vessels, in line with the seed-and-soil theory.44 Additionally, differences in the endothelial properties and structure of vascular beds of different organs contribute to the patterns of organotropism seen in metastasis.45 Therefore, using specific endothelial cells from targeted tissues, and replicating unique cell–cell and cell–ECM interactions to maintain the feature of specific vascular bed are essential to accurately recreate the extravasation process of tumor cells in the MPS platforms.
Confocal fluorescence microscopy is a predominant experimental technique for examining the interactions between tumor cells and vascular endothelium. Employing immunofluorescent staining or labeling on both endothelial and tumor cells provides valuable insights into how and to what extent different tumor cells penetrate endothelial barriers. This method allows for a detailed observation of the dynamic interplay between the tumor cells and the vascular structure.
2.3. Metastatic colonization at a distant organ
Metastatic colonization, the process where cancer cells spread and form new tumors, is notably inefficient.46 Most tumor cells perish during this process, with only a few managing to form significant tumors.47 This process involves several stages, including the ability of tumor cells to evade the immune system and other defense mechanisms in the distant organ.46 In their new environment, infiltrated tumor cells often face challenges, making them susceptible to immune system attacks.48 The unique immune cell compositions in each organ can affect how likely it is for that organ to develop visible metastasis.46 Therefore, understanding how metastasis targets specific organs requires studying how recapitulating tumor cells interact with the unique immune environments of these organs, which might be an important topic in MPS research.Additionally, the ability of tumor cells to establish themselves in a new location depends on finding a supportive niche.49 This involves interacting with the surrounding stroma cells and the ECM to activate pathways that promote growth and survival.49 Tumor cells also maintain their ability to initiate new tumors by retaining stem-like characteristics. The preference of cancer cells for specific organs is partly determined by whether the cellular and matrix components of an organ provide a supportive environment. Recapitulating these interactions between tumor cells and their new microenvironments is important to understand the underlying mechanisms of organotropism.
3. MPS recapitulating tumor metastasis
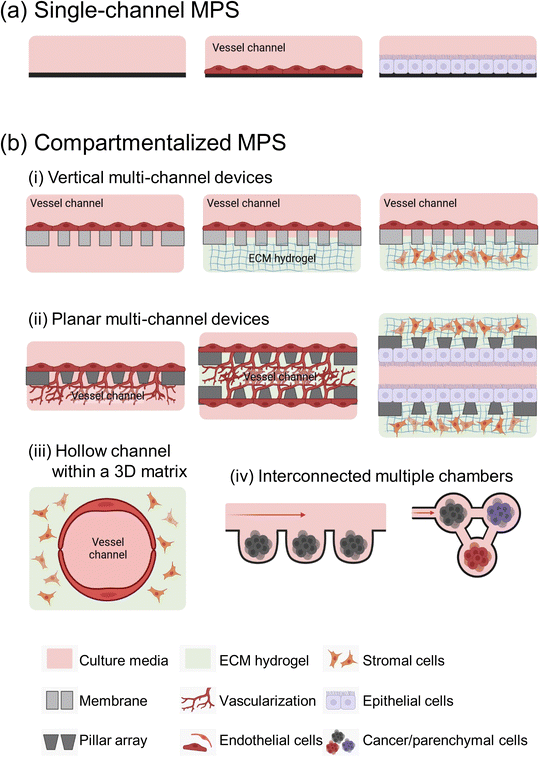
Microfluidic devices offer significant benefits in cancer metastasis research, particularly in its ability to control microstructures and fluid dynamics with precision.15 They support the growth of various cell types and accurately mimics tumor microenvironments (TMEs). MPS devices are generally classified into two main types: single-channel devices without separate compartments, and compartmentalized chips that include semipermeable membranes, pillar arrays, a hollow channel within hydrogels, and multi-chambers (Fig. 1).The complexity of MPS devices varies widely, tailored to the specific requirements of each metastasis study. The complexity of MPS devices is often dictated by the specific biological or pathological model being studied. For simpler studies, such as examining tumor cell migration and motility under well-defined conditions, a basic single-channel MPS device is often sufficient and more appropriate.
However, for more complex research areas like tumor metastasis involved in dynamic cell–cell interaction and the interplay between multiple organs, the need for more elaborate MPS devices arises. Crucially, in vitro replication of these interactions demands that MPS devices go beyond merely imitating the structural characteristics of organs. They must also emulate the functional aspects, including dynamic physiological processes like blood flow, the exertion of mechanical forces, and communication between different organs. This level of detail in modeling necessitates sophisticated designs in MPS devices.
It is important to note, however, that with increased complexity in MPS devices, there often comes a trade-off with aspects such as scalability, reproducibility, imaging and analysis capabilities, as well as the long-term viability and stability of the system.50
In this context, our focus is on MPS devices developed in the past five years, specifically engineered for cancer metastasis studies, which are comprehensively summarized in Table 1.
| Cancer type | Endothelial cells | Configuration | Application | Ref. |
|---|---|---|---|---|
| Single channel chip | ||||
| Lung cancer A549 cells | — | • Microchip with one side of the channel connected to the media reservoir (T-25 flask) and on the other side to a vacuum syringe pump | Flow-induced EMT characterization and effect of anti-cancer drugs | 37 |
| Tumor vessel | Human umbilical vein endothelial cells (HUVEC) | • Tumor vasculature MPS | Reproduction and characterization of the massive formation of thrombi and hemorrhage of tumor vessels | 52 |
| • Microfluidic channel with a serpentine morphology and rectangular cross-section with 200 μm × 50 μm (w × h) | ||||
| Breast cancer MCF-7 cells | HUVEC | • 3D spheroid-microvasculature MPS | Investigating the interaction between various tumor sizes and endothelial layer under a range of shear stresses | 51 |
| • Microfluidic straight channel with 600 μm × 300 μm × 25 mm (w × h × l) | ||||
| Breast cancer MCF-7 and MDA-MB-231 cells | HUVEC | • Vessel MPS | Evaluation of the role of interactions between cancer derived extracellular vesicles and platelets in pre-metastatic niche formation and tumor metastasis | 55 |
| • Microfluidic straight channel with 900 μm × 100 μm × 1 mm (w × h × l) | ||||
| Compartmentalized chip; vertical multi-channel | ||||
|---|---|---|---|---|
| Colon cancer HCT116 and HT29 cells | HUVEC | • Human colorectal cancer MPS | Analysis of tumor metastasis through cell intravasation and metabolic changes, exploration of novel cancer treatment strategies, and prediction of tumor metastasis potential and prognosis | 67 |
| • Upper (1 × 1 mm; w × h) and lower (1 × 0.2 mm; w × h) channels | ||||
| • Thin porous membrane; 50 μm thick with 7 μm diameter pores | ||||
| Breast cancer MCF-7 and MDA-MB-231 cells | Human hepatic sinusoidal endothelial cells | • Liver MPS | Investigating the role of breast cancer-derived extracellular vesicles in liver metastasis | 64 |
| • Channel with dimensions of 300 μm × 100 μm × 1 mm (w × h × l) | ||||
| • PDMS membrane; 8 μm thick with 8 μm diameter pores | ||||
| Breast cancer MDA-MB-231 cells and liver cancer HepG2 cells | Endothelial cells (Ea.hy 926) | • Dynamic tumor-vessel microsystem | Investigate different steps in the process of cancer metastasis and evaluate the effect of anticancer drugs on each step of tumor metastasis | 65 |
| • Three layers of PDMS plates with 30 mm diameter and 2 mm thicknesses | ||||
| • Polycarbonate membrane; 10 μm diameter pore | ||||
| Compartmentalized chip; planar multi-channel | ||||
|---|---|---|---|---|
| Breast cancer MDA-MB-231 cells | HUVEC | • Two-channel compartmentalized MPS | Real-time monitoring of the metastatic cascade | 78 |
| • Rectangular cross section; 210 μm × 50 μm × 2.7 cm (w × h × l) | ||||
| • Micropillar; 25 μm × 500 μm (w × l), 3 μm gap | ||||
| Breast cancer MCF-7, MDA-MB-231, SK-BR-3 and S-HBC cells | HUVEC and human lymphatic endothelial cells | • Lymph vessel-tissue–blood vessel MPS | Investigation of tumor invasion with interleukin-6 treatment which can induce EMT on breast cancers | 84 |
| • Three-channel microfluidic | ||||
| • Micropillar; three squared pillars (100 μm, h) | ||||
| Patient-derived GSCs | HUVEC | • Organotypic triculture MPS | Recapitulation of perivascular niche and study its effect on GSCs | 83 |
| • Inner tumor region bordered by two concentric semicircles serving as the stroma and vascular regions (diameters; 1, 2.5, and 3.5 mm, respectively) | ||||
| • Micropattern; hexagon and trapezoid pattern (200 μm, h) | ||||
| Liver cancer HepG2 cells | HUVEC and human dermal lymphatic endothelial cells | • Vascularized tumor spheroid MPS with a 3D perfusable vascular bed | Identification of reproduction of pathological characteristics of solid tumors, interactions between TMEs including formation of leaky tumor blood vessels, and evaluation of anti-angiogenic and anti-cancer drugs | 42 |
| • Micropattern; 150 μm height and 100 μm intervals | ||||
| Colon cancer SW620 cells | Induced pluripotent stem cell-derived endothelial cells | • Human induced pluripotent stem-cardiac-endothelial-tumor MPS | Simultaneous evaluation of cardiotoxicity and antitumor effect of drugs | 80 |
| • Three-chamber tissue | ||||
| Cervical cancer HeLa cells | HUVEC | • Extremely early stage (EES) on MPS | Replication of tumor progression at the EES and study of tumor behavior under the influence of endothelial cells and fibroblasts | 109 |
| • Central channel; 1.3 mm × 150.0 μm (w × h), lateral channel; 1.0 mm × 150.0 μm (w × h) | ||||
| • Micropattern; 100.0 μm (h) | ||||
| Compartmentalized chip; hollow channel | ||||
|---|---|---|---|---|
| Breast cancer MDA-MB-231 cells | HUVEC | • MPS of the solid tumor–vascular interface | Close recapitulation of the proliferative, migratory, angiogenic, and invasive properties of the tumor–vascular interface | 87 |
| • Hollow hydrogel channel (diameter, 350 μm) composed of collagen, Matrigel™ and fibrinogen | ||||
| Breast cancer MDA-MB-231 cells; bone tropic MDA-MB-231 and lung tropic MDA-MB-231 | Primary human dermal microvascular blood endothelial cells | • Engineered tumor–vascular network device | Organotrophic bone and lung metastasis by hydrogel invasion of breast cancers | 26 |
| • Hollow hydrogel channel (diameter, 350 μm) with collagen 1 hydrogel | ||||
| • ECM chamber with 20 mm3 volume | ||||
| Primary murine pancreatic cancer cells and human pancreatic cancer Panc-1 cells | HUVEC | • Biomimetic duct-blood vessel model | Observation of endothelial ablation | 86 |
| • Hollow hydrogel channel (diameter, 300 μm) with collagen 1 hydrogel | Confirmation of a critical role in activin-ALK7 signaling mediating endothelial excision | |||
| Lung cancer A549 cells | HUVEC | • 3D vascularized lung cancer MPS | Evaluation of tumor angiogenesis and drug efficacy | 19 |
| • Hollow hydrogel channel (diameter, 300 μm) with decellularized lung ECM hydrogels | ||||
| • ECM chamber in the shape of a rectangle with 20 mm × 25 mm × 2.5 mm (w × l × h) | ||||
| Glioblastoma U87 MG cells | HUVEC | • Tumor-vasculature MPS | Effect of distance between glioblastoma spheroids and blood vessels on vessel co-option induction | 110 |
| • Hollow hydrogel channel (diameter, 235 μm) with collagen 1 hydrogel | ||||
| • ECM chamber in the shape of a rectangle with 5 × 10 mm (w × l) | ||||
| Clear cell renal cell carcinoma | Both normal and tumor-associated endothelial cells | • Organotypic primary patient-specific blood vessel models | Determining the efficiency of restoration of vascular function, demonstrating the model's potential for single-patient clinical trials | 95 |
| • Hollow hydrogel channel (diameter, 350 μm) with collagen 1 hydrogel | ||||
| • Hexagonal chamber | ||||
| Breast cancer MDA-MB-231 cells | — | • TME-based MPS | Assessment of migration distance and matrix remodeling as well as crosstalk with human mammary fibroblasts or cancer-associated fibroblasts for breast cancers | 90 |
| • Hollow hydrogel channel (diameter, 280 μm) with collagen 1 hydrogel | ||||
| Compartmentalized chip; interconnected multiple chambers | ||||
|---|---|---|---|---|
| Liver cancer HepG2 cells and pancreatic cancer PANC-1 cells | HUVEC | • Vascular MPS | Fabrication of biomimetic vascular systems integrated with chamber-specific vascularized organs and prediction of multi-organ metastasis potential | 24 |
| • Leaf venation network with 150 μm depth | ||||
| Prostate cancer PC3 and C-2B cells | — | • Skeletal metastasis model | Integration and characterization of tumor and metastatic microenvironmental tissues and analysis of tumor chemotaxis | 98 |
| • Chamber; 6-well culture plates connected by large bore tubing | ||||
| Lung cancer A549 cells | — | • 3D culture multiorgan MPS | Hypoxia-induced cancer metastasis mechanisms | 107 |
| • Culture chambers; 5 mm diameter and 2 mm depth | Drug screening under a hypoxia environment on a multiorgan level | |||
| Neuroblastoma HTB-10 cells and Ewing sarcoma (ES) HTB-166 cells | — | • MPS integrating ES tumor and heart muscle interconnected by microfluidic circulation | ES tumor and cardiotoxicity evaluation of linsitinib, a novel anti-cancer drug for comparison with the results observed in the clinical trial | 18 |
3.1. Single-channel devices
The basic type of MPS, a single-channel MPS, has been a fundamental tool in metastasis research. Its simple design offers cost-efficiency, ease of fabrication, and is ideal for imaging. It is particularly useful in studying the EMT in various biomechanical cues, a crucial phase in early metastasis marked by changes in cell adhesion and migration. The Demirci group has extensively explored how flow-induced hydrodynamic shear stress influences EMT in various cancers, including esophageal cancer,35 ovarian cancer,36 and lung cancer.37 For example, they cultured cancer cells in the device and analyzed EMT biomarkers (E-cadherin and vimentin) through immunofluorescence in presence of fluid flow, which highlighted how flow-induced stress impacts EMT.37Furthermore, these single-channel systems can replicate tumor vasculature conditions. By designing microchannels that mimic the complex geometry of tumor vasculature, researchers have recreated abnormal blood flow patterns observed in tumors. This led to thrombosis formation in their MPS platform, which is also relevant to the process of metastasis.51
The type of device also facilitates the study of extravasation by mimicking interactions between CTC and vascular endothelium.37,51–55 It has been mostly used to examine cancer cell rolling and adhesion. Studies have explored how different conditions affect CTC–vasculature interactions, like how inflammation in brain microvasculature can hasten metastasis by increasing adhesive protein expression.56 Additionally, the devices are instrumental in studying CTC clusters not just singular CTCs, known for their higher metastatic potential due to greater stemness and plasticity.57 By examining these clusters' interactions with endothelial cells under fluid stress, researchers have gained insights into how shear stress affects CTC cluster expansion and endothelial cell polarity51 (Fig. 2a). However, there is a notable discrepancy between the size of CTC clusters (30–150 μm) used in experiments and those naturally found in the body, which consist of two or more nuclei.58 Using microfluidic systems that better mimic the smaller, natural clusters of circulating tumor cells (CTCs)59,60 could improve physiological relevance.
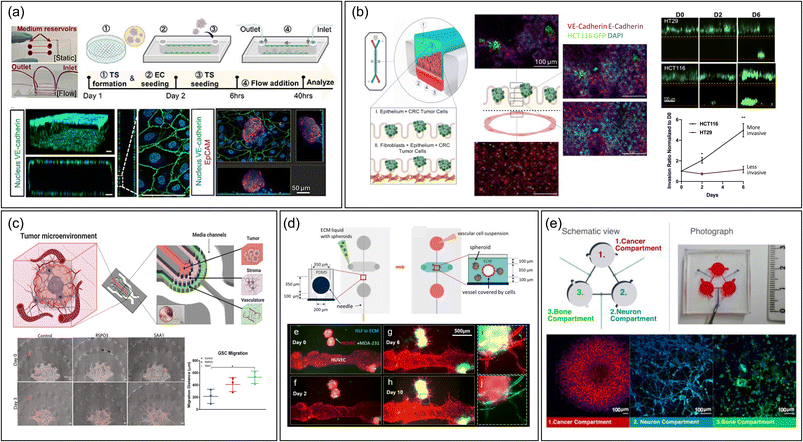 | ||
| Fig. 2 Representative MPS designs to study metastasis. (a) A single-channel device without compartments featuring the interaction between cancer spheroid and monolayered endothelial cells.51 The fabrication workflow is outlined as follows: (1) creating a tumor spheroid (TS), (2) introducing endothelial cells (EC) on day 1, (3) placing the TS in a single channel alongside an EC monolayer on day 2, (4) applying fluid flow, and (5) conducting analyses at 6 and 40 hours post TS seeding. Scale bar = 50 μm. Reproduced from ref. 51 with permission from IOP Publishing, copyright 2023. (b) A vertical multi-channel device compartmentalized by semipermeable PDMS membrane to study intravasation of colorectal cancer cells by monitoring cancer cells in upper epithelial channel migrate into the lower vascular channel chip containing features such as membranes.67 Scale bar = 100 μm. Reproduced from ref. 67 with permission from Elsevier, copyright 2021. (c) A planar multi-channel device compartmentalized by pillar arrays, having tumor, stromal, and vascular channels to recapitulate intravasation.83 Scale bar = 100 μm. Reproduced from ref. 83 with permission from Wiley, copyright 2022. (d) An MPS device having a vascular lumen within an ECM hydrogel embedding cancer spheroids to recapitulate angiogenesis and intravasation process.87 Scale bar = 500 μm. Reproduced from ref. 87 with permission from Springer Nature, copyright 2020. (e) Integrated multi-chamber device mirroring the migration of breast cancer driven by sympathetic activation on the interaction with bone cells.96 Scale bar = 100 μm. Reproduced from ref. 96 with permission from Elsevier, copyright 2022. Abbreviations: TS, Tumor spheroid; EC, Endothelial cell; CRC, Colorectal cancer; GSC, Glioma stem cell; HLF, Human lung fibroblast. | ||
While the single-channel MPS is effective in observing some metastatic features of cells, it falls short in mimicking complex cell–ECM interactions and intra- and extravasation events, pointing to the necessity for more advanced systems.
3.2. Compartmentalized devices
Research on individual tumor cells in culture has significantly advanced our understanding of tumors. However, more recent studies emphasize that MPSs, which better mimic the intricate physiology of tissues, can offer a more accurate representation of tumor cell behavior.29,61,62In this context, microfluidic devices with compartmentalized channels are particularly valuable.17 They facilitate the co-culture of diverse cell types in distinct areas, employing various techniques to authentically replicate the specific microenvironments of different organs with enhanced complexity. This quality renders them an exemplary model for examining tumor behavior in both original and distantly infiltrated tissues. Notably, these devices are instrumental in studying tumor metastasis, including processes like intra- and extravasation, which traditional 2D cultures or single-channel MPS devices cannot effectively replicate. These platforms utilize a variety of methods to create compartments that accommodate multiple cell types and matrix compositions. A thorough understanding of the characteristics and analytical techniques of these diverse platforms is essential for choosing the most suitable one for specific research objectives.
In these systems, porous membranes mimic the basement membrane—a specialized, dense, and porous layer of the ECM that separates and integrates tissues at their borders in the body. This interaction between cells and the ECM at the basement membrane is key to maintaining epithelial tissue's apical-basal polarity. Additionally, MPS devices can manipulate other conditions like air–liquid interfaces and fluidic shear stress, further promoting cell polarity.68
These MPS devices with integrated membranes effectively model the migration of tumor cells across epithelial barriers and their entry into the bloodstream (intravasation), as well as the exit of circulating cancer cells from the bloodstream and their invasion into specific organs (extravasation). The permeability porous membrane in these systems is crucial, dictating which biomolecules and cells can traverse the tissue barrier.69 The use of membranes having large pores (3–10 μm) are generally used in MPS not to hinder the motility of the migrating tumor cells, but to offer enough attachment area for cells forming tissue barriers.70
Porous polyethylene terephthalate (PET)71 or polycarbonate track-etched membranes,72 which are readily available on the commercial market, are commonly utilized as substitutes for the basement membrane. This is due to their availability of a diverse range of pore sizes and densities, making them suitable for this application. Another widely used material is polydimethylsiloxane (PDMS) membranes, allowing for pore size and density customization, stretchability, and seamless integration into PDMS-based MPS devices.73 PDMS membranes also offer better optical transparency than plastic membranes, enabling more effective monitoring of cell migration across tissue barriers. These membranes are often coated with basement membrane ECM proteins, such as collagen types I and IV, laminin, and fibronectin, or with animal-derived ECM mixtures like Matrigel™, to facilitate cell–ECM interaction and emulate biological signaling.68 Moreover, the inclusion of ultrathin, electrospun nanomembranes in MPS devices represents a significant advancement.74,75 These nanomembranes offer tissue-like stiffness and thickness, enhancing the realism of the model by mimicking the physical characteristics of the basement membrane.74,75
In a study examining colorectal cancer cell metastasis, an MPS with an upper channel for colon epithelium and a lower channel for endothelium, separated by a porous PDMS membrane, was utilized (Fig. 2b).67 This setup enabled monitoring the early stages of metastatic spread in various TMEs. The process of colorectal tumor cell intravasation was studied through confocal immunofluorescence analysis, observing how the seeded tumor cells penetrated the normal epithelial layer and migrated towards the endothelial channel.67 The design of this MPS, featuring open microchannels, allowed for the collection of effluents, which were then used to explore metabolic changes during metastasis. Although the device had only two channels, it effectively investigated the impact of stromal elements on tumor intravasation. This was achieved by additionally culturing cancer-associated fibroblasts beneath the colon epithelial layer, demonstrating the versatility and adaptability of the membrane-integrated MPS in metastasis research.67
Another study focused on breast cancer's premetastatic niche formation using human liver MPS.64 This system was designed with an upper channel for liver sinusoidal endothelium and a lower channel for liver epithelium. Differing from the colorectal cancer study,67 focusing on intravasation step, this research investigated the extravasation of breast cancer cells and their infiltration into the liver parenchyma, reflecting the organotropism characteristic of breast cancer. CTC extravasate through intracellular gaps in sinusoidal endothelial cells, known as fenestrae, and enter the subepithelial space of Disse.76 To more accurately mimic this breast cancer infiltration process, the epithelial channel was filled with an ECM hydrogel and fibroblasts, in addition to the hepatocyte epithelial layer,64 a methodology distinct from the colorectal cancer study.67 This platform demonstrated that extracellular vesicles derived from primary breast tumors promote metastasis by forming a premetastatic niche in the liver tissue.64
In a recent study, we explored how obesity influences the initial adhesion of tumors to the vasculature, a critical initial phase of extravasation using two-channel MPS device.77 Our MPS system was configured with an adipocyte channel, which was filled with an ECM hydrogel derived from adipose tissue and mature adipocytes. This was paired with an endothelium channel, separated by a porous PET membrane. Our findings demonstrated that circulating breast cancer cells adhered more effectively to the endothelium in an obese environment, recreated using cells and matrix materials derived from obese animals. Using the MPS device and confocal imaging analysis, we could reveal that obesity contributes to inflammation in microvessels, potentially enhancing cancer metastasis. However, it is important to note that the PET membrane used in this study had a pore size of 1 μm, which limited the observation of extravasation and transmigration of CTCs into the adipose tissue, indicating the importance of membrane porosity in metastasis study.
In summary, porous membranes offer a versatile means for compartmentalization in MPS. This feature allows for the effective evaluation of cancer cell motility within a well-structured tissue barrier and facilitates the study of cancer cell behavior in well-separated microenvironments recreated within the MPS device. As a result, these systems are a popular choice among researchers. However, it is noteworthy that the compartmentalization created by the membrane can increase the distance from the typical lens used in confocal microscopy. Additionally, the use of membranes with suboptimal optical properties can impede the effectiveness of detailed imaging analysis.
In one of its most basic forms, a two-channel planar MPS device has been utilized for real-time monitoring and analysis of the metastatic cascade in vitro.78 This device comprises two parallel channels separated by an array of rounded micropillars, effectively creating a permeable micromembrane. One channel functions as a vascular lumen, while the other serves as an extravascular channel, filled with a Matrigel™ hydrogel mimicking the stromal region of the tissue. The efficiency of intravasation is assessed by combining Matrigel™ with fluorescently labeled tumor cells, simulating malignant breast tumor tissue, and tracking the movement of tumor cells into the vascular channel under fluorescent microscopy. Conversely, the extravasation of CTC is replicated by flowing the CTCs in the vascular channel and monitoring their infiltration into the extravascular channel.78 The system's capability for real-time cell imaging in the MPS allows for precise analysis of cancer cell velocity under specific conditions, such as in an inflamed environment.
In a different study, researchers developed a three-channel MPS that simulates the structure of lymph vessels, tissues, and blood vessels, providing insights into cancer metastasis, particularly lymphatic dissemination.84 This MPS device features three parallel channels, delineated by rectangular micropatterned pillars: an open lymphatic vascular channel on the left, a central hydrogel-filled tissue channel, and an open microvascular channel on the right, specifically designed to study lymph node metastasis of breast cancer cells. The study highlighted the impact of the inflammatory cytokine IL-6. Researchers could monitor infused CTCs in the microvascular channel infiltrating the central tissue channel, then migrating towards the lymphatic vessel, in the presence of IL-6.
Another recent study introduced an MPS model that mimics the perivascular niche for examining the invasion of glioma stem cells (GSCs) through molecular interactions among GSCs, glial cells, and vascular cells in the brain (Fig. 2c).83 This model utilized U-shaped micropatterns to create three separate cell channels: a tumor channel with GSCs, a stroma channel containing glial cells, and a vascular channel. Additionally, there was a channel for media supply. Unlike the previously mentioned MPS design,84 this model also filled the microvascular channel with hydrogel, likely because the focus was on chemical interactions between multiple cell types rather than monitoring intravasation of tumor cells in a perfused vessel.83 Beyond confocal microscopy, the researchers employed single-cell RNA sequencing analysis by collecting cells from all channels, successfully identifying novel ligand–receptor pairs that drive the chemotactic invasion of GSCs, indicating the potential of single cell sequencing analysis to comprehensively understand the interaction of multi type of cells in the MPS.
In another innovative study, researchers highlighted the significance of accurately replicating the spatial distribution of different cell types in an MPS device, especially for mimicking cell–cell interaction-induced cytokine secretion and drug responses of metastases.85 They compared this approach to traditional transwell platforms. Their MPS model, designed to replicate the brain microenvironment, featured two channels: an open brain microvascular channel and an astrocyte channel embedded in hydrogel. At the side of those channels, media channel and tumor channel contained lung tumor spheroids within a collagen hydrogel, are located simulating brain metastases. The study revealed that specific cytokines were significantly elevated in tumor spheroids, accompanied by a global transcriptional shift when interacting with the brain microenvironment channels. This observation was in line with clinical data.85 In contrast, a transwell setup using the same types of cells did not show any notable changes. This finding underscores the advantage of using an MPS for understanding metastasis that involves multi-cell interactions, even though underlying molecular mechanism needs to be further investigated.
Overall, these pillar array-based multi-channel devices enable precise control of multicellular microenvironments by adjusting channel shapes and numbers, allowing for various analyses such as invasion, drug evaluation, interaction with metastatic niche providing a more accurate and detailed representation of the complex interplay between different cell types in metastatic processes compared to traditional in vitro systems.
The hollow channel enables the study of interactions between vascular endothelium, circulating cancer cells, and stromal cells within the hydrogel, facilitating observation of tumor cell intra/extravasation and tissue infiltration. For instance, the Beebe group has used this MPS design to examine how tumor cell behavior changes under various microenvironmental conditions.88,90,93–95 In one such study, a hollow vascular channel was positioned asymmetrically, allowing cells near the vessel to access nutrients and oxygen while those farther away experienced nutrient deprivation, hypoxia, and waste accumulation. This setup was used to study tumor metabolic vulnerabilities90 and NK cell exhaustion.94 Fluorescent labeling of each cell type simplified the observation of these processes in the MPS.
The Chen group developed an MPS with two hollow channels to simulate pancreatic cancer ducts and microvessels to efficiently recreate unique tumor-endothelium interaction in pancreatic ductal adenocarcinoma (PDAC).86 PDAC invades peripancreatic vasculature like many other tumors, but paradoxically, it exhibits hypo-vascularity. Their MPS platform allowed for the efficient observation of tumor cells from pancreatic cancer duct channel invading, and displacing vascular endothelium, referred as endothelial ablation.86 Furthermore, it allowed to reveal a key pathway in the endothelial ablation process, by using chemical inhibitors. Similarly, Lee group cultured tumor spheroids in ECM hydrogel alongside the hollow vascular channel created by needles and observed angiogenesis and vascular invasion through fluorescence staining analysis (Fig. 2d).87
In summary, the hollow vascular channel within an ECM hydrogel enhances the biological relevance of tumor studies, significantly improving cell–cell and cell–ECM interaction studies in metastasis research. However, a notable limitation of this system is that the perivascular region is not an open channel. This design aspect makes it challenging to analyze effluents from the vascular and perivascular regions, which can be crucial for a comprehensive understanding of the TME and its dynamics in metastasis.
For example, one study transformed a 6-well plate into a microfluidic system to study bone metastasis in prostate cancer.98 The larger well plates allowed for the cultivation of mouse xenograft tumor tissue, facilitating research on metastatic characteristics of patient-derived samples. Another innovation reported the photolithographic-free fabrication of millimeter-sized chambers for multi-cellular cultivation.99 This technique created controlled micro-patterns for uniform cell seeding and effectively isolated the chamber, enabling the study of tumor-fibroblast metastasis through fluorescent imaging analysis. Likewise, a three-chamber MPS device utilizing 3D printing effectively emulated the impact of sympathetic activation on the interaction between breast cancer cells and bone cells mediated by neurons (Fig. 2e).96 This platform relies on static diffusion between the three chambers to enable bidirectional communication, not incorporating fluid flow.96
The Mayorca-Guiliani team took a different approach, proposing a more in vivo-like vasculature formation by decellularizing and then recellularizing the blood vessels of intact mouse tissues.100 This model provided a precise means to monitor lung metastasis behavior of tumor cells and enabled the recreation of unique tissue microenvironments by co-culturing cells in original tissues. Furthermore, it offers a controllable, organ-specific ex vivo model that closely mimics in vivo metastasis processes across various tissues by inducing tumor metastasis cell signaling.
3.3. Hybrid compartmentalization approach
With the evolution of MPS, there has been a trend towards developing devices that merge the advantages of different MPS designs. For instance, Zhang group created a dual-chamber MPS, establishing separate chambers for primary and secondary tumor sites.97 The MPS integrated a PET membrane to efficiently compartmentalize the tumor sites and the vascular chamber, enabling the study of bone metastasis originating from primary liver cancer. Another innovative design combined microwells and membranes to investigate the progression and drug response of kidney tumor cells in the context of liver metastasis.101 In this setup, HepLL and Caki-1 cells were embedded within a liver-specific ECM in seven distinct microwells, each containing varying ratios of cells. The employment of a membrane to separate these compartments effectively mimicked the progression of liver-metastatic tumor cells, showcasing the potential of combining different MPS components.4. MPS recapitulating metastasis organotropism
The exact processes driving organotrophic metastasis are not fully understood yet, but they are believed to arise from a network of complex interactions. These include the molecular traits of cancer cells, each organ's microenvironment, the unique properties of the vascular bed in each organ, and the immune system.28,51,102,103 Unraveling these complex mechanisms is key to developing methods for predicting, preventing, and treating cancer metastasis.Organotropism has been extensively studied in animal models such as mice, rats, or chick embryos.104 These in vivo studies typically monitor the distribution of injected tumor cells into the bloodstream and their spread to various organs, shedding light on the patterns of metastasis in different types of tumors. However, these models often fall short in precisely controlling cellular behaviors, posing challenges in deciphering why cancer cells favor certain tissues over others for metastasis. MPS, particularly those modeling multiple organs, have been suggested as promising tools for unraveling the intricacies of metastatic organotropism. Although the number of such MPS studies remains limited, they have already begun to shed light on the complex mechanisms of how cancer cells choose their metastatic destinations.
The Lee group, for instance, developed an MPS with a hollow channel formed from human umbilical vein endothelial cells (HUVECs), surrounded by an ECM hydrogel containing parenchymal cells.26 This design enabled the study of extravasation in triple-negative breast cancer cells (TNBC), influenced by chemical signals from the parenchymal cells. Their research revealed that bone-like environments with osteoblasts and mesenchymal stem cells promote extravasation in bone-tropic TNBC cells, while lung-like environments are favored by lung-tropic TNBC cells. This MPS's comparative analysis has demonstrated that vascular permeability can be rapidly altered due to interactions between parenchymal and tumor cells, providing vital insights into tumor metastasis patterns and highlighting the role of organ-specific parenchymal cells in guiding the extravasation of organotrophic CTCs.26
Another research group developed two distinct MPSs to study extravasation and homing behavior of breast cancer cells distinctly25 (Fig. 3a). The first platform focused on invasion/chemotaxis with partitioned pillar arrays in parallel channels, comprising a homing and a tumor channel. The second, an extravasation platform, featured a homing channel adjacent to a vascular channel with an interior surface coated with laminin to create an intact endothelial monolayer. In this setup, lung, liver, and breast tissue-specific parenchymal cells were cultured in the homing channel, and the migration of breast cancer cells from the tumor or vascular channel was quantified using confocal fluorescence microscopy.
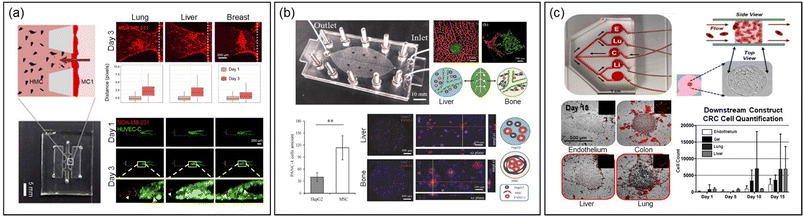 | ||
| Fig. 3 Exploring metastasis organotropism through the MPS technology. (a) MPSs featuring invasion/chemotaxis (upper) and extravasation (lower) to quantitatively assess infiltration and extravasation into specific tissues, mimicking the lung, liver and breast microenvironments. Reproduced from ref. 25 with permission from Wiley, copyright 2021. (b) A vascular MPS system, inspired by the structure of leaf veins and featuring two chambers simulating the liver and bone microenvironments, facilitates the validation of conducting comparative experiments for organ-specific metastasis studies on a single MPS platform. Reproduced from ref. 24 with permission from Wiley, copyright 2021. (c) An MPS with a single upper chamber for growing colorectal cancer spheroids, linked to downstream chambers simulating the lung, liver, and endothelium. The migration of cancer cells from the primary site to each chamber is monitored using fluorescence microscopy, allowing for the assessment of metastatic organ preference. Reproduced from ref. 23 with permission from Wiley, copyright 2018. | ||
A recent innovative MPS study involved a system with multiple chambers connected by microfluidic structures, inspired by leaf vein branching patterns (Fig. 3b).24 This design created complex architectures resembling the human cardiovascular system, linking vascularized organs in millimeter-sized chambers. In this system, pancreatic cancer cells were introduced to mimic organotropism metastasis, showing extravasation and infiltration into liver and bone chambers containing HepG2 cells and mesenchymal stem cells, respectively.
The Skardal group developed a simpler MPS with a single upper chamber for growing colorectal cancer spheroids, linked to downstream chambers housing liver, lung, and endothelial constructs in parallel (Fig. 3c).23 The tumor cells released from the spheroids migrated to these downstream chambers, demonstrating attachment, proliferation, and a notable preference for the lung.
5. Concluding and future perspectives
In this review, we have discussed the recent advancements in MPS as an in vitro platform for studying tumor metastasis and organotropism. MPS are highly effective in replicating the intricate and dynamic nature of cancer tissues, thus providing a valuable tool for exploring various aspects, including cancer mechanisms, the impact of treatments, and the screening of anti-cancer drugs. These assessments are anticipated to be pivotal in narrowing the gap between preclinical studies and clinical trials, potentially hastening the introduction of new therapeutic or diagnostic solutions. Significantly, in December 2022, the Food and Drug Administration endorsed alternative approaches to animal testing for drug approval, such as the use of MPS, showcasing a growing reliance on MPS in development and preclinical investigations.105Despite these advancements, MPS still confronts numerous biological and technical challenges. Biologically, there are difficulties in accurately simulating the complex TME, including the ECM and immune contexts, achieving tissue maturity, and capturing inter-individual variability. Technically, the field grapples with issues like intricate manufacturing processes, challenges in large-scale production, limitations in clearing and imaging technologies, and the absence of standardized analysis methodologies.
Recent MPS studies have successfully replicated the known patterns of metastasis organotropism by incorporating tissue-specific parenchymal cells. However, these models often do not fully represent the unique immune environments or endothelial characteristics of the tissues. A more comprehensive approach would involve using primary endothelial cells from the target organ to establish a tissue-specific vascular environment and co-culturing them with resident immune cells of that tissue. Enhancing the ECM environment to be tissue-specific, such as by using decellularized ECM hydrogel29 or fibroblast-derived matrices from the target tissue,106 would also add to the clinical relevance of MPS.
Presently, MPS research often employs homogeneous tumor cell lines; however, the reality of significant inter- and intra-patient variability greatly influences metastasis patterns61,62 (Fig. 4). Using patient-derived tumor organoids, which retain this heterogeneity, can significantly improve our understanding of tumor organotropism and advance the clinical application of MPS.16
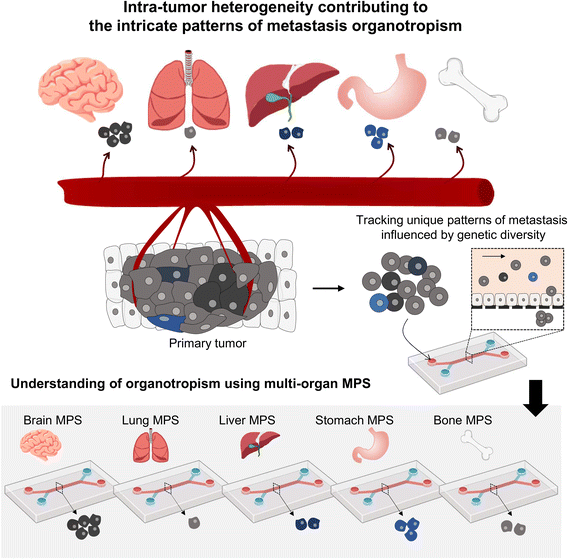 | ||
| Fig. 4 Investigating the metastasis organotropism mechanisms driven by genetic heterogeneity within a primary tumor by employing a multi-organ MPS platform. | ||
For a deeper analysis of tumor metastasis in multi-organ MPS, integrating omics technologies could provide groundbreaking insights. Omics analysis allows for the collection and examination of a wide range of molecular data, including gene expression, protein levels, and metabolites. This approach can reveal intricate details about the mechanisms of organotropism in metastatic tumors.42,67,96,107 By exploring specific gene expression changes linked to preferred metastatic sites or the role of proteins in metastasis, researchers can gain valuable insights. Integrating diverse omics data facilitates understanding the root causes and mechanisms behind metastatic tumors, contributing to the development of specific treatment and prevention strategies.108 Detailed characterizations of metastasized tumors in multi-organ MPS using omics data would enable comprehensive understanding, preventing, and treating metastatic organotropism.
Author contributions
HY composed the initial draft, JS and SWC provided revisions to that draft, and TEP supervised all work.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by a National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT (NRF-2022M3A91015716, 2020R1C1C1014753, and 2021R1A4A3030597).References
- I. Dagogo-Jack and A. T. Shaw, Nat. Rev. Clin. Oncol., 2018, 15, 81–94 CrossRef CAS PubMed.
- D. A. Lawson, K. Kessenbrock, R. T. Davis, N. Pervolarakis and Z. Werb, Nat. Cell Biol., 2018, 20, 1349–1360 CrossRef CAS PubMed.
- J. Fares, M. Y. Fares, H. H. Khachfe, H. A. Salhab and Y. Fares, Signal Transduction Targeted Ther., 2020, 5, 28 CrossRef PubMed.
- M. W. Pickup, J. K. Mouw and V. M. Weaver, EMBO Rep., 2014, 15, 1243–1253 CrossRef CAS PubMed.
- K. W. Hunter, R. Amin, S. Deasy, N.-H. Ha and L. Wakefield, Nat. Rev. Cancer, 2018, 18, 211–223 CrossRef CAS PubMed.
- G. S. Karagiannis, J. S. Condeelis and M. H. Oktay, Clin. Exp. Metastasis, 2018, 35, 269–284 CrossRef CAS PubMed.
- P. Bouchalova and P. Bouchal, Cancer Cell Int., 2022, 22, 394 CrossRef PubMed.
- M. E. Katt, A. L. Placone, A. D. Wong, Z. S. Xu and P. C. Searson, Front. Bioeng. Biotechnol., 2016, 4, 12 Search PubMed.
- D. Ferreira, F. Adega and R. Chaves, in Oncogenomics and Cancer Proteomics, ed. C. López-Camarillo and E. Aréchaga-Ocampo, IntechOpen, Rijeka, 2013, ch. 6, pp. 139–166, DOI:10.5772/53110.
- X.-L. Yang, Y. Shi, D.-D. Zhang, R. Xin, J. Deng, T.-M. Wu, H.-M. Wang, P.-Y. Wang, J.-B. Liu, W. Li, Y.-S. Ma and D. Fu, Mol. Ther.--Oncolytics, 2021, 21, 255–263 CrossRef CAS PubMed.
- B. Vikas and S. Anil, in Cell Growth, ed. B. Vikas and M. Fasullo, IntechOpen, Rijeka, 2019, ch. 7, p. 101, DOI:10.5772/intechopen.90226.
- M. A. Heinrich, A. M. R. H. Mostafa, J. P. Morton, L. J. A. C. Hawinkels and J. Prakash, Adv. Drug Delivery Rev., 2021, 174, 265–293 CrossRef CAS PubMed.
- L. Gómez-Cuadrado, N. Tracey, R. Ma, B. Qian and V. G. Brunton, Dis. Models Mech., 2017, 10, 1061–1074 CrossRef PubMed.
- I. W. Mak, N. Evaniew and M. Ghert, Am. J. Transl. Res., 2014, 6, 114–118 Search PubMed.
- D. E. Ingber, Nat. Rev. Genet., 2022, 23, 467–491 CrossRef CAS PubMed.
- M. C. Koyilot, P. Natarajan, C. R. Hunt, S. Sivarajkumar, R. Roy, S. Joglekar, S. Pandita, C. W. Tong, S. Marakkar, L. Subramanian, S. S. Yadav, A. V. Cherian, T. K. Pandita, K. Shameer and K. K. Yadav, Cell, 2022, 11(11), 1828 CrossRef CAS PubMed.
- M. Jouybar, C. M. de Winde, K. Wolf, P. Friedl, R. E. Mebius and J. M. J. den Toonder, Trends Biotechnol., 2023 DOI:10.1016/j.tibtech.2023.10.001.
- A. Chramiec, D. Teles, K. Yeager, A. Marturano-Kruik, J. Pak, T. Chen, L. Hao, M. Wang, R. Lock, D. N. Tavakol, M. B. Lee, J. Kim, K. Ronaldson-Bouchard and G. Vunjak-Novakovic, Lab Chip, 2020, 20, 4357–4372 RSC.
- S. Park, T. H. Kim, S. H. Kim, S. You and Y. Jung, Cancers, 2021, 13(16), 3930 CrossRef CAS PubMed.
- Z. Ao, H. Cai, Z. Wu, L. Hu, X. Li, C. Kaurich, M. Gu, L. Cheng, X. Lu and F. Guo, Theranostics, 2022, 12, 3628–3636 CrossRef CAS PubMed.
- Y. Nashimoto, R. Okada, S. Hanada, Y. Arima, K. Nishiyama, T. Miura and R. Yokokawa, Biomaterials, 2020, 229, 119547 CrossRef CAS PubMed.
- J.-Y. Shoji, R. P. Davis, C. L. Mummery and S. Krauss, Adv. Healthcare Mater., 2023, 2301067 CrossRef PubMed.
- J. Aleman and A. Skardal, Biotechnol. Bioeng., 2019, 116, 936–944 CrossRef CAS PubMed.
- M. Mao, H. P. Bei, C. H. Lam, P. Chen, S. Wang, Y. Chen, J. He and X. Zhao, Small, 2020, 16, 2000546 CrossRef CAS PubMed.
- B. Firatligil-Yildirir, G. Bati-Ayaz, I. Tahmaz, M. Bilgen, D. Pesen-Okvur and O. Yalcin-Ozuysal, Biotechnol. Bioeng., 2021, 118, 3799–3810 CrossRef CAS PubMed.
- T. J. Kwak and E. Lee, Biofabrication, 2021, 13, 015002 CrossRef PubMed.
- Y. Gao, I. Bado, H. Wang, W. Zhang, J. M. Rosen and X. H. Zhang, Dev. Cell, 2019, 49, 375–391 CrossRef CAS PubMed.
- W. Chen, A. D. Hoffmann, H. Liu and X. Liu, npj Precis. Oncol., 2018, 2, 4 CrossRef PubMed.
- X. Tian, M. E. Werner, K. C. Roche, A. D. Hanson, H. P. Foote, S. K. Yu, S. B. Warner, J. A. Copp, H. Lara, E. L. Wauthier, J. M. Caster, L. E. Herring, L. Zhang, J. E. Tepper, D. S. Hsu, T. Zhang, L. M. Reid and A. Z. Wang, Nat. Biomed. Eng., 2018, 2, 443–452 CrossRef CAS PubMed.
- Y. Liu and X. Cao, Cell Res., 2016, 26, 149–150 CrossRef CAS PubMed.
- T. Yu, C. Wang, M. Xie, C. Zhu, Y. Shu, J. Tang and X. Guan, Biomed. Pharmacother., 2021, 137, 111314 CrossRef CAS PubMed.
- V. Mittal, Annu. Rev. Pathol., 2018, 13, 395–412 CrossRef CAS PubMed.
- M. A. Nieto, R. Y. Huang, R. A. Jackson and J. P. Thiery, Cell, 2016, 166, 21–45 CrossRef CAS PubMed.
- M. S. Brown, K. E. Muller and D. R. Pattabiraman, Cancers, 2022, 14(5), 1138 CrossRef CAS PubMed.
- G. Calibasi Kocal, S. Güven, K. Foygel, A. Goldman, P. Chen, S. Sengupta, R. Paulmurugan, Y. Baskin and U. Demirci, Sci. Rep., 2016, 6, 38221 CrossRef CAS PubMed.
- I. Rizvi, U. A. Gurkan, S. Tasoglu, N. Alagic, J. P. Celli, L. B. Mensah, Z. Mai, U. Demirci and T. Hasan, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, E1974–E1983 CrossRef CAS PubMed.
- V. Mani, Z. Lyu, V. Kumar, B. Ercal, H. Chen, S. V. Malhotra and U. Demirci, Adv. Biosyst., 2019, 3, 1800223 CrossRef PubMed.
- S. Valastyan and R. A. Weinberg, Cell, 2011, 147, 275–292 CrossRef CAS PubMed.
- A. K. Shenoy and J. Lu, Cancer Lett., 2016, 380, 534–544 CrossRef CAS PubMed.
- J. A. Nagy, S. H. Chang, A. M. Dvorak and H. F. Dvorak, Br. J. Cancer, 2009, 100, 865–869 CrossRef CAS PubMed.
- K. B. Quencer, T. Friedman, R. Sheth and R. Oklu, Cardiovasc. Diagn. Ther., 2017, 7, S165–S177 CrossRef PubMed.
- J. Ahn, D.-H. Kim, D.-J. Koo, J. Lim, T.-E. Park, J. Lee, J. Ko, S. Kim, M. Kim, K.-S. Kang, D.-H. Min, S.-Y. Kim, Y. Kim and N. L. Jeon, Acta Biomater., 2023, 165, 153–167 CrossRef CAS PubMed.
- Q. Fu, Q. Zhang, Y. Lou, J. Yang, G. Nie, Q. Chen, Y. Chen, J. Zhang, J. Wang, T. Wei, H. Qin, X. Dang, X. Bai and T. Liang, Oncogene, 2018, 37, 6105–6118 CrossRef CAS PubMed.
- G. Follain, N. Osmani, A. S. Azevedo, G. Allio, L. Mercier, M. A. Karreman, G. Solecki, M. J. Garcia Leòn, O. Lefebvre, N. Fekonja, C. Hille, V. Chabannes, G. Dollé, T. Metivet, F. Hovsepian, C. Prudhomme, A. Pichot, N. Paul, R. Carapito, S. Bahram, B. Ruthensteiner, A. Kemmling, S. Siemonsen, T. Schneider, J. Fiehler, M. Glatzel, F. Winkler, Y. Schwab, K. Pantel, S. Harlepp and J. G. Goetz, Dev. Cell, 2018, 45, 33–52.e12 CrossRef CAS PubMed.
- S. Rajput, P. Kumar Sharma and R. Malviya, Med. Drug Discovery, 2023, 18, 100158 CrossRef CAS.
- J. Massagué and A. C. Obenauf, Nature, 2016, 529, 298–306 CrossRef PubMed.
- A. F. Chambers, A. C. Groom and I. C. MacDonald, Nat. Rev. Cancer, 2002, 2, 563–572 CrossRef CAS PubMed.
- J. Eyles, A. L. Puaux, X. Wang, B. Toh, C. Prakash, M. Hong, T. G. Tan, L. Zheng, L. C. Ong, Y. Jin, M. Kato, A. Prévost-Blondel, P. Chow, H. Yang and J. P. Abastado, J. Clin. Invest., 2010, 120, 2030–2039 CrossRef CAS PubMed.
- T. Oskarsson, E. Batlle and J. Massagué, Cell Stem Cell, 2014, 14, 306–321 CrossRef CAS PubMed.
- C. M. Leung, P. de Haan, K. Ronaldson-Bouchard, G.-A. Kim, J. Ko, H. S. Rho, Z. Chen, P. Habibovic, N. L. Jeon, S. Takayama, M. L. Shuler, G. Vunjak-Novakovic, O. Frey, E. Verpoorte and Y.-C. Toh, Nat. Rev. Methods Primers, 2022, 2, 33 CrossRef CAS.
- Y. Zhang, F. Jiang, Y. C. Zhao, A.-N. Cho, G. Fang, C. D. Cox, H. Zreiqat, Z. F. Lu, H. Lu and L. A. Ju, Biomed. Mater., 2023, 18, 055008 CrossRef PubMed.
- M. Llenas, R. Paoli, N. Feiner-Gracia, L. Albertazzi, J. Samitier and D. Caballero, Bioengineering, 2021, 8, 81 CrossRef CAS PubMed.
- M. A. U. Khalid, Y. S. Kim, M. Ali, B. G. Lee, Y.-J. Cho and K. H. Choi, Biochem. Eng. J., 2020, 155, 107469 CrossRef CAS.
- A. S. Piotrowski-Daspit, J. Tien and C. M. Nelson, Integr. Biol., 2016, 8, 319–331 CrossRef CAS PubMed.
- J. Kim, V. Sunkara, J. Kim, J. Ro, C.-J. Kim, E. M. Clarissa, S. W. Jung, H. J. Lee and Y.-K. Cho, Lab Chip, 2022, 22, 2726–2740 RSC.
- H. Xu, Z. Li, Y. Guo, X. Peng and J. Qin, Electrophoresis, 2017, 38, 311–319 CrossRef CAS PubMed.
- N. Aceto, M. Toner, S. Maheswaran and D. A. Haber, Trends Cancer, 2015, 1, 44–52 CrossRef PubMed.
- I. J. Fidler, Eur. J. Cancer, 1973, 9, 223–227 CrossRef CAS PubMed.
- X. Tang, X. Liu, P. Li, F. Liu, M. Kojima, Q. Huang and T. Arai, Anal. Chem., 2020, 92, 11607–11616 CrossRef CAS PubMed.
- J. Nilsson, M. Evander, B. Hammarström and T. Laurell, Anal. Chim. Acta, 2009, 649, 141–157 CrossRef CAS PubMed.
- A.-N. Cho, Y. Jin, Y. An, J. Kim, Y. S. Choi, J. S. Lee, J. Kim, W.-Y. Choi, D.-J. Koo, W. Yu, G.-E. Chang, D.-Y. Kim, S.-H. Jo, J. Kim, S.-Y. Kim, Y.-G. Kim, J. Y. Kim, N. Choi, E. Cheong, Y.-J. Kim, H. S. Je, H.-C. Kang and S.-W. Cho, Nat. Commun., 2021, 12, 4730 CrossRef CAS PubMed.
- H.-G. Yi, Y. H. Jeong, Y. Kim, Y.-J. Choi, H. E. Moon, S. H. Park, K. S. Kang, M. Bae, J. Jang, H. Youn, S. H. Paek and D.-W. Cho, Nat. Biomed. Eng., 2019, 3, 509–519 CrossRef CAS PubMed.
- D. Coradini, C. Casarsa and S. Oriana, Acta Pharmacol. Sin., 2011, 32, 552–564 CrossRef CAS PubMed.
- J. Kim, C. Lee, I. Kim, J. Ro, J. Kim, Y. Min, J. Park, V. Sunkara, Y.-S. Park, I. Michael, Y.-A. Kim, H. J. Lee and Y.-K. Cho, ACS Nano, 2020, 14, 14971–14988 CrossRef CAS PubMed.
- B. Jing, Y. Luo, B. Lin, J. Li, Z. A. Wang and Y. Du, RSC Adv., 2019, 9, 17137–17147 RSC.
- S. Hao, L. Ha, G. Cheng, Y. Wan, Y. Xia, D. M. Sosnoski, A. M. Mastro and S.-Y. Zheng, Small, 2018, 14, 1702787 CrossRef PubMed.
- C. Strelez, S. Chilakala, K. Ghaffarian, R. Lau, E. Spiller, N. Ung, D. Hixon, A. Y. Yoon, R. X. Sun, H.-J. Lenz, J. E. Katz and S. M. Mumenthaler, iScience, 2021, 24, 102509 CrossRef CAS PubMed.
- G. Salimbeigi, N. E. Vrana, A. M. Ghaemmaghami, P. Y. Huri and G. B. McGuinness, Mater. Today Bio, 2022, 15, 100301 CrossRef CAS PubMed.
- H. H. Chung, M. Mireles, B. J. Kwarta and T. R. Gaborski, Lab Chip, 2018, 18, 1671–1689 RSC.
- B. Choi, J.-W. Choi, H. Jin, H.-R. Sim, J.-H. Park, T.-E. Park and J. H. Kang, Biofabrication, 2021, 13, 045020 CrossRef CAS PubMed.
- J.-W. Choi, M. Seo, K. Kim, A. R. Kim, H. Lee, H.-S. Kim, C. G. Park, S. W. Cho, J. H. Kang, J. Joo and T.-E. Park, ACS Nano, 2023, 17, 8153–8166 CrossRef CAS PubMed.
- F. Yin, X. Zhang, L. Wang, Y. Wang, Y. Zhu, Z. Li, T. Tao, W. Chen, H. Yu and J. Qin, Lab Chip, 2021, 21, 571–581 RSC.
- J. Grant, A. Özkan, C. Oh, G. Mahajan, R. Prantil-Baun and D. E. Ingber, Lab Chip, 2021, 21, 3509–3519 RSC.
- J. W. Choi, J. Youn, D. S. Kim and T. E. Park, Biomaterials, 2023, 293, 121983 CrossRef CAS PubMed.
- J. Youn, H. Hong, W. Shin, D. Kim, H. J. Kim and D. S. Kim, Biofabrication, 2022, 14, 025010 CrossRef PubMed.
- T. Huu Hoang, M. Sato-Matsubara, H. Yuasa, T. Matsubara, L. T. T. Thuy, H. Ikenaga, D. M. Phuong, N. V. Hanh, V. N. Hieu, D. V. Hoang, H. Hai, Y. Okina, M. Enomoto, A. Tamori, A. Daikoku, H. Urushima, K. Ikeda, N. Q. Dat, Y. Yasui, H. Shinkawa, S. Kubo, R. Yamagishi, N. Ohtani, K. Yoshizato, J. Gracia-Sancho and N. Kawada, Sci. Adv., 2022, 8, eabo5525 CrossRef PubMed.
- H. Yoon, J. K. Seo and T.-E. Park, Acta Biomater., 2023, 159, 188–200 CrossRef CAS PubMed.
- H. Mollica, R. Palomba, R. Primavera and P. Decuzzi, ACS Biomater. Sci. Eng., 2019, 5, 4834–4843 CrossRef CAS PubMed.
- Y.-C. Toh, A. Raja, H. Yu and D. Van Noort, Bioengineering, 2018, 5(2), 29 CrossRef PubMed.
- K.-C. Weng, Y. K. Kurokawa, B. S. Hajek, J. A. Paladin, V. S. Shirure and S. C. George, Tissue Eng., Part C, 2019, 26, 44–55 CrossRef PubMed.
- M. S. Loeian, S. Mehdi Aghaei, F. Farhadi, V. Rai, H. W. Yang, M. D. Johnson, F. Aqil, M. Mandadi, S. N. Rai and B. Panchapakesan, Lab Chip, 2019, 19, 1899–1915 RSC.
- P. Zhang, X. Wu, G. Gardashova, Y. Yang, Y. Zhang, L. Xu and Y. Zeng, Sci. Transl. Med., 2020, 12, eaaz2878 CrossRef CAS PubMed.
- E. A. Adjei-Sowah, S. A. O'Connor, J. Veldhuizen, C. Lo Cascio, C. Plaisier, S. Mehta and M. Nikkhah, Adv. Sci., 2022, 9, 2201436 CrossRef CAS PubMed.
- H. Y. Cho, J. H. Choi, K. J. Kim, M. Shin and J. W. Choi, Front. Bioeng. Biotechnol., 2020, 8, 611802 CrossRef PubMed.
- H. Kim, J. K. Sa, J. Kim, H. J. Cho, H. J. Oh, D.-H. Choi, S.-H. Kang, D. E. Jeong, D.-H. Nam, H. Lee, H. W. Lee and S. Chung, Adv. Sci., 2022, 9, 2201785 CrossRef CAS PubMed.
- D.-H. T. Nguyen, E. Lee, S. Alimperti, R. J. Norgard, A. Wong, J. J.-K. Lee, J. Eyckmans, B. Z. Stanger and C. S. Chen, Sci. Adv., 2019, 5, eaav6789 CrossRef CAS PubMed.
- T. J. Kwak and E. Lee, Sci. Rep., 2020, 10, 20142 CrossRef CAS PubMed.
- J. M. Ayuso, R. Truttschel, M. M. Gong, M. Humayun, M. Virumbrales-Munoz, R. Vitek, M. Felder, S. D. Gillies, P. Sondel, K. B. Wisinski, M. Patankar, D. J. Beebe and M. C. Skala, OncoImmunology, 2019, 8, 1553477 CrossRef PubMed.
- Q. Liu, L. S. Mille, C. Villalobos, I. Anaya, M. Vostatek, S. Yi, W. Li, J. Liao, H. Wu, Y. Song, L. Xiong and Y. S. Zhang, Bio-Des. Manuf., 2023, 6, 373–389 CrossRef CAS.
- K. M. Lugo-Cintrón, M. M. Gong, J. M. Ayuso, L. A. Tomko, D. J. Beebe, M. Virumbrales-Muñoz and S. M. Ponik, Cancers, 2020, 12(5), 1173 CrossRef PubMed.
- N. Y. C. Lin, K. A. Homan, S. S. Robinson, D. B. Kolesky, N. Duarte, A. Moisan and J. A. Lewis, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 5399–5404 CrossRef CAS PubMed.
- L. L. Bischel, E. W. Young, B. R. Mader and D. J. Beebe, Biomaterials, 2013, 34, 1471–1477 CrossRef CAS PubMed.
- J. M. Ayuso, S. Rehman, M. Farooqui, M. Virumbrales-Muñoz, V. Setaluri, M. C. Skala and D. J. Beebe, Int. J. Mol. Sci., 2020, 21(23), 9075 CrossRef CAS PubMed.
- J. M. Ayuso, S. Rehman, M. Virumbrales-Munoz, P. H. McMinn, P. Geiger, C. Fitzgerald, T. Heaster, M. C. Skala and D. J. Beebe, Sci. Adv., 2021, 7, eabc2331 CrossRef CAS PubMed.
- M. Virumbrales-Muñoz, J. Chen, J. Ayuso, M. Lee, E. J. Abel and D. J. Beebe, Lab Chip, 2020, 20, 4420–4432 RSC.
- F. Conceição, D. M. Sousa, J. Loessberg-Zahl, A. R. Vollertsen, E. Neto, K. Søe, J. Paredes, A. Leferink and M. Lamghari, Mater. Today Bio, 2022, 13, 100219 CrossRef PubMed.
- F. Sharifi, O. Yesil-Celiktas, A. Kazan, S. Maharjan, S. Saghazadeh, K. Firoozbakhsh, B. Firoozabadi and Y. S. Zhang, Bio-Des. Manuf., 2020, 3, 189–202 CrossRef CAS.
- T. Osawa, W. Wang, J. Dai and E. T. Keller, Sci. Rep., 2019, 9, 14979 CrossRef PubMed.
- L. Zhao, T. Guo, L. Wang, Y. Liu, G. Chen, H. Zhou and M. Zhang, Anal. Chem., 2018, 90, 777–784 CrossRef CAS PubMed.
- M. Rafaeva, E. R. Horton, A. R. D. Jensen, C. D. Madsen, R. Reuten, O. Willacy, C. B. Brøchner, T. H. Jensen, K. W. Zornhagen, M. Crespo, D. S. Grønseth, S. R. Nielsen, M. Idorn, P. thorStraten, K. Rohrberg, I. Spanggaard, M. Højgaard, U. Lassen, J. T. Erler and A. E. Mayorca-Guiliani, Adv. Healthcare Mater., 2022, 11, 2100684 CrossRef CAS PubMed.
- Y. Wang, D. Wu, G. Wu, J. Wu, S. Lu, J. Lo, Y. He, C. Zhao, X. Zhao, H. Zhang and S. Wang, Theranostics, 2020, 10, 300–311 CrossRef CAS PubMed.
- X. Xie, S. Lian, Y. Zhou, B. Li, Y. Lu, I. Yeung and L. Jia, J. Controlled Release, 2021, 331, 404–415 CrossRef CAS PubMed.
- C. Wang, K. Xu, R. Wang, X. Han, J. Tang and X. Guan, J. Exp. Clin. Cancer Res., 2021, 40, 370 CrossRef CAS PubMed.
- A. Giacobbe and C. Abate-Shen, Trends Cancer, 2021, 7, 916–929 CrossRef CAS PubMed.
- J. J. Han, Artif. Organs, 2023, 47, 449–450 CrossRef PubMed.
- A. R. D. Jensen, E. R. Horton, L. H. Blicher, E. J. Pietras, C. Steinhauer, R. Reuten, E. M. Schoof and J. T. Erler, Cancers, 2021, 13(13), 3331 CrossRef CAS PubMed.
- L. Zheng, B. Wang, Y. Sun, B. Dai, Y. Fu, Y. Zhang, Y. Wang, Z. Yang, Z. Sun, S. Zhuang and D. Zhang, ACS Sens., 2021, 6, 823–832 CrossRef CAS PubMed.
- S. Chakraborty, M. I. Hosen, M. Ahmed and H. U. Shekhar, BioMed Res. Int., 2018, 2018, 9836256 Search PubMed.
- C. Li, S. Li, K. Du, P. Li, B. Qiu and W. Ding, ACS Appl. Mater. Interfaces, 2021, 13, 19768–19777 CrossRef CAS PubMed.
- J. Bae, M.-H. Kim, S. Han and S. Park, BioChip J., 2023, 17, 77–84 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2024 |