Open Access Article
Open Access ArticleDietary influences on urinary tract infections: unraveling the gut microbiota connection†
Yifan
Du‡
a,
Xiuyuan
Sui‡
a,
Yang
Bai‡
a,
Zhiyuan
Shi
a,
Bin
Liu
a,
Zeyuan
Zheng
a,
Zhengying
Zhang
a,
Yue
Zhao
a,
Jiqing
Wang
d,
Qian
Zhang
c,
Yuanhang
Zhu
b,
Qing
Liu
a,
Mingshan
Wang
a,
Huimin
Sun
*b and
Chen
Shao
 *a
*a
aDepartment of Urology, Xiang'an Hospital of Xiamen University, School of Medicine, Xiamen University, Xiamen, 361101, China. E-mail: cshao@xah.xmu.edu.cn
bCentral Laboratory, Xiang'an Hospital of Xiamen University, School of Medicine, Xiamen University, Xiamen, 361101, China
cDepartment of Endocrinology, Xiang'an Hospital of Xiamen University, School of Medicine, Xiamen University, Xiamen, 361000, China
dDepartment of Orthopedics, The First Affiliated Hospital of Xi'an Jiaotong University, Xi'an, 710061, China
First published on 18th September 2024
Abstract
This study employs Mendelian randomization to investigate the causal relationships between dietary factors, gut microbiota, and urinary tract infections (UTIs). Our analysis revealed statistically significant associations, including high alcohol intake, cheese, and oily fish consumption with UTI risk, as well as links between UTI risk and specific gut microbiota, such as Prevotellaceae, Butyrivibrio, Anaerotruncus, and Dorea. Additionally, we observed associations with inflammatory markers, including C-Reactive Protein and Interleukin-6. Although the observed effects of these dietary factors on UTI risk are minimal and may limit their clinical relevance, these findings can still hold significant implications at the population level in public health. This research offers novel insights into the interplay between diet, gut microbiota, and UTI risk, laying a foundation for future studies. Further research is warranted to validate these associations and to explore the underlying mechanisms and their broader impact on public health.
1. Introduction
Urinary tract infections (UTIs) stand as one of the most prevalent infections in both community and hospital settings. Approximately 12% of males and 40% of females experience at least one UTI in their lifetime.1 The high incidence rate of UTIs results in significant healthcare expenditures on a population scale. Unfortunately, UTIs often go undiagnosed, overdiagnosed, or inadequately diagnosed, leading to delayed treatment. UTIs encompass a spectrum of clinical phenotypes, including cystitis, pyelonephritis, prostatitis, urosepsis, and catheter-associated UTIs (CA-UTIs).2 Notably, UTIs are not only common but also characterized by a high recurrence rate. Recurrent UTIs (rUTIs) are defined by a frequency of at least two episodes within the preceding six months or three episodes within the past year. Acute UTIs predominantly manifest as uncomplicated cystitis and affect 50–80% of females within the general population. About a quarter of women experience frequent recurrences following an initial acute UTI. Emerging evidence underscores that susceptibility to rUTIs is, in part, mediated through the gut–bladder axis, involving disruptions in gut ecology and differential immune responses to bacterial colonization of the bladder. This has led to escalated antibiotic usage and the inadequacy of conventional treatment approaches, compounded by antibiotic resistance, elevating the urgency of addressing the impending threat of UTIs to human health.3 Thus, investigating the risk factors associated with UTIs and mitigating their occurrence holds paramount significance.UTIs typically result from the retrograde infection of bacteria from the fecal microbiota. Given that the composition of gut microbiota is substantially influenced by dietary choices, alterations in diet may potentially modulate the risk of UTIs. Prior case-control studies have indicated that the consumption of cranberry products and fermented dairy containing beneficial probiotic strains, such as Lactobacillus, effectively reduces the risk of recurrent UTIs.4,5 Additionally, prospective research has explored the association between vegetarian diets and UTIs.6 Establishing a link between diet and UTIs has been challenging.
The dietary components selected for this study—coffee, tea, alcohol, cheese, oily fish, poultry, pork, processed meat, non-oily fish, beef, and mutton—were chosen based on their hypothesized roles in modulating inflammation and immune responses, which are critical mechanisms in the pathogenesis of UTIs. These selections were guided by existing literature and preliminary data suggesting potential links between these dietary factors and inflammatory processes that may influence UTI risk. For example, alcohol consumption has been shown to impair immune function and increase gut permeability, leading to an elevated risk of bacterial translocation and infection.7 Cheese, a fermented dairy product rich in probiotics, has been associated with reduced inflammation and enhanced gut barrier function,8 which may protect against UTIs. Oily fish, high in omega-3 polyunsaturated fatty acids, has anti-inflammatory properties and may help maintain gut microbiota balance,9,10 reducing UTI risk. However, despite considering a broad range of dietary factors,11–13 our Mendelian randomization analysis revealed that only alcohol, cheese, and oily fish had statistically significant associations with UTI risk. This outcome highlights the importance of robust, data-driven methods like Mendelian randomization in identifying true causal relationships, minimizing the influence of confounding variables and reverse causation. The absence of significant associations for the other dietary factors suggests that, while these factors may have broader health effects as documented in the literature, they do not appear to be major contributors to UTI risk in the context of our study.
While randomized controlled trials (RCTs) have long been considered the gold standard for establishing causality in multicenter studies, most long-term nutritional epidemiological investigations rely on food frequency questionnaires to gauge food and nutrient consumption. Such an approach is susceptible to bias due to self-reported measurement errors. Furthermore, the widespread consumption of fortified foods and vitamin supplements adds complexity to assessing nutritional intake. Observational studies can introduce biases due to confounding factors and reverse causation, particularly when causal inferences are involved. Finally, RCTs have their own limitations related to ethical concerns, observation duration, and resource and cost constraints. These limitations can lead to measurement inaccuracies, making it challenging to conclusively link specific dietary habits to UTI risk. However, Mendelian randomization (MR) studies offer a promising approach to overcome these challenges.
MR is a method for assessing causal relationships between risk factors and diseases. It utilizes one or more genetic variants associated with the exposure of interest as instrumental variables to evaluate the association between the exposure and the outcome. As alleles are randomly allocated during conception, genetic variations remain unaffected by measurement biases or biases arising from reverse causation. Furthermore, MR offers a feasible approach for inferring correlations between specific dietary intake and diseases, utilizing the uniqueness of genotypes to investigate causal relationships between exposures and outcomes. It employs genetically correlated instrumental variables (IVs), closely related to the exposure, to mimic a randomized controlled setting. The MR design can mitigate potential residual confounding effects and counteract reverse causation biases. Leveraging MR-based research designs allows the investigation of exposures that cannot be subjected to randomized controlled trials. While the connection between diet and UTIs, as well as the relationship between gut microbiota and UTIs, has garnered considerable attention from researchers, there has been a lack of direct Mendelian randomization studies concerning the causal relationships between dietary factors, gut microbiota, and UTIs. Hence, we employ MR analysis to explore the associations between dietary factors and gut microbiota with UTIs. Through MR analysis, our objective is to identify the various associations between dietary habits and urinary tract infections and the impact of different gut microbiota on urinary tract infections. This research aims to provide novel insights into the prevention and treatment of urinary tract infections.
2. Materials and methods
2.1 Study design
The MR study design, as depicted in Fig. 2, adheres to established diagnostic criteria, including those outlined by the International Classification of Diseases, Tenth Revision (ICD-10) under code N39, which encompasses various urinary system disorders. This investigation leverages publicly accessible datasets from the UK Biobank, the FinnGen study, and prior Genome-Wide Association Studies (GWAS). Specifically, two datasets, the UK Biobank (ukb-b-8814) and FinnGen (finn-b-N14_URETHRAOTH), were meticulously chosen to mutually corroborate findings pertaining to urinary tract infection-related phenomena. All exposure-specific MR analyses can be located in the IEU OPEN GWAS PROJECT along with their respective datasets. We initiated our study by conducting MR analyses on dietary habits and urinary tract infections, subsequently subjecting the extracted dietary habits associated with urinary tract infections to further MR analysis in conjunction with levels of some commonly acknowledged urinary tract infection biomarkers. Finally, we conducted MR analysis to investigate the relationship between 208 types of gut microbiota and UTIs. And this study is reported following the Strengthening the Reporting of Observational Studies in Epidemiology Using Mendelian Randomization guidelines (STROBE-MR, S1 Checklist). Subsequently, we provided relevant findings pertaining to specific gut microbiota associated with UTIs, all of which have been previously reported in internationally authoritative journals, mitigating any concerns regarding database integrity (Fig. 1).2.2 Data sources
Analysis was conducted using publicly available datasets from The IEU Open GWAS project. Exposure-specific dietary factors were obtained from the UK Biobank and included coffee intake (ukb-b-5237), tea intake (ukb-b-6066), alcohol intake frequency (ukb-b-5779), cheese intake (ukb-b-1489), poultry intake (ukb-b-8006), pork intake (ukb-b-5640), processed meat intake (ukb-b-6324), oily fish intake (ukb-b-2209), non-oily fish intake (ukb-b-17627), beef intake (ukb-b-2862), and mutton intake (ukb-b-14179). Data related to urinary tract infections were sourced from the UK Biobank (ukb-b-8814) and the FinnGen biobank analysis (finn-b-N14_URETHRAOTH).Regarding urinary tract infection biomarkers, C-Reactive protein levels were sourced from MR-Base,14,15 This study measured serum CRP in mg L−1 using standard laboratory techniques and transformed the values by natural log. Individuals with autoimmune diseases, those taking immune-modulating agents (if this information was available), and those with CRP values 4 SD or more from the mean were excluded from all analyses. Neutrophil gelatinase-associated lipocalin and Procalcitonin levels were sourced from the Genomic Atlas of the Human Plasma Proteome, Interleukin-1-beta levels from the Genome-wide Association Study Identifies 27 Loci Influencing Concentrations of Circulating Cytokines and Growth Factors,16 and Interleukin-6, Interleukin-8, and Myeloperoxidase levels from the Genomic and Drug Target Evaluation of 90 Cardiovascular Proteins in 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 931 Individuals,17 Elevated erythrocyte sedimentation rate and abnormality of plasma viscosity data were obtained from the FinnGen biobank analysis. For these specific inflammatory biomarkers, all datasets excluded individuals with recent major illnesses (e.g., myocardial infarction, stroke, cancer, HIV, hepatitis B or C) and recent infections, as detailed in the cited literature.
931 Individuals,17 Elevated erythrocyte sedimentation rate and abnormality of plasma viscosity data were obtained from the FinnGen biobank analysis. For these specific inflammatory biomarkers, all datasets excluded individuals with recent major illnesses (e.g., myocardial infarction, stroke, cancer, HIV, hepatitis B or C) and recent infections, as detailed in the cited literature.
The 208 gut microbiota taxa used in our analysis originated from the MiBioGen consortium dataset,18 one of the largest and most comprehensive human microbiome GWAS to date. This dataset includes data from multiple cohorts across diverse populations, providing extensive coverage of gut microbiota diversity and their potential associations with various health outcomes. Initially comprising 211 species. Using a threshold of P < 1 × 10−5 to filter for suitable SNPs, three species were excluded due to the lack of suitable SNPs. Therefore, the actual number of species used in our analysis was 208. Definitions, inclusion criteria, and processing methods for all these data can be found on their respective official websites or citations and are not reiterated in this manuscript. Ethical approval was not required for the current analysis, as all incorporated GWAS data were publicly available and had already received approval from their respective ethics review boards.
2.3 Instrumental variable selection
We established a set of IVs for dietary factors and urinary tract infection markers, selecting SNPs with a genome-wide significance threshold of p < 5 × 10−8. We also applied an exclusion criterion, removing SNPs with an r2 > 0.001 (using a clumping window of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 kb) to account for linkage disequilibrium. To refine our SNP selection, we further filtered each SNP based on potential confounders related to age, weight, gender, ethnicity, as well as factors related to the outcome or potential confounding factors within the dietary factors using the PhenoScanner website.19 Subsequently, we computed the F-statistic to assess the strength of each individual SNP as an instrument. SNPs with an F-statistic greater than 10 were considered sufficiently strong to mitigate potential weak instrument bias, while SNPs with an F-statistic below 10 were excluded from the analysis.
000 kb) to account for linkage disequilibrium. To refine our SNP selection, we further filtered each SNP based on potential confounders related to age, weight, gender, ethnicity, as well as factors related to the outcome or potential confounding factors within the dietary factors using the PhenoScanner website.19 Subsequently, we computed the F-statistic to assess the strength of each individual SNP as an instrument. SNPs with an F-statistic greater than 10 were considered sufficiently strong to mitigate potential weak instrument bias, while SNPs with an F-statistic below 10 were excluded from the analysis.
2.4 MR analysis
The primary analytical method employed to calculate causal effects was the Inverse Variance Weighted (IVW) method. The IVW model stands as the most robust method for detecting causal relationships within two-sample MR analyses. Two additional approaches, including the Weighted Median and MR-Egger methods, were employed as sensitivity analyses. The Weighted Median method furnishes valid estimates if over 50% of the information originates from valid instrumental variables (IVs). The MR-Egger method serves to assess horizontal pleiotropy in the selected IVs. The Cochrane's Q statistic was utilized to detect heterogeneity among the chosen IVs. Should heterogeneity be present, a random-effects model was adopted; otherwise, a fixed-effects model was employed. Furthermore, leave-one-out sensitivity analyses were conducted to ascertain if the overall estimate was disproportionately influenced by individual SNPs. The estimation of the F-statistic quantified the strength of instrumental variables for each exposure; , where R2 represents the proportion of variance in the exposure explained by the genetic variants, N represents the sample size, and K represents the final number of instrumental variables after the selection process. All statistical analyses were executed using the “TwoSampleMR” package in R version 4.3.2.
, where R2 represents the proportion of variance in the exposure explained by the genetic variants, N represents the sample size, and K represents the final number of instrumental variables after the selection process. All statistical analyses were executed using the “TwoSampleMR” package in R version 4.3.2.
3. Results
3.1 The impact of dietary habits on urinary tract infections
Due to the inherent complexity of dietary factors, involving numerous confounding elements, we adopted a cross-validation approach by utilizing two databases to substantiate the authenticity and reliability of our conclusions. Employing the IVW method in conjunction with a comprehensive meta-analysis, it is noteworthy that many Confidence Intervals (CI) approach 1, a result of presenting results with two decimal places. Nonetheless, we assert that the combined IVW analysis yields meaningful results.The results demonstrate a negative correlation between processed meat intake (OR = 0.99; 95% CI, 0.98–1.00; p = 0.03), cheese intake (OR = 0.99; 95% CI, 0.99–1.00; p = 0.02), and oily fish intake (OR = 0.99; 95% CI, 0.99–1.00; p = 0.04) with urinary tract infections. Conversely, alcohol intake frequency is positively associated with an increased risk of urinary tract infections (OR = 1.00; CI, 1.00–1.01; p = 0.01). No evidence of horizontal pleiotropy or substantial heterogeneity was detected. Consistent results are observed in scatter plots and forest plots, as presented in ESI,† respectively. The leave-one-out sensitivity analysis, depicted in ESI,† suggests that the overall estimate remains unaffected by any individual SNP (Fig. 2).
3.2 Specific dietary habits and urinary tract infection markers
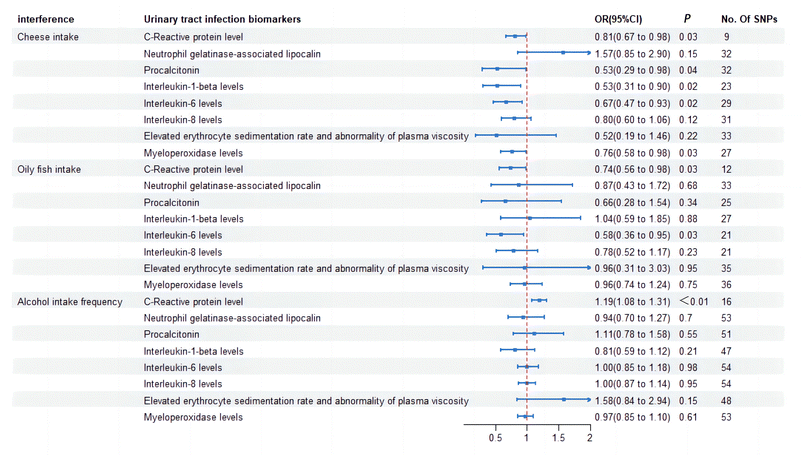
The IVW results indicate a positive association between alcohol intake and an increase in C-Reactive Protein (CRP) levels (OR = 1.19; 95%CI, 1.08–1.31; p < 0.001). Additionally, a reduction in cheese intake is observed to be associated with lower levels of CRP (OR = 0.810; 95%CI, 0.671–0.978; p = 0.03), procalcitonin (OR = 0.529; 95%CI, 0.286–0.978; p = 0.04), IL6 (OR = 0.665; 95%CI, 0.474–0.934; p = 0.02), and myeloperoxidase levels (OR = 0.756; 95%CI, 0.584–0.977; p = 0.03). Similarly, oily fish intake is associated with decreased levels of CRP (OR = 0.738; 95%CI, 0.557–0.977; p = 0.03) and IL6 (OR = 0.584; 95%CI, 0.360–0.945; p = 0.03). Consistent findings are mirrored in scatter plots, forest plots, funnel plots, and leave-one-out sensitivity analysis plots (ESI†) (Fig. 3).3.3 The relationship between different subtypes of intestinal flora and urinary tract infection
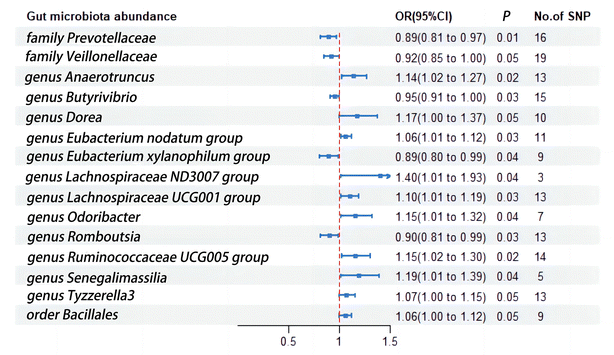
Given that urinary tract infections are often a result of retrograde infection from the gut microbiota, and the direct link between dietary habits and gut microbiota has been extensively reported, we proceeded to delve into the impact of various subtypes of gut microbiota on urinary tract infections. In this instance, we utilized the larger patient dataset and SNP count from the FinnGen database to shed light on the subgroups associated with urinary tract infections (Fig. 4). Family Prevotellaceae (OR = 0.89; 95%CI, 0.81–0.97; p = 0.01), Family Veillonellaceae (OR = 0.92; 95%CI, 0.85–1.00; p = 0.05), Genus Butyrivibrio (OR = 0.95; 95%CI, 0.91–1.00; p = 0.03), Genus Eubacterium xylanophilum group (OR = 0.89; 95%CI, 0.80–0.99; p = 0.03), and Genus Romboutsia (OR = 0.90; 95%CI, 0.81–0.99; p = 0.03) exhibited a negative correlation with urinary tract infections. Conversely, Genus Anaerotruncus (OR = 1.14; 95%CI, 1.02–1.27; p = 0.02), Genus Dorea (OR = 1.17; 95%CI, 1.00–1.37; p = 0.05), Genus Eubacterium nodatum group (OR = 1.06; 95%CI, 1.01–1.12; p = 0.03), Genus Lachnospiraceae ND3007 group (OR = 1.40; 95%CI, 1.01–1.93; p = 0.04), Genus Lachnospiraceae UCG001 group (OR = 1.10; 95%CI, 1.01–1.19; p = 0.03), Genus Odoribacter (OR = 1.15; 95%CI, 1.01–1.32; p = 0.04), Genus Ruminococcaceae UCG005 group (OR = 1.15; 95%CI, 1.02–1.30; p = 0.02), Genus Senegalimassilia (OR = 1.19; 95%CI, 1.01–1.39; p = 0.01), Genus Tyzzerella3 (OR = 1.07; 95%CI, 1.00–1.15; p = 0.05), and Order Bacillales (OR = 1.06; 95%CI, 1.00–1.12; p = 0.05) were associated with an increased risk of urinary tract infections (Fig. 4).4. Discussion
This study employs Mendelian randomization to investigate the causal relationships between dietary factors, gut microbiota, and UTI risk. Our analysis revealed statistically significant associations between certain dietary factors, such as high alcohol intake, cheese, and oily fish consumption, and UTI risk. However, the observed effect sizes, with ORs close to 1, suggest that these effects are small. It is crucial to interpret these findings cautiously, as the practical implications may be limited. While the MR approach provides a robust framework for causal inference by mitigating biases from confounding and reverse causation, it is important to recognize that statistical significance does not necessarily equate to clinical significance, especially when effect sizes are marginal. Despite the small effect sizes, these findings may still hold important implications, particularly in public health, where even small but widespread changes in dietary behaviors could lead to significant reductions in disease incidence.20–22 In this MR analysis, we assessed the causal relationships between dietary intake and gut microbiota species with susceptibility to UTIs. We observed potential protective effects of oily fish and cheese intake against UTIs, while high alcohol intake frequency appeared to increase the risk of UTIs. Additionally, we identified 15 associations between gut microbiota and UTIs.Numerous studies have confirmed that alcohol impairs the functionality of various components of the immune defense system.7,23,24 Prolonged alcohol consumption disrupts various aspects of acquired immune responses, including cell-mediated and humoral responses, rendering individuals more susceptible to viral and bacterial infections, as well as sterile inflammation. Existing research suggests that alcohol can alter the balance and interactions between the host immune system and the host microbiota to influence immune function. Alcohol consumption disrupts the gut barrier by increasing oxidative stress in the gut, leading to increased gut permeability and dysbiosis.25 This disruption can eventually result in the translocation of Gram-negative bacterial products, and the observed increase in CRP levels in our results aligns well with these findings. Moreover, as previously reported,26,27 alcohol consumption can lead to a decrease in the abundance of Bacteroidetes phyla (synonym Bacteroidota) bacteria and an increase in Firmicutes phylum (synonym Bacillota), specifically the Bacilli class. In our study, the protective effect of the family Prevotellaceae, which belongs to the Bacteroidetes phylum (Bacteroidota), against UTIs, and the increased risk associated with genera Anaerotruncus and Dorea (both of Firmicutes phylum: Bacilli class), corroborate this perspective. Therefore, the elevated frequency of alcohol consumption may enhance susceptibility to UTIs by affecting the immune system and altering the gut microbiota composition.
Oily fish exhibit a potential protective effect against UTIs. In comparison to white fish, oily fish are rich in omega-3 polyunsaturated fatty acids (PUFAs), specifically eicosapentaenoic acid and docosahexaenoic acid, both of which belong to the ω-3 class. The biochemical and physiological actions of these PUFAs largely depend on their conversion to 20-carbon or 22-carbon acids, followed by subsequent metabolism into bioactive lipid mediators such as prostaglandins, leukotrienes, lipoxins, and resolvins. Research indicates that omega-3 PUFAs have mitigating effects on various inflammation-related diseases. They achieve this by attenuating the activation of MAPK, NF-κB, activator protein-1, and oxidative stress pathways, or by enhancing the activation of PPARγ or GPR120, thus diminishing inflammation.9 Omega-3 PUFAs can act as alternative substrates for cyclooxygenase or lipoxygenase, impeding the conversion of arachidonic acid into pro-inflammatory eicosanoids, thereby reducing the production of inflammatory mediators. The reduction in IL-6 levels associated with oil fish intake in our findings further underscores this perspective. Research suggests that omega-3 PUFAs from fish oil can reduce the abundance of Firmicutes in animal models, corroborating our results.10 In terms of maintaining intestinal epithelial integrity, PUFAs influence gut inflammation either as precursors for anti-inflammatory eicosanoids synthesis or by regulating tight junction functionality to enhance gut integrity.28 When intestinal barrier function is compromised, resulting in heightened gut permeability and increased lipopolysaccharide (LPS) translocation, the subsequent postprandial endotoxemia results in mild systemic inflammation.29 Omega-3 PUFAs can alter the microbiota composition by inducing the production and secretion of intestinal alkaline phosphatase, thereby reducing the abundance of LPS-producing bacteria.30 The decrease in CRP in our results also supports this notion.
In addition to being rich in saturated fatty acids, cheese, as a dairy product, contains a diverse array of constituents including vitamins D, calcium, protein, probiotics, and bioactive peptides. Research indicates a close correlation between the Dietary Approaches to Stop Hypertension (DASH) diet pattern (inclusive of dairy products) and reduced inflammation. Studies in mouse models have revealed that dietary calcium, found in cheese, reduces the expression of inflammatory cytokines in adipocytes by inhibiting the formation of calcitriol. Dairy-derived proteins such as lactoferrin may also exhibit anti-inflammatory effects through the regulation of cytokine release, immune cell recruitment, and activation. Thus, while saturated fats might potentially elevate inflammation levels, dairy protein could exert a neutral or even beneficial impact on inflammation. Moreover, dairy products can modulate immune function within the gastrointestinal tract by interacting with the mucosal layer, enhancing gut barrier function, and stimulating immune cells. Cheese, categorized as a probiotic food, harbors an abundance of live microorganisms. Existing research indicates that certain specific bacterial strains possess the ability to inhibit inflammatory responses within the body.31 Clinical trials have demonstrated the beneficial effects of probiotic members, such as Lactobacillus and Bifidobacterium, which are prominent within cheese, on immunity and inflammation. Adequate cheese consumption can regulate the gut microbiota and confer beneficial effects on the host. A study has shown that supplementation with probiotics in cheese alleviates symptoms and prevents recurrent infections in UTI patients, aligning with our finding of a negative correlation between cheese intake and infection indicators,8 supporting the conclusion that cheese consumption has a protective effect against UTIs.
Dietary factors play a multifaceted role in influencing UTIs. Our focus has centered on investigating the impact of dietary factors on UTI biomarkers and the interplay between gut microbiota and UTIs. The human gut, harboring an estimated 100 trillion microorganisms, stands as one of the most densely colonized organs, endowing the host with a multitude of functionalities.32,33 Since the emergence of the gut–kidney axis concept proposed by Meijers in 2011, numerous clinical and animal model studies have affirmed the correlation between gut microbiota and various diseases. The gut–kidney axis describes the bidirectional relationship between the gut microbiota and renal function, wherein gut-derived metabolites such as short-chain fatty acids (SCFAs) and uremic toxins like indoxyl sulfate and p-cresyl sulfate can directly influence kidney health. These metabolites are either protective or deleterious depending on the state of the gut microbiota. For example, SCFAs, produced by the fermentation of dietary fibers, are known to exert anti-inflammatory effects and support renal function. In contrast, uremic toxins produced by certain gut bacteria can exacerbate renal injury and promote systemic inflammation. Conversely, impaired kidney function, often characterized by reduced clearance of these toxins, can lead to an accumulation of uremic solutes, further altering the gut microbiota composition and contributing to a vicious cycle of gut and kidney dysfunction. This interaction is crucial in understanding the systemic effects of gut microbiota on overall health, including the increased susceptibility to infections like UTIs in the context of dysbiosis and renal impairment.34 Within the framework of the gut–kidney axis theory, the bidirectional communication between gut microbiota and kidney health has been emphasized,35 suggesting that gut dysbiosis can lead to compromised kidney function and increased susceptibility to infections like UTIs.
Our study adds to this growing body of evidence by identifying specific dietary factors and gut microbiota taxa that are causally linked to UTI risk. For example, the protective effects observed from oily fish and cheese intake against UTIs can be attributed to their anti-inflammatory properties and ability to promote a healthy gut microbiota. Omega-3 PUFAs from oily fish are known to modulate inflammation by reducing the production of pro-inflammatory eicosanoids and promoting the synthesis of anti-inflammatory mediators. Similarly, the probiotics present in cheese, such as Lactobacillus and Bifidobacterium, have been shown to enhance gut barrier function and inhibit the colonization of uropathogens in the urinary tract. On the other hand, our findings that high alcohol intake frequency increases UTI risk are consistent with existing literature, which indicates that alcohol consumption impairs immune function and disrupts the gut barrier, leading to increased gut permeability, systemic inflammation, and the translocation of bacterial products such as LPS, which are known to exacerbate infection risk.
However, it is important to note that while our results suggest associations between high alcohol intake, cheese, and oily fish consumption and UTI risk, these dietary factors are not traditionally recognized as major risk factors for UTIs in the broader literature. Our findings should be viewed as exploratory, contributing to the ongoing discussion about the role of diet in UTI risk, but not definitive. Further studies, including those with a focus on clinical outcomes and mechanistic insights, are required to validate these associations and to better understand their potential impact on public health. The identified associations between gut microbiota and UTI risk further reinforce the importance of maintaining a balanced gut microbiota. For instance, the high relative abundance of beneficial taxa such as Romboutsia has been linked to a reduced risk of UTIs,36 likely due to their role in supporting gut barrier integrity and modulating local immune responses. Conversely, the presence of pathogenic or dysbiotic bacteria can compromise these defenses, increasing the likelihood of infection.
As we continue to refine our understanding of nutrient intake and the gut microbiota, the concept of the gut–kidney axis provides a valuable framework for exploring the complex interactions that influence UTI risk. This study highlights the potential of dietary modifications as a strategy to reduce the incidence and clinical burden of UTIs, particularly in high-risk populations such as individuals with congenital anomalies, diabetes-related UTIs, and kidney transplant recipients. By targeting both diet and gut microbiota, we can develop more effective preventive measures against UTIs, aligning with the broader goals of precision medicine and personalized healthcare.
5. Limitations
Several limitations must be acknowledged in the context of our study. Firstly, our investigation was confined to the European population, potentially limiting the generalizability of our findings to other ethnic cohorts. Secondly, our classification of gut microbiota at the genus level, while suitable for identifying broad population-level trends, may not fully capture the functional diversity within genera. This limitation could obscure more specific microbial interactions that occur at the species or strain level, which are crucial for understanding the precise mechanisms underlying urinary tract infections. Future research should aim to incorporate species-level or strain-level analyses to provide a more detailed understanding of these interactions.Additionally, the complex interplay of dietary factors in relation to urinary tract infections is manifold. Despite our emphasis on biomarkers and gut microbiota, these aspects may not comprehensively capture the full spectrum of potential effects. Real-world research is also susceptible to unidentified or unmeasured confounding factors. While these factors could potentially introduce variability in the results, the absence of pertinent data precludes their thorough control.
Finally, the primary objective of this article is not to advocate for immediate dietary recommendations for UTI patients but rather to offer novel insights into the potential connections between diet, gut microbiota, and UTI risk. These findings could serve as a foundation for future research and contribute to the development of targeted preventive strategies.
Author contributions
Yifan Du and Xiuyuan Sui were responsible for data acquisition and processing, and they performed statistical analyses and interpreted the results. Yang Bai and Yue Zhao contributed to the literature review and manuscript preparation. Zhiyuan Shi and Bin Liu played pivotal roles in data analysis, including statistical modeling and data interpretation. Zeyuan Zheng and Zhengying Zhang were responsible for crafting the introduction and results sections of the manuscript. Qian Zhang and Jiqing Wang was responsible for creating graphical illustrations and figures to enhance the presentation of the research findings. Chen Shao and Huimin Sun served as corresponding authors, overseeing the project, securing funding, and providing overall guidance. They also actively contributed to manuscript writing and revision.All authors collectively reviewed and approved the final manuscript for submission.
Data availability
The data utilized in this study are sourced from publicly available academic journals, books, and other published materials. All referenced data and citations can be accessed through their respective publications or databases. Detailed definitions, inclusion criteria, and processing methods for these data are available on the official websites or in the original publications and are not reiterated in this manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81972373), and the Medical Leading Talents of Xiamen City.References
- J. Kranz, R. Bartoletti, F. Bruyere, T. Cai, S. Geerlings, B. Koves, S. Schubert, A. Pilatz, R. Veeratterapillay, F. M. E. Wagenlehner, K. Bausch, W. Devlies, J. Horvath, L. Leitner, G. Mantica, T. Mezei, E. J. Smith and G. Bonkat, European Association of Urology Guidelines on Urological Infections: Summary of the 2024 Guidelines, Eur. Urol., 2024, 86(1), 27–41 CrossRef PubMed.
- R. A. Lee, R. M. Centor, L. L. Humphrey, J. A. Jokela, R. Andrews, A. Qaseem, P. Scientific Medical Policy Committee of the American College of, E. A. Akl, T. A. Bledsoe, M. A. Forciea, R. Haeme, D. L. Kansagara, M. Marcucci, M. C. Miller and A. J. Obley, Appropriate Use of Short-Course Antibiotics in Common Infections: Best Practice Advice From the American College of Physicians, Ann. Intern. Med., 2021, 174, 822–827 CrossRef PubMed.
- R. D. Klein and S. J. Hultgren, Urinary tract infections: microbial pathogenesis, host-pathogen interactions and new treatment strategies, Nat. Rev. Microbiol., 2020, 18, 211–226 CrossRef CAS PubMed.
- E. Harris, Updated Meta-analysis: Cranberry Products Reduced UTI Risk, J. Am. Med. Assoc., 2023, 329, 1730 Search PubMed.
- M. Beerepoot and S. Geerlings, Non-Antibiotic Prophylaxis for Urinary Tract Infections, Pathogens, 2016, 5(2), 36 CrossRef PubMed.
- Y. C. Chen, C. C. Chang, T. H. T. Chiu, M. N. Lin and C. L. Lin, The risk of urinary tract infection in vegetarians and non-vegetarians: a prospective study, Sci. Rep., 2020, 10, 906 CrossRef CAS PubMed.
- J. Calleja-Conde, V. Echeverry-Alzate, K. M. Buhler, P. Duran-Gonzalez, J. A. Morales-Garcia, L. Segovia-Rodriguez, F. Rodriguez de Fonseca, E. Gine and J. A. Lopez-Moreno, The Immune System through the Lens of Alcohol Intake and Gut Microbiota, Int. J. Mol. Sci., 2021, 22(14), 7485 CrossRef CAS PubMed.
- K. Mestrovic Popovic, P. Povalej Brzan, T. Langerholc and N. Marcun Varda, The Impact of Lactobacillus Plantarum PCS26 Supplementation on the Treatment and Recurrence of Urinary Tract Infections in Children-A Pilot Study, J. Clin. Med., 2022, 11(23), 7008 CrossRef PubMed.
- Y. Fu, Y. Wang, H. Gao, D. Li, R. Jiang, L. Ge, C. Tong and K. Xu, Associations among Dietary Omega-3 Polyunsaturated Fatty Acids, the Gut Microbiota, and Intestinal Immunity, Mediators Inflammation, 2021, 2021, 8879227 Search PubMed.
- H. N. Yu, J. Zhu, W. S. Pan, S. R. Shen, W. G. Shan and U. N. Das, Effects of fish oil with a high content of n-3 polyunsaturated fatty acids on mouse gut microbiota, Arch. Med. Res., 2014, 45, 195–202 CrossRef CAS PubMed.
- J. H. O'Keefe, S. K. Bhatti, H. R. Patil, J. J. DiNicolantonio, S. C. Lucan and C. J. Lavie, Effects of habitual coffee consumption on cardiometabolic disease, cardiovascular health, and all-cause mortality, J. Am. Coll. Cardiol., 2013, 62, 1043–1051 CrossRef PubMed.
- T. Ohishi, S. Goto, P. Monira, M. Isemura and Y. Nakamura, Anti-inflammatory Action of Green Tea, Antiinflamm. Antiallergy Agents Med. Chem., 2016, 15, 74–90 CrossRef CAS PubMed.
- S. Rohrmann, K. Overvad, H. B. Bueno-de-Mesquita, M. U. Jakobsen, R. Egeberg, A. Tjonneland, L. Nailler, M. C. Boutron-Ruault, F. Clavel-Chapelon, V. Krogh, D. Palli, S. Panico, R. Tumino, F. Ricceri, M. M. Bergmann, H. Boeing, K. Li, R. Kaaks, K. T. Khaw, N. J. Wareham, F. L. Crowe, T. J. Key, A. Naska, A. Trichopoulou, D. Trichopoulos, M. Leenders, P. H. Peeters, D. Engeset, C. L. Parr, G. Skeie, P. Jakszyn, M. J. Sanchez, J. M. Huerta, M. L. Redondo, A. Barricarte, P. Amiano, I. Drake, E. Sonestedt, G. Hallmans, I. Johansson, V. Fedirko, I. Romieux, P. Ferrari, T. Norat, A. C. Vergnaud, E. Riboli and J. Linseisen, Meat consumption and mortality–results from the European Prospective Investigation into Cancer and Nutrition, BMC Med., 2013, 11, 63 CrossRef PubMed.
- S. Ligthart, A. Vaez, U. Vosa, M. G. Stathopoulou, P. S. de Vries, B. P. Prins, P. J. Van der Most, T. Tanaka, E. Naderi, L. M. Rose, Y. Wu, R. Karlsson, M. Barbalic, H. Lin, R. Pool, G. Zhu, A. Mace, C. Sidore, S. Trompet, M. Mangino, M. Sabater-Lleal, J. P. Kemp, A. Abbasi, T. Kacprowski, N. Verweij, A. V. Smith, T. Huang, C. Marzi, M. F. Feitosa, K. K. Lohman, M. E. Kleber, Y. Milaneschi, C. Mueller, M. Huq, E. Vlachopoulou, L. P. Lyytikainen, C. Oldmeadow, J. Deelen, M. Perola, J. H. Zhao, B. Feenstra, S. LifeLines Cohort, M. Amini, C. I. W. Group, J. Lahti, K. E. Schraut, M. Fornage, B. Suktitipat, W. M. Chen, X. Li, T. Nutile, G. Malerba, J. Luan, T. Bak, N. Schork, M. F. Del Greco, E. Thiering, A. Mahajan, R. E. Marioni, E. Mihailov, J. Eriksson, A. B. Ozel, W. Zhang, M. Nethander, Y. C. Cheng, S. Aslibekyan, W. Ang, I. Gandin, L. Yengo, L. Portas, C. Kooperberg, E. Hofer, K. B. Rajan, C. Schurmann, W. den Hollander, T. S. Ahluwalia, J. Zhao, H. H. M. Draisma, I. Ford, N. Timpson, A. Teumer, H. Huang, S. Wahl, Y. Liu, J. Huang, H. W. Uh, F. Geller, P. K. Joshi, L. R. Yanek, E. Trabetti, B. Lehne, D. Vozzi, M. Verbanck, G. Biino, Y. Saba, I. Meulenbelt, J. R. O'Connell, M. Laakso, F. Giulianini, P. K. E. Magnusson, C. M. Ballantyne, J. J. Hottenga, G. W. Montgomery, F. Rivadineira, R. Rueedi, M. Steri, K. H. Herzig, D. J. Stott, C. Menni, M. Franberg, B. St Pourcain, S. B. Felix, T. H. Pers, S. J. L. Bakker, P. Kraft, A. Peters, D. Vaidya, G. Delgado, J. H. Smit, V. Grossmann, J. Sinisalo, I. Seppala, S. R. Williams, E. G. Holliday, M. Moed, C. Langenberg, K. Raikkonen, J. Ding, H. Campbell, M. M. Sale, Y. I. Chen, A. L. James, D. Ruggiero, N. Soranzo, C. A. Hartman, E. N. Smith, G. S. Berenson, C. Fuchsberger, D. Hernandez, C. M. T. Tiesler, V. Giedraitis, D. Liewald, K. Fischer, D. Mellstrom, A. Larsson, Y. Wang, W. R. Scott, M. Lorentzon, J. Beilby, K. A. Ryan, C. E. Pennell, D. Vuckovic, B. Balkau, M. P. Concas, R. Schmidt, C. F. Mendes de Leon, E. P. Bottinger, M. Kloppenburg, L. Paternoster, M. Boehnke, A. W. Musk, G. Willemsen, D. M. Evans, P. A. F. Madden, M. Kahonen, Z. Kutalik, M. Zoledziewska, V. Karhunen, S. B. Kritchevsky, N. Sattar, G. Lachance, R. Clarke, T. B. Harris, O. T. Raitakari, J. R. Attia, D. van Heemst, E. Kajantie, R. Sorice, G. Gambaro, R. A. Scott, A. A. Hicks, L. Ferrucci, M. Standl, C. M. Lindgren, J. M. Starr, M. Karlsson, L. Lind, J. Z. Li, J. C. Chambers, T. A. Mori, E. de Geus, A. C. Heath, N. G. Martin, J. Auvinen, B. M. Buckley, A. J. M. de Craen, M. Waldenberger, K. Strauch, T. Meitinger, R. J. Scott, M. McEvoy, M. Beekman, C. Bombieri, P. M. Ridker, K. L. Mohlke, N. L. Pedersen, A. C. Morrison, D. I. Boomsma, J. B. Whitfield, D. P. Strachan, A. Hofman, P. Vollenweider, F. Cucca, M. R. Jarvelin, J. W. Jukema, T. D. Spector, A. Hamsten, T. Zeller, A. G. Uitterlinden, M. Nauck, V. Gudnason, L. Qi, H. Grallert, I. B. Borecki, J. I. Rotter, W. Marz, P. S. Wild, M. L. Lokki, M. Boyle, V. Salomaa, M. Melbye, J. G. Eriksson, J. F. Wilson, B. Penninx, D. M. Becker, B. B. Worrall, G. Gibson, R. M. Krauss, M. Ciullo, G. Zaza, N. J. Wareham, A. J. Oldehinkel, L. J. Palmer, S. S. Murray, P. P. Pramstaller, S. Bandinelli, J. Heinrich, E. Ingelsson, I. J. Deary, R. Magi, L. Vandenput, P. van der Harst, K. C. Desch, J. S. Kooner, C. Ohlsson, C. Hayward, T. Lehtimaki, A. R. Shuldiner, D. K. Arnett, L. J. Beilin, A. Robino, P. Froguel, M. Pirastu, T. Jess, W. Koenig, R. J. F. Loos, D. A. Evans, H. Schmidt, G. D. Smith, P. E. Slagboom, G. Eiriksdottir, A. P. Morris, B. M. Psaty, R. P. Tracy, I. M. Nolte, E. Boerwinkle, S. Visvikis-Siest, A. P. Reiner, M. Gross, J. C. Bis, L. Franke, O. H. Franco, E. J. Benjamin, D. I. Chasman, J. Dupuis, H. Snieder, A. Dehghan and B. Z. Alizadeh, Genome Analyses of >200,000 Individuals Identify 58 Loci for Chronic Inflammation and Highlight Pathways that Link Inflammation and Complex Disorders, Am. J. Hum. Genet., 2018, 103, 691–706 CrossRef CAS PubMed.
- G. Hemani, J. Zheng, B. Elsworth, K. H. Wade, V. Haberland, D. Baird, C. Laurin, S. Burgess, J. Bowden, R. Langdon, V. Y. Tan, J. Yarmolinsky, H. A. Shihab, N. J. Timpson, D. M. Evans, C. Relton, R. M. Martin, G. Davey Smith, T. R. Gaunt and P. C. Haycock, The MR-Base platform supports systematic causal inference across the human phenome, eLife, 2018, 7, e34408 CrossRef PubMed.
- B. B. Sun, J. C. Maranville, J. E. Peters, D. Stacey, J. R. Staley, J. Blackshaw, S. Burgess, T. Jiang, E. Paige, P. Surendran, C. Oliver-Williams, M. A. Kamat, B. P. Prins, S. K. Wilcox, E. S. Zimmerman, A. Chi, N. Bansal, S. L. Spain, A. M. Wood, N. W. Morrell, J. R. Bradley, N. Janjic, D. J. Roberts, W. H. Ouwehand, J. A. Todd, N. Soranzo, K. Suhre, D. S. Paul, C. S. Fox, R. M. Plenge, J. Danesh, H. Runz and A. S. Butterworth, Genomic atlas of the human plasma proteome, Nature, 2018, 558, 73–79 CrossRef CAS PubMed.
- L. Folkersen, S. Gustafsson, Q. Wang, D. H. Hansen, A. K. Hedman, A. Schork, K. Page, D. V. Zhernakova, Y. Wu, J. Peters, N. Eriksson, S. E. Bergen, T. S. Boutin, A. D. Bretherick, S. Enroth, A. Kalnapenkis, J. R. Gadin, B. E. Suur, Y. Chen, L. Matic, J. D. Gale, J. Lee, W. Zhang, A. Quazi, M. Ala-Korpela, S. H. Choi, A. Claringbould, J. Danesh, G. Davey Smith, F. de Masi, S. Elmstahl, G. Engstrom, E. Fauman, C. Fernandez, L. Franke, P. W. Franks, V. Giedraitis, C. Haley, A. Hamsten, A. Ingason, A. Johansson, P. K. Joshi, L. Lind, C. M. Lindgren, S. Lubitz, T. Palmer, E. Macdonald-Dunlop, M. Magnusson, O. Melander, K. Michaelsson, A. P. Morris, R. Magi, M. W. Nagle, P. M. Nilsson, J. Nilsson, M. Orho-Melander, O. Polasek, B. Prins, E. Palsson, T. Qi, M. Sjogren, J. Sundstrom, P. Surendran, U. Vosa, T. Werge, R. Wernersson, H. J. Westra, J. Yang, A. Zhernakova, J. Arnlov, J. Fu, J. G. Smith, T. Esko, C. Hayward, U. Gyllensten, M. Landen, A. Siegbahn, J. F. Wilson, L. Wallentin, A. S. Butterworth, M. V. Holmes, E. Ingelsson and A. Malarstig, Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals, Nat. Metab., 2020, 2, 1135–1148 CrossRef CAS PubMed.
- A. Kurilshikov, C. Medina-Gomez, R. Bacigalupe, D. Radjabzadeh, J. Wang, A. Demirkan, C. I. Le Roy, J. A. Raygoza Garay, C. T. Finnicum, X. Liu, D. V. Zhernakova, M. J. Bonder, T. H. Hansen, F. Frost, M. C. Ruhlemann, W. Turpin, J. Y. Moon, H. N. Kim, K. Lull, E. Barkan, S. A. Shah, M. Fornage, J. Szopinska-Tokov, Z. D. Wallen, D. Borisevich, L. Agreus, A. Andreasson, C. Bang, L. Bedrani, J. T. Bell, H. Bisgaard, M. Boehnke, D. I. Boomsma, R. D. Burk, A. Claringbould, K. Croitoru, G. E. Davies, C. M. van Duijn, L. Duijts, G. Falony, J. Fu, A. van der Graaf, T. Hansen, G. Homuth, D. A. Hughes, R. G. Ijzerman, M. A. Jackson, V. W. V. Jaddoe, M. Joossens, T. Jorgensen, D. Keszthelyi, R. Knight, M. Laakso, M. Laudes, L. J. Launer, W. Lieb, A. J. Lusis, A. A. M. Masclee, H. A. Moll, Z. Mujagic, Q. Qibin, D. Rothschild, H. Shin, S. J. Sorensen, C. J. Steves, J. Thorsen, N. J. Timpson, R. Y. Tito, S. Vieira-Silva, U. Volker, H. Volzke, U. Vosa, K. H. Wade, S. Walter, K. Watanabe, S. Weiss, F. U. Weiss, O. Weissbrod, H. J. Westra, G. Willemsen, H. Payami, D. Jonkers, A. Arias Vasquez, E. J. C. de Geus, K. A. Meyer, J. Stokholm, E. Segal, E. Org, C. Wijmenga, H. L. Kim, R. C. Kaplan, T. D. Spector, A. G. Uitterlinden, F. Rivadeneira, A. Franke, M. M. Lerch, L. Franke, S. Sanna, M. D'Amato, O. Pedersen, A. D. Paterson, R. Kraaij, J. Raes and A. Zhernakova, Large-scale association analyses identify host factors influencing human gut microbiome composition, Nat. Genet., 2021, 53, 156–165 CrossRef CAS PubMed.
- M. A. Kamat, J. A. Blackshaw, R. Young, P. Surendran, S. Burgess, J. Danesh, A. S. Butterworth and J. R. Staley, PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations, Bioinformatics, 2019, 35, 4851–4853 CrossRef CAS PubMed.
- K. Bibbins-Domingo, G. M. Chertow, P. G. Coxson, A. Moran, J. M. Lightwood, M. J. Pletcher and L. Goldman, Projected effect of dietary salt reductions on future cardiovascular disease, N. Engl. J. Med., 2010, 362, 590–599 CrossRef CAS PubMed.
- G. Danaei, E. L. Ding, D. Mozaffarian, B. Taylor, J. Rehm, C. J. Murray and M. Ezzati, The preventable causes of death in the United States: comparative risk assessment of dietary, lifestyle, and metabolic risk factors, PLoS Med., 2009, 6, e1000058 CrossRef PubMed.
- M. R. Law and N. J. Wald, Risk factor thresholds: their existence under scrutiny, Br. Med. J., 2002, 324, 1570–1576 CrossRef CAS PubMed.
- T. N. Bukong, Y. Cho, A. Iracheta-Vellve, B. Saha, P. Lowe, A. Adejumo, I. Furi, A. Ambade, B. Gyongyosi, D. Catalano, K. Kodys and G. Szabo, Abnormal neutrophil traps and impaired efferocytosis contribute to liver injury and sepsis severity after binge alcohol use, J. Hepatol., 2018, 69, 1145–1154 CrossRef CAS PubMed.
- V. Bountziouka, C. P. Nelson, V. Codd, Q. Wang, C. Musicha, E. Allara, S. Kaptoge, E. Di Angelantonio, A. S. Butterworth, J. R. Thompson, E. M. Curtis, A. M. Wood, J. N. Danesh, N. C. Harvey, C. Cooper and N. J. Samani, Association of shorter leucocyte telomere length with risk of frailty, J. Cachexia Sarcopenia Muscle, 2022, 13, 1741–1751 CrossRef PubMed.
- E. Mutlu, A. Keshavarzian, P. Engen, C. B. Forsyth, M. Sikaroodi and P. Gillevet, Intestinal dysbiosis: a possible mechanism of alcohol-induced endotoxemia and alcoholic steatohepatitis in rats, Alcohol: Clin. Exp. Res., 2009, 33, 1836–1846 CrossRef CAS PubMed.
- Y. Chen, F. Yang, H. Lu, B. Wang, Y. Chen, D. Lei, Y. Wang, B. Zhu and L. Li, Characterization of fecal microbial communities in patients with liver cirrhosis, Hepatology, 2011, 54, 562–572 CrossRef PubMed.
- P. A. Engen, S. J. Green, R. M. Voigt, C. B. Forsyth and A. Keshavarzian, The Gastrointestinal Microbiome: Alcohol Effects on the Composition of Intestinal Microbiota, Alcohol. Res., 2015, 37, 223–236 Search PubMed.
- L. Costantini, R. Molinari, B. Farinon and N. Merendino, Impact of Omega-3 Fatty Acids on the Gut Microbiota, Int. J. Mol. Sci., 2017, 18(12), 2645 CrossRef PubMed.
- F. Laugerette, C. Vors, A. Geloen, M. A. Chauvin, C. Soulage, S. Lambert-Porcheron, N. Peretti, M. Alligier, R. Burcelin, M. Laville, H. Vidal and M. C. Michalski, Emulsified lipids increase endotoxemia: possible role in early postprandial low-grade inflammation, J. Nutr. Biochem., 2011, 22, 53–59 CrossRef CAS PubMed.
- K. Kaliannan, B. Wang, X. Y. Li, K. J. Kim and J. X. Kang, A host-microbiome interaction mediates the opposing effects of omega-6 and omega-3 fatty acids on metabolic endotoxemia, Sci. Rep., 2015, 5, 11276 CrossRef CAS PubMed.
- J. H. Kim, K. Kim and W. Kim, Cream Cheese-Derived Lactococcus chungangensis CAU 28 Modulates the Gut Microbiota and Alleviates Atopic Dermatitis in BALB/c Mice, Sci. Rep., 2019, 9, 446 CrossRef PubMed.
- R. E. Ley, D. A. Peterson and J. I. Gordon, Ecological and evolutionary forces shaping microbial diversity in the human intestine, Cell, 2006, 124, 837–848 CrossRef CAS PubMed.
- R. Sender, S. Fuchs and R. Milo, Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans, Cell, 2016, 164, 337–340 CrossRef CAS PubMed.
- B. Meijers, P. Evenepoel and H. J. Anders, Intestinal microbiome and fitness in kidney disease, Nat. Rev. Nephrol., 2019, 15, 531–545 CrossRef PubMed.
- B. K. Meijers and P. Evenepoel, The gut-kidney axis: indoxyl sulfate, p-cresyl sulfate and CKD progression, Nephrol., Dial., Transplant., 2011, 26, 759–761 CrossRef CAS PubMed.
- M. Magruder, E. Edusei, L. Zhang, S. Albakry, M. J. Satlin, L. F. Westblade, L. Malha, C. Sze, M. Lubetzky, D. M. Dadhania and J. R. Lee, Gut commensal microbiota and decreased risk for Enterobacteriaceae bacteriuria and urinary tract infection, Gut Microbes, 2020, 12, 1805281 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4fo03271c |
| ‡ Yifan Du, Xiuyuan Sui, and Yang Bai contributed equally to this article. |
| This journal is © The Royal Society of Chemistry 2024 |