Open Access Article
Open Access ArticleAdministering Lactiplantibacillus fermentum F6 decreases intestinal Akkermansia muciniphila in a dextran sulfate sodium-induced rat colitis model†
Qiuwen
He‡
abcd,
Tao
Zhang‡
abcd,
Weiqin
Zhang
abcd,
Cuijiao
Feng
abcd,
Lai-Yu
Kwok
abcd,
Heping
Zhang
abcd and
Zhihong
Sun
 *abcd
*abcd
aKey Laboratory of Dairy Biotechnology and Engineering, Ministry of Education, Inner Mongolia Agricultural University, Hohhot 010018, China. E-mail: sunzhihong78@163.com
bKey Laboratory of Dairy Products Processing, Ministry of Agriculture and Rural Affairs, Inner Mongolia Agricultural University, Hohhot 010018, China
cInner Mongolia Key Laboratory of Dairy Biotechnology and Engineering, Inner Mongolia Agricultural University, Hohhot 010018, China
dCollaborative Innovative Center for Lactic Acid Bacteria and Fermented Dairy Products, Ministry of Education, Inner Mongolia Agricultural University, Hohhot 010018, China
First published on 7th May 2024
Abstract
Probiotics are increasingly used to manage gut dysbiosis-related conditions due to their robust ability to manipulate the gut microbial community. However, few studies have reported that probiotics can specifically modulate individual gut microbes. This study demonstrated that administering the probiotic, Lactiplantibacillus fermentum F6, could ameliorate dextran sulfate sodium-induced colitis in a rat model, evidenced by the decreases in the disease activity index score, histopathology grading, and serum pro-inflammatory cytokine levels, as well as the increase in the serum anti-inflammatory cytokine levels. Shotgun metagenomics revealed that the fecal metagenomic of colitis rats receiving the probiotic intervention contained substantially fewer Akkermansia muciniphila than the dextran sulfate sodium group. Thus, the probiotic mechanism might be exerted by reducing specific gut microbial species associated with disease pathogenesis. A new paradigm for designing probiotics that manage diseases through direct and precise manipulation of gut microbes has been provided through this study.
1. Introduction
The close association between gut microbiota and human health has been demonstrated by a large body of correlational, observational, and fecal microbiota transplantation (FMT) studies conducted in the last decade examining alterations in its composition and function across a range of diseases using high-throughput multi-omics technologies.1–4 Metagenomics and metabolomics data from various distinct cohorts have shown changes in the gut microbiome and metabolome between people with the condition and healthy controls, as in the case of inflammatory bowel disease (IBD).5,6 IBD is a group of disorders characterized by chronic intestinal inflammation, including Crohn's disease and ulcerative colitis (UC). It is an example of digestive disease in that the gut microbiome and metabolome are perturbed7,8 and clinical remission has been achieved by FMT in some high-quality clinical trials.9–11 The encouraging clinical intervention outcomes suggest that the gut microbiome might be a crucial determinant of the onset and progression of the disease and that restoring a healthier gut microbiota could mitigate its symptoms, opening up new clinical management opportunities for IBD and other diseases resulting from gut dysbiosis for which there are currently no cure-all treatments.Akkermansia muciniphila (A. muciniphila) is a mucin-degrading bacterium that has often been detected in the gut of humans and animals with UC.12 Some studies have reported a reduction in the abundance of A. muciniphila,13–16 while others have observed the opposite trend,17–23 suggesting a potential negative or positive correlation with UC. More importantly, these correlations have been further confirmed by some interventional studies showing that the consumption of A. muciniphila can alleviate24,25 or aggravate26–28 disease process, indicating that it has a dichotomous role in UC. Regardless, attempts to maintain the homeostasis of A. muciniphila in the gut would provide new insights into disease prevention and treatment.
Probiotics are “live microorganisms which when administered in adequate amounts confer a health benefit on the host”.29 Evidence is accumulating that probiotics have become one of the most prevalent gut microbiome-management tools for a variety of gut-related disorders due to their robust capacity to modulate the gut microbial community.30 As reported in multiple studies, a common symptom-alleviating mechanism of probiotics on the disease is through modulating the gut microbiome.31,32 However, few studies have pinpointed the effect of probiotics on specific groups of gut microbes. Thus, this study hypothesized that probiotics can target and modulate specific gut bacterial taxa.
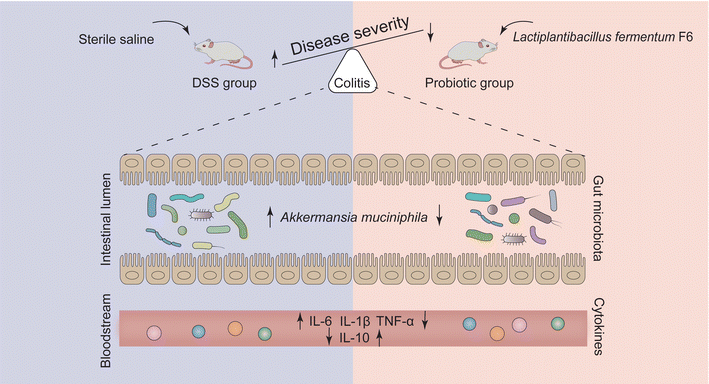
To confirm the hypothesis, we evaluated the modulatory effects of a probiotic, Lactiplantibacillus fermentum F6 (L. fermentum F6), on the gut microbiome in a dextran sulfate sodium (DSS)-induced rat colitis model. We found that probiotic administration to diseased rats could specifically modulate the abundance of intestinal A. muciniphila in the rats, which was accompanied by the remission of symptoms of UC. Our research provides a paradigm for probiotic-based precision therapy for gut-related diseases.
2. Materials and methods
2.1. Probiotic strain
The probiotic strain, L. fermentum F6, was originally isolated from traditional dairy products in Inner Mongolia, China. It is characterized by a high tolerance to simulated gastrointestinal juice and bile salts and a broad antibacterial activity.33 Packaged live probiotic powder of L. fermentum F6 (200 billion CFU per g) was provided by the Key Laboratory of Dairy Biotechnology and Engineering, Ministry of Education, Inner Mongolia Agricultural University, China.2.2. Construction of the DSS-induced colitis model and probiotic intervention
All experiments involving rats were approved by the Experimental Animal Ethics Committee of the Inner Mongolia Agricultural University (approval code: NND2021087), following the ARRIVE guidelines 2.0.34 Twenty-four 8-week-old specific pathogen-free male Wistar wild-type rats were obtained from Vital River Laboratories (Beijing, China). The rats were housed in a standard specific pathogen-free environment in the animal house of the Key Laboratory of Dairy Biotechnology and Engineering, Ministry of Education, Inner Mongolia Agricultural University. Every two rats were maintained in an individually ventilated cage (temperature, 22 ± 2 °C; relative humidity, 45 ± 10%; a 12 h light/dark cycle) with ad libitum access to water and food. After a 7-day acclimatization period, all rats were randomly assigned to three groups (n = 8 each group; ESI Fig. 1†): (1) control group: given drinking water for seven days, followed by daily oral administration of 2 mL sterile saline (Solarbio, Beijing, China) for 14 days; (2) DSS group: given drinking water containing 3% DSS (molecular mass 36–50 kDa, MP Biologicals, Solon, OH, USA) for seven days, followed by daily oral administration of 2 mL sterile saline for 14 days; (3) probiotic group: given drinking water containing 3% DSS for seven days, followed by daily oral administration of 2 mL sterile saline containing probiotics (4 × 109 CFU per day) for 14 days.The body weight of rats was recorded weekly throughout the animal trial. After two weeks of probiotic intervention, the fecal samples of rats were collected aseptically and stored immediately at −80 °C for subsequent metagenome analysis. Meanwhile, the parameters of weight loss, stool consistency, and fecal occult blood were monitored to determine the disease activity index (DAI) scores according to the guidelines listed in ESI Table 1.†![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 Rats were fasted for 12 h before they were sacrificed on day 22 by isoflurane (RWD Life Science, Shenzhen, China) administration. Blood and colonic tissues were collected for further processing and analyses.
35 Rats were fasted for 12 h before they were sacrificed on day 22 by isoflurane (RWD Life Science, Shenzhen, China) administration. Blood and colonic tissues were collected for further processing and analyses.
2.3. Histopathology evaluation
A segment of the distal colon was cut off, flushed with sterile saline, and then fixed in 4% paraformaldehyde (Solarbio, Beijing, China) at room temperature for 24 h. Paraformaldehyde-fixed colon tissues were embedded in paraffin and sectioned (4 μm thick). Paraffin sections were stained with hematoxylin and eosin (Servicebio, Wuhan, China) using standard protocols. Images were acquired using an upright optical microscope (Nikon Eclipse E100, Nikon, Tokyo, Japan) with an imaging system (Nikon DS-U3, Nikon, Tokyo, Japan). The histopathology of the colon tissues was graded according to the guidelines provided in ESI Table 2.†![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 36
36
2.4. Enzyme-linked immunosorbent assays
To obtain rat sera, the collected blood samples were centrifuged at 3000 rpm at 4 °C for 10 min after standing at room temperature for 60 min. The obtained sera were stored at −80 °C until enzyme-linked immunosorbent assays for measuring the levels of inflammatory cytokines, including interleukin (IL)-6, IL-10, IL-1β, and tumor necrosis factor-α (TNF-α) with commercial kits (Elabscience, Wuhan, China). The measurements were performed according to the manufacturer's instructions.2.5. Extraction of fecal DNA and whole-genome shotgun sequencing
Fecal metagenomic DNA was extracted from the frozen fecal samples using a QIAamp Fast DNA Stool Mini Kit (Qiagen, Hilden, Nordrhein-Westfalen, Germany) according to the manufacturer's instructions. The quality of the extracted DNA was assessed with a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and 1% agarose gel electrophoresis. Qualified DNA samples were stored at −80 °C before further analysis.Sequencing libraries were generated with a NEBNext Ultra II DNA Library Prep Kit (New England Biolabs, Ipswich, MA, USA) following the manufacturer's recommendations. The quality of the constructed DNA libraries was confirmed with a Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). Libraries were sequenced on a NovaSeq 6000 instrument (Illumina, San Diego, CA, USA). All samples were paired-end sequenced with a 150 bp read length to a targeted data size of 3 GB.
KneadData pipeline (https://huttenhower.sph.harvard.edu/kneaddata) was used to preprocess the raw reads, with its primary functions include filtering low-quality sequences via Trimmomatic,37 and removing host-contaminated sequences through Bowtie 2.38 The resulting clean data were taxonomically and functionally profiled using HUMAnN2 pipeline.39
2.6. Statistical analysis
All statistical analyses and graphical output of the data were achieved using R (v4.2.3); and the graphics were combined with Adobe Illustrator (v26.5.0). Data are expressed as mean ± standard deviation.3. Results
3.1. Administering L. fermentum F6 ameliorated DSS treatment-induced colitis in rats
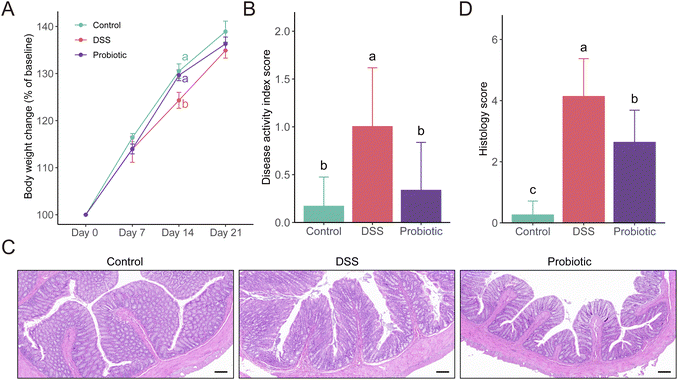
To assess the protective effects of L. fermentum F6 administration in UC, a DSS-induced colitis rat model was constructed (ESI Fig. 1†). The body weight of all three groups of rats increased as the intervention trial progressed. A significant difference was only seen at day 14 when the increase in body weight was higher in the control and probiotic groups compared with the DSS group (P < 0.05; Fig. 1A), suggesting that the probiotic intervention could prevent DSS-related body weight loss.The DAI score reflects the health conditions of the rats. We observed a significantly higher DAI score in the DSS group compared with the control and the probiotic groups on day 22 (P < 0.05; Fig. 1B). Such results were consistent with the colon histopathological examination and grading (P < 0.05; Fig. 1C and D). These results suggested that the probiotic intervention could reduce the severity of the disease.
3.2. Administering L. fermentum F6 reversed DSS treatment-induced inflammation
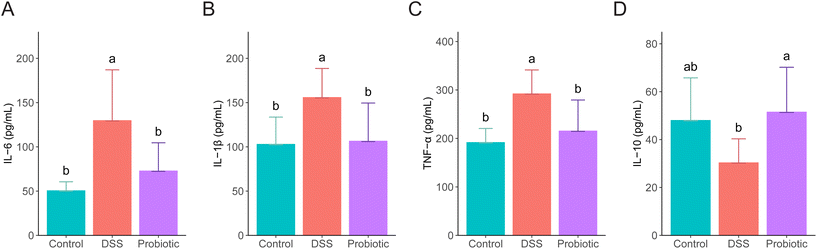
To evaluate the level of systemic inflammation of the rats, the serum concentrations of several pro- and anti-inflammatory cytokines were measured on day 22. The levels of all the measured pro-inflammatory cytokines (IL-6, IL-1β, and TNF-α) were significantly higher in the DSS group compared with the control and probiotic groups (P < 0.05; Fig. 2A–C). DSS treatment induced a reduced serum IL-10 level, which was significantly reversed by probiotic supplementation (P < 0.05; Fig. 2D).3.3. Administering L. fermentum F6 decreased the abundance of gut A. muciniphila in diseased rats
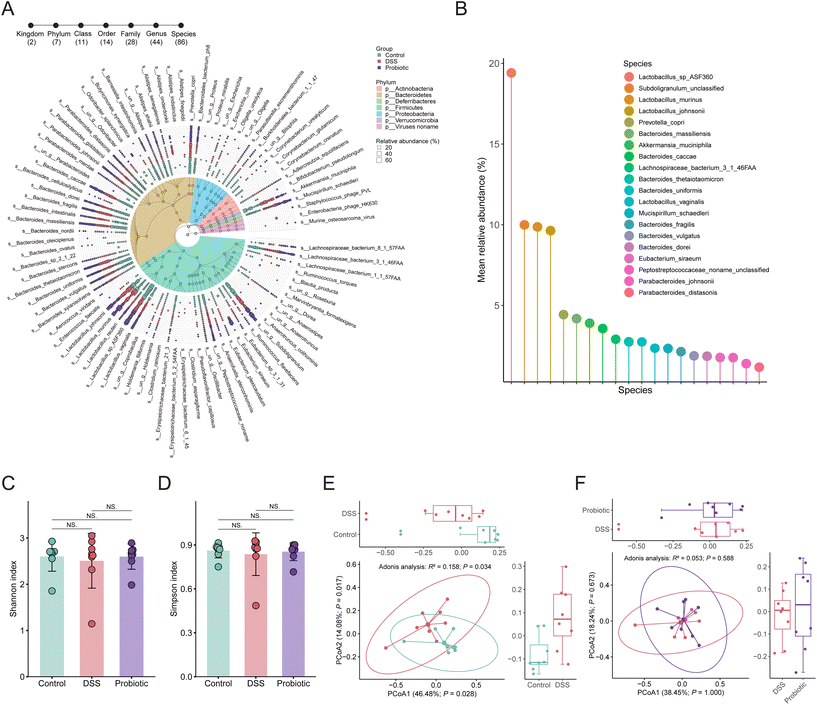
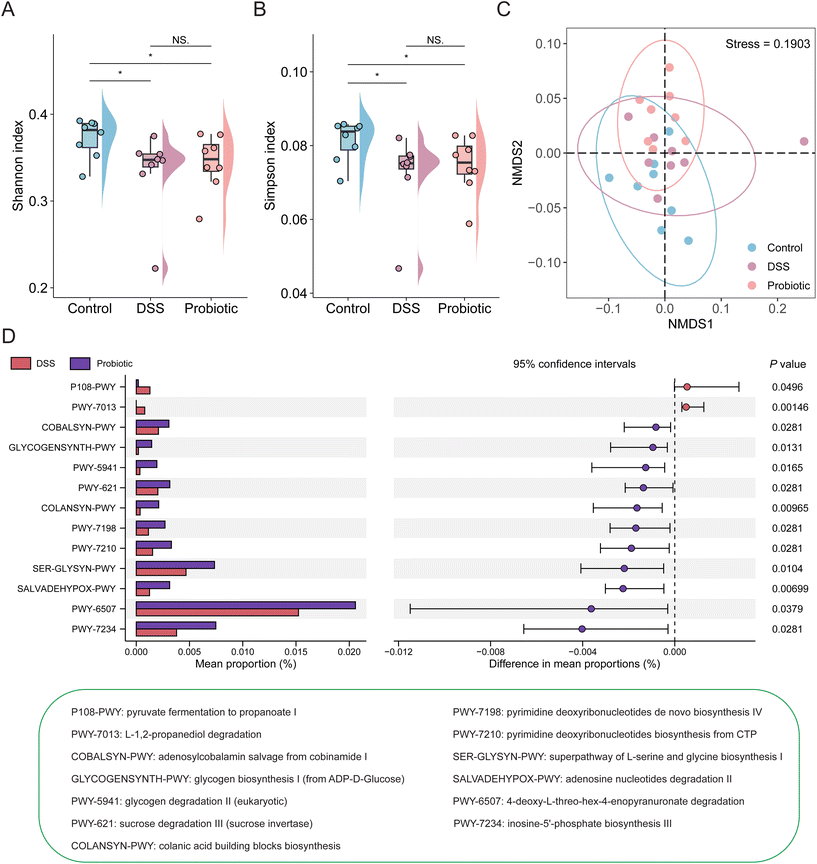
To investigate the changes in gut microbiota in association with colitis and probiotic intervention, a total of 24 stool samples from the three groups of rats were subjected to metagenomics sequencing analysis. The overall fecal metagenome dataset comprised two kingdoms, seven phyla, 11 classes, 14 orders, 28 families, 44 genera, and 86 species (Fig. 3A). The most dominant phyla were Firmicutes and Bacteroidetes (60.12% and 32.94%, respectively). There were 20 species with an average relative abundance greater than 1% across all samples, mainly including Lactobacillus sp. ASF360, Subdoligranulum unclassified, Lactobacillus murinus, Lactobacillus johnsonii, Prevotella copri, Bacteroides massiliensis, A. muciniphila, and Bacteroides caccae (Fig. 3B).We then assessed the alpha and beta diversity of the species-level fecal gut microbiota. Both the values of the Shannon and Simpson's diversity indices (representing the alpha diversity) were non-significantly lower in the DSS group compared with the control and probiotic groups (Fig. 3C and D), suggesting that the probiotic intake could help conserve the gut microbial diversity of DSS-treated rats. The beta diversity analysis was assessed through principal coordinate analysis using Bray–Curtis dissimilarity distance, revealing a pronounced difference in the gut microbial community structure between the control and DSS groups (Adonis test, R2 = 0.158, P = 0.034; Fig. 3E) but not between the DSS and probiotic group (Adonis test, R2 = 0.053, P = 0.588; Fig. 3F). These results suggested that the DSS treatment drastically affected the gut microbiota structure of the rats, which was not restored by probiotic intake.
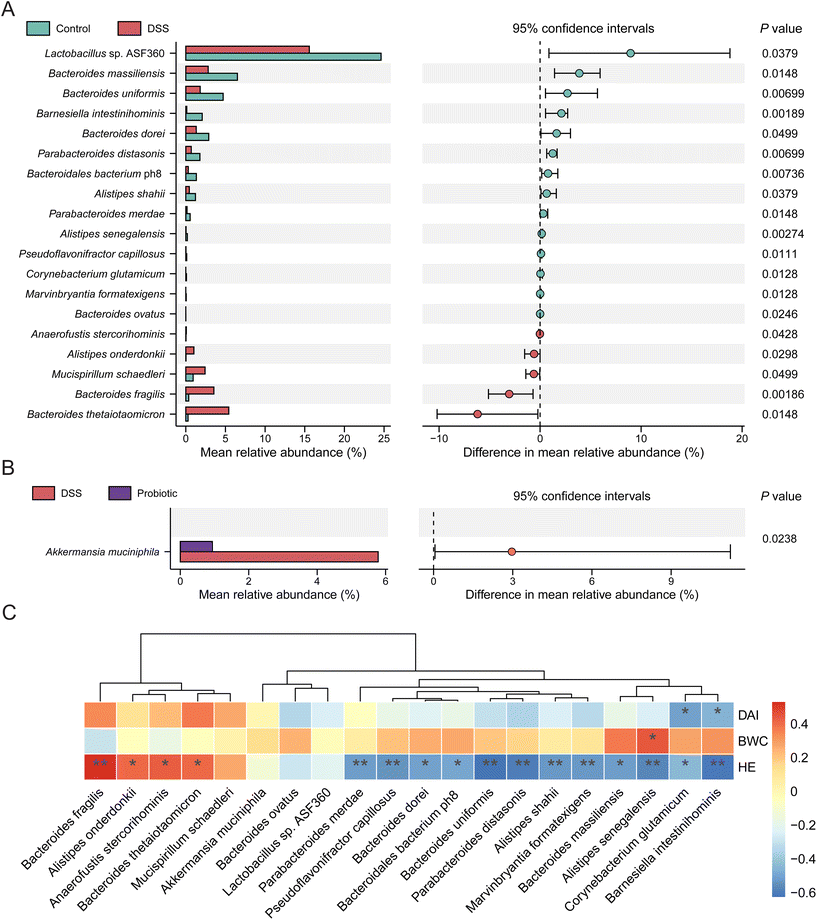
Given that the probiotic intervention failed to improve the overall alpha and beta diversity of gut microbiota disrupted by DSS treatment, we then asked whether the probiotics acted on specific gut microbes in UC remission. We first identified significant differential taxa between the control and DSS groups (P < 0.05, Wilcoxon rank-sum test), yielding two phyla, two classes, one order, five families, and seven genera (ESI Fig. 2†). Nineteen significant differentially abundant species were identified, 14 and five of which were enriched in the fecal microbiota of the control and DSS groups, respectively (Fig. 4A). Specifically, the fecal microbiota of the control group had significantly more Lactobacillus sp. ASF360, Bacteroides massiliensis, and Bacteroides uniformis but fewer Bacteroides thetaiotaomicron, Bacteroides fragilis, and Mucispirillum schaedleri compared with that of the DSS group (P < 0.05). Next, we identified the significant differentially abundant taxa between the DSS and probiotic groups, yielding one phylum, two classes, two orders, two families, and two genera (ESI Fig. 3†). At the species level, the abundance of one species, A. muciniphila, was significantly more in the fecal microbiota of the DSS group than in that of the probiotic group after probiotic intervention (Fig. 4B), indicating that administering L. fermentum F6 substantially lowered the intestinal abundance of this species in the diseased rats.
Spearman's correlation analysis was further performed to investigate whether these discriminatory species were associated with the colitis indices (including body weight change, DAI, and histopathology). Our results showed that 14 species enriched in the control group showed weak to moderate degree of negative correlations with DAI and histopathology grade, whereas five species enriched in the DSS group exhibited weak to moderate degree of positive correlations with DAI and histopathology grade (Fig. 4C). A. muciniphila showed a weak positive correlation with DAI (Fig. 4C). These results provide evidence that administering L. fermentum F6 did not drastically change the gut microbiota structure but fine-tuning the gut microbiota of diseased rats that flavored UC remission, accompanied by a drastic reduction in the intestinal abundance of A. muciniphila.
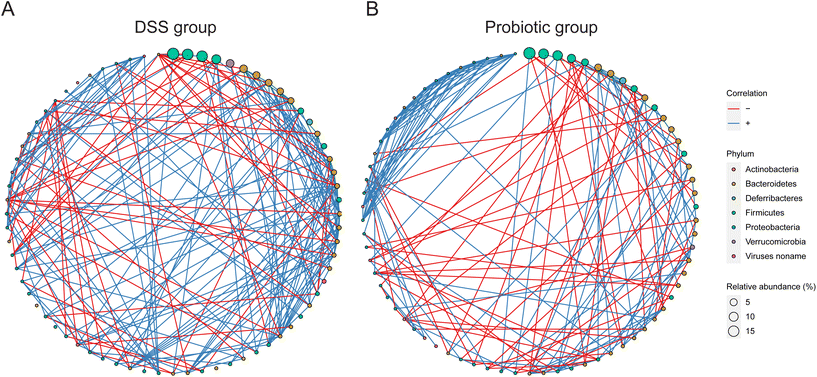
3.4. The gut microbiota interconnectedness was enhanced by the DSS treatment but reduced by the probiotic intervention
To investigate how the DSS treatment and probiotic intervention affect the interactions between the gut microbes, we then analyzed the gut microbiota interconnectedness by a co-occurrence network analysis. The fecal microbiota of the DSS group had the strongest interconnectedness among the three sample groups (DSS group: 246 correlations including 187 positive and 59 negative correlations; probiotic group: 227 correlations including 168 positive and 59 negative correlations; control group: 194 correlations including 160 positive and 34 negative correlations; Fig. 5A, B, and ESI Fig. 4,† respectively). These results suggested that the DSS treatment could enhance the gut microbiota interconnectedness, while such effect was reversed by the probiotic intervention.3.5. Administering L. fermentum F6 modulated the functional gut microbial metagenome
To investigate how the functional gut microbiota was associated with UC remission, we comparatively analyzed the fecal metagenomic potential of the three groups using the HUMAnN2 pipeline with default parameters. The functional fecal metagenome of both the DSS and probiotic groups had a significantly lower alpha diversity (represented by Shannon and Simpson's diversity indices) than the control group (Fig. 6A and B; P < 0.05), suggesting that the DSS treatment disrupted the diversity of MetaCyc pathways encoded in the fecal metagenome, which was not restored by the probiotic intervention. The beta diversity of the functional fecal metagenome was analyzed by non-metric multidimensional scaling (Bray–Curtis dissimilarity distance). On the score plot, the symbols representing the functional metagenome of the control and DSS groups (but not between the DSS and probiotic groups) showed a significant group-based clustering pattern (Adonis test of control and DSS groups: R2 = 0.190, P = 0.029; DSS and probiotic group: R2 = 0.039, P = 0.557; Fig. 6C). Such results suggested that the DSS treatment disrupted the functional potential of rats’ fecal metagenome, and such effect was not reversed by the probiotic intervention.We then investigated the changes in the distribution of specific MetaCyc pathways of the functional fecal metagenome of the three groups. Our results identified a total of 277 enriched MetaCyc pathways across all samples; and the top pathways were involved in adenosine ribonucleotides de novo biosynthesis (0.12%), starch degradation V (0.09%), and UMP biosynthesis (0.08%) (ESI Fig. 5†).
Three over-represented and 42 under-represented pathways were detected in the DSS group compared with the control group (ESI Fig. 6†). In addition, we identified 13 differential pathways between the DSS and probiotic groups (Fig. 6D), with 11 pathways enriched in the probiotic group, mainly involved in amino acid and carbohydrate metabolism. These results suggested that despite the probiotic intervention did not restore the functional metagenome of DSS-treated rats to the original healthy state, it did improve the functional potential of the intestinal microbiota.
4. Discussion
In the present study, we examined the impact of a probiotic, L. fermentum F6, on UC using a rat colitis model induced by DSS. We found that L. fermentum F6 administration could ameliorate the disease, as evidenced by the lower DAI, histopathology scores, and serum levels of pro-inflammatory cytokines but higher levels of anti-inflammatory cytokines. Even though administering L. fermentum F6 did not change the gut microbiota diversity, the intestinal abundance of A. muciniphila notably decreased in the probiotic-supplemented rats compared with the DSS-treated rats. Meanwhile, our correlation analysis revealed that A. muciniphila was likely involved in the development of UC. Our study provides intriguing insights into the pathogenesis of UC.Differences in the gut microbiota diversity metrics between healthy and colitis rodents have been widely reported. For example, when analyzed using multivariate analysis methods like principal coordinate analysis, experimental colitis mice induced by 2.5% or 5% DSS showed a distinct difference in the gut microbiota structure from healthy mice, and a lower alpha diversity (assessed by Chao 1, Ace, Shannon, and Simpson indices) was noted in diseased mice compared with the healthy ones.14,40 Similar outcomes were also observed in a rat model of UC induced by 4% DSS supplement.41 The common features between these studies are that DSS induction resulted in significantly reduced microbial community richness and evenness and altered gut microbiota structure. In our study, we observed obvious changes in the gut microbiota composition in DSS-treated rats compared with healthy controls, confirming an intestinal dysbiotic state in these rats. The identification of colitis-associated alterations in the gut microbiota would be helpful to further understand how the gut microbiota relates to the disease onset and pathogenesis.
In fact, some previous studies have used 16S rRNA gene sequencing to determine the composition of the gut microbiota in UC. For example, one study reported the enrichment in the genera Rikenellaceae_RC9_gut_group, Escherichia–Shigella, Romboutsia, Bacteroides, and Parabacteroides in colitis mice, whereas the genera Anaeroplasma, Ruminococcaceae_UCG-014, and Prevotellaceae_UCG-001 showed decreased abundances compared with the control group.42 Another study observed the under-representation of Lactobacillus, Ligilactobacillus, and Lachnospiraceae_NK4A136_group but over-representation in Prevotellaceae_UCG-001 and Bacteroides in DSS-induced mice relative to healthy mice.43 A third study found that DSS-treated mice had more Muribaculum, Akkermansia, and Escherichia–Shigella but fewer Coriobacteriaceae_UCG-002, Lactobacillus, and Parabacteroides compared with the healthy controls.17 The resolution of amplicon-based sequencing, however, is restricted to the genus level, making it difficult to collect information at the species or strain level, which is essential for shedding light on the disease pathogenesis. Thus, this work implemented higher-resolution shotgun metagenomics rather than 16S rRNA gene sequencing to track the changes in rats’ gut microbiota after DSS treatment and the probiotic intervention.
Specifically, the fecal microbiota of the DSS group had significantly more Bacteroides thetaiotaomicron, Bacteroides fragilis, Mucispirillum schaedleri, Alistipes onderdonkii, and Anaerofustis stercorihominis, but fewer Lactobacillus sp. ASF360, Bacteroides massiliensis, Bacteroides uniformis, Bacteroides dorei, Parabacteroides distasonis, and Bacteroidales bacterium ph8 than that of the control group. The species Mucispirillum schaedleri enriched in the DSS-induced rats might link to UC pathogenesis as a previous study have shown that Mucispirillum schaedleri could drive the development of Crohn's disease-like colitis in mice with severe immune deficiency.44 Moreover, consistent with previously reported studies,17,18,43 our study also confirmed an inverse correlation between the genus Lactobacillus and UC, which is worth further investigating in future studies. Parabacteroides distasonis is a species that is often associated with different health statuses. For example, it was diminished in patients with rheumatoid arthritis and hepatic fibrosis, and administering this bacterium could ameliorate disease pathogenesis in mouse models.45,46 On the other hand, Parabacteroides distasonis administration exerted beneficial effects in diseases such as type 2 diabetes47 and non-alcoholic steatohepatitis.48 Although our data revealed that Parabacteroides distasonis was diminished in DSS-induced rats, its impact on disease is unclear. Notably, several Bacteroides species exhibited opposite patterns of intestinal abundance in DSS-induced rats in this study, highlighting the importance and value of identifying microorganisms at a greater taxonomic resolution. The differential bacterial species identified in healthy and colitis rats deserve further attention in future studies to determine whether they are beneficial or harmful to the disease.
Given that gut dysbiosis is associated with UC, attempts to engineer gut microbiota to reach a premorbid state might open up new therapeutic avenues for managing UC. This notion has given rise to much research on manipulating the gut microbial community to ameliorate UC, especially with probiotic intervention.15,19,49–52 These studies showed that probiotic administration can beneficially modulate the host gut microbiota by promoting the beneficial microbes (such as Clostridium sensu stricto-1 and Butyricicoccus) while reducing the harmful ones (such as Escherichia–Shigella).52 A surprising observation in our study is that administering L. fermentum F6 specifically decreased the intestinal abundance of A. muciniphila, and the species was positively correlated with the severity of colitis. A diminished abundance of gut A. muciniphila was seen in various diseases,53 but some studies reported the opposite result (summarized in ESI Table 3†). For instance, some recent experimental evidence noted a marked increase in the intestinal abundance of A. muciniphila in DSS-induced colitis mice compared with healthy controls.17–23 In addition to UC, this phenomenon was also seen in some neurodegenerative disorders, such as multiple sclerosis54,55 and Parkinson's disease.56,57 Our study also observed an elevated level of gut A. muciniphila in DSS-treated rats, although no significant difference was seen between the control and DSS groups (ESI Fig. 7†). A. muciniphila is increasingly recognized as a next-generation probiotic,58 but its role in diseases including UC remains to be clarified. Overall, our study suggested that administering probiotics can modulate the gut microbiota composition, and the capacity to accurately regulate species that trigger or aggravate the disease would be a crucial basis for probiotic-based precision therapy for gut-related diseases.
Another interesting observation of this work was that the DSS-induced gut dysbiosis was accompanied not only by an abnormal shift of the gut microbiota but also an enhanced interconnectedness of the overall gut microbiota compared with that of the healthy rats. Moreover, this stronger interconnectedness among the gut microorganisms was abated by probiotic intake. The relationship between dysbiosis and the interconnectedness of the gut microbiota and their impact on diseases remain obscure and warrant further study.
On the one hand, even though we observed a positive association between A. muciniphila and the pathogenesis of colitis, we were unable to prove the causality of A. muciniphila in the disease, which might need to be validated in a separate experiment by exogenous supplementation of A. muciniphila to the DSS-treated rats with and without receiving the probiotic intervention. On the other hand, despite our seminal demonstration that probiotics could specifically regulate individual gut microbes, whether this function is generalizable in other colitis models or diseases requires further investigation. Nonetheless, our study offers valuable insights into target modulation of gut microbes of interest or disease causation to change the course of the disease (Fig. 7), which is an essential basis for precision probiotic therapies.
Data availability
The sequencing data generated in this study have been made available to the public with no restrictions at the NCBI Sequence Read Archive under BioProject accession number (PRJNA1007659).Author contributions
Q. H. and T. Z. contributed equally to this work. Q. H. designed the experiments, T. Z., W. Z., and C. F. conducted the animal experiments. T. Z. analyzed the results and wrote the manuscript. Q. H. and L. K. extensively revised the manuscript and provided advice. H. Z. and Z. S. supervised all data analysis. All authors read and approved the final manuscript.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
We are profoundly thankful to the National Natural Science Foundation of China (32325040, U22A20540), the National Key R&D Program of China (2022YFD21007002), and the Earmarked Fund for CARS36.References
- J. W. J. Lee, D. Plichta, L. Hogstrom, N. Z. Borren, H. Lau, S. M. Gregory, W. Tan, H. Khalili, C. Clish, H. Vlamakis, R. J. Xavier and A. N. Ananthakrishnan, Multi-omics reveal microbial determinants impacting responses to biologic therapies in inflammatory bowel disease, Cell Host Microbe, 2021, 29, 1294–1304 CrossRef CAS PubMed .e4.
- B. Tang, L. Tang, S. Li, S. Liu, J. He, P. Li, S. Wang, M. Yang, L. Zhang, Y. Lei, D. Tu, X. Tang, H. Hu, Q. Ouyang, X. Chen and S. Yang, Gut microbiota alters host bile acid metabolism to contribute to intrahepatic cholestasis of pregnancy, Nat. Commun., 2023, 14, 1305 CrossRef CAS PubMed.
- Y. Fan, R. K. Stoving, S. Berreira Ibraim, T. Hyotylainen, F. Thirion, T. Arora, L. Lyu, E. Stankevic, T. H. Hansen, P. Dechelotte, T. Sinioja, O. Ragnarsdottir, N. Pons, N. Galleron, B. Quinquis, F. Levenez, H. Roume, G. Falony, S. Vieira-Silva, J. Raes, L. Clausen, G. K. Telleus, F. Backhed, M. Oresic, S. D. Ehrlich and O. Pedersen, The gut microbiota contributes to the pathogenesis of anorexia nervosa in humans and mice, Nat. Microbiol., 2023, 8, 787–802 CrossRef CAS PubMed.
- B. D. Needham, M. Funabashi, M. D. Adame, Z. Wang, J. C. Boktor, J. Haney, W. L. Wu, C. Rabut, M. S. Ladinsky, S. J. Hwang, Y. Guo, Q. Zhu, J. A. Griffiths, R. Knight, P. J. Bjorkman, M. G. Shapiro, D. H. Geschwind, D. P. Holschneider, M. A. Fischbach and S. K. Mazmanian, A gut-derived metabolite alters brain activity and anxiety behaviour in mice, Nature, 2022, 602, 647–653 CrossRef CAS PubMed.
- E. A. Franzosa, A. Sirota-Madi, J. Avila-Pacheco, N. Fornelos, H. J. Haiser, S. Reinker, T. Vatanen, A. B. Hall, H. Mallick, L. J. McIver, J. S. Sauk, R. G. Wilson, B. W. Stevens, J. M. Scott, K. Pierce, A. A. Deik, K. Bullock, F. Imhann, J. A. Porter, A. Zhernakova, J. Fu, R. K. Weersma, C. Wijmenga, C. B. Clish, H. Vlamakis, C. Huttenhower and R. J. Xavier, Gut microbiome structure and metabolic activity in inflammatory bowel disease, Nat. Microbiol., 2019, 4, 293–305 CrossRef CAS PubMed.
- J. Lloyd-Price, C. Arze, A. N. Ananthakrishnan, M. Schirmer, J. Avila-Pacheco, T. W. Poon, E. Andrews, N. J. Ajami, K. S. Bonham, C. J. Brislawn, D. Casero, H. Courtney, A. Gonzalez, T. G. Graeber, A. B. Hall, K. Lake, C. J. Landers, H. Mallick, D. R. Plichta, M. Prasad, G. Rahnavard, J. Sauk, D. Shungin, Y. Vazquez-Baeza, R. A. White, 3rd, I. Investigators, J. Braun, L. A. Denson, J. K. Jansson, R. Knight, S. Kugathasan, D. P. B. McGovern, J. F. Petrosino, T. S. Stappenbeck, H. S. Winter, C. B. Clish, E. A. Franzosa, H. Vlamakis, R. J. Xavier and C. Huttenhower, Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases, Nature, 2019, 569, 655–662 CrossRef CAS PubMed.
- R. Pittayanon, J. T. Lau, G. I. Leontiadis, F. Tse, Y. Yuan, M. Surette and P. Moayyedi, Differences in Gut Microbiota in Patients With vs Without Inflammatory Bowel Diseases: A Systematic Review, Gastroenterology, 2020, 158, 930–946 CrossRef PubMed .e1.
- A. Vich Vila, S. Hu, S. Andreu-Sanchez, V. Collij, B. H. Jansen, H. E. Augustijn, L. A. Bolte, R. Ruigrok, G. Abu-Ali, C. Giallourakis, J. Schneider, J. Parkinson, A. Al-Garawi, A. Zhernakova, R. Gacesa, J. Fu and R. K. Weersma, Faecal metabolome and its determinants in inflammatory bowel disease, Gut, 2023, 72, 1472–1485 CrossRef PubMed.
- P. Moayyedi, M. G. Surette, P. T. Kim, J. Libertucci, M. Wolfe, C. Onischi, D. Armstrong, J. K. Marshall, Z. Kassam, W. Reinisch and C. H. Lee, Fecal Microbiota Transplantation Induces Remission in Patients With Active Ulcerative Colitis in a Randomized Controlled Trial, Gastroenterology, 2015, 149, 102–109 CrossRef PubMed .e6.
- S. Paramsothy, M. A. Kamm, N. O. Kaakoush, A. J. Walsh, J. van den Bogaerde, D. Samuel, R. W. L. Leong, S. Connor, W. Ng, R. Paramsothy, W. Xuan, E. Lin, H. M. Mitchell and T. J. Borody, Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial, Lancet, 2017, 389, 1218–1228 CrossRef PubMed.
- S. P. Costello, P. A. Hughes, O. Waters, R. V. Bryant, A. D. Vincent, P. Blatchford, R. Katsikeros, J. Makanyanga, M. A. Campaniello, C. Mavrangelos, C. P. Rosewarne, C. Bickley, C. Peters, M. N. Schoeman, M. A. Conlon, I. C. Roberts-Thomson and J. M. Andrews, Effect of Fecal Microbiota Transplantation on 8-Week Remission in Patients With Ulcerative Colitis: A Randomized Clinical Trial, J. Am. Med. Assoc., 2019, 321, 156–164 CrossRef PubMed.
- M. Zheng, R. Han, Y. Yuan, Y. Xing, W. Zhang, Z. Sun, Y. Liu, J. Li and T. Mao, The role of Akkermansia muciniphila in inflammatory bowel disease: Current knowledge and perspectives, Front. Immunol., 2023, 13, 1089600 CrossRef PubMed.
- S. L. James, C. T. Christophersen, A. R. Bird, M. A. Conlon, O. Rosella, P. R. Gibson and J. G. Muir, Abnormal fibre usage in UC in remission, Gut, 2015, 64, 562–570 CrossRef CAS PubMed.
- X. Huang, J. Hu, H. Zhang, J. Li, X. Zhu, Y. Liu, Y. Liang and Y. Mei, Clostridium butyricum and Chitooligosaccharides in Synbiotic Combination Ameliorate Symptoms in a DSS-Induced Ulcerative Colitis Mouse Model by Modulating Gut Microbiota and Enhancing Intestinal Barrier Function, Microbiol. Spectrum, 2023, 11, e0437022 CrossRef PubMed.
- Y. Wu, A. Li, H. Liu, Z. Zhang, C. Zhang, C. Ma, L. Zhang and J. Zhang, Lactobacillus plantarum HNU082 alleviates dextran sulfate sodium-induced ulcerative colitis in mice through regulating gut microbiome, Food Funct., 2022, 13, 10171–10185 RSC.
- K. Sheng, Y. Xu, X. Kong, J. Wang, X. Zha and Y. Wang, Probiotic Bacillus cereus Alleviates Dextran Sulfate Sodium-Induced Colitis in Mice through Improvement of the Intestinal Barrier Function, Anti-Inflammation, and Gut Microbiota Modulation, J. Agric. Food Chem., 2021, 69, 14810–14823 CrossRef CAS PubMed.
- Z. Zhou, W. He, H. Tian, P. Zhan and J. Liu, Thyme (Thymus vulgaris L.) polyphenols ameliorate DSS-induced ulcerative colitis of mice by mitigating intestinal barrier damage, regulating gut microbiota, and suppressing TLR4/NF-κB-NLRP3 inflammasome pathways, Food Funct., 2023, 14, 1113–1132 RSC.
- H. Lu, M. Shen, Y. Chen, Q. Yu, T. Chen and J. Xie, Alleviative effects of natural plant polysaccharides against DSS-induced ulcerative colitis via inhibiting inflammation and modulating gut microbiota, Food Res. Int., 2023, 167, 112630 CrossRef CAS PubMed.
- H. Gao, Y. Li, J. Xu, X. Zuo, T. Yue, H. Xu, J. Sun, M. Wang, T. Ye, Y. Yu and Y. Yao, Saccharomyces boulardii protects against murine experimental colitis by reshaping the gut microbiome and its metabolic profile, Front. Microbiol., 2023, 14, 1204122 CrossRef PubMed.
- K. Jiang, D. Wang, L. Su, X. Liu, Q. Yue, B. Li, K. Li, S. Zhang and L. Zhao, Structural characteristics of locust bean gum hydrolysate and its alleviating effect on dextran sulfate sodium-induced colitis, Front. Microbiol., 2022, 13, 985725 CrossRef PubMed.
- Y. Y. Huang, Y. P. Wu, X. Z. Jia, J. Lin, L. F. Xiao, D. M. Liu and M. H. Liang, Lactiplantibacillus plantarum DMDL 9010 alleviates dextran sodium sulfate (DSS)-induced colitis and behavioral disorders by facilitating microbiota-gut-brain axis balance, Food Funct., 2022, 13, 411–424 RSC.
- Y. Huang, Y. Zheng, F. Yang, Y. Feng, K. Xu, J. Wu, S. Qu, Z. Yu, F. Fan, L. Huang, M. Qin, Z. He, K. Nie and K. F. So, Lycium barbarum Glycopeptide prevents the development and progression of acute colitis by regulating the composition and diversity of the gut microbiota in mice, Front. Cell. Infect. Microbiol., 2022, 12, 921075 CrossRef CAS PubMed.
- Y. Hu, X. Jin, F. Gao, T. Lin, H. Zhu, X. Hou, Y. Yin, S. Kan and D. Chen, Selenium-enriched Bifidobacterium longum DD98 effectively ameliorates dextran sulfate sodium-induced ulcerative colitis in mice, Front. Microbiol., 2022, 13, 955112 CrossRef PubMed.
- X. Bian, W. Wu, L. Yang, L. Lv, Q. Wang, Y. Li, J. Ye, D. Fang, J. Wu, X. Jiang, D. Shi and L. Li, Administration of Akkermansia muciniphila Ameliorates Dextran Sulfate Sodium-Induced Ulcerative Colitis in Mice, Front. Microbiol., 2019, 10, 2259 CrossRef PubMed.
- S. Qu, L. Fan, Y. Qi, C. Xu, Y. Hu, S. Chen, W. Liu, W. Liu and J. Si, Akkermansia muciniphila Alleviates Dextran Sulfate Sodium (DSS)-Induced Acute Colitis by NLRP3 Activation, Microbiol. Spectrum, 2021, 9, e0073021 CrossRef PubMed.
- S. S. Seregin, N. Golovchenko, B. Schaf, J. Chen, N. A. Pudlo, J. Mitchell, N. T. Baxter, L. Zhao, P. D. Schloss, E. C. Martens, K. A. Eaton and G. Y. Chen, NLRP6 Protects IL10−/− Mice from Colitis by Limiting Colonization of Akkermansia muciniphila, Cell Rep., 2017, 19, 733–745 CrossRef CAS PubMed.
- S. Qu, Y. Zheng, Y. Huang, Y. Feng, K. Xu, W. Zhang, Y. Wang, K. Nie and M. Qin, Excessive consumption of mucin by over-colonized Akkermansia muciniphila promotes intestinal barrier damage during malignant intestinal environment, Front. Microbiol., 2023, 14, 1111911 CrossRef PubMed.
- Q. Liu, W. Lu, F. Tian, J. Zhao, H. Zhang, K. Hong and L. Yu, Akkermansia muciniphila Exerts Strain-Specific Effects on DSS-Induced Ulcerative Colitis in Mice, Front. Cell. Infect. Microbiol., 2021, 11, 698914 CrossRef CAS PubMed.
- C. Hill, F. Guarner, G. Reid, G. R. Gibson, D. J. Merenstein, B. Pot, L. Morelli, R. B. Canani, H. J. Flint, S. Salminen, P. C. Calder and M. E. Sanders, Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic, Nat. Rev. Gastroenterol. Hepatol., 2014, 11, 506–514 CrossRef PubMed.
- M. E. Sanders, D. J. Merenstein, G. Reid, G. R. Gibson and R. A. Rastall, Probiotics and prebiotics in intestinal health and disease: from biology to the clinic, Nat. Rev. Gastroenterol. Hepatol., 2019, 16, 605–616 CrossRef PubMed.
- F. Zhao, Z. Guo, L. Y. Kwok, Z. Zhao, K. Wang, Y. Li, Z. Sun, J. Zhao and H. Zhang, Bifidobacterium lactis Probio-M8 improves bone metabolism in patients with postmenopausal osteoporosis, possibly by modulating the gut microbiota, Eur. J. Nutr., 2023, 62, 965–976 CAS.
- H. Lan, W. H. Liu, H. Zheng, H. Feng, W. Zhao, W. L. Hung and H. Li, Bifidobacterium lactis BL-99 protects mice with osteoporosis caused by colitis via gut inflammation and gut microbiota regulation, Food Funct., 2022, 13, 1482–1494 RSC.
- Y. Bao, Y. Zhang, Y. Zhang, Y. Liu, S. Wang, X. Dong, Y. Wang and H. Zhang, Screening of potential probiotic properties of Lactobacillus fermentum isolated from traditional dairy products, Food Control, 2010, 21, 695–701 CrossRef CAS.
- N. Percie du Sert, V. Hurst, A. Ahluwalia, S. Alam, M. T. Avey, M. Baker, W. J. Browne, A. Clark, I. C. Cuthill, U. Dirnagl, M. Emerson, P. Garner, S. T. Holgate, D. W. Howells, N. A. Karp, S. E. Lazic, K. Lidster, C. J. MacCallum, M. Macleod, E. J. Pearl, O. H. Petersen, F. Rawle, P. Reynolds, K. Rooney, E. S. Sena, S. D. Silberberg, T. Steckler and H. Wurbel, The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research, PLos Biol., 2020, 18, e3000410 CrossRef CAS PubMed.
- S. N. Murthy, H. S. Cooper, H. Shim, R. S. Shah, S. A. Ibrahim and D. J. Sedergran, Treatment of dextran sulfate sodium-induced murine colitis by intracolonic cyclosporin, Dig. Dis Sci., 1993, 38, 1722–1734 CrossRef CAS PubMed.
- L. A. Dieleman, M. J. Palmen, H. Akol, E. Bloemena, A. S. Pena, S. G. Meuwissen and E. P. Van Rees, Chronic experimental colitis induced by dextran sulphate sodium (DSS) is characterized by Th1 and Th2 cytokines, Clin. Exp. Immunol., 1998, 114, 385–391 CrossRef CAS PubMed.
- A. M. Bolger, M. Lohse and B. Usadel, Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics, 2014, 30, 2114–2120 CrossRef CAS PubMed.
- B. Langmead and S. L. Salzberg, Fast gapped-read alignment with Bowtie 2, Nat. Methods, 2012, 9, 357–359 CrossRef CAS PubMed.
- E. A. Franzosa, L. J. McIver, G. Rahnavard, L. R. Thompson, M. Schirmer, G. Weingart, K. S. Lipson, R. Knight, J. G. Caporaso, N. Segata and C. Huttenhower, Species-level functional profiling of metagenomes and metatranscriptomes, Nat. Methods, 2018, 15, 962–968 CrossRef CAS PubMed.
- M. Zhao, P. Xia, X. Zhang, H. He and T. Hou, Selenium-containing soybean peptides ameliorate intestinal inflammation and modulate gut microbiota dysbacteriosis in DSS-induced ulcerative colitis mice, Food Funct., 2023, 14, 6187–6199 RSC.
- B. Ran, C. E. Guo, Y. Zhang, C. Han, T. Cao, H. Huang, Z. Geng and W. Li, Preventive effect of Chinese dwarf cherry [Cerasus humilis (Bge.) Sok.] fermentation juice on dextran sulfate sodium-induced ulcerative colitis rats through the regulation of IgA and the intestinal immune barrier, Food Funct., 2022, 13, 5766–5781 RSC.
- Z. Mei, X. Huang, H. Zhang, D. Cheng, X. Xu, M. Fang, J. Hu, Y. Liu, Y. Liang and Y. Mei, Chitin derivatives ameliorate DSS-induced ulcerative colitis by changing gut microbiota and restoring intestinal barrier function, Int. J. Biol. Macromol., 2022, 202, 375–387 CrossRef CAS PubMed.
- R. Yang, Y. Wang, S. Mehmood, M. Zhao, X. Yang, Y. Li, W. Wang, J. Chen and Q. Jia, Polysaccharides from Armillariella tabescens mycelia mitigate DSS-induced ulcerative colitis via modulating intestinal microbiota in mice, Int. J. Biol. Macromol., 2023, 245, 125538 CrossRef CAS PubMed.
- R. Caruso, T. Mathes, E. C. Martens, N. Kamada, A. Nusrat, N. Inohara and G. Nunez, A specific gene-microbe interaction drives the development of Crohn’s disease-like colitis in mice, Sci. Immunol., 2019, 4, eaaw4341 CrossRef CAS PubMed.
- H. Sun, Y. Guo, H. Wang, A. Yin, J. Hu, T. Yuan, S. Zhou, W. Xu, P. Wei, S. Yin, P. Liu, X. Guo, Y. Tang, Y. Yan, Z. Luo, M. Wang, Q. Liang, P. Wu, A. Zhang, Z. Zhou, Y. Chen, Y. Li, J. Li, J. Shan and W. Zhou, Gut commensal Parabacteroides distasonis alleviates inflammatory arthritis, Gut, 2023, 72, 1664–1677 CrossRef CAS PubMed.
- Q. Zhao, M. Y. Dai, R. Y. Huang, J. Y. Duan, T. Zhang, W. M. Bao, J. Y. Zhang, S. Q. Gui, S. M. Xia, C. T. Dai, Y. M. Tang, F. J. Gonzalez and F. Li, Parabacteroides distasonis ameliorates hepatic fibrosis potentially via modulating intestinal bile acid metabolism and hepatocyte pyroptosis in male mice, Nat. Commun., 2023, 14, 1829 CrossRef CAS PubMed.
- D. Liu, S. Zhang, S. Li, Q. Zhang, Y. Cai, P. Li, H. Li, B. Shen, Q. Liao, Y. Hong and Z. Xie, Indoleacrylic acid produced by Parabacteroides distasonis alleviates type 2 diabetes via activation of AhR to repair intestinal barrier, BMC Biol., 2023, 21, 90 CrossRef CAS PubMed.
- W. Wei, C. C. Wong, Z. Jia, W. Liu, C. Liu, F. Ji, Y. Pan, F. Wang, G. Wang, L. Zhao, E. S. H. Chu, X. Zhang, J. J. Y. Sung and J. Yu, Parabacteroides distasonis uses dietary inulin to suppress NASH via its metabolite pentadecanoic acid, Nat. Microbiol., 2023, 8, 1534–1548 CrossRef CAS PubMed.
- Y. Ni, Y. Zhang, L. Zheng, N. Rong, Y. Yang, P. Gong, Y. Yang, X. Siwu, C. Zhang, L. Zhu and Z. Fu, Bifidobacterium and Lactobacillus improve inflammatory bowel disease in zebrafish of different ages by regulating the intestinal mucosal barrier and microbiota, Life Sci., 2023, 324, 121699 CrossRef CAS PubMed.
- Q. Lin, W. J. Hao, R. M. Zhou, C. L. Huang, X. Y. Wang, Y. S. Liu and X. Z. Li, Pretreatment with Bifidobacterium longum BAA2573 ameliorates dextran sulfate sodium (DSS)-induced colitis by modulating gut microbiota, Front. Microbiol., 2023, 14, 1211259 CrossRef PubMed.
- T. Wang, C. Shi, S. Wang, Y. Zhang, S. Wang, M. Ismael, J. Zhang, X. Wang and X. Lü, Protective Effects of Companilactobacillus crustorum MN047 against Dextran Sulfate Sodium-Induced Ulcerative Colitis: A Fecal Microbiota Transplantation Study, J. Agric. Food Chem., 2022, 70, 1547–1561 CrossRef CAS PubMed.
- L. Ma, Q. Shen, W. Lyu, L. Lv, W. Wang, M. Yu, H. Yang, S. Tao and Y. Xiao, Clostridium butyricum and Its Derived Extracellular Vesicles Modulate Gut Homeostasis and Ameliorate Acute Experimental Colitis, Microbiol. Spectrum, 2022, 10, e0136822 CrossRef PubMed.
- P. D. Cani, C. Depommier, M. Derrien, A. Everard and W. M. de Vos, Akkermansia muciniphila: paradigm for next-generation beneficial microorganisms, Nat. Rev. Gastroenterol. Hepatol., 2022, 19, 625–637 CrossRef PubMed.
- S. Jangi, R. Gandhi, L. M. Cox, N. Li, F. von Glehn, R. Yan, B. Patel, M. A. Mazzola, S. Liu, B. L. Glanz, S. Cook, S. Tankou, F. Stuart, K. Melo, P. Nejad, K. Smith, B. D. Topcuolu, J. Holden, P. Kivisakk, T. Chitnis, P. L. De Jager, F. J. Quintana, G. K. Gerber, L. Bry and H. L. Weiner, Alterations of the human gut microbiome in multiple sclerosis, Nat. Commun., 2016, 7, 12015 CrossRef CAS PubMed.
- E. Cekanaviciute, B. B. Yoo, T. F. Runia, J. W. Debelius, S. Singh, C. A. Nelson, R. Kanner, Y. Bencosme, Y. K. Lee, S. L. Hauser, E. Crabtree-Hartman, I. K. Sand, M. Gacias, Y. Zhu, P. Casaccia, B. A. C. Cree, R. Knight, S. K. Mazmanian and S. E. Baranzini, Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 10713–10718 CrossRef CAS PubMed.
- F. Zhang, L. Yue, X. Fang, G. Wang, C. Li, X. Sun, X. Jia, J. Yang, J. Song, Y. Zhang, C. Guo, G. Ma, M. Sang, F. Chen and P. Wang, Altered gut microbiota in Parkinson's disease patients/healthy spouses and its association with clinical features, Parkinsonism Relat. Disord., 2020, 81, 84–88 CrossRef PubMed.
- A. H. Tan, C. W. Chong, S. Y. Lim, I. K. S. Yap, C. S. J. Teh, M. F. Loke, S. L. Song, J. Y. Tan, B. H. Ang, Y. Q. Tan, M. T. Kho, J. Bowman, S. Mahadeva, H. S. Yong and A. E. Lang, Gut Microbial Ecosystem in Parkinson Disease: New Clinicobiological Insights from Multi-Omics, Ann. Neurol., 2021, 89, 546–559 CrossRef CAS PubMed.
- Q. Zhai, S. Feng, N. Arjan and W. Chen, A next generation probiotic, Akkermansia muciniphila, Crit. Rev. Food Sci. Nutr., 2019, 59, 3227–3236 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4fo00462k |
| ‡ These authors contributed equally: Qiuwen He and Tao Zhang. |
| This journal is © The Royal Society of Chemistry 2024 |