Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A magnetic microneedle to isolate single immunomagnetically labeled cells†
Michiel
Stevens
 *a,
Philip
Harder
b and
Leon W. M. M.
Terstappen
*a,
Philip
Harder
b and
Leon W. M. M.
Terstappen
 ac
ac
aDepartment of Medical Cell BioPhysics, Faculty of Science and Technology, University of Twente, Enschede, The Netherlands. E-mail: m.stevens@utwente.nl
bUniversity of Applied Sciences and Arts, Bielefeld, Germany
cDepartment of General, Visceral and Pediatric Surgery, Heinrich-Heine University, University Hospital Düsseldorf, Düsseldorf, Germany
First published on 6th December 2023
Abstract
Immunomagnetic enrichment of cell populations from bodily fluids followed by immunofluorescent labeling is an established sample preparation method often used for the detection and enumeration of rare cells such as circulating tumor cells. For a detailed analysis of the heterogeneous characteristics of these cells, the cells need to be retrieved individually. Although several technologies are available to obtain 100% pure cells either individually or in bulk, these are often expensive, have low specificity, or suffer from high cell losses, either inherent to the technology or caused by sample transfer into special chips. To solve this issue, we introduce the magnetic micro-needle approach, which allows for the isolation of immunomagnetically labeled target cells by the use of a magnetized microneedle directly from glass slides. The magnetic microneedle approach makes use of the already present magnetic labeling used for enrichment, while the glass-slide-based open sample container allows for easy and loss-free sample loading. Additionally, the system facilitates not only the isolation but also the precise placement of cells. As the used parts are low cost, the technology provides researchers with an affordable and efficient method to pick up and isolate, as well as specifically place magnetically labeled cells from enriched fractions, thereby enabling the researchers to isolate or analyze these rare cells in more detail.
Introduction
The isolation of intact single cells is an important tool in biological research. For those applications where an abundance of cells is available often techniques such as fluorescence-activated cell sorting (FACS) or limited dilution are used. However, when cells are rare, such as circulating tumor cells (CTCs), there is an increased appreciation of the information that can be attained from single, intact cells.1–3 For this purpose, multiple commercially available single-cell isolation techniques exist, including the laser dissection approach as well as more recently developed techniques such as the CellCelector,4 DEP-Array,5 and Puncher systems.6 Additionally, multiple research groups have shown proof of principle work on chip-based single-cell isolation and/or direct characterization strategies.7–11 While some of these technologies can work with thousands to millions of cells as inputs, in the case of rare cells an initial enrichment step is needed in order to achieve a sufficient sample purity. Immunomagnetic enrichment by the FDA-cleared CellSearch system is frequently used for enrichment, after which the immunomagnetically enriched and fluorescently labeled cells are obtained from the cartridges. The loss of cells during sample transfer and the following isolation procedure is however a serious constraint for samples containing only a few CTCs.The key element in these positive immunomagnetic enrichment procedures is the binding of a magnetic label to the cells of interest. As a result, these cells can be manipulated using magnetic fields. This characteristic is mostly used to hold all the labeled cells in place using large external magnetic configurations, allowing for the separation of bound cells from unbound cells, or to remove liquid during staining procedures. When however a local magnetic field is used, individual magnetic particles can be selectively manipulated, as shown by Timonen and Grzybowski, who used a small electromagnetic needle in order to pick and place single magnetic particles.12 Similarly, local magnetic fields have been used to selectively attract magnetic beads bound to cells as a tool to measure cell flexibility.13 Moving single cells by using magnetic clusters manipulated by external magnetic fields has also been done.14 The DEP-Array, CellCelector, or Puncher systems do not make use of the magnetic label present on the target cells and require sample transfer prior to single-cell isolation. Here, we show the use of a magnetic microneedle for the pick-up, isolation, and precise placement of immunomagnetically labeled CTCs.
Methods
Healthy donor blood samples
Whole blood samples were collected in CellSave preservative tubes (Menarini Silicon Biosystems, Bologna, Italy) from anonymized healthy volunteers at the TechMed Centre Donor Service of the University of Twente. From all volunteers informed consent was obtained and the blood collection procedure was approved by the local Medical Research Ethics Committee (METC Twente; Registration number K11-23). This research does not fall within the scope of the Dutch Medical Research Involving Human Subjects Act.Cell lines
PC3 (CRL-1435) and LNCaP (CRL-1740) were purchased from the ATCC (Manassas, VA, USA) while the PC3-9 cell line was kindly provided by Immunicon (Huntingdon Valley, PA, USA). The PC3-9 cell line was tested to be a subclone of PC3 using STR-profiling. All cell lines were cultured in RPMI1640 (Lonza, Basel, Switzerland) supplemented with 10% FBS (Sigma-Aldrich, St. Louis, MO, USA) and 1% penicillin/streptomycin (Lonza, Basel, Switzerland). Upon reaching 70–80% confluence, they were trypsinized using 0.05% trypsin-EDTA (Gibco, Waltham, MA, USA) and fixated using either CellSave (Menarini, Bologna, Italy) fixative or 1% formaldehyde. To measure the level of EpCAM expression the cells were stained using anti-EpCAM (Vu1d9)-PE (Sigma-Aldrich, St. Louis, MO, USA). The PE intensity was measured and quantified using a FACS Aria II flow cytometer (BD, Franklin Lakes, USA) and the BD Quantibrite™ Beads PE Fluorescence Quantitation Kit (BD). We used prostate cancer cell lines with different amounts of the target antigen EpCAM, with PC3 having a measured mean expression of 7300 EpCAM antigens per cell, PC3-9 a measured mean of 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 EpCAM antigens per cells, and LNCaP with a measured mean of 463
200 EpCAM antigens per cells, and LNCaP with a measured mean of 463![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 EpCAM antigens per cells.
900 EpCAM antigens per cells.
Sample holder
For the identification, magnetic pick-up and isolation of cells a custom sample holder was used consisting of a glass slide onto which a 3-D printed part was glued, resulting in four open chambers; three chambers that can be used for sample deposition and one from which the single cell aspiration can take place. After sample placement, the cells were allowed to settle for 20 min after which the chambers were connected by the addition of buffer.Experimental setup
The setup used for single cell pick up and aspiration consists of an inverted fluorescence microscope (Nikon, Minato, Japan) with a controllable X, Y, Z stage (ASI, Eugene, USA), a LED light source (Lumencor, Beaverton, USA) and CCD camera (Hamamatsu, Shizuoka, Japan). To move the magnetic microneedle into proximity of the cell of interest and subsequently perform cell pickup, the solid magnetic microneedle is mounted onto an XYZ-micromanipulator (Eppendorf, Hamburg, Germany), which is attached to the microscope. To attach the magnetic microneedle to the micromanipulator, a flexible adapter was designed and 3D printed. The aspiration pipet was placed into a custom 3D printed holder and fixed onto the microscope stage to retain its position relative to the sample holder during movement. Using Labview control of the XY stage, the XY position of the stage at which the pipet tip used for cell aspirations was in view was saved. After the pickup of the target cell, the magnetic microneedle was automatically moved to the location of the aspiration pipet tip to enable cell extraction. The actual setup is shown in Fig. 1, with in panel A the system overview, in panel B a closer view of the sample holder, the aspiration pipet mounted for cell extraction, and the Luer lock dispensing tip into which the magnetic microneedle is placed. In the zoom-in on panel C the magnetic microneedle can be seen.Magnetic labeling
Unless stated otherwise, tumor cells were magnetically labelled by CellSearch ferrofluid (Menarini Silicon Biosystems) directed against EpCAM. To assess the possibility of using smaller or larger magnetic particles, cells were labelled with Miltenyi CD326 (EpCAM) MicroBeads (Miltenyi, Bergisch Gladbach, Germany) directed against EpCAM or first labeled with the anti-EpCAM antibody Vu1D9 coupled to biotin and subsequently incubated with Dynabeads MyOne C1 streptavidin-coated magnetic beads (ThermoFisher, Waltham, USA).Magnetic microneedle
Although any ferro-, ferri- or even para-magnetic material would be possible, for our experiments we used as a magnetic microneedle solid pins made of ferritic stainless steel (Austerlitz INSECT PINS, Slavkov u Brna, Czech Republik), normally used for the preparation of insect displays. They are 12 mm long and 100 μm in diameter, with one of the ends being sharpened into a 12.5 μm diameter tip end. For easy handling and attachment into the manipulation system, the magnetic microneedles were fitted into 22 ga × 0.25 in Luer dispensing tips (Instech, Plymouth Meeting, US) by bending the blunt end of the magnetic microneedles into an angle before inserting into the dispensing tips, causing the microneedles to remain stuck.In our experiments, we magnetized the used magnetic microneedles prior to use, by placing them in between opposing permanent N45 magnets. The resulting permanent magnetization allowed us to perform the pickup and transfer without the use of an external magnet. The maximum remnant magnetism of the used magnetic microneedles was determined to be 14.5 μA m2 by vibrating sample magnetometer (VSM). The magnetic microneedles could also be magnetized using electromagnets, as well as at the time of pickup. In the latter case, the (electro-)-magnet is placed in close proximity to the magnetic microneedle within the setup.
Aspiration pipet
After pick up, the target cells are either placed in a different location or transferred to the opening of a standard plastic 10 μL pipet tip for aspiration. For this, pipet tips were placed onto a 10 μL pipet, which was then mounted into a custom holder on the microscope stage. The pipet was set to a 2 μL volume and prefilled with PBS. During the cell transfer, the pipet stop was pressed down manually. After the magnetic microneedle was positioned directly in front of the pipet tip opening the pipet stop was released, causing the cell to be aspirated into the pipet tip.Bead size
To test the ability of cell pick up after the magnetic enrichment of cells using commercially available beads with different sizes, PC3 and LNCaP cells were magnetically labeled with Miltenyi CD326 (EpCAM) MicroBeads, CellSearch ferrofluid, and Dynabeads C1 enrichment beads, having a diameter of 60 nm, 200 nm, and 1 μm respectively. Magnetically labeled cells were enriched from the buffer using an iMag magnetic separator (BD) and placed onto the sample holder, after which the pickup of at least 20 cells was tested.Blood sample spiking, enrichment, and isolation
For a more realistic evaluation of the system, cells from the LnCAP cell line were fixed using CellSave preservative and in three independent experiments cells were precisely spiked into 7.5 mL of blood from healthy volunteers as described previously.15 The samples were then diluted to 14 mL using CellSearch dilution buffer and centrifuged at 800g for 10 min. Then, the plasma was aspirated and the remaining ∼6 mL was supplemented with CellSearch dilution buffer to 10 mL, after which CellSearch 150 μL ferrofluid and 150 μL capture enhancement reagents (Menarini) were added. The samples were incubated in a magnet for 8 times 3 min, where the samples were mixed by inversion between each 3 min incubation step. After incubation, the samples were magnetically separated using our previously described magnetic flow-through system.16 The enriched samples, containing enriched tumor cells as well as some non-specifically co-enriched leukocytes were stained using the CellSearch staining reagents and placed onto the custom sample holder. The number of successfully enriched LNCaP and PC3 cells was determined manually, after which the cells were relocated and single cells were picked up using the magnetic microneedle. After transfer to the isolation pipet, the cells were aspirated into a 10 μL pipet using a 2 μL volume and transferred to a separate glass microscope slide for confirmation of successful isolation. Here it was also checked if only the target cell and no contaminating leukocytes were isolated.Isolation from high cell-density samples
To test the ability of the system to pick up single cells from increasingly dense cell populations, we mixed magnetically labeled PC3-9 and LNCaP cells at a ratio of 100 to 1. LNCaP cells were stained using CellTracker Orange prior to mixing to facilitate identification, while all cells were stained with Hoechst. Increasing concentrations of this sample were placed onto a sample holder, after which it was attempted to pick up 20 randomly selected single LNCaP cells without any contaminating PC3-9 cells.Cell placement and transfer
To test the proof-of-principle of cell transfer and placement, magnetically labeled LNCaP or PC3-9 cells were placed onto a sample holder, and 10 randomly selected cells were attempted to pick up, retract into the holder, and transferred to a 30-well microscope slide. On a multiwell slide, the cells were placed into the middle of a single well.Results
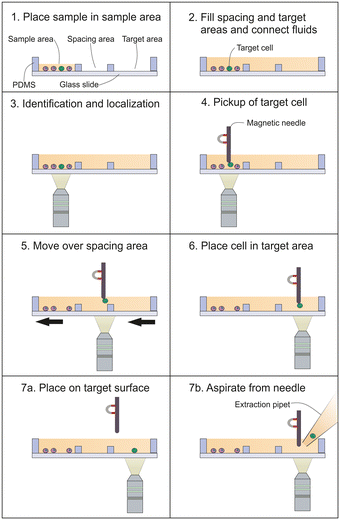
Principle of magnetic microneedle single cell transfer and isolation
Fig. 2 depicts the magnetic micro-needle cell transfer principle as performed in our proof of principle experiments. The immunomagnetically enriched cells are placed on a glass surface and allowed to sediment (1). Next, additional buffer was added to the sample holder, causing the fluid in the different areas to become connected. In this way, a fluid connection is established to the target area. In our setup, an empty area was present between the sample and target area to further separate the sample and target areas and prevent contamination of the target area with unwanted cells (2). The positions of the target cells (green) and non-target cells (purple) are identified by microscopy (3). A magnetic microneedle is positioned above the target cell and by lowering the microneedle it is brought into proximity of the cell, causing the cell to attach to the magnetic microneedle (4). Next, the cell is lifted from the surface through the solution and transferred to the target location (5). The cell is placed on the target location (6). Then, either the cell is released from the magnetic microneedle onto the surface, by quickly moving it in order to allow the inertial-, drag- and surface adhesion forces to overcome the magnetic and adhesion force to the microneedle (7a), or the cell is brought into proximity of a pipet tip and aspirated directly from the microneedle (7b).Magnetic bead size
The force a magnetic bead exerts is dependent on the applied magnetic field together with the resulting magnetic moment of the bead, which generally increases with its size. Magnetic beads used for rare cell separation range from 60 nm to 2.8 μm and we have therefor tested the ability of the magnetic microneedle to pick-up magnetically labelled cells across different bead sizes. Our results show that the current magnetic microneedle can be used successfully to pick up cells enriched from buffer using the 1 μm diameter Dynabead C1 beads, 200 nm diameter CellSearch ferrofluid, while also the 60 nm large Miltenyi cell enrichment particles resulted in 100% successful pickup when using the high EpCAM expressing LNCaP cell line. When using the very low EpCAM expressing PC3 cells, Dynal bead labelling still allowed 100% of the enriched cells to be picked up, while CellSearch and Miltenyi labeling resulted in respectively 97% and 63% of the enriched cells being successfully picked up. A complete overview of the number of (successful) pickup attempts can be found in ESI† Table S1. It is worth noting that the distance at which the cell is visibly attracted to the microneedle is dependent on the magnetic labeling, causing cells to be attracted from several tens of μm when labeled with the Dynabeads, while attraction is only visible in the last 1–2 μm when labelled with the small Miltenyi magnetic beads.Single-cell isolation from enriched blood samples
To test the efficiency of the magnetic microneedle single-cell isolation system in combination with tumor cell enrichment from whole blood, in three independent experiments an exact number of cells from the LNCaP cell line (average 21, range 19 to 25) were spiked into blood samples obtained from healthy donors. After immunomagnetic enrichment, the resulting samples, containing the enriched tumor cells as well as a background of co-enriched white blood cells, were placed onto the sample holder to determine recovery, before isolating the cells using the magnetic microneedle isolation setup.Although after enrichment on average 81% of the spiked LNCaP cells were successfully identified on the sample holder, the lack of automated identification and relocation in our setup in combination with bleaching of the identifying fluorophore, resulted in not all initially identified tumor cells being relocated for potential isolation. Of on average 87% of the originally identified cells isolation could attempted. Cell extraction was defined as the combination of pickup, transfer to the aspiration pipet, and aspiration into the pipet. As shown in Fig. 3, all of these steps were successful in on average 98% of the attempted cases. Of these extracted cells on average 89% could be relocated in the 2 μL droplet on a separate glass slide, wherein 95% of these cases only the single target cell was detected. In total, 83% of the single-cell isolation attempts resulted in the successful extraction and relocation of the uncontaminated target cell on the examination slide.
In the total process, including the enrichment, the loss of cells that could not be relocated due to bleaching as well as the single cell isolation and relocation, 58% of the originally spiked cells could be relocated as single cells in a 2 μL droplet. A complete overview of the success rate for each step can be found in ESI† Table S2.
To examine if this proof-of-principle setup is also already capable of isolating very low-expressing cells, the very low EpCAM-expressing PC3 cells were spiked into healthy donor blood and enriched in the same way, resulting in 12% recovery.
Although in this experiment 86% of the recovered cells were successfully picked up as single cells, even the minimal transfer speed of 1 mm s−1 that could be set in our setup resulted in the loss of these cells during transfer to the aspiration pipet.
Isolation of single cells among a high-density of non-target cells
As the density of the cells present in the sample will influence the efficiency to selectively pick target cells we created samples with increasing cell densities, being approximately 400, 800, 1600, and 3200 cells per mm2. With an average cell diameter 16 μm, this results in a surface occupancy of 8%, 16%, 32%, and 64% respectively. From these samples, consisting of PC3-9 and LNCaP cells in a 100 to 1 ratio, single LNCaP cells were isolated using the magnetic microneedle system. In Fig. 4A the resulting success rate and transfer times are shown. The success rate is here defined as the percentage of cells that could be picked up without any contaminating non-target cells. The results show that transfer efficiency decreases at higher cell densities, while the transfer time increases (Fig. 4B), both due to the more challenging pickup of only the single target cell. This indicates that to maintain sufficient efficiency, the sample holder should be large enough to avoid excessive cell densities. Although the surface coverage can reach up to 64% while still retaining an above 50% success rate, for most applications the surface coverage should be limited to 20% or less.Cell placement
After aspiration from the magnetic microneedle, the cell of interest can be placed into a tube or plate for further analysis or single cell culture. As however the cell can be precisely placed using the magnetic microneedle, it is also possible to deposit the cell in a specific position within the same sample container, as schematically shown in panel 7A of Fig. 2. To demonstrate this ability, an example is shown in Fig. 5, where four single cells in a low cell density sample (∼20 cells per mm2) are picked up from their original location and placed into a single file line surrounding an already present cell on the target location in the same sample.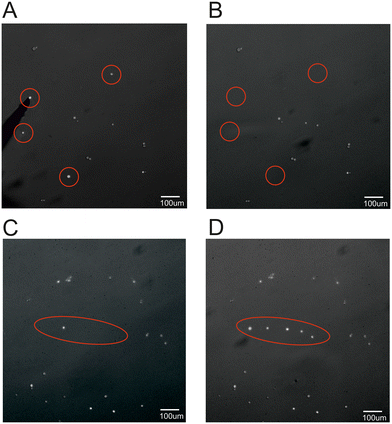 | ||
| Fig. 5 Fluorescent image of A) before and B) after magnetic pickup showing the magnetic microneedle on the left side of image A. Target location before (C) and after (D) cell placement. | ||
Cell transfer
For the magnetic needle to reach the desired position for cell placement, it has to remain within the liquid during the transportation of the cell from the pickup to the target location. If removed from the liquid, the surface tension dislodges the cell, preventing it from being transported to a different container. In most applications however, the cell needs to be placed onto or into a different sample format. As shown, this can be achieved by aspirating the cell from the microneedle into a pipet tip. This however eliminates the possibility of precise placement of the cells as demonstrated in Fig. 5. To circumvent this limitation, we have tested a microneedle retraction system that is capable of retracting the microneedle into a syringe, allowing for transport between different sample carriers, shown schematically in Fig. 6A. Here, the magnetic microneedle is mounted into a holder that protrudes from a syringe-like container (panel i). After picking up the cell, the magnetic microneedle is retracted within the syringe-like container (panel ii). When the syringe is then removed from the sample area, the liquid within is transferred along, thereby keeping the microneedle and attached cell contained in the liquid. After transfer to the target area/liquid, the microneedle is protruded again and the cell can be placed into a specific position as described previously (panel iii). This way the cell can be transferred to a different type of sample holder or liquid, without the need for the cell and microneedle to overcome the surface tension. Additionally, the cell can be moved at large speeds without being subjected to large drag forces. In Fig. 6B the proof-of-principle device used to test this approach is shown while Fig. 6C shows an example of a relocated cell placed onto a microwell slide.Using this proof-of-principle device, the pickup, transfer, and placement of magnetically labeled LNCaP cells was successful in 90% of attempts, while the lower EpCAM expressing PC3-9 cells resulted in a reduction in successful transfer and placement to 50% of attempts.
Discussion
Here we introduce a magnetic microneedle to isolate single magnetically labeled cells or place these in a desired location.Although different bead sizes can be used, the efficiency of the method is dependent on the expression level of the targeted antigen, which determines the number of beads bound per cell. An increase in magnetic attraction force is likely needed to successfully isolate the low EpCAM-expressing cells seen in patient samples.15 As in this proof-of-principle study, we used simple steel insect pins as magnetic microneedles, we expect an increase in magnetic attraction can be achieved by optimization of the used material and microneedle shape. Similarly, although automation will also likely aid in the stability of cell pickup, an improvement in needle shape and material leading to an even more localized magnetic field will likely be needed to achieve successful pickup in samples with very high cell densities. To pick up cells without any EpCAM expression another surface antigen such as PSMA or Her-2 could be targeted.
Although we have demonstrated the principle of the magnetic microneedle approach for isolation of single CTC isolation, for routine use further automation of imaging, target cell recognition, cell pick-up, and aspiration of the target cells is needed. Essential for magnetic microneedle single-cell isolation is an inverted microscope system with an X–Y-stage onto which the sample holder can be mounted together with an XYZ-translation stage for the magnetic microneedle. A large benefit of the magnetic microneedle above some other single-cell isolation approaches such as the VyCAP Puncher or Menarini DEP-Array is that no expensive chips/cartridges are needed, as the sample holder consists of a simple chamber on a glass slide.
A constraint of picking up single cells with the magnetic microneedle is the cell density, which should not exceed approximately 400 cells per mm2. However, the surface of the chamber in which we deposited immunomagnetically enriched cells is divided into three cell deposition areas each measuring approximately 200 mm2 implying that the deposited cell number per area should not exceed 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells to ensure efficient pickup, which is high compared to the 10
000 cells to ensure efficient pickup, which is high compared to the 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 30
000 to 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells that fit into a VyCAP Puncher or DEP-array chip or the 50
000 cells that fit into a VyCAP Puncher or DEP-array chip or the 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 that can be loaded onto the CellCelector.4–6 For CTC enrichment the most commonly used method is CellSearch, which after the immunomagnetic CTC enrichment from 7.5 mL of blood has a nucleated cell count ranging from 433–384
000 that can be loaded onto the CellCelector.4–6 For CTC enrichment the most commonly used method is CellSearch, which after the immunomagnetic CTC enrichment from 7.5 mL of blood has a nucleated cell count ranging from 433–384![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 015, with 27% of samples having a nucleated cell count greater than 100
015, with 27% of samples having a nucleated cell count greater than 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000.17 The chamber used for CTC analysis in CellSearch has a surface of 2.7 × 33 = 89 mm2. With a nucleated count of 100
000.17 The chamber used for CTC analysis in CellSearch has a surface of 2.7 × 33 = 89 mm2. With a nucleated count of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, the surface is completely covered with cells, impeding the identification of individual cells. The surface of the chamber we used has three cell deposition areas each having a 2.2-fold larger surface area, allowing for the placement of in total 240
000, the surface is completely covered with cells, impeding the identification of individual cells. The surface of the chamber we used has three cell deposition areas each having a 2.2-fold larger surface area, allowing for the placement of in total 240![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells. Although this ensures a sufficiently low cell density for most samples, it also increases the time needed for scanning and target identification. Depending on the sample size, only one or two of the chambers can be used to reduce the scan time.
000 cells. Although this ensures a sufficiently low cell density for most samples, it also increases the time needed for scanning and target identification. Depending on the sample size, only one or two of the chambers can be used to reduce the scan time.
After the enrichment of tumor cells from blood and loading of the sample onto the holder, it takes approximately 20 min for the cells to settle onto the glass slide. The subsequent identification and isolation of 20 cells using the current magnetic microneedle isolation system takes approximately 1.5 to 2 hours. The majority of this time consists of the manual identification and relocation, which are separated to ensure all cells are identified before bleaching occurs. Automated scanning, identification, and relocation could therefore vastly reduce this time, considering that the manual isolation procedure including pick up, transfer and aspiration takes on average only 60 to 90 seconds per cell. This is slower than the VyCAP Puncher system (1 cell per second6) or the ALS CellCelector (20 seconds per cell4), but it is faster than the Menarini DEP-Array system (107 seconds for a single cell5). As an example, if through automation the scanning and identification could be reduced to 20 min, the entire procedure to isolate 20 single tumor cells could, including sample holder loading, be performed in about an hour.
Although both the VyCAP Puncher as well as Menarini DEP-Array indicate a high recovery of cells once detected in the cartridge or chip, the loading of these often results in high cell loss, with a minimal loss of 23% due to dead volume for the DEP-Array, while small cells will pass through the pores of the VyCAP chip. The simple sample holder in combination with a pick-and-place methodology shows the most resemblance to the ALS CellCelector, where the main advantage of the here presented method is the ability to not only precisely pick up cells in relatively high-density samples, but also place these onto specific target locations for further characterization.
The possibility of isolating a single CTC has already led to several applications, including genetic, epigenetic, transcriptomic, and proteomic analysis of a single CTC, which can also enable the discovery of novel drugs by exploring the content of the CTCs. This analysis can also be performed after the extraction of CTCs using the magnetic microneedle approach. However, due to the placement capability, other types of analysis involving the secretion profile or electrophysiological analysis of these cells can also be undertaken. This makes the magnetic microneedle method a versatile approach for researchers interested in the multifaceted and innovative analysis of single cells retrieved from magnetically enriched samples.
Conclusion
A novel magnetic microneedle system is presented capable of efficiently picking up single magnetically labeled cells for single-cell isolation or precise placement. Although the method's efficiency is dependent on the expression level of the target antigen, the magnetic microneedle can pick up cells enriched using a range of magnetic labeling beads and allows efficient pick up from samples placed at concentrations up to 400 cells per mm2. The picked-up cells can be isolated by simple pipet aspiration directly from the magnetic microneedle or placed onto specific positions by rapid movement of the magnetic microneedle. When precise placement onto a surface in a different container is needed, the magnetic microneedle and cell can be subtracted into a syringe to facilitate transfer. Overall, the low-cost magnetic microneedle approach is a versatile method for the isolation and placement of magnetically enriched cells, allowing the potential applications for their study to be increased.Author contributions
Conceptualization, LWMMT; investigation, MS & PH; methodology, MS & LWMMT; supervision, LWMMT; visualization, MS; writing – original draft, MS & LWMMT; writing – review and editing, MS, PH & LWMMT;Conflicts of interest
MS, PH and LWMMT have filed a patent application on the here-introduced method.References
- S. B. Lim, W. Di Lee and J. Vasudevan, et al. , npj Precis. Oncol., 2019, 3, 23 CrossRef PubMed.
- C. P. Wu, P. Wu, H. F. Zhao, W. L. Liu and W. P. Li, DNA Cell Biol., 2018, 37, 78–89 CrossRef PubMed.
- L. Keller and K. Pantel, Nat. Rev. Cancer, 2019, 19, 553–567 CrossRef PubMed.
- C. Nelep and J. Eberhardt, Cytometry, Part A, 2018, 93, 1267–1270 CrossRef PubMed.
- M. Di Trapani, N. Manaresi and G. Medoro, Cytometry, Part A, 2018, 93, 1260–1266 CrossRef PubMed.
- M. Stevens, L. Oomens, J. Broekmaat, J. Weersink, F. Abali, J. Swennenhuis and A. Tibbe, Cytometry, Part A, 2018, 93, 1255–1259 CrossRef PubMed.
- H. Pei, L. Li, Z. Han, Y. Wang and B. Tang, Lab Chip, 2020, 20, 3854–3875 RSC.
- Y. Song, T. Tian, Y. Shi, W. Liu, Y. Zou, T. Khajvand, S. Wang, Z. Zhu and C. Yang, Chem. Sci., 2017, 8, 1736–1751 RSC.
- J. Lamanna, E. Y. Scott, H. S. Edwards, M. D. Chamberlain, M. D. M. Dryden, J. Peng, B. Mair, A. Lee, C. Chan, A. A. Sklavounos, A. Heffernan, F. Abbas, C. Lam, M. E. Olson, J. Moffat and A. R. Wheeler, Nat. Commun., 2020, 11, 1–13 Search PubMed.
- M. Zhang, Y. Zou, X. Xu, X. Zhang, M. Gao, J. Song, P. Huang, Q. Chen, Z. Zhu, W. Lin, R. N. Zare and C. Yang, Nat. Commun., 2020, 11, 1–13 Search PubMed.
- Y. Wang, X. Wang, T. Pan, B. Li and J. Chu, Lab Chip, 2021, 21, 3695–3706 RSC.
- J. V. I. Timonen and B. A. Grzybowski, Adv. Mater., 2017, 29, 1603516 CrossRef PubMed.
- Z. Cenev, H. Zhang, V. Sariola, A. Rahikkala, D. Liu, H. A. Santos and Q. Zhou, Adv. Mater. Technol., 2018, 3, 1–9 Search PubMed.
- L. Zhu, W. Huang, F. Yang, L. Yin, S. Liang, W. Zhao, L. Mao, X. Yu, R. Qiao and Y. Zhao, Adv. Biosyst., 2019, 3, 1–11 Search PubMed.
- A. Mentink, K. T. Isebia, J. Kraan and L. W. M. M. Terstappen, Sci. Rep., 2023, 1–11 Search PubMed.
- M. Stevens, P. Liu, T. Niessink, A. Mentink, L. Abelmann and L. Terstappen, Diagnostics, 2021, 11, 1020 CrossRef PubMed.
- S. de Wit, L. L. Zeune, T. J. N. Hiltermann, H. J. M. Groen, G. van Dalum and L. W. M. M. Terstappen, Cancers, 2018, 10, 1–16 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3lc00920c |
| This journal is © The Royal Society of Chemistry 2024 |