Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Development of a self-contained microfluidic chip and an internet-of-things-based point-of-care device for automated identification of respiratory viruses†
Huynh Quoc
Nguyen‡
 ,
Van Dan
Nguyen‡
,
Vu Minh
Phan
and
Tae Seok
Seo
,
Van Dan
Nguyen‡
,
Vu Minh
Phan
and
Tae Seok
Seo
 *
*
Department of Chemical Engineering (BK21 FOUR Integrated Engineering Program), Kyung Hee University, Yongin, 17104, South Korea. E-mail: seots@khu.ac.kr; Fax: +82 31 204 8114; Tel: +82 31 201 3676
First published on 28th March 2024
Abstract
The COVID-19 pandemic greatly impacted the in vitro diagnostic market, leading to the development of new technologies such as point-of-care testing (POCT), multiplex testing, and digital health platforms. In this study, we present a self-contained microfluidic chip integrated with an internet-of-things (IoT)-based point-of-care (POC) device for rapid and sensitive diagnosis of respiratory viruses. Our platform enables sample-to-answer diagnostics within 70 min by automating RNA extraction, reverse transcription-loop-mediated isothermal amplification (RT-LAMP), and fluorescence detection. The microfluidic chip is designed to store all the necessary reagents for the entire diagnostic assay, including a lysis buffer, a washing buffer, an elution buffer, and a lyophilized RT-LAMP cocktail. It can perform nucleic acid extraction, aliquoting, and gene amplification in multiple reaction chambers without cross-contamination. The IoT-based POC device consists of a Raspberry Pi 4 for device control and data processing, a CMOS sensor for measuring fluorescence signals, a resistive heater panel for temperature control, and solenoid valves for controlling the movement of on-chip reagent solutions. The proposed device is portable and features a touchscreen for user control and result display. We evaluated the performance of the platform using 11 clinical respiratory virus samples, including 5 SARS-CoV-2 samples, 2 influenza A samples, and 4 influenza B samples. All tested clinical samples were accurately identified with high specificity and fidelity, demonstrating the ability to simultaneously detect multiple respiratory viruses. The combination of the integrated microfluidic chip with the POC device offers a simple, cost-effective, and scalable solution for rapid molecular diagnosis of respiratory viruses in resource-limited settings.
The COVID-19 pandemic has had a profound impact on the in vitro diagnostic market in a number of ways.1 In particular, real-time reverse transcriptase-polymerase chain reaction (RT-PCR) assays, which are considered to be the most accurate and trustworthy for diagnosis, have seen an upsurge in demand that created challenges for supply chains and logistics,2,3 while new customer segments, such as public health authorities, employers, schools, and consumers, needed faster and more accessible testing solutions. The RT-PCR testing has become more difficult due to constraints on cost and turnaround time as well as a shortage of supplies. This challenge has accelerated the development and implementation of new technologies, including POC testing, multiplex testing, and digital health platforms.
The development of POC molecular devices makes it possible for the quick, sensitive, and low-cost identification of COVID-19 infection at the clinic level.4–7 Some of the most popular POC molecular testing for COVID-19 detection includes lateral flow assays (LFAs),8,9 loop-mediated isothermal amplification (LAMP)6,10 and CRISPR-based methods.11–13 These approaches have different advantages and limitations in terms of sensitivity, specificity, speed, cost and ease of use. LFAs are widely used for COVID-19 diagnosis because they are fast, easy to use, and low-cost.14 However, they have critical limitations of low sensitivity and specificity, which means that early infections that have low viral loads can be missed.15,16 Therefore, lateral flow assays are always used with caution and eventually they are interlinked with other testing methods such as RT-PCR to confirm the results and avoid misdiagnosis. In contrast to the LFA kits, RT-LAMP exhibits superior sensitivity and specificity, thereby enabling the detection of low viral loads. Compared to RT-PCR, RT-LAMP is a nucleic acid amplification assay that is quick, sensitive, specific, and low-cost. This technique has the capability to yield results within a timeframe of 15 to 60 min, without necessitating thermal cycling and sophisticated equipment. RT-LAMP works by first converting the SARS-CoV-2 RNA into DNA with a reverse transcriptase enzyme, and then amplifying targeted regions of the viral genome with a Bst polymerase enzyme and specifically designed primers. This amplification process produces a huge quantity of amplicons, which can be identified using a variety of techniques, including color change,17 fluorescence,18 and turbidity.19 Furthermore, its cost-effectiveness and scalability make it conducive to large-scale testing and screening, particularly in environments with limited resources. However, sample pretreatment and purification are usually required due to the negative effect of the inhibitors or impurities on the reaction efficiency. It may only be capable of detecting a few variations or co-infections due to its limited multiplexing capabilities.20
CRISPR-based methods for viral detection are a new approach to the accurate detection and treatment of viruses such as SARS-CoV-2. They use CRISPR–Cas systems like Cas12 and Cas13 for diagnosis of viral RNA.4,11 Although CRISPR-based methods for viral detection reduce the turnaround time, the drawback of the CRISPR-based methods for viral detection is the complexity of designing appropriate crRNAs that are limited to target regions in proximity to a protospacer adjacent motif (PAM). Mutations in the viral PAM sequence may disable recognition by the Cas protein as the virus evolves.12 In addition, most CRISPR–Cas diagnostic kits require an amplification step such as PCR, RPA, or LAMP before the CRISPR–Cas assay to enhance the detection sensitivity.7 This prerequisite complicates the whole procedure when a POC device for the CRISPR–Cas assay is developed. There are several reports showing the possibility of detecting with CRISPR/Cas without pre-amplification steps; however, a separate sample preparation step is needed.21–23
Recently, POC devices have presented a highly promising alternative to diagnose viral disease directly at the location where patients receive care.11,15,24–26 These devices offer rapid and dependable results while requiring minimal user involvement. However, they still lack suitable sample preparation methods. Among the diverse array of POC technologies, microfluidics-based devices have garnered significant interest owing to their inherent advantages in terms of miniaturization, integration, automation, and the ability to perform multiplexed assays simultaneously.27–29 The utilization of microfluidics in POC devices holds immense potential in revolutionizing virus detection and improving healthcare outcomes.
In this paper, we present a novel and cost-effective POC diagnostic platform that utilizes a self-contained microfluidic chip and an IoT-based POC device for respiratory virus detection. The all-in-one microfluidic chip contains all the necessary reagents for the assay and enables complete sample pretreatment using magnetic bead-based extraction. It also allows for simultaneous RT-LAMP assay and fluorescence detection in four reaction chambers, enabling the detection of multiple respiratory viruses, including SARS-CoV-2, influenza A, and influenza B, within a single device in just 70 minutes. To validate the high performance for real-world scenarios, we analyzed 11 clinical samples with high specificity and sensitivity.
Materials and methods
Chemicals and reagents
A MagMAX™ viral/pathogen nucleic acid isolation kit was purchased from Applied Biosystems™ (USA), which included a magnetic bead solution, a binding buffer solution, a proteinase K solution, and a washing I solution. A LavaLAMP RNA component kit was obtained from Lucigen (UK), consisting of a 10× LavaLAMP™ RNA buffer, LavaLAMP™ RNA enzyme, and 100 mM MgSO4. A dNTP mix (10 mM each) was purchased from Thermo Fisher Scientific (USA). A 10× SYBR dye solution was ordered from Invitrogen (USA), and the LAMP primers were synthesized by Macrogen (Korea). Heat-inactivated SARS-CoV-2 was obtained from ATCC (VR-1986HK™, Lot # 70036071, USA). Three mm-thick PMMA sheets were ordered from Acrytal (Korea), and a pressure-sensitive adhesive (PSA) film was purchased from HJ-Bioanalytik GmbH (Cat. 900360, Germany). Paraffin wax (ASTM D 87) with a melting point of 58–62 °C was from Sigma (USA). A super-hydrophobic reagent was delivered from Ultratech (USA).Preparation of an RT-LAMP assay
The MagMAX™ viral/pathogen nucleic acid isolation kit provided magnetic beads, a bead binding solution, a proteinase K solution, and a washing I solution. The lysis solution was prepared by mixing 530 μl of the bead binding solution, 20 μl of the magnetic bead solution, and 10 μl of the proteinase K solution. A washing II solution (80% ethanol) was prepared using ACS-graded ethanol. Nuclease-free water was used to elute the RNA from the magnetic beads. For the RT-LAMP reaction mix in one run on a chip, the components included 5.0 μl of a 10× LavaLAMP™ RNA buffer, 2.0 μl of LavaLAMP™ RNA enzyme, 2.5 μl of 100 mM MgSO4, and 4.0 μl of dNTP mix. The LAMP primers (F3, B3, FIP, BIP, LF, and LB) targeting genes of SARS-CoV-2, influenza A, and influenza B were adopted from previous reports.10 Information about the primers and target sequences is provided in Table S1.† For reference, the RT-LAMP reaction in a 200 μL tube was also conducted using a commercial qPCR machine (CFX Connect, BioRad, USA). The volume of the RT-LAMP mixture was 10 μL, comprising 5 μl of reaction mixture (RM), 0.4 μl of enzyme mixture (EM), 0.4 μl of SYBR green, 1.8 μl of nuclease-free water, and 1 μl of the RNA sample. The one-step RT-LAMP reaction proceeded at 65 °C for 60 min, and the fluorescence intensity of the RT-LAMP mixture was measured at 1 min intervals.Preparation of a self-contained microfluidic chip
The self-contained microfluidic chip was designed using Cut2D software (Vectric, USA) and then etched onto a 3.0 mm-thick PMMA sheet using a CNC machine (Tinyrobo30, Korea). The microfluidic circuits, including microchannels, chambers, and manifolds, were patterned on one side of the sheet. After fabrication, the chips underwent a cleaning process, which involved sonicating them in a detergent solution, followed by rinsing with 70% ethanol and DNase-free water. The chips were then sterilized for 30 min under UV light. Paraffin wax with a melting point of 58–62 °C was placed in the wax valves near the reaction chambers. The passive valves of the chip were coated with a super-hydrophobic reagent by applying 3 μL onto each valve and allowing it to dry for 30 min. Next, 2.4 μL of the primer mix was dropped into each reaction chamber and left to dry for 1 hour at room temperature. Subsequently, 0.5 μL of SYBR green was loaded into each reaction chamber, followed by the loading of the RT-LAMP reaction mix into the mixing chambers. The microfluidic chips were rapidly transferred to a −80 °C freezer and kept there for 2 hours before being placed in a vacuum freeze-dryer for 8 hours. After the drying process, the microfluidic chip was promptly removed and sealed on the front side using a PSA film to prevent the dried reagents from being exposed to humidity. Finally, the washing solutions and the elution solution were loaded into their designated chambers through the holes in the backside, and the backside of the chip was sealed with a PSA film. For each microfluidic chip, 50 μL of a lysis solution, 80 μL of a washing I solution, 100 μL of a washing II solution, and 50 μL of nuclease-free water were loaded. For molecular diagnostics, 50 μL of the sample was introduced into the lysis chamber.Hardware components of the IoT-based POC device
The structure of the device was created using Autodesk 360 Fusion, and it was printed using a Formlabs 3B+ SLA 3D printer with Clear Resin V4. The outer covers, on the other hand, were printed using an FDM printer (Raise3D E2, USA) and PLA filaments. The central processing unit of the device was a Raspberry Pi 4. High torque servos (TowerPro, MG996R) and solenoid valves were used to control the operation of the microfluidic chip and the direction of liquid flow. The device included three resistive heaters that were etched onto a copper-clad laminate sheet using a CNC machine. The temperature was monitored using a microcontroller (Arduino Nano) with three thermistors and three MOSFETs (IRLB3813).For real-time fluorescence detection, the device incorporated a high-intensity light-emitting diode (5 mW) with an excitation wavelength of 488 nm (488T-60 DS5, Q-LINE, China), a bandpass filter (525 nm CWL, dia. 12.5 mm, 70 nm bandwidth, Edmund Optics, USA), and a Sony IMX219 8 megapixel CMOS sensor. User control and data display were facilitated through a 5 inch 800 × 480 TFT Raspberry Pi DSI Touchscreen. Additionally, an uninterruptible power supply (UPS HAT) (B)5V-5A from Waveshare (Hong Kong) was utilized to ensure continuous power supply to the device. The total cost of the hardware was less than $500.
Software development
The JavaScript-based in-house software was developed using the Node-RED programming tool, an IoT framework, to control the input and output pins of the Raspberry Pi 4. Node-RED facilitates remote control and monitoring of the device through wireless and internet-based channels. To implement PID control for the heater, the Arduino IDE was used to write the code in C++ for the Arduino Nano. Communication between the Raspberry Pi and Arduino Nano was established via a serial port. The fluorescence signal generated by each reaction chamber was captured using a CMOS sensor, and the cumulative green intensities in a 200 × 200 pixel region surrounding each reaction chamber were calculated. The intensity data were then fitted to a 5-parameter log-logistic (5PLL) curve using eqn (1). The 5PLL model is a well-established statistical model commonly employed to analyze quantitative PCR data for determining the threshold cycle of an amplification reaction. This is accomplished by fitting a log-logistic function to the fluorescence intensities versus the reaction time. Our device utilizes a customized algorithm to estimate all the model parameters. The algorithm iteratively refines the parameter estimates over 1000 iterations to identify the set of parameters that produce the lowest root-mean-square deviation. The parameters required by the 5PLL model include the inflection point (P[0]), lower asymptote (P[1]), upper asymptote (P[2]), slope of the inflection point (P[3]), and maximum reaction rate (P[4]). Once these parameters have been estimated, the threshold time can be calculated using eqn (2). In our experimental setup, the threshold value was defined as 5 times the standard deviation of the fluorescence values. | (1) |
 | (2) |
Results and discussion
Design of the self-contained microfluidic chip
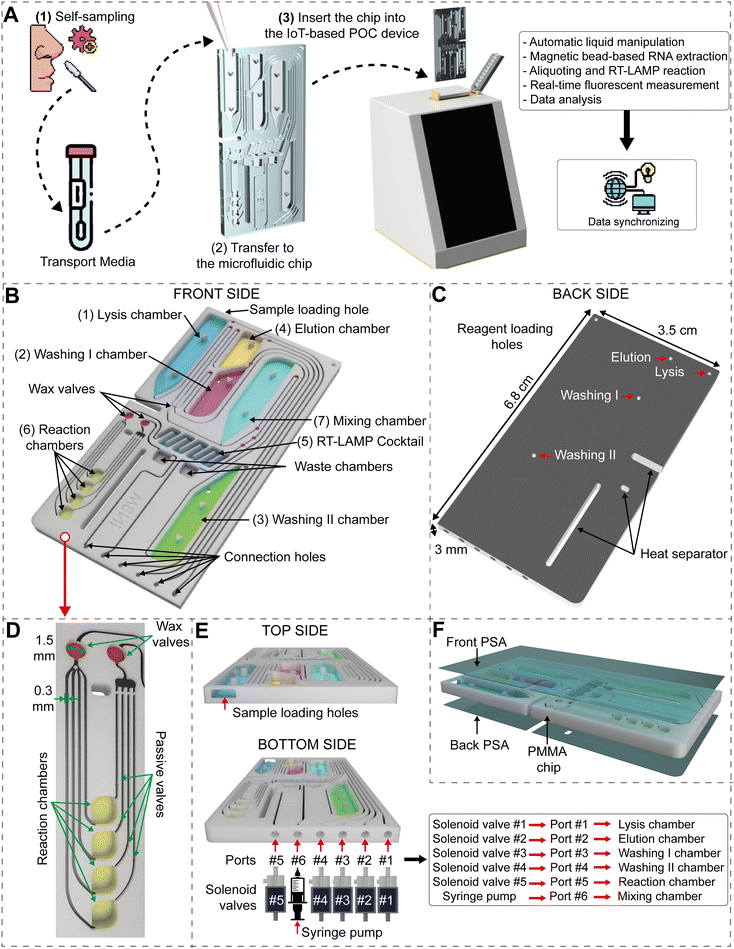
Our study focused on the development of not only a fully self-contained microfluidic chip but also a dedicated point-of-care (POC) device designed to streamline the diagnostic process from sample collection to result acquisition. With our platform, viral molecular diagnosis can be simplified into three straightforward steps (Fig. 1A). Firstly, users collect a sample themselves using a nasopharyngeal swab and deposit it into a transport medium. Secondly, a small portion (50–75 μl) of the medium is transferred onto the self-contained microfluidic chip. Finally, the microfluidic chip is inserted into the developed POC device. The remaining diagnostic process is automatically carried out within the POC device, including sequential and precise liquid manipulation, magnetic bead capture, regional temperature control, fluorescence detection, and data analysis. Moreover, our device is equipped with embedded IoT features, enabling seamless data sharing and updates through an internet connection.To ensure simplicity of use, the fully self-contained microfluidic chip was specifically designed to store all the necessary reagents for the complete diagnostic assay (refer to Fig. 1B). The chip consists of seven functional chambers, where chambers (1) to (4) are intended for storing reagents in the liquid form, while chambers (5) and (6) contain dried reagents. The mixing chamber (7) is left empty (Fig. 1B). The reagents include (1) a lysis solution, (2) a washing I solution, (3) a washing II solution, (4) an elution solution for RNA extraction, (5) lyophilized RT-LAMP cocktails, and (6) fluorescent dyes and primers for the amplification and detection of the viral RNA content. The microfluidic chip has dimensions of 6.8 cm (height) × 3.5 cm (width) × 3.0 mm (thickness) (Fig. 1C). On the front side of the microfluidic chip, reagent chambers were etched onto the PMMA sheet with a depth of 2.5 mm.
The on-chip reagent preparation process is detailed in the Preparation of a self-contained microfluidic chip section. To ensure an equal distribution of the RT-LAMP reaction solution among the four reaction chambers, an aliquoting structure was implemented. Each reaction chamber was designed to specifically target a different respiratory virus. This approach facilitated accurate compartmentalization of the reaction solution for precise and targeted diagnostic testing. The aliquoting structure started with a normally open circular wax valve (1.5 mm in diameter and 1.5 mm in depth) followed by four distinct microchannels. These microchannels led to reaction chambers with dimensions of 2.5 mm × 2.8 mm × 1.5 mm (height × width × depth). The reaction chambers were followed by passive valves, which were coated with a hydrophobic material. The passive valves created a higher resistance for the solution to flow through, so that the RT-LAMP reaction solution was completely filled in before reaching another open circular wax valve (Fig. 1D). The two wax valves were initially in the normally open state. When heated to the wax melting point, the wax would melt and flow, effectively blocking the microchannel. This mechanism served to prevent evaporation of the reaction solution and minimize the risk of cross-contamination during the RT-LAMP reaction. While all the necessary reagents were preloaded into the microfluidic chip, only the sample was loaded shortly before the diagnosis. This was done through a sample loading hole located on the top side of the microfluidic chip (Fig. 1E). The bottom side of the microfluidic chip featured six vertically fabricated connection holes in the PMMA sheet, with a depth of 2 mm. These connection holes were designed to align with a series of solenoid valves and a syringe pump (Fig. 1E). When the microfluidic chip was connected to the diagnostic device, the lysis, elution, washing I, washing II, and reaction chambers were aligned with solenoid valves #1, #2, #3, #4, and #5, respectively, via the corresponding connection holes and microchannels. The mixing chamber, on the other hand, was connected to a syringe pump. Both the front side and the back side of the microfluidic chip were sealed using a PSA film (Fig. 1F). This sealing ensured proper containment and prevention of any leakage during the diagnostic process.
Design and construction of the IoT-based diagnostic device
Next, we developed a dedicated POC device with the goal of automating the operation of the microfluidic chip and providing faster and more convenient diagnostic results compared to traditional laboratory-based tests. The device is portable with a size of 12 cm × 13 cm × 10 cm (length × height × width) (Fig. 2A). Despite its compact size, the device comprises several key components. These include a touchscreen for user control and result display, a 3D-printed scaffold for component alignment and positioning of the microfluidic chip, a servo motor to control the syringe pump, a vibration motor for efficient bead mixing, a CMOS sensor to measure the fluorescence signal from the reaction chambers, a Raspberry Pi 4 as the device controlling unit and for data computing, an Arduino Nano serving as the heat controlling unit, and a resistive heater panel to regulate the temperature of the microfluidic chip during lysis and RT-LAMP reactions. Additionally, the device incorporates an electromagnet to capture magnetic beads during RNA purification, solenoid valves for controlling the movement order of preloaded reagent solutions, and a power regulator to ensure stable voltage supply for the device's operation. Fig. 2B shows the fabricated and assembled POC device with wires connected. To firmly attach the microfluidic chip to the POC device, a PDMS layer was integrated, creating a flexible interface between the bottom side of the chip and the scaffold for the solenoid valves (Fig. 2C). Since the size of the chip was smaller than the five solenoid valves, the internal channels were patterned in the scaffold to match the connection holes of the chip with each solenoid valve. The flexible and sticky properties of the PDMS layer provided air-tight linkage between the chip and the scaffold (Fig. 2C). Once the microfluidic chip was securely placed inside the POC device, its front side would be in direct contact with the front side of the heat panel. This arrangement ensured alignment between the lysis chamber, mixing chamber, normally open wax valves, and reaction chamber with the lysis heater, electromagnet, wax heater, and reaction heater, respectively (Fig. 2D). To enable simultaneous measurement of the fluorescence signals from the four reaction chambers during the RT-LAMP reaction, an optical module was integrated into the region of the reaction chamber (Fig. 2F). This optical module consisted of a CMOS camera serving as a detector, a bandpass filter with a central wavelength of 525 nm to selectively transmit the desired fluorescence signal, and a 488 nm LED used as the excitation light source.Operation of the microfluidic chip using the IoT-based diagnostic device
Prior to the experiment, thorough cleaning and sterilization of the microfluidic chip were carried out. Briefly, the wax was inserted into the star-shaped junction between the lysis, washing I chamber, washing II chamber, elution chamber, and mixing chamber. Additionally, two circular holes of the aliquoting structure were also filled with wax (Fig. 3A). The star-shaped wax valve was initially filled completely to create a normally closed wax valve. When the heat was applied and the wax melted, the blocking wax could be pushed away so that the air and liquid flowed (i.e., the wax valve was open). On the other hand, the circular wax valves, with the wax loaded in the deeper etched circular chamber, were initially open because the underlying microchannel was not filled with the wax, allowing the passage of air and liquid. However, when the wax reached its melting point (65 °C), it was fully melted and blocked the underlying microchannels. As the temperature dropped, the wax was solidified again, resulting in the complete closure of the microchannels (i.e., the wax valve was closed).The lyophilized RT-LAMP cocktail was stored in the first half of a zigzag channel within the microfluidic chip (Fig. 3A). The specific primers and fluorescent dye were located in the four reaction chambers. Once these components were loaded, the front side of the microfluidic chip was sealed using a PSA film. Other solutions required for the diagnostic process were loaded into their designated chambers through the holes on the backside of the chip. The backside of the chip was then sealed with another layer of PSA film (Fig. 1C and 3A). During the diagnosis, the sample to be tested was loaded into the lysis chamber and sealed with a PSA film on the top side of the chip. When the chip was inserted into the POC device, a lid was used to secure and firmly press the chip in place. The lid was secured with a screw to ensure a tight connection between the microfluidic chip and the POC device (Fig. 3B).
Fig. 3C and Video S1† provide a visual representation of the automated steps involved in on-chip RNA extraction and RT-LAMP assay preparation using the IoT diagnostic device. (1) The first step of the diagnosis involved the lysis reaction, which lasted for 3 min. During this step, the sample was subjected to lysis facilitated by the vibration motor operating at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 RPM and the lysis heater maintained at a temperature of 65 °C, as recommended by the MagMax RNA extraction kit. The combination of vibration and heat helped to facilitate lysis of the sample, while the released viral RNAs were captured on the magnetic beads. Furthermore, the gradual increase in temperature resulted in the melting and subsequent opening of the star-shaped wax valve. (2) In the second step, the mixing process was performed. To initiate the mixing, the syringe connected to the mixing chamber was slowly withdrawn, creating a negative pressure within the mixing chamber. Simultaneously solenoid valve #1, which was connected to the top end of the lysis chamber, was opened. As a result, the sample solution present in the lysis chamber (indicated by its blue color) was forced to move from the lysis chamber into the mixing chamber. During this step, the electromagnet behind the mixing chamber was activated, causing the magnetic beads to adhere to the walls of the mixing chamber. (3) In the third step, bead capture was executed to separate the magnetic beads from the lysis solution. The direction of the syringe pump was reversed to push back, causing the sample solution to be pushed back from the mixing chamber to the lysis chamber. Meanwhile the electromagnet remained activated to immobilize the magnetic beads, ensuring that they remained attached to the walls of the mixing chamber. (4) The subsequent step involved adding the washing I solution (indicated by its purple color) to the mixing chamber. This was accomplished by opening solenoid valve #3 and operating the syringe pump in withdrawal mode. The vibration motor was activated to facilitate the agitation and washing of the beads in the washing I solution. During this step, the electromagnet was turned off to ensure proper washing of the beads. (5) In the fifth step, another round of bead capture was carried out. The electromagnet was activated, causing the magnetic beads to be separated from the washing I solution. The washing I solution was then moved back to the washing I chamber using the push mode of the syringe pump. Steps (6) and (7) involved washing the magnetic beads with the washing II solution (indicated by its green color), following a similar procedure to steps (4) and (5). In this case, solenoid valve #4 was activated instead of solenoid valve #3 to allow the washing II solution to be transferred to the mixing chamber. (8) In the eighth step, the elution solution (indicated by its yellow color) was transferred to the mixing chamber to release the RNAs that were attached to the magnetic beads into the solution. This was achieved by opening solenoid valve #2 and the withdrawal mode of the syringe, allowing the elution solution to flow into the mixing chamber. (9) In the next step, the RT-LAMP mixing process took place. By operating the syringe pump in the pushing mode and activating solenoid valve #5, the elution solution containing the released RNAs from the previous step was directed to the zigzag chamber. At this step, the eluted solution only filled half of the zigzag chamber, where the lyophilized RT-LAMP cocktail was located. To properly rehydrate the lyophilized RT-LAMP cocktail and ensure uniform distribution of the components within the reaction mixture, the elution solution was held in place within the chamber for a duration of 2 min. During this time, the vibration motor was activated, providing agitation and promoting the effective mixing of the reagents. This step allowed for optimal mixing and enhanced the efficiency and accuracy of the subsequent RT-LAMP amplification process. In step (10), the aliquoting of the RT-LAMP mixture was performed using the pushing mode of the syringe pump. The RT-LAMP reaction solution was transferred from the mixing chamber into the aliquoting structure, effectively filling all four reaction chambers. Once the RT-LAMP mixture was distributed into the reaction chambers, the wax heater was activated and heated to 80 °C. This temperature caused the normally open wax valves to quickly melt, resulting in the closure of the reaction chambers. This sealing process was crucial to prevent evaporation during the RT-LAMP reaction, ensuring the integrity and accuracy of the reaction conditions throughout the amplification process.
000 RPM and the lysis heater maintained at a temperature of 65 °C, as recommended by the MagMax RNA extraction kit. The combination of vibration and heat helped to facilitate lysis of the sample, while the released viral RNAs were captured on the magnetic beads. Furthermore, the gradual increase in temperature resulted in the melting and subsequent opening of the star-shaped wax valve. (2) In the second step, the mixing process was performed. To initiate the mixing, the syringe connected to the mixing chamber was slowly withdrawn, creating a negative pressure within the mixing chamber. Simultaneously solenoid valve #1, which was connected to the top end of the lysis chamber, was opened. As a result, the sample solution present in the lysis chamber (indicated by its blue color) was forced to move from the lysis chamber into the mixing chamber. During this step, the electromagnet behind the mixing chamber was activated, causing the magnetic beads to adhere to the walls of the mixing chamber. (3) In the third step, bead capture was executed to separate the magnetic beads from the lysis solution. The direction of the syringe pump was reversed to push back, causing the sample solution to be pushed back from the mixing chamber to the lysis chamber. Meanwhile the electromagnet remained activated to immobilize the magnetic beads, ensuring that they remained attached to the walls of the mixing chamber. (4) The subsequent step involved adding the washing I solution (indicated by its purple color) to the mixing chamber. This was accomplished by opening solenoid valve #3 and operating the syringe pump in withdrawal mode. The vibration motor was activated to facilitate the agitation and washing of the beads in the washing I solution. During this step, the electromagnet was turned off to ensure proper washing of the beads. (5) In the fifth step, another round of bead capture was carried out. The electromagnet was activated, causing the magnetic beads to be separated from the washing I solution. The washing I solution was then moved back to the washing I chamber using the push mode of the syringe pump. Steps (6) and (7) involved washing the magnetic beads with the washing II solution (indicated by its green color), following a similar procedure to steps (4) and (5). In this case, solenoid valve #4 was activated instead of solenoid valve #3 to allow the washing II solution to be transferred to the mixing chamber. (8) In the eighth step, the elution solution (indicated by its yellow color) was transferred to the mixing chamber to release the RNAs that were attached to the magnetic beads into the solution. This was achieved by opening solenoid valve #2 and the withdrawal mode of the syringe, allowing the elution solution to flow into the mixing chamber. (9) In the next step, the RT-LAMP mixing process took place. By operating the syringe pump in the pushing mode and activating solenoid valve #5, the elution solution containing the released RNAs from the previous step was directed to the zigzag chamber. At this step, the eluted solution only filled half of the zigzag chamber, where the lyophilized RT-LAMP cocktail was located. To properly rehydrate the lyophilized RT-LAMP cocktail and ensure uniform distribution of the components within the reaction mixture, the elution solution was held in place within the chamber for a duration of 2 min. During this time, the vibration motor was activated, providing agitation and promoting the effective mixing of the reagents. This step allowed for optimal mixing and enhanced the efficiency and accuracy of the subsequent RT-LAMP amplification process. In step (10), the aliquoting of the RT-LAMP mixture was performed using the pushing mode of the syringe pump. The RT-LAMP reaction solution was transferred from the mixing chamber into the aliquoting structure, effectively filling all four reaction chambers. Once the RT-LAMP mixture was distributed into the reaction chambers, the wax heater was activated and heated to 80 °C. This temperature caused the normally open wax valves to quickly melt, resulting in the closure of the reaction chambers. This sealing process was crucial to prevent evaporation during the RT-LAMP reaction, ensuring the integrity and accuracy of the reaction conditions throughout the amplification process.
Evaluation of the molecular diagnostic performance on the IoT-based POC device
The performance of the proposed platform was evaluated using three respiratory viruses: SARS-CoV-2, influenza A, and influenza B. The four reaction chambers within the microfluidic chip were pre-coated with specific LAMP primer sets. These primer sets targeted different genes, including (1) the 18S rRNA gene of humans, (2) the As1e gene of SARS-CoV-2, (3) the Neuraminidase gene of influenza A, and (4) the Nonstructural I gene of influenza B. The amplification curves and fluorescence images of the reaction chambers were obtained for each test and are shown in Fig. 4A. When testing a sample of SARS-CoV-2 (103 copies per μl), the amplification curves and corresponding fluorescence signals for the 18S rRNA gene of humans and the As1e gene of SARS-CoV-2 were detected in the first two chambers of the chip. Similarly, when testing an influenza A sample (103 pfu mL−1), the fluorescence signals for the 18S rRNA gene of humans and the Neuraminidase gene of influenza A were observed in the first and third chambers, respectively. Likewise, for an influenza B sample (103 pfu mL−1), the device detected fluorescence signals for the 18S rRNA gene of humans in the first chamber and the Nonstructural I gene of influenza B in the fourth chamber. These results confirm the successful identification of the specific viruses in their respective chambers without any cross-contamination, highlighting the effectiveness and accuracy of the microfluidic chip for diagnostic purposes.The limit-of-detection (LOD) for the three respiratory viruses, SARS-CoV-2, influenza A, and influenza B, was determined using the developed platform. For SARS-CoV-2, the concentration range analyzed was from 1 × 102 to 1 × 105 copies per μL. Within this range, the platform successfully identified the presence of SARS-CoV-2 with threshold times ranging from 33.7 min to 12.2 min (top panel of Fig. 4B). For influenza A, the virus was detectable in a concentration range of 1 × 102 to 1 × 105 pfu ml−1, with threshold times ranging from 48.1 min to 20.6 min (middle panel of Fig. 4B). Similarly, for influenza B, the platform could detect the virus within a concentration range of 1 × 102 to 1 × 105 pfu ml−1, with threshold times ranging from 44.9 min to 15.2 min (bottom panel of Fig. 4B). Therefore, the LOD was determined to be 1 × 102 copies per μL for SARS-CoV-2, 1 × 102 pfu ml−1 for influenza A, and 1 × 102 pfu ml−1 for influenza B on the developed POC device. These findings demonstrate the sensitivity of the platform in detecting low concentrations of the target viruses.
Validation of the proposed POC platform for the clinical sample test
To assess the practical application of our POC platform, we conducted tests on 11 clinical respiratory virus samples obtained from patients, including 5 samples of SARS-CoV-2, 2 samples of influenza A, and 4 samples of influenza B. The positivity of these clinical samples was confirmed by Kyung Hee University Hospital at Gangdong (Seoul, Korea) using a commercial qPCR machine. Fig. 5 illustrates the threshold times recorded for each reaction chamber after testing the clinical samples. Among the six SARS-CoV-2 samples, the fluorescence signal was detected within the threshold time of 14.3 min in the chambers containing the 18S rRNA gene, and within the threshold time of 25.2 min in the chambers containing the As1e gene. However, no signal was observed in the chambers with primer sets targeting influenza A and B. For the two influenza A samples, only the 18S rRNA human gene and Neuraminidase I gene were detected at an early threshold time of 9.2 min. Similarly, the four influenza B samples only exhibited fluorescence signals in the chambers containing the 18S rRNA human gene and the Nonstructural I gene of influenza B, as expected. It is worth noting that the negative control, which contained water instead of a viral sample, showed no signals in any of the reaction chambers, indicating the absence of false positive results in our diagnostic platform. The entire process, from sample loading to result acquisition, was completed within 70 min. These results validate the high fidelity of our system for the simultaneous detection of respiratory viruses, as well as the high specificity of the RT-LAMP primers employed. | ||
| Fig. 5 Validation of the practical applications of the proposed POC platform using 11 clinical respiratory virus samples. | ||
Conclusions
The study aimed to develop a fully self-contained microfluidic chip and an IoT-based POC device to streamline the diagnostic process for viral molecular diagnosis. The microfluidic chip was specifically designed to store all the necessary reagents for the diagnostic assay, including lyophilized RT-LAMP cocktails, fluorescent dyes, and primers. An aliquoting structure was implemented to ensure equal distribution of the RT-LAMP reaction solution into four reaction chambers, each targeting a specific respiratory virus. The microfluidic chip was integrated with the POC device, which featured various components such as a touchscreen for user control, a 3D-printed scaffold for precise alignment, a servo motor for controlled fluid manipulation, a vibration motor for mixing, a CMOS sensor for fluorescence detection, a Raspberry Pi for device control and data processing, an Arduino Nano for heat control, a resistive heater panel for precise temperature regulation, an electromagnet for bead capture, solenoid valves for fluid movement control, and a power regulator for stable voltage supply. Additionally, the IoT functionality of the device allowed for real-time data monitoring, data storage, and data transfer via the internet. The diagnostic performance of the developed platform was assessed by testing three respiratory viruses: SARS-CoV-2, influenza A, and influenza B. The system demonstrated the ability to accurately detect and identify each virus without any cross-contamination between the reaction chambers. The LOD for each virus indicated high sensitivity, allowing for the detection of low viral concentrations. The platform was further validated using 11 clinical respiratory virus samples, including SARS-CoV-2, influenza A, and influenza B samples. The results showed accurate detection and confirmation of these viruses within a short turnaround time of 70 min. The combination of the self-contained microfluidic chip and the POC device provided a simple, efficient, and accurate method for viral molecular diagnosis, which is applicable for effective disease management and control. In addition, our platform needs low power consumption and can be operated with a built-in battery (Video S2†). Thus, we believe that our advanced diagnostic system can be applied for on-site respiratory virus screening anytime, anywhere, and by anyone.Author contributions
All authors have given approval to the final version of the manuscript.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the Ministry of Health and Welfare of South Korea (HI22C0426), the Engineering Research Center of Excellence Program of the National Research Foundation of Korea (2021R1A5A6002853), the National Research Foundation of Korea (NRF), and the Ministry of Science and ICT (MSIT) (2020R1A2C1003960). We are grateful to Kyung Hee University Hospital for the donation of the clinical samples.References
- M. Kyhlstedt and S. W. Andersson, Health Policy Technol., 2020, 9, 126–128 CrossRef PubMed.
- W. Feng, A. M. Newbigging, C. Le, B. Pang, H. Peng, Y. Cao, J. Wu, G. Abbas, J. Song, D. B. Wang, M. Cui, J. Tao, D. L. Tyrrell, X. E. Zhang, H. Zhang and X. C. Le, Anal. Chem., 2020, 92, 10196–10209 CrossRef CAS PubMed.
- L. Lanser, R. Bellmann-Weiler, K. W. Öttl, L. Huber, A. Griesmacher, I. Theurl and G. Weiss, Infection, 2021, 49, 555–557 CrossRef CAS PubMed.
- J. P. Broughton, X. Deng, G. Yu, C. L. Fasching, V. Servellita, J. Singh, X. Miao, J. A. Streithorst, A. Granados, A. Sotomayor-Gonzalez, K. Zorn, A. Gopez, E. Hsu, W. Gu, S. Miller, C.-Y. Pan, H. Guevara, D. A. Wadford, J. S. Chen and C. Y. Chiu, Nat. Biotechnol., 2020, 38(7), 870–874 CrossRef CAS PubMed.
- M. J. Flynn, O. Snitser, J. Flynn, S. Green, I. Yelin, M. Szwarcwort-Cohen, R. Kishony and M. B. Elowitz, medRxiv, 2020, preprint, DOI:10.1101/2021.03.02.21252704.
- R. Sivakumar, V. P. Dinh and N. Y. Lee, Lab Chip, 2021, 21, 700–709 RSC.
- Q. Song, X. Sun, Z. Dai, Y. Gao, X. Gong, B. Zhou, J. Wu and W. Wen, Lab Chip, 2021, 21, 1634–1660 RSC.
- Y. Zhou, Y. Wu, L. Ding, X. Huang and Y. Xiong, TrAC, Trends Anal. Chem., 2021, 145, 116452 CrossRef CAS PubMed.
- K. Dighe, P. Moitra, M. Alafeef, N. Gunaseelan and D. Pan, Biosens. Bioelectron., 2022, 200, 113900 CrossRef CAS PubMed.
- H. Q. Nguyen, H. K. Bui, V. M. Phan and T. S. Seo, Biosens. Bioelectron., 2022, 195, 113655 CrossRef CAS PubMed.
- B. Casati, J. P. Verdi, A. Hempelmann, M. Kittel, A. G. Klaebisch, B. Meister, S. Welker, S. Asthana, S. Di Giorgio, P. Boskovic, K. H. Man, M. Schopp, P. A. Ginno, B. Radlwimmer, C. E. Stebbins, T. Miethke, F. N. Papavasiliou and R. Pecori, Nat. Commun., 2022, 13, 1–11 Search PubMed.
- M. Patchsung, K. Jantarug, A. Pattama, K. Aphicho, S. Suraritdechachai, P. Meesawat, K. Sappakhaw, N. Leelahakorn, T. Ruenkam, T. Wongsatit, N. Athipanyasilp, B. Eiamthong, B. Lakkanasirorat, T. Phoodokmai, N. Niljianskul, D. Pakotiprapha, S. Chanarat, A. Homchan, R. Tinikul, P. Kamutira, K. Phiwkaow, S. Soithongcharoen, C. Kantiwiriyawanitch, V. Pongsupasa, D. Trisrivirat, J. Jaroensuk, T. Wongnate, S. Maenpuen, P. Chaiyen, S. Kamnerdnakta, J. Swangsri, S. Chuthapisith, Y. Sirivatanauksorn, C. Chaimayo, R. Sutthent, W. Kantakamalakul, J. Joung, A. Ladha, X. Jin, J. S. Gootenberg, O. O. Abudayyeh, F. Zhang, N. Horthongkham and C. Uttamapinant, Nat. Biomed. Eng., 2020, 4, 1140–1149 CrossRef CAS PubMed.
- A. Ramachandran, D. A. Huyke, E. Sharma, M. K. Sahoo, C. Huang, N. Banaei, B. A. Pinsky and J. G. Santiago, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 29518–29525 CrossRef CAS PubMed.
- N. Taleghani and F. Taghipour, Biosens. Bioelectron., 2021, 174, 112830 CrossRef CAS PubMed.
- A. Yakoh, U. Pimpitak, S. Rengpipat, N. Hirankarn, O. Chailapakul and S. Chaiyo, Biosens. Bioelectron., 2021, 176, 112912 CrossRef CAS PubMed.
- M. Yüce, E. Filiztekin and K. G. Özkaya, Biosens. Bioelectron., 2021, 172, 112752 CrossRef PubMed.
- H. Q. Nguyen, V. D. Nguyen, H. Van Nguyen and T. S. Seo, Sci. Rep., 2020, 10, 1–10 CrossRef PubMed.
- Y. Cao, L. Wang, L. Duan, J. Li, J. Ma, S. Xie, L. Shi and H. Li, Sci. Rep., 2017, 7, 13394 CrossRef PubMed.
- N. Tomita, Y. Mori, H. Kanda and T. Notomi, Nat. Protoc., 2008, 3, 877–882 CrossRef CAS PubMed.
- X. Zhu, X. Wang, L. Han, T. Chen, L. Wang, H. Li, S. Li, L. He, X. Fu, S. Chen, M. Xing, H. Chen and Y. Wang, Biosens. Bioelectron., 2020, 166, 112437 CrossRef CAS PubMed.
- H. Shinoda, Y. Taguchi, R. Nakagawa, A. Makino, S. Okazaki, M. Nakano, Y. Muramoto, C. Takahashi, I. Takahashi, J. Ando, T. Noda, O. Nureki, H. Nishimasu and R. Watanabe, Commun. Biol., 2021, 4, 476 CrossRef CAS PubMed.
- A. Parinaz Fozouni, S. Son, M. Díaz de Leó Derby, J. A. Doudna, D. A. Fletcher, M. Ott, P. Fozouni, G. J. Knott, C. N. Gray, M. V. D'Ambrosio, C. Zhao, N. A. Switz, G. Renuka Kumar, S. I. Stephens, D. Boehm, C.-L. Tsou, J. Shu, A. Bhuiya, M. Armstrong, A. R. Harris, P.-Y. Chen, J. M. Osterloh, A. Meyer-Franke, B. Joehnk, K. Walcott, A. Sil, C. Langelier, K. S. Pollard, E. D. Crawford, A. S. Puschnik, M. Phelps, A. Kistler and J. L. DeRisi, Cell, 2021, 184, 323–333 CrossRef PubMed.
- H. Shinoda, T. Iida, A. Makino, M. Yoshimura, J. Ishikawa, J. Ando, K. Murai, K. Sugiyama, Y. Muramoto, M. Nakano, K. Kiga, L. Cui, O. Nureki, H. Takeuchi, T. Noda, H. Nishimasu and R. Watanabe, Commun. Biol., 2022, 5, 473 CrossRef CAS PubMed.
- H. Van Nguyen, V. D. Nguyen, H. Q. Nguyen, T. H. T. Chau, E. Y. Lee and T. S. Seo, Biosens. Bioelectron., 2019, 141, 111466 CrossRef PubMed.
- V. D. Nguyen, H. Q. Nguyen, K. H. Bui, Y. S. Ko, B. J. Park and T. S. Seo, Biosens. Bioelectron., 2022, 195, 113632 CrossRef CAS PubMed.
- G. Cao, J. Kong, Z. Xing, Y. Tang, X. Zhang, X. Xu, Z. Kang, X. Fang and M. Guan, Anal. Chim. Acta, 2018, 1024, 123–135 CrossRef CAS PubMed.
- S. J. Oh, B. H. Park, J. H. Jung, G. Choi, D. C. Lee, D. H. Kim and T. S. Seo, Biosens. Bioelectron., 2016, 75, 293–300 CrossRef CAS PubMed.
- K. Kadimisetty, J. Song, A. M. Doto, Y. Hwang, J. Peng, M. G. Mauk, F. D. Bushman, R. Gross, J. N. Jarvis and C. Liu, Biosens. Bioelectron., 2018, 109, 156–163 CrossRef CAS PubMed.
- S. Arshavsky-Graham and E. Segal, in Advances in Biochemical Engineering/Biotechnology, Springer Science and Business Media Deutschland GmbH, 2022, vol. 179, pp. 247–265 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Primer sequences for amplifying the target genes of SARS-CoV-2, influenza A, and influenza B and the human 18S rRNA gene (words). Video S1. Liquid manipulation on the POCT device (.mp4). Video S2. POCT heaters operating on a portable battery (.mp4). See DOI: https://doi.org/10.1039/d3lc00933e |
| ‡ Huynh Quoc Nguyen and Dan Van Nguyen contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |