Open Access Article
Open Access ArticleOral administration of cranberry-derived exosomes attenuates murine premature ovarian failure in association with changes in the specific gut microbiota and diminution in ovarian granulosa cell PANoptosis†
Zeyu
Cui‡
a,
Te
Liu‡
 *a,
Yichao
Wen‡
a,
Weihao
Li‡
b,
Jianghong
Xu
c,
Yingjuan
Chen
c,
Danping
Chen
c and
Ying
Zhu
c
*a,
Yichao
Wen‡
a,
Weihao
Li‡
b,
Jianghong
Xu
c,
Yingjuan
Chen
c,
Danping
Chen
c and
Ying
Zhu
c
aShanghai Geriatric Institute of Chinese Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai 200031, China. E-mail: liute1979@shutcm.edu.cn; Fax: +86-21-64720010; Tel: +86-21-64720010
bHandan Municipal Centre for Disease Prevention and Control, Hebei 056008, China
cDepartment of Gynaecology, Jingan Hospital of Traditional Chinese Medicine, Shanghai, China
First published on 12th November 2024
Abstract
Background: Although a high-fat and high-sugar diet (HFHS) can induce ovarian insufficiency and premature ovarian failure (POF)—making the treatment difficult—plant-derived exosome-like nanovesicles manifest numerous therapeutic effects on various diseases. Purpose: To explore the therapeutic effects and the molecular biology mechanism of exosomes derived from Vaccinium macrocarpon Ait (cranberry) (Va-exos) in the treatment of murine HFHS-POF. Methods: The exosomes from cranberry (Va-exos) were isolated, purified and fed to HFHS-POF model mice. The pathological changes in ovaries, livers, intestines were detected by H&E and Masson staining. The 16s rRNA-seq technique was used to investigate the changes in the gut microbiota and microecology. The mRNA and protein expressions of PANoptosis and their phosphorylation levels in ovarian granulosa cells were detected by qPCR and western blot. Results: Pathological examination showed that Va-exos not only significantly alleviated the symptoms of POF in model mice but also improved the intestinal barrier function and inhibited the production of inflammatory factors. The high-throughput sequencing results of 16s rRNA indicated that the relative abundances of Akkermansia and Allobaculum microorganisms in the intestines of the Va-exos group of mice significantly increased, while the relative abundances of uncultured-bacterium_f-Muribaculaceae, Dubosiella, and uncultured-bacterium_f-Lachnospiraceae microorganisms were significantly reduced. The FCM test results indicated that Va-exos significantly reduced necrosis, apoptosis, and accumulation of reactive oxygen species in ovarian granulosa cells (OGCs) of the HFHS POF mice. Finally, both qPCR and western-blot analyses indicated that Va-exos significantly attenuated the expression levels of key regulatory factors in the PANoptosis of OGCs in HFHS POF mice. Conclusion: We confirmed that oral administration of cranberry-derived exosomes attenuated murine POF by modulating the gut microbiota and decreasing ovarian granulosa cell PANoptosis.
1. Introduction
Premature ovarian failure (POF) refers to a disease in women that occurs before the age of 40 and is characterized by amenorrhea, infertility, low estrogen levels, high concentrations of gonadotropins, and a lack of mature ovarian follicles. POF is one of the common causes of female infertility and is closely related to endocrine and metabolic disorders. Many studies have revealed that a high-fat and high-sugar (HFHS) diet elevates the risk of obesity, tumors, and cardiovascular disease,1–6 seriously affecting the ovarian function and oocyte quality.1–6 Our previous research also confirmed that the HFHS diet induced aging, cell death, and POF-like manifestations in mouse ovarian granulosa cells (mOGCs) by inducing histone H2A·X-specific site-phosphorylation modifications and Lin28/YY2 deficiency.7 Therefore, it is particularly important to investigate POF therapy that targets the regulation of glucose and lipid metabolism.Exosomes are extracellular nanoscale vesicles with a diameter of 30–150 nm; they show a complete membrane structure, and they are principally responsible for intercellular material transport and information transmission. In recent years, scholars have found that—like the exosomes secreted by animal cells—plants secrete exosomes that also contain RNA, lipids, and proteins. These exosomes are not only endocytosed by plant cells but are also directly incorporated by animal or human cells and serve as cellular messengers to mediate intercellular communication and regulate the host gene expression.8–12 Plant-derived exosome-like nanomedicines, therefore, exhibit distinct advantages in the treatment of certain diseases and in drug delivery and are considered to possess broad pharmaceutical applications.8–12
Vaccinium macrocarpon Ait (cranberry) is a herbaceous plant of the Rhododendron family and the Vaccinium genus, with spherical berries that are purplish-red in color.5,13–16 Cranberries contain numerous active substances, with high concentrations of hyperoside, pentahydroxyflavonoid, proanthocyanidins, ellagic acid, phenolic acid, resveratrol, and myricetin.5,13–16 Many studies have indicated that cranberry not only manifests significant antioxidant, anti-tumor, antiviral, anti-infective, and other functions but also protects the liver and gastrointestinal systems and relieves oral and gastric ulcers.5,13–16 However, there have been no reports on whether cranberry or cranberry-derived exosomes significantly improve POF symptoms.
PANoptosis is an inflammatory programmed cell death pathway activated by a specific trigger, and it is regulated by the PANoptosome complex (see below). It integrates the key features of cellular pyroptosis, apoptosis, and/or necroptosis, but it cannot be characterized solely by any one of these modes of cell death.17–21 PANoptosis is regulated by a cascade of upstream receptors and molecular signals that assemble into a polymeric complex called the PANoptosome.20,22,23 The PANoptosome, as a molecular scaffold, allows for the coupling and binding of key molecules involved in the processes of pyroptosis, cellular apoptosis, and necroptosis.17–19,21 After sensing pathogenic or heterologous antigenic components, sensor proteins mediate the assembly of proteins such as RIPK3, RIPK1, CASP8, and FADD into PANoptosome complexes, thereby inducing the onset of PANoptosis.17–19,21 In addition, the activation of the PANoptosome is also inhibited by TAK1, PSTIP2, SHARPIN, HOIP, HOIL-1, and A20.17–19,21 An increasing number of studies have indicated that PANoptosis is not only a unique phenomenon of infectious diseases but may also facilitate tumor treatment. However, the understanding of PANoptosis is still in its early stages, and there have been no reports as to whether POF induces PANoptosis.
In accordance with the above evidence, the aim of this study was to determine the molecular mechanisms underlying HFHS diet-induced PANoptosis and POF in mouse OGCs. We also explored the potential therapeutic effects of exosomes derived from cranberries on HFHS-induced murine POF via the regulation of the gut microbiota so as to inhibit the release of inflammatory factors and OGC PANoptosis.
2. Materials and methods
2.1 Isolation and enrichment of cranberry-derived exosomes
As per previous reports,8–12 fresh cranberries were incubated with cellulase (3 g per 100 ml) and pectinase (0.2 g per 100 ml) for 12 h after washing to remove sludge, and then their weight was recorded. Subsequently, the cranberry juice mixture was centrifuged at 2800g for 20 min. The supernatant was then centrifuged to remove plant fibers (850g, 40 min, 4 °C) and concentrated to approximately 100 ml using a hollow-fiber module. The concentrated sample was placed in an ultra-clean ultracentrifuge tube after 7 ml of 0.971 M sucrose was added to the bottom. The sucrose layer was collected and diluted in sterile phosphate-buffered saline (PBS) after centrifugation at 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 90 min at 4 °C, and cranberry sucrose was removed using an ultrafiltration tube. The cranberry-derived exosomes were ultimately resuspended in PBS, and the newly extracted exosomes were quantified based on the protein concentration using a BCA protein quantification kit (Beyotime, Shanghai, China).
000g for 90 min at 4 °C, and cranberry sucrose was removed using an ultrafiltration tube. The cranberry-derived exosomes were ultimately resuspended in PBS, and the newly extracted exosomes were quantified based on the protein concentration using a BCA protein quantification kit (Beyotime, Shanghai, China).
2.2 HFHS-POF model construction and the cranberry-derived exosome treatment
As per previously published methods,6,7,24 10-week-old female C57BL/6 mice (n = 24) were purchased from the Experimental Animal Centre at Shanghai University of Traditional Chinese Medicine. The mice were randomly allocated to three groups, with eight mice per group: the blank control-group (WT) mice were fed with regular feed but without any intervention; the cranberry-derived exosome-treated (Va-exos) mice were fed with high-fat feed (8 g kg−1), with a daily gavage of 200 μl of 30% high fructose and 100 μl of cranberry-derived exosomes at a concentration of 1 × 106 particles each; and the HFHS-POF model group mice (Ctrl) were fed a high-fat diet (8 g kg−1) and were orally administered 200 μl of 30% high fructose daily, along with an equal volume of physiological saline once each. Each group of mice was continuously treated for 2 months. This study was approved by the Ethics Committee of the Shanghai Geriatric Institute of Chinese Medicine (SHAGESYDW202009), and all experiments complied with the regulations of the National Science and Technology Commission of China on the use of experimental animals.2.3 Isolation and culture of mOGCs
Briefly, murine ovarian tissues were isolated under sterile conditions and placed in PBS at 4 °C as in our previous study.7,24 The tissues were minced, and 2.0 ml of hyaluronidase (0.1%, Sigma-Aldrich, St Louis, MO, USA) was added to the tissues for 1 min of digestion at 37 °C. The tissue suspension was gently pipetted, and 200 μl of fetal calf serum (Gibco, Gaithersburg, MD, USA) was added to the suspension to terminate digestion; the suspension was then filtered through a 200-mesh cell strainer. Next, 5.0 ml of PBS was added to the filtrate and all components were mixed well, followed by centrifugation at 1500 rpm for 5 min at 10 °C. The supernatant was discarded, and the pellet was re-suspended in 5.0 ml of PBS, followed by centrifugation at 1500 rpm for 5 min at 10 °C. The supernatant was ultimately discarded.2.4 Blood lipid testing
We collected peripheral blood serum from each group of mice and followed the instructions of the reagent kits to determine the levels of total cholesterol (TC), triglyceride (TG), low-density lipoprotein (LDL), and high-density lipoprotein (HDL) in serum using a chemiluminescence method.2.5 ELISA
We collected peripheral blood from each group of mice and centrifuged the blood samples at a speed of 3000 rpm for 10 min. We then collected the serum and used enzyme-linked immunosorbent assay (ELISA) kits to determine the concentrations of serum hormones (estradiol-17β [E2], follicle-stimulating hormone [FSH], and anti-Müllerian hormone [AMH]). Briefly, we prepared serum samples and diluted standards and added the prepared samples and the corresponding concentrations of the diluted standards to the coated ELISA plate and reacted them at 37 °C for 30 min, washed the ELISA plate three times, added the antibody reaction solution and reacted the mixture at 37 °C for 30 min, rinsed the ELISA plate three times, added chromogenic solutions A and B and reacted them at 37 °C for 30 min, added termination solution, and read the OD values within 15 min.2.6 H&E staining
Tissue samples were dehydrated with an ascending series of alcohol concentrations (75%, 85%, 95%, and anhydrous alcohol) for 4 h, cleared with xylene for 1.5 h, immersed in paraffin for 4.5 h, and embedded to form wax tissue blocks. The blocks were cut into paraffin sections at a thickness of 4 μm using a slicer and subjected to xylene dewaxing for 10 min and a descending alcohol gradient (anhydrous ethanol, 95% ethanol, 85% ethanol, and 75% ethanol; 5 min each). After removing xylene, cell nuclei and cytoplasm were stained with hematoxylin and eosin, and neutral gum was used to seal the sections. Images were observed and acquired under a microscope.2.7 Masson staining
Tissue samples were dehydrated with an alcohol gradient (75%, 85%, 95%, and anhydrous alcohol) for 4 h, cleared with xylene for 1.5 h, immersed in paraffin for 4.5 h, and embedded to form wax tissue blocks. The blocks were cut into paraffin sections at a thickness of 4 μm using a slicer, and the paraffin sections were subjected to xylene dewaxing for 10 min and an alcohol gradient (anhydrous ethanol, and 95%, 85%, and 75% ethanol; 5 min each). We washed the sections twice with PBS after removing xylene. Subsequently, Masson's reagent kit was used to stain the sections with hematoxylin for 5 min, followed by washing with PBS and then differentiation with acidic ethanol for 10 s. Washing with PBS was followed by 3 min of reaction until a color change was observed, followed by washing with PBS; this was followed by 10 min of staining with Ponceau S, 5 min of staining with phosphomolybdic acid, 5 min of staining with toluidine blue, and 1 min of reaction with 1% glacial acetic acid. Dehydration and sealing were again conducted, followed by observation and image acquisition using a light microscope.2.8 Immunohistochemical stain assay
Briefly, all fresh tissues were soaked in 4% paraformaldehyde (Sigma-Aldrich, St Louis, USA) at room temperature and fixed for 30 min. Tissues were exposed to ethanol gradient dehydration, paraffin embedding, sectioning at a thickness of 4 μm, and immersion in xylene for dewaxing. Tissue sections were sealed with immunohistochemical blocking solution (Beyotime Biotechnology Co., Ltd, Zhejiang, China) at 37 °C for 30 min. We discarded the blocking solution and added an immunohistochemical cleaning solution (Beyotime Biotechnology Co., Ltd) to clean the sections at room temperature three times for 5 min each. Then, primary antibodies (Mouse anti-IL-1β (3A6) mAb (#12242), Rabbit anti-IL-6 (D5W4V) XP® mAb (#12912), CST, MA, USA) were added and incubated at 37 °C for 45 min, and an immunohistochemical cleaning solution (Beyotime Biotechnology Co., Ltd) was used to clean slides at room temperature three times 5 min each. Secondary antibodies (Goat Anti-Mouse IgG H&L (Alexa Fluor® 555) preadsorbed (ab150118) and Goat Anti-Rabbit IgG H&L (Alexa Fluor® 488) (ab150077), Abcam, MA, USA) were added and incubated at 37 °C for 45 min, and an immunohistochemical cleaning solution (Beyotime Biotechnology Co., Ltd) was added to clean the slides at room temperature three times 5 min each. Finally, an immunofluorescence blocking solution was added (Sigma-Aldrich, St Louis, USA) for encapsulation.2.9 Gut microbial analysis
Fresh fecal samples were collected during the final 5 experimental days for gut microbial analysis as described in a previous study.25 Bacterial genomic DNA was extracted from frozen samples stored at −80 °C, and the V3 and V4 regions of the 16S rRNA gene were amplified by PCR using specific bacterial primers (forward primer, 5′-ACTCCTACGGGAGGCAGCA-3′; reverse primer, 5′-GGACTACHVGGGTWTCTAAT-3′). High-throughput pyrosequencing of the PCR products was performed on an Illumina MiSeq platform at Biomarker Technologies Co., Ltd (China). The raw paired-end reads from the original DNA fragments were merged using FLASH32 and assigned to each sample according to their unique barcodes. The QIIME (version 1.8.0) UCLUST software was used based on 97% sequence similarity, and the tags were clustered into operational taxonomic units (OTUs). The alpha-diversity index was evaluated using the Mothur software (version, v.1.30), and to compare the diversity indices among samples, the number of sequences in each sample was standardized. Analyses included OTU rank, rarefaction, and Shannon curves; and the Shannon, Chao1, Simpson, and abundance-based coverage indicator (ACE) indices were calculated. For beta-diversity analysis, heatmaps of redundancy analysis (RDA)-identified key OTUs, principal coordinate analysis (PCoA), non-metric multidimensional scaling (NMDS), and unweighted pair-group method with arithmetic mean (UPGMA) were obtained using QIIME. The line discriminant analysis (LDA) effect size (LEfSe) method was used for the quantitative analysis of biomarkers in each group. Briefly, LEfSe analysis, an LDA threshold >4, a non-parametric factorial Kruskal–Wallis rank-sum test, and an unpaired Wilcoxon rank-sum test were conducted to identify the most differentially abundant taxa.2.10 Annexin V/PI staining and flow-cytometric detection
Briefly, 200 μL of annexin V binding buffer (Invitrogen) and 5 μL of FITC annexin V solution (Invitrogen) were added to each group of moGCs and stained at room temperature for 15 min. Then, 2 μL of PI staining solution (Invitrogen) was added and cells were stained at room temperature for 5 min. We ultimately analyzed the fluorescence signals for FITC and PI using the flow cytometer's FL1 and FL2 channels and counted them.2.11 Cytokine array assay
A mouse cytokine array (Abcam) designed to detect 22 cytokines was employed in accordance with the manufacturer's instructions. Briefly, OGC proteins in each group of mice were incubated with the membrane at 4 °C overnight, and the analysis was performed with a chemiluminescent western-blot assay using biotinylated detector antibodies and streptavidin horseradish peroxidase (HRP). The targets of this ELISA-like array were granulocyte-colony stimulating factor (GCSF), granulocyte-macrophage colony stimulating factor (GM-CSF), interleukin (IL)-2, IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12 p40/p70, IL-12 p70, IL-13, and IL-17, interferon gamma (IFN-γ), monocyte chemoattractant proteins 1 and 5 (MCP-1 and MCP-5), regulated on activation, normal T cell expressed and secreted (RANTES), stem cell factor (SCF), soluble tumor necrosis receptor factor 1 (sTNFR1), tumor necrosis factor alpha (TNF-α), thrombopoietin, and vascular endothelial growth factor (VEGF). Results were analyzed using the ImageJ software (NIH, USA).2.12 RNA extraction, reverse-transcription reaction, and qPCR assay
We extracted total RNA from each group of cells according to the instructions provided with the TRIzol reagent (Invitrogen). After DNase I (Sigma Aldrich) treatment, total RNA was quantified using a ReverTra Ace-αFirst Strand cDNA Synthesis Kit (TOYOBO) and reverse-transcribed to generate cDNA. qRT-PCR was conducted in a RealPlex4 real-time PCR detection system (Eppendorf Co., Ltd, Germany) using a SyBR Green RealTime PCR Master MIX (TOYOBO) as the fluorescent dye for nucleic acid amplification. qRT-PCR involves 40 amplification cycles: first, denaturation at 95 °C for 15 s, then annealing at 58 °C for 30 s, and finally primer template extension at 72 °C for 42 s. We applied the 2−ΔΔCt calculation method to determine the relative expression level of genes as follows: ΔCt = Ct_genes − Ct_18srRNA; ΔΔCt = ΔCt_all_groups − ΔCt_control_group. The mRNA expression levels were corrected based on the expression level of 18s rRNA. The lists of qPCR primer sequences were as follows: MLKL-FP: 5′-AATTGTACTCTGGGAAATTGCCA-3′; MLKL-RP: 5′-TCTCCAAGATTCCGTCCACAG-3′; IL1B-FP: 5′-GAAATGCCACCTTTTGACAGTG-3′; IL1B-RP: 5′-TGGATGCTCTCATCAGGACAG-3′; Ki67-FP: 5′-CAAGGCGAGCCTCAAGAGATA-3′; Ki67-RP: 5′-TGTGCTGTTCTACATGCCCTG-3′; P16-FP: 5′-CGCAGGTTCTTGGTCACTGT-3′; P16-RP: 5′-TGTTCACGAAAGCCAGAGCG-3′; P21-FP: 5′-CCTGGTGATGTCCGACCTG-3′; P21-RP: 5′-CCATGAGCGCATCGCAATC-3′; P53-FP: 5′-GCGTAAACGCTTCGAGATGTT-3′; P53-RP: 5′-TTTTTATGGCGGGAAGTAGACTG-3′; GSDMD-FP: 5′-CCATCGGCCTTTGAGAAAGTG-3′; GSDMD-RP: 5′-ACACATGAATAACGGGGTTTCC-3′; Casp3-FP: 5′-ATGGAGAACAACAAAACCTCAGT-3′; Casp3-RP: 5′-TTGCTCCCATGTATGGTCTTTAC-3′; Casp9-FP: 5′-TCCTGGTACATCGAGACCTTG-3′; Casp9-RP: 5′-AAGTCCCTTTCGCAGAAACAG-3′; Casp7-FP: 5′-CGGAATGGGACGGACAAAGAT-3′; Casp7-RP: 5′-CTTTCCCGTAAATCAGGTCCTC-3′; BCL2-FP: 5′-GTCGCTACCGTCGTGACTTC-3′; BCL2-RP: 5′-CAGACATGCACCTACCCAGC-3′; BAX-FP: 5′-TGAAGACAGGGGCCTTTTTG-3′; BAX-RP: 5′-AATTCGCCGGAGACACTCG-3′; XIAP-FP: 5′-CGAGCTGGGTTTCTTTATACCG-3′; XIAP-RP: 5′-GCAATTTGGGGATATTCTCCTGT-3′; RREB1-FP: 5′-CCCACTAAGATGTGACATTTGCT-3′; RREB1-RP: 5′-GCAGGAATCGAAGGGTTGTTCT-3′; MAZ-FP: 5′-GCCCCAGTTGCATCTGTCTT-3′; MAZ-RP: 5′-CTTCGGAGGTTGTAGCCGTT-3′; RIP3-FP: 5′-TGGGCCTGCTAAGATGGCT-3′; RIP3-RP: 5′-CTGCCAGAGTGTGGATTTGGT-3′; 18S rRNA-F: CAGCCACCCGAGATTGAGCA; 18S rRNA-F: TAGTAGCGACGGGCGGTGTG.2.13 Western blot analysis
We added 300 μL of cellular lysate to each group of cell samples, placed them on ice for 20 min to lyse, and centrifuged them at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min. We aspirated the supernatant, added 60 μL of 6× SDS loading buffer, and boiled the mixture at 100 °C for 10 min. Subsequently, the protein samples were separated using SDS-PAGE and transferred onto a PVDF membrane. We sealed the PVDF membrane with 5% bovine serum albumin at room temperature and added primary antibodies (Mouse Reactive PANoptosis Antibody Sampler Kit (#70934), Rabbit anti-GAPDH (14C10) mAb (#2118), CST, MA, USA), incubated at 37 °C for 45 min, and rinsed four times with TBST solution for 15 min each time. We added secondary antibodies (Goat anti-Rabbit IgG H&L (HRP) (ab97051) and Goat anti-Mouse IgG H&L (HRP) (ab6789), Abcam, MA, USA), incubated at 37 °C for 45 min, and washed four times with TBST solution for 15 min each time. Finally, the ECL luminescent substrate was added to detect protein bands.
000g for 10 min. We aspirated the supernatant, added 60 μL of 6× SDS loading buffer, and boiled the mixture at 100 °C for 10 min. Subsequently, the protein samples were separated using SDS-PAGE and transferred onto a PVDF membrane. We sealed the PVDF membrane with 5% bovine serum albumin at room temperature and added primary antibodies (Mouse Reactive PANoptosis Antibody Sampler Kit (#70934), Rabbit anti-GAPDH (14C10) mAb (#2118), CST, MA, USA), incubated at 37 °C for 45 min, and rinsed four times with TBST solution for 15 min each time. We added secondary antibodies (Goat anti-Rabbit IgG H&L (HRP) (ab97051) and Goat anti-Mouse IgG H&L (HRP) (ab6789), Abcam, MA, USA), incubated at 37 °C for 45 min, and washed four times with TBST solution for 15 min each time. Finally, the ECL luminescent substrate was added to detect protein bands.
2.14 Nanoparticle tracking analysis
By using a dark-field microscope (NS500; Nanosight, Amesbury, UK) equipped with a 45 mW 405 nm laser and an electron multiplying charge-coupled device, we tracked the Brownian motion of cranberry-derived exosome single particles.2.15 Statistical analysis
Each experiment was performed at least three times, and data are shown as the mean ± standard error where applicable. Using the one-way ANOVA with Bonferroni test, significant interactions, differences and main effects for all statistical tests were analyzed (α = 0.05). A P value less than 0.05 was considered to be statistically significant.3. Results
3.1 Cranberry-derived exosomes significantly improve lipid metabolism abnormalities and POF symptoms in HFHS-POF model mice
To investigate whether Va-exos improve the condition of HFHS-POF model mice, Va-exos were isolated and enriched using gradient-density ultracentrifugation (Fig. 1A). Transmission electron microscopy (TEM) and Nanoparticle Tracking Analysis (NAT) results indicated that Va-exos exhibited a typical exosomal morphology and that their particle sizes ranged from 60 nm to 280 nm (Fig. 1B and C). The HFHS-POF model mice were subsequently fed Va-exos. After treatment, the HFHS-POF model group (Ctrl) mice were found to be obese, with dull hair color and significant hair loss, while the Va-exos group mice exhibited bright black fur and normal body posture (Fig. 1D). The pathological diagnostic results of H&E staining showed that the interstitial spaces in the ovaries increased, granulosa cell staining deepened, and atretic follicles were more common in the Ctrl group of mice. However, the ovarian tissue structure of the Va-exos group mice was tight, with a normal morphology of granulosa cells and more normal follicles (Fig. 1E). The blood lipid test results showed that the HDL of the Ctrl group was significantly lower than that of the WT group, while the LDL was significantly higher than that of the WT group, and the HDL and LDL of the Va-exos group mice were exactly opposite to those of the Ctrl group mice, showing significant improvement (Fig. 1F). Follicle counting showed that the proportion of blocked follicles in the Ctrl group mice was significantly higher than that in the WT group, while the proportion of blocked follicles in the Va-exos group mice was significantly lower than that in the Ctrl group (Fig. 1G). Finally, the ELISA test results showed that the levels of AMH and E2 in the peripheral blood of Ctrl group mice were significantly lower than those of the WT group, while the levels of AMH and E2 in the peripheral blood of the Va-exos group mice were significantly higher than those of the Ctrl group mice (Fig. 1H). The level of FSH in the peripheral blood of the Ctrl group mice was also significantly higher than that of the WT group, while the level of FSH in the peripheral blood of the Va-exos group mice was significantly lower than that of the Ctrl group (Fig. 1H). The above results all suggest that feeding HFHS-POF model mice with Va-exos significantly improved lipid-metabolism abnormalities, ovarian reserve, and sex hormone levels in the mice.3.2 Cranberry-derived exosomes significantly improve the structural abnormalities of liver and colorectal tissues in HFHS-POF model mice and mitigate the release of inflammatory factors
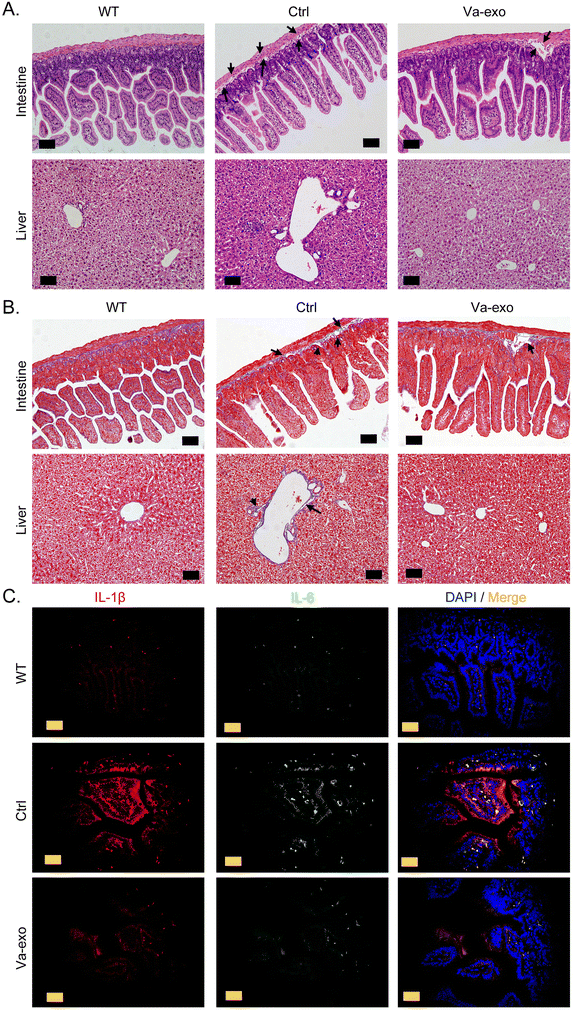
The H&E staining results indicated that the colorectal tissue structure of the Ctrl group mice was incomplete, with a loose arrangement of epithelial cells, obvious damage to the mucosal layer, an abnormal and disorderly arrangement of intestinal glands, an increased gap between glands and the basement membrane, and the presence of partial inflammatory swelling (Fig. 2A). However, the arrangement of glands in the colorectal tissue of Va-exos group mice was significantly improved, with a tight connection between the glands and the basement membrane and a relatively complete tissue structure (Fig. 2A). The Ctrl group mice showed moderate vacuolar lesions in liver cells, with some liver cells showing degeneration and necrosis; liver sinusoids showing partial dilation; an abnormal vascular structure in the liver; and deeply stained liver cells (Fig. 2A). However, the liver cells of the Va-exos group were arranged radially along the central vein and the cells appeared relatively healthy, with no obvious degeneration or necrosis observed; and the intrahepatic blood vessels and liver tissue structures were intact (Fig. 2A). In addition, Masson staining revealed that the collagen layer between the intestinal gland and the basement membrane of the Ctrl group had mostly disappeared, with a large gap visible; and liver vascular endothelial fibrosis was significant (Fig. 2B). The Va-exos group mice manifested abundant and tightly connected collagen in the intestinal basement membrane and a complete liver structure (Fig. 2B). Finally, the results of tissue immunofluorescence staining indicated that high concentrations of inflammatory factors IL-1β and IL-6 were present around the colonic tissue of mice in the Ctrl group, while the levels of IL-1β and IL-6 were significantly reduced around the colonic tissue of the mice in the Va-exos group (Fig. 2C and Fig. S1†). Therefore, our results suggested that cranberry-derived exosomes significantly improved the structure of mouse liver and colorectal tissues and reduced the production of inflammatory factors.3.3 Cranberry-derived exosomes significantly alter the quantity of the gut microbiota in HFHS POF model mice
High-throughput sequencing was conducted on the 16S rRNA V3 + V4 region of microorganisms found in the feces of WT, Ctrl, and Va-exos groups of mice to evaluate the composition and specific distribution of the gut microbiota. After sequencing, there were 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 885
885![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 010 paired-end reads among 24 samples, and there were 7
010 paired-end reads among 24 samples, and there were 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 698
698![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 785 clean tags after paired-end read alignment and filtering. For each sample, there were at least 72
785 clean tags after paired-end read alignment and filtering. For each sample, there were at least 72![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 439 clean tags and 320
439 clean tags and 320![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 783 clean tags on average. Using the QIIME (version 1.8.0) UCLUST software and based on 97% sequence similarity, tags were clustered into OTUs. In terms of the OTU number, the Va-exos group reflected significantly more OTUs than the Ctrl and WT groups (Fig. 3A). Subsequently, Venn-diagram statistics from the three OTU groups revealed a total of 518 OTUs (Fig. 3B). Next, we conducted an alpha-diversity index evaluation on the OTUs of the three groups (Fig. 3C–E). Our analysis indicated that there were significant differences between the OTUs of the Ctrl group and the WT group in Shannon and Simpson index curves, while the OTUs of the Va-exos group and the Ctrl group showed significant differences in Chao1 and ACE index curves (Fig. 3F–I). Preliminary test results suggested that cranberry-derived exosomes significantly altered the quantity of the gut microbiota in HFHS-POF model mice.
783 clean tags on average. Using the QIIME (version 1.8.0) UCLUST software and based on 97% sequence similarity, tags were clustered into OTUs. In terms of the OTU number, the Va-exos group reflected significantly more OTUs than the Ctrl and WT groups (Fig. 3A). Subsequently, Venn-diagram statistics from the three OTU groups revealed a total of 518 OTUs (Fig. 3B). Next, we conducted an alpha-diversity index evaluation on the OTUs of the three groups (Fig. 3C–E). Our analysis indicated that there were significant differences between the OTUs of the Ctrl group and the WT group in Shannon and Simpson index curves, while the OTUs of the Va-exos group and the Ctrl group showed significant differences in Chao1 and ACE index curves (Fig. 3F–I). Preliminary test results suggested that cranberry-derived exosomes significantly altered the quantity of the gut microbiota in HFHS-POF model mice.
3.4 Cranberry-derived exosomes significantly alter the distribution and diversity of the gut microbiota in HFHS-POF model mice
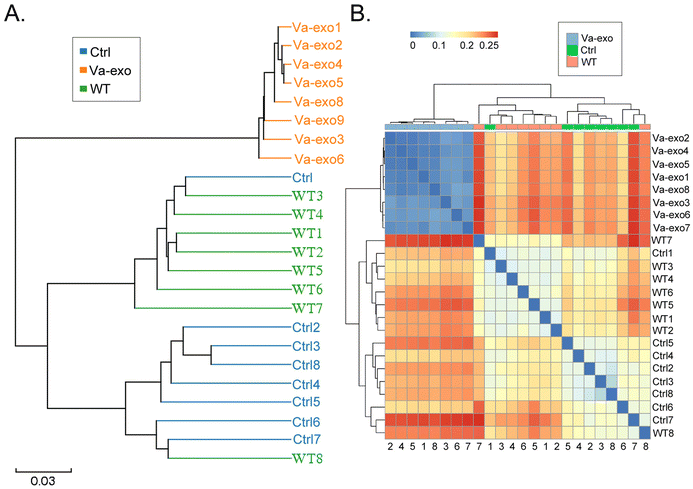
By comparing the representative sequences of OTUs with the microbial reference database, each OTU was classified into a species, and the community composition of each sample was then analyzed. At different taxonomic levels (kingdom, phylum, class, order, family, genus, and species), the QIIME software was used to generate species-abundance tables, and R language tools were used to construct the community structure of samples at different taxonomic levels. Phylum-level analysis showed that the relative abundance of microorganisms belonging to Verrucomicrobia and Proteobacteria in the gut of the Va-exos group mice was significantly increased compared to that of the Ctrl group (Fig. 4A). Further analysis of microbial differences at the genus and species levels revealed that at the genus level, the relative abundances of Akkermansia and Allobaculum microorganisms in the gut of Va-exos group mice were significantly augmented compared to those of the Ctrl group, while the relative abundances of uncultured_bacterium_f_Muribaculaceae, Dubosiella, and uncultured_bacterium_f_Lachnospiraceae microorganisms were significantly reduced (Fig. 4B). Finally, we analyzed the various species of microorganisms present. Compared to the Ctrl group, the relative abundances of uncultured_bacterium_g_Akkermansia, uncultured_bacterium_g_Allobaculum, and uncultured_bacterium_g_Bifidobacterium in the gut of Va-exos group mice were significantly elevated, while the relative abundances of uncultured_bacterium_g_Dubosiella and uncultured_bacterium_f_Muribaculaceae were significantly diminished (Fig. 4C).Subsequently, we used the Bray–Curtis algorithm to analyze the inter-group differences in microbial communities among the three groups, with the analyses primarily consisting of PCoA, principal component analysis (PCA), and NMDS. The results of these analyses showed that the microbial communities in the three groups exhibited significant differences in community distribution (Fig. 4D and E; ESI Table S2†). The results of the UPGMA hierarchical clustering analysis suggested that the intestinal microbiota in the Ctrl and WT groups had high sequence similarity and relatively close genetic backgrounds (Fig. 5A and B). By combining the UPGMA clustering tree with a histogram, we could demonstrate the characteristics of the microbial communities in each group. Our comprehensive analysis showed that in the Va-exos group, microorganisms such as those of the Allobaculum genus, Bifidobacterium genus, and Akkermansia genus were the dominant taxa; while the Dubosiella genus and uncultured_bacterium_f_Lachnospiraceae_NK4A136_group genera exhibited low abundances. However, in the Ctrl group, the microbial distribution results were contrary to those in the Va-exos group (Fig. 6A–C), and this result was consistent with the species abundance table generated by the QIIME software. In addition, we used the LEfSe method to identify high-dimensional biomarkers in the intestinal microbiota of each group; and with the LDA score set to 4.0, LDA scores greater than 4 were considered to be important biomarkers. As shown in the cladogram analysis and LDA score-distribution results, the number of microorganisms in the g__Allobaculum, s__uncultured_bacterium_g_Allobaculum, f__Bifidobacterium, g__Bifidobacterium, and o__Bifidobacterium genera rose significantly in the intestinal microbiota of mice in the Va-exos group. However, the number of microorganisms in the uncultured_bacterium_f_Lachnospiraceae family fell sharply in the intestinal microbiota of mice in the Va-exos group, and the distribution of these two microbial communities was reversed in the Ctrl group (Fig. 7).
3.5 Cranberry-derived exosomes significantly alter the expression distribution and diversity of 16S functional genes and metabolic-signaling pathways in the gut microbiota of HFHS-POF model mice
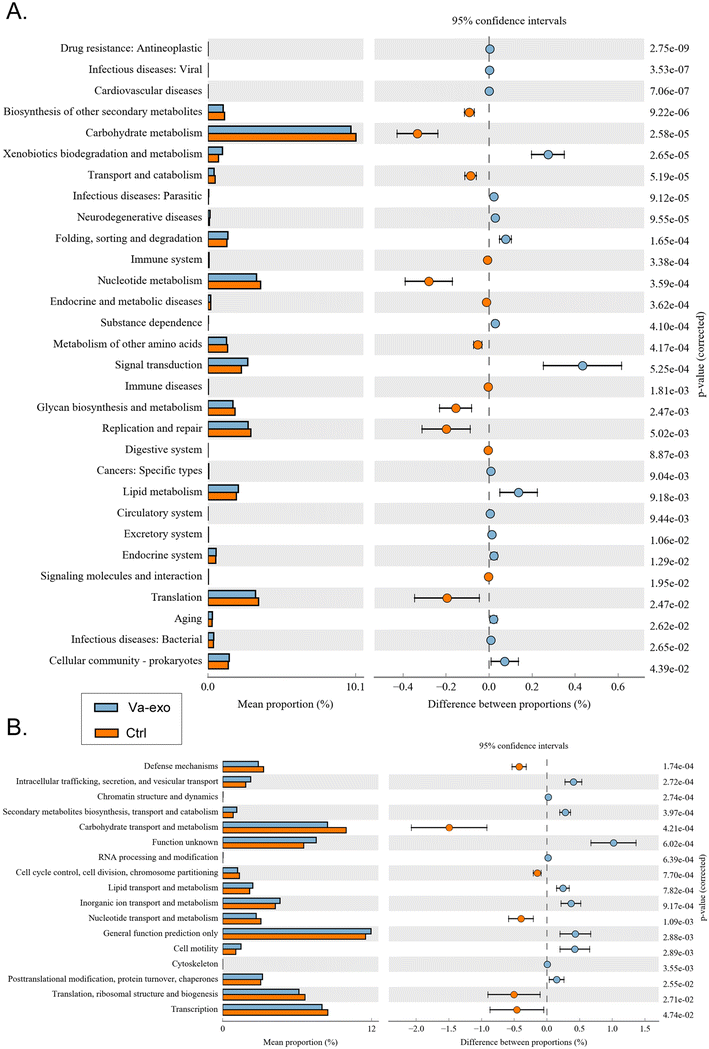
Differential analysis of KEGG-metabolic pathways can reveal differences in the functional genes of microbial communities between different groups of samples, thus allowing the exploration of metabolic alterations that occur and subsequent microbial adaptation to environmental changes. Our analysis showed that compared with the Ctrl group, the intestinal microbial community of the Va-exos group improved xenobiotic biodegradation and metabolism (metabolism), signal transduction (environmental information processing), lipid metabolism (metabolism), and other metabolic pathways, while reducing carbohydrate metabolism (metabolism), nucleotide metabolism (metabolism), and glycan biosynthesis and metabolism (metabolism) (Fig. 8A).The distributions and abundances of homologous protein clusters in the intestinal microbiota of mice can be analyzed through the database clusters of orthologous genes/groups of proteins (COG) database. Using COGs, our data depicted an abundance of proteins related to cellular processes and signaling, such as intracellular trafficking, secretion, and vesicular transport and cell motility, and proteins related to metabolism, such as inorganic ion transport and metabolism; and the analysis revealed that these proteins were significantly elevated in the Va-exos group relative to the Ctrl group (Fig. 8B). In contradistinction, the abundances of proteins related to metabolism, such as carbohydrate transport and metabolism and nucleotide transport and metabolism, and proteins related to information storage and processing, such as translation, ribosomal structure, and biogenesis, were significantly attenuated (Fig. 8B). These data showed that cranberry-derived exosomes significantly altered the expression distribution and diversity of 16S functional genes and metabolic-signaling pathways in the intestinal microbiota of HFHS-POF model mice.
3.6 Cranberry-derived exosomes significantly improve PANoptosis in ovarian granulosa cells in HFHS-POF model mice
The FCM results showed that the proportion of apoptotic and necrotic mOGCs in the ovarian tissues of the Ctrl group was significantly higher than that of the WT group, while the proportion of apoptotic and necrotic mOGCs in the ovarian tissues of Va-exos mice was significantly lower than that in Ctrl group mice (Fig. 9A). FCM detection also showed that ROS levels in the ovarian tissues of Va-exos mice were significantly lower than those in Ctrl group mice (Fig. 9B). Cytokine microarray results showed that the levels of cytokines IL-2, IL-3, IL-12 p40p70, VEGF, and thrombopoietin in the mOGCs of Va-exos mice were statistically greater than those of the Ctrl group (Fig. 9C). Subsequently, qPCR showed that the expression levels of cell-proliferation inhibitory factors (p16, p21, p53, and Bax) and pan-apoptosis family members (MLKL, IL1B, GSDMD, Casp3, Casp7, RREB1, MAZ, and RIP3) in the mOGCs of Ctrl group mice were significantly higher than the levels of the WT group, while the mRNA expression levels of these genes in the mOGCs of the Va-exos group mice were significantly lower than those of the Ctrl group (Fig. 9D). Immunoblot analysis also showed that the expression and phosphorylation levels of pan-apoptotic proteins in the mOGCs of Ctrl group mice were significantly higher than those of the WT group, while the expression and phosphorylation levels of pan-apoptotic proteins of the Va-exos group were lower than those of the Ctrl group (Fig. 9E). Collectively, these experimental results suggested that cranberry-derived exosomes significantly improved the PANoptosis of OGCs in HFHS-POF model mice by inhibiting ROS release and increasing cytokine expression.4. Discussion
In this investigation, we systematically explored the therapeutic effects of exosomes (a type of extracellular vesicle [EV]) from cross-species sources on murine POF by modulating the microecological structure of the intestinal flora and the release of inflammatory factors. EVs are natural nano-sized particles secreted by cells that possess a double-membrane structure and diameters ranging from 40 nm to 1000 nm,26,27 are key mediators of paracrine signaling in cells, and transport internal genetic materials such as mRNA, ncRNA, and proteins as well as bioactive proteins to target organs so as to exert biological effects.26,27 EVs also constitute long-distance intercellular communication in the body26,27 and are currently divided into three main categories: exosomes, microvesicles, and apoptotic bodies.26,27 Although an increasing number of studies have indicated that exosomes comprise a promising cell therapy drug,28,29 some reports have shown that exosomes from plant sources also contain various components such as RNA, lipids, and proteins—similar to the case of animal exosomes.10,28,29 Exosomes from plant sources can be ingested or otherwise incorporated by plants, animals, bacteria, and even human cells, and they can also exert antioxidant, anti-inflammatory, and tissue-regenerative effects.8,10–12 Hwang et al. demonstrated that yam-derived exosome-like nanovesicles could stimulate murine osteoblast formation and prevent osteoporosis by activation of the BMP-2/p-p38-dependent Runx2 pathway.8 Liu et al. reported that garlic-derived exosomes improved the symptoms of nonalcoholic fatty liver disease in high-fat diet-fed mice via macrophage–hepatocyte crosstalk and that they played a role in PFKFB3 expression.9 Ou et al. indicated that Catharanthus roseus (L.) Don leaf-derived exosome-like nanovesicles strongly stimulated the secretion of TNF-α, activated the NF-κB-signaling pathway, and elevated the expression of the hematopoietic function-related transcription factor PU.1 both in vitro and in vivo.10 Deng et al. also reported that broccoli-derived nanoparticles suppressed murine colitis by activating AMP-activated protein kinase expression in dendritic cells.12 All of the aforementioned studies confirmed that exosome-like nanovesicles derived from plants exhibit potential therapeutic and immunoregulatory effects on diseases. Inspired by these research reports, we herein focused on whether Vaccinium macrocarpon Ait (cranberry)-derived exosome-like nanovesicles (Va-exos) also exerted significant therapeutic or ameliorative effects on POF symptoms. First, pathological examination of ovarian tissue, peripheral blood hormone levels, and blood lipids all confirmed that Va-exos significantly improved HFHS-POF and blood lipid levels in mice, presumably due to the flavonoids and antioxidant properties of cranberry itself.5,13–16 Second, when we evaluated digestive organs such as the liver and intestine, we found that Va-exos improved the intestinal barrier and ameliorated liver injury. We speculate that the effects of Va-exos are principally due to their ability to attenuate the release of inflammatory cytokines (especially IL-1β and IL-6) from immune cells in the intestinal mucosa. Third, we demonstrated that HFHS significantly induced PANoptosis in mOGCs, and this may constitute one of the primary mechanisms underlying POF induction in mice. However, after oral administration of Va-exos to HFHS-POF, we ascertained that PANoptosis in mOGCs was significantly improved. Since the state of mOGCs directly affects the overall functioning of the ovary, and the onset of POF is largely related to the status of the OGCs,30,31 we hypothesize that Va-exos play a therapeutic role by improving PANoptosis in mOGCs.Since the interventional route used in this study was oral administration, the microecology of the intestinal flora would reflect a significant role. In fact, the interactions between the gut microbiota and the human body have various effects.32–36 Increasing numbers of studies have reported that the gut microbiota (gut ecology) affects normal physiological and biochemical functions and is associated with various diseases in mammals, including gynaecological diseases.32–36 Guo et al. found that there were significant differences in the composition and distribution of the gut microbiota between a letrozole-induced mouse model of polycystic ovary syndrome (PCOS) and healthy controls.32 The levels of Lactobacillus, Ruminococcus, and Clostridium were lower, while the levels of Prevotella were significantly higher in the PCOS group than those in the control group.32 Yuan et al. found significant differences between mice in the endometriosis and mock groups, where the reduction in Bacteroidetes levels was particularly significant.33 In addition, an imbalance in the gut ecology causes an abnormal increase in the blood oestrogen levels, stimulating the growth of endometriotic lesions and the pathology of cyclic bleeding.34,35 Hence, the gut microbiota appears to be closely associated with the occurrence and outcomes of gynaecological disorders.36 In addition, our previous studies also confirmed that oral administration of fisetin regulated the distribution and content of Akkermansia and Lachnospiraceae microorganisms in the gut flora of POF mice, reduced the number of specific T lymphocyte subsets and the concentration of IL-12 released in peripheral blood, and reduced inflammation levels—achieving therapeutic effects in POF.25 Therefore, we have reason to believe that oral administration of Va-exos will also improve the structure and distribution of the intestinal flora microecology in HFHS-POF mice, achieving regulatory effects on the “gut–ovary axis”. The results of high-throughput sequencing of 16S rRNA confirmed that the relative abundances of Akkermansia and Allobaculum microorganisms in the mouse gut were significantly increased in the Va-exos group compared to those in the Ctrl group, while the relative abundances of uncultured_bacterium_f_Muribaculaceae, Dubosiella, and uncultured_bacterium_f_Lachnospiraceae microorganisms were significantly decreased. At the species level, we discerned that the relative abundances of uncultured_bacterium_g_Akkermansia, uncultured_bacterium_g_Allobaculum, and uncultured_bacterium_g_Bifidobacterium were increased in the mouse gut in the Va-exos group compared to those in the Ctrl group, while the relative abundances of uncultured_bacterium_g_Dubosiella and uncultured_bacterium_f_Muribaculaceae were decreased. Many investigators have indicated that Allobaculum is an important short-chain fatty acid (SCFA) producer in the human gut, and SCFAs are the ultimate metabolites of gut microorganisms that inhibit the growth of pathogenic bacteria and promote the proliferation of beneficial bacteria by reducing intestinal pH, thereby improving the intestinal microenvironment.37–40 Therefore, Allobaculum is critical to maintaining the integrity of the intestinal barrier and constructing the intestinal immune system, and it is a protective flora in the gut. The diminution in the number of Allobaculum bacteria in the gut is also closely correlated with constipation, colitis, obesity, heavy-metal poisoning, and many other diseases.37–40 Bifidobacterium is an anaerobic Gram-positive bacillus that often bifurcates at its terminus;37–40 it is an important component of the physiological microbiota in the intestines of humans and animals, and it is also the second largest group of bacteria found in breastfed infants.37–40 Research shows that obesity, diabetes, allergies, and other diseases are all related to the reduction in Bifidobacteria at all stages of life.37–40 Lachnospiraceae is a family of Gram-positive bacteria that are anaerobic, spore forming, and motile.37–40 The relative abundance of Lachnospiraceae in patients with enteritis, intestinal tumors, liver steatosis, glycolipid metabolic diseases, and diabetic pregnancies is also significantly increased.37–40
In this study, although we have suggested that oral administration of Va-exos could improve the symptoms of HFHS-POF mice via achieving regulatory effects on the “gut–ovary axis” and the structure and distribution of the intestinal flora microecology, there are still some flaws. Firstly, we were unable to transplant the gut microbiota. Secondly, we did not conduct a thorough analysis of the content of Va-exos. Therefore, the efficacy and target of Va-exos on HFHS-POF mice revealed also have certain limitations in this study. The above two directions will be the focus of our future study.
5. Conclusions
In summary, we herein confirmed that oral administration of cranberry-derived exosomes attenuated murine POF by modulating the gut microbiota and inhibiting ovarian granulosa cell PANoptosis (Fig. 10). | ||
| Fig. 10 Oral administration of cranberry-derived exosomes attenuates murine POF by modulating the gut microbiota and inhibiting ovarian granulosa cell PANoptosis. | ||
Abbreviations
| HFHS | High-fat and high-sugar |
| POF | Premature ovarian failure |
| Cranberry | Vaccinium macrocarpon Ait |
| OGCs | Ovarian granulosa cells |
| TC | Total cholesterol |
| TG | Triglyceride |
| LDL | Low-density lipoprotein |
| HDL | High-density lipoprotein |
| ELISA | Enzyme-linked immunosorbent assay |
| E2 | Estradiol-17β |
| FSH | Follicle-stimulating hormone |
| AMH | Anti-Müllerian hormone |
| ACE | Abundance-based coverage indicator |
| RDA | Redundancy analysis |
| PCoA | Principal coordinate analysis |
| NMDS | Non-metric multidimensional scaling |
| UPGMA | Unweighted pair-group method with arithmetic mean |
| LDA | Line discriminant analysis |
| LEfSe | Effect size |
| GCSF | Granulocyte-colony stimulating factor |
| HRP | Streptavidin horseradish peroxidase |
| GM-CSF | Granulocyte-macrophage colony stimulating factor |
| IFN-γ | Interferon gamma |
| MCP-1/5 | Monocyte chemoattractant proteins 1/5 |
| RANTES | Regulated on activation, normal T cell expressed and secreted |
| SCF | Stem cell factor |
| sTNFR1 | Soluble tumor necrosis receptor factor 1 |
| TNF-α | Tumor necrosis factor alpha |
| VEGF | Vascular endothelial growth factor |
| TEM | Transmission electron microscopy |
| NAT | Nanoparticle Tracking Analysis |
| COG | Genes/groups of proteins |
Author contributions
Zeyu Cui, Te Liu, Yichao Wen, and Weihao Li performed the majority of the experiments in the study. Jianghong Xu, Yingjuan Chen, Danping Chen, and Ying Zhu contributed to the analysis of the experimental data. Te Liu contributed to the study design and manuscript writing and provided experimental funding support. All authors read and approved the final manuscript.Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank LetPub (https://www.letpub.com) for its linguistic assistance during the preparation of this manuscript. This work was supported by a grant from the National Natural Science Foundation of China (no. 82274569) and the Clinical Research Special Fund of the Shanghai Municipal Health Commission (202240223).References
- G. Liu, F. Shi, U. Blas-Machado, Q. Duong, V. L. Davis, W. G. Foster and C. L. Hughes, Ovarian effects of a high lactose diet in the female rat, Reprod., Nutr., Dev., 2005, 45, 185–192 CrossRef PubMed.
- L. Tappy and K. A. Le, Metabolic effects of fructose and the worldwide increase in obesity, Physiol. Rev., 2010, 90, 23–46 CrossRef CAS PubMed.
- N. Wang, L. L. Luo, J. J. Xu, M. Y. Xu, X. M. Zhang, X. L. Zhou, W. J. Liu and Y. C. Fu, Obesity accelerates ovarian follicle development and follicle loss in rats, Metab., Clin. Exp., 2014, 63, 94–103 CrossRef CAS PubMed.
- Y. Wu, Z. Zhang, X. Liao and Z. Wang, High fat diet triggers cell cycle arrest and excessive apoptosis of granulosa cells during the follicular development, Biochem. Biophys. Res. Commun., 2015, 466, 599–605 CrossRef CAS PubMed.
- Z. Fu, D. Liska, D. Talan and M. Chung, Cranberry Reduces the Risk of Urinary Tract Infection Recurrence in Otherwise Healthy Women: A Systematic Review and Meta-Analysis, J. Nutr., 2017, 147, 2282–2288 CrossRef CAS PubMed.
- M. D. Goncalves, C. Lu, J. Tutnauer, T. E. Hartman, S. K. Hwang, C. J. Murphy, C. Pauli, R. Morris, S. Taylor, K. Bosch, S. Yang, Y. Wang, J. Van Riper, H. C. Lekaye, J. Roper, Y. Kim, Q. Chen, S. S. Gross, K. Y. Rhee, L. C. Cantley and J. Yun, High-fructose corn syrup enhances intestinal tumor growth in mice, Science, 2019, 363, 1345–1349 CrossRef CAS PubMed.
- T. Liu, F. Jing, P. Huang, Z. Geng, J. Xu, J. Li, D. Chen, Y. Zhu, Z. Wang, W. Huang and C. Chen, Thymopentin alleviates premature ovarian failure in mice by activating YY2/Lin28A and inhibiting the expression of let-7 family microRNAs, Cell Proliferation, 2021, 54, e13089 CrossRef CAS PubMed.
- J. H. Hwang, Y. S. Park, H. S. Kim, D. H. Kim, S. H. Lee, C. H. Lee, S. H. Lee, J. E. Kim, S. Lee, H. M. Kim, H. W. Kim, J. Kim, W. Seo, H. J. Kwon, B. J. Song, D. K. Kim, M. C. Baek and Y. E. Cho, Yam-derived exosome-like nanovesicles stimulate osteoblast formation and prevent osteoporosis in mice, J. Controlled Release, 2023, 355, 184–198 CrossRef CAS PubMed.
- J. Liu, W. Li, Y. Bian, X. Jiang, F. Zhu, F. Yin, L. Yin, X. Song, H. Guo and J. Liu, Garlic-derived exosomes regulate PFKFB3 expression to relieve liver dysfunction in high-fat diet-fed mice via macrophage-hepatocyte crosstalk, Phytomedicine, 2023, 112, 154679 CrossRef CAS PubMed.
- X. Ou, H. Wang, H. Tie, J. Liao, Y. Luo, W. Huang, R. Yu, L. Song and J. Zhu, Novel plant-derived exosome-like nanovesicles from Catharanthus roseus: preparation, characterization, and immunostimulatory effect via TNF-alpha/NF-kappaB/PU.1 axis, J. Nanobiotechnol., 2023, 21, 160 CrossRef CAS PubMed.
- O. Urzi, M. Cafora, N. R. Ganji, V. Tinnirello, R. Gasparro, S. Raccosta, M. Manno, A. M. Corsale, A. Conigliaro, A. Pistocchi, S. Raimondo and R. Alessandro, Lemon-derived nanovesicles achieve antioxidant and anti-inflammatory effects activating the AhR/Nrf2 signaling pathway, iScience, 2023, 26, 107041 CrossRef CAS PubMed.
- Z. Deng, Y. Rong, Y. Teng, J. Mu, X. Zhuang, M. Tseng, A. Samykutty, L. Zhang, J. Yan, D. Miller, J. Suttles and H. G. Zhang, Broccoli-Derived Nanoparticle Inhibits Mouse Colitis by Activating Dendritic Cell AMP-Activated Protein Kinase, Mol. Ther., 2017, 25, 1641–1654 CrossRef CAS PubMed.
- B. V. Nemzer, F. Al-Taher, A. Yashin, I. Revelsky and Y. Yashin, Cranberry: Chemical Composition, Antioxidant Activity and Impact on Human Health: Overview, Molecules, 2022, 27, 1503–1509 CrossRef CAS PubMed.
- M. Mantzorou, A. Zarros, G. Vasios, S. Theocharis, E. Pavlidou and C. Giaginis, Cranberry: A Promising Natural Source of Potential Nutraceuticals with Anticancer Activity, Anti-Cancer Agents Med. Chem., 2019, 19, 1672–1686 CrossRef CAS PubMed.
- H. Shmuely, I. Ofek, E. I. Weiss, Z. Rones and Y. Houri-Haddad, Cranberry components for the therapy of infectious disease, Curr. Opin. Biotechnol., 2012, 23, 148–152 CrossRef CAS PubMed.
- C. C. Neto, Cranberry and blueberry: evidence for protective effects against cancer and vascular diseases, Mol. Nutr. Food Res., 2007, 51, 652–664 CrossRef CAS PubMed.
- Y. Wang and T. D. Kanneganti, From pyroptosis, apoptosis and necroptosis to PANoptosis: A mechanistic compendium of programmed cell death pathways, Comput. Struct. Biotechnol. J., 2021, 19, 4641–4657 CrossRef CAS PubMed.
- S. Christgen, M. Zheng, S. Kesavardhana, R. Karki, R. K. S. Malireddi, B. Banoth, D. E. Place, B. Briard, B. R. Sharma, S. Tuladhar, P. Samir, A. Burton and T. D. Kanneganti, Identification of the PANoptosome: A Molecular Platform Triggering Pyroptosis, Apoptosis, and Necroptosis (PANoptosis), Front. Cell. Infect. Microbiol., 2020, 10, 237 CrossRef CAS PubMed.
- P. Samir, R. K. S. Malireddi and T. D. Kanneganti, The PANoptosome: A Deadly Protein Complex Driving Pyroptosis, Apoptosis, and Necroptosis (PANoptosis), Front. Cell. Infect. Microbiol., 2020, 10, 238 CrossRef CAS PubMed.
- X. Sun, Y. Yang, X. Meng, J. Li, X. Liu and H. Liu, PANoptosis: Mechanisms, biology, and role in disease, Immunol. Rev., 2024, 321, 246–262 CrossRef CAS PubMed.
- R. K. S. Malireddi, S. Kesavardhana and T. D. Kanneganti, ZBP1 and TAK1: Master Regulators of NLRP3 Inflammasome/Pyroptosis, Apoptosis, and Necroptosis (PAN-optosis), Front. Cell. Infect. Microbiol., 2019, 9, 406 CrossRef CAS PubMed.
- B. Sundaram, N. Pandian, R. Mall, Y. Wang, R. Sarkar, H. J. Kim, R. K. S. Malireddi, R. Karki, L. J. Janke, P. Vogel and T. D. Kanneganti, NLRP12-PANoptosome activates PANoptosis and pathology in response to heme and PAMPs, Cell, 2023, 186, 2783–2801 CrossRef CAS PubMed.
- S. Lee, R. Karki, Y. Wang, L. N. Nguyen, R. C. Kalathur and T. D. Kanneganti, AIM2 forms a complex with pyrin and ZBP1 to drive PANoptosis and host defence, Nature, 2021, 597, 415–419 CrossRef CAS PubMed.
- T. Liu, J. Lin, C. Chen, X. Nie, F. Dou, J. Chen, Z. Wang and Z. Gong, MicroRNA-146b-5p overexpression attenuates premature ovarian failure in mice by inhibiting the Dab2ip/Ask1/p38-Mapk pathway and gammaH2A.X phosphorylation, Cell Proliferation, 2021, 54, e12954 CrossRef CAS PubMed.
- J. Lin, X. Nie, Y. Xiong, Z. Gong, J. Chen, C. Chen, Y. Huang and T. Liu, Fisetin regulates gut microbiota to decrease CCR9(+)/CXCR3(+)/CD4(+) T-lymphocyte count and IL-12 secretion to alleviate premature ovarian failure in mice, Am. J. Transl. Res., 2020, 12, 203–247 CAS.
- G. van Niel, G. D'Angelo and G. Raposo, Shedding light on the cell biology of extracellular vesicles, Nat. Rev. Mol. Cell Biol., 2018, 19, 213–228 CrossRef CAS PubMed.
- D. K. Jeppesen, Q. Zhang, J. L. Franklin and R. J. Coffey, Extracellular vesicles and nanoparticles: emerging complexities, Trends Cell Biol., 2023, 33, 667–681 CrossRef CAS PubMed.
- J. J. Lai, Z. L. Chau, S. Y. Chen, J. J. Hill, K. V. Korpany, N. W. Liang, L. H. Lin, Y. H. Lin, J. K. Liu, Y. C. Liu, R. Lunde and W. T. Shen, Exosome Processing and Characterization Approaches for Research and Technology Development, Adv. Sci., 2022, 9, e2103222 CrossRef PubMed.
- C. He, S. Zheng, Y. Luo and B. Wang, Exosome Theranostics: Biology and Translational Medicine, Theranostics, 2018, 8, 237–255 CrossRef CAS PubMed.
- S. J. Chon, Z. Umair and M. S. Yoon, Premature Ovarian Insufficiency: Past, Present, and Future, Front. Cell Dev. Biol., 2021, 9, 672890 CrossRef PubMed.
- R. W. Rebar, Premature ovarian failure, Obstet. Gynecol., 2009, 113, 1355–1363 CrossRef PubMed.
- Y. Guo, Y. Qi, X. Yang, L. Zhao, S. Wen, Y. Liu and L. Tang, Association between Polycystic Ovary Syndrome and Gut Microbiota, PLoS One, 2016, 11, e0153196 CrossRef PubMed.
- M. Yuan, D. Li, Z. Zhang, H. Sun, M. An and G. Wang, Endometriosis induces gut microbiota alterations in mice, Hum. Reprod., 2018, 33, 607–616 CrossRef CAS PubMed.
- R. Flores, J. Shi, B. Fuhrman, X. Xu, T. D. Veenstra, M. H. Gail, P. Gajer, J. Ravel and J. J. Goedert, Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study, J. Transl. Med., 2012, 10, 253 CrossRef CAS PubMed.
- J. M. Baker, L. Al-Nakkash and M. M. Herbst-Kralovetz, Estrogen-gut microbiome axis: Physiological and clinical implications, Maturitas, 2017, 103, 45–53 CrossRef CAS PubMed.
- O. Koren, J. K. Goodrich, T. C. Cullender, A. Spor, K. Laitinen, H. K. Backhed, A. Gonzalez, J. J. Werner, L. T. Angenent, R. Knight, F. Backhed, E. Isolauri, S. Salminen and R. E. Ley, Host remodeling of the gut microbiome and metabolic changes during pregnancy, Cell, 2012, 150, 470–480 CrossRef CAS PubMed.
- N. Zmora, J. Suez and E. Elinav, You are what you eat: diet, health and the gut microbiota, Nat. Rev. Gastroenterol. Hepatol., 2019, 16, 35–56 CrossRef CAS PubMed.
- G. Vrancken, A. C. Gregory, G. R. B. Huys, K. Faust and J. Raes, Synthetic ecology of the human gut microbiota, Nat. Rev. Microbiol., 2019, 17, 754–763 CrossRef CAS PubMed.
- X. Jiang and X. Hu, Data Analysis for Gut Microbiota and Health, Adv. Exp. Med. Biol., 2017, 1028, 79–87 CrossRef PubMed.
- P. P. Ahern and K. J. Maloy, Understanding immune-microbiota interactions in the intestine, Immunology, 2020, 159, 4–14 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4fo03468f |
| ‡ These authors contributed equally to this work and shared the first authorship. |
| This journal is © The Royal Society of Chemistry 2024 |